Abstract
We examined the prevalence of interactions between pairs of short chromosomal regions from one species (Solanum habrochaites) co-introgressed into a heterospecific genetic background (Solanum lycopersicum). Of 105 double introgression line (DIL) families generated from a complete diallele combination of 15 chromosomal segments, 39 (∼38%) showed evidence for complex epistasis in the form of genotypic and/or allelic marker transmission distortion in DIL F2 populations.
INTRINSIC postzygotic isolation (environment-independent hybrid inviability and sterility) is often due to deleterious genetic interactions between loci that have functionally diverged during the evolution of new species [i.e., “Dobzhansky–Muller” incompatibilities (DMIs)] (Coyne and Orr 2004). Most standard models of this process assume that individual DMIs are due to pairwise genetic interactions (one in each diverging lineage) and that each DMI contributes additively to the expression of hybrid incompatibility between diverging species (some models can relax the first assumption; Turelli and Orr 2000; Orr and Turelli 2001; Welch 2004). Nonetheless, if interspecific epistasis is more complex, there could be important consequences for the temporal accumulation of species barriers, and the number of loci required to complete speciation (Turelli et al. 2001; Kondrashov 2003; Welch 2004). For example, if epistasis between different conspecific loci is generally synergistic (i.e., if the combined effect of two conspecific loci is greater than expected on the basis of their individual effects on hybrid incompatibility), fewer DMIs will be required for the expression of complete reproductive isolation, with a correspondingly shorter time to speciation. [Conceptually similar expectations were first developed in the context of the epistasis among deleterious recessive loci causing inbreeding depression (Kondrashov 1984).] Epistasis among more than two loci, therefore, could be fundamentally important in determining patterns and rates of evolution of isolation between diverging species. Nonetheless, the prevalence and nature of interactions between more than two loci from one or both species involved in a hybridization (i.e., “complex epistasis”), and their effects on hybrid incompatibility, is poorly understood empirically.
The goal of this study was to assess evidence for genetic interactions between different chromosomal regions from one species when introgressed together pairwise in the background of a second species (Figure 1). Genetic interactions influencing hybrid incompatibility were detected by measuring the degree to which the genetic composition of F2 populations deviated from Mendelian expectations [i.e., transmission ratio distortion (TRD)]. We selected 15 chromosomal regions for inclusion in the study (Table 1), drawing from a set of near-isogenic lines (NILs) previously developed between two plant species in the genus Solanum section Lycopersicon (the tomato clade). Each NIL contains a unique short chromosomal region from the wild species Solanum habrochaites (SH) introgressed into the otherwise isogenic genetic background of the domesticated tomato, S. lycopersicum (SL) (Monforte and Tanksley 2000; see also Moyle and Graham 2005 for a previous summary). Note that some of the NILs used here are known to contain QTL for hybrid incompatibility that acts at later stages of development (partial pollen and/or seed sterility); however, the detection of TRD at early postzygotic embryonic stages in this study appears to be unrelated to whether one or both introgressions contain loci for these later acting incompatibilities (see Table 1 and below).
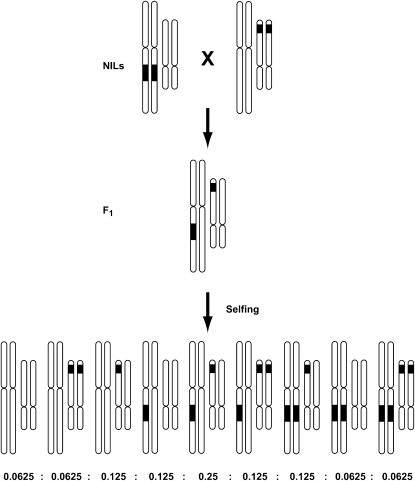
Figure 1.—
Schematic of approach to generating double introgression lines. Solid bars within a chromosome represent introgressed S. habrochaites (SH) chromosomal regions in an (open) isogenic S. lycopersicum (SL) background. Crosses were carried out by emasculating maternal plants in the bud and hand-pollinating with freshly collected pollen from the paternal parent. For each of 105 DIL families, we germinated 120 F2 seeds, extracted DNA (96-well format, phenol chloroform extractions), and genotyped each individual for two marker locations, one at each introgressed region represented in that DIL family. In cases where <120 individuals were available for a DIL family, we genotyped all available F2's. Total experimental size = 12,048 × 2 markers = 24,096. The detection of an SH allele at a marker location indicates the presence of the SH introgression at that location. Because markers are codominant, homozygous and heterozygous genotypes can be distinguished. Included are the expected genotypic ratios in an F2 population for each DIL family, if marker transmission is Mendelian.
TABLE 1.
NILs used to generate double introgression lines
| LA number | QTL status | Chromosome location | Introgression length (cM) | % genome |
|---|---|---|---|---|
| LA3975 | None | 3 | 12.1 | 0.0096 |
| LA3968 | None | 12 | 14.1 | 0.0112 |
| LA3964 | None | 10 | 22.5 | 0.0179 |
| LA3957 | None | 9 | 44.8 | 0.0356 |
| LA3947 | None | 6 | 8.6 | 0.0068 |
| LA3956 | Pollen | 9 | 57.4 | 0.0456 |
| LA3935 | Pollen | 4 | 53 | 0.0421 |
| LA3950 | Pollen | 7 | 33.8 | 0.0268 |
| LA3963 | Pollen | 10 | 30.3 | 0.0241 |
| LA3948 | Pollen | 7 | 50.4 | 0.0400 |
| LA3931 | Seed | 4 | 18.7 | 0.0149 |
| LA3939 | Seed | 5 | 25.8 | 0.0205 |
| LA3943 | Seed | 5 | 34 | 0.0270 |
| LA3915 | Seed | 1 | 34.8 | 0.0276 |
| LA3977 | Seed | 4 | 19 | 0.0151 |
LA number, seed accession identifier (see tgrc.ucdavis.edu). QTL status, five NILs contained pollen sterility QTL and five contained seed sterility QTL previously identified in the genomewide survey of hybrid incompatibility between these two species (Moyle and Graham 2005). The remaining five NILs had no detected effects on hybrid pollen and seed fertility but had introgression lengths (centimorgans) comparable to the sterility QTL NIL set. Note that segregation distortion detected here is unlikely to be systematically due to the direct effects of pollen or seed sterility QTL because: all known sterility QTL cause only partial sterility; lines carrying seed sterility QTL were preferentially used as pollen parents, and vice versa; our crossing methodology maximizes potential seed set (i.e., copious pollen is transferred and crosses are repeated multiple times to obtain sufficient seed); and, in only 4 of 31 cases where individual SH introgressions were significantly underrepresented in F2 populations (see text), was a parental line known to be partially sterile for the relevant parental function (i.e., seed sterility for maternal alleles, pollen sterility for paternal alleles).
To generate lines with two introgressed regions [double introgression lines (DILs)], a complete diallele cross was performed to combine each introgression with every other introgression (Figure 1), for a total of 105 unique pairwise combinations of the 15 regions. In one of the NIL–NIL combinations, no crosses produced viable seed despite being performed at least 20 times using both reciprocal directions. Heterozygote F1 DILs from each of the 104 remaining NIL–NIL combinations were selfed to generate F2 seeds (“DIL family”), and up to 120 F2's in each DIL family were grown and genotyped for two markers, one located in each introgressed region. Species-specific codominant PCR-based markers were used to diagnose the presence of each introgression (SH vs. SL alleles) (see supplemental Table 1), for a total experimental size of 104 DIL families × 120 F2's × 2 markers = 24,960. Figure 1 shows the expected genotypic ratios in an F2 population for each DIL family, if transmission is Mendelian.
Evidence for complex epistasis among genotypes:
Our a priori hypothesis was that, if epistatic interactions affecting the expression of hybrid incompatibility were operating between chromosomal regions, evidence for this would most likely be seen as underrepresentation of individuals that are homozygous for both introgressions. This expectation was based on data indicating that hybrid incompatibility loci generally act recessively (Coyne and Orr 2004), as they do in this and other Solanum species crosses (Moyle and Nakazato 2008), and therefore that incompatibility effects should primarily be seen in individuals with homozygous introgressions. In each F2 DIL family we performed χ2 tests to assess whether individuals in the double homozygote genotypic class were more or less frequent than expected under Mendelian segregation ratios (e.g., 0.0625 or 7.5 per 120 individuals, assuming 50:50 allele ratios at each locus). Because significant deviations in the double homozygote class might also be due to under- or overrepresented homozygotes at only one of the introgressions, rather than exclusively due to interactions between the two introgressions, in our χ2 tests we conditioned the expected frequency of double homozygotes on the observed allele frequencies at both SH loci in each F2 genotyped DIL population [i.e., expected frequency = (qlocus1)2 × (qlocus2)2]. Conditioning on observed allele frequencies in each F2 population also has the effect of testing specifically for genotypic distortion, by accounting for any allelic distortion present in each DIL F2 population.
We detected a significant deviation in this genotypic class in 8 of 104 lines for which there were data (Table 2). In these cases, we infer that epistasis between the two introgressed regions was necessary for Mendelian distortion in the double homozygote class. In six of these cases, SH double homozygotes were underrepresented in the corresponding DIL F2 populations, as expected if carriers of both of these chromosomal regions are preferentially inviable due to incompatible interactions with the SL background. These interactions are presumably expressed at early stages of reproductive development (i.e., early postzygotic embryonic stages in the F2's). In two cases, F2 populations had higher than expected numbers of double SH homozygotes. Because our test for distortion in double homozygotes is fairly weak, none of the detected deviations survive correction for multiple testing, even in the three DIL families where no double SH homozygotes were detected. However, due to biological nonindependence in this experiment (each NIL is represented in 14 DIL families) it is unclear how to formally correct for multiple testing; we have conservatively used a Bonferroni correction (Sokal and Rohlf 1995). For the 3 families in which we detected no double SH homozygotes, we preformed additional crosses to assess biological repeatability of our initial result. In all cases, SH double homozygous genotypes were also underrepresented in this second round of crosses and genotyping; in one case, we failed to generate any individuals in this genotypic class (data not shown).
TABLE 2.
Double introgression line F2 families showing significant under- or overrepresentation of SH double homozygote genotypes
| DIL family | NIL 1 | NIL 2 | χ2 | P-value | Direction |
|---|---|---|---|---|---|
| 36 | LA3935 | LA3956 | 5.109 | 0.024 | Under |
| 37 | LA3950 | LA3956 | 4.309 | 0.038 | Under |
| 44 | LA3948 | LA3950 | 7.175 | 0.007 | Under |
| 62 | LA3935 | LA3931 | 4.205 | 0.040 | Over |
| 69 | LA3964 | LA3939 | 4.051 | 0.044 | Under |
| 108 | LA3964 | LA3977 | 4.144 | 0.042 | Under |
| 110 | LA3947 | LA3977 | 4.275 | 0.039 | Under |
| 118 | LA3939 | LA3977 | 4.722 | 0.030 | Over |
Direction, under- or overrepresentation of double SH homozygotes. Under a χ2 test with 1 d.f. we can detect a significant underrepresentation only when we find two or fewer double homozygotes in 120 genotyped individuals. Therefore, this test is relatively conservative with respect to detecting underrepresented double homozygotes. Supplemental Table 2 gives results for all pairwise and single tests of allelic and genotypic distortion.
Evidence for complex epistasis among alleles:
Our test for underrepresented double introgression homozygotes (above) does not detect allelic distortion that occurs prior to creation of F2 genotypes (i.e., prior to fertilization). However, because angiosperm pollen expresses its own haploid genome during fertilization (Frova and Pe 1997), competitive superiority of some alleles over others at postmating, prefertilization stages could also result in allelic distortions in segregating populations. We detected 13 DIL families that showed significant allelic distortion at one or both single introgressions (P < 0.05, Fisher's exact test) and/or at both introgressions considered together (P < 0.05, 2 × 2 χ2 test). In eight of these cases, the relevant locus also showed genotypic distortion (either in double homozygotes or in single introgressions; see below); we attribute these cases to selection for or against SH genotypes rather than alleles. In the five remaining cases, distorted ratios might be due to prefertilization processes involving allelic selection; all show significant deviations in introgression combinations (one also shows deviations at one of the introgressions). Interestingly, introgression combination deviations appear to be in opposite directions in all five cases (i.e., at locus 1, SH alleles are underrepresented; at locus 2, SH alleles are overrepresented). This observation might suggest that negative gametic interactions between conspecific (SH) alleles are producing distortions in the transmission of these alleles in the biological context of a second (SL) species.
Evidence for DMIs at single chromosomal locations:
Finally, we evaluated evidence for significant genotypic distortion at each SH introgression individually, using χ2 tests for significant over- or underrepresentation of SH homozygotes within each DIL family (expected segregation ratio of 3:1 at each locus). Of 104 lines, 31 showed segregation distortion at one or both individual introgressions. Each of these cases can be explained by a deleterious interaction (i.e., DMI) between this locus and one or more loci in the SL genetic background. In no case did an introgression show significant segregation distortion independent of the 14 other introgressions with which it was paired in this experiment (i.e., no SH introgression was over- or underrepresented in every pairwise combination). This result suggests that the expression of deleterious hybrid interactions at these loci is sensitive to the conspecific genetic background in which an introgression is found, even when there is no genotypic or allelic distortion at the interacting locus.
Underlying genetic mechanisms:
Across the whole experiment, 39 DIL families (37.5%) showed significant (P < 0.05) genotypic or allelic segregation distortion for at least one introgression segment (supplemental Table 1). In most cases where segregation was distorted (35/39), SH alleles are underrepresented. Overall, these cases show a diversity of mechanisms for complex epistasis, ranging from direct evidence (i.e., specific underrepresentation of double introgression homozygotes) through to more indirect indicators of genetic interactions (i.e., underrepresentation in only some genetic backgrounds). These data indicate that complex epistasis is detectable but not overwhelmingly pervasive between these species, at least at the developmental stages that could be assessed here. Nonetheless, the frequency with which we detect evidence for complex epistasis suggests that it could be an influential factor in the temporal accumulation of species barriers, and the number of loci required to complete speciation. For example, our results suggest that in many cases greater than one allelic substitution per lineage is required for the expression of hybrid incompatibility phenotypes at early developmental stages in this species pair.
Note that the genomic regions evaluated in this analysis might represent multiple linked genes, each of which potentially contributes to expression of transmission ratio distortion. Strictly speaking, therefore, our analysis evaluates the prevalence of interactions between chromosomal regions in different parts of the genome; detected interactions might be pairwise or higher order between any individual loci that are contained within individual introgressions. The fine-scale structure of loci contained within currently identified genomic regions can be directly evaluated in the future via fine mapping and eventual positional cloning.
In terms of our a priori hypothesis (underrepresentation of double SH homozygotes), we found only modest evidence of complex epistasis. The direction of results, however, suggests that complex epistasis more frequently acts to enhance the expression of hybrid incompatibility (i.e., greater viability selection against individuals carrying two chromosomal regions). Some evidence suggests that these synergistic epistatic interactions might be common in interspecific crosses. For example, interspecific male sterility QTL in Drosophila show evidence of complex epistasis such that several individual genomic regions appear to be simultaneously required for the expression of some male sterility phenotypes (e.g., Wu et al. 1996; other evidence is reviewed in Coyne and Orr 2004). If synergistic epistasis is common for hybrid incompatibility, although >1 substitution might be required to observe incompatibility phenotypes, once substitutions have begun to accumulate substantial hybrid incompatibility could be expressed rapidly between two diverging species. Interestingly, a previous analysis of complex epistasis for yield-related traits in a similar tomato cross (Eshed and Zamir 1996) found the opposite result; introgression combinations showed less-than-additive effects on traits like fruit size, perhaps marking a difference in the underlying genetic basis of these different classes of traits. Conversely, in the two cases where SH double homozygotes were overrepresented, we hypothesize that these genotypes must have conferred greater pollen competitive advantage and/or greater early postfertilization viability in comparison to gametes or individuals carrying one or no introgressions at these regions. This is intriguing and suggests that co-introgression contributes to reproductive heterosis in these cases.
Formally, interactions involving more than two loci that influence hybrid incompatibility could be of two types. First, more than two loci (i.e., more than one locus from each species) could be required to express a single DMI (“complex conspecific epistasis”; Coyne and Orr 2004, p. 307). Complex conspecific epistasis (CCE) could be due to biochemical redundancy in developmental pathways, so that multiple losses or changes in function in one or both lineages are simultaneously required to express a DMI (Coyne and Orr 2004, pp. 275–276). Alternatively, higher-order interactions might also occur between loci involved in different DMIs [i.e., “complex heterospecific epistasis” (CHE)]. That is, conspecific loci underlying different DMIs further interact with each other to amplify or suppress the expression of hybrid incompatibility. For example, two different mutations might each have a deleterious individual effect within the same developmental pathway, but their pairwise combination might suppress these deleterious effects (as has been observed, for example, with double deletion strains in yeast; Jasnos and Korona 2007). Both forms of epistasis might be more likely where gene duplicates are common in the relevant developmental pathways (i.e., leading to functional redundancy; Coyne and Orr 2004, p. 275) and/or where particular developmental or biochemical pathways are frequently involved in the expression of a class of hybrid incompatibilities. Indeed, the principle that genes that show nonadditive effects in combination are likely to be acting in the same biochemical or physiological pathways is routinely used to infer functional interactions between these loci (e.g., Koornneef et al. 1991). The moderate evidence for complex epistasis we have detected here might therefore suggest that any underlying DMIs are distributed among several different developmental pathways.
Finally, where we have detected evidence for complex epistasis here, we are presently unable to distinguish complex conspecific from heterospecific epistasis (CCE vs. CHE). However, this study is a prelude to an equivalent analysis of complex epistasis for hybrid male (pollen) and female (seed) fertility. Because we have constructed our DILs using lines that both carry and do not carry known hybrid incompatibility QTL that act at these later stages of fertility (Table 1 and supplemental Table 2), this ongoing work can assess the frequency with which complex epistasis preferentially involves QTL with main effects vs. no main effects on hybrid sterility.
Acknowledgments
We thank M. Fryska and R. Burns for assistance with plant germination and cultivation, C. Muir, M. W. Hahn, and K. Montooth for helpful suggestions and/or comments on the manuscript. This work was supported by National Science Foundation (Division of Environmental Biology) grant 0532097 (to L.C.M.).
References
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Eshed, Y., and D. Zamir, 1996. Less-than-additive epistatic interactions of quantitative trait loci in tomato. Genetics 143 1807–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frova, C., and M. E. Pe, 1997. Gene expression during pollen development, pp. 31–48 in Sexual Plant Reproduction, edited by M. Cresti and A. Tiezzi. Springer, Berlin.
- Jasnos, L., and R. Korona, 2007. Epistatic buffering of fitness loss in yeast double deletion strains. Nat. Genet. 39 550–554. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1984. Deletrious mutations as an evolutionary factors. I. The advantage of recombination. Genet. Res. 44 199–217. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., 2003. Accumulation of Dobzhansky-Muller incompatibilities with a spatially structured population. Evolution 57 151–153. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., C. J. Hanhart and J. H. Van Der Veen, 1991. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229 57–66. [DOI] [PubMed] [Google Scholar]
- Monforte, A. J., and S. D. Tanksley, 2000. Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome 43 803–813. [PubMed] [Google Scholar]
- Moyle, L. C., and E. B. Graham, 2005. Genetics of hybrid incompatibility between Lycopersicon esculentum and L. hirsutum. Genetics 169 355–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle, L. C., and T. Nakazato, 2008. Comparative genetics of hybrid incompatibility: sterility in two Solanum species crosses. Genetics 179 1437–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and M. Turelli, 2001. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55 1085–1094. [DOI] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1995. Biometry, Ed. 3. W. H. Freeman, New York.
- Turelli, M., N. H. Barton and J. A. Coyne, 2001. Theory and speciation. Trends Ecol. Evol. 16 330–343. [DOI] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 2000. Dominance, epistasis and the genetics of postzygotic isolation. Genetics 154 1663–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, J. J., 2004. Accumulating Dobzhansky-Muller incompatibilities: reconciling theory and data. Evolution 58 1145–1156. [DOI] [PubMed] [Google Scholar]
- Wu, C. I., N. A. Johnson and M. F. Palopoli, 1996. Haldane's rule and its legacy: Why are there so many sterile males? Trends Ecol. Evol. 11 281–284. [DOI] [PubMed] [Google Scholar]