Abstract
Loss of Abf2p, an abundant mitochondrial nucleoid-associated protein, results in increased mitochondrial frameshifts and direct-repeat mediated deletions but has no effect on the rate of mitochondrial point mutations. The instability of repeated sequences in this strain may be linked to the loss of mitochondrial DNA in abf2-Δ strains.
A functional mitochondrial genome is essential for the survival of eukaryotic cells. Several inherited neurodegenerative disease syndromes arise as a result of mitochondrial mutation. Deletions and rearrangements of the mitochondrial DNA (mtDNA) are easily detected and have been found in a variety of tissue types in aging mammals. Accumulation of deletions in mtDNA of mice lacking the proofreading domain of the mitochondrial polymerase, Polγ, has been linked to precocious aging of these animals (Vermulst et al. 2008), underscoring the importance of maintaining mitochondrial genome stability. Similar mutations are also observed in mitochondria of S. cerevisiae. When yeast cells are grown on a fermentable carbon source such as glucose, nonrespiring, “petite” variants arise that form smaller colonies than respiring cells (Dujon 1981). Analysis of deletion junctions in mtDNA from petite cells suggests that at least some mutations arise by recombination between directly repeated sequences (Dujon 1981).
mtDNA is organized into nucleoids, structures that are more DNase resistant than naked DNA (Williamson 1976; Miyakawa et al. 1987, 1995; Newman et al. 1996). Each nucleoid consists of multiple mitochondrial genomes and tightly associated proteins. In yeast, the most extensively studied nucleoid-associated protein is Abf2p, which shows homology to HMG (high mobility group) proteins that bend DNA and are involved in nuclear transcription and chromatin packaging. ABF2 is a nuclear gene whose deletion gives rise to rapid loss of mtDNA when cells are grown on a fermentable carbon source, although cells can maintain mtDNA when grown in media that selects for mitochondrial gene expression (Diffley and Stillman 1991). Yeast abf2-Δ strains can be rescued by both the Escherichia coli HU protein and the human homolog of Abf2p, h-mTFA, suggesting broad conservation of at least some functions (Megraw and Chae 1993; Parisi et al. 1993). Chromatin immunoprecipitation experiments have demonstrated that Abf2p binds most regions of the mtDNA, with a higher affinity for GC-rich sequences. In addition, there is a decreased Abf2p to DNA ratio in glycerol grown cells relative to those grown in dextrose. This decrease correlates with the increased sensitivity of nucleoids to DNase I treatment, suggesting that the genome is more loosely condensed as the availability of Abf2p declines (Kucej et al. 2008).
Studies have implicated Abf2p in histone-like DNA packaging, replication, and in the localization of other mitochondrial proteins to nucleoids (Newman et al. 1996; Zelenaya-Troitskaya et al. 1998; Friddle et al. 2004). In addition, a role for Abf2p in mtDNA recombination has been proposed. In crosses, mitochondrial recombinants were obtained approximately fivefold less frequently when both parents were deleted for ABF2 than in wild-type crosses. This suggests a role for Abf2p in promotion of recombination events; however, this analysis is complicated by the fact that altered mtDNA and protein mixing during mating was observed in these strains (Zelenaya-Troitskaya et al. 1998). The significant changes to mitochondrial nucleoid structure and organization observed in abf2-Δ strains, even under conditions where mtDNA was retained, prompted us to test whether mutations accumulate in the mitochondrial genome under these conditions.
As previously reported, abf2-Δ strains rapidly lose mitochondrial function after cells are shifted to medium containing a fermentable carbon source. In our strain background, 81% of abf2-Δ cells are petite after ∼18 generations of growth in dextrose, as compared to 0.35% in the wild-type control (Table 1). MtDNA can be maintained in abf2-Δ strains when grown in medium containing a nonfermentable carbon source; however, it is not known whether the mitochondrial genome displays higher rates of mutation under these conditions. To determine the role of Abf2p in preventing genome instability under conditions in which mitochondrial genome retention is selected, we examined the effect of an ABF2 deletion on a broad spectrum of mitochondrial mutations. Using erythromycin, we are able to measure rates of point mutation accumulation, as erythromycin resistance is conferred by point mutations in the mitochondrially encoded 21S rRNA gene. As shown in Table 1, there is no effect of deletion of ABF2 on the rate of mitochondrial base substitutions.
TABLE 1.
Rates of mutation in the mitochondrial genome
| % petites | Rate of eryR | Rate of microsatellite instability | Rate of direct repeat-mediated deletion | |
|---|---|---|---|---|
| Wild-type | 0.35 | 2.0 × 10−8 | 6.5 × 10−7 | 0.7 × 10−4 |
| abf2-Δ | 81 | 2.1 × 10−8 (1.1-fold) | 4.0 × 10−6 (6-fold, P = 0.01) | 3.6 × 10−4 (5-fold, P < 0.0001) |
The percentage of petite cells was determined by selecting respiring cells in glycerol medium (YPG), then isolating independent colonies on synthetic dextrose medium to release selection. For each experiment, at least 20 independent colonies were resuspended in sterile water and appropriate dilutions were plated on YPG + 0.1% dextrose. Petite and grande colonies were scored after 4 days incubation at 30°. The rate of erythromycin resistance (eryR) was determined as described (Mookerjee and Sia 2006). To measure the rate of microsatellite instability, independent colonies containing the reporter were grown with glycerol selection for 3 days at 30°, resuspended in sterile water, and appropriate dilutions were plated on YPG to determine the number of respiring cells, and synthetic dextrose medium lacking arginine (SD −Arg), to select for reversion of the ARG8m frameshift. Arg+ colonies were scored after 7 days at 30°. Rates of DRMD were determined similarly: strains were grown for 2 days on SD −Arg to select the reporter; independent colonies were isolated on SD medium and allowed to grow for 3 days at 30°. Cells from 20 independent colonies were resuspended in sterile water and appropriate dilutions plated on YPG and SD −Arg. Recombinants were scored on YPG after 3 days at 30°. All experiments were performed at least three times. All rates were calculated by the method of the median (Lea and Coulson 1949), and two-tailed P-values were calculated using an unpaired t-test performed by Instat3 for Macintosh (GraphPad Software).
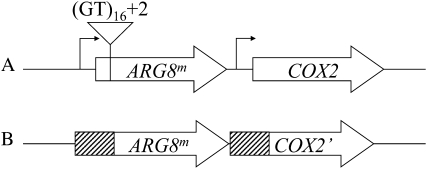
Microsatellite sequences are sensitive indicators of genomic instability, and mitochondrial microsatellite instability can also be monitored in yeast using a previously constructed reporter (Kalifa and Sia 2007). This reporter makes use of a synthetic mitochondrial gene, derived from the S. cerevisiae nuclear ARG8 gene, whose encoded protein functions within the mitochondria in the biosynthesis of arginine. ARG8 has been recoded to reflect the codon usage of a mitochondrial gene, and this synthetic gene is called ARG8m. Insertion of ARG8m into the mitochondrial genome as a translational fusion with a mitochondrially encoded gene complements a chromosomal arg8-Δ allele, resulting in strains that are phenotypically Arg+ (Steele et al. 1996). Microsatellite sequences consisting of poly(GT) repeats were introduced into the mitochondrial genome, interrupting the reading frame of ARG8m and creating frameshift mutations that will be translated in the +2 reading frame, designated (GT)16+2∷arg8m. The microsatellite reporter has been inserted 5′ of COX2 in the mitochondrial genome, and the cells containing this reporter maintain respiratory function, allowing us to select for functional mitochondria and retention of the reporter in the abf2-Δ cells (Figure 1A). We find that deletion of ABF2 results in a sixfold increase in the rate of microsatellite instability (Table 1).
Figure 1.—
Reporters to measure the stability of repetitive sequences in mtDNA. (A) The microsatellite reporter. The open arrows indicate the ARG8m and COX2 coding sequences. The location of the site used for insertion of the microsatellite sequence in the ARG8m coding sequence is indicated. (B) The mitochondrial direct repeat-mediated deletion reporter. The striped boxes represent the 96 bases of COX2 sequence 3′ of the ATG start. The open arrows indicate the ARG8m coding sequence, including the ATG start fused in-frame with the COX2 sequences, and the remainder of the COX2 sequence, respectively. Recombination between the repeated sequences restores the functional COX2 gene.
To determine whether loss of Abf2p results in a general destabilization of repetitive DNA, we made use of a reporter strain that allows us to measure the rate at which direct repeat-mediated deletions (DRMDs) occur (Phadnis et al. 2005). This strain consists of 96 base pairs of directly repeated COX2 sequence flanking the ARG8m reporter gene integrated at the COX2 locus in the mitochondrial genome (Figure 1B). Cells with the intact reporter are prototrophic for arginine and respiration deficient, due to the interrupted COX2 gene. DRMD events at this site restore an active COX2 locus, thereby restoring respiration competence. The rate of these events can be estimated by selecting recombinants on a nonfermentable carbon source. Previously, it had been hypothesized that Abf2p plays a role, perhaps directly, in mitochondrial recombination (Macalpine et al. 1998). However, we find a significant increase in mitochondrial DRMDs in the abf2-Δ cells (Table 1), indicating that these events are suppressed in the presence of Abf2p. These results do not support a model in which Abf2p is important for promoting mtDNA recombination at directly-repeated sequences in yeast cells during mitosis.
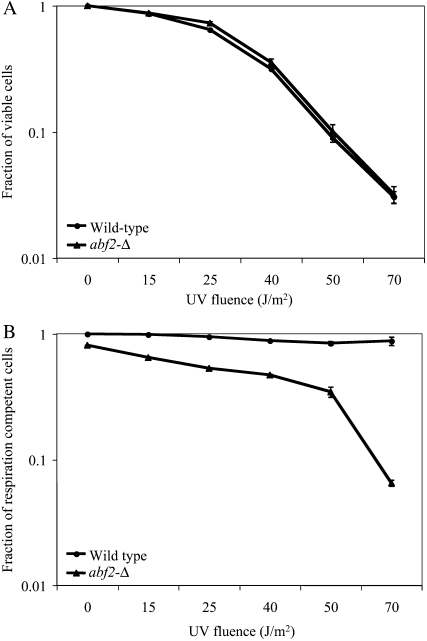
To determine whether mtDNA in the strain lacking Abf2p was more vulnerable to DNA damage, we tested the UV sensitivity of the abf2-Δ strain for respiratory function. An increase in petite cells was observed in an otherwise wild-type strain carrying an abf2-Δ mutation relative to the wild-type strain after UV exposure; however, no significant difference in viability was observed. Therefore, mitochondrial function is specifically affected in the mutant (Figure 2), suggesting that Abf2p may be involved in protection or recovery of mtDNA from UV-induced damage.
Figure 2.—
Viability and respiration loss of yeast strains following exposure to UV light. (A) Fraction of the total number of cells that are viable. (B) Fraction of viable cells that are respiration competent. For each experiment, yeast cells were grown to saturation in YPG medium. Dilutions were plated on YPG + 0.1% dextrose plates in duplicate. Plates were exposed to 0–70 J/m2 UV light and incubated for 4 days at 30° in the dark to prevent repair by photolyase. Plates were then scored for viability and respiring and nonrespiring cells. The percentage of nonrespiring cells averaged over 2–3 independent experiments is shown. Error bars indicate the standard error of the mean.
We conclude that although the mitochondrial genome is maintained under selective conditions in the abf2-Δ strain, alteration of the nucleoid structure in these mutants results in increased mutagenesis and increased sensitivity to DNA damage. Previous studies of RECA3 and MSH1 mutants in Arabidopsis, and MSH1 in yeast, have demonstrated that these nuclear-encoded proteins suppress mitochondrial recombination at short direct repeats. In yeast, however, these mutants, unlike the abf2-Δ cells, also display increased rates of point mutation accumulation (Mookerjee and Sia 2006; Shedge et al. 2007), suggesting that the suppression by Abf2p may be mechanistically distinct.
The instability of repeated sequences in the abf2-Δ strain may be linked to the high rate of respiration loss observed in this strain. Because the mitochondrial genome is completely lost in these strains, ABF2 deletion likely results in improper regulation of mtDNA replication or segregation under fermentative conditions. Mitochondrial nucleoid organization is altered in response to growth conditions, and the stable maintenance of mtDNA is dependent on enzymes normally involved in metabolism (Aco1p) and amino acid biosynthesis (Ilv5p), which appear to directly link growth conditions to changes in nucleoid structure (Zelanaya-Troitskaya et al. 1995; Chen et al. 2005). The overexpression of Aco1p and Ilv5p can suppress the loss of mtDNA in the abf2-Δ cells and may likewise suppress the instability of repetitive mtDNA.
Acknowledgments
This work was sponsored by National Science Foundation grant MCB0543084.
References
- Chen, X. J., X. Wang, B. A. Kaufman and R. A. Butow, 2005. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307 714–717. [DOI] [PubMed] [Google Scholar]
- Diffley, J. F., and B. Stillman, 1991. A close relative of the nuclear, chromosomal high-mobility group protein Hmg1 in yeast mitochondria. Proc. Natl. Acad. Sci. USA 88 7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon, B., 1981. Mitochondrial genetics and functions, pp. 505–635 in Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance, edited by J. N. Strathern, E. W. Jones and J. R. Broach. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Friddle, R. W., J. E. Klare, S. S. Martin, M. Corzett, R. Balhorn et al., 2004. Mechanism of DNA compaction by yeast mitochondrial protein Abf2p. Biophys. J. 86 1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa, L., and E. A. Sia, 2007. Analysis of Rev1p and Polζ in mitochondrial mutagenesis suggests an alternative pathway of damage tolerance. DNA Repair 6 1732–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucej, M., B. Kucejova, R. Subramanian, X. J. Chen and R. A. Butow, 2008. Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J. Cell Sci. 121 1861–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49 264–285. [DOI] [PubMed] [Google Scholar]
- MacAlpine, D. M., P. S. Perlman and R. A. Butow, 1998. The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc. Natl. Acad. Sci. USA 95 6739–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw, T. L., and C.-B. Chae, 1993. Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J. Biol. Chem. 268 12758–12763. [PubMed] [Google Scholar]
- Miyakawa, I., S. Nobundo, K. Shigeyuki, N. Soichi and K. Tsuneyoshi, 1987. Isolation of morphologically intact mitochondrial nucleoids from the yeast, Saccharomyces cerevisiae. J. Cell Sci. 88 431–439. [DOI] [PubMed] [Google Scholar]
- Miyakawa, I., S. Fumoto, T. Kuroiwa and N. Sando, 1995. Characterization of DNA-binding proteins involved in the assembly of mitochondrial nucleoids from the yeast, Saccharomyces cerevisiae. Plant Cell Physiol. 36 1179–1188. [PubMed] [Google Scholar]
- Mookerjee, S. A., and E. A. Sia, 2006. Overlapping contributions of Msh1p and putative recombination proteins Cce1p, Din7p, and Mhr1p in large-scale recombination and genome sorting events in the mitochondrial genome of Saccharomyces cerevisiae. Mutat. Res. 595 91–106. [DOI] [PubMed] [Google Scholar]
- Newman, S. M., O. Zelenaya-Troitskaya, P. S. Perlman and R. A. Butow, 1996. Analysis of mitochondrial DNA nucleoids in wild-type and a mutant strain of Saccharomyces cerevisiae that lacks the mitochondrial HMG-box protein Abf2p. Nucleic Acids Res. 24 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M. A., B. J. Xu and D. A. Clayton, 1993. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol. 13 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis, N., R. A. Sia and E. A. Sia, 2005. Analysis of repeat-mediated deletions in the mitochondrial genome of Saccharomyces cerevisiae. Genetics 171 1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedge, V., M. Arrieta-Montiel, A. C. Christensen and S. A. Mackenzie, 2007. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell 19 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele, D. F., C. A. Butler and T. D. Fox, 1996. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. USA 93 5253–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermulst, M., J. Wanagat, G. C. Kujoth, J. H. Bielas, P. S. Rabinovitch et al., 2008. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat. Genet. 40 392–394. [DOI] [PubMed] [Google Scholar]
- Williamson, D. H., 1976. Packaging and recombination of mitochondrial DNA in vegetatively growing cells, pp. 99–115 in Genetics, Biogenesis and Bioenergetics of Mitochondria, edited by W. Bandlow, R. J. Schweyen, D. Y. Thomas, T. K. Wolf and F. Kaudewitz. Walter de Gruyter, Berlin.
- Zelanaya-Troitskaya, O., P. S. Perlman and R. A. Butow, 1995. ILV5 encodes a bifunctional mitochondrial protein involved in branched chain amino acid biosynthesis and maintenance of mitochondrial DNA. EMBO J. 14 3268–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenaya-Troitskaya, O., S. M. Newman, K. Okamoto, P. S. Perlman and R. A. Butow, 1998. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics 148 1763–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]