Abstract
In many physiological and disease processes, TGF-β usurps branches of MAP kinase pathways in conjunction to Smads to induce apoptosis and epithelial to mesenchymal transition, but the detailed mechanism of how a MAP kinase cascade is activated by TGF-β receptors is not clear. We report here that TRAF6 is specifically required for the Smad-independent activation of JNK and p38 and its carboxyl TRAF homology domain physically interacts with TGF-β receptors. TGF-β induces K63-linked ubiquitination of TRAF6, and promotes association between TRAF6 and TAK1. Our results indicate that TGF-β activates JNK and p38 through a mechanism similar to that operating in the interleukin-1β/Toll-like receptor pathway.
INTRODUCTION
Transforming growth factor-β (TGF-β) exerts its diverse functions through multiple intracellular signaling pathways on a wide array of cellular processes (Massagué et al., 2000). In the classically described signaling pathway, TGF-β acts through a complex of type I (TβRI) and type II (TβRII) transmembrane receptors to induce phosphorylation of Smad proteins, leading to their accumulation in the nucleus to regulate target gene expression (Feng and Derynck, 2005; Massagué et al., 2005). In many instances, ligand-bound TGF-β receptors also activate JNK and p38 of mitogen-activated protein (MAP) kinase pathways independent of Smads to induce apoptosis during normal physiological processes and in tumor suppression or to promote epithelial to mesenchymal transition (EMT) at the onset of metastasis (Derynck and Zhang, 2003; Moustakas and Heldin, 2005). However, how TGF-β receptors activate JNK and p38 in this so-called Smad-independent pathway remains to be determined.
The MAP kinase pathways consist of cascades of sequentially acting protein kinases that ultimately activate nuclear transcription factors by phosphorylation in response to extracellular cues (Chang and Karin, 2001). The p38 and JNK MAP kinases are at the tertiary layer of a kinase cascade, in which they are activated by the MAP kinase kinases (MKKs), specifically the MKK3/6 or MKK4, respectively. Further upstream, MKKs are activated by the MAP kinase kinase kinases (MAP3Ks); in case of MKK3/6 and MKK4, the TGF-β-activated kinase 1 (TAK1) is one of the activating MAP3Ks. Vertebrate TAK1 was identified based on the ability of a mouse cDNA to substitute for a latent MAP3K of S. cerevisiae in the yeast mating pheromone responses and the cloned kinase was found to activate and be activated by TGF-β signaling (Yamaguchi et al., 1995). The physiological significance of TAK1 in TGF-β signaling was revealed in recent studies of mouse embryonic fibroblasts (MEFs) in which the TAK1 gene was inactivated through homologous recombination (Shim et al., 2005; Jadrich et al., 2006). It was demonstrated that TAK1 is absolutely required for the TGF-β-induced JNK and NFκB activation (Shim et al. 2005). TAK1 was also discovered independently in a protein complex required for the activation of JNK and the I-kappa B kinase (IKK) complex that situates at a critical node of an intracellular signaling network controlling a master transcription factor, NF-κB, in response to tumor necrosis factor α (TNF–α), interleukin 1- β (IL-1β), or ligands for Toll-like receptors (TLRs) (Chen, 2005). Activation of JNK and IKK by these cytokines requires the kinase activity of TAK1, which is activated by TRAF2 or TRAF6, two RING domain ubiquitin ligases that catalyze the synthesis of polyubiquitin chains linked via a lysine at position 63 (K63). The ubiquitin ligase activity is essential for TRAF’s function, as a mutant TRAF6 lacking the RING domain was found unable to activate MAPKs in response to IL-1β and LPS in TRAF6-deficient cells (Kobayashi et al., 2001). Unlike the type of polyubiquitin chains formed via lysine-48 (K48) that normally target proteins for degradation in the proteasomes, the K63-linked polyubiquitin chains serve as a regulatory signal and provide a scaffold for the assembly of protein kinase complexes to mediate their activation (Laine and Ronai, 2005; Adhikari et al., 2007).
In light of the critical roles of TRAF E3 ligases in TAK1 activation, we asked if a similar mechanism also operates in the Smad-independent pathway of TGF-β signaling. Our investigation demonstrated that the ligand-activated TGF-β receptor complex binds and induces K63 polyubiquitination of TRAF6, which is specifically required for activating the downstream JNK and p38 by TGF-β.
RESULTS AND DISCUSSION
TRAF6 is essential to the TGF-β-induced activation of JNK and p38
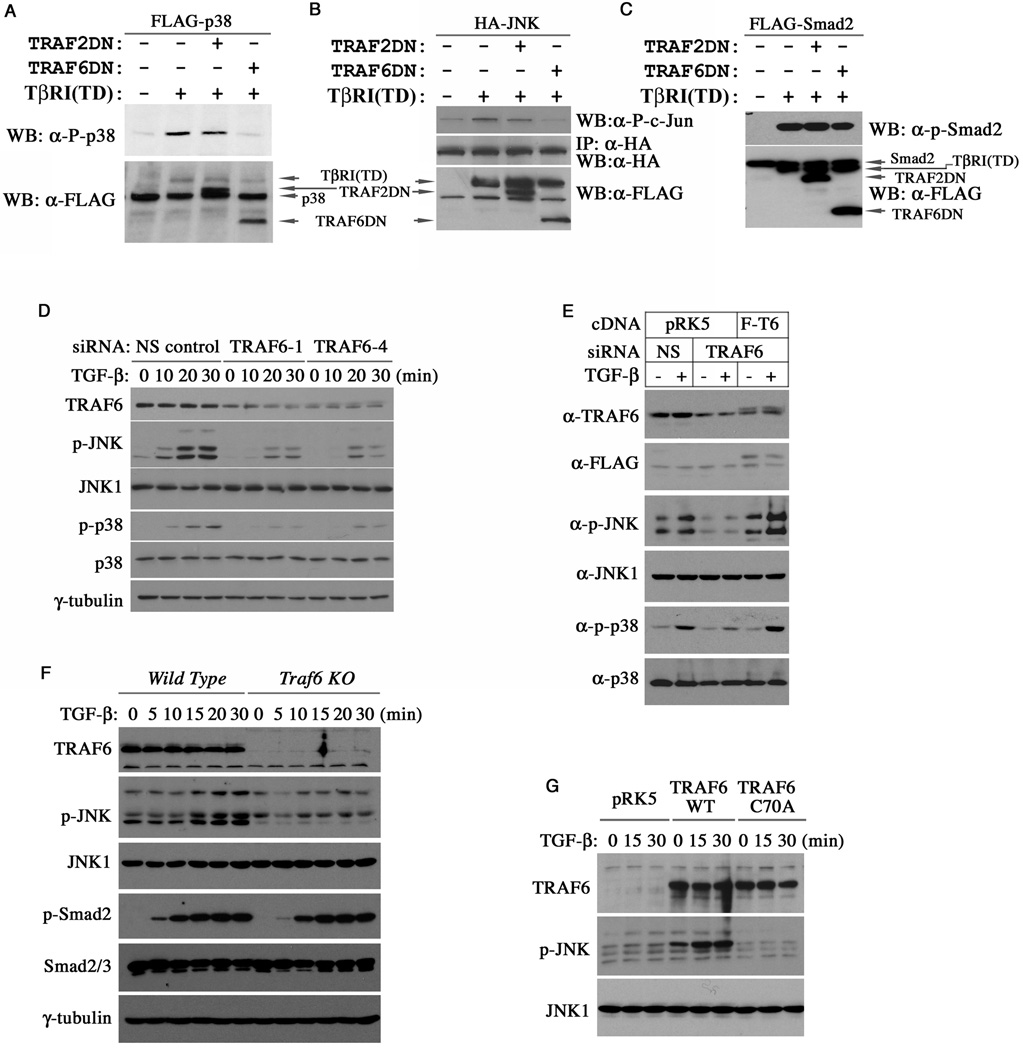
Several TRAF proteins including TRAF2 and TRAF6 consist of the highly conserved RING ubiquitin ligase domain in the amino terminus followed by a zinc finger domain, a coiled-coil domain and a carboxyl TRAF homology domain that interacts with the ligand activated receptors of various pathways (Chung et al., 2002). To determine if TRAF2 or TRAF6 is required for the TGF-β-induced p38 and JNK activation, we co-expressed a FLAG-tagged p38 along with dominant negative mutants of TRAF2 or TRAF6 lacking the RING domain in RIBL17 mink lung epithelial cells, which are deficient in TβRI. As we showed previously (Yu et al., 2002), adding back a constitutively active receptor mutant, TβRI(TD), to these cells efficiently activated p38, as evident by immunoblotting with phospho-p38 specific antibodies (Figure 1A). However, phosphorylation of p38 induced by TβRI(TD) was severely curtailed by a TRAF6 deletion mutant (TRAF6DN) lacking the RING domain but not by a similar TRAF2 mutant (TRAF2DN), implying a specific requirement of the endogenous TRAF6 function. A similar result was observed on the activation of JNK, which was assayed in vitro on a recombinant GST-c-Jun substrate following immunoprecipitation of the HA-tagged JNK from transfected RIBL17 cells (Figure 1B). In contrast, neither TRAF2DN nor TRAF6DN had any effect on the TβRI(TD)-induced Smad2 phosphorylation (Figure 1C). To determine if TRAF6 is required for the TGF-β-induced activation of JNK and p38 in vivo, we resorted to RNAi to silence TRAF6 expression in AML12 cells, a line of mouse hepatocytes, in which TGF-β is able to induce a rapid activation of p38 and JNK (Perlman et al., 2001; Liao et al., 2001). In this experiment, activation of both p38 and JNK was strongly inhibited by two different siRNAs against TRAF6 (Figure 1D). The specificity of these siRNA experiments was confirmed in a “rescue” experiment, in which a FLAG-tagged human TRAF6 cDNA that does not contain target sequences of the siRNA was reintroduced into AML12 cells. This human TRAF6 cDNA completely reversed the inhibition of the mouse TRAF6-specific siRNA on the ability of TGF-β to activate JNK and p38 (Figure 1E). The requirement of TRAF6 was also investigated in TRAF6-deficient MEFs, in which TGF-β failed to activate JNK whereas a robust activation was observed in wild type MEFs (Figure 1F). Once again, reintroducing wild type TRAF6 to the TRAF6-deficient MEFs restored the TGF-β-induced JNK activation to the extent that it actually generated a higher basal level of phospho-JNK, possibly due to the effect of overexpression (Figure 1G). Conversely, a mutant TRAF6 with a highly conserved cysteine residue in the RING domain being replaced by an alanine (C70A) had no effect in this experiment (Figure 1G), indicating that the integrity of the RING domain is essential.
Figure 1. TRAF6 is required for the TGF-β-induced activation of JNK and p38.
(A–C) Effects of TRAF2 and TRAF6 mutants on the activation of exogenous FLAG-p38, HA-JNK or FLAG-Smad2 by the constitutively active TβRI(TD) in R1BL17 cells. The levels of phosphorylated and total p38 were analyzed by Western blot analyses (A), and the activity of immunoprecipitated HA-JNK was determined in an in vitro kinase reaction using GST-c-Jun as the substrate (B). Only the TRAF6 mutant inhibited the activation of both p38 and JNK. Neither TRAF2 nor TRAF6 affected Smad2 phosphorylation (C). (D) Effect of the TRAF6 RNAi on the activation of endogenous JNK and p38 by TGF-β. AML12 cells were treated with TGF-β as indicated two days after siRNA transfection. The levels of phospho- or total JNK and p38 in cell lysates were analyzed by Western blots. (E) Rescuing JNK and p38 activation in TRAF6-siRNA transfected AML12 cells with wild type human TRAF6 cDNA. TGF-β treatment was for 30 minutes. (F) Western analyses showing absent JNK activation but unabated Smad2 activation by TGF-β in TRAF6-deficient MEFs. (G) Restoration of the TGF-β-induced JNK activation in TRAF6-deficient MEFs with wild type TRAF6 but not the RING domain mutant TRAF6(C70A).
TRAF6 is physically associated with activated TGF-β receptors
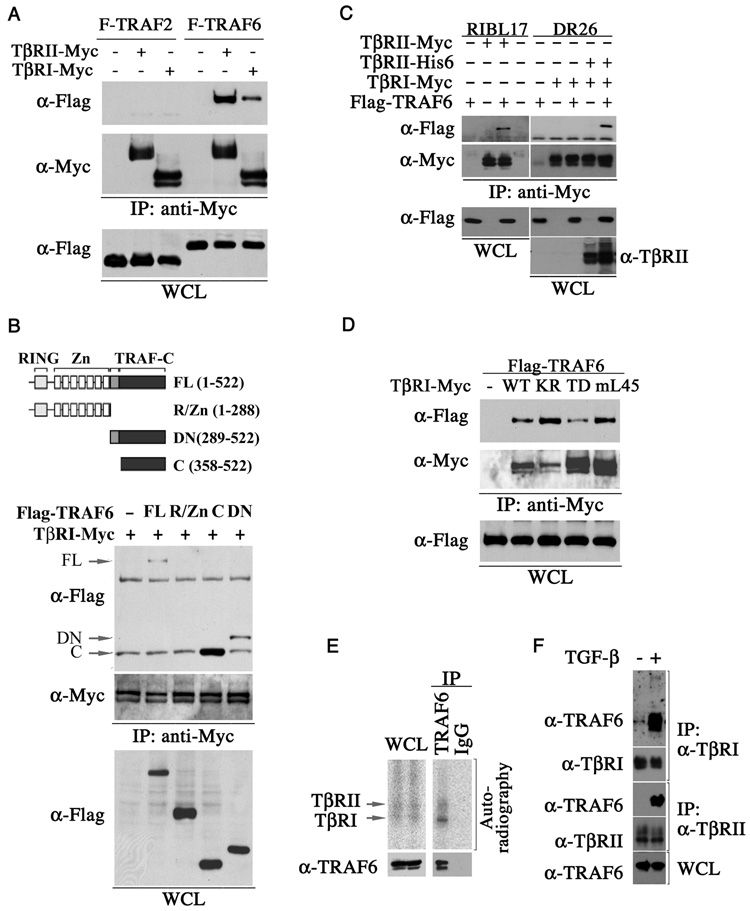
In the signaling by pro-inflammatory cytokines, TRAFs are recruited to the ligand-activated receptor complexes (Chung et al, 2002). We therefore asked if TRAF6 binds to TGF-β receptors by co-immunoprecipitation experiments in HEK293 cells. Our results showed that TRAF6 indeed was capable of binding to both TβRI and TβRII, with slightly higher affinity towards TβRII (Figure 2A). In contrast, TRAF2 lacks such binding activity (Figure 2A). Deletion mapping experiments indicated that TRAF6 binds to TGF-β receptors via the TRAF homology domain at the carboxyl terminus (Figure 2B, supplemental figure 1), in line with the interaction between TRAFs and other cytokine receptors (Chung et al, 2002). Because the two TGF-β receptors can complex together even in the absence of ligand when they are over-expressed (Feng et al., 1996), we then re-examined this interaction in TβRI-deficient RIBL17 or TβRII-deficient DR26 cells to determine if TRAF6 binds to individual receptors or to receptor complexes as a whole. We were able to bring down the FLAG-tagged TRAF6 using anti-Myc antibodies against the Myc-tagged TβRII in RIBL17 cells, but not with antibodies against the Myc-tagged TβRI in DR26 cells (Figure 2C). However, when both TβRII and TβRI were expressed in DR26 cells, TRAF6 was found to bind Myc-TβRI (Figure 2C, right panel). Superficially, this result suggests that TRAF6 interacts directly with TβRII while its interaction with TβRI is mediated through TβRII. However, when co-expressed with a series of type I receptor mutants, TRAF6 exhibited affinities to TβRI in a descending order from the kinase defective mutant, TβRI(KR), to the wild type, and to the constitutively active mutant, TβRI(TD) (Figure 2D), implying a role of TβRI in the binding. A parsimonious explanation of our binding data would be a model in which TRAF6 recognizes an activated receptor complex consisted of a homo- or heterocomplex of two types of receptors but the interaction to the heterocomplex is likely to be transitory as TRAF6 was dissociated upon activation or became trapped to the catalytically inactive TβRI mutant (Figure 2D). The fact that TβRII can become activated in a homocomplex, whereas TβRI can only be activated by TβRII is consistent with the higher affinity of TRAF6 to TβRII (Figure 2A) and the role of the kinase activity of TβRI in binding (Figure 2D). Previously, we showed that p38 and JNK can be activated by a mutant type I receptor, TβRImL45, which has lost the ability to interact with Smads due to mutations in the L45 loop of the kinase domain (Yu et al., 2002). This mutant receptor was still capable of binding to TRAF6 (Figure 2D), consistent with its retained activity in activating p38 and JNK.
Figure 2. TRAF6 specifically interacts with the TGF-β receptors.
(A) Immunoprecipitation and Western analyses of specific interaction between TGF-β receptors and TRAF6 but not TRAF2 in transfected HEK293 cells. (B) Deletion mapping of the TβRI binding domain in TRAF6. A schematic representation of various deletion constructs was shown. (C) Interaction between TRAF6 and TGF-β receptors in TβRI-deficient RIBL17 or TβRII-deficient DR26 cells. (D) Interaction between TRAF6 and wild-type (WT), kinase-deficient (KR), constitutively active (TD) or Smad-binding defective (mL45) TβRI receptors in HEK293 cells. Note the increase of TRAF6 in the TβRI(KR) immunocomplex, indicative of a higher binding affinity. (E) Binding of TRAF6 to the ligand-occupied TGF-β receptor complexes on cell surface. AML12 cells were affinity-labeled with [125I] TGF-β1 and crosslinked with DSS. Lysates were subjected to anti-TRAF6 immunoprecipitation followed by autoradiography to detect TRAF6 bound receptors. (F) TGF- β stimulates interaction between endogenous TRAF6 and TβRI or TβRII. AML12 cells were crosslinked with formaldehyde, and the receptor-bound TRAF6 were analyzed by Western blot following immunoprecipitation.
To determine if TRAF6 indeed interacts with the TGF-β receptor complex, we labeled AML12 cell surface proteins with [125I] TGF-β in the presence of disuccinimidyl suberate (DSS), a membrane permeable crosslinker. Immunoprecipition of TRAF6 brought down both [125I] TGF-β-labeled type I and type II receptors as evident in autoradiogram (Figure 2E). In a separate experiment, we treated AML12 cells with formaldehyde, another membrane permeable crosslinking agent, and detected endogenous TRAF6 in either anti-TβRI or anti-TβRII immunocomplexes upon TGF-β stimulation (Figure 2F).
TGF-β promotes K63 ubiquitination of TRAF6 and interaction with TAK1
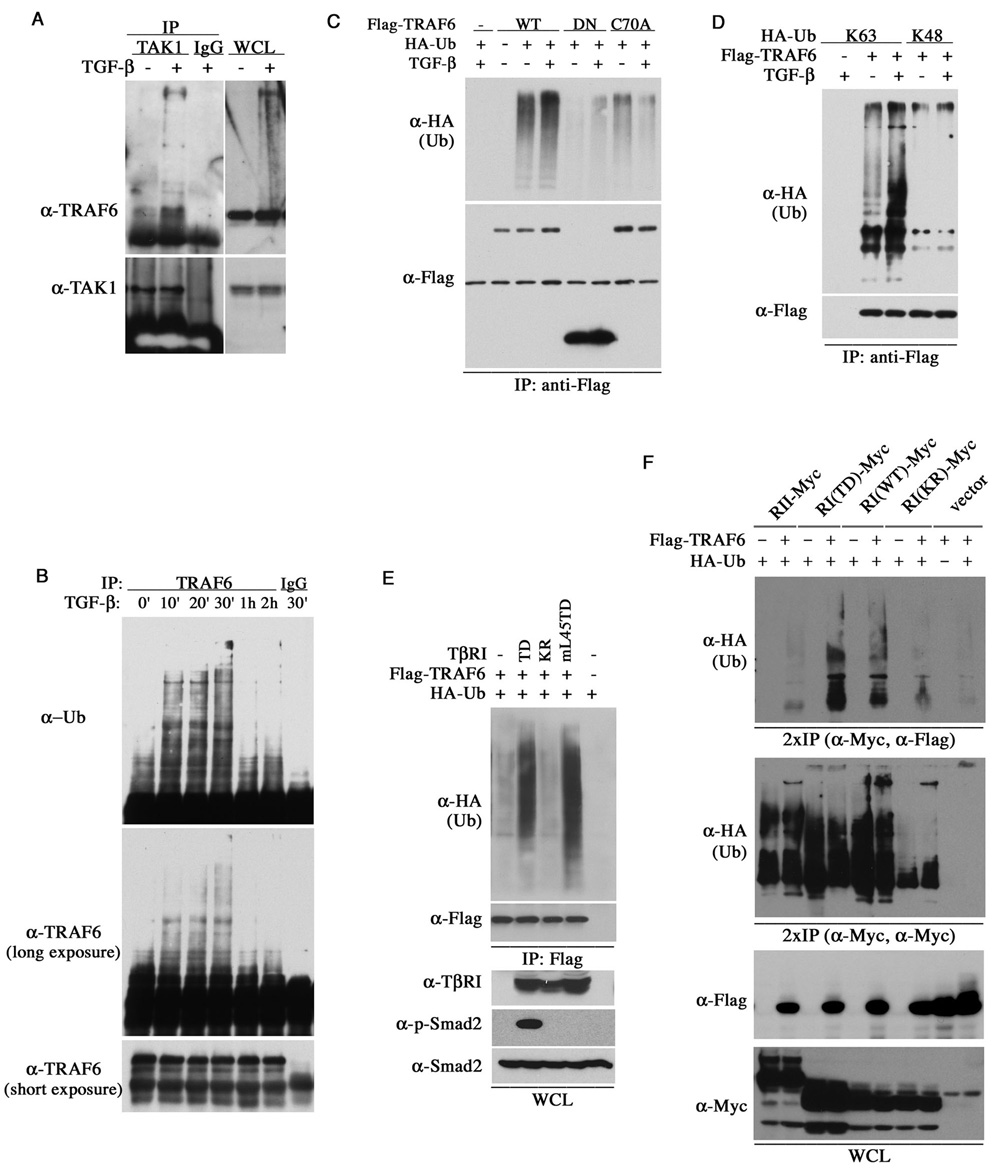
In the IL-1R/TLR signaling, TRAF6 itself undergoes K63-linked polyubiquitination, which is essential for the interaction with TAK1 and the activation of JNK and p38 (Wang et al., 2001). We observed an enrichment of TRAF6 in the anti-TAK1 immunocomplex and a ladder of high molecular weight TRAF6 in AML12 cells under TGF-β treatment (Figure 3A). Small amount of high molecular weight TRAF6 was also detected in the anti-TβRI immunocomplex (Figure 2F), suggesting that TGF-β may promote TRAF6 ubiquitination. Indeed, TRAF6 became polyubiquitinated in AML12 cells within 30 minutes of TGF-β treatment, but the level of polyubiquitinated TRAF6 abruptly dropped after one hour (Figure 3B), reflecting a transitory nature of this modification. This time course is consistent with the kinetics of JNK and p38 activation induced by TGF-β (Figure 1D). TGF-β also enhanced ubiquitination of the exogenous, FLAG-tagged TRAF6 but not TRAF6DN or TRAF6(C70A) (Figure 3C), indicating that the RING domain is required for its ubiquitination. To determine the type of ubiquitin chain linkage, we co-expressed TRAF6 with ubiquitin mutants that had all lysines but K63 or K48 replaced with arginine, and found that only the K63- but not K48-ubiquitin mutant allowed ubiquitination of TRAF6, which was enhanced by TGF-β (Figure 3D).
Figure 3. TGF-β promotes K63 polyubiquitin chain modification of TRAF6.
(A) Interaction between the endogenous TRAF6 and TAK1 in AML12 cells as determined by immunoprecipitation with anti-TAK1 antibody, followed by anti-TRAF6 Western blot. The TGF-β treatment was for 30 minutes and the total rabbit IgG was used as a control. (B) TGF-β–induced ubiquitination of endogenous TRAF6 in AML12 cells. Cells were lyzed in RIPA buffer after treated with TGF-β for the indicated duration. The amount of ubiquitinated TRAF6 was visualized by anti-Ub and anti-TRAF6 blots after immunoprecipitation. (C) Requirement of the RING E3 ligase domain in TRAF6 ubiquitination. AML12 cells tranfected with plasmids encoding HA-ubiquitin, Flag-TRAF6 or its mutants were lyzed in RIPA buffer after treated with TGF-β for 30 min. Ubiquitination of transfected TRAF6 was detected using anti-HA antibody after immunoprecipitation with the FLAG antibody. (D) TGF-β promotes the formation of K63- but not K48-linked polyubiquitin chain on TRAF6. AML12 cells were transfected with plasmids encoding TRAF6 and either K63 or K48 only HA-ubiquitin, and treated with TGF-β for 30 min. Ubiquitinated TRAF6 was visualized as in (C). (E) Smad-independent ubiquitination of TRAF6 in RIBL17 cells. The level of ubiquitinated TRAF6 was detected as in (C), and the levels of transfected TβRI, phospho-Smad2 and Smad2 in total lysates were shown at the bottom. (F) Association of the ubiquitinated TRAF6 with TGF-β receptors in HEK293 cells. The myc-tagged receptors were isolated from total cell lysates by immunoprecipitation, boiled in a SDS buffer, and then the resuspended proteins were reprecipitated with either FLAG or Myc antibodies prior to Western analyses.
To determine if the TGF-β-induced ubiquitination of TRAF6 is a direct result of receptor activation, we examined the impact of various type I receptor mutants on this reaction in RIBL17 cells. Our results showed that only the constitutively active TβRI(TD) and mL45(TD) mutants supported the ubiquitination of TRAF6, while the kinase deficient TβRI(KR) did not (Figure 3E). This indicate that the kinase activity of TβRI, therefore activation of the receptor complex, is essential for TRAF6 ubiquitination whereas Smad binding is not. This notion is consistent with our previously reported requirement of type I receptor kinase activity but not Smad activity in the activation of p38 and JNK (Yu et al., 2002; Itoh et al., 2003). Finally, to demonstrate the presence of receptor-bound, ubiquitinated TRAF6, we first isolated the Myc-tagged receptors from transfected HEK293 cells, and then after disrupting the immunocomplexes with a SDS buffer we re-precipitated the receptor-bound FLAG-tagged TRAF6. Following this procedure, we detected ubiquitinated TRAF6 that was specifically associated with the constitutively active or wild type type I receptor, but not TβRII alone or the kinase-deficient TβRI(KR) (Figure 3F). In a control experiment, we found that the various TGF-β receptors used in this experiment were also ubiquitinated, but their ubiquitination was unaffected by the co-expressed TRAF6 (Figure 3F).
TRAF6 functions specifically in the Smad-independent branch of TGF-β signaling
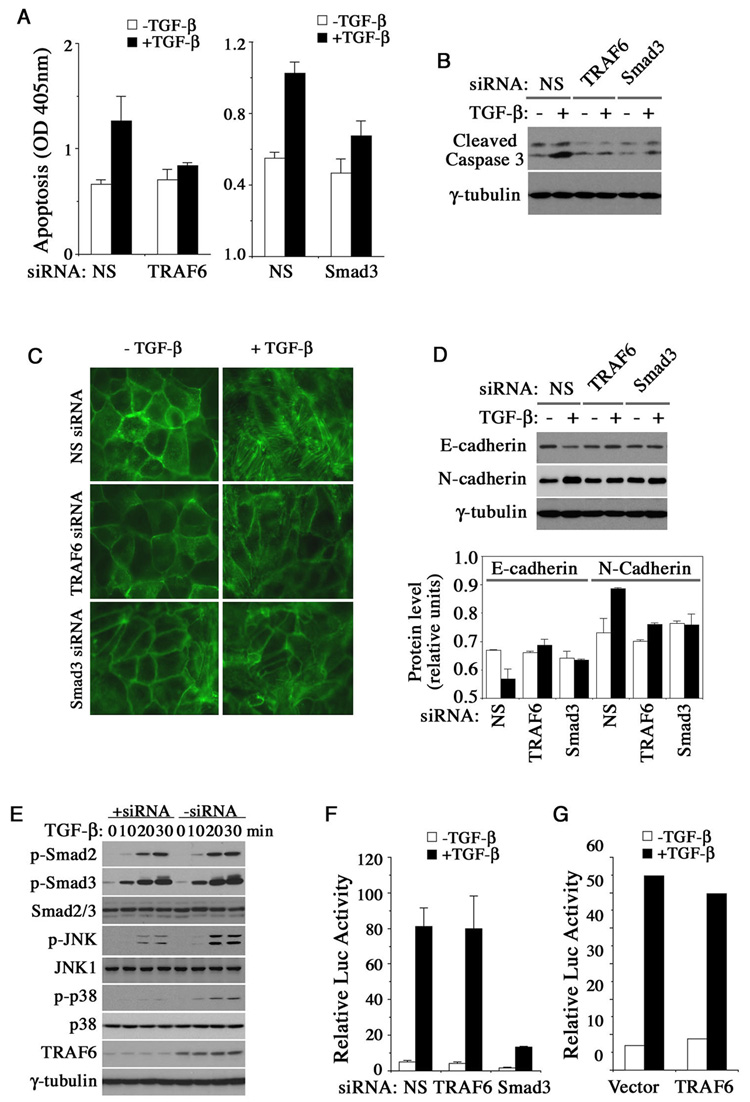
In many physiological functions of TGF-β, such as apoptosis and EMT, inputs from both Smad and MAP kinase pathways are simultaneously required to generate a full-fledged signaling response (Zavadil et al., 2001; Yu et al., 2002; Valcourt et al., 2005; Yang et al., 2006). We therefore investigated if TRAF6 acts in TGF-β-mediated apoptosis and EMT responses by RNAi-mediated gene silencing with a TRAF6 specific siRNA. Treating AML12 cells with TGF-β led to apoptosis, a typical TGF-β response that requires dual pathway activation. These cells were protected from death (Figure 4A, left panel) and caspase-3 activation (Figure 4B) when TRAF6 expression was knocked down. Knocking-down Smad3 expression had a similar protective effect to AML12 cells (Figure 4A, right panel and Figure 4B), in keeping with our previous observation that the TGF-β-induced apoptosis requires both Smad3-mediated transcriptional responses and p38 activation (Yang et al., 2006). The role of TRAF6 on TGF-β-induced EMT was examined in NMuMG mouse mammary epithelial cells whose actin cytoskeleton appeared in a regular cobble stone-like shape at cell-cell junctions (Figure 4C). Following 48 hours of TGF-β treatment, this regular pattern gave away to a spindle-shaped structure of actin stress fibers, accompanied by a downregulation of E-cadherin and an upregulation of N-cadherin (Figure 4C and 4D). Once again, silencing either TRAF6 or Smad3 in NMuMG cells blocked the EMT (Figure 4C and 4D). However, RNAi-mediated silencing of TRAF6 failed to alter Smad2 and Smad3 phosphorylation (Figure 4E), nor did it change the response of a Smad-specific luciferase reporter, (CAGA)12-Luc, to TGF-β (Figure 4F). Overexpression of TRAF6 also did not change the (CAGA)12-Luc response (Figure 4G). These results are in agreement with the observation that loss of TAK1 does not affect Smad activation or Smad-dependent transcriptional responses (Shim et al. 2005).
Figure 4. TRAF6 is required for the TGF-β-induced apoptosis and EMT.
(A–B) Effect of transfected TRAF6 and Smad3 siRNA on the apoptosis of AML12 cells. (A) Cell death was measured with the Cell Death Elisa kit after treating cells with or without TGF-β for 24 hours. (B) Caspase-3 activation was analyzed by Western blot. (C–D) Effect of transfected TRAF6 and Smad3 siRNA on EMT in NMuMG cells after treated with or without TGF-β for 48 hours. The cells were fixed and stained for F-actin (C) or subjected to Western analysis of E-cadherin and N-cadherin expression (D). Quantitations of E-cadherin and N-cadherin levels relative to γ-tubulin are shown at bottom, open bar: −TGF-β; solid bar: +TGF-β. (E) TGF-β-induced Smad2 and Smad3 activation in AML12 cells in the presence or absence of transfected TRAF6 siRNA. (F, G) (CAGA)12-Luc reporter assay of TGF-β responses in Hep3B cells that were transfected with siRNAs or TRAF6 expression plasmid. The ligand treatment was for 24 hours. All bar graphs are displayed as mean ± standard deviation.
In summary, we have identified an essential role of TRAF6 in the TGF-β induced activation of p38 and JNK by perturbing endogenous TRAF6 function with dominant negative mutants or siRNAs, and by restoring JNK activation in TRAF6-deficient cells using wild type but not mutant TRAF6 cDNA (Figure 1). We showed that TRAF6 binds to the ligand-occupied receptor complex in vivo, and examined the nature of this interaction in transfected cells (Figure 2). We further demonstrated that TGF-β receptors induces the K63 polyubiquitination of TRAF6 when they are associated in a complex (Figure 3), and TRAF6 has a clear role in TGF-β-induced apoptosis and EMT independent of Smads (Figure 4).
Because TRAF6 is a critical activator of TAK1 that functions upstream as the MAP3K of p38 and JNK, our finding connects a missing link between the ligand activated receptors and the MAP kinase branch of Smad-independent TGF-β signaling. Previously, another RING E3 ligase, XIAP, was shown to have the ability to activate TAK1/JNK by interacting with TGF-β receptors and TAB1, a constituent of the TAK1 kinase complex (Yamaguchi et al., 1999; Birkey Reffey et al., 2001). However, the L45 loop of the type I receptor kinase domain, which we showed as being dispensable for p38/JNK activation, was actually known to be required for XIAP binding (Itoh et al. 2003), and the TGF-β-induced JNK activation was found to proceed normally in TAB1 null MEFs (Shim et al., 2005). Moreover, a more recent report indicated that XIAP plays a negative role in the TGF-β-induced JNK activation and apoptosis by targeting TAK1 for degradation in the proteosomes (Kaur et al., 2005). Our results also reveal an essential requirement for the kinase activity of TβRI in TRAF6 ubiquitination and activation, although the significance of this requirement is not clear at the moment since TRAF6 itself was not phosphorylated by TβRI (data not shown). Finally, although the TRAF6 activity described here is independent of Smad, the resulting activated p38 and JNK still function in conjunction with the Smad-dependent pathway to regulate downstream cellular responses to TGF-β Such a mechanism could allow a fine-tuning of the level of TGF-β signaling in determining the ultimate nature of cell fate in response to this cytokine.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used: phospho-Smad2 (465/467), phospho-p38, p38, phospho-JNK, JNK1, Caspase-3 (Cell signaling Technology); TRAF6 (H274), TRAF6 (D10), TAK1 (M579), c-Myc (9E10), TβRII (L21), TβRI (V22) (Santa Cruz Biotech); phospho-Smad3 (423/425) (Rockland); E-cadherin, N-cadherin, Smad2/3, TβRI, TβRII (BD Biosciences), Flag (M2), Flag-POD (Sigma), γ-tubulin (Invitrogen); HA (HA11) (Covance); ubiqutin (Biomol); Myc-POD, HA-HRP (Roche).
Cell Culture and transfection
HEK293, AML12, Hep3B and NMuMG cells were obtained from the American Type Culture Collection, mutant mink lung epithelial cells, RIBL17 and DR26, were provided by Dr. J. Massagué (Cárcamo et al., 1994), and TRAF6-deficient MEFs (Lomaga et al., 1999) were obtained from Dr. Tak Mak. Whenever used, TGF-β1 (PeproTech) was added to serum-starved cells at 4 ng/ml in medium containing 0.2% FBS. Transfection of HEK293 and RIBL17 cells was carried out using Fugene 6 (Roche), while AML12 cells using Lipofectamine 2000 (Invitrogen), DR26 cells using nucleofector (Amaxa). Oligofectamine (Invitrogen) was used in all siRNA transfections.
Immunoblotting, immunoprecipitation, in vitro kinase assay
For Western and immunoprecipitation experiment, cells were lysed in the NP-40 lysis buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and a cocktail of protease and phosphatase inhibitors. The JNK kinase assay, affinity labeling cell surface TGF-β receptors with [125I]-TGF-β1(NEN), regular and sequential immunoprecipitation experiments were described previously (Chen and Derynck, 1994; Yu et al., 2002). In order to detect transient interaction between endogenous TRAF6 and TGF-β receptors, AML12 cells were crosslinked with 1% formaldehyde before harvesting. After sonicating cell lysates, TGF-β-receptors were immunoprecipitated with biotinylated goat-anti-mouse TβRI or TβRII antibodies (R&D Systems) absorbed on streptavidin-coupled magnetic beads (Promega). In TRAF6 ubiquitination experiments, cells were harvested in the RIPA buffer containing 50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% sodium deoxycholate, 0.2% SDS with protease and phosphatase inhibitors.
Apoptosis assay
Prior to assay, AML12 cells were maintained in DMEM/F12- 0.2% FBS with or without TGF-β1 treatment for 24 hours. Apoptosis was quantified with the Cell Death Detection ELISA assay kit (Roche), which quantitatively determines DNA fragmentation.
EMT and fluorescence staining
To visualize EMT, NMuMG cells were grown on coverslips in 12-well plates. After 48 hours of TGF-β treatment in 10%FBS containing medium, cells were fixed in 4.5% paraformaldehyde and permeabilized with 0.2% Triton X-100. Actin cytoskeleton was stained with fluorescin-conjugated-phalloidin (Molecular Probe).
Transcription reporter assay
(CAGA)12-Luc (Dennler et al., 1998) was kindly provided by P. ten Dijke. All experiments were performed in triplicates as described previously (Yu et al., 2002), and the data were presented as average ± SD.
Expression plasmids and siRNAs
Supplementary Material
ACKNOWLEDGEMENTS
We are in debt to Drs. D. Goeddel, Y. Zheng, P. ten Dijke, Tak Mak and J. Massagué for providing various plasmids and cell lines. This research was supported by the intramural research program of the Center for Cancer Research, National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adhikari A, Xu M, Chen ZJ. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene. 2007;26:3214–3226. doi: 10.1038/sj.onc.1210413. [DOI] [PubMed] [Google Scholar]
- Birkey Reffey S, Wurthner JU, Parks WT, Roberts AB, Duckett CS. X-linked inhibitor of apoptosis protein functions as a cofactor in transforming growth factor-β signaling. J. Biol. Chem. 2001;276:26542–26549. doi: 10.1074/jbc.M100331200. [DOI] [PubMed] [Google Scholar]
- Cárcamo J, Weis FMB, Ventura F, Wieser R, Wrana JL, Attisano L, Massagué J. Type I receptors specify growth-inhibitory and transcriptional responses to transforming growth factor β and activin. Mol. Cell. Biol. 1994;14:3810–3821. doi: 10.1128/mcb.14.6.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signaling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Chen RH, Derynck R. Homomeric interactions between Type II transforming growth factor-β receptors. J. Biol. Chem. 1994;269:22868–22874. [PubMed] [Google Scholar]
- Chen ZJ. Ubiquitin signaling in the NF-kB pathway. Nat. Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JY, Park YC, Ye H, Wu H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 2002;115:679–688. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Ligand-independent activation of transforming growth factor (TGF) β signaling pathways by heteromeric cytoplasmic domains of TGF-β receptors. J. Biol. Chem. 1996;271:13123–13129. doi: 10.1074/jbc.271.22.13123. [DOI] [PubMed] [Google Scholar]
- Feng X-H, Derynck R. Specificity and versatility in TGF-beta signaling through Smads. Annu Rev Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Itoh S, Thorikay M, Kowanetz M, Moustakas A, Itoh F, Heldin C-H, ten Dijke P. Elucidation of Smad requirement in transforming growth factor-β type I receptor-induced responses. J. Biol. Chem. 2003;278:3751–3761. doi: 10.1074/jbc.M208258200. [DOI] [PubMed] [Google Scholar]
- Jadrich JL, O’Connor MB, Coucouvanis E. The TGF beta activated kinase TAK1 regulates vascular development in vivo. Development. 2006;133:1529–1541. doi: 10.1242/dev.02333. [DOI] [PubMed] [Google Scholar]
- Kaur S, Wang F, Venkatraman M, Arsura M. X-linked inhibitor of apoptosis (XIAP) inhibits c-Jun N-terminal kinase 1 (JNK1) activation by transforming growth factor β1 (TGF-β1) and through ubiquitin-mediated proteosomal degradation of the TGF-β1-activated kinase 1 (TAK1) J. Biol. Chem. 2005;280:38599–38608. doi: 10.1074/jbc.M505671200. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Kadono Y, Naito A, Matsumoto K, Yamamoto T, Tanaka S, Inoue J. Segregation of TRAF6-mediated signaling pathways clarifies its role in osteoclastogenesis. EMBO J. 2001;20:1271–1280. doi: 10.1093/emboj/20.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A, Ronai Z. Ubiquitin chains in the ladder of MAPK signaling. Sci. STKE. 2005;2005(281) doi: 10.1126/stke.2812005re5. ref5. [DOI] [PubMed] [Google Scholar]
- Liao JH, Chen JS, Chai MQ, Zhao S, Song JG. The involvement of p38 MAPK in transforming growth factor β1-induced apoptosis in murine hepatocytes. Cell Res. 2001;11:89–94. doi: 10.1038/sj.cr.7290072. [DOI] [PubMed] [Google Scholar]
- Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, et al. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable Disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin C-H. Non-Smad TGFβ signals. J.Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- Perlman R, Schiemann WP, Brooks MW, Lodish HF, Weinberg RA. TGF-β-induced apoptosis is mediated by the adapter protein Daxx that facilitates JNK activation. Nat. Cell Biol. 2001;3:708–714. doi: 10.1038/35087019. [DOI] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, Lee KY, Bussey C, Steckel M, Tanaka N, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcourt U, Kowanetz M, Niimi H, Heldin C-H, Moustakas A. TGF-β and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol. Biol. Cell. 2005;16:1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Nagai S, Ninomiya-Tsuji J, Nishita M, Tamai K, Irie K, Ueno N, Nishida E, Shibuya H, Matsumoto K. XIAP, a cellular member of the inhibitor of apoptosis protein family, links the receptors to TAB1-TAK1 in the BMP signaling pathway. EMBO J. 1999;18:179–187. doi: 10.1093/emboj/18.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YA, Zhang GM, Feigenbaum L, Zhang YE. Smad3 reduces susceptibility to hepatocarcinoma by sensitizing hepatocytes to apoptosis through downregulation of Bcl-2. Cancer Cell. 2006;9:445–457. doi: 10.1016/j.ccr.2006.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Hebert MC, Zhang YE. TGF-β receptor-activated p38 MAP kinase mediates Smad-independent TGF-β responses. EMBO J. 2002;21:3749–3759. doi: 10.1093/emboj/cdf366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J, Bitzer M, Liang D, Yang YC, Massimi A, Kneitz S, Piek E, Bottinger EP. Genetic programs of epithelial cell plasticity directed by transforming growth factor-β. Proc. Natl. Acad. Sci. USA. 2001;98:6686–6691. doi: 10.1073/pnas.111614398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.