Abstract
Streptococcus pneumoniae normally resides in the human nasopharynx in a nondisease state. In response to unknown triggers this organism can descend to the lower respiratory tract and/or invade the bloodstream. Regulation and activation of virulence genes play essential roles in this process of disease development. Characterization of S. pneumoniae regulatory networks has been a recent area of interest, but despite inroads little is known about regulation of virulence genes in this pathogen. A putative transcriptional regulator in S. pneumoniae, mgrA, which exhibits homology to the virulence gene activator mga of group A streptococcus, was previously identified as a regulator that is required for development of pneumonia in a murine model. In this study we confirmed that mgrA plays a role in both nasopharyngeal carriage and pneumonia. Transcriptional profiling by microarray technology was used to show that mgrA acts as a repressor of the previously characterized rlrA pathogenicity islet. This is manifested phenotypically by a decrease in adherence to epithelial cells in tissue culture since the rlrA pathogenicity islet contains genes mediating adherence.
Streptococcus pneumoniae is a major human pathogen in all age groups worldwide (14). It accounts for over one million deaths annually in children under the age of 5 years alone, is the most common cause of bacterial meningitis in children under 2 years old in the United States, and is a leading cause of bacteremia, otitis media, and sinusitis (18, 25). Despite the fact that it is capable of causing invasive disease, this bacterium usually exists as a benign colonizer on respiratory tract mucosal surfaces. The transition from colonization of the nasopharyngeal mucosa to invasive disease is likely to involve adaptive responses in the bacterium and coordinate expression of virulence factors. Although characterization of pneumococcal regulatory networks has been an area of interest recently (16, 19, 26), there is still much more to be discovered, particularly with respect to regulation of pneumococcal genes important for virulence. Identification of the virulence-associated regulatory networks may lead to new therapeutic strategies designed to inhibit the disease process by affecting the expression of virulence genes or to new vaccine strategies based on antigens encoded within regulons.
Two-component signal transduction systems and other putative transcriptional regulators have been identified by sequence analysis and signature-tagged mutagenesis (STM) screening (12, 13, 22, 29). These systems and regulators are likely to play roles in the ability of S. pneumoniae to adapt to changing environments. In three STM studies the workers identified factors that are essential for bacterial survival in murine models of infection (8, 13, 22). Among these factors were genes predicted to code for proteins with regulatory functions. One screening analysis showed that many of these factors are specific for particular host environments, suggesting that they are important in adaptation to the different niches that the bacteria encounter during host infection or colonization (8).
One putative transcriptional regulator that has been identified, encoded by sp1800 and designated MgrA, is an orthologue of Mga from group A streptococcus (GAS), which activates several virulence genes in that organism (17, 21, 24, 28). Given its homology to mga and its identification in the STM screening analysis, we hypothesized that mgrA is important for virulence gene regulation in pneumococcus. In this study we confirmed that mgrA plays a role in lung infection and nasopharyngeal colonization in a murine model. Microarray gene expression and RNase protection assays (RPAs) were used to identify genes whose expression is altered in mutants lacking or overexpressing mgrA. Finally, transcriptional regulation of the genes identified was correlated to phenotypic changes in adherence to tissue culture monolayers.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The bacterial strains, plasmids, and primers used in this study are listed in Tables 1 and 2. The parental strain used for all S. pneumoniae genetic manipulations was AC353, a streptomycin-resistant (Smr) derivative of serotype 4 strain TIGR4 (8, 9). S. pneumoniae strains were grown in Todd-Hewitt broth containing 5% yeast extract (THY) and supplemented with Oxyrase (5 μl ml−1) and 0.8% maltose as indicated below. The antibiotic concentrations used in this study were as follows: 100 μg of streptomycin per ml, 4 μg of chloramphenicol per ml, and 200 μg of spectinomycin per ml for S. pneumoniae; and 100 μg of ampicillin per ml, 100 μg of spectinomycin per ml, and 10 μg of chloramphenicol per ml for Escherichia coli. The primers used in this study are listed in Table 2. PCRs were performed in reaction buffer containing 1× Taq reaction buffer (Promega), each deoxynucleoside triphosphate at a concentration of 250 μM, each primer at a concentration of 1 μM, and either Taq alone or a 10:1 mixture of Taq and Pfu DNA polymerases. The reaction conditions consisted of 25 cycles of 95°C for 30 s, 45 to 53°C for 30 s, and 72°C for 30 s/kb of DNA, followed by 10 min at 72°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Reference(s) or source |
|---|---|---|
| E. coli DH5α | F− Δ(lacZYA-argF)U169 recA1 endA1 hsdR1 supE44 thi-1 gyrA96 relA1 | 7, 11 |
| S. pneumoniae strains | ||
| AC353 | TIGR4 Smr derivative | 8 |
| AC1213 | rlrA::magellan2 Smr Cmr | 9 |
| AC1272 | mgrA::magellan2 Smr Cmr | This study |
| AC1478 | malM-rlrA-cat-malP Smr Cmr | 9 |
| AC1481 | malM-mgrA-cat-malP Smr Cmr | This study |
| AC1500 | ΔmgrA Smr Spcr | This study |
| STM206 | mgrA 5′-UTR::magellan2 Smr Cmr | 8 |
| Plasmids | ||
| pGEM-T | Cloning vector; Apr | Promega |
| pCR-Script | Cloning vector; Apr | Stratagene |
| Amp SK(+) | ||
| pEMCat | Contains magellan2; Apr/Cmr | 3 |
| PEMSpc/pR412 | Contains magellan5; Apr Spcr | 15 |
| pAC1000 | S. pneumoniae suicide vector | 9 |
| pCH84 | pAC1000 malm-rlrA-cat-malP; Smr Cmr | 9 |
| pAC1472 | pAC1000 malM-mgrA-cat-malP; Smr Cmr | This study |
| pAC1499 | pAC1000 ΔmgrA′::aad9′; Cmr Spcr | This study |
| pAC1279 | pGEM-T rlrA RPA probe; Apr | 9 |
| pAC1280 | pGEM-T rrgA RPA probe; Apr | 9 |
| pAC1281 | pGEM-T rrgB RPA probe; Apr | 9 |
| pAC1285 | pGEM-T srtD RPA probe; Apr | 9 |
| pCH207 | pGEM-T mgrA RPA probe; Apr | This study |
TABLE 2.
Sequences of primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| 180AF1 | GTTTGGATCCTTTAAAATTAAAAATATAATCTT |
| 1800R8 | GCATGCCATGGTTAAGAAGAATGAAAAATCAAG |
| DN1800 | GGATGCATGCCAAAAATAACAAAAAAAC |
| 180SRX | GGGACTCGAGATGAAACACAAGGAATGGCA |
| SPCFN | CATGCCATGGTCTAGAACTAGTGGATCCC |
| SPCRS | CGTTGCATGCTTATAATTTTTTTAATCTG |
| MALMFX | CCCTCGAGTGAAAGCTATCGTGAGCAATT |
| MALMRP | CCGAGCTCAAGATCTGGATCCTTATTTCTTTAAATCTACC |
| MALPF2 | CCTCTAGAGAGCATGCGACAATAATCAGGAGACAAC |
| MALPRP | CCGCGGCTCGAGTTCAAGAGGCCATTTTTCAAG |
| PCATF1 | CCCGGTCTAGAGTCGACGGTATCGATAAGCT |
| PCATR1 | CCGGCGCATGCTTATAAAAGCCAGTCATTAG |
| SP1800F | CGCGGATCCAAAGGAGAATCATCATGAGAGATTTATTATCTAAAAAAAG |
| SP1800R | CCCTCTAGATTACTCATCTAATCGAATAAA |
Construction of mgrA deletion strain.
AC1500 was constructed by replacing 1,299 bp of the coding sequence of mgrA (corresponding to bp 180 to the stop codon) with the Spcr gene (aad9) and its promoter amplified from pEMSpc, previously designated pR412 (15). DNA fragments containing the region 5′ to the mgrA gene and ending 180 bp into the coding sequence of the gene and the region 3′ to the mgrA gene were PCR amplified from AC353 by using two primer pairs, primers 180AF1 and 1800R8 and primers DN1800 and 180SRX. The aad9 gene and its promoter, conferring spectinomycin resistance to E. coli and S. pneumoniae, was PCR amplified from pEMSpc by using primers SPCFN and SPCRS. Each of these fragments was subcloned separately into pCR-Script Amp SK(+) (Stratagene) and subsequently inserted into pAC1000 to obtain the allelic exchange vector pAC1499. To generate AC1500, pAC1499 was linearized by digestion with XhoI, and the gel-purified fragment was transformed into naturally competent AC353 as previously described (4). The double recombination event was selected for by plating on medium containing spectinomycin and was confirmed by PCR and DNA sequencing.
Construction of an MgrA-overexpressing strain.
To construct a strain that overexpresses mgrA from an inducible promoter, the coding sequence of the mgrA gene was introduced into the S. pneumoniae maltose locus downstream of malM (1, 23). To do this, DNA fragments containing the 3′ end of the malM gene and the 5′ end of the malP gene were PCR amplified from AC353 with primers MALMFX and MALMRP and with primers MALPF2 and MALPRP, respectively. The cat gene, conferring chloramphenicol resistance (Cmr) in both E. coli and S. pneumoniae, was PCR amplified from pAC1000 with primers PCATF1 and PCATR1, and the coding sequence of the mgrA gene was PCR amplified from AC353 with primers SP1800F and SP1800R. In the latter case the Shine-Dalgarno sequence of the S. pneumoniae rpoB gene was engineered into the forward primer sequence to allow a high level of translation efficiency of mgrA at the maltose locus. Each of these fragments was subcloned separately into pCR-Script Amp SK (+) (Stratagene) and subsequently inserted into pAC1000 to obtain pAC1472. The final construct contained the 3′ malM sequence and the 5′ malP sequence flanking the mgrA coding sequence and the cat gene. To generate AC1481, the S. pneumoniae strain overexpressing mgrA, pAC1472 was linearized by digestion with XhoI, and the gel-purified fragment was transformed into naturally competent AC353. The double recombination event was selected for by plating on medium containing chloramphenicol and was confirmed by PCR and DNA sequencing.
Animal competition assays.
For all animal infections 6- to 10-week-old female Swiss Webster mice (Taconic Laboratories) were used. Prior to competition experiments magellan2 insertion mutations were backcrossed into AC353 as described previously (8). Bacteria were prepared for inoculation as follows. Mutant and wild-type (AC353) strains were grown separately overnight on blood agar plates with appropriate antibiotics and resuspended in THY, and the optical density was adjusted to an appropriate value for the input dose. Mutant and wild-type strains were mixed at a 1:1 ratio prior to inoculation. In all experiments mice were lightly anesthetized with methoxyflurane prior to inoculation and were sacrificed by CO2 asphyxiation at the end of the infection.
For lung infection 40 μl (approximately 1 × 107 CFU) of the mutant-wild type mixture was inoculated intranasally. The infection was allowed to proceed for 44 h, and then the lungs were aseptically removed and mechanically homogenized and the bacteria were recovered from the homogenate. Approximately 5 × 105 bacteria were inoculated for intraperitoneal infection, and bacteria were recovered from the bloodstream by cardiac puncture after 20 h. To assess nasopharyngeal carriage, 10 μl (approximately 1 × 108 CFU) of a mutant-wild type mixture was inoculated intranasally, and bacteria were recovered after 7 days by washing the nasopharynx with 400 μl of sterile phosphate-buffered saline (PBS). The input ratio of mutant bacteria to wild-type bacteria inoculated into the mice was determined as described below. In parallel for all infections, the same mutant-wild type mixture was inoculated into 10 ml of THY supplemented with streptomycin (50 μg ml−1) and Oxyrase (5 μg ml−1) and grown to the mid-log or late log phase to assess in vitro growth.
For each mouse experiment and for in vitro growth experiments, the ratio of the mutant bacteria recovered to the wild-type bacteria recovered was determined by plating recovered bacteria on Trypticase soy agar (TSA) blood plates supplemented with streptomycin and subsequently replica plating colonies on TSA blood plates containing either streptomycin and streptomycin-chloramphenicol or streptomycin and streptomycin-spectinomycin. In vivo competition indices were calculated by determining the ratio of the mutant bacteria recovered to the wild-type bacteria recovered from each animal adjusted by the input ratio.
RNA purification and labeling.
Total RNA was isolated from 5-ml aliquots of S. pneumoniae grown in THY supplemented with Oxyrase (5 μl ml−1) and 0.8% maltose. Cells were pelleted at 4°C and snap frozen. Bacterial pellets were thawed on ice and treated with 3 mg of lysozyme ml−1 in 100 μl of PBS for 5 min at room temperature. RNA was isolated with a Qiagen RNeasy kit used in accordance with the recommendations of the manufacturer (Qiagen). Samples were treated on the column with DNase I (Qiagen), as recommended by the manufacturer.
Probe synthesis and hybridization.
RNA was converted to cDNA in 20-μl reaction mixtures by combining 1 μg of RNA and 0.5 μg of random hexamers (Amersham), heating the mixtures to 65°C for 10 min, and then snap cooling the mixtures on ice. The following preparations were then added: 2 μl of 0.1 M dithiothreitol, 0.5 μl containing each of the deoxynucleoside triphosphates at a concentration of 10 mM, 4 μl of 5× RT buffer (Invitrogen), and 1 μl (200 U) of Superscript II (Gibco BRL). The mixture was incubated at 42°C for 150 min. RNA was hydrolyzed with 1 μl of 1 M NaOH at 65°C for 10 min and neutralized with 1 μl of 1 M HCl. Samples were purified with a Qia-quick PCR column (Qiagen) used according to the manufacturer's instructions and were eluted with 40 μl of elution buffer. Amino-allyl dUTP was incorporated into the cDNA samples as follows. For each sample, 40 μl of the eluted DNA was incubated for 5 min at 99°C and then for 5 min on ice. Five microliters of 10× random octamer buffer (1550-2; New England Biolabs), 3 μl of a deoxynucleoside triphosphate-dUTP mixture (3 mM dGTP, 3 mM dATP, 3 mM dCTP, 1.8 mM amino-allyl dUTP [A-0410; Sigma-Aldrich], 1.2 mM dTTP), and 2 μl of Exo− Klenow fragment (NEB) were added, and the mixture was incubated for 150 min at 37°C and then stored at 4°C overnight. Free amines were removed with a Qia-quick PCR purification kit (Qiagen), and the eluted sample was dried under vacuum. Samples were resuspended in 4.5 μl of distilled H2O and incubated with either 1 μM Cy3 or 1 μM Cy5 monofunctional reactive dye (Amersham) for 1 h at room temperature in the dark. The time point samples were incubated with Cy5, and the reference samples were incubated with Cy3. Reference samples were prepared by reverse transcribing a pool of RNA comprised of equimolar aliquots of RNA isolated from wild-type (AC353) bacteria at each optical density. The reactions were quenched with 4.5 μl of 4 M hydroxylamine for 15 min at room temperature, and then each Cy5-labeled sample was mixed with a Cy3-labeled reference sample. The unincorporated dye was removed with a Qia-quick PCR purification kit, and the probes were eluted with 40 μl of elution buffer and were dried under vacuum. For hybridization, the samples were resuspended in a solution containing 11.3 μl of Tris-EDTA (pH 7.5), 1 μl of a 10-mg ml−1 solution of yeast tRNA, 2.25 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 0.45 μl of 10% sodium dodecyl sulfate. The mixture was heated to 99°C for 2 min and immediately centrifuged for 2 min at the maximum speed. The probe was applied to a microarray, which is described elsewhere (http://falkow.stanford.edu/whatwedo/supplementarydata/pub7/MicroarrayDesign.pdf), and was incubated for at least 24 h at 60°C.
Data analysis.
Arrays were scanned by using a GenePix 4000A scanner (Axon Instruments, Union City, Calif.), and images were analyzed with the GenePix Pro 3.0 software. Microarray data were stored in the Stanford Microarray Database (6, 27) and are available at http://genome-www.stanford.edu/microarray. The data were filtered to remove poor-quality measurements (for example, spots affected by scratches on the array were not considered), and the red/green ratios were log2 transformed. Genes for which reliable measurements were obtained for over 80% of the arrays in the resulting data set were selected and organized by hierarchical clustering by using the CLUSTER program and were viewed in TREEVIEW (5). A statistical analysis was performed by using Significance Analysis of Microarrays (30).
RPAs.
Template DNA for the generation of riboprobes was PCR amplified with two primer sets, primers 1800PF2 and 1800PR and primers RPOBF3 and RPOBR2. The resulting products were purified with a Qia-quick PCR purification kit, subsequently cloned into pGEM-T (Promega), and confirmed by PCR performed with both a T7 or SP6 primer and a primer specific for the cloned insert. These plasmids were used as templates for generation of riboprobes for the detection of mgrA and rpoB mRNA. Riboprobes for detecting rlrA, rrgA, rrgB, and srtD mRNA were generated as previously described (9). Synthesized probes were purified on a 4% denaturing polyacrylamide gel containing 7 M urea. RPAs were carried out as described by the manufacturer with an RPA11 kit (Ambion) in triplicate with at least three independently isolated RNA samples. The protected fragments were visualized by exposing each gel to a phosphorimaging screen (Kodak) and were analyzed with a Storm 860 scanner and the IQMac V 1.2 imaging software. The relative amount of each protected fragment in each assay mixture was normalized to the amount of rpoB protected RNA in each lane.
Adherence assays.
A549 cells, a human lung epithelial cell line, were grown according to American Type Culture Collection guidelines and were seeded into 24-well tissue culture plates 48 h prior to the binding assays. S. pneumoniae cells were grown to the mid-exponential phase (optical density at 600 nm [OD600], 0.3 to 0.4) and washed once with PBS, the OD600 was adjusted to 0.3 with PBS, and the preparation was diluted 1:10 with Ham's F12K tissue culture medium containing 10% fetal bovine serum. Aliquots (350 μl) were added to semiconfluent monolayers of A549 cells at an multiplicity of infection of 10:1. Bacteria were incubated with A549 cells for 30 min at 37°C in 6% CO2, and then the culture fluid was removed from each well and the monolayers were washed three times with PBS (pH 7.4) to remove nonadherent bacteria. Epithelial cells were detached from the wells by treatment with 200 μl of 0.25% trypsin-1 mM EDTA and were lysed by addition of 800 μl of ice-cold 0.025% Triton X-100. Appropriate dilutions were plated on TSA blood plates containing streptomycin to count the number of bacteria that adhered to the eukaryotic cells. The titer of adherent bacteria for each strain was compared to the input titer of the strain, and the percentage of adherent bacteria was determined for the strain. All experiments were performed in quadruplicate, and each experiment was replicated three times on different days.
RESULTS
Identification of mgrA, an orthologue of GAS mga, as a virulence factor in S. pneumoniae.
An STM screening of TIGR4 resulted in identification of several putative transcriptional regulators as regulators that are essential for lung infection in a murine model (8). One of these transcriptional regulators, encoded by the sp1800 gene (designated mgrA [Mga-like repressor A] here), is 51% similar and 25% identical to the multiple gene regulator Mga of GAS. The region that includes mgrA was identified twice in the pneumococcal STM screening analysis. One transposon insertion mapped to the middle of the 1,479-bp open reading frame, and a second transposon insertion mapped to a noncoding region 300 bp upstream of the predicted start site of mgrA (mgrA 5′ untranslated region). The mutants with the transposon insertion mutations were backcrossed into the wild-type strain, and their phenotypes were confirmed in competition assays with the wild type in three murine models of infection (namely, nasopharyngeal carriage, bacteremia, and lung infection). The mutants were attenuated for both nasopharyngeal carriage and lung infection but were as virulent as or slightly more virulent than the wild type in the bacteremia model (Table 3).
TABLE 3.
Virulence in mice of mrgA mutant strains as assessed by competition assays
| Strain | Relevant genotype and phenotype | Geometric mean in vivo competition indexa
|
||
|---|---|---|---|---|
| Lung infection | Carriage | Bacteremia | ||
| AC1500 | ΔmgrA Smr Spcr | 0.26 (12) | 0.034 (6) | 2.1 (8) |
| AC1272 | mgrA::magellan2 Smr Cmr | 0.47 (8) | 0.58 (6) | NDb |
| STM206 | mgrA 5′-UTR::magellan2 Smr Cmr | <0.023 (4) | 0.009 (6) | 0.56 (3) |
The geometric mean of the in vivo competition index was determined. The numbers in parentheses are the numbers of animals infected in the experiments. For the experiments in which no mutant bacteria were recovered from an animal the number 1 was used as the numerator when the in vivo ratio for the animal was determined; thus, the in vivo mean competition index is less than the calculated value. The values in boldface type are significantly different (P < 0.05, as determined by the Student two-tailed t test).
ND, not determined.
Construction of a deletion strain and a strain overexpressing mgrA.
Two additional mutants were constructed; one (AC1500) had a deletion-insertion of an Spcr gene in place of mgrA, and the other, a merodiploid strain (AC1481), had a second copy of the gene inserted between malM and malP of the maltose operon. In the latter strain mgrA is under control of the maltose promoter, which allows overexpression of mgrA when the organism is grown in maltose. These strains were used to assess whether there were any genes under control of MgrA. The phenotype of the mgrA deletion mutant was also assessed in competition assays with the wild type in the three murine models of infection. Similar to the transposon insertion mutants, the deletion strain was attenuated for both nasopharyngeal carriage and pneumonia but not for bacteremia (Table 3).
Unlike mga, mgrA does not appear to regulate neighboring genes.
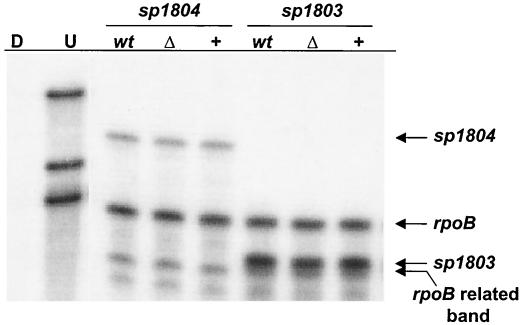
In GAS mga lies directly upstream of and reads in the same direction as some of the genes that it regulates, including the M protein and C5a peptidase genes (17, 21, 24, 28). In contrast, the arrangement around mgrA on the S. pneumoniae serotype 4 genome is different. mgrA is approximately 450 bp upstream of and is divergently transcribed from a cluster of five small open reading frames encoding putative hypothetical proteins having unknown functions and downstream of a gene encoding a putative LacI family transcriptional regulator (sp1799). The mgrA gene itself has a predicted transcriptional terminator directly after the stop codon, and therefore, transcription would not be predicted to read through to the neighboring transcriptional regulator. To test whether mgrA regulates transcription of the cluster of downstream genes, RPAs were performed by using probes for two putative genes in the cluster, sp1803 and sp1804. In these experiments RNA was harvested from AC1500 and AC1481 grown in 0.8% maltose to the mid-exponential phase (OD600, 0.4). A probe for the rpoB gene, which codes for the β subunit of RNA polymerase, was used to probe the same RNA and served as a loading control for each experiment. Although both sp1803 and sp1804 were transcriptionally active, there was no detectable difference in the level of either transcript in either mutant, which is consistent with the hypothesis that mgrA does not affect transcription of these genes under the in vitro conditions tested (Fig. 1).
FIG. 1.
RPA to analyze the mRNA levels of sp1803 and sp1804 in wild-type strain AC353 (lanes wt), mgrA deletion strain AC1500 (lanes Δ), and mgrA-overexpressing strain AC1481 (lanes +). Riboprobes for sp1803, sp1804, and rpoB were generated and hybridized to 10 μg of S. pneumoniae RNA from the three strains. RNA was harvested from cells grown in 0.8% maltose to an OD600 of 0.4. Lanes U and D contained undigested riboprobes and riboprobes digested by RNase in the absence of S. pneumoniae RNA, respectively.
mgrA acts as a transcriptional repressor of the rlrA pathogenicity islet.
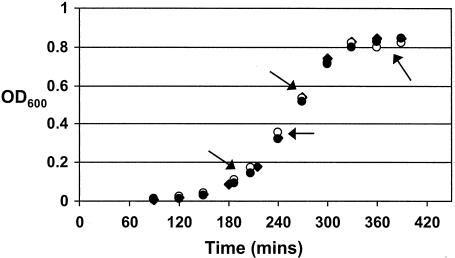
cDNA microarray analysis of the gene expression patterns of the mgrA deletion and mgrA-overexpressing strains (AC1500 and AC1481, respectively) was used in a further attempt to identify genes regulated by MgrA. To examine the hypothesis that MgrA may not be active at all phases of growth, RNA was harvested from pelleted cells at four separate times on the growth curve (early exponential, mid-exponential, late exponential, and early stationary phases). Later times were not assessed because of the level of mRNA degradation and autolysis (data not shown). All strains had similar growth kinetics in vitro (Fig. 2). The vast majority of genes were expressed at comparable levels in the two strains. A subset of genes that had different profiles in the two mutant strains at one or two times were not considered further.
FIG. 2.
Growth curves for the wild-type strain AC353 (⧫), the mgrA deletion mutant (•), and the mgrA-overexpressing strain (○) cultured statically in THY supplemented with Oxyrase (5 μl ml−1) and 0.8% maltose. Aliquots were removed for RNA extraction at four different times, indicated by the arrows (the equivalent OD600 values were as follows: early log phase, ∼0.2; mid-log phase, ∼0.4; late log phase, ∼0.6; and stationary phase, ∼0.8).
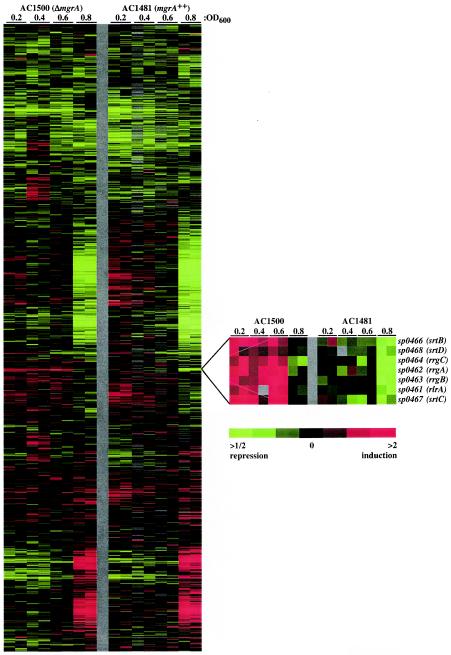
One set of genes was notable because the transcripts were more abundant in the ΔmgrA strain than in the mgrA-overexpressing strain at all the times assessed, suggesting that MgrA is involved in repressing these genes (Fig. 3 and Table 4). These genes were the genes in the rlrA pathogenicity islet, some of which have been characterized previously as virulence factors required for lung infection and nasopharyngeal carriage (9).
FIG.3.
Gene expression profiling of non-MgrA-expressing strain AC1500 and MgrA-overexpressing strain AC1481 by spotted DNA microarray analysis. The data are measures of relative gene expression at the culture optical densities indicated and represent the quotient of hybridization of the fluorescent cDNA probe prepared from the AC1500 or AC1481 sample and hybridization of a reference pool. Red and green represent high and low experimental sample/reference ratios, respectively (see scale bar). Gray indicates technically inadequate or missing data. The columns represent arrays, and the rows represent genes. See Materials and Methods for experimental details. Data for all of the arrays used in this study are available elsewhere (6).
TABLE 4.
Average expression of rlrA islet genes in AC1500, an mgrA deletion strain, compared to expression in AC1481, an mgrA-overexpressing straina
| TIGR4 open reading frameb | Gene | Fold difference in expression for the time course at an OD600 of:
|
|||
|---|---|---|---|---|---|
| 0.2 | 0.4 | 0.6 | 0.8 | ||
| SP0461 | rlrA | 2.0 | 1.6 | 2.8 | 2.3 |
| SP0462 | rrgA | 2.4 | 2.1 | 3.0 | 3.6 |
| SP0463 | rrgB | 2.2 | 1.9 | 3.5 | 2.8 |
| SP0464 | rrgC | 1.8 | 1.7 | 3.1 | 2.5 |
| SP0466 | srtB | 2.0 | 2.8 | 2.7 | 2.8 |
| SP0467 | srtC | 1.7 | 2.7 | 4.4 | 2.4 |
| SP0468 | srtD | 2.4 | 1.7 | 2.3 | 1.5 |
The log red/green ratios from replicate arrays were averaged at each time point. For each OD600, the average log2 value for each AC1481 sample was subtracted from the corresponding log2 AC1500 value. The results are expressed as the fold difference in expression between the two strains. Data from all of the arrays used in this study are available elsewhere (6).
Designations of the open reading frames in the TIGR4 sequence which are on the microarray.
Confirmation of this phenotype was obtained by performing RPAs with RNA isolated from three independent sets of cultures and assessing two time points in the growth curve for each culture (mid-exponential phase [OD600, 0.4] and late exponential phase [OD600, 0.6]).
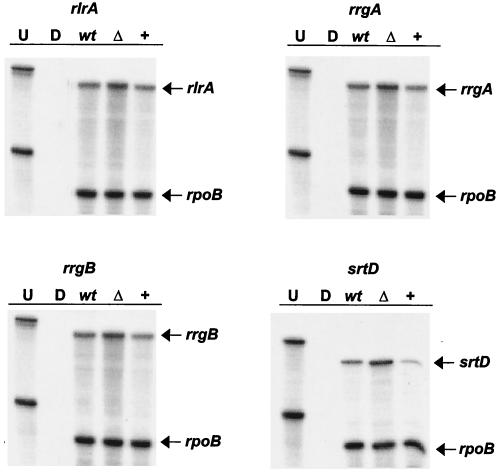
The islet contains four promoters that drive transcription of seven genes (8). Probes for a gene in each transcriptional unit (namely, rlrA, rrgA, rrgB, and srtD), as well for rpoB, were synthesized. Again, rpoB was used as a loading control for each experiment. All four of the islet genes tested were up-regulated in the mgrA deletion strain and down-regulated in the mgrA-overexpressing strain at both times, and the levels were similar to the levels that were revealed by the microarray experiments, which is consistent with the hypothesis that MgrA acts as a transcriptional repressor of these genes (Fig. 4). The level of each islet gene message was two- to threefold higher in the mgrA deletion strain and at least twofold lower in the mgrA-overexpressing strain than in the wild type at all of the times tested. Together, these data are consistent with the hypothesis that MgrA acts as a transcriptional repressor of the islet genes. MgrA may act at each promoter or, alternatively, may alter directly only the expression of rlrA, a transcriptional activator, which subsequently affects the expression of the six other islet genes.
FIG. 4.
RPAs to analyze the mRNA levels of a gene in each transcriptional unit in the rlrA islet in wild-type strain AC353 (lanes wt), mgrA deletion strain AC1500 (lanes Δ), and mgrA-overexpressing strain AC1481 (lanes +). Riboprobes for each gene, as well as rpoB, were generated and hybridized to 10 μg of S. pneumoniae RNA from the three strains. RNA was harvested from cells grown in 0.8% maltose to an OD600 0.4 or 0.6. Transcript levels were comparable at either time point. The results obtained with an OD600 of 0.4 are shown. Lanes U and D contained undigested riboprobes and riboprobes digested by RNase in the absence of S. pneumoniae RNA, respectively.
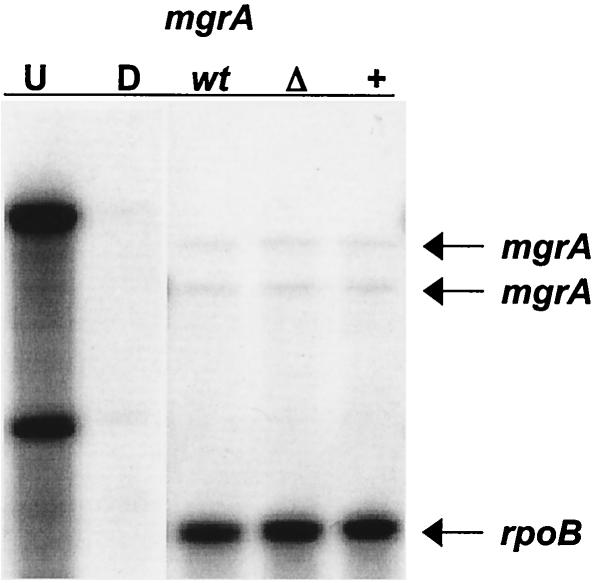
RlrA exhibits homology to RofA and Nra of GAS (9). In GAS Nra acts as a transcriptional repressor of mga, while Mga activates nra transcription (20). To test the possible role of RlrA in regulation of mgrA in S. pneumoniae serotype 4, the transcript levels of mgrA in the wild type and in strains overexpressing rlrA or with rlrA inactivated were assessed. RPAs were performed by probing for mgrA in RNA harvested from wild-type strain AC353, strain AC1213, a strain that harbors a transposon insertion in rlrA, and strain AC1278, a strain that overexpresses rlrA from the maltose-inducible promoter. rpoB was used as a loading control. As shown in Fig. 5, there was no detectable difference between the amount of mgrA message in the AC1213 or AC 1278 strain and the amount in wild-type strain AC353. Two mRNA species were detected with the mgrA riboprobe in all strains, which is consistent with the hypothesis that the gene has two transcriptional start sites, as the riboprobe for mgrA was complementary to RNA overlapping the 5′ untranslated mRNA and the coding sequence of the gene. Neither transcript was altered in the rlrA mutant strains, suggesting that there is a regulatory circuit different from that observed for the homologues in GAS.
FIG. 5.
RPA to analyze the mgrA mRNA levels in wild-type strain AC353 (lane wt), rlrA deletion strain AC1213 (lane Δ), and rlrA-overexpressing strain AC1278 (lane +). Riboprobes for mgrA and rpoB were generated and hybridized to 10 μg of S. pneumoniae RNA from the three strains. RNA was harvested from cells grown in 0.8% maltose to an OD600 of 0.4. Lanes U and D contained undigested riboprobes and riboprobes digested by RNase in the absence of S. pneumoniae RNA, respectively.
Biological consequences of MgrA regulation.
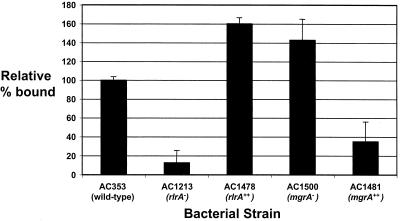
The islet genes rlrA, rrgA, and srtD have been shown previously to be required for lung infection and/or nasopharyngeal carriage in mice (8). This phenotype is similar to that seen in the mgrA deletion strain, which is contrary to what one would predict given that mgrA acts as a transcriptional repressor of the islet genes. Three of the rlrA islet genes, rrgA, rrgB, and rrgC, are predicted to code for cell wall-anchored proteins that, based on homology to microbial surface components recognizing adhesive matrix molecules (9), may be important for bacterial adhesion to mucosal surfaces. To determine if the level of transcriptional derepression of the islet genes seen in the mgrA deletion mutant corresponded to a measurable biological consequence in the bacterial cell, the abilities of the wild type, rlrA mutants AC1213 and AC1278, and mgrA mutants AC1500 and AC1481 to adhere to lung epithelial cell line A549 cells were analyzed. Bacteria that were grown in vitro were added to epithelial cells at a multiplicity of infection of 10:1. After 30 min, nonadherent bacteria were removed by washing, and the adherent bacteria were liberated and counted. The four mutant strains exhibited either increased or decreased adherence compared to the adherence of the wild type depending on the nature of the mutation. The transposon insertion in the rlrA gene reduced binding of S. pneumoniae to A549 cells to 10% of wild-type binding, and conversely, overexpression of rlrA increased binding to 160% of the wild-type binding. Overexpression of mgrA reduced binding to 30% of the wild-type binding, and deletion of mgrA increased binding to 140% of the wild-type binding (Fig. 6).
FIG. 6.
Adherence of S. pneumoniae to A549 lung epithelial cells. Mid-exponential-phase S. pneumoniae cells (1 × 106 CFU) were added to A549 cells at a multiplicity of infection of 10 and incubated at 37°C for 30 min. The percentages of the input bacteria that adhered to the monolayers after three washes with PBS were determined. The adherence of mutant strains is expressed relative to the adherence of the wild type. Representative results from one of four independent experiments are shown. The assay was performed in quadruplicate in each experiment, and the error bars indicate the standard deviations.
These data show that the alteration in islet gene expression caused by MgrA seen in vitro results in a change in the adherence phenotype of the pneumococcal strain. However, this in vitro phenotypic change does not appear to correlate with the in vivo phenotype of reduced virulence.
DISCUSSION
The mgrA gene was initially identified by STM screening as a gene that is essential for lung infection by S. pneumoniae serotype 4 (8). We confirmed here that this gene is also required for wild-type levels of nasopharyngeal colonization in a murine model but appears to have only a minimal effect on the ability of the bacteria to cause bacteremia when the mucosal surfaces are bypassed by direct intraperitoneal inoculation of bacteria.
The two transposon insertion mutants from the STM screening and the mgrA deletion strain obtained in this work had slightly different phenotypes in competition assays. The transposon insertion 300 bp upstream of the start site had the greatest effect on virulence. RPA data (data not shown) obtained by probing for the mgrA transcript in the three mutants showed that there was no detectable transcript in either the deletion mutant or the mutant with the transposon inserted into the coding sequence, but a transcript larger than the transcript seen in the wild-type strain was detectable in the mutant with the insertion upstream of the mgrA start site. It is possible that in the latter strain the transposon insertion leads to disregulation of mgrA due to readthrough transcription from the transposon or due to effects on protein binding to the mgrA promoter region. The hypothesized inappropriate gene expression may be more detrimental than an actual gene knockout. Alternatively, we have not excluded the possibility that the upstream insertion also has effects on the putative open reading frame in the opposite direction, SP1801.
MgrA exhibits amino acid sequence homology to Mga, which activates the transcription of numerous virulence genes involved in GAS pathogenesis. Many of these genes encode surface-associated factors, such as M family proteins, C5a peptidase, and a secreted inhibitor of complement (21). These three genes are adjacent to mga in the GAS genome, but Mga is also involved in regulating transcription of distant genes.
Using RPAs, we determined that MgrA in S. pneumoniae does not regulate the transcription of neighboring putative genes in vitro. To determine if MgrA influences pneumococcal gene transcription, like Mga, we compared the transcriptional profiles of two strains, one having a deletion of mgrA and one overexpressing mgrA, over time by microarray analysis. Our data indicated that MgrA may act as a transcriptional repressor of genes in the rlrA pathogenicity islet, a previously identified element that contains virulence genes (9). All the genes in the islet were identified in our screening analysis as genes that exhibit a consistent and statistically significant change in expression (at least a twofold change) at all growth times assayed. The variation in expression of these genes appeared to be the only difference between our mutant strains. Genes in this islet had previously been found to be important for establishing nasopharyngeal carriage and lung infection in murine models, and at least three genes in the islet are thought to code for surface-associated factors (8). Interestingly, the rlrA pathogenicity islet has only been found in a minority of S. pneumoniae serotype strains, yet we found mgrA in all serotypes in which we looked for it (data not shown). It seems unlikely, therefore, that the primary role of this regulator is to repress islet gene expression. It would be interesting to perform similar microarray experiments with serotypes that do not possess the pathogenicity islet. Perhaps there are other loci found only in particular serotypes that are under the control of MgrA.
One hypothesis to explain the failure to identify more target genes is that the conditions chosen were not suitable for complete or appropriate activation of MgrA and did not mimic the relevant host environment. The in vivo data for mice are consistent with this hypothesis. Strains having a deletion of mgrA have a decreased ability to cause lung infection and nasopharyngeal colonization despite their increased ability to adhere to lung epithelial cells in vitro. It is possible that a different set of genes is regulated by MgrA in response to the in vivo environment experienced by the pneumococci in the mouse and under vitro conditions. This could lead to expression of different surface factors and different phenotypes with respect to adhesion to host cells.
It has previously been shown that the timing of expression of virulence factors is crucial to disease progression. Activation or expression of factors at the wrong time may actually be detrimental to the bacteria and lead to loss of function instead of gain of function (2). This may explain the attenuation in carriage and lung infection seen in the mgrA deletion strain, in which we expected that expression of the islet genes was at least partially deregulated.
The fact that the rlrA islet is not widely conserved among pneumococcal serotypes suggests that it may have been acquired by horizontal transfer. This suggestion is supported by the fact that the islet is flanked by two IS1167 insertion sequences allowing integration of the element into the genome that occurred by homologous recombination or a transposition event. Data presented here show that the element has not only inserted into the genome but has also integrated into at least one regulatory network in S. pneumoniae serotype 4. The phenomenon of regulators encoded outside a pathogenicity island regulating genes encoded in the island is well described (10) and is one of many mechanisms important in bacterial evolution that enable bacteria to adapt to new environments and maintain a survival advantage.
Acknowledgments
We thank S. Falkow for his generous support of this work.
This work was supported by Wellcome Trust Research Training Fellowship 061036 to C.J.H., by NRSA grant 1F32 A151859-01 to E.A.J., by a grant from the CHIRON Vaccines IRIS Research Center, Siena, Italy, by the Center for Gastroenterology Research on Absorptive and Secretory Processes, and by NIH grant AI52374 to A.C.
REFERENCES
- 1.Acebo, P., C. Nieto, M. A. Corrales, M. Espinosa, and P. Lopez. 2000. Quantitative detection of Streptococcus pneumoniae cells harbouring single or multiple copies of the gene encoding the green fluorescent protein. Microbiology 146:1267-1273. [DOI] [PubMed] [Google Scholar]
- 2.Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611-620. [DOI] [PubMed] [Google Scholar]
- 3.Akerley, B. J., E. J. Rubin, A. Camilli, D. J. Lampe, H. M. Robertson, and J. J. Mekalanos. 1998. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl. Acad. Sci. USA 95:8927-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bricker, A. L., and A. Camilli. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172:131-135. [DOI] [PubMed] [Google Scholar]
- 5.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gollub, J., C. A. Ball, G. Binkley, J. Demeter, D. B. Finkelstein, J. M. Herbert, T. Hernandez-Boussard, H. Jin, M. Kaloper, J. C. Maltese, M. Schroeder, P. O. Brown, D. Botstein, and G. Sherlock. 2003. The Standford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 31:94-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 8.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1405. [PMC free article] [PubMed] [Google Scholar]
- 9.Hava, D. L., C. J. Hemsley, and A. Camilli. 2003. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J. Bacteriol. 185:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaper, J. B., and J. Hacker. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 11.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 12.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 13.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 14.Leowiski, J. 1986. Mortality from acute respiratory infections in children under 5 years of age: global estimates. World Health Stat. 39:138-144. [PubMed] [Google Scholar]
- 15.Martin, B., M. Prudhomme, G. Alloing, C. Granadel, and J. P. Claverys. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 38:867-878. [DOI] [PubMed] [Google Scholar]
- 16.Mascher, T., D. Zahner, M. Merai, N. Balmelle, A. B. De Saizieu, and R. Hakenbeck. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J. Bacteriol. 185:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McIver, K., and R. L. Myles. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol. Microbiol. 43:1591-1601. [DOI] [PubMed] [Google Scholar]
- 18.Musher, D. M. 1992. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis and treatment. Clin. Infect. Dis. 14:801-809. [DOI] [PubMed] [Google Scholar]
- 19.Peterson, S., R. T. Cline, H. Tettelin, V. Sharov, and D. A. Morrison. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 182:6192-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pobielski, A., M. Woischnik, B. A. Leonard, and K. H. Schmidt. 1999. Characterisation of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 21.Pobielski, A., M. Woischnik, B. Pohl, and K. H. Schmidt. 1996. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med. Microbiol. Immunol. 185:171-181. [DOI] [PubMed] [Google Scholar]
- 22.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puyet, A., and M. Espinosa. 1993. Structure of the maltodextrin-uptake locus of Streptococcus pneumoniae. Correlation to the Escherichia coli maltose regulon. J. Mol. Biol. 230:800-811. [DOI] [PubMed] [Google Scholar]
- 24.Rasmussen, M., A. Edén, and L. Björck. 2000. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect. Immun. 68:6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuchat, A., K. Robinson, and J. D. Wenger. 1997. Bacterial meningitis in the United States in 1995. N. Engl. J. Med. 337:970-976. [DOI] [PubMed] [Google Scholar]
- 26.Sebert, M. E., L. M. Palmer, M. Rosenberg, and J. N. Weiser. 2002. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terao, Y., S. Kawabata, E. Kunitomo, J. Murakami, I. Nakagawa, and S. Hamada. 2001. Fba, a novel fibronectin-binding protien from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol. Microbiol. 42:75-86. [DOI] [PubMed] [Google Scholar]
- 29.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 30.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]