Abstract
There is significant evidence linking nucleocytoplasmic transport to cell cycle control. The budding yeast, Saccharomyces cerevisiae, serves as an ideal model system for studying transport events critical to cell cycle progression because the nuclear envelope remains intact throughout the cell cycle. Previous studies linked the classical nuclear localization signal (cNLS) receptor, importin-α/Srp1, to the G2/M transition of the cell cycle. Here, we utilize two engineered mutants of importin-α/Srp1 with specific molecular defects to explore how protein import affects cell cycle progression. One mutant, Srp1-E402Q, is defective in binding to cNLS cargoes that contain two clusters of basic residues termed a bipartite cNLS. The other mutant, Srp1-55, has defects in release of cNLS cargoes into the nucleus. Consistent with distinct in vivo functional consequences for each of the Srp1 mutants analyzed, we find that overexpression of different nuclear transport factors can suppress the temperature-sensitive growth defects of each mutant. Studies aimed at understanding how each of these mutants affects cell cycle progression reveal a profound defect at the G1 to S phase transition in both srp1-E402Q and srp1-55 mutants as well as a modest G1/S defect in the temperature-sensitive srp1-31 mutant, which was previously implicated in G2/M. We take advantage of the characterized defects in the srp1-E402Q and srp1-55 mutants to predict candidate cargo proteins likely to be affected in these mutants and provide evidence that three of these cargoes, Cdc45, Yox1, and Mcm10, are not efficiently localized to the nucleus in importin-α mutants. These results reveal that the classical nuclear protein import pathway makes important contributions to the G1/S cell cycle transition.
THE compartmentalized transport of macromolecules, including proteins and RNAs, into and out of the nucleus is a highly regulated process essential for all eukaryotic cells. Bidirectional movement of these macromolecules controls cell growth through coordinating nuclear and cytoplasmic aspects of gene expression (Moll et al. 1991; Beg et al. 1992; Sidorova et al. 1995; Briscoe et al. 1996). The orchestration of the cell cycle is one of the most complex processes that cells must undergo, requiring coordination of numerous cytoplasmic and nuclear events. Many previous studies have uncovered links between cell cycle control and nuclear transport (Moll et al. 1991; Pines and Hunter 1991; Loeb et al. 1995; David-Pfeuty et al. 1996), but how these two cellular processes control and influence one another is not yet understood in detail.
The nuclear envelope provides a physical mechanism for regulation of numerous events that contribute to cell cycle transitions. In higher eukaryotic cells, the nuclear envelope breaks down during mitosis, allowing for redistribution of macromolecules between the nucleus and the cytoplasm (Burke and Ellenberg 2002; Hetzer et al. 2005). Despite this transient disappearance of the barrier separating the nucleus and the cytoplasm, there are numerous protein transport events that occur during stages of the cell cycle in which the nuclear envelope remains intact. For example, critical regulators such as cyclin A, cyclin B1, and the tumor suppressor p53 are transported in and out of the nucleus during phases of the cell cycle in which the nuclear envelope is intact (Pines and Hunter 1991; David-Pfeuty et al. 1996; Middeler et al. 1997). Cyclin A is transported into the nucleus during S phase (Pines and Hunter 1991) and cyclin B1 is transported to the nucleus at the beginning of mitosis before the nuclear envelope breaks down (Pines and Hunter 1991). p53 enters the nucleus at the early mid-G1 phase of the cell cycle (David-Pfeuty et al. 1996). These cases are examples where regulated transport into the nucleus adds an extra level of control over the activity of these critical regulatory proteins.
Many of the cargo proteins that contribute to control of the cell cycle are likely to be targeted to the nucleus through a classical nuclear localization signal (cNLS) (Lange et al. 2007). The classical NLS consists of a sequence of basic amino acids in a single cluster (monopartite) or two clusters separated by a nonconserved amino acid linker (bipartite) (Kalderon et al. 1984; Robbins et al. 1991). cNLS cargo recognition and transport is mediated by a soluble heterodimeric protein receptor composed of an adapter, importin/karyopherin-α, which recognizes the cNLS cargo in the cytoplasm and a carrier, importin/karyopherin-β, which targets the complex to the nuclear pore complex (NPC) for transport (Görlich et al. 1995; Bayliss et al. 2000; Liu and Stewart 2005). Significant evidence has accumulated to support the idea that rates of import into the nucleus are largely determined by interaction between the NLS cargo and the NLS receptor (Hodel et al. 2006; Timney et al. 2006; Riddick and Macara 2007), making recognition of the NLS cargo by the NLS receptor essentially the rate-limiting step in the process of nuclear protein import.
Numerous studies have provided a detailed molecular understanding of how the import receptor, importin-α, recognizes cNLS-containing cargoes (Conti et al. 1998; Kobe 1999). Importin-α consists of three functional domains (see Figure 1A). The N-terminal region contains an importin-β binding (IBB) domain that interacts with importin-β (Görlich et al. 1996; Weis et al. 1996). The IBB domain also contains an internal NLS-like sequence or auto-inhibitory motif that regulates cNLS cargo binding and facilitates cNLS cargo release in the nucleus (Kobe 1999; Harreman et al. 2003b). The central region of importin-α, which contains 10 armadillo repeat motifs (ARM), constitutes the NLS binding pocket (Conti et al. 1998; Conti and Kuriyan 2000; Fontes et al. 2000). A portion of the N-terminal IBB domain in cooperation with the C-terminal domain of importin-α contains a binding site for the export receptor, Cse1/CAS (Hood and Silver 1998; Solsbacher et al. 1998; Schroeder et al. 1999), which is required for recycling importin-α back to the cytoplasm after the cNLS cargo is released in the nucleus (Gilchrist et al. 2002; Gilchrist and Rexach 2003; Matsuura and Stewart 2004).
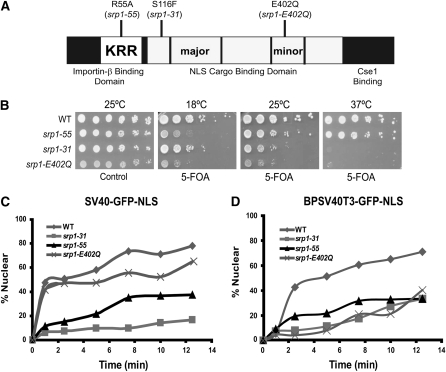
Figure 1.—
Functional analysis of importin-α mutants in vivo. (A) Schematic of importin-α with the three major domains (IBB; NLS cargo-binding domain with major and minor NLS binding pockets; Cse1 binding domain) indicated. The position of the auto-inhibitory NLS-like sequence (54KRR56) is also indicated. The approximate location as well as the amino acid change for each of the srp1 mutant alleles employed in the study is shown. (B) Growth of each srp1 mutant was assessed at 18°, 25°, and 37°. Each mutant was covered with a wild-type SRP1 URA3 plasmid (pAC876) and analyzed by serial dilution and spotting on control plates (where the wild-type SRP1 plasmid is maintained) or on 5-FOA plates (where the wild-type SRP1 plasmid is lost). Plates were incubated at the indicated temperatures for 3–7 days. (C) Kinetic nuclear import assay for monopartite SV40-GFP NLS and (D) bipartite BPSV40T3-GFP NLS import reporters. The initial import rates for the cNLS import reporters were analyzed using a kinetic import assay as described in materials and methods (Shulga et al. 1996). Cultures were grown to early mid-log phase at 25° and then shifted to the nonpermissive temperature (37° for srp1-31 and srp1-E402Q cells and 18° for srp1-55 cells). Initial import kinetics was measured by determining the percentage of cells showing nuclear accumulation of the cNLS reporter at a given time. For scoring, 2-μl samples were removed every 2.5 min. Cells were scored as “nuclear” if both the nucleus was brighter than the surrounding cytoplasm and a nuclear–cytoplasmic boundary was visible. At least 100 cells were counted at each time point. Results are plotted as the percentage of cells showing nuclear cNLS reporter signal vs. time for wild type (WT) (♦), srp1-31(▪), srp1-55 (▴), and srp1-E402Q (X) cells.
The classical import pathway consists of four key steps (Stewart 2007). First, the trimeric import complex consisting of importin-β, importin-α, and the NLS-containing cargo is assembled in the cytoplasm where the cargo is recognized by importin-α. Second, the import complex is targeted to the nuclear pore by importin-β and the trimeric complex is translocated through the NPC (Bayliss et al. 2000; Liu and Stewart 2005). Third, the complex is disassembled in the nucleus following binding of the small GTPase Ran in its GTP-bound form to importin-β, which triggers the release of the cNLS cargo and delivery into the nucleus (Vetter et al. 1999; Lee et al. 2005). Finally, the import receptors are recycled to the cytoplasm for another round of protein import with importin-α exported to the cytoplasm by Cse1/CAS in complex with RanGTP (Matsuura and Stewart 2004).
There is a long history suggesting that nuclear transport factors play key roles in regulating the cell cycle. Many of the original hints come from defects in cell cycle progression associated with mutations in nuclear transport factors (Nishitani et al. 1991; Sazer and Nurse 1994; Loeb et al. 1995; Gruss et al. 2001; Nachury et al. 2001). For example, the first evidence for involvement of Ran in the cell cycle was the observation that cultured cells containing a conditional allele of RCC1, the Ran guanine nucleotide exchange factor (RanGEF), prematurely entered mitosis in the presence of unreplicated DNA (Nishitani et al. 1991). Furthermore, in a conditional allele of RanGEF in fission yeast cells, the mutant cells progress through one round of DNA replication and mitosis before arresting at the mitosis-interphase transition of the cell cycle due to the failure of chromatin decondensation (Sazer and Nurse 1994). Subsequent studies have revealed that RanGTP regulates the activity of several mitotic proteins primarily by modulating interactions with import factors (Sazer and Dasso 2000; Gruss et al. 2001; Nachury et al. 2001; Wiese et al. 2001; Du et al. 2002).
Importin-α orthologs in the budding yeast, Saccharomyces cerevisiae, and the fission yeast, Schizosaccharomyces pombe, have also been linked to the cell cycle (Yano et al. 1994; Loeb et al. 1995; Umeda et al. 2005). Interestingly, a conditional allele of S. cerevisiae importin-α (Srp1), srp1-31, has been reported to arrest at the G2/M stage of the cell cycle (Loeb et al. 1995). This mutant also shows defects in nuclear protein import (Loeb et al. 1995), suggesting that the cell cycle delay could be due to the inability to import a nuclear protein required to traverse the G2/M transition. However, since this mutant of importin-α was identified in a genetic screen, the molecular defects that lead to either the cell cycle delay or the impaired nuclear import are not clear (Yano et al. 1994; Loeb et al. 1995). The amino acid substitution in the srp1-31 protein variant is a serine-to-phenylalanine change at position 116. This change lies within the first of the ARM domains that compose the cNLS binding pocket (see Figure 1A); however, serine 116 is located outside the cNLS binding pocket, making it unclear how this amino acid change affects the function of importin-α.
Therefore, the goal of this study is to understand how specific engineered amino acid changes that cause defects in importin-α cNLS cargo binding and release into the nucleus affect cell cycle progression. Results of this analysis reveal an important role for the classical nuclear import pathway in the G1/S transition of the cell cycle, suggesting that key cargoes containing bipartite cNLS motifs need to be imported to the nucleus to allow cells to properly enter S phase and replicate DNA.
MATERIALS AND METHODS
Yeast strains and plasmids:
All chemicals were obtained from Sigma or USBiological unless otherwise noted. All DNA manipulations were performed by standard protocols (Sambrook and Russell 2001), and all media were prepared by standard procedures (Adams et al. 1997). All yeast strains and plasmids used in this study are listed in Tables 1 and 2.
TABLE 1.
S. cerevisiae strains used in this study
| Name | Strains | Genotype | References |
|---|---|---|---|
| Wild type | pSY580 (ACY192) | MATaura3-52 leu2Δ1 trp1 | Winston et al. (1995) |
| Wild type | ACY247 | MATa/α ura3-52 leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 ade2/ADE2 ade3/ADE3 lys2/LYS2 trp1/TRP1 | Jones et al. (2000) |
| srp1-55 | ACY641 | MATaura3-52 his3Δ200 leu2Δ1 trp1 ade2 srp1-55∷LEU2 | Harreman et al. (2003a) |
| srp1-55 | ACY642 | MATα ura3-52 his3Δ200 leu2Δ1 trp1 srp1-55∷LEU2 | Harreman et al. (2003a) |
| srp1-E402Q | ACY1560 | MATaura3-52 his3Δ200 leu2Δ1 trp1 ade2 lys2 srp1-E402Q∷LEU2 | This study |
| srp1-31 | ACY1561 | MATα ura3-52 leu2Δ1 trp1 his3Δ200 lys2 | This study |
| srp1-31 | ACY1562 | MATaura3-52 leu2Δ1 trp1 | This study |
| Cdc45-GFP | YLR103C (ACY1886) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Huh et al. (2003) |
| srp1-31-Cdc45-GFP | ACY1889 | MATaleu2Δ0 met15Δ0 ura3Δ0 trp1 | This study |
| srp1-55-Cdc45-GFP | ACY1890 | MATamet15Δ0 ura3Δ0 | This study |
| Mcm10-GFP | YIL150C (ACY1887) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Huh et al. (2003) |
| srp1-31-Mcm10-GFP | ACY1891 | MATaleu2Δ0 met15Δ0 ura3Δ0 | This study |
| srp1-55-Mcm10-GFP | ACY1892 | MATamet15Δ0 ura3Δ0 | This study |
| Yox1-GFP | YML027W (ACY1888) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Huh et al. (2003) |
| srp1-31-Yox1-GFP | ACY1893 | MATaleu2Δ0 met15Δ0 ura3Δ0 | This study |
| srp1-55-Yox1-GFP | ACY1894 | MATamet15Δ0 ura3Δ0 | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | References |
|---|---|---|
| pRS406 | LEU2, integration, AMPR | Kahana et al. (1995) |
| pRS424 | 2μ, TRP1, AMPR | Sikorski and Hieter (1989) |
| pAC592 | RSL2, 2μ, TRP1, AMPR | Harreman et al. (2003a) |
| pAC876 | SRP1, CEN, URA3, AMPR | Harreman et al. (2003b) |
| pAC1059 | pMET25-BPSV40T3-NLS-GFP-GFP, CEN, URA3, AMPR | Hodel et al. (2006) |
| pAC1065 | pMET25-SV40-NLS-GFP-GFP, CEN, URA3, AMPR | Hodel et al. (2006) |
| pAC1303 | CSE1, 2μ, TRP1, AMPR | Harreman et al. (2003a) |
| pAC1344 | TUB1-GFP, integration, URA3, AMPR | Straight and Murray (1997) |
| pAC1354 | SRP1, CEN, TRP1, AMPR | Harreman et al. (2003a) |
| pAC1385 | NUP2, 2μ, TRP1, AMPR | Harreman et al. (2003a) |
| pAC1999 | E402Q-SRP1, LEU2, integrating, AMPR | This study |
| p305.2 | ARS305 CEN5 URA3 | Friedman et al. (1996) |
| pARS1 | ARS1 CEN5 URA3 | Friedman et al. (1996) |
| p12 | ARS1412 CEN5 URA3 | Friedman et al. (1996) |
To generate mutants of importin-α that could be directly compared to one another, each mutant was integrated into the same strain background as the previously generated allele srp1-55 (ACY641) (Harreman et al. 2003a). The E402Q importin-α mutant replaced the wild-type copy of importin-α. To integrate E402Q importin-α, the E402Q mutation was subcloned into the open reading frame of the LEU2 integrating plasmid, pRS406 (Sikorski and Hieter 1989), to create srp1-E402Q (pAC1999). E402Q importin-α was then integrated at the endogenous SRP1 locus by linearization of the srp1-E402Q (pAC1999) plasmid and transformation into the S288C wild-type diploid cells (ACY247). Transformants that grew on plates lacking leucine were selected for further analysis. The presence of the E402Q importin-α mutation was confirmed by PCR and sequencing. The heterozygous diploid was then sporulated and tetrads were dissected to generate the haploid srp1-E402Q (ACY1560). This integration strategy is designed to make E402Q importin-α the only copy of importin-α expressed in the haploid strain.
Although the srp1-31 (ACY639) mutant already existed in an S288C background, the mutant was further backcrossed to an S288C wild-type strain (PSY580) (Harreman et al. 2003a). The heterozygous diploid strain was sporulated and tetrads were dissected to generate srp1-31 haploids (ACY1561 and ACY1562).
To generate cells in which microtubules could be visualized directly with GFP, TUB1-GFP (pAC1344) was integrated at the URA3 locus as described previously (Straight and Murray 1997). The Cdc45, Mcm10, and Yox1 proteins were visualized by monitoring the localization of the previously described integrated C-terminal GFP fusion proteins: Cdc45-GFP (YLR103C), Mcm10-GFP (YIL150C), or Yox1-GFP (YML027W) (Huh et al. 2003). Each of these strains was crossed to either the srp1-31 (ACY1561) or the srp1-55 (ACY642) mutant. The resulting diploid strains were sporulated and tetrads were dissected to generate srp1 mutant strains expressing Cdc45-GFP, Yox1-GFP, or Mcm10-GFP (Table 1).
In vivo functional analysis:
The function of importin-α variants in vivo was assessed by examining the growth of the integrated alleles srp1-31, srp1-55, and srp1-E402Q. As a control, each mutant was covered with a wild-type SRP1 URA3 plasmid (pAC876) to ensure that conditional phenotypes were complemented prior to the growth assays. Single colonies were grown to saturation in liquid culture, serially diluted (1:10), and spotted on minimal medium plates as a control or on fluoroorotic acid (5-FOA) plates. The drug 5-FOA eliminates the URA3 plasmid-encoded wild-type SRP1 (pAC876) to reveal the phenotype of the mutants (Boeke et al. 1987). Plates were incubated at the indicated temperatures for 3–7 days.
Immunoblot analysis:
Immunoblot analysis was performed using standard methods (Towbin et al. 1979). Cultures were grown to log phase in yeast extract peptone dextrose (YEPD) media at 25° and then shifted to the indicated temperature. Cells were then harvested by centrifugation and washed twice in water and once in PBSMT [100 mm KH2PO4, pH 7.0, 15 mm (NH4)2SO4, 75 mm KOH, 5 mm MgCl2, 0.5% Triton X-100]. Cells were subsequently lysed in PBSMT with protease inhibitors (0.5 mm phenylmethylsulfonyl fluoride, 3 μg/ml each of aprotinin, leupeptin, chymostatin, and pepstatin) by glass bead lysis. Equal amounts of total protein were resolved by SDS–PAGE and immunoblotted with polyclonal anti-importin-α antibody (1:5000 dilution) raised against recombinant GST–importin-α followed by anti-rabbit secondary antibody (1:5000 dilution).
NLS–GFP import assay:
The NLS–GFP import assay was performed as described previously (Shulga et al. 1996; Hodel et al. 2006). Cells were resuspended in glucose-containing synthetic media pre-equilibrated to 25° (permissive), 37° (srp1-31, srp1-E402Q), or 18° (srp1-55). For scoring, 2-μl samples were removed every 2.5 min and images were collected through a GFP-optimized filter (Chroma Technology) using an Olympus BX60 epifluorescence microscope. Cells were scored as “nuclear” if both the nucleus was brighter than the surrounding cytoplasm and a nuclear-cytoplasmic boundary was visible. At least 100 cells were counted at each time point.
Microscopy:
Direct fluorescence microscopy was performed to localize GFP fusion proteins in live cells. For all experiments, cultures were also labeled with Hoechst dye (1 μg/ml) to visualize DNA and confirm the location of the nucleus. The localization of GFP fusion proteins was monitored by directly viewing the GFP signal in living cells through a GFP-optimized filter (Chroma Technology) using an Olympus BX60 epifluorescence microscope equipped with a Photometrics Quantix digital camera. For localization of candidate cargoes, cells expressing Cdc45-GFP, Mcm10-GFP, or Yox1-GFP were grown to log phase at the permissive temperature and then shifted to the nonpermissive temperature for 3 hr.
High-copy suppressor analysis:
For high-copy suppressor analysis, high-copy plasmids (2μ TRP1) encoding the nuclear transport factors importin-β (pAC592), Cse1 (pAC1303), and Nup2 (pAC1385) were transformed into srp1-55 (ACY641), srp1-E402Q (ACY1560), and srp1-31 (ACY1561) cells covered by an SRP1 URA3 maintenance plasmid (pAC876). Genetic suppression was assessed by growing single colonies in liquid culture to saturation, serially diluting (1:10), and spotting on minimal medium plates as a control or on 5-FOA plates. Plates were incubated at the indicated temperatures for 3–6 days.
Cell cycle arrest and release:
For cell cycle studies, cultures were synchronized by treatment with α-factor or hydroxyurea. For arrest with α-factor, cells were grown to early mid-log phase (OD600 0.25–0.35) in YEPD media at 25°. Cell cycle arrest was accomplished by pelleting the cells, washing with YEPD (pH 3.9), and resuspending in fresh YEPD (pH 3.9) containing a 1:500 dilution of 5 mg/ml of α-factor (Sigma) followed by a 90-min incubation at 25°. Additional α-factor (1:1000 dilution) was added every 30 min. Cells were released from α-factor arrest by pelleting the cells, washing twice with YEPD, and resuspending in fresh YEPD for release at the indicated temperatures for an additional 3 hr (Swaminathan et al. 2007).
For hydroxyurea arrest, cells were arrested with α-factor as described above. Then cells were pelleted, washed twice with YEPD, and resuspended in YEPD containing 200 mm hydroxyurea. Cultures were incubated for an additional 2 hr at 25°. Cells were then pelleted, washed twice with YEPD, and resuspended in fresh YEPD for release at the indicated temperatures for an additional 3 hr (Swaminathan et al. 2007).
Flow cytometry analysis:
Cells were prepared for flow cytometry analysis by staining with propidium iodide (Sazer and Sherwood 1990). Briefly, asynchronous or synchronous cultures were ethanol fixed overnight at 4°, washed, and resuspended in 50 mm sodium citrate. Cells were then treated with 0.1 mg/ml RNase A for 2 hr at 37° and 10 mg/ml of Proteinase K for 1 hr at 50°. Cells were stained with 8 μg/ml of propidium iodide. Each sample was analyzed with a FACSVantage SE (Becton Dickinson, Franklin Lakes, NJ). Data were analyzed using FloJo 7.2.2 software.
Plasmid loss:
Plasmid loss was determined in wild type or mutant cells containing plasmids with an early (ARS305)-, middle (ARS1)-, or late (ARS1412)-firing autonomously replicating sequence (ARS) (Friedman et al. 1996). Cells were grown to log phase after which at least 200 cells were plated on nonselective YEPD plates. Colonies that grew on YEPD plates were then replica plated to ura− selective plates to determine plasmid loss. The percentage of plasmid loss was calculated as 100 − (the ratio of colonies on the ura− selective plate/colonies on the YEPD plate) × 100. The experiment was performed in triplicate.
Statistical analysis:
Statistical methods were employed to determine if a significant difference was observed in the percentage of plasmid loss in the srp1 mutant cells as compared to wild-type cells. Data were analyzed using a one-way analysis of variance (ANOVA) for origin of replication followed by a Dunnett's multiple comparison test using Graph Pad Prism 3.0. The significance level (α) was set at 0.05 for all statistical tests. If the calculated P-value was <α, then the difference in plasmid loss was reported as being statistically significant.
RESULTS
To examine links between the classical nuclear protein import pathway and cell cycle progression, we exploited two mutants of importin-α with specific molecular defects. The srp1-E402Q mutant alters a critical glutamic acid residue in the minor pocket of the cNLS cargo-binding domain to glutamine (Conti et al. 1998; Fontes et al. 2000). This conservative amino acid substitution at position 402 creates the conditional allele of importin-α, srp1-E402Q, which causes a decrease in bipartite cNLS cargo binding in vitro and affects the steady-state localization of a bipartite NLS cargo in vivo (Leung et al. 2003).
As a complement to the analysis of the NLS cargo-binding pocket, a variant of importin-α that affects cargo delivery was also employed. A conserved NLS-like sequence within the N-terminal IBB domain of importin-α, 54KRR56, is essential for the auto-inhibitory function of importin-α (Harreman et al. 2003b). An arginine-to-alanine substitution at position 55 within this auto-inhibitory sequence (KRR → KAR) creates a conditional allele of importin-α, srp1-55, which specifically affects cargo delivery/release into the nucleus (Harreman et al. 2003a).
Functional analysis of srp1 mutants:
To compare the consequences of defects in cargo binding and cargo release in vivo, we generated alleles of the srp1-55 and srp1-E402Q importin-α mutants that could be directly compared to one another (see materials and methods). As a control, we also generated the previously characterized srp1-31 mutant (Loeb et al. 1995) in the same genetic background. As an initial characterization and comparison of these mutants, we analyzed their growth at various temperatures. To ensure that equal numbers of cells were grown and spotted, the srp1-31, srp1-55, and srp1-E402Q mutants were transformed with a wild-type SRP1 plasmid. To assay growth, 10-fold serial dilutions of the samples were spotted on control plates where wild-type SRP1 is maintained or on 5-FOA plates where the plasmid encoding wild-type SRP1 is lost. In comparison to wild-type cells, srp1-55 mutant cells show a growth defect at 18° as previously reported (Harreman et al. 2003a). In contrast, srp1-31 and srp1-E402Q mutants show growth defects at 37° (Figure 1B). Immunoblot analysis indicates no significant change in the level of any of the Srp1 mutant proteins at the nonpermissive temperatures as compared to wild-type importin-α, indicating that the growth defects observed are not due simply to loss of the essential importin-α protein (data not shown).
NLS–GFP import assay:
Prior studies examined the cargo-binding properties of the srp1-55 and srp1-E402Q variants in vitro as well as their impact on the steady-state localization of cNLS cargo in vivo (Harreman et al. 2003a; Leung et al. 2003). To further characterize the impact of each of these alleles as well as the srp1-31 mutant on nuclear protein import, we used a semiquantitative kinetic NLS–GFP import assay that assesses the initial rate of NLS cargo import (Shulga et al. 1996). For this assay, import of both a monopartite SV40 and a bipartite BPSV40T3 NLS cargo was examined. The SV40 and BPSV40T3 cargoes were selected for the NLS–GFP import assay because both bind to importin-α with a similar affinity (∼10 nm) and, importantly, the bipartite cargo is engineered such that productive binding to importin-α absolutely depends on the basic cluster of amino acids that binds to the minor NLS binding pocket (Hodel et al. 2001). Experiments were carried out at the nonpermissive temperatures in live cells as described in materials and methods. In comparison to wild-type cells, the srp1-31 and srp1-55 mutants showed a decrease in the initial rate of nuclear import of the monopartite cNLS cargo SV40-GFP (Figure 1C). As expected, there was no observed change in the initial import rate of the monopartite cargo in srp1-E402Q cells (Figure 1C) because the E402Q amino acid substitution decreases bipartite cargo binding without impairing binding to monopartite cNLS cargo (Leung et al. 2003). However, all three mutants, srp1-31, srp1-55, and srp1-E402Q, show a decrease in the initial rate of nuclear import of the bipartite cNLS cargo BPSV40T3-GFP (Figure 1D). These results confirm that cells expressing each variant of importin-α have defects in cNLS protein import.
Importin-α variants differ in their in vivo molecular defects:
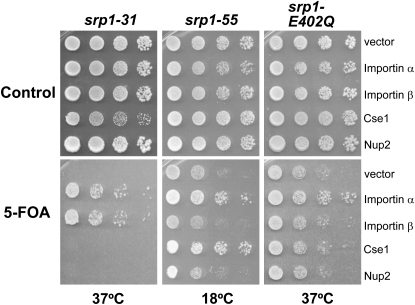
Although all of the importin-α mutant cells showed defects in the kinetic NLS import assay, we hypothesized that the different engineered amino acid changes in each importin-α variant should impair nuclear import through distinct mechanisms. To address this hypothesis, we tested whether each of the importin-α mutants could be suppressed by overexpression of several nuclear transport factors with the assumption that suppression by specific factors could provide information about what interactions are compromised in vivo. For this analysis, we examined overexpression of importin-β, Cse1, and the nuclear pore protein, Nup2. Importin-β interacts with the IBB domain of importin-α to target the import complex to the nuclear pore (Görlich et al. 1996; Weis et al. 1996). Cse1 is the export receptor for recycling importin-α to the cytoplasm (Hood and Silver 1998; Solsbacher et al. 1998) and both Cse1 and Nup2 facilitate cargo delivery in the nucleus (Matsuura et al. 2003; Matsuura and Stewart 2004; Liu and Stewart 2005). As controls, each srp1 mutant was also transformed with a vector alone or with a wild-type importin-α plasmid. To ensure equal growth and spotting, each mutant was transformed with wild-type SRP1 on a URA3 plasmid. Cultures were grown to saturation, serially diluted, and then spotted either on control plates where the SRP1 maintenance plasmid is retained or on plates containing 5-FOA where the maintenance plasmid is lost but the overexpression plasmids are retained (Figure 2). The 5-FOA plates were incubated at the nonpermissive temperature as previously determined (see Figure 1B) for each mutant. Interestingly, we find that the temperature-sensitive growth defect of srp1-31 cells is suppressed by overexpression of importin-β (Figure 2). As previously observed (Harreman et al. 2003a), the cold-sensitive growth defect of srp1-55 mutant cells is suppressed by overexpression of the nuclear export factor, Cse1 (Figure 2). In contrast, the temperature-sensitive growth defect of srp1-E402Q cells is not suppressed by any of the nuclear transport factors (Figure 2). Taken together, these results suggest that the in vivo defects that underlie diminished protein import in each of these mutants are likely to be distinct from one another.
Figure 2.—
High-copy suppressor analysis. High-copy plasmids encoding the nuclear transport factors, importin-β (pAC592), Cse1 (pAC1303), and Nup2 (pAC1385), were transformed into srp1-55, srp1-E402Q, and srp1-31 cells, which also contained a wild-type SRP1 URA3 plasmid (pAC876). As a control, each mutant was also transformed with vector alone (pRS424) or with wild-type SRP1 plasmid (pAC1354). Genetic suppression was assessed by serially diluting and spotting on control plates (where the wild-type SRP1 plasmid is retained) or on 5-FOA plates (where the wild-type SRP1 plasmid is lost). Plates were incubated at the indicated temperatures for 3–6 days. Suppression was scored as enhanced growth at the nonpermissive temperature as compared to the vector control.
Mutants of importin-α affect cell cycle progression:
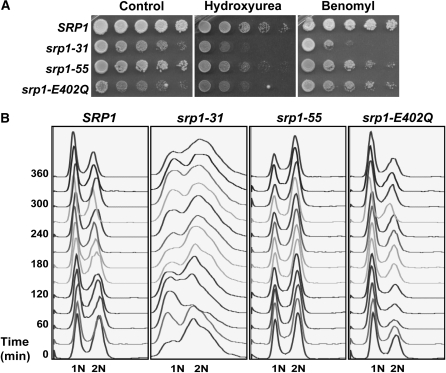
To probe the link between importin-α and cell cycle events, we exploited these srp1 mutants that affect the classical nuclear protein import pathway through distinct mechanisms. As a first step to determining whether all defects in importin-α impair G2/M of the cell cycle as previously described for the srp1-31 mutant (Loeb et al. 1995), each importin-α mutant was spotted on plates containing either hydroxyurea, which inhibits DNA replication (Mitchison and Creanor 1971), or benomyl, which blocks mitosis prior to the onset of anaphase (Li and Murray 1991). Consistent with previous reports, the srp1-31 mutant shows sensitivity to growth on benomyl, suggesting a defect in G2/M of the cell cycle (Figure 3A) (Loeb et al. 1995). In contrast, the srp1-55 and srp1-E402Q mutant cells show no obvious sensitivity to benomyl. Surprisingly, in comparison to wild-type cells, srp1-31, srp1-55, and srp1-E402 mutants all show sensitivity to hydroxyurea, suggesting a defect in processes critical to the G1/S cell cycle transition, including DNA replication, DNA repair, and/or checkpoint function (Figure 3A).
Figure 3.—
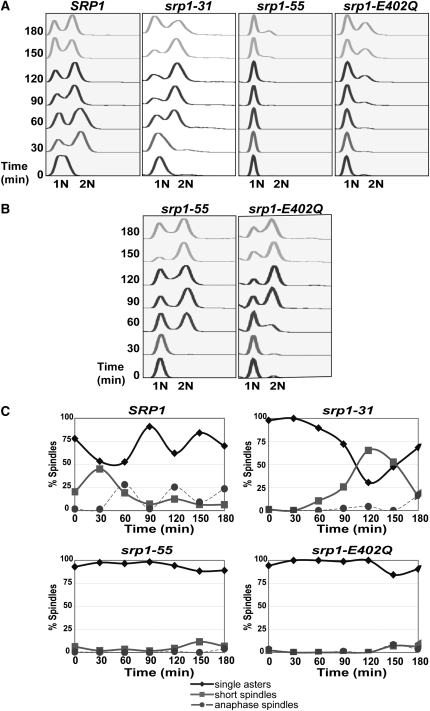
Analysis of cell cycle defects in srp1 mutants. (A) Analysis of cell growth on hydroxyurea and benomyl plates. To determine whether any of the srp1 mutant alleles show hypersensitivity to growth on plates containing hydroxyurea or benomyl, cultures were grown to saturation at the permissive temperature and then serially diluted and spotted on control YEPD plates, YEPD containing 100 mm hydroxyurea, or YEPD plates containing 10 mg/ml benomyl. Plates were incubated at the permissive temperature for 3–7 days. (B) Analysis of cell cycle progression in unsynchronized cells. Wild-type (SRP1) and mutant (srp1-31, srp1-55, and srp1-E402Q) cells were grown to early mid-log phase at the permissive temperature and then shifted to the nonpermissive temperature (37° for srp1-31 and srp1-E402Q cells and 18° for srp1-55 cells). Samples were collected every 30 min for 6 hr for flow cytometry to analyze DNA content. The positions of unreplicated DNA (1N) and replicated DNA (2N) are indicated below the graphs.
To begin to assess how the cell cycle is affected in each of the srp1 mutants, the DNA content of each mutant was measured in an asynchronous cell population using flow cytometry (Figure 3B). Samples were analyzed at 30-min intervals over 6 hr at both permissive and nonpermissive temperatures. The cells are scored as having 1N (G1) or 2N (G2/M) DNA content. All importin-α mutants show a distribution of 1N and 2N DNA content that is indistinguishable from wild-type (SRP1) cells at the permissive temperature (data not shown). In wild-type (SRP1) cells, the distribution of 1N and 2N DNA content is unchanged relative to the permissive temperature. Consistent with previous results (Loeb et al. 1995), srp1-31 mutant cells show some accumulation of cells in the G2/M phase of the cell cycle as compared to the G1 phase. As with srp1-31 cells, srp1-55 cells show some increase in cells with 2N DNA content as compared to wild-type cells (SRP1), which is consistent with previous analysis of an asynchronous population of these cells (Harreman et al. 2003a). In contrast to the srp1-31 and srp1-55 mutants, srp1-E402Q mutant cells show an increase in the population of cells with 1N DNA content as compared to wild-type (SRP1) cells. While these results are consistent with impaired cell cycle transitions due to compromised classical nuclear protein import, defects at multiple points in the cell cycle are not readily uncovered by analysis of asynchronous cultures.
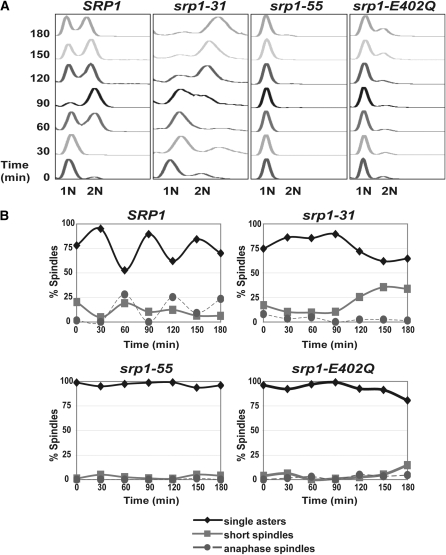
Both sensitivity to hydroxyurea and analysis of DNA content for the srp1-E402Q mutant suggest a role for importin-α in the G1/S cell cycle transition. To assess the DNA content of synchronized cultures, cells were arrested in late G1 phase of the cell cycle with the mating-type pheromone, α-factor (Siede et al. 1989; Siede and Friedberg 1990), and then analyzed by flow cytometry over time following release from α-factor. Samples were analyzed at 30-min intervals over 3 hr at both permissive and nonpermissive temperatures. All importin-α mutants show wild-type progression at the permissive temperature (data not shown). As expected, wild-type cells progress through the cell cycle over the time course of the experiment at all temperatures. As previously reported, srp1-31 mutant cells display a defect in progression through G2/M (Figure 4A) (Loeb et al. 1995). The srp1-31 mutant also shows a slight defect in the G1/S phase transition, which is consistent with growth sensitivity to hydroxyurea. Interestingly, both srp1-55 and srp1-E402Q mutant cells show a profound defect in the G1/S transition (Figure 4A). These results link importin-α and hence the classical nuclear import pathway to the G1/S transition of the cell cycle as well as the previously reported G2/M transition.
Figure 4.—
Analysis of cell cycle progression in cells synchronized in G1 phase of the cell cycle. (A) Wild-type (SRP1) and mutant (srp1-31, srp1-55, and srp1-E402Q) cells were grown to early mid-log phase at the permissive temperature and then arrested at late G1 phase with α-factor. Samples were then released to the nonpermissive temperature (37° for srp1-31 and srp1-E402Q cells and 18° for srp1-55 cells). Samples were collected every 30 min for 3 hr for flow cytometry to analyze DNA content. The positions of unreplicated DNA (1N) and replicated DNA (2N) are indicated below the graphs. (B) Spindle morphology. Wild-type (SRP1) and mutant (srp1-31, srp1-55, and srp1-E402Q) cells expressing Tub1-GFP to visualize microtubules were treated as described above in A. Spindles were visualized by examining integrated Tub1-GFP signal by direct fluorescence microscopy. Results are plotted as the percentage of spindles in the total cell population scored as single asters (no spindles, ♦); short spindles (spindle not extending to the duplicated nucleus of the daughter cell, ▪); or anaphase spindles (spindle extending to the duplicated nucleus, •) vs. time.
Using the synchronization method employed for flow cytometry, the spindle morphology of each mutant following the shift to the nonpermissive temperature was assessed over time (Figure 4B). The percentage of spindle formation was defined as containing the following: single asters (no spindles), short spindles (spindle not extending to the duplicated nucleus of the daughter cell), or anaphase spindles (spindle extending to the duplicated nucleus) (Goh and Surana 1999). In comparison to wild-type cells, srp1-31 cells show an increased number of short spindles and a decrease in the relative number of anaphase spindles after 3 hr at the nonpermissive temperature (Figure 4B). In contrast, srp1-55 and srp1-E402Q mutant cells show a constant level of single asters over time, which correlates with a defect in the G1/S phase transition where spindle formation has not yet begun (Figure 4B).
Since a profound delay at the G1/S transition was observed in srp1-55 and srp1-E402Q mutant cells, we wanted to determine whether these cells were defective not only in transition but also in progression through S phase. To examine progression through S phase, the mutant cells were arrested in G1 using α-factor at the permissive temperature and then released into media containing hydroxyurea to synchronize cells in early S phase. After 2 hr, cells were released at either the permissive or the nonpermissive temperature. Samples were then analyzed by flow cytometry. At the permissive temperature, wild-type and all mutant cells progress through the cell cycle in a comparable manner (data not shown). However, when mutants are released to the nonpermissive temperature, both srp1-55 and srp1-E402Q cells progress slowly through S phase in comparison to wild-type cells (Figure 5A). This experiment also provides further evidence that srp1-31 cells have defects in S phase progression as well as in G2/M as they also progress through S phase more slowly than wild-type cells (Figure 5A). To ensure that both srp1-55 and srp1-E402Q cells were able to release from α-factor arrest, the DNA content of these mutant cells was also monitored at the permissive temperature (Figure 5B). Flow cytometry analysis shows that both srp1-55 and srp1-E402Q cells can release from α-factor arrest and enter the cell cycle at the permissive temperature, providing evidence that these cells, when first synchronized with α-factor and then released into media containing hydroxyurea, should be synchronized in early S phase. We also employed spindle morphology analysis for each mutant as independent confirmation of the cell cycle delays observed in the srp1-31, srp1-55, and srp1-E402Q cells (Figure 5C). As observed for the α-factor arrest, both srp1-55 and srp1-E402Q cells showed nearly 100% of cells with single asters throughout the time course of the experiment. This spindle morphology is consistent with the failure to enter the cell cycle over the time course of the experiment. As suggested by the flow cytometry data, the srp1-31 cells show a delay in spindle formation relative to wild-type (SRP1) cells, but spindles do begin to form and elongate in this mutant after ∼90 min [compare to ∼30 min for wild-type (SRP1) cells].
Figure 5.—
Analysis of cells synchronized in S phase of the cell cycle. (A) Wild-type (SRP1) and mutant (srp1-31, srp1-55, and srp1-E402Q) cells were grown to early mid-log phase at the permissive temperature and then arrested at late G1 phase with α-factor. Samples were then released into hydroxyurea at the permissive temperature and incubated for 2 hr. Finally, the cells synchronized in early S phase were released to the nonpermissive temperature (37° for srp1-31 and srp1-E402Q cells and 18° for srp1-55 cells). Samples were collected every 30 min for 3 hr for analysis by flow cytometry to analyze DNA content. The positions of unreplicated DNA (1N) and replicated DNA (2N) are indicated below the graphs. (B) Mutant (srp1-31 and srp1-E402Q) cells were grown to early mid-log phase at the permissive temperature and then arrested at late G1 phase with α-factor. Samples were then released to the permissive temperature to determine if the cells are released from α-factor arrest. Samples were collected every 30 min for 3 hr for flow cytometry to analyze DNA content. The positions of unreplicated DNA (1N) and replicated DNA (2N) are indicated below the graphs. (C) Spindle morphology. Wild-type (SRP1) and mutant (srp1-31, srp1-55, and srp1-E402Q) cells expressing Tub1-GFP to visualize microtubules were treated as described above in A. Spindles were visualized by examining integrated Tub1-GFP signal by direct fluorescence microscopy. Results are plotted as the percentage of spindles in the total cell population scored as single asters (no spindles, ♦), short spindles (spindle not extending to the duplicated nucleus of the daughter cell, ▪), or anaphase spindles (spindle extending to the duplicated nucleus, •) vs. time.
Results indicate that all of the importin-α mutants examined show defects in the G1/S transition and in S phase progression. As an initial approach to understanding how srp1 mutants might affect DNA replication, we utilized a plasmid loss assay (Friedman et al. 1996; Dohrmann and Sclafani 2006). This assay monitors loss of plasmids containing an ARS, which function as replication origins in S. cerevisiae (Dohrmann and Sclafani 2006). Although defects in many cell cycle pathways can contribute to chromosome loss, mutants with general DNA replication defects exhibit loss regardless of the origin of replication used in the plasmid, whereas mutants specific for replication initiation often exhibit differential loss dependent on the nature of the replication origin (Maine et al. 1984; Hartwell and Smith 1985; Palmer et al. 1990).
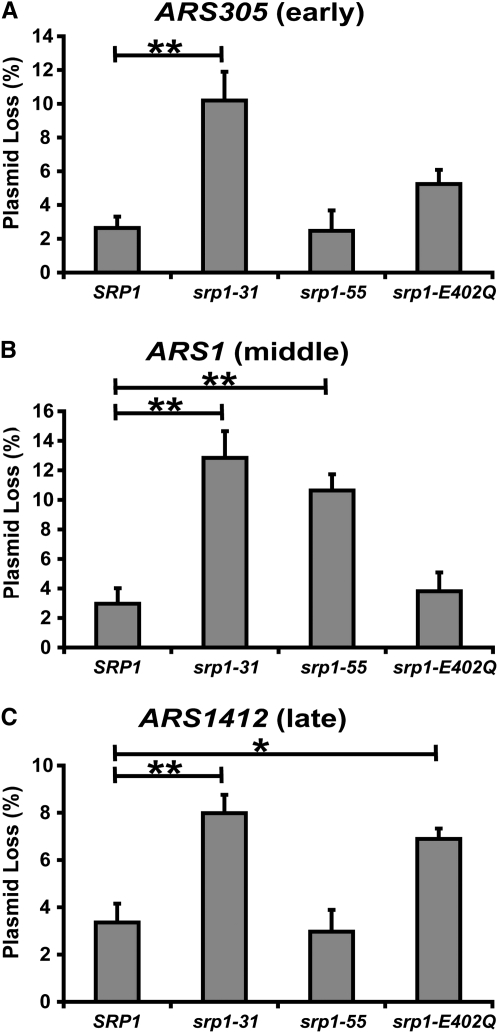
Therefore, we compared the rate of loss of plasmids containing an ARS that fires early in S phase (ARS305), in the middle of S phase (ARS1), or late in S phase (ARS1412) (Friedman et al. 1996). All results are compared to wild-type (SRP1) cells (Figure 6). In comparison to wild-type (SRP1) cells, both the srp1-31 and srp1-E402Q mutant cells exhibit plasmid loss of the early firing origin of replication, ARS305. As described in materials and methods, statistical analysis reveals a significant increase in plasmid loss in the srp1-31 mutant as compared to the wild-type (SRP1) cells (**P < 0.01) (Figure 6A). We also observed increased plasmid loss in srp1 mutant cells with ARS1, which fires in the middle of S phase. Statistical analysis shows a significant increase in plasmid loss in both the srp1-31 (**P < 0.01) and the srp1-55 (**P < 0.01) mutant cells (Figure 6B). Finally, the srp1-31 and srp1-E402Q mutants show an increase in plasmid loss for the late-firing origin of replication, ARS1412. Statistical analysis reveals a significant increase in plasmid loss for both srp1-31 (**P < 0.01) and srp1-E402Q (*P < 0.05) mutant cells as compared to wild-type (SRP1) cells (Figure 6C). To rule out changes within the wild-type control, plasmid loss for early, middle-, and late-firing replication origins was compared in wild-type cells. This comparative analysis shows no significant difference in plasmid loss between the early, middle, and late replication origins in the wild-type cells (P = 0.8417) (data not shown). Interestingly, the srp-31 mutant shows a significant increase in plasmid loss at the early, middle-, and late-firing origins of replication, suggesting a general chromosome loss phenotype. The plasmid loss defect in the srp1-55 mutant suggests a specific defect in the middle-firing origin of replication, ARS1. In contrast, the srp1-E402Q mutant shows a specific defect in the late-firing origin of replication, ARS1412. Together, these results indicate that these srp1 alleles have distinct effects on chromosome loss, which may result from differential import of key cell cycle proteins.
Figure 6.—
Analysis of plasmid loss. To determine if srp1 mutant cells have defects in DNA replication, a plasmid loss assay was performed. Plasmid loss was determined in cells containing plasmids with early (ARS305)-, middle (ARS1)-, or late (ARS1412)-firing ARSs as described in materials and methods. Results are plotted as the mean percentage of plasmid loss for (A) ARS305 (early)-, (B) ARS1 (middle)-, or (C) ARS1412 (late)-firing ARS sequences for each srp1 mutant (srp1-31, srp-55, and srp1-E402Q) as well as for wild-type (SRP1) control cells. Experiments were performed in triplicate. Standard deviations in the data are indicated by the error bars. A one-way ANOVA followed by a Dunnett's multiple comparison test using an α-value of 0.05 was used to determine the significance of the results. Significant differences between samples corresponding to the P-values of *P < 0.05 and **P < 0.01 are indicated by the lines above the bars.
Bipartite cNLS candidates involved in G1/S of the cell cycle:
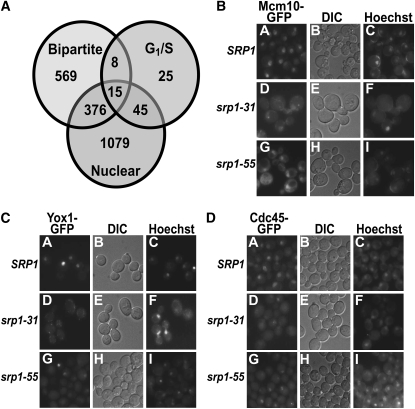
While it is likely that numerous import cargoes are affected in the srp1 mutant cells, we wanted to determine whether there were key candidates most likely to be affected. To identify candidate cargoes whose import could be impaired in the srp1-55 and srp1-E402Q cells, a bioinformatic approach was taken to identify proteins containing a putative bipartite cNLS that are involved in G1/S of the cell cycle and are localized to the nucleus (Figure 7A). To identify the entire complement of candidate bipartite cNLS cargoes, the PSORT II algorithm for predicting bipartite cNLS motifs (Nakai and Horton 1999) was used to query the S. cerevisiae GenBank (Benson et al. 2006). Of 5850 proteins, 968 contain a predicted bipartite cNLS. We then used the PSORTII algorithm to query the pool of 1515 nuclear or nucleolar proteins as defined by localization of GFP fusion proteins (Huh et al. 2003). Of these 1515 nuclear/nucleolar proteins, 391 contain a predicted bipartite cNLS (Huh et al. 2003). As a preliminary approach to identifying a subset of these proteins linked to the G1/S cell cycle transition, we searched for genes containing the terms G1, S, or G1/S in their Gene Ontology definition (http://www.geneontology.org/). Through this initial approach, we identified 93 proteins implicated in G1/S of the cell cycle. The overlap between these three data sets identifies 15 proteins that contain a predicted bipartite cNLS, localize to the nucleus, and are already implicated in the G1/S transition (Figure 7A). These 15 candidate proteins that could be affected in cells that are defective in nuclear import of cargoes containing bipartite cNLS are listed in Table 3.
Figure 7.—
Candidate bipartite proteins implicated in G1/S of the cell cycle are not efficiently targeted to the nucleus in importin-α mutants. (A) The PSORT II algorithm for bipartite cNLSs was used to query three sets of data: the 5850 proteins in the S. cerevisiae GenBank (bipartite) (Benson et al. 2006), the 1515 proteins localized to either the nucleus or the nucleolus in the global yeast GFP-fusion library (nuclear) (Huh et al. 2003), and 93 proteins implicated in G1/S of the cell cycle based on the Gene Ontology definition (G1/S) (http://www.geneontology.org/). The overlap between these three data sets identifies 15 proteins that contain a predicted bipartite cNLS, localize to the nucleus, and are already implicated in G1/S of the cell cycle. Data from this analysis are presented as a Venn diagram. (B–D) Wild-type (SRP1) and srp1 mutant (srp1-31, srp1-55) cells expressing Mcm10-GFP (B), Yox1-GFP (C), or Cdc45-GFP (D) were analyzed by direct fluorescence microscopy. Cultures were grown to log phase at the permissive temperature and then shifted to the nonpermissive temperature (37° for srp1-31 cells and 18° for srp1-55 cells) for 3 hr. Wild-type cells (SRP1) were analyzed following shifts to both 37° and 18° and identical results were obtained with each of the GFP fusion proteins. Results are shown for the wild-type cells shifted to 37°. Corresponding DIC and Hoechst DNA-staining images are shown.
TABLE 3.
Bipartite cNLS candidates involved in G1/S
| Protein | Function |
|---|---|
| Bck2 | Molecular function unknown |
| Cdc40 | RNA splicing factor activity |
| Cdc45 | DNA replication |
| Mcm10 | DNA replication and chromatin binding |
| Rsc3 | DNA binding |
| Swi5 | DNA replication and transcriptional activator activity |
| Tos4 | Transcription factor activity |
| Tye7 | Transcription factor activity |
| Pog1 | RNA POL II transcription factor activity |
| Taf1 | RNA POL II transcription factor activity |
| Taf13 | RNA POL II transcription factor activity |
| Taf14 | RNA POL II transcription factor activity |
| Yap5 | RNA POL II transcription factor activity |
| Yhp1 | DNA binding |
| Yox1 | DNA binding |
As an initial test of whether the nuclear import of any of these candidate cargoes is impaired in the importin-α mutants, we selected three candidates that had previously been visualized as nuclear GFP fusion proteins: Cdc45, Mcm10, and Yox1 (Huh et al. 2003). Cdc45 is involved in DNA replication and remains nuclear throughout the S. cerevisiae cell cycle (Moir et al. 1982; Hopwood and Dalton 1996). Cdc45 interacts with the minichromosome maintenance proteins, which regulate DNA replication (Hopwood and Dalton 1996). Mcm10 is a constitutively nuclear chromatin-associated protein that plays a role in DNA replication initiation by recruiting both the Mcm2-7 complex and DNA polymerase-α to DNA replication origins (Burich and Lei 2003; Ricke and Bielinsky 2004; Zhu et al. 2007). Yox1 is a homeodomain transcriptional repressor that regulates the expression of genes that are critical for the G1/S transition (Pramila et al. 2002; Braun and Breeden 2007). We examined the localization of Mcm10-GFP, Yox1-GFP, and Cdc45-GFP in wild-type (SRP1), srp1-31, and srp1-55 cells (Figure 7, B–D). At the permissive temperature, Mcm10-GFP, Yox1-GFP, and Cdc45-GFP are localized to the nucleus in all cells examined (data not shown). Following a shift to the nonpermissive temperature, both srp1-31 and srp1-55 cells show a decrease in nuclear localization of Mcm10-GFP (Figure 7B) and Yox1-GFP (Figure 7C) in comparison to wild-type (SRP1) cells. Interestingly, Cdc45-GFP remains nuclearly localized in srp1-55 cells but is mislocalized to the cytoplasm in srp1-31 cells (Figure 7D).
DISCUSSION
Results of this study define an important role for the NLS receptor, importin-α, in DNA replication. Specifically, we report that two mutants of importin-α cause a profound delay in the G1/S cell cycle transition. Although previous studies described defects in G2/M associated with other importin-α mutants, srp1-31 (Loeb et al. 1995) and srp1-55 (Harreman et al. 2003a), this is the first report demonstrating that mutations in the NLS receptor and hence defects in the classical protein import pathway impair the G1/S transition.
Our study also reveals that the srp1-31 mutant shows a delay in both the G2/M and G1/S cell cycle transitions, although the G1/S delay for the srp1-31 cells is not as profound as observed for srp1-55 and srp1-E402Q cells. The G2/M cell cycle defect observed for the srp1-31 mutant is consistent with previous work (Loeb et al. 1995) and also with hypersensitivity to benomyl. Genetic analysis indicated that the temperature-sensitive growth defect of srp1-31 is suppressed by the overexpression of the nuclear import receptor, importin-β. This finding suggests that the amino acid change in the srp1-31 variant protein could impair importin-β binding. As interaction between importin-α and -β is critical to targeting cargoes to the nuclear pore for import (Stewart 2007), decreased interaction with importin-β could lead to profound defects in nuclear import of critical cargoes required for cell cycle transitions.
This study is distinct from the previous analysis of the srp1-31 allele because the importin-α variants employed here have well-characterized molecular defects. The Srp1-E402Q mutant protein is impaired in binding to cargoes that contain a bipartite cNLS (Leung et al. 2003). The Srp1-55 protein shows defects in cargo delivery into the nucleus (Harreman et al. 2003a). We find that cells that express each of these importin-α variants as the sole cellular copy of importin-α have defects in the G1/S cell cycle transition. The finding that importin-α variants with defects in cargo binding and cargo delivery cause similar cell cycle defects may be surprising; however, the most logical interpretation of this result may be that both variants are defective in the effective nuclear import of cargo proteins that contain a classical bipartite NLS. This conclusion is fairly obvious for the srp1-E402Q protein, which has an amino acid substitution in a key residue of the minor NLS-binding pocket that makes contact only with cargoes that contain a bipartite cNLS and, in fact, shows only defects in the import of bipartite cNLS cargoes (Conti et al. 1998; Leung et al. 2003). In the case of the srp1-55 protein, the auto-inhibitory, and hence the inherent cargo release mechanism, is impaired (Harreman et al. 2003a). However, in cells, there are additional factors that facilitate cargo release, including the export receptor, Cse1, and the nuclear pore protein, Nup2 (Solsbacher et al. 1998, 2000; Gilchrist et al. 2002). Indeed, overexpression of Cse1 suppresses the growth defect of the srp1-55 mutant. Possibly, cargoes that bind tightly to the NLS receptor may be most affected in vivo when the cargo delivery function inherent to importin-α is impaired. These tight-binding cargoes are most likely to be the cargoes that contain a bipartite cNLS as bipartite cargoes typically bind with a stronger affinity to the NLS receptor than monopartite cargoes (Hodel et al. 2001). Thus, both srp1-55 and srp1-E402Q cells may, logically, be most impaired in nuclear delivery of bipartite cNLS cargo proteins.
Although srp1-55 and srp1-E402Q mutant cells show similar defects in the cell cycle, the plasmid loss assay provides further insight into the molecular defect. The plasmid loss assay indicates that a defect in cNLS cargo binding (srp1-E402Q) affects DNA replication late in S phase. Similarly, the plasmid loss assay suggests that a defect in cNLS cargo release (srp1-55) also affects DNA replication but in the middle of S phase. Furthermore, as with srp1-31 cells, our results suggest that srp1-55 cells are impaired in both the G1/S and the G2/M cell cycle transitions. Interestingly, these results suggest that different defects in the NLS receptor importin-α are likely to affect the import of distinct complements of nuclear import cargoes, including some that may be uniformly affected and some that may be distinct for each mutant. Presumably, which cargoes are affected in each mutant depends on both cargo recognition in the cytoplasm and release into the nucleus. Other factors that effect recognition and release could also influence the phenotype of these mutants.
The use of variants of importin-α with specific molecular defects allowed us to identify a number of candidate proteins that may be affected in these mutants. For three of these candidate proteins, Mcm10, Yox1, and Cdc45, we show that nuclear localization is differentially impaired in the importin-α mutants. While Mcm10 and Yox1 are mislocalized in each importin-α mutant examined, Cdc45 is mislocalized to the cytoplasm only in srp1-31 cells. Overall, these results show that different defects in importin-α affect distinct cargoes. In the future, genetic approaches that exploit the conditional growth phenotypes of these srp1 mutants may be useful in identifying additional cargo proteins affected in these mutants, including some that may be specific for each mutant.
Collectively, our data demonstrate a molecular role for the nuclear localization signal receptor, importin-α, during the G1/S stage of the cell cycle. This finding adds to our previous understanding of the link between nuclear transport and cell cycle progression as previously only defects in the G2/M transition had been linked to mutants in the nuclear import receptor. Future studies will be aimed at identifying key cargoes that must enter the nucleus to mediate these critical cell cycle transitions.
Acknowledgments
We are very grateful to Deanna Koepp for her help with cell cycle studies as well as her comments on the manuscript. We thank Ryan Mills for his help with bioinformatics analysis as well as members of the Corbett laboratory for helpful suggestions. This work was supported by a National Institutes of Health (NIH) grant to A.H.C. K.F.P. was supported by an NIH and Facilitating Academic Careers in Engineering and the Sciences supplement.
References
- Adams, A., D. E. Gottschling, C. A. Kaiser and T. Stearns, 1997. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bayliss, R., A. H. Corbett and M. Stewart, 2000. The molecular mechanism of transport of macromolecules through nuclear pore complexes. Traffic 1 448–456. [DOI] [PubMed] [Google Scholar]
- Beg, A. A., S. M. Ruben, R. I. Scheinman, S. Haskill, C. A. Rosen et al., 1992. I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: a mechanism for cytoplasmic retention. Genes Dev. 6 1899–1913. [DOI] [PubMed] [Google Scholar]
- Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell and D. L. Wheeler, 2006. GenBank. Nucleic Acids Res. 34 D16–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. D., J. Truehart, G. Natsoulis and G. Fink, 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154 164–175. [DOI] [PubMed] [Google Scholar]
- Braun, K. A., and L. L. Breeden, 2007. Nascent transcription of MCM2-7 is important for nuclear localization of the minichromosome maintenance complex in G1. Mol. Biol. Cell 18 1447–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe, J., F. Kohlhuber and M. Muller, 1996. JAKs and STATs branch out. Trends Cell Biol. 6 336–340. [DOI] [PubMed] [Google Scholar]
- Burich, R., and M. Lei, 2003. Two bipartite NLSs mediate constitutive nuclear localization of Mcm10. Curr. Genet. 44 195–201. [DOI] [PubMed] [Google Scholar]
- Burke, B., and J. Ellenberg, 2002. Remodelling the walls of the nucleus. Nat. Rev. Mol. Cell Biol. 3 487–497. [DOI] [PubMed] [Google Scholar]
- Conti, E., and J. Kuriyan, 2000. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure 8 329–338. [DOI] [PubMed] [Google Scholar]
- Conti, E., M. Uy, L. Leighton, G. Blobel and J. Kuriyan, 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94 193–204. [DOI] [PubMed] [Google Scholar]
- David-Pfeuty, T., F. Chakrani, K. Ory and Y. Nouvian-Dooghe, 1996. Cell cycle-dependent regulation of nuclear p53 traffic occurs in one subclass of human tumor cells and in untransformed cells. Cell Growth Differ. 7 1211–1225. [PubMed] [Google Scholar]
- Dohrmann, P. R., and R. A. Sclafani, 2006. Novel role for checkpoint Rad53 protein kinase in the initiation of chromosomal DNA replication in Saccharomyces cerevisiae. Genetics 174 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Q., L. Taylor, D. A. Compton and I. G. Macara, 2002. LGN blocks the ability of NuMA to bind and stabilize microtubules: a mechanism for mitotic spindle assembly regulation. Curr. Biol. 12 1928–1933. [DOI] [PubMed] [Google Scholar]
- Fontes, M. R., T. Teh and B. Kobe, 2000. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 297 1183–1194. [DOI] [PubMed] [Google Scholar]
- Friedman, K. L., J. D. Diller, B. M. Ferguson, S. V. Nyland, B. J. Brewer et al., 1996. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev. 10 1595–1607. [DOI] [PubMed] [Google Scholar]
- Gilchrist, D., and M. Rexach, 2003. Molecular basis for the rapid dissociation of nuclear localization signals from karyopherin alpha in the nucleoplasm. J. Biol. Chem. 278 51937–51949. [DOI] [PubMed] [Google Scholar]
- Gilchrist, D., B. Mykytka and M. Rexach, 2002. Accelerating the rate of disassembly of karyopherin.cargo complexes. J. Biol. Chem. 277 18161–18172. [DOI] [PubMed] [Google Scholar]
- Goh, P. Y., and U. Surana, 1999. Cdc4, a protein required for the onset of S phase, serves an essential function during G(2)/M transition in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 5512–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich, D., S. Kostka, R. Kraft, C. Dingwall, R. A. Laskey et al., 1995. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 5 383–392. [DOI] [PubMed] [Google Scholar]
- Görlich, D., P. Henklein, R. A. Laskey and E. Hartmann, 1996. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15 1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Gruss, O. J., R. E. Carazo-Salas, C. A. Schatz, G. Guarguaglini, J. Kast et al., 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104 83–93. [DOI] [PubMed] [Google Scholar]
- Harreman, M. T., P. E. Cohen, M. R. Hodel, G. J. Truscott, A. H. Corbett et al., 2003. a Characterization of the auto-inhibitory sequence within the N-terminal domain of importin alpha. J. Biol. Chem. 278 21361–21369. [DOI] [PubMed] [Google Scholar]
- Harreman, M. T., M. R. Hodel, P. Fanara, A. E. Hodel and A. H. Corbett, 2003. b The auto-inhibitory function of importin alpha is essential in vivo. J. Biol. Chem. 278 5854–5863. [DOI] [PubMed] [Google Scholar]
- Hartwell, L. H., and D. Smith, 1985. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 110 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer, M. W., T. C. Walther and I. W. Mattaj, 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21 347–380. [DOI] [PubMed] [Google Scholar]
- Hodel, M. R., A. H. Corbett and A. E. Hodel, 2001. Dissection of a nuclear localization signal. J. Biol. Chem. 276 1317–1325. [DOI] [PubMed] [Google Scholar]
- Hodel, A. E., M. T. Harreman, K. F. Pulliam, M. E. Harben, J. S. Holmes et al., 2006. Nuclear localization signal receptor affinity correlates with in vivo localization in Saccharomyces cerevisiae. J. Biol. Chem. 281 23545–23556. [DOI] [PubMed] [Google Scholar]
- Hood, J. K., and P. A. Silver, 1998. Cse1p is required for export of Srp1p/importin-alpha from the nucleus in Saccharomyces cerevisiae. J. Biol. Chem. 273 35142–35146. [DOI] [PubMed] [Google Scholar]
- Hopwood, B., and S. Dalton, 1996. Cdc45p assembles into a complex with Cdc46p/Mcm5p, is required for minichromosome maintenance, and is essential for chromosomal DNA replication. Proc. Natl. Acad. Sci. USA 93 12309–12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425 686–691. [DOI] [PubMed] [Google Scholar]
- Jones, A. L., B. B. Quimby, J. K. Hood, P. Ferrigno, P. H. Keshava et al., 2000. SAC3 may link nuclear protein export to cell cycle progression. Proc. Natl. Acad. Sci. USA 97 3224–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana, J. A., B. J. Schnapp and P. A. Silver, 1995. Kinetics of spindle pole body separation in budding yeast. Proc. Natl. Acad. Sci. USA 92 9707–9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon, D., B. L. Roberts, W. D. Richardson and A. E. Smith, 1984. A short amino acid sequence able to specify nuclear location. Cell 39 499–509. [DOI] [PubMed] [Google Scholar]
- Kobe, B., 1999. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 6 388–397. [DOI] [PubMed] [Google Scholar]
- Lange, A., R. E. Mills, C. J. Lange, M. Stewart, S. E. Devine et al., 2007. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282 5101–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. J., Y. Matsuura, S. M. Liu and M. Stewart, 2005. Structural basis for nuclear import complex dissociation by RanGTP. Nature 435 693–696. [DOI] [PubMed] [Google Scholar]
- Leung, S. W., M. T. Harreman, M. R. Hodel, A. E. Hodel and A. H. Corbett, 2003. Dissection of the karyopherin alpha nuclear localization signal (NLS)-binding groove: functional requirements for NLS binding. J. Biol. Chem. 278 41947–41953. [DOI] [PubMed] [Google Scholar]
- Li, R., and A. W. Murray, 1991. Feedback control of mitosis in budding yeast. Cell 66 519–531. [DOI] [PubMed] [Google Scholar]
- Liu, S. M., and M. Stewart, 2005. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-beta homologue, Kap95p. J. Mol. Biol. 349 515–525. [DOI] [PubMed] [Google Scholar]
- Loeb, J. D., G. Schlenstedt, D. Pellman, D. Kornitzer, P. A. Silver et al., 1995. The yeast nuclear import receptor is required for mitosis. Proc. Natl. Acad. Sci. USA 92 7647–7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine, G. T., P. Sinha and B. K. Tye, 1984. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics 106 365–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, Y., and M. Stewart, 2004. Structural basis for the assembly of a nuclear export complex. Nature 432 872–877. [DOI] [PubMed] [Google Scholar]
- Matsuura, Y., A. Lange, M. T. Harreman, A. H. Corbett and M. Stewart, 2003. Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 22 5358–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middeler, G., K. Zerf, S. Jenovai, A. Thulig, M. Tschodrich-Rotter et al., 1997. The tumor suppressor p53 is subject to both nuclear import and export, and both are fast, energy-dependent and lectin-inhibited. Oncogene 14 1407–1417. [DOI] [PubMed] [Google Scholar]
- Mitchison, J. M., and J. Creanor, 1971. Induction synchrony in the fission yeast. Schizosaccharomyces pombe. Exp. Cell Res. 67 368–374. [DOI] [PubMed] [Google Scholar]
- Moir, D., S. E. Stewart, B. C. Osmond and D. Botstein, 1982. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties and pseudoreversion studies. Genetics 100 547–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, T., G. Tebb, U. Surana, H. Robitsch and K. Nasmyth, 1991. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66 743–758. [DOI] [PubMed] [Google Scholar]
- Nachury, M. V., T. J. Maresca, W. C. Salmon, C. M. Waterman-Storer, R. Heald et al., 2001. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104 95–106. [DOI] [PubMed] [Google Scholar]
- Nakai, K., and P. Horton, 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24 34–36. [DOI] [PubMed] [Google Scholar]
- Nishitani, H., M. Ohtsubo, K. Yamashita, H. Iida, J. Pines et al., 1991. Loss of RCC1, a nuclear DNA-binding protein, uncouples the completion of DNA replication from the activation of cdc2 protein kinase and mitosis. EMBO J. 10 1555–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, R. E., E. Hogan and D. Koshland, 1990. Mitotic transmission of artificial chromosomes in cdc mutants of the yeast, Saccharomyces cerevisiae. Genetics 125 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines, J., and T. Hunter, 1991. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol. 115 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila, T., S. Miles, D. GuhaThakurta, D. Jemiolo and L. L. Breeden, 2002. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 16 3034–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke, R. M., and A. K. Bielinsky, 2004. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell 16 173–185. [DOI] [PubMed] [Google Scholar]
- Riddick, G., and I. G. Macara, 2007. The adapter importin-alpha provides flexible control of nuclear import at the expense of efficiency. Mol. Syst. Biol. 3 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, J., S. M. Dilworth, R. A. Laskey and C. Dingwall, 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64 615–623. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. Russell, 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sazer, S., and M. Dasso, 2000. The ran decathlon: multiple roles of Ran. J. Cell Sci. 113(Pt. 7): 1111–1118. [DOI] [PubMed] [Google Scholar]
- Sazer, S., and P. Nurse, 1994. A fission yeast RCC1-related protein is required for the mitosis to interphase transition. EMBO J. 13 606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer, S., and S. W. Sherwood, 1990. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97(Pt. 3): 509–516. [DOI] [PubMed] [Google Scholar]
- Schroeder, A. J., X. H. Chen, Z. Xiao and M. Fitzgerald-Hayes, 1999. Genetic evidence for interactions between yeast importin alpha (Srp1p) and its nuclear export receptor, Cse1p. Mol. Gen. Genet. 261 788–795. [DOI] [PubMed] [Google Scholar]
- Shulga, N., P. Roberts, Z. Gu, L. Spitz, M. M. Tabb et al., 1996. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 135 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova, J. M., G. E. Mikesell and L. L. Breeden, 1995. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol. Biol. Cell 6 1641–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siede, W., and E. C. Friedberg, 1990. Influence of DNA repair deficiencies on the UV sensitivity of yeast cells in different cell cycle stages. Mutat. Res. 245 287–292. [DOI] [PubMed] [Google Scholar]
- Siede, W., G. W. Robinson, D. Kalainov, T. Malley and E. C. Friedberg, 1989. Regulation of the RAD2 gene of Saccharomyces cerevisiae. Mol. Microbiol. 3 1697–1707. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher, J., P. Maurer, F. R. Bischoff and G. Schlenstedt, 1998. Cse1p is involved in export of yeast importin alpha from the nucleus. Mol. Cell. Biol. 18 6805–6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher, J., P. Maurer, F. Vogel and G. Schlenstedt, 2000. Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha. Mol. Cell. Biol. 20 8468–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, M., 2007. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8 195–208. [DOI] [PubMed] [Google Scholar]
- Straight, A. F., and A. W. Murray, 1997. The spindle assembly checkpoint in budding yeast. Methods Enzymol. 283 425–440. [DOI] [PubMed] [Google Scholar]
- Swaminathan, S., A. C. Kile, E. M. MacDonald and D. M. Koepp, 2007. Yra1 is required for S phase entry and affects Dia2 binding to replication origins. Mol. Cell. Biol. 27 4674–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timney, B. L., J. Tetenbaum-Novatt, D. S. Agate, R. Williams, W. Zhang et al., 2006. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 175 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin, H., T. Staehelin and J. Gordon, 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some application. Proc. Natl. Acad. Sci. USA 76 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda, M., S. Izaddoost, I. Cushman, M. S. Moore and S. Sazer, 2005. The fission yeast Schizosaccharomyces pombe has two importin-α proteins, Imp1p and Cut15p, which have common and unique functions in nucleocytoplasmic transport and cell cycle progression. Genetics 171 7–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter, I. R., A. Arndt, U. Kutay, D. Görlich and A. Wittinghofer, 1999. Structural view of the Ran-importin β interaction at 2.3 Å resolution. Cell 97 635–646. [DOI] [PubMed] [Google Scholar]
- Weis, K., U. Ryder and A. I. Lamond, 1996. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 15 1818–1825. [PMC free article] [PubMed] [Google Scholar]
- Wiese, C., A. Wilde, M. S. Moore, S. A. Adam, A. Merdes et al., 2001. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science 291 653–656. [DOI] [PubMed] [Google Scholar]
- Winston, F., C. Dollard and S. L. Ricupero-Hovasse, 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11 53–55. [DOI] [PubMed] [Google Scholar]
- Yano, R., M. L. Oakes, M. M. Tabb and M. Nomura, 1994. Yeast Srp1p has homology to armadillo/plakoglobin/beta-catenin and participates in apparently multiple nuclear functions including the maintenance of the nucleolar structure. Proc. Natl. Acad. Sci. USA 91 6880–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W., C. Ukomadu, S. Jha, T. Senga, S. K. Dhar et al., 2007. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 21 2288–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]