Abstract
Small noncoding regulatory RNAs (sRNAs) play a key role in regulating the expression of many genes in Escherichia coli and other bacteria. Many of the sRNAs identified in E. coli bind to mRNAs in an Hfq-dependent manner and stimulate or inhibit translation of the mRNAs. Several sRNAs are regulated by well-studied global regulators. Here, we report characterization of the CyaR (RyeE) sRNA, which was previously identified in a global search for sRNAs in E. coli. We demonstrated that CyaR is positively regulated by the global regulator Crp under conditions in which cyclic AMP levels are high. We showed by using microarray analysis and Northern blotting that several genes are negatively regulated by CyaR, including ompX, encoding a major outer membrane protein; luxS, encoding the autoinducer-2 synthase; nadE, encoding an essential NAD synthetase; and yqaE, encoding a predicted membrane protein with an unknown function. Using translational lacZ fusions to yqaE, ompX, nadE, and luxS, we demonstrated that the negative regulation of these genes by CyaR occurs at the posttranscriptional level and is direct. Different portions of a highly conserved 3′ region of CyaR are predicted to pair with sequences near the ribosome binding site of each of these targets; mutations in this sequence affected regulation, and compensatory mutations in the target mRNA restored regulation, confirming that there is direct regulation by the sRNA. These results provide insight into the mechanisms by which Crp negatively regulates genes such as luxS and ompX and provide a link between catabolite repression, quorum sensing, and nitrogen assimilation in E. coli.
Noncoding regulatory RNAs posttranscriptionally regulate gene expression in bacteria, archaea, and eukaryotes. The posttranscriptional regulation of gene expression by noncoding regulatory RNAs called small noncoding regulatory RNAs (sRNAs) in Escherichia coli is a model system for understanding this mechanism of regulating gene expression in bacteria. In E. coli, ∼100 sRNAs have been identified (10, 66, 71). Many of the genes encoding sRNAs were identified because they were located in a conserved intergenic region and flanked by a recognizable promoter and/or Rho-independent terminator (2, 66). These sRNAs are typically ∼100 bp long, and many of them have been shown to bind to the RNA chaperone Hfq (71).
Hfq is a member of the Sm-like family of RNA binding proteins and was first identified by Kajitani and Ishihama as the factor required for replication of the Qβ phage (29). Later, Tsui et al. found that an insertion mutation in hfq caused diverse pleiotropic phenotypes (61). Hfq has subsequently been shown to catalyze the binding of sRNAs to a complementary site in the targeted mRNAs (for a review, see reference 63).
Binding of an sRNA to a targeted mRNA frequently results in inhibition of translation and coupled degradation of the mRNA and the sRNA. One of the best-characterized examples of this mechanism of gene regulation involves the regulation of the sodB mRNA by the RyhB sRNA; Massé et al. have shown that expression of RyhB sRNA leads to coupled degradation of RyhB and the mRNA encoding sodB by RNase E (37). In other cases, the binding of an sRNA to a targeted mRNA leads to stabilization of the mRNA and stimulation of translation. The best-characterized example of this outcome is the regulation of the rpoS mRNA by the sRNA DsrA; DsrA pairing disrupts secondary structure in the rpoS mRNA that would otherwise block ribosome binding (for a review, see reference 35).
Many of the sRNAs that have been identified have been shown to be regulated by a specific global regulator. RyhB is regulated by the Fur repressor (38), RprA is regulated by the RcsC/RcsD/RcsB phosphorelay (34), OmrA and OmrB are regulated by the EnvZ/OmpR two-component system (21), and MicA and RybB are regulated by σE (16, 27, 50, 59). Thus, many different classes of transcriptional regulators regulate sRNAs, and it may be that most global regulators include at least one sRNA as part of the regulon. Here, we report that expression of the CyaR sRNA (previously designated RyeE) is regulated by the global regulator Crp and that activation of cyaR expression by Crp occurs under conditions in which cyclic AMP (cAMP) levels are known to be high. Two other groups of workers have recently reported regulation of CyaR by Crp (26, 49) in agreement with our findings. Moreover, we show here that the CyaR sRNA affects the expression of a number of genes; some of these genes, including ompX and luxS, were previously shown to be negatively regulated by Crp. We also demonstrate that the negative regulation of luxS by CyaR leads to a decrease in autoinducer-2 production by E. coli. This sRNA has been renamed CyaR to reflect its regulation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All E. coli K-12 strains used in this study are derivatives of strain MG1655. All strains and plasmids used in this study are listed in Table 1. The primers and 5′-biotinylated probes used in this study (Table 2) were supplied by Integrated DNA Technologies, Inc. Transduction was performed using phage P1vir as described by Miller (42). The ΔcyaR strain NRD345 and the ΔluxS strain NRD381 were generated by lambda Red recombinase-mediated gene replacement using the PCR product generated from template plasmid pKD3 and using the ryeEKO For and Rev primers and the luxSKO For and Rev primers, respectively. The lsrACDBFG deletion harbored by strain NRD389 was created by lambda Red recombinase-mediated gene replacement using the PCR product generated with the template plasmid pKD4 and the lsrACDBFGKO For and Rev primers.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant features | Reference or source |
|---|---|---|
| Strains | ||
| BB170 | V. harveyi luxN::Tn5 | 6 |
| CV600 | MG1655 ΔlacX74 Δcrp::cat | C. Vanderpool |
| CV8000 | MG1655 ΔlacX74imm21 PcyaR-lacZ (positions −76 to 64) | C. Vanderpool |
| DJ480 | MG1655 ΔlacX74 | D. Jin, NCI |
| DJ624 | MG1655 ΔlacX74mal::lacIq | D. Jin, NCI |
| MG1655 | Wild type | Lab strain collection |
| MG1225 | MG1655 ΔlacX74mal::lacIq λRS PBAD::ompX::lacZ | M. Guillier |
| NRD345 | MG1655 ΔcyaR::cat | This study |
| NRD348 | MG1655 ΔlacX74imm21 PcyaR-lacZ (positions −41 to 64) | This study |
| NRD351 | MG1655 ΔlacX74imm21 PcyaR-lacZilvD::Tn10 ΔcyaA | CV8000 + P1(SG2201) |
| NRD352 | MG1655 ΔlacX74imm21 PcyaR-lacZ (positions −76 to 64) Δcrp::cat | CV8000 + P1(CV600) |
| NRD359 | MG1655 ΔlacX74mal::lacIq ΔcyaR::cat | DJ624 + P1(NRD345) |
| NRD356 | MG1655 Δcrp::cat | MG1655 + P1(CV600) |
| NRD377 | MG1655 ΔlacX74mal::lacIq λRS PBAD::ompX::lacZ ΔcyaR::cat | MG1225 + P1(NRD345) |
| NRD381 | MG1655 ΔluxS::cat | This study |
| NRD389 | MG1655 ΔlsrACDBFG::kan | This study |
| NRD390 | MG1655 ΔlacX74mal::lacIq ΔlsrACDBFG::kan | DJ624 + P1(NRD389) |
| NRD392 | MG1655 ΔlacX74mal::lacIq ΔlsrACDBFG::kan ΔcyaR::cat | NRD390 + P1(NRD345) |
| NRD399 | mal::lacIq ΔaraBAD lacI′::PBAD::nadE′-lacZ | This study |
| NRD400 | mal::lacIq ΔaraBAD lacI′::PBAD::yqaE′-lacZ | This study |
| NRD402 | MG1655 ΔlacX74mal::lacIq ΔlsrACDBFG::kan ΔluxS::cat | NRD390 + P1(NRD381) |
| NRD406 | mal::lacIq ΔaraBAD lacI′::PBAD::luxS(T-7A)′-lacZ | This study |
| NRD407 | mal::lacIq ΔaraBAD lacI′::PBAD::nadE(T-3A C-2T A-1G)′-lacZ | This study |
| NRD408 | mal::lacIq ΔaraBAD lacI′::PBAD::yqaE(T7A C9G)′-lacZ | This study |
| NRD410 | mal::lacIq ΔaraBAD lacI′::PBAD::nadE(T-3A C-2T A-1G)′-lacZ ΔryeE::cat | NRD407 + P1(NRD345) |
| NRD411 | mal::lacIq ΔaraBAD lacI′::PBAD::yqaE(T7A C9G)′-lacZ ΔryeE::cat | NRD408 + P1(NRD345) |
| NRD413 | mal::lacIq ΔaraBAD lacI′::PBAD::luxS′-lacZ | This study |
| NRD415 | mal::lacIq ΔaraBAD lacI′::PBAD::luxS(T-7A)′-lacZ ΔryeE::cat | NRD406 + P1(NRD345) |
| PM1205 | mal::lacIq ΔaraBAD lacI′-PBAD::cat sacB::lacZ | Pierre Mandin, NIH |
| SG2201 | ilvD::Tn10 Δcya | Lab strain collection |
| Top10 | φ80lacZΔM15 ΔlacX74recA1 | Invitrogen |
| Plasmids | ||
| pBR-plac | Ampr; araBAD promoter-based expression vector having a pBR322 origin | 21 |
| pNRD405 | AatII-EcoRI cyaR-containing fragment cloned into the same sites in pBR-plac | This study |
| pNRD407 | A44T site-directed mutation in cyaR in pNRD405 | This study |
| pNRD408 | T47A site-directed mutation in cyaR in pNRD405 | This study |
| pNRD409 | T50A site-directed mutation in cyaR in pNRD405 | This study |
| pNRD410 | G38C, G39A, and A40T site-directed mutations in cyaR in pNRD405 | This study |
| pKD46 | Ampr; RepA101(Ts); λ exo, γ, and β expressed from an araBAD promoter | 12 |
TABLE 2.
Primers and probes used in this study
| Primer or probe | Sequence (5′-3′) |
|---|---|
| Primers | |
| c14Bam | CCGGGGATCCGATTACACAGGCTAAGGAGG |
| c14Eco | CACGGAATTCCGATTATTTTTCCCGGATGG |
| deeplac | CGGGCCTCTTCGCTA |
| EcopBAD2 | CGACGAATTCTCCCGCCATTCAGAGAAGAAACC |
| luxS(T-7A) For | CGGAGGAGGCTAAATGCCGTTGTTAGATAG |
| luxS(T-7A) Rev | CTATCTAACAACGGCATTTAGCC |
| luxSKO For | GAAAGAGTTCAGAAAATTTTTAAAAAAATTACCGGAGGTGGCTAAGTGTAGGCTGGAGCTGCTTC |
| luxSKO Rev | TGAACTGGCTTTTTTCAATTAATTGTGAAGATAGTTTACTGACTACATATGAATATCCTCCTTAG |
| lsrACDBFGKO For | AATACATTTGTTCAAAACTCACCTGCAAAACTGAACGGGGGAAATGTGTAGGCTGGAGCTGCTTC |
| lsrACDBFGKO Rev | CGTCAGCGCGTTACCCCAATTGTAGATAAAATTGATAAATTCGCCCATATGAATATCCTCCTTAG |
| M13For(−20) | GTAAAACGACGGCCAG |
| nadE(T-3A C-2T A-1G)-lacZ For | CTTTTTATCGCAACTCTCTACTGTTTCTCCATCTTTTCTGTCTGGAGGGGTATGATGACATTGCAACAAGTCG |
| ompXSma | CGACTCCCGGGCAGTGCTGAAAGACATGCAATTTTTTTCATAACCACCTC |
| pBAD-DNA-RACE | ACCTGACGCTTTTTATCGCAACTCTCTACTGTTTCTCCAT |
| pBAD-luxS4-lacZ | AACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGTATGATCGACTGTGAAGCTATC |
| pBAD-nadE-lacZ For | ACCTGACGCTTTTTATCGCAACTCTCTACTGTTTCTCCATCTTTTCTGTCTGGAGGGGTTC |
| pBAD-nadE-lacZ Rev | AACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACTTGTTGCAATGTCATTGAACCC |
| pBADompX | CCAGATTATTCCTAATCACCAGACTAATGATTCCATCAATCCTGGATGGAGAAACAGTAGAGAGTTGCG |
| pBAD-yqaE-lacZ For | ACCTGACGCTTTTTATCGCAACTCTCTACTGTTTCTCCATGCTTAAGAGATAAAATCTCTTTTTAAACAA |
| pBAD-yqaE-lacZ Rev | AACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGATGACGATTCTCCAGAAACC |
| pBR-placRyeE For | GCGTGACGTCGCTGAAAAACATAACCCATAAAATGCTAG |
| pBR-placRyeE Rev | GAGAATTCCCTTTTATTTCATTGTATTACGCGTAAAAAATAAGC |
| pBAD-X-lacZ For | GCGGATCCTACCTGACGC |
| RACE_nadE1 | GTTGTTGCAATGTCATTGAAC |
| RACE_yqaE2 | CCAAGCAGTGTCAACAGAA |
| RACE _ryeE1 | GGAGGTGGTTCCTGGTACAG |
| RACE_universal | GATATGCGCGAATTCCTGTAGAACGAACACTAGAAGAAA |
| ryeEKO For | GGAAAATTCTTAGAAACCGATCACATACAGCTGCATTTATTAAGGGTGTAGGCTGGAGCTGCTTC |
| ryeEKO Rev | GAAATAAATCCTTTTATTTCATTGTATTACGCGTAAAAAATAAGCCATATGAATATCCTCCTTAG |
| ryeEA44T For | GCTAGCTGTACCAGGAACCTCCTCCTTAGCCTGTGTAATCTC |
| ryeEA44T Rev | GATATTACACAGGCTAAGGAGGAGGTTCCTGGTACAGCTAGC |
| ryeET47A For | CTAGCTGTACCAGGAACCACCACCTTAGCCTGTGTAATCTCC |
| ryeET47A Rev | GGAGATTACACAGGCTAAGGTGGTGGTTCCTGGTACAGCTAG |
| ryeET50A For | CTGTACCAGGAACCACCTCCATAGCCTGTGTAATCTCCC |
| ryeET50A Rev | GGGAGATTACACAGGCTATGGAGGTGGTTCCTGGTACAG |
| ryeEG38C-G39A-A40T For | CCCATAAAATGCTAGCTGTACCACATACCACCTCCTTAGCCTGTG |
| ryeEG38C-G39A-A40T Rev | CACAGGCTAAGGAGGTGGTATGTGGTACAGCTAGCATTTTATGGG |
| yqaE(T7A C9G)-lacZ rev | GTTTTCCCAGTCACGACGTTGTAAAACGACGATGACGATTCTCCACATACCCATATGTACTCCCTATAAG |
| Probes | |
| cfaprobe1 | CCACCAGCCATCCATATAACTTTCGCCTAACCCCA |
| yqaEprde1 | CCCAACGGAACCCTTTACCGAGCAGCACGCCGAGC |
| flu-probe | CCCTGCAGGAGTGCATCGCCTTCACGGATGGTCAGGGTATCG |
| luxSprobe1 | TCCCTCTTTCTGGCATCACTTCTTTGTTCGGCACG |
| mscLprobe1 | CCCAGAGGAGGCATGATGATATCGGCAACCAGTGA |
| nadEprobe1 | GGTTAATACGCGATCCGGTTGAATAAAGGCAATGG |
| ompXprobe1 | TACGGAAGTACCTGCGGTGAAAGCCAGAACTGCGG |
| ryeEprobe1 | TGGTTCCTGGTACAGCTAGCATTTTATGGGTTATG |
| uspGprobe1 | CGGACATGTTGTTTAATGCGGGAAGGATCGATGGT |
lacZ translational fusions to yqaE, nadE, and luxS that were under control of the araBAD promoter were generated using a procedure developed by P. Mandin, NIH (P. Mandin and S. Gottesman, unpublished data). First, the transcription start sites of yqaE, nadE, and luxS were determined by 5′ rapid amplification of cDNA ends (5′ RACE) as described below. Primers pBAD-yqaE-lacZ For and pBAD-yqaE-lacZ Rev, primers pBAD-nadE-lacZ For and pBAD-nadE-lacZ rev, and primers pBAD-luxS4-lacZ and pBAD-DNA-RACE were then used to PCR amplify from cDNA or genomic DNA the sequence from the transcription start site to the first few codons of yqaE, nadE, and luxS, respectively. The ends of the PCR products had sequences that were homologous to the araBAD promoter at one end and homologous to lacZ at the other end. Each PCR product was inserted into the chromosome of strain PM1205 (Table 1) by lambda Red recombinase-mediated gene replacement, resulting in translational lacZ fusions under transcriptional control of the araBAD promoter.
lacZ translational fusions to nadE and yqaE that harbored point mutations in the translational initiation region were constructed by amplifying the nadE′- and yqaE′-lacZ translational fusions from NRD399 and NRD400, respectively, using primers carrying the mutations [primers nadE(T-3A C-2T A-1G)-lacZ For and deeplac or primers pBAD-X-lacZ For and yqaE(T7A C9G)-lacZ rev]. The PCR products were then recombined into the chromosome of PM1205 as described above to generate strains NRD407 [nadE(T-3A C-2T A-1G)′-lacZ fusion] and NRD408 [yqaE(T7A C9G)′-lacZ fusion]. The luxS′-lacZ translational fusion that had a point mutation in the translational initiation region was constructed by first amplifying two overlapping pieces of this region using primers luxS(T-7A) For and deeplac and primers pBAD-X-lacZ For and luxS(T-7A) Rev. The two PCR fragments were then joined by overlap extension PCR, and the resulting PCR product was recombined into the chromosome of strain PM1205 to generate strain NRD406.
Strain CV8000, which harbors a cyaR-lacZ transcriptional fusion that contains the full-length promoter of cyaR (long fusion), was obtained from Carin K. Vanderpool. The cyaR-lacZ fusion harbored by strain CV8000 was generated by amplifying a portion of the cyaR gene and the promoter upstream of cyaR from E. coli K-12 genomic DNA using the c14Eco and c14Bam primers and cloned into pRS1553 (54) digested with EcoRI and BamHI. Strain MG1655 was transformed with this plasmid, and the transformants were then infected with λRS468 phage (54) to generate recombinant phages harboring a cyaR-lacZ transcriptional fusion. Strain DJ480 was infected with these recombinant λ phages, and single lysogens were isolated (54).
Strain MG1225 carrying the ompX′-lacZ translational fusion was acquired from Maude Guillier and was constructed as follows. The araBAD promoter was amplified by PCR from pBAD24 using the EcopBAD2 and pBADompX primers. The resulting PCR product and the ompXSma primer were used to amplify a sequence of E. coli K-12 strain MG1655 genomic DNA containing the first 10 codons of ompX and the 241 nucleotides directly upstream of the ompX start codon. The resulting PCR product was cloned into pRS414 (54) digested with EcoRI and SmaI. Recombinant lambda phages containing the lacZ translational fusion to ompX were generated, and the lacZ fusion was inserted into the chromosome of strain DJ480 as described above.
Plasmid pNRD405 was constructed by PCR amplification of cyaR from strain MG1655 using the pBR-placRyeE For and pBR-placRyeE Rev primers and subsequently cloning the PCR product into the pBR-plac vector digested with AatII and EcoRI. cyaR site-directed mutants with A44T, T47A, T50A, and G38C G39A A40T mutations were constructed using a QuikChange II site-directed mutagenesis kit (Stratagene), template plasmid pNRD405, and primers ryeEA44T For and ryeEA44T Rev, primers ryeET47A For and ryeET47A Rev, primers ryeET50A For and ryeET50A Rev, and primers ryeEG38C-G39A-A40T For and ryeEG38C-G39A-A40T Rev, respectively. All of the cyaR site-directed mutants were confirmed by sequencing.
Culture media and growth conditions.
All MG1655 derivatives were grown in liquid medium or agar plates containing Lennox broth or in M63 minimal medium supplemented with vitamin B1 at a final concentration of 0.001% and glucose or glycerol at a final concentration of 0.2%. In some instances, LB medium was supplemented with glucose or arabinose at a final concentration of either 0.2 or 0.01%. Antibiotics were added to LB liquid medium or agar plates at the following concentrations: tetracycline, 25 mg liter−1; chlorampenicol, 25 mg liter−1; and ampicillin, 100 mg liter−1. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to cultures at a final concentration of 100 μM.
β-Galactosidase assays.
All strains were grown overnight at 37°C in LB liquid medium. Each overnight culture was then diluted 200-fold in fresh LB liquid medium or LB liquid medium supplemented with glucose and incubated at 37°C. β-Galactosidase assays were performed as described by Miller (42) using samples removed from the cultures during log phase (optical density at 600 nm [OD600], 0.3 to 0.4), late log phase (OD600, 1.0 to 1.2), and stationary phase (OD600, 3.2 to 5.2). All β-galactosidase assays were performed at least twice.
Northern blotting.
For determination of the effects of cyaR expression on the abundance of potential target mRNAs, overnight cultures of NRD359/pBR-plac or NRD359/pNRD405 (plac-cyaR+) grown in LB liquid medium containing ampicillin were diluted 200-fold in fresh medium and incubated at 37°C. A 700-μl sample was removed from each culture when the OD600 was 0.5. IPTG was then added to each culture, and additional samples were removed 2, 5, 10, and 15 min after the addition of IPTG. RNA was extracted from the samples using the hot phenol method described by Massé et al. (37).
For examination of the role of Crp in the expression of cyaR, an overnight culture of strain MG1655 or NRD356 grown in LB liquid medium was diluted 200-fold in fresh LB medium and incubated at 37°C. When each culture reached log phase or late log phase, a 700-μl sample was removed from the culture and the RNA was extracted as described above.
To determine the effect of expression of CyaR from its native promoter on the abundance of the yqaE, ompX, nadE, and luxS mRNAs, overnight cultures of strain DJ624 or NRD359 grown in LB liquid medium were diluted 200-fold in fresh LB medium and incubated at 37°C. Samples (700 μl) were removed from log-, late-log-, and stationary-phase cultures of each strain, and the RNA was extracted from each sample as described above.
Northern blot analysis of CyaR expression was performed by fractionating 3 μg of RNA on a Bio-Rad Criterion 10% Tris-borate-EDTA (TBE)-urea polyacrylamide gel that was prerun for 30 min at 55 V and subsequently run at 55 V for 3 h in 1× TBE. The fractionated RNA was then transferred to a 0.2-μm nylon membrane (Whatman Nytran N) by electroblotting at 200 mA for 2 h in 0.5× TBE. The RNA was cross-linked to the nylon membrane by using UV irradiation. The membrane was then hybridized with 5′-biotinylated ryeEprobe1 in ULTRAhyb solution (Ambion) at 42°C, and the CyaR sRNA was detected using a Brightstar Biotect kit (Ambion).
Northern blot analysis of ompX, nadE, yqaE, luxS, mscL, uspG, cfa, and flu expression was performed by fractionating 10 μg of total mRNA on a 1.2% agarose gel that was prerun at 12 V/cm for at least 5 min and subsequently run at 5 V/cm for 2 h in 1× morpholinepropanesulfonic acid (MOPS). After capillary transfer of the RNA to a 0.45-μm nylon membrane (Whatman Nytran SuPerCharge), the membrane was hybridized with ompXprobe1, nadEprobe1, yqaEprobe1, luxSprobe1, mscLprobe1, uspGprobe1, cfaprobe1, or flu-probe, and the mRNA was detected as described above.
5′ RACE.
5′ RACE was performed by using the method described by Argaman et al. (2), with the following modifications. One microgram of RNA ligated to the 5′ universal RNA adapter (5′-GAU AUG CGC GAA UUC CUG UAG AAC GAA CAC UAG AAG AAA-3′) or the 5′ pBAD-DNA-RACE RNA adapter (ACC TG CGC TTT TTA TCG CAA CTC TCT ACT GTT TCT CCA T) was reverse transcribed by using Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions and the pBAD-luxS4-lacZ, RACE-nadE1, RACE-yqaE, or RACE-ryeE1 primer and then treated with RNase H. The cDNA was then amplified by PCR using the pBAD-luxS4-lacZ, RACE-nadE1, RACE-yqaE, or RACEryeE1 primer and the RACEUniversal or pBAD-DNA-RACE primer. The PCR products were then TOPO cloned into the pCR4-TOPO vector (Invitrogen), and the resulting plasmids were transformed into Top10 cells. At least five transformants harboring plasmids containing the amplified cDNA from the tobacco acid pyrophosphatase-treated RNA were sequenced using the M13For(−20) primer supplied with the TOPO cloning kit (Invitrogen).
Microarray analysis.
The cyaR deletion strain NRD359 harboring the vector pBR-plac or plasmid pNRD405, which contains cyaR under control of a lac promoter, was grown overnight at 37°C in LB liquid medium containing ampicillin. Each strain was then diluted 200-fold in fresh medium and incubated at 37°C until the OD600 was 0.5. IPTG was then added to each culture to a final concentration of 100 μM. After 15 min of induction with IPTG, a sample was taken from each culture, and the RNA was extracted by the hot phenol method as described above. The RNA was treated with Turbo DNase (Ambion), and the RNA was then recovered as described above for 5′ RACE. The labeled cDNA was then prepared and hybridized to an E. coli Genome 2.0 array (Affymetrix) according to the manufacturer's instructions.
Analyses of the microarray data were performed as described by Massé et al., except that genes that were not present (P > 0.05) in three or more of the cDNA samples or had an average difference of less than twofold under the two conditions used for the two microarray experiments were eliminated from further consideration. The genes whose expression patterns were not reproducible in the two microarray experiments (i.e., genes that did not show a change in expression in the same direction under the two conditions in both microarray experiments) were also eliminated. Finally, the data for genes that gave a low signal (<300) in all of the microarrays were also removed from the data set. Of the remaining genes (Table 3), those with the greatest change and the most reproducible signal intensities (yqaE, ompX, nadE, and luxS) were analyzed further by performing Northern blot analysis as described above. Other genes with smaller changes in abundance that were of interest to us were also examined by Northern blotting (uspG, mscL, and cfa), but the changes were not significant and these genes were not studied further.
TABLE 3.
Genes regulated by CyaR
| Gene | Locus | Expt 1
|
Expt 2
|
Avg change (fold) | Gene product | ||||
|---|---|---|---|---|---|---|---|---|---|
| Level when CyaR was not expresseda | Level when CyaR was expresseda | Change (fold) | Level when CyaR was not expresseda | Level when CyaR was expresseda | Change (fold) | ||||
| uspG | b0607 | 951.8 | 647.5 | −1.47 | 1,232.6 | 448.8 | −2.75 | −2.1 | Universal stress protein |
| modC | b0765 | 297.3 | 263.8 | −1.13 | 451.2 | 111.8 | −4.04 | −2.6 | Component of a molybdenum ABC-type transporter |
| ompX | b0814 | 31,805.2 | 4,250.6 | −7.48 | 19,716 | 2,519.9 | −7.82 | −7.7 | Outer membrane protein with unknown function |
| gsiB | b0830 | 1,507.9 | 447 | −3.37 | 565.1 | 428.5 | −1.32 | −2.35 | Component of a predicted glutathione ABC-type transporter |
| nadE | b1740 | 2,776.2 | 450.9 | −6.16 | 3,628.7 | 767.8 | −4.73 | −5.4 | NAD+ synthase |
| yobD | b1820 | 388.8 | 176.5 | −2.20 | 458.8 | 229.7 | −2.00 | −2.1 | Conserved inner membrane protein with unknown function |
| yeeD | b2012 | 3,063.7 | 772.8 | −3.96 | 182.6 | 170.5 | −1.07 | −2.5 | Conserved predicted protein |
| gabD | b2661 | 531.3 | 222.6 | −2.39 | 649 | 340.6 | −1.91 | −2.1 | Succinic semialdehyde dehydrogenase |
| ygaU | b2665 | 1,307 | 864.6 | −1.51 | 1,377.9 | 455.3 | −3.03 | −2.3 | Protein with unknown function |
| yqaE | b2666 | 3,254.6 | 286.7 | −11.3 | 2,304.7 | 390.2 | −5.91 | −8.6 | Conserved predicted membrane protein |
| luxS | b2687 | 6,105.7 | 1,961.7 | −3.11 | 5,417.7 | 1,589.3 | −3.41 | −3.3 | Autoinducer-2 synthase |
| cysN | b2751 | 2,528.7 | 211.1 | −12.0 | 127.3 | 24 | −5.30 | −8.6 | Subunit of sulfate adenylyltransferase |
| cysH | b2762 | 2,125.2 | 133.2 | −16.0 | 131.4 | 27 | −4.87 | −10.4 | Phosphoadenylyl sulfate reductase |
| mscL | b3291 | 2,135.8 | 1,015.1 | −2.10 | 2,005.3 | 1,029.4 | −1.95 | −2.0 | Mechanosensitive channel that relieves pressure on the membrane |
| ibpB | b3686 | 3,952.1 | 438.7 | −9.00 | 3,387.6 | 2,764.3 | −1.23 | −5.1 | Heat shock protein |
| ibpA | b3687 | 5,866.2 | 1,651.1 | −3.55 | 7,108.9 | 5,687.2 | −1.25 | −2.4 | Heat shock protein |
| sbp | b3917 | 1,454.4 | 192.9 | −7.54 | 78 | 14.1 | −5.53 | −6.5 | Sulfate binding protein involved in sulfate import |
| ryeA | b4432 | 3,807.7 | 2,131 | −1.79 | 5,880.6 | 2,309.7 | −2.55 | −2.2 | sRNA |
| sfuC | b0066 | 97.8 | 147.8 | 1.51 | 120.3 | 339.3 | 2.82 | 2.2 | Thiamine transporter |
| ykgC | b0304 | 119.7 | 160.3 | 1.34 | 103.3 | 345.3 | 3.34 | 2.3 | Predicted oxidoreductase |
| fepA | b0584 | 467.8 | 1,247.3 | 2.67 | 1,360.1 | 1,997 | 1.47 | 2.1 | Outer membrane receptor and transport protein for enterobactin |
| fepC | b0588 | 146.1 | 491.2 | 3.36 | 543 | 647.4 | 1.19 | 2.3 | Fe3+-enterobactin ABC-type transporter |
| flu | b2000 | 279 | 11,245 | 40.3 | 277.4 | 9,614 | 34.7 | 37.0 | Outer membrane protein involved in autoaggregation |
| yeeR | b2001 | 151 | 1,897.1 | 12.6 | 130.6 | 1,461.4 | 11.2 | 12.0 | Predicted membrane protein |
| yeeS | b2002 | 9 | 598.7 | 66.5 | 45.3 | 377.7 | 8.34 | 37.4 | Predicted DNA repair protein |
Units are Affymetrix-reported values for gene signal.
The expression of three genes (yeeR, yeeS, and flu) appeared to increase substantially upon cyaR overexpression in the microarray experiments (Table 3). These three genes are adjacent to each other in the E. coli genome and are transcribed in the same direction. Reexamination of the stocks of strains NRD359/pBR-plac and NRD359/pNRD405 used in the microarray experiments described above demonstrated that these two strains were dominated by different phase variants of flu, providing an explanation for the substantial changes in expression. Northern blotting of overnight cultures of NRD359/pBR-plac and NRD359/pNRD405 that were in the same phase with respect to flu expression (i.e., there was not rapid settling of cells in the cultures) demonstrated that there was no difference in flu expression in the presence and in the absence of CyaR expression. The negative regulation of yqaE, ompX, nadE, and luxS by CyaR was observed regardless of whether the two strains were in the same phase with respect to flu expression.
Autoinducer-2 assays.
Autoinducer-2 assays were performed as described by Surette and Bassler (55) and Wang et al. (65). Strains NRD359 and NRD392 harboring pBR-plac or pNRD405 were grown overnight at 37°C in LB liquid medium containing ampicillin. Cells from 1 ml of each overnight culture were then pelleted, resuspended in 1 ml of fresh LB medium, and diluted and grown at 30°C in fresh LB medium containing IPTG at a final concentration of 100 μM. Samples were removed every hour, and a cell-free culture filtrate was prepared. Cell-free culture filtrates were stored at −20°C overnight.
In duplicate, 10 μl of a cell-free culture filtrate or fresh LB medium was added to a Corning Costar 3912 flat-bottom assay plate. Ninety microliters of AB medium containing Vibrio harveyi diluted 5,000-fold from an overnight culture was added to each well. The 96-well microplate was then incubated at 30°C, and bioluminescence was measured 2 h after inoculation and then every 0.5 h for an additional 3 h.
RESULTS
Northern blotting previously showed that CyaR sRNA was expressed well in LB liquid medium but was expressed poorly in liquid M63 minimal medium supplemented with glucose and vitamin B1 (66). We confirmed this expression pattern using strain CV8000, a lac derivative of E. coli K-12 strain MG1655 harboring a lacZ transcriptional fusion to the promoter region of cyaR (positions −74 to 64) (Fig. 1). Consistent with the Northern blotting performed by Wassarman et al. (66), expression of cyaR was fivefold higher in a late-log-phase culture (OD600, 1.0 to 1.2) of strain CV8000 grown in LB liquid medium (254 Miller units) than in a late-log-phase culture grown in liquid M63 minimal medium (54 Miller units).
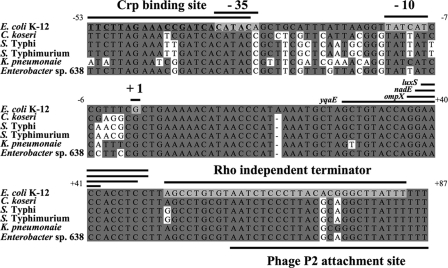
FIG. 1.
Alignment of cyaR and the cyaR promoter region of several gammaproteobacteria. DNA sequence identity is indicated by dark gray shading. The predicted −10 region and −35 region of the cyaR promoter, the Rho-independent terminator, and the transcription start site mapped by 5′ RACE are indicated by light gray shading. The regions of CyaR predicted to pair with the yqaE, ompX, nadE, and luxS mRNAs are indicated by bars above the regions. The sequence of the predicted CRP binding site is indicated by bold type and is underlined. C. koseri, Citrobacter koseri; K. pneumoniae, Klebsiella pneumoniae.
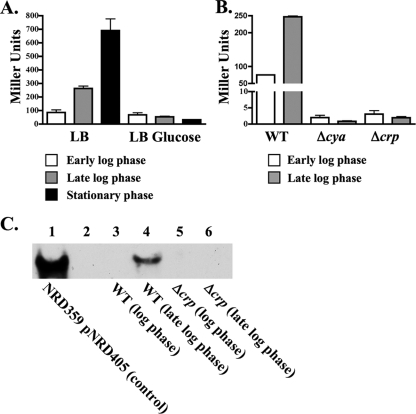
One possible source of the difference between expression of cyaR in LB liquid medium and expression of cyaR in M63 minimal medium containing glucose might be the difference in glucose levels. This possibility was tested by comparing the β-galactosidase activities of samples of cells taken from log-phase, late-log-phase, and stationary-phase cultures of CV8000 grown in LB medium or LB medium supplemented with glucose at a final concentration of 0.2%. The levels of expression of cyaR were similar for the cultures of CV8000 grown in M63 minimal medium containing vitamin B1 and glucose and in LB liquid medium containing glucose in both the log phase (61 and 67 Miller units) and the late log phase (54 and 53 Miller units). However, the level of expression of cyaR was significantly higher in cultures of CV8000 grown in LB medium than in cultures grown in LB medium containing glucose in the late log phase (5-fold higher; 263 and 53 Miller units) and the stationary phase (22-fold higher; 691 and 32 Miller units) (Fig. 2A).
FIG. 2.
Comparison of CyaR expression under different conditions. (A) β-Galactosidase assays of strain CV8000, which contains a lacZ transcriptional fusion to the cyaR promoter, grown at 37°C in LB liquid medium with or without glucose. β-Galactosidase assays were performed using samples taken from the cultures during log phase, late log phase, or stationary phase. (B) β-Galactosidase assays of strain CV8000 (WT) or derivatives of strain CV8000 harboring a cya deletion (NRD351) or a crp deletion (NRD352) grown at 37°C in LB liquid medium. β-Galactosidase assays were performed using samples taken from cultures of each strain during log phase or late log phase. The data in panels A and B are averages of three independent experiments. (C) Northern blot analysis of total RNA from a culture of strain NRD359/pNRD405 grown in LB liquid medium containing ampicillin and IPTG (control) (lane 1), from log-phase and late-log-phase cultures of wild-type strain MG1655 (WT) (lanes 3 and 4, respectively), or from log-phase and late-log-phase cultures of the Δcrp strain NRD345 (lanes 5 and 6, respectively) grown in LB liquid medium at 37°C. Five times more RNA was loaded in lanes 5 and 6 (Δcrp) than in lanes 3 and 4 (wild type). The ryeEprobe1 was used to detect RNA encoding cyaR.
Since cyaR expression was subject to a “glucose effect” in the experiments described above, we hypothesized that cyaR expression might be activated by cAMP receptor protein (Crp) in the presence of cAMP. In E. coli, glucose import prevents activation of cAMP synthesis by adenylate cyclase, which is encoded by the cya gene. In the absence of transport of glucose into E. coli, adenylate cyclase is activated, resulting in the synthesis of cAMP, which then binds to Crp (for a review, see reference 51). With cAMP bound, Crp is able to bind upstream of specific promoters, leading to activation of gene expression. To test whether cyaR expression is dependent on Crp and cAMP, isogenic derivatives of CV8000 carrying either a cya or crp deletion were constructed and tested to determine expression of the reporter fusion during growth in LB broth. Expression of cyaR was drastically reduced (<1% of wild-type expression) in late-log-phase cultures of strains having either Δcya or Δcrp mutations (227 Miller units for a wild-type strain versus 0.7 and 1.8 Miller units for its Δcya and Δcrp derivatives, respectively) (Fig. 2B).
We also examined the abundance of the CyaR sRNA in log-phase and late-log-phase cultures of MG1655 and its Δcrp derivative by Northern blotting. While the CyaR sRNA was easily detected by Northern blotting in cells from a late-log-phase culture of the parent strain, strain MG1655, it was not detected in cells of the crp mutant strain NRD356 (Fig. 2C, compare lanes 4 and 6), even though five times more RNA was loaded into the lanes containing Δcrp strain NRD356 (lanes 5 and 6) than into the lanes containing MG1655 (lanes 3 and 4).
Analysis of the cyaR promoter.
To further characterize cyaR and regulation of its transcription, we mapped the transcription start site of cyaR by using 5′ RACE. Six of the seven independently cloned cDNAs of cyaR had the same transcription start site (Fig. 1). Based on the identified transcription start site and the predicted Rho-independent terminator, we predicted that the cyaR sRNA is 84 bp long. A comparison of the region encompassing cyaR and its predicted promoter in several gammaproteobacteria revealed a conserved −10 sequence and a highly conserved sequence that overlaps the predicted −35 region of the promoter (Fig. 1). This highly conserved sequence is centered at position −42.5 and has a high level of homology to the consensus sequence for Crp binding sites (5′-AAATGTGATCTAGATCACATTT-3′; the bases conserved in the Crp binding site upstream of cyaR are underlined) (15, 17, 47). To confirm that this conserved putative Crp binding site is required for cyaR expression, we constructed a lacZ transcriptional fusion to the cyaR promoter identical to the fusion constructed in CV8000, except that it lacked the region upstream of the Crp binding site of cyaR (positions −76 to −54) and the Crp binding site of cyaR was changed from 5′-TTCTTAGAAACCGATCACATAC-3′ to 5′-GGATCGGAATTCGATCACATAC-3′ (the bases that were changed are underlined). Cells carrying this reporter fusion expressed β-galactosidase at levels that were 18- and 27-fold lower than the levels expressed by cells with the wild-type fusion during log phase (3.3 versus 60 Miller units) and late log phase (7.8 versus 206 Miller units), respectively. The low level of residual activity of this mutant promoter, compared to the very low levels of expression in a strain with crp deleted, may have been due to the presence of the unaltered second half of the Crp binding site and the presence of three conserved residues in the 5′ end of the mutant Crp binding site (5′-GGATCGGAATTCGATCACATAC-3′; conserved residues are underlined). Our results are consistent with direct Crp/cAMP regulation of ryeE, which was renamed cyaR to reflect this regulation. Similar results have recently been reported by Johansen et al. for E. coli (27) and by Papenfort et al. for Salmonella enterica serovar Typhimurium (49).
Identification of the targets of the CyaR sRNA.
As mentioned above, Hfq-binding sRNAs recognize specific mRNAs by base pairing. The binding of an sRNA to an mRNA results in either inhibition of translation and coupled degradation of the sRNA and targeted mRNA or an increase in translation and an increase in message stability. To identify genes that are either negatively or positively regulated by the CyaR sRNA, we used a microarray-based approach similar to that described by Massé et al. (39) and Guillier and Gottesman (20) to examine the effects of CyaR expression. Total genome expression was compared in ΔcyaR strain NRD359 carrying a vector control (pBr-plac) and ΔcyaR strain NRD359 carrying cyaR controlled by plac; samples were collected from log-phase cultures after 15 min of IPTG induction.
In the microarray experiments, the expression of 18 genes decreased on average at least twofold, and the expression of 7 genes increased at least twofold, not counting cyaR itself. Four of the genes whose mRNA levels decreased upon CyaR expression (cysH, cysN, spb, and yeeD) were expressed at very low levels in one of the two experiments and were not considered further. The reason for the difference in basal expression levels between experiments is not known, but we noted that many cys genes were expressed much better in experiment 1 than in experiment 2. Of the remaining 15 genes, yqaE, ompX, nadE, and luxS are four of the five genes showing the largest decreases in expression upon cyaR expression. The fifth gene, ibpB, and the upstream gene ibpA, both encoding heat shock proteins, were not examined further; the decreases in expression of these genes may suggest that there is a gradual decrease in expression of the heat shock regulon in response to CyaR overexpression, since other genes involved in the heat shock response were also downregulated, although to a lesser extent (see Table S1 in the supplemental material).
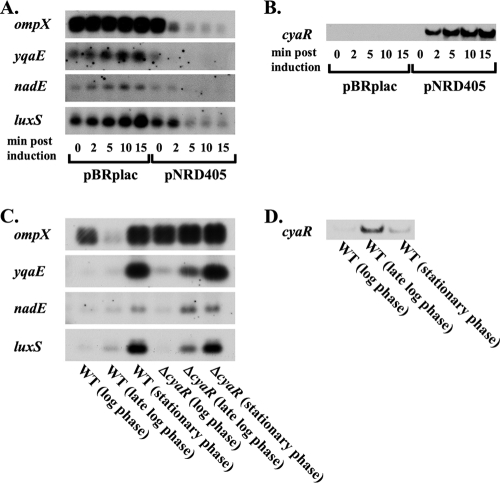
To confirm the negative regulation of yqaE, ompX, nadE, and luxS by CyaR, we examined the expression of these genes under conditions similar to those used for the microarrays, except that RNA was isolated from the cultures at additional time points (0, 2, 5, 10, and 15 min after IPTG was added to cultures of NRD359/pBR-plac and NRD359/pBR-cyaR [pNRD405]). The levels of mRNAs for ompX, yqaE, and nadE clearly decreased or the mRNAs disappeared within 2 min upon induction of CyaR expression, and the levels of mRNA for luxS showed a marked decrease within 5 min upon induction of CyaR expression (Fig. 3A), consistent with the hypothesis that there is direct regulation of the stability of these mRNAs by CyaR. As expected, cyaR expression was not induced until after IPTG was added, and the abundance of the CyaR sRNA continued to increase after IPTG induction (Fig. 3B).
FIG. 3.
Northern blot analysis of ompX, nadE, yqaE, and luxS expression in the presence or absence of cyaR expression. (A and B) The cyaR deletion strain NRD359 harboring an empty vector (pBR-plac) or a plasmid containing cyaR under a lac-based promoter (pNRD405) was grown at 37°C in LB liquid medium containing ampicillin to an OD600 of 0.5, and a sample of each culture was removed. IPTG was then added to each culture, and additional samples were removed from each culture 2, 5, 10, and 15 min after the addition of IPTG. RNA was extracted by the hot phenol method, fractionated on a 1.2% agarose gel (A) or a 10% TBE-urea gel (B), and transferred to a nylon membrane. 5′-Biotinylated probes were used to detect mRNA-encoding genes showing negative regulation by cyaR in the microarray experiments whose results are shown in Table 3, including ompX, yqaE, nadE, and luxS (A) or the CyaR sRNA (B). (C and D) Strain DJ624 (WT) and ΔcyaR strain NRD359 (ΔcyaR) were grown at 37°C in LB liquid medium, and samples were removed from each culture at log phase (at an OD600 between 0.3 and 0.4), late log phase (at an OD600 between 0.9 and 1.2), and stationary phase (at an OD600 more than 5.0; 6 h postinoculation). RNA was extracted, fractionated, and transferred to a nylon membrane as described above. 5′-Biotinylated probes were used to detect ompX, yqaE, nadE, luxS and ompA (loading control [data not shown]) (C) or the CyaR sRNA (D). Based on the RNA ladder run alongside the mRNA samples for the Northern blots shown in panel A, the mRNAs encoding ompX, nadE, yqaE, and luxS are estimated to be approximately 900 and 600 bp (ompX), 1,000 bp (nadE), 300 bp (yqaE), and 700 bp (luxS) long, respectively, indicating that all of the transcripts detected encoded only a single gene. Two ompX mRNAs that were different sizes were detectected because ompX is transcribed from two different promoters located 170 bp apart (40).
Northern blotting was also performed under these conditions for three other genes of interest, mscL, uspG, and cfa, whose expression decreased 2.0-, 2.1-, and 1.9-fold upon cyaR expression in the microarray experiments described above. However, we did not detect decreases in mscL, uspG, or cfa expression upon overexpression of cyaR (data not shown).
The four mRNAs confirmed by Northern blotting, the ompX, yqaE, nadE, and luxS mRNAs, were selected for further study. The functions of these mRNAs are discussed briefly here, but no obvious common theme was suggested by the results for this group of genes. OmpX and YqaE are both membrane proteins, a class of proteins that many other sRNAs have been found to regulate (8, 11, 21, 27, 43, 50, 59, 62, 64). ompX, whose expression was reduced almost eightfold upon CyaR expression, encodes an outer membrane protein with an unknown function that is not essential (40), and yqaE, whose expression was reduced almost ninefold upon CyaR expression, is predicted to encode a conserved inner membrane protein. Another gene that was expressed at much lower levels after CyaR expression was nadE. nadE is an essential gene and encodes an ammonia-dependent NAD synthetase, which synthesizes NAD from nicotinic adenine dinucleotide (24, 53, 69). Finally, luxS, which encodes the autoinducer-2 synthase involved in quorum sensing, was also significantly downregulated upon CyaR expression. Autoinducer-2-mediated quorum sensing has a significant influence on the expression of many genes in E. coli, including genes involved in biofilm formation and motility (14, 18, 33).
Consistent with regulation of CyaR by cAMP and Crp, several of the downregulated transcripts (including ompX and luxS) have previously been shown to be negatively regulated by Crp (19, 65, 72). LuxS was previously shown by Wang et al. (65) to be negatively regulated by crp, but it lacked a sequence upstream of the coding sequence for luxS that was able to bind CRP in vitro. Thus, the regulation by CyaR explains this discrepancy.
For the mRNAs whose levels increased after CyaR expression, the most dramatic increases (flu, yeeR, and yeeS) were found to be due to phase variation and not CyaR (see Materials and Methods). For other mRNAs there were modest changes or relatively low levels of expression were observed, and these mRNAs were not analyzed further.
CyaR expression results in decreases in the abundance of yqaE, ompX, nadE, and ompX mRNAs during late log phase in wild-type E. coli cells.
In the microarray and Northern blot experiments described above, target mRNAs were identified and confirmed to be negatively regulated by CyaR under CyaR overexpression conditions (compare Fig. 2, lane 1 [1 μg of RNA isolated from NRD359/pNRD405 expressing cyaR for 15 min], to lane 4 [3 μg of RNA isolated from a late-log-phase culture of MG1655]). To assess the effect of CyaR expression from its native promoter on the abundance of these mRNAs, we compared the levels of yqaE, ompX, nadE, and luxS mRNAs in cells taken from log-phase, late-log-phase, and stationary-phase cultures of a cyaR+ strain, DJ624, and its ΔcyaR derivative NRD359 by Northern blotting. Consistent with our observation that CyaR expression is highest during late log phase, the levels of all four of these mRNAs were much lower in strain DJ624 than in its ΔcyaR derivative during late log phase, while no significant differences were observed during the log and stationary phases (Fig. 3C). No significant differences in the levels of the ompA mRNA, which was used as a control in these experiments, were observed between the strains in any growth phase (data not shown).
The negative regulation of yqaE, ompX, luxS, and nadE by CyaR occurs at the posttranscriptional level and is direct.
To determine whether yqaE, luxS, and nadE were negatively regulated by CyaR at the posttranscriptional level, we constructed translational lacZ fusions to these genes and placed these fusions under the control of a different promoter, the araBAD promoter. Replacing the normal promoter of yqaE, luxS, and nadE with the araBAD promoter allowed us to decouple posttranscriptional regulation of these genes from transcriptional regulation and to control the expression of the fusions by altering the concentration of arabinose in the medium. A strain harboring a lambda prophage carrying a lacZ translational fusion to ompX was obtained from Maude Guillier and was used to examine whether ompX was negatively regulated by CyaR. The ompX-′lacZ translational fusion in this strain was also under the control of the araBAD promoter, but it still had its native proximal promoter; the two promoters of ompX were previously determined to be at positions −220 and −50 relative to the start codon (40).
Since we did not want to affect the posttranscriptional regulation of yqaE, luxS, and nadE by altering the 5′ end of the mRNAs that normally encode these genes, we mapped the 5′ end of each of the mRNAs by using 5′ RACE and then constructed the fusions in such a way as to allow transcription from the araBAD promoter to begin at the mapped position 1 of the genes. The transcription start sites of yqaE, luxS, and nadE were mapped to positions −57, −83, and −22 relative to the initiation of translation, respectively. Surprisingly, the transcription start site for luxS is within the divergent gene for MicA, an sRNA that negatively regulates expression of the major outer membrane protein OmpA (62). As a result, the first 15 bp of the luxS mRNA is complementary to the first 15 bp of the MicA sRNA, an unusual arrangement.
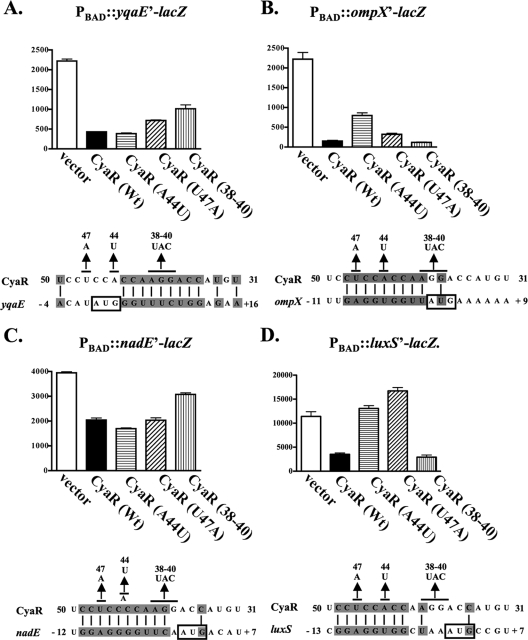
lacZ translational fusions to yqaE, luxS, and nadE were constructed and placed in the chromosome under control of the araBAD promoter such that transcription began at the mapped transcriptional start site for these genes. As mentioned above, the ompX translational lacZ fusion is under the control of its native proximal promoter. The fusion genes were expressed in a ΔaraBAD ΔcyaR strain harboring either an empty vector (pBR-plac) or a plasmid expressing CyaR from a lac promoter (pNRD405), and the amounts of translated fusion proteins were determined by β-galactosidase activity assays. Expression of CyaR from the plasmid reduced the translation of yqaE, nadE, luxS, and ompX 4.7-, 1.7-, 5.2-, and 14.3-fold, respectively (Fig. 4).
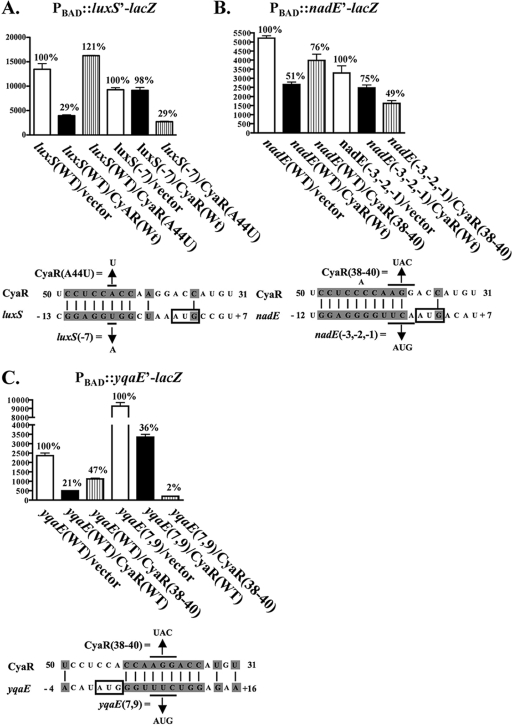
FIG. 4.
β-Galactosidase assays of yqaE-lacZ, ompX-lacZ, nadE-lacZ, and luxS-lacZ translational fusions in the presence or absence of wild-type or mutant CyaR expression. Overnight cultures of (A) NRD400 (yqaE-lacZ fusion), (B) NRD377 (ompX-lacZ fusion), (C) NRD399 (nadE-lacZ fusion), or (D) NRD413 (luxS-lacZ fusion) harboring pBR-plac (vector), pNRD405 (wild-type cyaR [Wt]), pNRD407 (cyaR A44T), pNRD408 (cyaR T47A), or pNRD410 (cyaR G38C G39A A40T [CyaR (38-40]) were diluted 200-fold in fresh LB medium containing IPTG (100 μM), ampicillin (100 mg liter−1), and arabinose (0.01%) or (in the case of NRD377) no arabinose. These strains were then incubated at 37°C for 6 h. The OD600 of each culture was determined, and β-galactosidase assays were performed as described in Materials and Methods. The data are the averages of three independent experiments. Below the graphs showing the results of the β-galactosidase activity assays are diagrams showing the predicted pairing between CyaR and yqaE (A), ompX (B), nadE (C), or luxS (D) mRNA. The start codon of yqaE, ompX, nadE, and luxS is indicated by a box, and the base substitutions in CyaR are indicated above the CyaR sequence.
The sequence of CyaR and the 5′ end of mRNAs encoding yqaE, nadE, luxS, and ompX were examined, and possible regions of base pairing were determined (see Fig. S1 in the supplemental material). CyaR is predicted to base pair with ompX, luxS, and nadE mRNAs in a region that includes the ribosome binding site of these genes, whereas CyaR is predicted to base pair with the yqaE-encoding mRNA immediately downstream of the start codon for yqaE. Based on these predictions of base pairing, several site-directed cyaR mutants were constructed by using QuikChange site-directed mutagenesis and the template plasmid pNRD405. The resulting mutants with the A44U, U47A, and U50A single mutations and the G38C G39A A40U triple mutation in CyaR were then tested to determine their abilities to negatively regulate the translational fusions as described above. The U50A change was not predicted to disrupt base pairing with any of the four targets (see Fig. S1 in the supplemental material), and, consistent with this prediction, a plasmid expressing this mutant CyaR was able to downregulate all four fusions (see Fig. S2 in the supplemental material). The A44U substitution in CyaR reduced the negative regulation of ompX and eliminated the negative regulation of luxS (Fig. 4B and 4D). Papenfort et al. recently reported that an A44U substitution in CyaR also reduces the ability of this molecule to negatively regulate ompX in S. enterica serovar Typhimurium (49). The U47A substitution in CyaR also eliminated the ability of CyaR to negatively regulate luxS, but it had less effect on ompX (Fig. 4B and 4D). Both the A44U and U47A CyaR mutants were still able to negatively regulate the yqaE and nadE mRNAs (Fig. 4A and 4C). In contrast, the G38C G39A A40U triple substitution (Fig. 4) reduced the ability of CyaR to negatively regulate yqaE and nadE expression and had no effect on the ability of CyaR to negatively regulate luxS and ompX expression. Since each of the substitutions in CyaR (A44U, U47A, or G38C G39A A40U) reduced the expression of only a subset of the four mRNAs regulated by CyaR, these mutations in CyaR most likely acted by disrupting base pairing between CyaR and the affected mRNAs rather than by reducing active CyaR levels. However, it is also worth noting that these mutations did not totally eliminate regulation, except regulation of luxS.
To confirm that the A44U substitution in CyaR disrupts the base pairing between CyaR and the luxS mRNA, a mutant with a compensatory mutation in the luxS-lacZ translational fusion was prepared and assessed as described above. As predicted, the compensatory mutation in the luxS-lacZ fusion restored the ability of the A44U CyaR mutant and eliminated the ability of wild-type CyaR to negatively regulate luxS expression (Fig. 5A), consistent with our model for pairing. A mutation in ompX that would have compensated for the A44U substitution in CyaR was not constructed, since Papenfort et al. recently reported that a compensatory mutation in the predicted pairing region in ompX restored the ability of the A44U CyaR mutant to negatively regulate ompX in S. enterica serovar Typhimurium (49).
FIG. 5.
β-Galactosidase assays measuring the effect of compensatory mutations in the translation initiation regions of luxS, nadE, and yqaE on restoration of negative regulation by CyaR. Overnight cultures of (A) NRD413 (luxS-lacZ fusion), NRD415 [luxS (T-7A)-lacZ fusion], (B) NRD399 (nadE-lacZ fusion), NRD410 [nadE (T-3A; C-2T; A-1G)-lacZ fusion], (C) NRD400 (yqaE-lacZ fusion), or NRD411 [yqaE (T7A;C9G)-lacZ fusion] harboring pBR-plac (vector), pNRD405 (wild-type cyaR), pNRD407 (cyaR A44T), or pNRD410 (cyaR G38C G39A A40T [CyaR(38-40)]) were diluted 200-fold in fresh LB liquid medium containing IPTG (100 μΜ), ampicillin (100 mg liter−1), and arabinose (0.01%). After the strains were incubated at 37°C for 6 h, the OD600 of each culture was determined, and β-galactosidase assays were performed as described in Materials and Methods. The variant of the lacZ translational fusion examined and the variant of CyaR expressed, if any (bases changed in the lacZ translational fusion or in cyaR are indicated), are indicated on the x axis. The percentage of β-galactosidase activity relative to the activity of the strain harboring the empty vector is indicated above each bar. Below the graphs showing the results of the β-galactosidase assays are diagrams showing the predicted pairing between CyaR and the mRNA encoding luxS (A), nadE (B), or yqaE (C) and the base substitutions in cyaR and the compensatory base changes made in the mRNAs encoding the genes. The start codon of each gene is indicated by a box. The results are averages of two independent experiments. Wt and WT, wild type.
Compensatory mutations for the G38C G39A A44U mutant CyaR were also constructed in the predicted pairing region of the nadE and yqaE mRNAs. As predicted, these compensatory mutations increased the ability of the G38C G39A A44U mutant CyaR and decreased the ability of the wild-type CyaR to negatively regulate the nadE-lacZ and yqaE-lacZ fusions (Fig. 5B and C). In fact, the triple mutation and compensating mutations led to regulation of yqaE that was tighter than the regulation observed for the wild-type molecule (Fig. 5C).
Expression of CyaR results in a decrease in autoinducer production.
Since expression of CyaR results in a dramatic decrease in the translation of luxS encoding autoinducer-2 synthase (Fig. 4D), we examined the effect of CyaR expression on the production of autoinducer-2 by E. coli. Autoinducer-2 levels are measured by collecting culture filtrates from the producer strain and measuring their abilities to activate expression of the lux genes in V. harveyi (55).
Strain NRD359, a derivative of strain DJ624 that harbors a cyaR deletion, was transformed with pBR-plac or pNRD405, which transcribes cyaR from a lac promoter. The transformed strains were grown at 30°C, and culture filtrates were prepared from each culture every hour after inoculation as described in Materials and Methods. The relative amounts of autoinducer-2 in culture filtrates were then determined by measuring the amount of light produced by V. harveyi strain BB170, a response to the concentration of autoinducer-2 present in the culture filtrate (56).
As NRD359/pBR-plac and NRD359/pNRD405 approached late log phase (4 h postinoculation), autoinducer-2 could be detected in the cell-free culture filtrates of these strains, and the autoinducer-2 levels peaked at 5 h postinoculation (Fig. 6A and 6B). When the peak autoinducer-2 levels were present, the levels were 2.8-fold lower in the strain expressing cyaR (NRD359/pNRD405) than in the strain lacking cyaR (NRD359/pBR-plac) (Fig. 6B), consistent with regulation of luxS by CyaR. These results were obtained in two independent experiments.
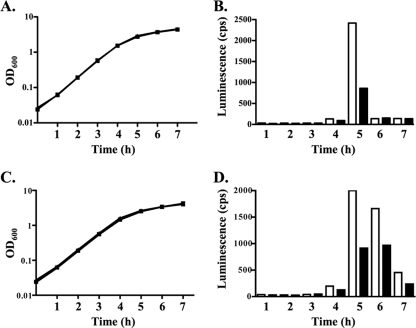
FIG. 6.
Comparison of autoinducer-2 production in the presence and absence of CyaR expression. (A and C) Growth at 30°C of the ΔcyaR strain NRD359 (A) or the ΔlsrACDBFG ΔcyaR strain NRD392 (C) harboring the vector pBR-plac (squares) or the cyaR-expressing plasmid pNRD405 (triangles). (B and D) Autoinducer-2 production by NRD359/pBR-plac (open bars) and NRD359/pNRD405 (filled bars) (B) or by NRD392 pBR-plac (open bars) and NRD392/pNRD405 (filled bars) (D). Overnight cultures of strains NRD359/pBR-plac, NRD359/pNRD405, NRD392/pBR-plac, and NRD392/pNRD405 grown in LB medium containing ampicillin were pelleted, the supernatants were removed, and the cells were resuspended in LB medium. The washed cells were then diluted 200-fold in LB medium containing IPTG and incubated at 37°C. Every hour after inoculation the OD600 of each culture was determined and samples were removed. Cell-free culture filtrates were then prepared, and the relative amounts of autoinducer-2 were determined as described in Materials and Methods. Each experiment was performed twice, and the data are representative of the two experiments.
To confirm this significant difference in autoinducer-2 production in the presence and absence of CyaR expression, we constructed strain NRD392, a derivative of strain DJ624 that harbors an lsrACDBFG deletion and a cyaR deletion. lsrACDBFG encodes a transporter that mediates the uptake of autoinducer-2 from the medium; the transcription of lsrACDBFG is induced by autoinducer-2 (57). Deletion of lsrACDBFG reduces the uptake of autoinducer-2 by E. coli (57) and thus allows higher levels of autoinducer-2 to remain in the culture medium for a longer period of time. We transformed NRD392 with pBR-plac or pNRD405 and grew the resulting strains at 30°C. We collected cell-free culture filtrates and measured the autoinducer-2 levels with the V. harveyi BB170 reporter strain as described above.
At late log phase, autoinducer-2 could be detected in the culture filtrates of strains NRD392/pBRplac and NRD392/pNRD405 (Fig. 6C and 6D), and at 5 h postinoculation the autoinducer-2 levels were 2.2-fold lower in the culture filtrates of the strain expressing cyaR (NRD392/pNRD405) than in the culture filtrates of the strain lacking cyaR (NRD392/pBR-plac). Moreover, a significant difference in autoinducer levels between the culture filtrates of the strain expressing cyaR and the culture filtrates of the strain lacking cyaR was still observed 7 h after inoculation. A similar difference in autoinducer levels between strains NRD359/pBR-plac and NRD359/pNRD405 was observed in an independent experiment (data not shown).
We also examined the production of autoinducer-2 in a wild-type strain of E. coli (DJ624) and a ΔcyaR derivative of this strain by growing these strains at 30°C in LB medium, preparing cell-free culture filtrates, and measuring the level of autoinducer-2 as described above. No significant difference in autoinducer-2 production between the wild-type strain and its ΔcyaR derivative was found (data not shown). This may not be surprising given the transient expression of CyaR in E. coli strains grown in LB medium (Fig. 3C).
DISCUSSION
We showed that the expression of the RyeE sRNA, renamed CyaR here, is activated by Crp under conditions in which cAMP levels are high and that CyaR directly negatively regulates the expression of several genes, including yqaE, ompX, nadE, and luxS, at the posttranscriptional level.
Crp is one of the best-studied regulatory proteins. It has been shown that when cAMP is present, Crp activates the expression of numerous genes which are primarily involved in the transport and catabolism of sugars and amino acids (19, 72, 73). In E. coli, the synthesis of cAMP by adenylate cyclase is inhibited by import of the preferred catabolite glucose, and the levels of cAMP and therefore the DNA binding activity of Crp are high when poor carbon sources are available and low when glucose is available (25). Consistent with this pattern, expression of a cyaR-lac fusion (Fig. 2A) is increased in LB medium compared to the expression in LB medium containing glucose, and expression is completely dependent upon both cya (for synthesis of cAMP) and crp (Fig. 2B and C). Additionally, upstream of cyaR and overlapping the predicted −35 region of the cyaR promoter in E. coli K-12 is a conserved sequence similar to the consensus sequence of a Crp binding site; in our experiments, alteration of this Crp binding site resulted in a drastic decrease in expression from the cyaR promoter. In parallel experiments with Salmonella, the cyaR promoter was shown to directly bind Crp, further supporting the hypothesis that there is direct regulation by cAMP and Crp (49). Another sRNA, Spot 42, is negatively regulated by cAMP and Crp (44), and Spot 42 downregulates expression of galK. This has been interpreted as a mechanism for adjusting expression of Gal epimerase, which is needed for cell wall synthesis, relative to expression of Gal kinase, which is needed only for galactose metabolism, when glucose rather than galactose is used by cells as a carbon source.
CyaR is an Hfq-binding RNA, and other sRNAs have been shown to both positively and negatively regulate gene expression by pairing with specific mRNA targets. The results of microarray experiments, Northern blotting, and experiments with translational fusions all are consistent with the hypothesis that CyaR negatively regulates ompX, yqaE, nadE, and luxS at a posttranscriptional level by pairing with the mRNA. As mentioned above, several of these genes that have been shown to be negatively regulated by CyaR, including luxS and ompX, have also been shown to be negatively regulated by Crp (19, 65). Our results are consistent with these findings and provide a mechanism for the negative regulation of these genes via Crp activation of CyaR.
A large number of outer membrane proteins have been shown to be subject to regulation by Hfq-binding small RNAs (8, 11, 20, 27, 43, 50, 59, 62, 64). Some of this regulation serves as a feedback regulatory mechanism for the sigma E regulon (27, 50, 59), while in other cases (20) it is unclear what the functions of the regulatory effects are. OmpX, an outer membrane protein with an unknown function, had been suspected to be regulated by an sRNA, because the level of ompX mRNA increases significantly in an hfq mutant (22). Our work and recently described work of Papenfort et al. (49) and Johansen et al. (26) clearly confirmed this and demonstrated that CyaR regulates OmpX in both E. coli and Salmonella, thus leading to downregulation of OmpX in the presence of a poor carbon source or under low-glucose conditions.
ompX has a long untranslated leader, and the primary region of pairing is predicted to be positions −2 to −9 relative to the translation start site of ompX (see Fig. S1 in the supplemental material). A translational fusion containing 241 nucleotides upstream of the start site and 30 nucleotides of the ompX coding region was downregulated sixfold when CyaR was overexpressed from a lac promoter (Fig. 4B). Therefore, the 5′ end of ompX extending from the transcription start site to the 10th codon of the gene is sufficient for regulation; this region contains the predicted pairing region. The results of experiments carried out by Papenfort et al. to examine CyaR regulation of OmpX in Salmonella (49) are consistent with this predicted pairing. The A44U mutation in CyaR previously shown by Papenfort et al. to disrupt pairing between CyaR and OmpX in Salmonella had similar effects in E. coli (Fig. 4B).
OmpX belongs to a family of outer membrane proteins that have been implicated in promoting bacterial adhesion and entry into and survival in mammalian cells. While the data for E. coli are not compelling (deletion of ompX did not affect adherence to human HEp-2 cells and indirectly stimulated adherence [40, 48]), in Enterobacter cloacae overexpression of OmpX increased invasion of rabbit illeal tissue (13). OmpX of E. cloacae has 83% protein identity to E. coli OmpX, and the proposed binding site for cyaR upstream of ompX is conserved in E. cloacae. Therefore, it seems reasonable to predict that the regulation of OmpX by CyaR has a role in downregulating adherance under low-glucose conditions. This could be part of a starvation response, for instance, in which cells should move to new environments in search of nutrients.
Another target of CyaR that is also involved in the behavior of bacteria at cell surfaces and during infection is luxS. LuxS synthesizes autoinducer-2 (56), a signal molecule used in quorum sensing in E. coli and a wide range of bacteria (5, 52). In E. coli, autoinducer-2 influences the expression of many genes, including genes involved in transport and processing of the autoinducer, in motility, and in biofilm development (14, 18, 33, 65). Autoinducer-2 has been shown to increase the mass and thickness of biofilms (18, 33). LuxS activity also impacts methionine metabolism, since homocysteine, a precursor of methionine, is generated in the reaction catalyzed by LuxS that produces autoinducer-2 (52).
The luxS mRNA has a long 5′ untranslated region that begins 83 bp upstream of the AUG start codon. CyaR has a region of base pairing with the luxS 5′ untranslated region that overlaps the ribosome binding site and involves the same region of CyaR used for ompX regulation; this region is conserved in the luxS leader (see Fig. S1 in the supplemental material). Consistent with this, the A44U mutation that disrupts pairing between CyaR and the ompX mRNA also disrupts pairing between CyaR and the luxS mRNA (Fig. 4D), and a compensatory mutation in the pairing region of the luxS mRNA restores negative regulation by the mutant form of CyaR (Fig. 5A). The effect of downregulation of luxS mRNA by CyaR overexpression is a decrease in the amount of autoinducer-2 produced by E. coli (Fig. 6B and 6D). In E. coli, autoinducer-2 simulates biofilm formation. Thus, as observed for ompX, it seems possible that downregulation of luxS under glucose-limiting conditions may help cells decrease biofilm formation, increase planktonic behavior, and move in search of nutrients.
The sequence encoding the long 5′ end of the luxS mRNA extends into micA. As a result, the luxS mRNA and the MicA sRNA share 15 bp of complementary sequence. Thus, the sigma E-regulated promoter of micA lies within the sequence encoding the luxS mRNA, and at least some of the promoter of luxS must lie within micA. A possible consequence of this arrangement is that expression from the promoter of one of these genes may interfere with expression of the other gene. Another potential consequence of this arrangement is that the 15-bp complementarity between these genes may facilitate the posttranscriptional regulation of luxS by MicA. It is interesting that the same region of MicA was suggested by Johansen et al. to base pair with the ompX mRNA, which these authors identified as a target of negative regulation by MicA and which is negatively regulated by CyaR (26).
Another confirmed target, nadE, is an essential gene whose product uses ammonia to catalyze the last step in NAD synthesis (24, 53, 69). A sequence immediately upstream of nadE that has the potential to pair with CyaR was found; however, this sequence is not well conserved in other closely related enterobacteria, including S. enterica serovar Typhimurium and S. enterica serovar Typhi (Fig. 1), even though the region of CyaR that pairs with this sequence is absolutely conserved in these species (Fig. 1). Thus, nadE may not be a target of negative regulation by CyaR in these other species of enterobacteria. Nonetheless, the results of our experiments with the translational lacZ fusion to nadE demonstrated that the regulation of nadE by CyaR is posttranscriptional and is direct (Fig. 5B). None of the other genes in the pathway for NAD biosynthesis or recycling (nadC, nadD, nadA, nadB, and nadR) showed any significant effect of CyaR overproduction in our arrays, suggesting that it is not NAD synthesis per se, but possibly limiting NadE ammonia use under limiting C or energy source conditions, that is the physiological basis for this regulation. This is not the first example of a protein involved in nitrogen metabolism that is regulated by catabolite repression. Crp was previously shown to decrease the expression of glutamine synthetase (36, 60), which catalyzes the synthesis of glutamine from glutamate and ammonia, and to activate the expression of GlnHPQ, a high-affinity glutamine transporter (36). Possibly because nadE is an essential gene, the downregulation of nadE by CyaR is less dramatic than the downregulation seen with other targets (Fig. 5). While it is unusual for targets of these small RNAs to be essential, one essential gene, ftsZ, has previously been shown to be regulated by the small RNA DicF (58).
The fourth gene for which there is strong and reproducible regulation by CyaR is the gene encoding YqaE, a highly conserved predicted membrane protein with an unknown function. In plants, this gene has been found to be a stress-induced gene that is expressed at higher levels at low temperatures and under salt stress and dehydration conditions (41). There is some evidence that in E. coli yqaE may be regulated by RpoS; the level of expression of yqaE is higher in a wild-type strain than in an rpoS mutant when the organisms are challenged with a hyperosmotic shock (nearly fivefold higher) or a decrease in the pH of the growth medium (threefold higher) (67). There is a substantial amount of pairing between CyaR and the region immediately downstream of the start codon of yqaE in E. coli that is highly conserved (see Fig. S1 in the supplemental material). This pairing between CyaR and YqaE has been confirmed (Fig. 5C).
As mentioned above, the sequences upstream of ompX, nadE, and luxS that are capable of pairing with CyaR overlap the ribosome binding sites of these genes, while the predicted pairing region of yqaE is immediately downstream of its start codon. The extent of base pairing between CyaR and the translation initiation regions of ompX, luxS, nadE, and yqaE is substantial, and the base pairing should efficiently block ribosome binding to these mRNAs when the level of CyaR expression is high.
The base pairing tests whose results are shown in Fig. 4 and 5 provided some hints about the complexity and redundancy of regulation by sRNAs of the sort examined here. The lengths of the predicted pairing “seed” (continuous pairing section) are 10 bp for nadE, 9 bp for yqaE and ompX, and 8 bp for luxS. Disrupting the seed by a single nucleotide change did not change regulation for nadE (Fig. 4C), changed regulation modestly for ompX (Fig. 4B), and changed regulation drastically for luxS (Fig. 4D and 5A). Even a change of 3 nucleotides led to only a partial release of regulation for yqaE (Fig. 4A and 5C) and nadE (Fig. 5B). While all of these experiments were done under CyaR overproduction conditions and it is possible that CyaR can pair elsewhere on the mRNA, it seems clear that there is not an absolute requirement for extensive seed pairing. Why luxS is particularly sensitive to single nucleotide changes remains to be explored. The results of further in vivo pairing tests should help in the development of a more complete understanding of sRNA function.
As mentioned above, Crp has been shown to affect the expression of many genes. Most of the genes that have been shown to be directly regulated by Crp are involved in the transport or catabolism of sugars or amino acids. Interestingly, all of the genes identified here as genes that are indirectly regulated by Crp via the CyaR sRNA are involved in processes other than sugar and amino acid transport and catabolism. The genes that are indirectly negatively regulated by Crp via the CyaR sRNA are involved in or associated with other cellular processes, such as adherance (ompX), NAD biosynthesis (nadE), and quorum sensing (luxS). The function of the predicted membrane protein encoded by yqaE remains unknown.
While E. coli utilizes Crp to directly regulate transport and catabolism of sugars and amino acids and utilizes CyaR to indirectly regulate other processes via Crp, other species of nonenteric gammaproteobacteria, such as Pseudomonas aeruginosa and Pseudomonas putida, divide the regulation of these different processes between two different regulatory proteins, Vfr and Crc. P. aeruginosa Vfr is 91% similar to the E. coli Crp protein, binds cAMP, and binds to a DNA sequence that is similar to, but significantly different from, the consensus Crp binding sites (30). Vfr directly regulates the expression of genes involved in the production of exotoxin A, protease production, quorum sensing, type IV pilus formation, and twitching motility (1, 7, 30, 68). On the other hand, Crc is responsible for repressing the expression of genes involved in the catabolism of nonideal catabolites, when ideal catabolites, including tricarboxylic acid intermediates, are present (23, 45, 46, 70). Crc has been shown to repress the expression of genes by a cAMP-independent mechanism.
Why does E. coli regulate some genes directly by Crp and other genes indirectly by Crp via the CyaR sRNA? Certainly most of the genes directly regulated by Crp are positively regulated, while the genes described here are negatively regulated; it may be easier to evolve negative regulation by this activator protein via an sRNA intermediate. Another possibility is that this division of labor allows E. coli to rapidly decrease expression of a subset of genes by actively destroying the mRNAs, which may be particularly important for genes with longer mRNA half-lives, although this possibility has not been specifically investigated for these targets. Another possibility is that the process of regulating genes through CyaR sRNA provides E. coli with another way of fine-tuning the expression of its target genes in response to different factors.
cyaR as a phage attachment site.
The cyaR gene of E. coli and its upstream binding site for Crp is highly conserved in other enterobacteria (49), and it is downstream of a conserved gene that is designated yegQ in E. coli and Salmonella. However, Papenfort et al. noted that the gene downstream of cyaR varies (49). The variation downstream is likely the result of a second function of the cyaR gene, as a phage attachment site. As originally noted by Wassarman et al. (66), the 3′ end of the small RNA overlaps an attachment site for phage P2, and E. coli K-12 has a fragment of a cryptic prophage at this site. While the att site is clearly conserved (Fig. 1), the prophage is not present in some enterobacteria, while a large number of prophage genes are present in other enterobacteria (for instance, avian pathogeneic E. coli [28]). In at least one strain, a P2 prophage also has introduced a restriction/modification gene cluster at this location (32). Many tRNAs serve as phage attachment sites, as does tmRNA (ssrA) (9, 31), and recently the sRNA RyeB was reported to serve as an integration site for phages in S. enterica serovar Typhimurium (4). With the addition of CyaR, it is clear that sRNAs, like tRNAs, are bacteriophage attachment sites.
It has been suggested that a conserved sequence and/or structure associated with tRNAs may be an attractive conserved site for phage lysogenization (9, 31). Certainly, the conservation of cyaR in a broad range of enterobacterial species suggests that phages related to P2 may make use of this site in all of these species.
Supplementary Material
Acknowledgments
We thank B. L. Bassler for providing the V. harveyi strain and the protocol for autoinducer-2 detection. We thank P. Mandin for providing the strain and protocol for constructing translational lacZ fusions, advice, and editorial comments. We also thank C. Vanderpool and M. Guillier for providing strains. We are grateful to K. Moon, P. Milanesio, M. Guillier, N. Majdalani, and G. Storz for their advice, as well as their editorial comments.
This work was supported by the Intramural Research Program of the NIH National Cancer Institute Center for Cancer Research.
Footnotes
Published ahead of print on 31 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. H. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1793928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11941-950. [DOI] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Balbontín, R., N. Figueroa-Bossi, J. Casadesus, and L. Bossi. 2008. Insertion hot spot for horizontally acquired DNA within a bidirectional small-RNA locus in Salmonella enterica. J. Bacteriol. 1904075-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 1794043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9773-786. [DOI] [PubMed] [Google Scholar]
- 7.Beatson, S. A., C. B. Whitchurch, J. L. Sargent, R. C. Levesque, and J. S. Mattick. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 1843605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossi, L., and N. Figueroa-Bossi. 2007. A small RNA downregulates LamB maltoporin in Salmonella. Mol. Microbiol. 65799-810. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, A. 2003. Prophage insertion sites. Res. Microbiol. 154277-282. [DOI] [PubMed] [Google Scholar]
- 10.Chen, S., E. A. Lesnik, T. A. Hall, R. Sampath, R. H. Griffey, D. J. Ecker, and L. B. Blyn. 2002. A bioinformatic based approach to discover small RNA genes in the Escherichia coli genome. BioSystems 65157-177. [DOI] [PubMed] [Google Scholar]
- 11.Chen, S., A. Zhang, L. B. Blyn, and G. Storz. 2004. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 1866689-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Kort, G., A. Bolton, G. Martin, J. Stephen, and J. A. M. van de Klundert. 1994. Invasion of rabbit ileal tissue by Enterobacter cloacae varies with the concentration of OmpX in the outer membrane. Infect. Immun. 624722-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 1835239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebright, R. H., Y. W. Ebright, and A. Gunasekera. 1989. Consensus DNA site for the Escherichia coli catabolite activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res. 1710295-10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figueroa-Bossi, N., S. Lemire, D. Maloriol, R. Balbontin, J. Casadesus, and L. Bossi. 2006. Loss of Hfq activates the σE-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 62838-852. [DOI] [PubMed] [Google Scholar]
- 17.Gaston, K., B. Chan, A. Kolb, J. Fox, and S. Busby. 1988. Alterations in the binding site of the cyclic AMP receptor protein at the Escherichia coli galactose operon regulatory region. Biochem. J. 253809-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez Barrios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gosset, G., Z. Zhang, S. Nayyar, W. A. Cuevas, and M. H. Saier, Jr. 2004. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J. Bacteriol. 1863516-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillier, M., and S. Gottesman. 2006. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 59231-247. [DOI] [PubMed] [Google Scholar]
- 21.Guillier, M., S. Gottesman, and G. Storz. 2006. Modulating the outer membrane with small RNAs. Genes Dev. 202338-2348. [DOI] [PubMed] [Google Scholar]
- 22.Guisbert, E., V. A. Rhodius, N. Ahuja, E. Witkin, and C. A. Gross. 2007. Hfq modulates the σE-mediated envelope stress response and the σ32-mediated cytoplasmic stress response in Escherichia coli. J. Bacteriol. 1891963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hester, K. L., J. Lehman, F. Najar, L. Song, B. A. Roe, C. H. MacGregor, P. Hager, P. V. Phibbs, Jr., and J. R. Sokatch. 2000. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 1821144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes, K. T., B. M. Olivera, and J. R. Roth. 1988. Structural gene for NAD synthetase in Salmonella typhimurium. J. Bacteriol. 1702113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishizuka, H., A. Hanamura, T. Kunimura, and H. Aiba. 1993. A lowered concentration of cAMP receptor protein caused by glucose is an important determinant for catabolite repression in Escherichia coli. Mol. Microbiol. 10341-350. [DOI] [PubMed] [Google Scholar]
- 26.Johansen, J., M. Eriksen, B. Kallipolitis, and P. Valentin-Hansen. 2008. Down-regulation of outer membrane proteins by non-coding RNAs: unravelling the cAMP-CRP- and σE-dependent CyaR-ompX regulatory case. J. Mol. Biol. 3831-9. [DOI] [PubMed] [Google Scholar]
- 27.Johansen, J., A. A. Rasmussen, M. Overgaard, and P. Valentin-Hansen. 2006. Conserved small non-coding RNAs that belong to the σE regulon: role in down-regulation of outer membrane proteins J. Mol. Biol. 3641-8. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, T. J., S. Kariyawasam, Y. Wannemuehler, P. Mangiamele, S. J. Johnson, C. Doetkott, J. A. Skyberg, A. M. Lynne, J. R. Johnson, and L. K. Nolan. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 1893228-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajitani, M., and A. Ishihama. 1991. Identification and sequence determination of the host factor gene for bacteriophage QB. Nucleic Acids Res. 191063-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanack, K. J., L. J. Runyen-Janecky, E. P. Ferrell, S. J. Suh, and S. E. H. West. 2006. Characterization of DNA-binding specificity and analysis of binding sites of Pseudomonas aeruginosa global regulator, Vfr, a homologue of the Escherichia coli cAMP receptor protein. Microbiology 1523485-3496. [DOI] [PubMed] [Google Scholar]
- 31.Kirby, J. E., J. E. Trempy, and S. Gottesman. 1994. Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J. Bacteriol. 1762068-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kita, K., H. Kawakami, and H. Tanaka. 2003. Evidence for horizontal transfer of the EcoT38I restriction-modification gene to chromosomal DNA by the P2 phage and diversity of defective P2 prophages in Escherichia coli TH38 strains. J. Bacteriol. 1852296-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, J., C. Attila, L. Wang, T. K. Wood, J. J. Valdes, and W. E. Bentley. 2007. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J. Bacteriol. 1896011-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majdalani, N., D. Hernandez, and S. Gottesman. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46813-826. [DOI] [PubMed] [Google Scholar]
- 35.Majdalani, N., C. K. Vanderpool, and S. Gottesman. 2005. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 4093-113. [DOI] [PubMed] [Google Scholar]
- 36.Mao, X. J., Y. X. Huo, M. Buck, A. Kolb, and Y. P. Wang. 2007. Interplay between CRP-cAMP and PII-Ntr systems forms novel regulatory network between carbon metabolism and nitrogen assimilation in Escherichia coli. Nucleic Acids Res. 351432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massé, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 172374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 994620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massé, E., C. K. Vanderpool, and S. Gottesman. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 1876962-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mecsas, J., R. Welch, J. W. Erickson, and C. A. Gross. 1995. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J. Bacteriol. 177799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medina, J., R. Catalá, and J. Salinas. 2001. Developmental and stress regulation of RCI2A and RCI2B, two cold-inducible genes of Arabidopsis encoding highly conserved hydrophobic proteins. Plant Physiol. 1251655-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Mizuno, T., M. Y. Chou, and M. Inouye. 1984. A unique mechanism regulating gene expression: translation inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. USA 811966-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Møller, T., T. Franch, C. Udesen, K. Gerdes, and P. Valentin-Hansen. 2002. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 161696-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morales, G., J. F. Linares, A. Beloso, J. P. Albar, J. L. Martinez, and F. Rojo. 2004. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 1861337-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreno, R., A. Ruiz-Manzano, L. Yuste, and F. Rojo. 2007. The Pseudomonas putida Crc global regulator is an RNA binding protein that inhibits translation of the AlkS transcriptional regulator. Mol. Microbiol. 64665-675. [DOI] [PubMed] [Google Scholar]
- 47.Morita, T., K. Shigesada, F. Kimizuka, and H. Aiba. 1988. Regulatory effect of a synthetic CRP recognition sequence placed downstream of a promoter. Nucleic Acids Res. 167315-7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otto, K., and M. Hermansson. 2004. Inactivation of ompX causes increased interactions of type 1 fimbriated Escherichia coli with abiotic surfaces. J. Bacteriol. 186226-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papenfort, K., V. Pfeiffer, A. Sonawane, J. C. D. Hinton, and J. Vogel. 2008. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol. Microbiol. 68890-906. [DOI] [PubMed] [Google Scholar]
- 50.Papenfort, K., V. Pfeiffer, F. Mika, S. Lucchini, J. C. D. Hinton, and J. Vogel. 2006. σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 621674-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saier, M. H., Jr. 1998. Multiple mechanisms controlling carbon metabolism in bacteria. Biotechnol. Bioeng. 58170-174. [DOI] [PubMed] [Google Scholar]
- 52.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing molecule. Mol. Microbiol. 41463-476. [DOI] [PubMed] [Google Scholar]
- 53.Schneider, B. L., and L. J. Reitzer. 1998. Salmonella typhimurium nit is nadE: defective nitrogen utilization and ammonia-dependent NAD synthetase. J. Bacteriol. 1804739-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 5385-96. [DOI] [PubMed] [Google Scholar]
- 55.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 957046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 961639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42777-793. [DOI] [PubMed] [Google Scholar]
- 58.Tétart, F., and J. P. Bouché. 1992. Regulation of the expression of the cell-cycle gene ftsZ by DicF antisense RNA. Division does not require a fixed number of FtsZ molecules. Mol. Microbiol. 6615-620. [DOI] [PubMed] [Google Scholar]
- 59.Thompson, K. M., V. A. Rhodius, and S. Gottesman. 2007. σE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 1894243-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tian, Z. Y., Q. S. Li, M. Buck, A. Kolb, and Y. P. Wang. 2001. The CRP-cAMP complex and downregulation of the glnAp2 promoter provides a novel regulatory linkage between carbon metabolism and nitrogen assimilation in Escherichia coli. Mol. Microbiol. 41911-924. [DOI] [PubMed] [Google Scholar]
- 61.Tsui, H. C., H. C. Leung, E., and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1335-49. [DOI] [PubMed] [Google Scholar]
- 62.Udekwu, K. I., F. Darfeuille, J. Vogel, J. Reimegard, E. Holmqvist, and E. G. Wagner. 2005. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 192355-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valentin-Hansen, P., M. Eriksen, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 511525-1533. [DOI] [PubMed] [Google Scholar]
- 64.Vanderpool, C. K., and S. Gottesman. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 541076-1089. [DOI] [PubMed] [Google Scholar]
- 65.Wang, L., Y. Hashimoto, C. Y. Tsao, J. J. Valdes, and W. E. Bentley. 2005. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 1872066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 151637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weber, J., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 1871591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.West, S. E. H., A. K. Sample, and L. J. Runyen-Janecky. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 1767532-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willison, J. C., and G. Tissot. 1994. The Escherichia coli efg gene and the Rhodobacter capsulatus adgA gene code for NH3-dependent NAD synthetase. J. Bacteriol. 1763400-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolff, J. A., C. H. MacGregor, R. C. Eisenberg, and P. V. Phibs. 1991. Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J. Bacteriol. 1734700-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, A., K. M. Wassarman, C. Rosenow, B. C. Tjaden, G. Storz, and S. Gottesman. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 501111-1124. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, Z., G. Gosset, R. Barabote, C. S. Gonzalez, W. A. Cuevas, and M. H. Saier, Jr. 2005. Functional interactions between the carbon and iron utilization regulators, Crp and Fur, in Escherichia coli. J. Bacteriol. 187980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng, D., C. Constantinidou, J. L. Hobman, and S. D. Minchin. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 325874-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.