Abstract
Caulobacter crescentus has become the predominant bacterial model system to study the regulation of cell cycle progression. Stage specific processes such as chromosome replication and segregation, and cell division are coordinated with the development of four polar structures: the flagellum, pili, stalk, and holdfast. The production, activation, localization, and proteolysis of specific regulatory proteins at precise times during the cell cycle culminate in the ability of the cell to produce two physiologically distinct daughter cells. We examine the recent advances that have enhanced our understanding of the mechanisms of temporal and spatial regulation that occur during cell cycle progression.
1. Introduction
Decades of work in eukaryotic biology have shown that many types of cells undergo programmed cell cycles, which consist of a series of invariant steps. The transitions from one stage to the next are mediated by complex regulatory networks that lead to the ordered production, localization, and activation of proteins for critical cellular events such as chromosome replication, organelle development, and cell division. The aquatic bacterium Caulobacter crescentus undergoes a programmed developmental cycle that requires integrated regulatory networks that may rival those of eukaryotic cells. Thus, C. crescentus is an excellent model system for studying cellular differentiation (Brun and Janakiraman, 2000).
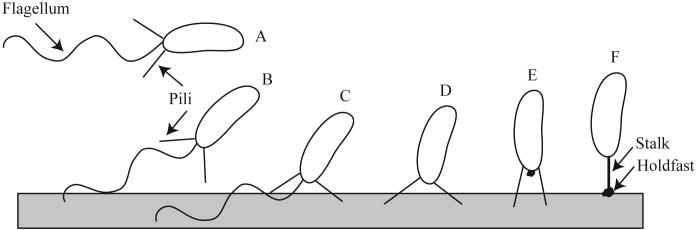
Progression through the cell cycle results in a sequential series of changes in cellular morphology and requires the coordination of processes including chromosome replication, chromosome segregation, polar morphogenesis, cell growth, and cell division. Unlike many prokaryotes, C. crescentus replicates its chromosome only once during the cell division cycle such that the G1, S, and G2 phases are readily distinguishable (Fig. 1). The cell division cycle is tightly coupled to a series of morphological transitions resulting in the formation of two distinct cell types, a motile swarmer cell and a sessile stalked cell. The swarmer cell has a polar flagellum and pili and is incapable of chromosome replication. After a gap period (G1) equivalent to about one-third of the cell cycle, the swarmer cell differentiates into a stalked cell. Cellular differentiation involves ejection of the flagellum, retraction of the pili, and synthesis of a stalk with an adhesive holdfast at the same pole that previously contained the flagellum. At the onset of S phase, the stalked cell initiates chromosome replication, cell division, and synthesis of a flagellum at the pole opposite the stalk. Flagellum rotation is activated just prior to cell separation. After cell division, pili are synthesized on the new swarmer cell and the stalked cell undergoes a new round of chromosome replication and cell division.
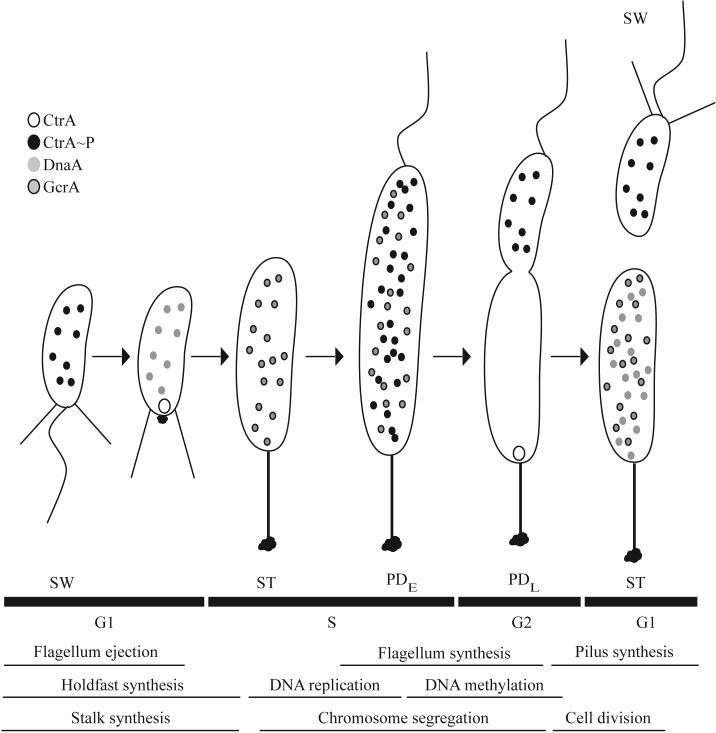
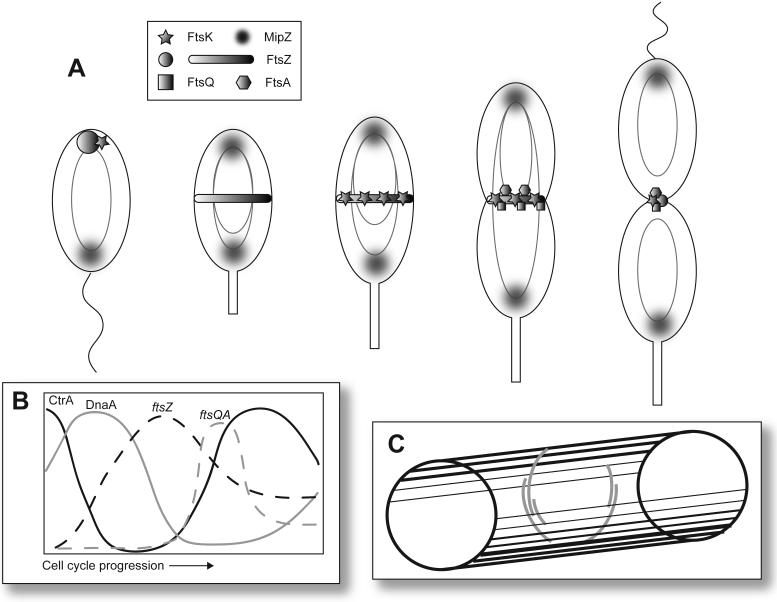
Figure 1.
The C. crescentus cell cycle and the localization of the major regulators CtrA, DnaA, and GcrA. The cell cycle, including the swarmer (SW), stalked (ST), early predivisional (PDE), late predivisional (PDL), and progeny cells, is depicted. The stages of the cell division cycle and cell cycle events are shown below the cell cycle schematic. G1 is the pre-synthesis gap period, S is the DNA synthesis period, and G2 is the post-synthesis gap period. Phosphorylated CtrA (CtrA∼P) is found in the swarmer cell. As the cell undergoes the swarmer to stalked cell differentiation, CtrA is recruited to the flagellar pole where it is proteolyzed. DnaA is then synthesized and leads to the production of GcrA, the dominant regulator in the stalked cell. In the predivisional cell, GcrA is responsible for the activation of ctrA transcription. CtrA is phosphorylated and represses transcription of gcrA. In the stalked compartment, CtrA is subject to proteolysis at the stalked pole and both GcrA and DnaA accumulate in the new stalked cell. In the swarmer compartment, phosphorylated CtrA is present and blocks chromosome replication in the new swarmer cell.
Many genes and gene products important for cell cycle and developmental regulation have been identified in a number of genetic screens. The ability to synchronize the cells and monitor changes in the transcriptome and proteome as cells proceed through the cell cycle has led to the identification of additional genes and proteins that are important for developmental regulation. In this review, we will highlight recent advances in the study of C. crescentus that have enhanced our understanding of the complex regulatory circuits that are required for cell cycle progression. We will examine the roles of several regulatory elements and discuss how they influence polar development, maintenance of cell shape, and cell division.
2. Global Regulation
Most bacteria respond to environmental and physiological changes by using complex global regulatory mechanisms to adjust the transcription levels of specific genes. The numerous combinations of interactions amongst cis- and trans-acting elements lead to an extensive web of transcriptional regulation that controls critical events such as cell division and cell differentiation.
A compilation of earlier studies indicated that the transcription of 72 single genes varies as a function of the cell cycle, including many genes involved in the regulation of the cell cycle and cell differentiation (Laub et al., 2000). The availability of the genome sequence of C. crescentus (Nierman et al., 2001) resulted in the development of methods for global identification of genes that control the cell cycle. The initial global transcription analysis found that 19% of the genome (553 genes) was comprised of genes whose RNA levels varied throughout the cell cycle (Laub et al., 2000). The validity of this approach is supported by the fact that the 72 genes that were previously identified as being cell cycle regulated are found within the set of temporally regulated transcripts.
The use of global transcriptional profiling has provided a tantalizing glimpse into the molecular mechanism(s) involved in cell cycle control (Laub et al., 2000). Several insights have been made from this work, which allow us to begin to address the question of how C. crescentus regulates cell cycle progression at the molecular level. The observation that peak expression of cell cycle regulated genes occurs immediately before or coincident with the event requiring the gene product has led to the notion of “just-in-time” transcription that may allow the cell to grow and differentiate more efficiently. The roles of several regulatory elements known to be important in the cell cycle progression of C. crescentus are discussed below.
2.1 Coordination between Oscillating Global Regulators
At least three transcriptional regulators, CtrA, GcrA, and DnaA modulate the distinctive cell cycle and morphological changes that occur during the C. crescentus life cycle. Each regulator has a unique pattern of expression that is coincident with its function in the cell (Fig. 1; for review, see (Holtzendorff et al., 2006)). Phosphorylated CtrA, a response regulator protein, is found in the swarmer cell where it binds to the origin of replication and inhibits chromosome replication (Quon et al., 1998). In the swarmer cell, CtrA also represses ftsZ, which is required for cell division, and podJ, which is required for polar development. CtrA is rapidly degraded by proteolysis as the cell transitions from a swarmer cell to a stalked cell and DnaA is stablized. DnaA is an essential activator of chromosome replication (Gorbatyuk and Marczynski, 2001) and acts as a transcriptional regulator during early S phase, leading to the synthesis of GcrA (Hottes et al., 2005). In the stalked cell, DnaA is responsible for the activation of both ftsZ and podJ. During early S phase, GcrA is the dominant regulator and activates genes required for chromosome replication, cell elongation, and polar development by an unknown mechanism (Holtzendorff et al., 2004). As the cell transitions from the S phase to the G2 phase, CtrA transcription is activated by GcrA. Newly synthesized CtrA is phosphorylated and in turn represses gcrA. Recruitment of CtrA to the stalked pole in the stalked compartment of the predivisional cell leads to dephosphorylation and proteolysis of CtrA. The decrease in active CtrA level relieves repression of gcrA and allows GcrA to accumulate. Meanwhile, CtrA remains at a high level in the swarmer compartment of the predivisional cell and blocks gcrA transcription and chromosome replication. The sequential expression of the CtrA/DnaA/GcrA regulators drives the cell cycle progression of C. crescentus and allows the formation of two distinct progeny cells following cell division. The specific roles of each regulator in cell cycle progression are described below.
2.1.1 CtrA
In C. crescentus CtrA is an essential response regulator that serves as a major regulatory protein. CtrA contains a DNA-binding domain and recognizes specific sequences that are found upstream of many cell cycle controlled promoters (Laub et al., 2002; Quon et al., 1996). CtrA acts either as an activator or repressor for the transcription of a number of cell cycle regulated genes. The CtrA regulon includes genes whose products are required for flagellum assembly and activation, pili biogenesis, holdfast synthesis, DNA methylation, chromosome replication, and cell division. The level of ctrA transcription is influenced by two promoters, the methylation state of the DNA, and the presence of activators (Domian et al., 1997; Quon et al., 1996; Reisenauer and Shapiro, 2002). CtrA activity is controlled by cellular localization, proteolysis, and phosphorylation (Biondi et al., 2006a; Domian et al., 1997; Iniesta et al., 2006; McGrath et al., 2006; Ryan et al., 2004; Ryan et al., 2002). As a result, active CtrA appears only at precise times in the C. crescentus cell cycle, including the swarmer and predivisional cells.
2.1.1a CtrA transcription
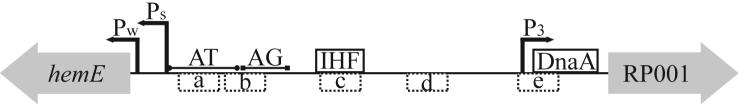
The expression of ctrA is controlled by two promoters, which are active at different times in the cell cycle (Domian et al., 1999). P1 is a relatively weak promoter located 122 basepairs (bps) upstream of the ctrA translation start site and is active only in the stalked cell. The P1 promoter contains a GAnTC methylation site and is activated soon after chromosome replication, when the chromosome is hemi-methylated (Marczynski, 1999; Reisenauer and Shapiro, 2002; Stephens et al., 1996). GcrA activates ctrA expression leading to the production of phosphorylated CtrA (CtrA∼P) (Holtzendorff et al., 2004). In predivisional cells, CtrA∼P activates the transcription of ccrM, which encodes the adenine DNA methyltransferase that methylates the newly replicated DNA (Quon et al., 1996). When the chromosome is fully methylated, the P1 promoter is repressed allowing minimal ctrA expression prior to a new round of chromosome replication (Domian et al., 1999). P2 is a stronger promoter located 65 bps upstream of the ctrA translational start site and is activated by CtrA∼P in the predivisional cell. Thus, both promoters are subject to feedback control by CtrA; P1 is repressed and P2 is activated. Combined with regulated phosphorylation and proteolysis of CtrA (see sections 2.1.1b and 2.1.1c), the feedback control of the ctrA promoters results in the appearance of active CtrA during specific times in the cell cycle (Fig. 1).
2.1.1b CtrA localization and proteolysis
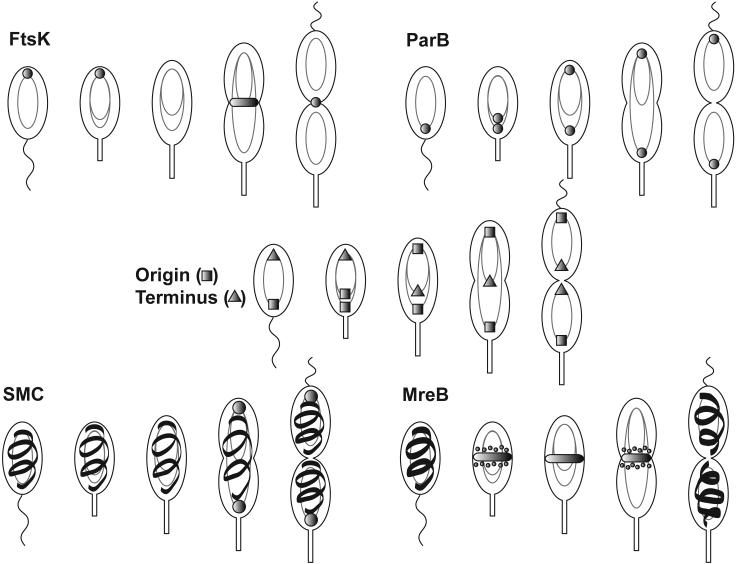
In addition to transcriptional control, proteolysis plays an important role in the presence of active CtrA. Following the production of CtrA, the predivisional cell becomes compartmentalized and CtrA is degraded from the stalked cell compartment, but not from the swarmer cell compartment prior to cell division (Domian et al., 1997). The daughter swarmer cell inherits CtrA, but the daughter stalked cell does not. CtrA is degraded as the swarmer cell undergoes differentiation into a stalked cell and reappears in the early predivisional cell (Fig. 2).
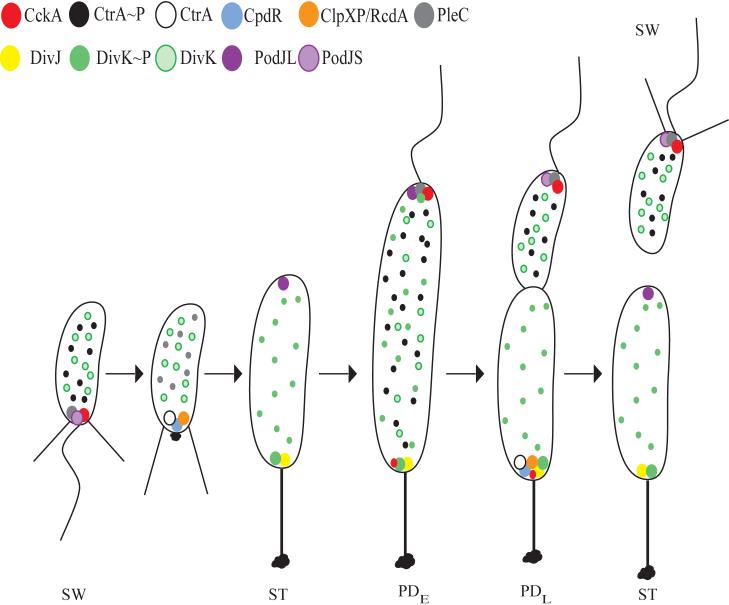
Figure 2.
Localization of proteins affecting CtrA phosphorylation and proteolysis during the C. crescentus cell cycle. All stages of the cell cycle, including the swarmer (SW), stalked (ST), early predivisional (PDE), late predivisional (PDL), and progeny cells, are shown. In the swarmer cell, PodJS is localized at the flagellar pole and is responsible for the localization of PleC. PleC dephosphorylates DivK∼P leading to the accumulation of delocalized DivK. CckA is also localized to the flagellar pole and controls CtrA prosphorylation (CtrA∼P), leading to a block in chromosome replication. As the cell undergoes the swarmer to stalked cell differentiation, PodJ is degraded, releasing PleC from the pole. CpdR is localized to the flagellated pole and recruits both CtrA and ClpXP/RcdA to the pole, resulting in the proteolysis of CtrA. In the stalked cell, PodJL is synthesized and localizes to the incipient swarmer pole. DivJ is localized to the stalked pole where it recruits and phosphorylates DivK, leading to accumulation of DivK∼P at the pole and in the cytoplasm. In the early predivisional cell, PodJL recruits PleC to the incipient swarmer pole where PleC dephosphorylates DivK∼P. Meanwhile, DivJ is localized to the stalked pole where DivK is phosphorylated. Due to the opposing actions of PleC and DivJ, DivK and DivK∼P are found in the cytoplasm. In the swarmer compartment of the late predivisional cell, the periplasmic protease PerP is responsible for the cleavage of PodJL to PodJS at the incipient flagellar pole. CckA and PleC remain at the flagellar pole and CtrA is phosphorylated. The absence of DivJ and the presence of PleC in the swarmer compartment lead to the accumulation of unphosphorylated DivK. In the stalked compartment of the predivisional cell, CpdR and ClpXP/RcdA join DivJ and DivK∼P at the stalked pole, resulting in the degradation of CtrA and accumulation of DivK∼P. In the new stalked cell, PodJL is synthesized, CtrA is completely degraded, and CpdR and ClpXP/RcdA are no longer polarly localized.
Proteolysis of CtrA is mediated by the essential ClpXP protease (Jenal and Fuchs, 1998). Depletion of either ClpX or ClpP leads to the stabilization of CtrA and arrest of the cell cycle prior to chromosome replication. Since CtrA is not degraded, it is presumed to remain bound to the origin of replication, preventing the initiation of chromosome replication. Even though clpX and clpP are constitutively expressed and ClpXP is present throughout the cell cycle, the activity of ClpXP must be modulated since CtrA is degraded only in the swarmer cell and the stalked compartment of the late predivisional cell.
CtrA degradation is controlled by two determinants within the CtrA protein (Ryan et al., 2002). One determinant is present at the C-terminus of CtrA. A proteolytically stable form of CtrA is generated by deleting the last three C-terminal amino acid residues or by modification of the last two residues from alanines to aspartates (Domian et al., 1997). The second determinant is located within the first 56 residues of CtrA. The N-terminus of CtrA is comprised of the receiver domain, including the critical aspartate residue that is phosphorylated by the CckA-ChpT-CtrA phosphorelay as described in section 2.1.1.c (Biondi et al., 2006a). Neither the expression of the receiver domain nor expression of the C-terminus of CtrA alone is sufficient for cell cycle degradation (Domian et al., 1997; Ryan et al., 2002). The phosphorylation state of the receiver domain modulates activity (see section 2.1.1c), but does not affect proteolysis (Ryan et al., 2002). Fusion of the receiver domain to the 15 C-terminal amino acid residues is sufficient for cell cycle regulated proteolysis of CtrA (Ryan et al., 2002). Fusion of the receiver domain and the 15 C-terminal residues of CtrA to yellow fluorescent protein (YFP) is sufficient to confer cell cycle dependent degradation to YFP. A series of five hybrid proteins generated using the receiver domain of CtrA homologs from other bacterial genera and the 15 C-terminal residues of CtrA were expressed in C. crescentus. Four of the hybrid proteins were degraded with a temporal pattern similar to native CtrA, indicating that they share a proteolytic signal. Ten candidate residues for a proteolytic signal were identified by a sequence alignment and nine of these residues are predicted to be located on one surface of CtrA. It has been hypothesized that some or all of these residues may form a binding pocket for an activating protein that stimulates degradation of CtrA by ClpXP (Ryan et al., 2002).
CtrA localizes to the incipient stalked pole in cells undergoing the swarmer to stalked cell differentiation and to the stalked pole in the stalked compartment of predivisional cells, just prior to its degradation (Fig. 2) (Ryan et al., 2004; Ryan et al., 2002). CtrA proteolysis in the stalked cell compartment of the predivisional cell occurs only after a diffusion barrier forms (Judd et al., 2003). The delay in proteolysis until after compartmentalization enables the production of two distinct daughter cells with very different levels of CtrA. These results indicate that there may be a regulatory link between compartmentalization and the initiation of CtrA proteolysis. In this model, polar localization is a prerequisite for proteolysis (Fig. 2). This is consistent with the hypothesis that an activating protein promotes the interaction of CtrA with ClpXP at a specific time and place.
The notion of an activating protein for degradation of CtrA by ClpXP is supported by the observation that CtrA and ClpX are found in a polar complex with a third protein, RcdA (McGrath et al., 2006). The localization pattern of RcdA matches that of both CtrA and ClpX; all three proteins appear at the same pole in cells undergoing the swarmer to stalked cell differentiation and in the stalked compartment of pre-divisional cells (Fig. 2). In cells lacking rcdA, ClpX localization still occurs, but polar CtrA foci are not observed and CtrA remains delocalized throughout the cell cycle. The half-life of CtrA in cells lacking rcdA is 2.6 times greater than the half-life of CtrA in wild-type cells. When cells expressing RcdA are depleted of ClpX, RcdA no longer forms polar foci. These results suggest that ClpX localizes to the pole prior to RcdA and that RcdA recruits CtrA to the cell pole. Therefore, the dynamic localization of this complex is responsible for the temporal control of CtrA proteolysis.
RcdA is not absolutely required for an interaction between ClpX and CtrA; complexes of non-polar ClpX-CtrA can be isolated from the cell (McGrath et al., 2006). These complexes are presumed to be non-functional in vivo as CtrA is not degraded in an rcdA mutant; however, in vitro studies using recombinant C. crescentus ClpXP indicate that an activating protein is not required for degradation of CtrA by ClpXP (Chien et al., 2007). In vitro studies indicate that ClpXP directly recognizes the C-terminal tag of CtrA. The N-terminal domain required for localization and proteolysis in vivo is dispensable for in vitro degradation. The rates of CtrA degradation by ClpXP are unchanged by the addition of RcdA, suggesting that RcdA is unlikely to act as a substrate-specific protease for degradation. Kinetic analyses indicate that the rate of unassisted CtrA degradation by ClpXP is sufficient to account for the observed rate of intracellular CtrA degradation. These results show that the N-terminal domain of CtrA and RcdA are not required for the activation of CtrA proteolysis in the in vitro environment.
The requirement for RcdA to mediate CtrA proteolysis in vivo, but not in vitro, suggests that there are likely to be additional factors involved in controlling CtrA proteolysis. One possibility is that there are unidentified inhibitors of ClpXP in the swarmer and predivisional cells (Chien et al., 2007). Since the level of ClpXP is limiting compared with the level of CtrA, competition between CtrA and other higher priority substrates could provide a mechanism for inhibition. This is an attractive model since a specific inhibitor is not required. Another possibility is that CtrA may interact with a protein that protects it from degradation in swarmer and predivisional cells. The inhibitor may interact with CtrA and protect CtrA from proteolysis by masking the C-terminal recognition motif either directly or indirectly. In this scenario, RcdA recruitment of CtrA to the pole may result in release of the protease inhibitor. In vitro studies conducted in the absence of the relevant inhibitor may suggest that RcdA is not required for efficient proteolysis. The disparity between the in vivo and in vitro results with regard to the role of RcdA in CtrA proteolysis clearly indicates that the regulation of CtrA proteolysis is likely to involve additional factors.
How is the timing of CtrA degradation controlled despite the presence of ClpXP through all stages of the cell cycle? To answer this question we turn our attention to the single-domain response regulator, CpdR, which mediates the localization and activity of the ClpXP protease (Iniesta et al., 2006). CtrA is stabilized in a cpdR deletion strain, indicating that CpdR is required for CtrA proteolysis. CpdR exhibits the same localization pattern as CtrA, ClpXP, and RcdA throughout the cell cycle suggesting that CpdR may be a localization factor (Fig. 2). Indeed, CpdR forms a complex with ClpX and is required for the specific localization of ClpXP, and subsequently RcdA and CtrA, to the pole at specific times in the cell cycle. Consistent with previous results, localization of both ClpXP and CtrA is a prerequisite for proteolysis. Proper localization of ClpXP requires the non-phosphorylated form of CpdR, which is produced as the cell undergoes the transition from swarmer to stalked cell and in the stalked compartment of the predivisional cell. Interestingly, CpdR phosphorylation is controlled by the same phosphorelay responsible for the phosphorylation of CtrA. Following the localization of ClpXP to the appropriate pole, the ClpXP/RcdA/CtrA complex forms and CtrA is degraded. In the predivisional cell, CtrA is stabilized following phosphorylation of CpdR by the CckA-ChpT phosphorelay, which is also responsible for the phosphorylation of CtrA (see section 2.1.1c). Additional research is likely to elucidate the mechanistic roles of CpdR and RcdA in restricting the proteolysis of CtrA by ClpXP to specific locations and times in the cell cycle.
Localization of CtrA to the flagellar pole and proteolysis of CtrA at this pole during swarmer to stalked cell differentiation is influenced by an additional single-domain response regulator, DivK (Hung and Shapiro, 2002). DivK is required for CtrA proteolysis (Hung and Shapiro, 2002) and is localized to the same pole during the swarmer to stalked cell differentiation, but does not recruit CtrA to the pole (Jacobs et al., 2001). A signaling pathway is responsible for controlling the phosphorylation state of DivK and contributes to the regulation of CtrA proteolysis (see section 2.4.1b). The histidine kinases, DivJ and PleC, which are responsible for the phosphorylation and dephosphorylation of DivK respectively, affect the degradation of CtrA in the stalked compartment of the predivisional cell (Judd et al., 2003; Matroule et al., 2004; Ryan et al., 2004). Accumulation of phosphorylated DivK prevents the phosphorylation of CpdR (Biondi et al., 2006a) (see section 2.1.1c for more detail), allowing localization of ClpXP and subsequent proteolysis of CtrA. It will be of interest to determine if this signaling cascade also influences the recruitment of CtrA by RcdA.
2.1.1c CtrA phosphorylation
In addition to the temporal expression of ctrA and the temporal degradation of CtrA, the activity of CtrA is modulated by the phosphorylation state of the protein. CtrA is phosphorylated in vivo and a non-phosphorylatable mutant of CtrA does not complement the lethality of a ctrA deletion when present at low-copy (Quon et al., 1996), but is partially active when present at high copy (Domian et al., 1997). Furthermore, phosphorylated CtrA has a higher binding affinity for CtrA-regulated promoters and CtrA binding sites in the origin of replication than non-phosphorylated CtrA (Reisenauer et al., 1999; Siam and Marczynski, 2000). Binding of phosphorylated CtrA to its binding sites in the origin of replication has been shown to promote cooperative binding of phosphorylated CtrA at adjacent binding sites (Siam and Marczynski, 2000). These results suggest that phosphorylation is required for CtrA to act as a transcriptional regulator.
While several kinases have been identified as playing a role in controlling the phosphorylation state of CtrA, only the hybrid histidine kinase/response regulator, CckA is required for phosphorylation of CtrA in vivo (Fig. 2) (Jacobs et al., 1999). The phosphorylation of CckA correlates with that of CtrA (Jacobs et al., 2003). Like ctrA, cckA is an essential gene. Temperature sensitive alleles of ctrA and cckA cause remarkably similar phenotypes and result in the differential regulation of a similar set of genes, suggesting that CckA modulates the activity of CtrA (Jacobs et al., 2003; Jacobs et al., 1999). Interestingly, unlike CtrA, CckA is found at constant levels throughout the cell cycle (Jacobs et al., 1999). How does CckA modulate CtrA levels if it is always present? The key to answering this question is to examine the transient polar localization of CckA in predivisional cells. CckA localizes predominantly to the flagellated pole of predivisional cell and swarmer cells; this localization correlates with the timing of CckA and CtrA phosphorylation (Jacobs et al., 1999). A deletion of the transmembrane domain of CckA is lethal and indicates that membrane anchoring is required for the essential activity of CckA, presumably mediating the phosphorylation of CtrA. When CckA lacking the transmembrane domain is fused to green fluorescent protein (GFP) and expressed in cells containing a wild-type copy of CckA, GFP does not preferentially localize to the nascent flagellar pole of predivisional cells. Taken together, these results suggest that the proper translocation of CckA to the membrane and to the pole is required for the proper temporal activation of CtrA. In addition to the role in CtrA phosphorylation, CckA stabilizes CtrA (Jacobs et al., 2003), indicating that CckA contributes to the control of CtrA by multiple mechanisms.
Does CckA phosphorylate CtrA directly or indirectly? Hybrid kinases can directly transfer the phosphoryl group to their cognate response regulators. Alternatively, a phosphorelay cascade may lead to indirect phosphorylation of the response regulator. Phosphorelay typically involves passing the phosphoryl group from the kinase domain to the receiver domain of the hybrid kinase, then onto a histidine phosphotransferase (Hpt), and finally to the cognate response regulator. In order to address this question, phosphotransfer profiling was performed using the purified kinase domain of CckA as the phosphoryl-donor and 50 purified response regulators as candidate phosphoryl-acceptors (Biondi et al., 2006a). CckA phosphorylates its own receiver domain as well as three other response regulators, but not CtrA, indicating that phosphorelay is likely necessary for phosphorylation of CtrA by CckA. Although no Hpt proteins had been identified in the genome sequence of C. crescentus (Nierman et al., 2001), a series of structural and functional criteria identified a candidate Hpt protein, ChpT, responsible for the phosphorelay from CckA to CtrA (Biondi et al., 2006a). Depletion of chpT generates a phenotype similar to temperature sensitive alleles of ctrA and cckA and the gene expression patterns of all three strains are highly correlated, suggesting that ChpT is the intermediate protein required for the phosphorylation of CtrA (Biondi et al., 2006a). Biochemical evidence confirmed that ChpT acts as an Hpt and established the presence of a CckA-ChpT-CtrA phosphorelay. CckA is the only kinase capable of phosphorylating ChpT. ChpT in turn phosphorylates both CtrA and CpdR. CpdR controls the activity of the ClpXP protease, which ultimately is responsible for the degradation of CtrA (see section 2.1.1b). These findings explain how CckA leads to both the activation and stabilization of CtrA: phosphorelay from CckA-ChpT-CtrA activates CtrA by phosphorylation and the phosphorelay from CckA-ChpT-CpdR leads to the production of phosphorylated CpdR and prevents the localization of ClpXP and the subsequent proteolysis of CtrA.
The histidine kinase DivJ and response regulator DivK have also been implicated in controlling the phosphorylation state of CtrA (Wu et al., 1998). The level of active phosphorylated CtrA is increased in a divJ deletion background, indicating that rather than mediating phosphorylation of CtrA, DivJ normally reduces the level of phosphorylated CtrA (Pierce et al., 2006). Suppressors of divJ, including cckA mutants, compensate for the loss of divJ by reducing the amount of CtrA∼P, but not total CtrA. A temperature sensitive allele of divK grown at the nonpermissive temperature has increased levels of active CtrA, due in part to the reduction in CtrA proteolysis (Hung and Shapiro, 2002). Accumulation of phosphorylated DivK results in delocalization of CckA, preventing the phosphorylation of CtrA and CpdR (Biondi et al., 2006a). These observations are consistent with a role of DivK in reducing the levels of CtrA∼P by preventing CckA from mediating the phosphorylation of CtrA and by allowing unphosphorylated CpdR to accumulate, leading to the degradation of any existing pools of CtrA∼P. Although transcription of divK is regulated by CtrA and peaks late in the predivisional cell, DivK is a stable protein and is found throughout the cell cycle with only a slight increase in late predivisional cells (Hecht et al., 1995; Jacobs et al., 2001). Thus, the phosphorylation of DivK late in the cell cycle leads to the inhibition of CckA and CtrA, allowing the cell cycle to reset.
The tyrosine kinase DivL has also been implicated in a signal transduction pathway leading to the phosphorylation of CtrA (see section 2.4.1d) (Wu et al., 1999). In vitro studies suggest that DivL phophorylates CtrA, but not DivK (Wu et al., 1999) and a conditional mutation in divL results in reduced levels of phosphorylated CtrA (Pierce et al., 2006). Taken together, these results suggest that DivL may regulate CtrA independently of the DivJK phosphorelay. This is supported by the observation that DivL does not impact CtrA proteolysis (Reisinger et al., 2007), suggesting that the mechanism of action differs from that of DivK, which utilizes the CckA-ChpT phosphorelay to impact both CtrA phosphorylation and proteolysis. It is equally possible that DivL functions within the DivJK pathway since a yeast-two-hybrid screen identified DivL as an interacting partner for DivK, which suggests that there may be some crosstalk between the DivJK and DivL phosphorelay pathways (Ohta and Newton, 2003). Two additional histidine kinases, CckN and CckO, were identified as interacting partners for DivK in the screen, but it remains unclear if interactions between DivK and DivL, CckO, or CckN impact the phosphorylation of CtrA or even if the interactions are relevant in vivo.
2.1.1d The CtrA regulon
The extensive regulation of CtrA transcription, localization, degradation, and phosphorylation, along with the essentiality of the ctrA gene, demonstrate its importance in modulating cell cycle regulated gene expression. The initial description of CtrA indicated that CtrA regulates promoters of the class II flagellar genes, fliQ and fliL, and the ccrM methyltranferase gene (Quon et al., 1996). FliQ and other flagellar proteins are required early in the predivisional cell when flagellum biosynthesis is initiated, whereas CcrM is required late in the predivisional cell to fully methylate newly replicated DNA. Additional studies have identified several genes that are regulated by CtrA. CtrA positively regulates the expression of the major chemotaxis operon in predivisional cells and three additional genes involved in flagellar filament production (Jones et al., 2001; Leclerc et al., 1998). In the predivisional cell, CtrA activates the transcription of pilA, which encodes the major pilin subunit, and leads to the synthesis of pili on the daughter swarmer cell (Skerker and Shapiro, 2000). Transcription of the early cell division gene ftsZ is repressed by CtrA in swarmer cells (Kelly et al., 1998). In contrast, CtrA activates the transcription of the late cell division genes ftsQ and ftsA (Wortinger et al., 2000). These results suggest that CtrA influences the transcription of a diverse array of genes throughout the cell cycle.
Microarray experiments indicate that CtrA regulates the transcription of nearly a quarter of all the cell cycle regulated genes (Laub et al., 2000). CtrA influences the transcription of 144 genes; however, only 55 genes (with an additional 40 genes in potential operons) are directly regulated by CtrA (Laub et al., 2002; Laub et al., 2000). Genes that are directly regulated by CtrA were identified by combining microarray analysis and genome-wide location analysis, which maps the in vivo CtrA binding sites using a modification of chromatin-immunoprecipitation (Laub et al., 2002). Among the 95 genes that are directly regulated by CtrA, 29 are repressed and 66 are activated by CtrA (Laub et al., 2002). Most of the genes that are repressed are maximally expressed during swarmer to stalked cell differentiation, coincident with CtrA proteolysis and clearing form the cells. Conversely, most of the genes that are activated by CtrA are maximally expressed after the accumulation of CtrA in predivisional cells.
CtrA directly regulates at least 14 regulatory genes, including operons encoding ten two-component signal transduction proteins and two sigma factors. rpoN, which encodes the alternative sigma factor, σ54, is activated by CtrA and is required for both flagellum and stalk formation, as well as proper cell division (Brun and Shapiro, 1992). sigT, which encodes another alternate sigma factor, is repressed by CtrA, leading to maximal expression of sigT during the swarmer to stalked cell transition suggesting that SigT may regulate gene expression early in the cell cycle (Laub et al., 2002; Laub et al., 2000). CtrA also activates additional regulators of unknown function. A number of regulatory genes, including the response regulators cheY, cheYII, cheYIII, and the histidine kinase cheA, are activated by CtrA and are known to affect polar morphogenesis (Laub et al., 2002). CtrA-dependent genes required for polar morphogenesis encode proteins required for the biosynthesis and activation of flagella, production of chemotaxis machinery, pili biogenesis and holdfast synthesis (see section 3) (Laub et al., 2002). Each of these processes occurs in the swarmer compartment of the predivisional cell in which CtrA binds the origin of replication, demonstrating the importance of CtrA in the formation of the non-replicative, motile daughter swarmer cell.
CtrA directly regulates essential cell processes including DNA methylation and cell division (Laub et al., 2002). The genes for DNA methylation include ccrM and metK, which encodes the S-adenosylmethionine synthetase that produces the substrate used for CcrM-dependent DNA methylation. CtrA influences the transcription of genes required for cell division initiation and progression by repressing ftsZ and activating ftsA, ftsQ, ftsW, and ftsI. The proteins encoded by these genes are part of a core set of proteins that localize to the division plane where they perform specific functions in cell division (see section 4.3). The remaining 39 genes directly regulated by CtrA have no known function (Laub et al., 2002).
Most of the genes positively regulated by CtrA are activated in the predivisional cell however; the genes are not transcribed at the exact same time. For example, fliQ is activated earlier in the predivisional cell than ccrM (Reisenauer et al., 1999). The differential temporal regulation of gene expression occurs because phosphorylated CtrA has different affinities for its binding site (Reisenauer et al., 1999). Disruption of an inverted repeat sequence within the ccrM promoter, which left the CtrA binding site intact, did not affect the timing of transcription initiation, but significantly reduced the amount of transcript indicating that an accessory factor may act as activator (Reisenauer et al., 1999).
Differences in the ability of CtrA to recognize the CtrA binding site have been attributed to specific features of the CtrA binding site including the specific CtrA binding site sequence, the distance between the TTAA elements, and the strength of the downstream promoter (Ouimet and Marczynski, 2000). Promoters strongly bound by CtrA contain the consensus sequence TTAA - N7-TTAA, followed by a poor match to the consensus -10 region (gCTANAWC). In this case, the transcription of the downstream gene is cell cycle regulated. When the upstream TTAA element contains mutations, there is a moderate decrease in transcription levels and transcription remains cell cycle regulated. When the spacing between the TTAA elements is changed by even a single base pair, there is a significant decrease in transcription levels and transcription remains cell cycle regulated. In contrast, when the downstream TTAA element contains mutations, there is a drastic decrease in transcription levels and transcription is no longer cell cycle regulated. When both TTAA elements contain mutations, the presence of a strong -10 promoter element restores cell cycle transcription. The pilA promoter contains four CtrA binding sites (Skerker and Shapiro, 2000) indicating that the number of CtrA binding sites may also impact CtrA binding affinity and affect the timing and levels of transcription. Naturally occurring promoters in C. crescentus contain mutations in the upstream TTAA element (fliQ) and the downstream TTAA element (ccrM and fliF) indicating that differences in the CtrA binding site contribute to the sequential activation of genes throughout the cell cycle (Ouimet and Marczynski, 2000).
2.1.2 GcrA
The GcrA protein is conserved exclusively among the alphaproteobacteria, but does not contain any known functional motifs and little is known about the mechanism of regulation by this protein (Holtzendorff et al., 2004). gcrA expression is cell cycle controlled and GcrA is essential and serves as a second global cell cycle regulator.
2.1.2a GcrA transcription
The pattern of gcrA transcription and subsequent protein production is strikingly out of phase with that of ctrA (Fig. 1). The maximal level of gcrA transcription occurs in the stalked cell where CtrA levels are low and gcrA transcript levels decrease in the predivisional cell as ctrA expression is reactivated (Collier et al., 2006; Holtzendorff et al., 2004). The reciprocal oscillation of CtrA and GcrA suggested that these two proteins might regulate the transcription of one another. Indeed, gcrA transcription is negatively regulated by CtrA whereas GcrA is required to activate the P1 promoter of ctrA (Holtzendorff et al., 2004). Phosphorylated CtrA binds directly to the promoter of gcrA and the level of gcrA transcription is increased in temperature sensitive mutants of CtrA and CckA at the restrictive temperature, confirming a role for CtrA and CckA in the repression of gcrA transcription (Holtzendorff et al., 2004). Mutagenesis of the CtrA binding site in the gcrA promoter does not eliminate the cell cycle regulation of gcrA expression (Collier et al., 2006). This observation suggests that the proteolysis of the CtrA repressor is not the only regulatory element controlling gcrA expression.
DnaA has been shown bind to the gcrA promoter and induces gcrA transcription (Collier et al., 2006; Hottes et al., 2005). Mutation of the DnaA box in the gcrA promoter or depletion of DnaA leads to a decrease in gcrA transcription (Collier et al., 2006). When the gcrA promoter is mutated such that it is either CtrA-independent or DnaA-independent, temporal regulation of gene expression is maintained (Collier et al., 2006). When the gcrA promoter is mutated to be both CtrA-independent and DnaA-independent, the temporal regulation is attenuated (Collier et al., 2006). The maximum change in the transcriptional activity throughout the cell cycle is 1.5 fold for cells with the CtrA- and DnaA-independent promoter compared with five fold for cells with the wild type promoter. These results indicate that the temporal regulation of gcrA transcription is directly mediated by both CtrA-dependent repression and DnaA-dependent activation. Since the cell cycle regulation of the gcrA promoter is not completely abolished when both the CtrA and DnaA binding sites are compromised, additional regulatory elements may also contribute to gcrA regulation. For example, full methylation of the gcrA promoter has been proposed to partially repress gcrA transcription (Collier et al., 2006).
2.1.2b GcrA proteolysis
The regulation of gcrA transcription is not the only mechanism for regulating the level of GcrA in the cells. When gcrA is transcribed constitutively in cells, GcrA abundance varies throughout the cell cycle (Collier et al., 2006). GcrA accumulates only in the stalked and predivisional cells (Fig. 1). The half-life of GcrA in stalked cells is about 40 minutes whereas the half-life of GcrA in swarmer cells is about 10 minutes. These results suggest that GcrA is subject to cell cycle regulated proteolysis, but the mechanism of GcrA proteolysis remains unknown.
2.1.2c GcrA regulon
Using microarray analysis, it was determined that GcrA depletion altered the gene expression of 125 genes, including 49 cell cycle regulated genes (Holtzendorff et al., 2004). Furthermore, chromatin immunoprecipitation assays indicate that GcrA interacts with promoter sequences, including the ctrA promoter (Holtzendorff et al., 2004). GcrA could interact directly with the promoter DNA or alternatively, could interact with a protein bound to the promoter DNA. Since GcrA lacks any detectable functional motifs, if an interaction with DNA occurs it is likely to be via a novel mechanism (Crosson et al., 2004). If GcrA does not interact directly with DNA, the mechanism of transcriptional regulation by this protein is likely to be novel.
The GcrA-regulated genes are involved in a vast array of functions including motility, polar development, cell wall biogenesis, amino acid metabolism and transport, chromosome replication, repair, and recombination. Given the maximal expression of gcrA in the stalked cell, when chromosome replication is initiated, it is not surprising that GcrA regulates a number of genes involved in DNA metabolism. The regulation of gcrA transcription and the initiation of chromosome replication share a number of common features (Collier et al., 2006). Both the C. crescentus origin of replication (Cori) and the gcrA promoter are repressed by CtrA binding, activated by DnaA binding, and contain DNA methylation sites suggesting that similar mechanisms couple the initiation of gcrA transcription and the initiation of chromosome replication (Collier et al., 2006; Holtzendorff et al., 2004; Marczynski and Shapiro, 2002). This is consistent with the observation that GcrA represses genes encoding chromosome replication initiation factors but activates genes encoding proteins involved in the progression of chromosome replication and segregation (Holtzendorff et al., 2004).
2.1.3 DnaA
DnaA is an essential bacterial chromosome replication initiation factor. In Escherichia coli, DnaA binds to a specific binding site (the DnaA box) in the origin of replication and unwinds the two DNA strands allowing the replication machinery to assemble on each DNA strand (for review of this process, see (Messer, 2002)). In C. crescentus, DnaA binds to a DnaA box in Cori (Marczynski and Shapiro, 2002), and depletion of DnaA leads to a block in chromosome replication and cell division (Gorbatyuk and Marczynski, 2001). The transition from swarmer to stalked cell is not affected by the depletion of DnaA and all stalked cells become filamentous. This observation indicates that transcription and protein synthesis processes remain intact; however, transcription of a subset of genes was inhibited, which suggests that DnaA may have an additional regulatory role. Indeed, DnaA has been shown to be a global transcriptional regulator (Hottes et al., 2005) that is subject to proteolysis at specific times in the cell cycle (Fig. 1) (Gorbatyuk and Marczynski, 2005).
2.1.3a DnaA transcription
Unlike CtrA and GcrA, transcription of DnaA does not appear to be dynamically regulated. Transcription of DnaA occurs throughout the cell cycle, but reaches a maximal level (two-fold higher than the minimal level) just prior to the swarmer to stalked cell transition (Zweiger and Shapiro, 1994). Transcription of dnaA has been shown to be DnaA-dependent, although the absence of a DnaA box in the promoter region suggests that the autoregulation of dnaA transcription may be indirect (Hottes et al., 2005).
2.1.3b DnaA proteolysis
DnaA synthesis occurs throughout the cell cycle, with a two-fold increase in the swarmer cells; however, DnaA is not a stable protein and it is targeted for proteolysis during the cell cycle (Gorbatyuk and Marczynski, 2005). DnaA is subject to proteolysis by ClpP throughout the cell cycle, but proteolysis occurs two-fold faster in swarmer cells. DnaA degradation is not reduced when ClpA is absent or ClpX is inactive. The presentation of DnaA to the ClpP protease likely requires an unidentified ATP-dependent chaperone or other specificity factor. The combination of DnaA synthesis and proteolysis ensures that newly synthesized DnaA is present as the cell undergoes the swarmer to stalked cell transition and initiates chromosome replication (Fig. 1).
2.1.3c The DnaA regulon
In addition to its role as an initiator of chromosome replication, DnaA has been shown to be a transcriptional regulator (Hottes et al., 2005). Using microarray analysis, transcription of 40 genes expressed during the swarmer to stalked cell differentiation was shown to be DnaA-dependent. Thirteen of these genes have putative DnaA boxes in the promoter region, indicating that DnaA directly regulates them. These genes encode nucleotide biosynthesis enzymes, chromosome replication machinery components, GcrA, the polar localization factor PodJ, and the cell division protein FtsZ. Gel-shift assays demonstrated that DnaA binds to the gcrA, podJ, and ftsZ promoters confirming the role of DnaA as a transcriptional regulator. The dual role of DnaA as a chromosome replication initiator and transcriptional regulator of components for chromosome replication, polar development, and cell division allows the coordination of multiple processes that are necessary for proper cell cycle progression (Hottes et al., 2005).
2.2 Sigma Factors
The three global regulators described above account for regulation of roughly 30% of the cell cycle regulated genes, indicating that additional levels of regulation must mediate cell cycle progression in C. crescentus. One possibility is that sigma factors may alter gene expression throughout the cell cycle. The genome sequencing of C. crescentus revealed the presence of 16 putative extracytoplasmic function sigma factors, which typically lead to changes in gene expression in response to periplasmic or extracellular stimuli (Nierman et al., 2001). Only six of the sigma factors, rpoD (σ73), rpoN (σ54), and rpoH (σ32), sigF (σF), sigT (σT), and sigU (σU) have been studied previously (Alvarez-Martinez et al., 2006; Alvarez-Martinez et al., 2007; Brun and Shapiro, 1992; Malakooti and Ely, 1995; Reisenauer et al., 1996; Wu and Newton, 1996). Elucidation of the roles of the remaining sigma factors is likely to aid in understanding the complex regulation of gene expression throughout the cell cycle.
2.2.1 σ73 - the principal sigma factor
rpoD is constitutively expressed throughout the cell cycle and encodes the principal sigma factor, σ73 (Malakooti and Ely, 1995). σ73 recognizes a consensus promoter sequence, TTGaCgS (n10-14) GCtANAWC, which is found in the promoter region of a number of biosynthetic and housekeeping genes (Malakooti and Ely, 1995; Malakooti et al., 1995). The consensus sequence for this sigma factor has been confirmed recently (see section 2.3; (McGrath et al., 2007)). σ73 also recognizes E. coli σ70-dependent promoters (Malakooti and Ely, 1995; Wu et al., 1997). This observation is interesting since the two sigma factors do not share a consensus sequence (Malakooti and Ely, 1995; Wu et al., 1997). Although the −35 region of σ73 promoter is similar to the consensus −35 region recognized by E. coli, σ70, the −10 region of the σ73 promoter is not recognizably similar to −10 region of the E. coli σ70 promoter. In addition, the space between the −35 and −10 regions is smaller in the σ73 promoters, when compared to the σ70 promoters. These observations suggest that σ73 has less promoter specificity than σ70.
2.2.2 σ32 - the heat shock sigma factor
Following heat shock, a transient increase in both rpoH transcription and σ32 protein levels is observed in C. crescentus (Reisenauer et al., 1996; Wu and Newton, 1996). The rpoH promoter has two promoter elements; P1 is dependent on σ73 and P2 is autoregulated by σ32, which recognizes the following consensus sequence, TNNCNCCCTTGAA (Wu and Newton, 1997). Transcription from P2 increases in response to heat shock. Heat shock also influences the expression of about 20 genes, including clpXP and genes that encode molecular chaperones (Gomes et al., 1986; Osteras et al., 1999). Transcription of clpP, which encodes an essential protease, is enhanced by heat shock, whereas transcriptionof clpX, which encodes an ATP-dependent chaperone, is repressed by heat shock (Osteras et al., 1999). The activation of clpP transcription by σ32 may explain why σ32 cannot be inactivated, even at low temperatures (Osteras et al., 1999; Reisenauer et al., 1996). One of the chaperones regulated by σ32, DnaK, has been shown to negatively regulate the heat shock response (da Silva et al., 2003). After heat shock, cells express a high level of DnaK and the level of σ32 is transiently high; however, the heat shock proteins are not induced. This result suggests that DnaK inhibits the activity of σ32 and may stimulate its degradation. It has been proposed that competition between σ73 and σ32 for RNA polymerase may be responsible for the downregulation of the heat shock response during the recovery phase (da Silva et al., 2003).
2.2.3 σ54 - the alternative sigma factor for polar development
The σ54 - RNA polymerase holoenzyme recognizes the consensus TGGCNCCGNCCTTGCA promoter and requires activator proteins in order initiate transcription (Brun and Shapiro, 1992; McGrath et al., 2007). In C. crescentus, the transcription of σ54 is cell cycle regulated, with an increase in expression just prior to both flagellum biosynthesis and stalk biosynthesis (Brun and Shapiro, 1992). rpoN mutants are non-motile, stalkless, and display aberrant cell division indicating that σ54 regulates genes involved in flagellum and stalk biosynthesis, as well as cell division (Brun and Shapiro, 1992). Indeed, σ54 is specifically responsible for the transcription of the class III and IV flagellar genes (Anderson et al., 1995).
2.2.3a σ54 activators
In the absence of an activator protein, the σ54-RNA polymerase holoenzyme is unable to form an open complex and transcription is not activated. σ54 activator proteins typically belong to the NtrC-family of response regulators and function as transcriptional activators in the phosphorylated state. The genome sequence of C. crescentus revealed the presence of four possible activators based on homology to the highly conserved central domain of NtrC (Nierman et al., 2001). In E. coli, σ54 has been shown to interact specifically with the threonine residue of the GAFTGA motif present in σ54 activator proteins (Bordes et al., 2004). The two activators which have been characterized, FlbD and TacA, have the complete GAFTGA motif that is required for the interaction with σ54. The remaining two possible activators do not have the complete motif required for interacting with σ54 and have not been characterized. Identifying the function of the remaining two activators and determining if they interact with σ54 may further elucidate the role of the alternative sigma factor, σ54, in polar development and cell division.
FlbD is required for the transcription of the class III and class IV flagellar genes in the swarmer compartment of the predivisional cell (see section 3.1.1a; (Ramakrishnan and Newton, 1990)). FlbD is present throughout the cell cycle, but is phosphorylated exclusively in the predivisional cells when class III flagellar transcription is initiated (Wingrove et al., 1993). The kinase responsible for the phosphorylation of FlbD has not been identified. In addition to activating late flagellar genes, FlbD represses early flagellum assembly genes, indicating the ability of this transcriptional regulator to affect multiple steps in flagellum assembly (Fig. 3). Mutants in flbD are not defective in stalk biosynthesis, indicating that another activator interacts with σ54 to regulate stalk synthesis.
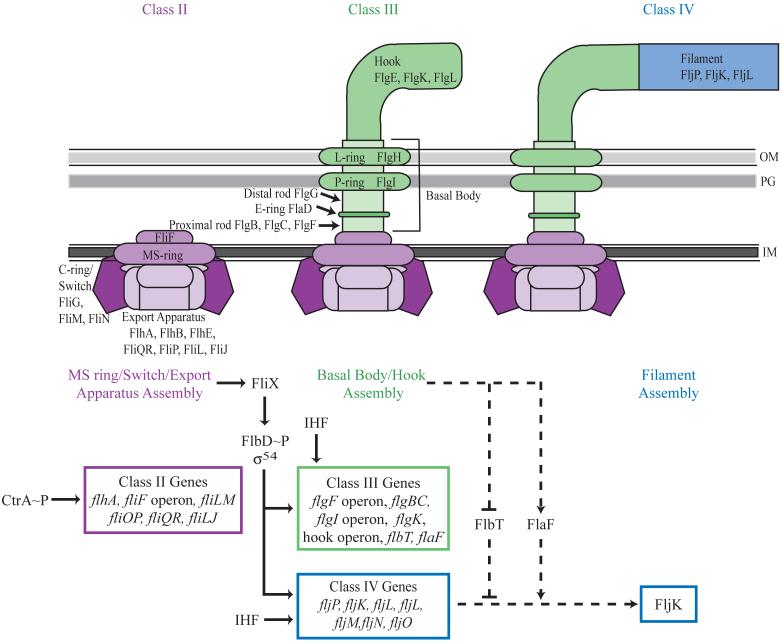
Figure 3.
Transcriptional regulation, translational regulation, and assembly of flagellar proteins. The presence of phosphorylated CtrA (CtrA∼P) leads to the transcription of the class II flagellar genes, which include flbD, which is part of the fliF operon. FlbD, aσ54-dependent transcriptional activator, is repressed by FliX until assembly of the class II flagellar proteins, including the export apparatus, C-ring/switch, and MS-ring, is complete. The assembly of class II flagellar proteins is detected by FliX through an unknown mechanism and leads to the activation of FlbD by phosphorylation in a FliX dependent manner. FlbD phosphorylation results in the transcriptional activation of the class III and IV flagellar genes. Maximal transcription of class III and IV flagellar genes also requires IHF. The class III flagellar proteins are then assembled to form the basal body and hook of the flagellum. Although the class IV genes are transcribed, they are subject to post-transcriptional regulation by FlbT and FlaF, which prevent translation of at least one class IV flagellin message, fljK, until assembly of the basal body and hook is complete. Following assembly of the basal body and hook, fljK transcripts are stabilized by FlaF. The presence of FljK, the major flagellin required for motility, enables filament assembly. With the addition of the filament, flagellum assembly is complete. Solid lines depict pathways involved in transcriptional control and dashed lines indicate pathways for translational control. IM, inner membrane; PG, peptidoglycan; OM, outer membrane.
TacA was initially identified in an effort to find σ54 activators by PCR amplification of conserved domains (Marques et al., 1997). The TacA promoter contains a CtrA binding site and the transcription of tacA is temporally regulated; maximal transcription of tacA occurs in the predivisional cell (Biondi et al., 2006b; Marques et al., 1997). Following chromosome replication initiation in the stalked cell, CtrA activates the transcription of rpoN and perhaps tacA; however CtrA binding to the tacA promoter has not been demonstrated. TacA is phosphorylated through a phosphorelay cascade (Biondi et al., 2006b). ShkA, a hybrid histidine kinase, autophosphorylates and transfers the phosphoryl group to its own receiver domain. From there, the phosphoryl group is transferred onto ShpA, an Hpt, which transfers the phosphoryl group to TacA. Phosphorylated TacA interacts with the RNA polymerase-σ54 holoenzyme to enable the transcription of genes required for the regulation of stalk biogenesis, including StaR, a regulator of stalk length (see section 3.3.3). It remains to be determined if TacA activity is spatially restricted.
2.2.4 σF - alternative sigma factor for oxidative stress during stationary phase
σF is not required during normal growth, however σF is specifically required for a response to oxidative stress during stationary phase (Alvarez-Martinez et al., 2006). In response to oxidative stress during stationary phase, σF activates the transcription of eight genes, including those which encode methioine sulfoxide reductase, superoxide dismutase, and carbonic anhydrase, which are involved in protection against oxidative stress. The function of the remaining proteins in the σF regulon remains unknown. Interestingly, transcription of sigF is reduced as the cell goes into stationary phase, however σF protein levels increase in stationary phase. In exponential cells, σF is degraded either directly or indirectly by the protease FtsH. During stationary phase, σF is less susceptible to degradation by FtsH. It has been suggested that an accessory protein may interact with σF during exponential phase and lead to increased degradation of this sigma factor (Alvarez-Martinez et al., 2006).
2.2.5 σT - alternative sigma factor for osmotic and oxidative stress
Transcription of sigT occurs specifically during the swarmer to stalked cell differentiation and is regulated by CtrA (Laub et al., 2002; Laub et al., 2000). Transcription of sigT is induced by growth in minimal media, heavy metal stress, and osmotic stress (Alvarez-Martinez et al., 2007; Hottes et al., 2004; Hu et al., 2005). σT is not essential under normal growth conditions however, σT is essential for survival under conditions of osmotic or oxidative stress (Alvarez-Martinez et al., 2007). Interestingly, sigT transcription is not induced under oxidative stress. This observation suggests that the normal levels of σT present in the cell are sufficient for protection against oxidative stress. The σT regulon includes genes predicted to encode proteins involved in the biosynthesis or structure of the cell envelope, stress response, and electron transfer suggesting the σT is involved in mediating a general stress response and cell envelope functions. The σT regulon revealed that σT regulates its own transcription, as well as two additional sigma factors, sigR and sigU. This observation indicates that σT likely initiates a regulatory cascade allowing the indirect regulation of additional genes.
2.2.6 σU - alternative sigma factor which may be involved in stress response
The transcription of sigU is positively regulated by σT and as a result, sigU is induced by growth in minimal media, heavy metal stress, and osmotic stress (Alvarez-Martinez et al., 2007; Hottes et al., 2004; Hu et al., 2005). σU is not required for growth under normal condition or under any environmental conditions tested thus far (Alvarez-Martinez et al., 2007). It has been proposed that σT activates sigU leading to the regulation of another distinct set of genes perhaps required for resistance to an unidentified growth condition (Alvarez-Martinez et al., 2007). Determining the function of this sigma factor, as well as the remaining sigma factors predicted by the genome sequence, may help identify genes with specific functions cell survival, stress response, or perhaps in cell cycle progression.
2.3 Promoter Architecture
Using an Affymetrix array, promoter motifs were identified by searching upstream of the transcriptional start sites of fourteen gene clusters comprised of genes that are coexpressed throughout the cell cycle of C. crescentus (McGrath et al., 2007). Fourteen promoter motifs were identified within ten different groups of coexpressed genes. Seven of the fourteen promoters had been previously described including a σ54-dependent promoter and a CtrA-dependent promoter. The remaining seven promoter motifs are found in gene clusters that are expressed at particular times in the cell cycle. This type of analysis demonstrates that there are likely to be more unknown regulatory elements that effect cell cycle progression. For example, one motif, cc_9, with a consensus sequence of GACACNNTGTCGCA, was identified upstream of a CtrA binding site in a set of genes maximally expressed early in predivisional cells. It is likely that the element that binds to cc_9 impacts the transcription of genes involved in polar development.
2.4 Two-Component Signal Transduction Proteins
In addition to global regulators and sigma factors, two component signal transduction proteins are known to dramatically impact cell cycle progression. The genome sequence of C. crescentus revealed the presence of 105 genes encoding two-component signal transduction proteins including 34 histidine kinases, 44 response regulators, and 27 hybrid histidine kinase/response regulators (Nierman et al., 2001). About a third of the histidine kinases are located adjacent to response regulators suggesting that these comprise functional pairs of signal transduction proteins. While two-component signal transduction proteins are typically involved in mediating responses to environmental changes, a number of the two-component proteins have been shown to function in cell cycle regulation in C. crescentus.
A systematic deletion analysis has shown that at least 39 of the two-component genes are required for cell cycle progression, growth, or morphology and nine of the genes are essential for cell survival (Skerker et al., 2005). Potential cell cycle or cell growth regulatory genes were identified from mutants with decreased motility in swarm agar and an increase in generation time. This strategy identified divJ, flbD, and tacA, which encode proteins previously known to be involved in cell cycle progression. Two regulators, ShkA and CpdR, were identified and have since been shown to play significant roles in regulation of cell cycle progression. A single deletion of any of seven regulators, including PhoB and six uncharacterized regulators, results in a phenotype indicative of prolonged swimming suggesting that these regulators may play a role in the swarmer to stalked cell differentiation. This observation suggests that there is much more to be learned about the complex regulation of cell cycle progression
2.4.1 Subcellular localization of signal transduction proteins
While global regulators and sigma factors primarily contribute to the temporal regulation of gene expression, the signal transduction proteins are typically involved in the activation of cognate genes or proteins. The spatial organization of signal transduction proteins in C. crescentus plays an important role in preparing each compartment of the predivisional cell for the formation of distinct cell types (Fig. 2).
2.4.1a CckA - the mediator of CtrA phosphorylation
As described in section 2.1.1 c, the histidine kinase, CckA, is necessary for the phosphorylation and activation of CtrA via the CckA-ChpT phosphorelay. In the predivisional cell, CckA is localized to the nascent swarmer pole leading to increased levels of CtrA∼P that block chromosome replication in the new swarmer cell (Fig. 2; (Jacobs et al., 1999)). Conversely, the absence of CckA in the stalked compartment of the predivisional cell prevents the phosphorylation of CtrA and allows a new round of chromosome replication to be initiated in the new stalked cell. Thus, the dynamic localization of CckA contributes to the formation of distinct daughter cells.
2.4.1b PleC-DivJ-DivK-PleD Multicomponent System
The histidine kinases, PleC and DivJ, are localized to opposite cell poles in the predivisional cells (Fig. 2; (Wheeler and Shapiro, 1999)) and have opposing actions on the response regulators, DivK and PleD. DivJ phosphorylates both DivK and PleD whereas PleC acts as a phosphatase for DivK∼P and prevents the formation of PleD∼P, perhaps by acting as a phosphatase although the mechanism has not been experimentally determined (Aldridge et al., 2003; Hecht and Newton, 1995). DivJ is responsible for the dynamic localization of DivK throughout the cell cycle (Fig. 2; (Lam et al., 2003)). In the swarmer cell, the presence of PleC at the flagellated pole results in low levels of DivK∼P, which stabilizes CckA and allows CtrA∼P to remain bound to Cori thereby preventing chromosome replication in the new swarmer cell following cell division (Biondi et al., 2006a). As the cell undergoes the swarmer to stalked cell differentiation, PleC delocalizes and DivJ is synthesized and binds to the stalked pole. DivJ recruits DivK to the stalked pole and the kinase activity of DivJ causes the level of DivK∼P to increase and DivK∼P localizes to the flagellar pole of the predivisonal cell (Lam et al., 2003), which leads to the delocalization of CckA and prevents the phosphorylation of CtrA (Fig. 2). The remaining pools of CtrA are dephosphorylated and degraded, allowing the initiation of chromosome replication to occur (Biondi et al., 2006a). As the predivisional cell is formed, DivK∼P is targeted to both the flagellar and stalked poles (Lam et al., 2003). In the predivisional cell, PleC is localized to the swarmer pole and dephosphorylates DivK. The unphosphorylated DivK freely diffuses in the cytoplasm to the stalked pole where it is phosphorylated again by DivJ (Matroule et al., 2004). The localization and activities of both PleC and DivJ at opposite cell poles result in a rapid exchange of DivK and DivK∼P at the cell poles and result in bipolar localization of DivK∼P (Matroule et al., 2004). Following cell division, DivK is completely delocalized in the swarmer cell compartment and the swarmer to stalked cell transition begins (Matroule et al., 2004). The difference in the level of DivK∼P in each of the daughter cells contributes to the distinct physiologies of the new stalked and swarmer cells.
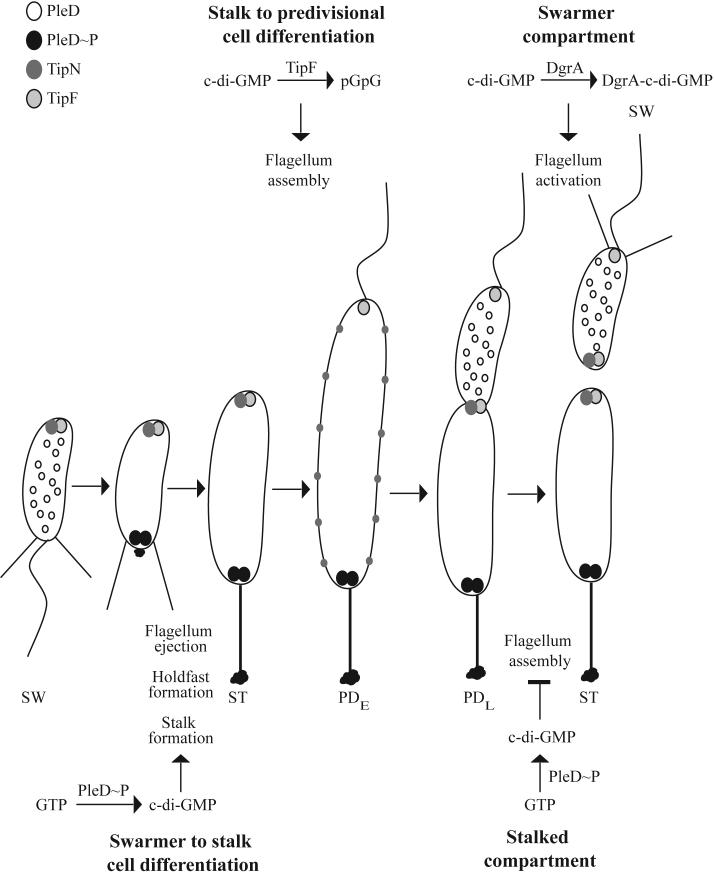
Localization of DivJ and PleC also impact the phosphorylation state and activity of PleD, a response regulator that contains a diguanylate cyclase domain. PleD∼P is required for ejection of the flagellum and elongation of the stalk during the swarmer to stalked cell differentiation and for preventing flagellum activation in the predivisional cell (Aldridge and Jenal, 1999). The presence of PleC in the new swarmer cell prevents the phosphorylation of PleD and premature ejection of the flagellum. As the cell undergoes the swarmer to stalked cell transition, DivJ is localized to the incipient stalked pole allowing the phosphorylation of PleD, which is then sequestered at the pole, resulting in subsequent ejection of the flagella and elongation of the stalk (Aldridge et al., 2003; Paul et al., 2004). Proper temporal control of holdfast synthesis during the swarmer to stalked cell differentiation is also dependent on PleD∼P (Levi and Jenal, 2006). PleD∼P has di-guanylate cyclase activity capable of converting two molecules of guanosine triphosphate (GTP) into cyclic-diguanosine monophosphate (c-di-GMP) (Paul et al., 2004). Since activated PleD∼P is found only at the stalked pole, and c-di-GMP can serve as a second messenger, it has been proposed that production of c-di-GMP may serve as a signal for polar development (Jenal and Malone, 2006). The structure of PleD suggests that the efficient production of c-di-GMP requires PleD dimerization and that feedback inhibition by the product is likely to limit the concentration of c-di-GMP in the cell (Chan et al., 2004; Wassmann et al., 2007). Indeed, biochemical analyses have shown that PleD is activated by dimerization and that dimerization occurs when the protein is phosphorylated (Paul et al., 2007). It will be of particular interest to determine the specific role of c-di-GMP signaling in the development of the stalked pole in C. crescentus.
2.4.1c PodJ - the localization factor
Localization is a critical element for spatial regulation by some two-component systems. Little is known about how CckA and DivJ are localized to the stalked pole, but a localization factor, PodJ, has been identified for PleC (Hinz et al., 2003; Viollier et al., 2002b). PodJ is subject to complex regulation throughout the cell cycle and impacts swarming motility and the formation of pili and holdfast. podJ transcription is repressed by CtrA, activated directly by DnaA, and activated, either directly or indirectly, by GcrA (Crymes et al., 1999; Holtzendorff et al., 2004; Hottes et al., 2005). As a result of this transcriptional regulation, podJ is transcribed following the swarmer to stalked cell differentiation and full length PodJ (PodJL) is then localized to the incipient flagellar pole of the predivisional cell (Hinz et al., 2003; Viollier et al., 2002b). PodJL is required for the localization of both PleC and CpaE, a protein required for pili assembly, to the incipient flagellar pole (Fig. 2; (Hinz et al., 2003; Viollier et al., 2002b)). Following cell division, PodJL, is processed into a short form (PodJS) by the periplasmic protease, PerP (Chen et al., 2006). PodJS remains localized at the flagellar pole following cell division and is required for chemotaxis and holdfast formation, presumably due to a role in localizing proteins required for these functions (Fig. 2; (Hinz et al., 2003; Lawler et al., 2006; Viollier et al., 2002b)). During the swarmer to stalked cell transition, PodJS is cleaved from the membrane by the protease MmpA and degraded completely by an unknown protease prior to the synthesis of new PodJL (Chen et al., 2005).
How can PodJ influence pili formation, holdfast formation, swarming motility, and PleC localization? PodJ has been shown to be a modular protein with functional domains that are important for specific functions (Lawler et al., 2006). PodJ has a single transmembrane domain with the N-terminus in the cytoplasm and the C-terminus in the periplasm (Viollier et al., 2002b). The cytoplasmic portion of PodJ contains three coiled-coil domains that are typically involved in protein-protein interactions and has been shown to be required for holdfast production and swarming motility (Crymes et al., 1999; Lawler et al., 2006; Smith et al., 2003). The periplasmic portion of PodJ is required for pili formation and contains three tetratricopeptide repeats, which are also involved in mediating protein-protein interactions, and a peptidoglycan binding domain (Crymes et al., 1999; Lawler et al., 2006; Viollier et al., 2002b). The region immediately preceding the transmembrane domain is necessary for the localization of PodJ to the correct pole throughout the cell cycle, whereas the region immediately following the transmembrane domain is required for localization of PleC (Lawler et al., 2006). Some PodJ mutants that are not able to localize PleC can still produce holdfast indicating that PleC does not have to localize to be active. In contrast, it has been suggested that PodJ localization is required for proper function (Lawler et al., 2006). The necessity of polar localization for proper function is likely to be a common feature for some of the cell cycle regulated proteins in C. crescentus.
2.4.1d DivL - an essential tyrosine kinase
DivL, a tyrosine kinase, is required for cell survival and impacts cell cycle progression of C. crescentus (Wu et al., 1999). DivL autophosphorylates on a tyrosine residue and is capable of passing the phosphoryl group to CtrA in vitro (Wu et al., 1999). Reduced levels of phosphorylated CtrA are observed in conditional mutants of divL (Pierce et al., 2006). Together these results indicate that DivL is involved in the activation of CtrA.
DivL is dynamically localized throughout the cell cycle (Sciochetti et al., 2005). Following cell division, DivL is absent from both the swarmer cell and the stalked cell. Shortly after cell division, DivL appears in the stalked cell and is localized to the pole opposite the stalk. The swarmer cell does not acquire DivL until after differentiation into the stalked cell. As the predivisional cell is formed, DivL remains localized at the incipient flagellated pole and is occasionally observed at the stalked pole. The polar foci of DivL are dispersed late in the predivisional cell, prior to cell division. The polar localization of DivL is dependent, either directly or indirectly, on the presence of DivJ. However, the kinase activity of DivL is not dependent on its polar localization (Sciochetti et al., 2005).
Interestingly, it is not clear if the kinase activity of DivL is important for its function in cell cycle progression. Mutation of the tyrosine residue in DivL results in a phenotype that is much less severe than DivL depletion, suggesting that DivL has a kinase independent function in cell cycle regulation (Reisinger et al., 2007). DivL affects CtrA phosphorylation, but not proteolysis suggesting that DivL acts independently from the CckA-ChpT phosphorelay. One model is that DivL may protect CtrA∼P rather than acting as a CtrA kinase (Reisinger et al., 2007). Irrespective of the mechanism of promoting CtrA phophorylation, DivL has another function in cell cycle control since deletion of divL in a strain expressing a phosphomimetic ctrA allele results in defects in chromosome replication and cell division. DivL has also been shown to modulate the localization and phosphorylation of DivK (Reisinger et al., 2007). DivL may promote DivK localization by regulating DivJ localization and kinase activity or by direct interactions with DivK at the flagellar pole, or both. Thus, kinase independent activities of DivL are likely to be responsible for the modulation of both CtrA and DivK. It remains to be determined if the kinase independent activities of DivL are dependent on its localization.
3. Polar Structure Biogenesis and Function
The regulatory mechanisms described above function primarily to impart polarity to the predivisional cell and allow the formation of two different types of daughter cells - the swarmer cell and the stalked cell. The polar structures found on each cell type are distinct; the swarmer cell contains a flagellum and pili whereas the stalked cell has an extension of the cell membranes and peptidoglycan layer called a stalk or prostheca and a polysaccharide containing adhesin at the tip of the stalk called a holdfast. The holdfast is found in both swarmer and stalked cells, but primarily serves to attach the stalked and predivisional cell to a surface. The role and biosynthesis of each of the polar structures is discussed below.
3.1 The Flagellum
C. crescentus produces a single polar flagellum that is responsible for fast and efficient swimming in swarmer cells (Li and Tang, 2006). The regulation of flagellum biosynthesis and assembly has been studied extensively and has been the subject of a number of recent reviews (Aldridge and Hughes, 2002; England and Gober, 2001; Gober and England, 2000; Jenal, 2000). The following sections focus on the most recent advances that have enhanced our understanding of the most characterized aspect of C. crescentus polar development, flagellum biogenesis.
3.1.1 Biosynthesis and assembly of flagella
The biosynthesis of flagella is a complex process, requiring a vast array of structural, regulatory and force generating proteins. In C. crescentus, this process is temporally and spatially constrained by a number of molecular mechanisms including both transcriptional and translational regulation.
3.1.1a Transcriptional and translational regulation of flagellar genes
The temporal transcriptional regulation of about 50 flagellar genes is mediated by a complex hierarchy that is tied to both cell cycle progression and assembly of the flagellum (Fig. 3; for review see (Gober and England, 2000)). CtrA∼P is responsible for the transcription of the class II flagellar genes in stalked cells following the initiation of chromosome replication. The class II flagellar genes encode the MS ring, flagellar switch, and flagellum export apparatus, which are the first flagellar components to be assembled. Class II flagellar genes also encode regulatory proteins including the transcriptional activator FlbD, the transacting factor FliX, and σ54, which impact the transcription of the class III and IV flagellar genes.
Maximal transcription of a subset of class III and IV flagellar genes also requires the sequence specific binding protein integration host factor (IHF) (Muir and Gober, 2005). In E. coli, binding of IHF induces a bend into DNA which brings RNA polymerase bound to the promoter into close proximity with transcriptional activators bound at distant site (Hoover et al., 1990). It has been proposed that IHF-induced bending of DNA may enhance the interaction of FlbD with other molecules of FlbD to promote oligomerization, which is thought to enhance the ability FlbD to isomerize RNA polymerase from a closed complex to an open complex (Muir and Gober, 2005).
The transcription of the class III and IV flagellar genes does not occur until assembly of the class II flagellar proteins is complete (Fig. 3). Prior to class II protein assembly, the trans-acting factor FliX interacts directly with FlbD, preventing FlbD from binding to enhancer sequences and subsequent σ54-dependent transcription of the class III and IV flagellar genes (Dutton et al., 2005; Muir and Gober, 2001; Muir and Gober, 2002; Muir and Gober, 2004; Muir et al., 2001). Once assembly of the class II flagellar proteins is complete, FliX inhibition of FlbD is relieved, FlbD is phosphorylated by an unknown kinase, and transcription of the class III and IV flagellar genes is initiated (Muir and Gober, 2001; Muir and Gober, 2002; Muir et al., 2001; Wingrove et al., 1993). It remains unclear how FliX senses the completion of assembly. The class III genes encode structural components of the flagella including the basal body, L ring, and P ring. The assembly of the class III structural components into the basal body hook complex requires the lytic transglycosylase, PleA (Viollier and Shapiro, 2003). pleA mutants are non-motile and lack flagella. Thus, it has been proposed that PleA is responsible for the hydrolysis of peptidoglycan at the swarmer pole, allowing the penetration of structural components of the flagellum through the peptidoglycan layer.
In addition to structural components of the basal body hook complex, the class III flagellar genes include flbT, which encodes a regulatory protein responsible for translational control of the class IV flagellar genes encoding the flagellins (Anderson and Gober, 2000; Mangan et al., 1999). In mutants unable to assemble the hook complex, flagellin mRNA is present at low levels due to messenger instability. When a flbT mutation is introduced into a strain which cannot assemble the hook complex, flagellin mRNA is stabilized indicating that FlbT is a negative regulator of flagellin expression (Mangan et al., 1999). The mechanism of FlbT repression of flagellin gene expression was explored using the fljK transcript, which encodes one of the major flagellins (Anderson and Gober, 2000). Studies using translational fusions revealed that only 63 nucleotides of the 5' untranslated region and the first 14 codons of fljK mRNA are necessary for FlbT-mediated repression. This region of the fljK mRNA is predicted to fold into two different secondary structures, including one structure that is likely to be unfavorable for translation since the ribosome binding site is blocked by base pairing. Using cell extracts, FlbT was shown to bind to a loop structure of the fljK mRNA, likely stabilizing the secondary structure that prevents translation. Based on these observations, it has been suggested that in the absence of a complete basal body hook complex, FlbT binds to flagellin mRNA and blocks translation leading to destabilization of the flagellin mRNA (Fig.3; (Anderson and Gober, 2000)). Following the assembly of the class III flagellar proteins into the basal body hook structure, FlbT repression is relieved by an unknown mechanism and fljK transcripts are stabilized leading to translation of the flagellins and assembly of the filament (Fig. 3). It remains unclear how the completion of basal body hook assembly is detected by FlbT.
The stabilization of flagellin mRNA requires FlaF, a protein required for motility. The flaF gene is directly downstream from the flbT gene and is required for fljK transcript stabilization (Llewellyn et al., 2005). In the absence of flaF, the basal body hook complex is assembled, but flagellin mRNAs are not translated. This observation indicates that FlaF is involved in the transition between completion of the basal body hook complex assembly and the initiation of flagellin translation and filament assembly. It has been proposed that throughout the cell cycle, the regulation of flagellin translation and filament assembly is controlled by the opposing actions of FlbT, which destabilizes flagellin mRNA, and FlaF, which stabilizes flagellin mRNA (Llewellyn et al., 2005). Both flbT and flaF are conserved among swimming bacteria belonging to the alphaproteobacteria indicating that this regulatory mechanism, which delays flagellin translation and filament assembly until after the completion of the basal body hook complex assembly, may constitute a conserved checkpoint in flagellum assembly.
3.1.1b Spatial control of flagellum assembly
How is the location of flagellum assembly determined? To answer that question, it is necessary to first consider how polarity is determined in the cell (Fig. 4). TipN, a “birth scar” protein, marks polarity in new daughter cells (Huitema et al., 2006; Lam et al., 2006). TipN is found exclusively at the new pole in both the swarmer and stalked cells and marks the site of future flagellum assembly. TipN remains at the new pole as the flagellum is assembled in the predivisional cell. Following the biosynthesis of the flagellum, TipN is briefly delocalized before localizing to the division site (Huitema et al., 2006; Lam et al., 2006). Following cell division, TipN localization serves as a molecular beacon, marking the new pole of the daughter cell.
Figure 4.
Cyclic-di-GMP signaling throughout the C. crescentus cell cycle. Each stage of the cell cycle, including the swarmer (SW), stalked (ST), early predivisional (PDE), late predivisional (PDL), and progeny cells, is shown. In the swarmer cell PleD is delocalized. As the cell undergoes the swarmer to stalked cell differentiation, dimers of PleD∼P are localized to the flagellar pole leading to the production of cyclic-di-GMP. The increase in c-di-GMP levels promotes flagellum ejection, holdfast formation, and stalk formation. Flagellum assembly in the early predivisional cell is achieved by a reduction in c-di-GMP levels through degradation into linear diguanylate (pGpG) by TipF and occurs at the pole marked by the presence of TipN. After flagellum biosynthesis, TipN is briefly delocalized. In the late predivisional cell, just prior to cell division, flagellum activation occurs as c-di-GMP levels decrease due to binding by proteins such as DgrA. TipF is localized to the pole opposite the flagellum throughout the cell cycle, until the onset of cell division, when both TipN and TipF localize to the mid-cell and mark the new pole following cell separation.
TipN is required for proper localization of TipF, a c-di-GMP phosphodiesterase protein, required for flagellum assembly (Huitema et al., 2006). In cells lacking TipN, the flagellum is assembled, but is frequently misplaced; however, in cells lacking TipF, the flagellum fails to assemble. These results suggest that presence of TipF, but not its localization to the pole, is required for flagellum assembly and that the interaction of TipN and TipF is responsible for assembly of the flagellum specifically at the new pole. Mutational analysis indicates that the TipF phosphodiesterase domain, which likely degrades c-di-GMP, is required for its function in flagellum assembly (Huitema et al., 2006). This result suggests that reducing the level of c-di-GMP may facilitate flagellum assembly.
3.1.2 Flagellum biosynthesis is coupled to cell division
Assembly of the class II flagellar structures is required for proper cell division; mutants in class II flagellar genes result in the formation of filamentous cells (Gober et al., 1995; Yu and Shapiro, 1992; Zhuang and Shapiro, 1995). Such a checkpoint is logical, delaying cell division until the assembly of the flagellum has initiated on the swarmer pole of the predivisional cell ensures that the swarmer cell will have a functional flagellum. Two links between flagellum biogenesis and cell division have recently been uncovered. First, the regulation of FlbD activity by FliX influences the transcription of late flagellar genes and the completion of cell division (Muir et al., 2005). Negative regulation of FlbD activity by FliX is responsible for the cell division defect in class II flagellar genes. Identifying the genes activated by σ54 and FlbD will reveal if FlbD is required for the transcription of any cell division genes. Second, once the predivisional cell is mature and the flagellum has been assembled, TipN and then TipF are localized to the division site (Huitema et al., 2006; Lam et al., 2006). The localization of TipN and TipF at the division site depends on FtsZ ring formation and constriction suggesting that these proteins may also have a role in cell division (Huitema et al., 2006; Lam et al., 2006).
3.1.3 Flagellum activation
Cell division defects result in nonmotile cells that have an assembled flagellum indicating that assembly is not sufficient for flagellum rotation and function (Degnen and Newton, 1972; Muir et al., 2005; Ohta et al., 1997; Quardokus and Brun, 2003). FliL is required for flagellum rotation, but is not part of the flagellar transcriptional hierachy and is not a component of the assembled flagellum (Jenal et al., 1994). The mechanism of motor control by FliL remains unknown, but recent results suggest that the level of the second messenger, c-di-GMP, may modulate motility and specifically reduce the levels of FliL (Fig 4; (Aldridge et al., 2003; Christen et al., 2007). Motility is blocked in C. crescentus cells with an increased level of c-di-GMP (Christen et al., 2007). The block in motility is caused by a loss of flagellum function and can be bypassed in a dgrA mutant (Christen et al., 2007). DgrA is a member of the diguanylate receptor proteins and has a high affinity for binding c-di-GMP. Cells lacking DgrA are motile in the presence of high levels of c-di-GMP and have reduced levels of FliL. However, the levels of other motility proteins including FliM, which is cotranscribed with FliL, are not affected. This result indicates that changes in FliL protein levels are likely to be due to translational regulation or decreased protein stability. The simplest model to explain these results is that when DgrA is bound to c-di-GMP, FliL is repressed and cannot promote flagellum rotation (Fig. 4).
3.1.4 Ejection of the flagellum
During the swarmer to stalked cell transition, the flagellum is ejected prior to the formation and elongation of the stalk. The ejected flagellum is largely intact, containing the filament, the hook, and part of the rod, indicating that the breakpoint occurs at the junction between the MS ring and the rod (Kanbe et al., 2005). Indeed, flagellum ejection coincides with the degradation of FliF, which forms the MS-ring that anchors the axial components of the flagellum to the cell (Aldridge and Jenal, 1999). Biochemical evidence suggests that the MS-ring, but not the rod, of C. crescentus flagella are particularly susceptible to protease activity (Kanbe et al., 2005). The ClpAP protease recognizes the hydrophobic tail of FliF and is responsible for the degradation of FliF during the swarmer to stalked cell differentiation (Grunenfelder et al., 2003; Grunenfelder et al., 2004).
The response regulator, PleD is also involved in the regulation of flagellum ejection. In a pleD mutant, FilF is stabilized and the flagellum is not ejected (Aldridge and Jenal, 1999). Furthermore, the GGDEF (Gly-Gly-Asp-Glu-Phe) output domain of PleD is responsible for the production of c-di-GMP and is required for flagellum ejection (Fig. 4; (Aldridge and Jenal, 1999) (Paul et al., 2004). This result suggests that c-di-GMP signaling mediated by PleD is responsible for the degradation of the MS-ring. Notably, the only flagellar protein required for efficient removal of FliF prior to flagellum ejection is FliL. FliL is known to be modulated by DgrA when bound c-di-GMP (Aldridge and Jenal, 1999; Christen et al., 2007). The mechanism by which PleD and c-di-GMP trigger the degradation of FliF remains unknown. Determining the interactions among proteins that generate, bind, and degrade c-di-GMP and a subset of flagellar proteins will lead to a greater understanding of the temporal regulation of flagellar assembly, activation, and ejection.
3.1.5 Function of the flagellum
C. crescentus live in oligotrophic aquatic environments and the ability to swim is certainly important for tracking nutrients and cell survival. Despite structural similarities between the flagella of E. coli and C. crescentus, C. crescentus swims nearly 10 times more efficiently than E. coli (Li and Tang, 2006). The increase in swimming efficiency may reflect an adaptation to low nutrient environments (Li and Tang, 2006).
The flagellum of C. crescentus also plays a role in the establishment and escape from biofilms under hydrodynamic conditions (Entcheva-Dimitrov and Spormann, 2004). C. crescentus forms biphasic biofilms consisting of a monolayer biofilm interspersed with large mushroom-like structures. Swimming motility mediated by the flagellum enhances the initial attachment event leading to eventual biofilm formation (Fig. 5; (Bodenmiller et al., 2004; Entcheva-Dimitrov and Spormann, 2004)). In a monolayer biofilm formed under hydrodynamic conditions, cell division produces new swarmer cells that swim away from the established biofilm (Entcheva-Dimitrov and Spormann, 2004). The release of swarmer cells from the biofilm can be viewed as a dispersal mechanism, allowing the establishment of new biofilms in other locations. In a mushroom-like biofilm structure, which is comprised primarily of clonal growth arising from established microcolonies, the flagellum is involved retaining new swarmer cells within the three dimensional structure and may function as an adhesin (Entcheva-Dimitrov and Spormann, 2004). The dual function of the flagellum in the two types of biofilm formations is intriguing and suggests that the role of the flagellum in C. crescentus may not be confined to cell motility.
Figure 5.
Attachment of a C. crescentus cell to a surface. The stages of initial attachment are shown, A-F. The surface for attachment is shown as a grey box and the polar structures are labeled. Contact with the surface is mediated by flagellar motility, which overcomes repulsive forces (A-B). The flagellum enhances the initial attachment of C. crescentus to the surface (C). Following flagellum ejection, the interaction of the pili with the surface may properly position the cell prior to a more permanent attachment (D). As the swarmer cell differentiates into a stalk cell, the holdfast is exported and the pili are hypothesized to retract (E). The biosynthesis of the stalk brings the holdfast in contact with the surface (F). The holdfast is responsible for the remarkably strong adhesion of C. crescentus to surfaces.
3.2 The Pili
Immediately after cell division, Flp (fimbrial low-weight molecular protein) pili are synthesized at the flagellar pole of the swarmer cell. Like the flagellum, the biosynthesis of the pili is temporally and spatially restricted and subject to transcriptional regulation. Despite the similarity in the timing and location of flagellum and pilus biogenesis, these two processes share few regulatory elements. Whereas the flagellar assembly machinery is similar to type III secretion machinery, the pilus assembly machinery more closely resembles the type II Tad (tight adherence) macromolecular transport system, indicating that these two structures are assembled independently from one another (Skerker and Shapiro, 2000; Tomich et al., 2007). In this section, an overview of the most recent advances regarding the regulation, assembly, and function of the pili is provided.
3.2.1 Transcription of the pili genes
DNA microarray experiments have shown that the genes required for pilus biosynthesis are maximally transcribed just prior to the assembly of the pili at the flagellar pole of the swarmer cell (Laub et al., 2000). The genes encoding the pilus secretion machinery, cpaBCDEF, are transcribed and then the cpaA gene that encodes the prepilin peptidase is transcribed. Transcription of the pilA gene, which encodes the major protein subunit in the pilus filament, occurs last. The transcription of pilA is under the control of CtrA, linking pilus biogenesis to cell cycle progression (Skerker and Shapiro, 2000). When the transcription of pilA is under the control of a constitutive promoter, pili are assembled prematurely in predivisional cells. This observation indicates that the temporal regulation of pilA transcription by CtrA prevents premature pilus biogenesis.
3.2.2 Assembly of the pili
How are the pili assembled at the proper time and place? PodJL binds to the future site of pilus biogenesis and serves as a localization factor for the pilus assembly protein, CpaE (Viollier et al., 2002b). The presence of CpaE at the incipient swarmer cell pole triggers the localization of the pilus secretion channel, CpaC, to the same pole (Viollier et al., 2002a). The localization of CpaC is required for pore formation and subsequent pilus assembly. The formation of the CpaC secretion channel depends on the presence of PleA, a transglycosylase, indicating that localized peptidoglycan hydrolysis is required for the formation of the channel (Viollier and Shapiro, 2003). This is consistent with the observation that pleA mutants lack pili. Following cell division, PilA accumulates and is polymerized to form the filament, which is secreted through the CpaC pore, thus completing pilus biogenesis (Skerker and Shapiro, 2000; Viollier et al., 2002a).
3.2.3 Function of the pili
The Flp pili enhance the initial binding of C. crescentus cells to a surface (Fig. 5; (Bodenmiller et al., 2004; Entcheva-Dimitrov and Spormann, 2004). Although, retraction of Flp pili has never been demonstrated in any bacterium, it is presumed that the C. crescentus pili retract since pili or pilin does not accumulate in the media following loss of pili from the cell surface (Lagenaur and Agabian, 1977). Further support for pili retraction is provided by the observation that shortly after the addition of phage, the phage can be visualized along the pili by TEM; however when more time is allowed, the phage are observed only at the pole, suggesting that the pili have retracted (Skerker and Shapiro, 2000). As the swarmer cell differentiates into a stalked cell, retraction of the pili is followed by the synthesis of the stalk with the adhesive holdfast at the tip. It has been proposed that the initial attachment mediated by the pili may properly position the cell and allow a more permanent surface attachment following contact between the holdfast and the surface (Bodenmiller et al., 2004; Janakiraman and Brun, 1999).
The pili are also involved in the maintenance of complex biofilms (Bodenmiller et al., 2004; Entcheva-Dimitrov and Spormann, 2004). On coverslips, cpaA mutants fail to form dense monolayer biofilms compared to wild-type cells after overnight incubation (Bodenmiller et al., 2004). Under hydrodynamic conditions, a deletion mutant lacking genes for pilus assembly and for the pilin subunits forms loose microcolonies, but does not develop the dense mushroom-like structures formed by wild-type cells within 96 hours (Entcheva-Dimitrov and Spormann, 2004). These results suggest that pili may function in mediating cell-to-cell contact and function in biofilm maturation.
3.3 The Stalk
The synthesis of a polar stalk as the swarmer cells differentiates to form a stalked cell results in a striking change in cellular morphology. Following flagellum ejection, stalk biosynthesis occurs at the same pole, culminating in the extension of a stalk one-fifth the diameter of the cell body. The synthesis of the stalk is temporally and spatially constrained, indicating that this process must be regulated throughout the cell cycle. In this section, the structure, biosynthesis, regulation and function of the stalks of C. crescentus will be discussed.
3.3.1 Structure of the stalk
The stalk is a true extension of the cell envelope, containing the inner and outer membranes, peptidoglycan, and periplasmic space, but is seemingly devoid of DNA, ribosomes, and cytoplasmic material (Ireland et al., 2002; Poindexter and Bazire, 1964; Wagner et al., 2006). The stalk contains crossbands, intermittent electron dense rings perpendicular to the long axis, which appear to be synthesized once during the cell cycle, and can be used to determine the age of a stalked cell (Poindexter and Staley, 1996). Crossbands have been proposed to compartmentalize or stabilize the stalk (Jones and Schmidt, 1973).
3.3.2 Stalk biosynthesis
Stalk elongation occurs as new cell envelope material is incorporated at the junction between the cell body and the stalk (Aaron et al., 2007; Schmidt and Stanier, 1966; Seitz and Brun, 1998; Smit and Agabian, 1982). The biosynthesis and elongation of the stalk is dependent on at least three proteins, which are also required for elongation of the cell body. Depletion or inactivation of penicillin binding protein 2 (PBP2), RodA, or MreB leads to defects in both cell body elongation and stalk elongation (Divakaruni et al., 2007; Seitz and Brun, 1998; Wagner et al., 2005). RodA or MreB depletion generates bulbous or short wide stalks at the proper pole (see section 4.1.1; Fig. 8; (Wagner et al., 2005)). Cells depleted, and then repleted for RodA and MreB have multiple mislocalized stalks indicating that the presence of ectopic poles (Wagner et al., 2005). Similar results were obtained when cells were briefly exposed to the PBP2 inhibitor mecillinam. The phenotype of multiple mislocalized stalks demonstrates that PBP2, RodA and MreB contribute to cell polarity and stalk localization (Wagner et al., 2005).
Figure 8.
The C. crescentus origin of replication. The region required for chromosome replication is shown as a thick black line and is flanked by the hemE and RP001 genes which are depicted as grey arrows that indicate polarity. The black arrows indicate RNA start sites for the transcription promoters, PW, PS, and P3. The AT and purine (AG) rich regions required for chromosome replication are shown as labeled thin solid bars. The binding sites of IHF and DnaA are shown in solid boxes. The CtrA binding sites are shown in dashed boxes and are labeled with the site designation, a-e.
The FtsH protease is also required for stalk elongation, but is not required for cell body elongation (Fischer et al., 2002). Depletion of ftsH leads to filamentous cells with either no stalk or with short stalks at the proper pole. Some of the short stalks contain up to four adjacent crossbands indicating a defect in stalk elongation. The role of FtsH in stalk elongation has not been elucidated; however, FtsH does not appear to modulate the activity σ54. One possibility is that FtsH may control the activity of one or more of the σ54 activators. The observation that depletion of FtsH results in filamentous cells indicates that FtsH directly or indirectly contributes to cell division. Proper stalk biogenesis also requires the cell division protein FtsZ, which is responsible for the formation of crossbands in the stalk ((Divakaruni et al., 2007); see section 4.2.2 for a description of FtsZ). FtsZ controls peptidoglycan at the division site; however, the requirement of FtsZ for crossband synthesis indicates that FtsZ may be required for multiple forms of peptidoglycan synthesis. FtsH and FtsZ impact both stalk biosynthesis and cell division indicating that the two processes may be coupled.
Since the stalk is found only at a specific location and is considerably narrower than the cell body, there must be stalk-specific proteins or mechanisms that determine the location and size of the stalk. Thus far, all mutations that impact stalk biosynthesis are pleiotropic in nature and often also impact cell division. Identification and characterization of proteins that are required for stalk biosynthesis specifically will illuminate the processes which allow a local extension of the cell membrane. Since stalks are not required for cell survival, the study of stalk biosynthesis is an extremely tractable experimental system. The study of stalk biosynthesis is expected to reveal basic principles that apply to other morphological changes (Wagner and Brun, 2007).
3.3.3 Regulation of biogenesis and stalk length
Mutations in a number of regulators involved in cell-cycle progression, including CtrA, CckA, PleC, PleD, σ54, and TacA, result in defects in stalk biogenesis (Aldridge and Jenal, 1999; Biondi et al., 2006b; Brun and Shapiro, 1992; Jacobs et al., 2003; Jacobs et al., 1999; Quon et al., 1996; Wang et al., 1993). The exact role of CckA and PleC in mediating stalk biosynthesis is unclear. One possibility is that the stalk defect of CckA and PleC is indirect and related to the role of these proteins in modulating CtrA activity, which is responsible for coupling stalk biogenesis to cell cycle progression. CtrA is responsible for the precisely timed transcription of rpoN and tacA (Laub et al., 2002; Laub et al., 2000). TacA is activated by phosphorylation and then binds to enhancer sequences, enabling σ54-dependent gene expression for stalk biogenesis (Biondi et al., 2006b). A phosphorelay cascade involving the hybrid histidine kinase, ShkA, and the Hpt protein, ShpA, mediates the activation of TacA. The signal responsible for initiating the ShkA-ShpA-TacA phosphorelay remains unknown.
Microarray analysis identified 30 candidate genes downstream of TacA (Biondi et al., 2006b). One of the TacA- σ54 targets is predicted to encode StaR, a putative transcriptional factor of the Cro/CI family, suggesting that a transcriptional cascade may control stalk biosynthesis (Biondi et al., 2006b). Although deletion of staR does not eliminate stalks, the stalks are considerably shorter than wild type. Determination of which genes are regulated by StaR may lead to increased understanding of how stalk length is regulated. Since a tacA deletion generates stalkless cells and a staR deletion does not, it is likely that additional downstream targets of TacA- σ54 contribute to the control of stalk biosynthesis.
While stalk biogenesis seems to be controlled largely by cues mediating cell cycle progression, stalk length is known to be impacted by the extracellular environment of the cell. In the natural, nutrient-depleted environment of C. crescentus, the stalks are remarkably long, up to 20 times the length of the cell body (Brun and Janakiraman, 2000; Poindexter, 1964). Limitation of organic phosphate or inactivation of the genes required for phosphate uptake leads to a dramatic increase in the rate of stalk elongation (Gonin et al., 2000; Schmidt and Stanier, 1966). By analogy with work in E. coli (Wanner, 1996), a model for the role of the Pho regulon in phosphate uptake and stalk elongation of C. crescentus has been proposed (Gonin et al., 2000). The PstSCAB proteins comprise a high affinity phosphate transporter that controls the autophosphorylation of the histidine kinase, PhoR. When phosphate is in excess, PhoR remains unphosphorylated. Under phosphate limiting conditions, PhoR is released from the Pst complex, autophosphorylates, and then transfers the phosphoryl group to PhoB, its cognate response regulator. Phosphorylated PhoB then directly or indirectly activates the expression of genes required for stalk elongation.
3.3.4 Function of the stalk
A number of possible functions have been attributed to the stalk of C. crescentus, including adhesion and nutrient acquisition. The localization of the adhesive holdfast to the distal tip of the stalk is often taken as an indication that the stalk may play a role in adhesion. Several lines of evidence indicate that there is an adhesion-independent function for the stalk. First, stalk and holdfast synthesis are not coupled. Stalk synthesis occurs after holdfast synthesis and does not depend on the presence of the holdfast (Bodenmiller et al., 2004; Levi and Jenal, 2006). Similarly, holdfast synthesis does not depend on stalk synthesis (Janakiraman and Brun, 1999). Second, in closely related organisms such as Asticaccaulis excentricus and A. biprosthecum, the holdfast is associated with the cell body and not the stalk (Poindexter, 1964). Lastly, the swarmer cell, which lacks a stalk, initiates the adhesion process (see figure 5; (Bodenmiller et al., 2004; Levi and Jenal, 2006). These observations suggest that there is a selective advantage for stalk synthesis, independent of adhesion.
The observation that stalks are considerably longer in nutrient depleted environments suggests that stalk elongation may provide a means to increase cellular surface area and nutrient uptake ability (Gonin et al., 2000; Poindexter, 1964). A number of recent results have provided evidence in support of a role for the stalk in nutrient uptake. First, in diffusion-limited environments, like the natural oligotrophic environment where C. crescentus lives, the rate of nutrient uptake should be proportional to the length of the cell, rather than to the surface area of the cell (Wagner et al., 2006). Thus, the elongation of the stalk rather than the entire cell body is likely to be bioenergetically favorable because synthesizing a long thin cell envelope extension requires less energy than elongating the cell by the same length. Second, stalks purified from C. crescentus are capable of transporting and hydrolyzing an organic phosphate-containing molecule (Wagner et al., 2006). Isolated stalks of A. biprosthecum are also able to transport glucose and all 20 amino acids (Larson and Pate, 1976; Tam and Pate, 1985). Thirdly, the predicted functions of the proteins associated with the stalk suggest a role for the stalk in nutrient uptake (Ireland et al., 2002; Wagner et al., 2006). A large number of outer membrane transporters and periplasmic binding proteins for a variety of nutrients, including phosphate, are found in the stalk (Ireland et al., 2002; Wagner et al., 2006). Each of these observations provides support for a model that the stalk of C. crescentus is a morphological adaptation that enables survival in an oligotrophic environment (for review see (Wagner and Brun, 2007)).
3.4 The Process of Adherence and the Holdfast
Many bacteria possess the ability to adhere to surfaces. What are the potential advantages of adherence? It may be bioenergetically favorable for bacteria to attach to a surface and absorb passing nutrients rather than to actively swim in pursuit of nutrients. This may be because nutrients as well as the enzymes the bacteria need to assimilate the nutrients tend to be concentrated near solid surfaces in a dilute aqueous environment (ZoBell, 1937). In addition, the association of the bacteria with larger extracellular structures, such as other organisms or surfaces of ships and pipes, may provide a physical barrier and prevent predation (Costerton et al., 1987). Adhesion mediates the formation of biofilms, in which bacteria are more resistant to antibiotics and are better protected from the environment, including changes in pH, desiccation, and shear forces in an aqueous environment (Fux et al., 2005; Jefferson, 2004; Stoodley et al., 2002).
C. crescentus, like many bacterial organisms, can vigorously adhere to both biotic and abiotic surfaces (Poindexter, 1964). While the pili and the flagellum facilitate the initial process of adherence and biofilm maturation, it is the holdfast, a polarly secreted adhesin comprised of both proteins and polysaccharides, which mediates a more permanent attachment (Fig. 5; see sections 3.1.5 and 3.2.5). C. crescentus cells are often associated with each other in large groups exclusively via their holdfasts, a structure that is referred to as a rosette. During early studies, C. crescentus was isolated based on its ability to adhere to glass slides and an extracellular structure at the tip of the stalk, termed holdfast, was hypothesized to be responsible for this attachment (Henrici and Johnson, 1935; Poindexter and Bazire, 1964). Identification and characterization of several groups of holdfast mutants determined that the holdfast contains acidic polysaccharides (Umbreit and Pate, 1978). Synthesis and export of the C. crescentus holdfast is cell cycle regulated (Janakiraman and Brun, 1999). Here we will discuss the process of adherence in C. crescentus and the role of the holdfast. We will also examine the properties and regulation of the holdfast, as well as the holdfasts of other prosthecate bacteria.
3.4.1 Role of polar structures in adherence and biofilm formation
For C. crescentus, several components are necessary for the initial stages of attachment, including flagella, pili and motility (Fig. 5)(Bodenmiller et al., 2004; Levi and Jenal, 2006). To initiate an association with a surface, the cell must overcome repulsive forces, possibly including hydrophobic and electrostatic forces (Marshall, 1972; ZoBell, 1943). Freshwater bacteria, such as C. crescentus, may have a relatively hydrophobic cell surface and therefore the aqueous phase at the water-solid interface may serve as a barrier to attachment (Marshall, 1972). Both the bacterial cell surface and most attachment surfaces carry a net negative charge leading to repulsion due to the electrostatic forces. Brownian motion is not sufficient to overcome these forces (Marshall, 1972); however motility via the flagellum likely aids in overcoming the repulsive forces. The initial attachment may be facilitated directly by both the flagella and pili, which help to orient the cell properly for more permanent adherence that is only achieved with the presence of the holdfast (Bodenmiller et al., 2004; Merker and Smit, 1988). After the early stages of attachment, C. crescentus can create a more complex biofilm structure (Entcheva-Dimitrov and Spormann, 2004; Smit et al., 2000). The biofilm begins with microcolonies that develop into mushroom shaped structures and a monolayer biofilm forms between these structures. The holdfast is the most important structure for proper biofilm formation although the pili and flagella were also found to be critical for the proper development and maintenance of the mushroom structures within the biofilm (Entcheva-Dimitrov and Spormann, 2004).
3.4.2 Holdfast composition and properties
The C. crescentus holdfast is comprised of both protein and polysaccharide. N-acetyl glucosamine (NAG) was identified as a component of the holdfast polysaccharide based on the ability of the holdfast to bind wheat germ agglutinin (WGA) lectin and on the sensitivity of the holdfast to chitinase and lysozyme, which are specific for NAG polymers (Merker and Smit, 1988). Lysozyme and chitinase cleave the β1-4 linkages of between N-acetyl-D-glucosamine residues in polysaccharides, indicating that some of the linkages in the C. crescentus holdfast are β1-4 linkages. NAG polymers that function as adhesins have been identified in other bacteria including E. coli (PGA) (Wang et al., 2004) and Staphylococcus sp. (PIA) (Cramton et al., 1999; Mack et al., 1996). The PGA and PIA adhesins of E. coli and Staphylococcus sp. are linear polymers of β1-6 NAG (Wang, 2004). PIA and PGA are important for biofilm formation and ultrastructure and the PIA adhesin is also important for the pathogenicity of Staphylococcus sp. through protection from the innate immune response (Cramton et al., 1999; Götz, 2002; Vuong et al., 2004; Wang et al., 2004). In addition to NAG, holdfast associated proteins may play an integral role in adherence since one group of proteins associated with the holdfast, the holdfast attachment proteins (HfaA, HfaB, and HfaD, see section 3.4.3b), have similarity to the curli proteins of E. coli, which are involved in bacterial attachment.
Recent biophysical analysis of the holdfast has shown that it is an elastic, gel-like substance, with impressive adhesive properties. Initial indications that the holdfast had strong adhesive properties came from the failure of laser tweezers with a maximum working force on the order of 10 pN to detach single C. crescentus cells from a glass surface (Tsang et al., 2006). In order to determine the force of adhesion of single C. crescentus cells, a micromanipulation method was developed. Cells were allowed to attach to thin flexible pipettes whose force constants had been determined by atomic force microscopy (AFM). A suction pipette was used to grab the cell body and pull it in a direction perpendicular from the flexible pipette. The force required to break the cell-to-pipette contact was determined by measuring the displacement of the flexible pipette at the time the contact was broken. The force of adhesion of individual cells ranged from 0.11 μN to 2.26 μN with an average of 0.59 μN (Tsang et al., 2006). The large variation in force of adhesion is thought to be due to the variation in the size of holdfasts, the different breaking points, and the angle of pulling. Cells attached to a glass surface were subjected to a strong jet of water and examination of a glass surface by AFM following cell detachment indicated that the contact between the cells and the surface broke most often within the stalk or at the stalk-to-holdfast junction. Since the holdfast-to-surface junction remained intact, the force calculated for single cell adhesion is an underestimate of the force of holdfast adhesion. The holdfast spreads over a larger area (∼411 nm) than the diameter of the stalk tip (∼119 nm), therefore the stress endured by the holdfast was calculated using finite element analysis, an engineering method used to determine stress in mechanical systems. This analysis revealed that the maximum stress at the holdfast-surface interface was 68 N/mm2, indicating that holdfast covering a 1 cm2 surface could theoretically hold a weight of ∼700 kg, making the holdfast the strongest biological adhesin described thus far (Tsang et al., 2006). Interestingly, such a force is in the right range to resist the force caused by the passage of an air-liquid interface, which can be up to 200 nN for a microscopic object. C. crescentus cells may be subjected to such forces when they are on surfaces that are hit by waves in an aquatic environment.
Individual C. crescentus cells adhered to a surface are constrained by both the stalk and the holdfast. Microscopic observation of attached cells in liquid medium shows that the attached cells are subject to Brownian motion that slightly displaces the cells, resulting in a deformation of the stalk and/or holdfast. This deformation causes a restoring force, which returns the cells to their original equilibrium position. Mathematical analysis of this behavior has been used to study the biophysical properties of the holdfast (Alipour-Assiabi et al., 2006; Li et al., 2005). After displacement of a cell by Brownian motion, the restoring force would be expected to result from the properties of both the holdfast and the stalk. The restoring force was determined by measuring the displacement of individual cells using optical microscopy and the force constant of the holdfast and stalk assembly was determined by fitting the displacement distribution to a modification of the Boltzmann equation, which typically describes the distribution of particles in a fluid (Li et al., 2005). If deformation of the stalk contributes significantly to the elastic nature of the stalk-holdfast assembly, the force constant would depend on stalk length since the ease of bending an object increases with its length. The analysis showed no correlation between the force constant and stalk length, indicating that the stalk is stiff as compared to the holdfast and that the force constant is a measure of the elasticity of the holdfast. A similar fluctuation analysis of the displacement of attached cells using a higher frequency of image recording was used to study pairs of cells attached to a surface by a shared holdfast (Alipour-Assiabi et al., 2006). This analysis showed that in some pairs, the elastic coupling between the two cells was stronger than their respective coupling to the surface, suggesting that the two cells attached to each other at the swarmer cell stage and subsequent holdfast synthesis allowed a strong crosslinking of the two cells, perhaps mediated by their respective Hfa proteins (see section 4.3.3b). In other pairs, one cell was tightly attached to the surface, whereas the other cell was coupled more tightly to the first cell than to the surface, suggesting that the cells attached to each other after holdfast synthesis, resulting in a less efficient melding of the two holdfasts.
Digestion of the holdfast with lysozyme the force constant of attached cells, indicating that the NAG polymer is important for the elastic properties of the holdfast (Li et al., 2005). Furthermore, lysozyme digestion drastically reduced the force of adhesion of the holdfast, such that cells were aspired by the suction pipette without causing a displacement of the flexible pipette in cell pulling experiments (Tsang et al., 2006). Notably, the adhesion force was still sufficient to resist detachment using laser tweezers with a force of 10 pN, suggesting that components other than NAG polymers also contribute to the force of adhesion of the holdfast.
3.4.3 Genetics of holdfast synthesis and attachment in C. crescentus
Several loci involved in the biosynthesis, regulation and anchoring of the holdfast in C. crescentus have been identified. Four classes of holdfast mutants have been characterized by transposon and UV mutagenesis: I) cells that make no holdfast, II) cells that make reduced levels of holdfast, III) cells that make holdfast that does not remain attached to the cell, and IV) cells with pleiotropic mutations that result in holdfast and polar development defects (Mitchell and Smit, 1990; Ong et al., 1990; Smith et al., 2003).
3.4.3a Holdfast secretion and biosynthesis loci
The mutations in the class I and II holdfast mutants that produce no holdfast or reduced amounts of holdfast, respectively, reside within the holdfast export and biosynthesis locus. Holdfast export is predicted to be accomplished by the products of three adjacent genes, hfsD (CC2432), hfsA (CC2431), and hfsB (CC2430) (Smith et al., 2003). The genes involved in holdfast polysaccharide biosynthesis are adjacent to those involved in holdfast export and are comprised of hfsC (CC2429), hfsE, hfsF, hfsG, and hfsH (CC2425-28) (Fig. 6B; E. Toh, H. Kurtz and Y. Brun, in preparation). hfsA, hfsB and hfsC are transcribed convergently with the predicted biosynthesis genes, while hfsD is transcribed divergently from hfsA, hfsB and hfsC (Fig. 6B) (Smith et al., 2003).
Figure 6.
Organization of the hfa and hfs loci in C. crescentus, Caulobacter sp. K31, M. maris and O. alexandrii. Each arrow represents a gene and the direction of transcription for that gene. Cc, C. crescentus CB15; K31, Caulobacter sp. K31; Mm, M. maris MCS10; Oa, Oceanicaulis alexandrii HTCC2633 (A) Comparison of the holdfast attachment loci for C. crescentus, M. maris and O. alexandrii. The holdfast attachment locus is composed of hfaA, hfaB, hfaD and hfaC. The corresponding homologs for hfaA (black), hfaB (dark grey) and hfaD (light grey) in Caulobacter sp. K31, M. maris and O. alexandrii are shown below the hfa locus for C. crescentus. The gene numbers are indicated in the arrow representing the gene for Caulobacter sp. K31, M. maris and O. alexandrii. Predicted open reading frames surrounding the hfa loci are represented with white arrows. There is a conserved rhomboid protease (rhom) upstream of many of the hfa loci. M. maris has two hfa loci. HfaC is not present in any of the other hfa loci. (B) Comparison of the holdfast biosynthesis loci from C. crescentus, Caulobacter sp. K31, M. maris and O. alexandrii. The holdfast biosynthesis and transport loci are comprised of hfsE, hfsF, hfsG, hfsH and then hfsD, hfsA, hfsB and hfsC. The gene numbers for each gene are indicated in the arrow representing the gene for Caulobacter sp. K31, M. maris and O. alexandrii. Both M. maris and O. alexandrii have additional genes that may function in holdfast biosynthesis, which are shown as white arrows. Caulobacter sp. K31 has additional genes within the holdfast biosynthesis and secretion loci (shown in white), but their suggested functions do not appear to be related to polysaccharide biosynthesis. gt indicates a predicted glycosyl transferase; mt, is a predicted mannosyl transferase; gh, a predicted glycosyl hydrolase; celD indicates a gene involved in cellulose biosynthesis. The gene function assignments are based on the annotations of TIGR or JGI and blastp analysis. O. alexandrii has a homolog to hfsE that is located elsewhere in the genome as indicated by the gene number.
Deletions of components of the holdfast export locus, hfsD, hfsA or hfsB, result in complete loss of holdfast polysaccharide synthesis. Based on sequence homology, holdfast polysaccharide export likely occurs by a wzy-dependent mechanism similar to group I capsule biosynthesis of E. coli (Fig. 7) (Whitfield, 2006). HfsD is a 25-kDa lipoprotein (Smith and Brun, unpublished) with similarity to the Wza secretin of E. coli, an octomeric outer membrane lipoprotein required for Type I capsule polysaccharide secretion (Drummelsmith and Whitfield, 2000). In the type I capsule export system of E. coli, Wza and Wzc form a complex that spans the inner membrane, the periplasm, and the outer membranes (Collins et al., 2007). HfsA is a predicted 55-kDa protein with similarity to Wzc, a member of the membrane periplasmic auxiliary (MPA-1) family of polysaccharide transport proteins (Paulsen et al., 1997; Smith et al., 2003). In E. coli, Wzc functions as a tetramer (Collins et al., 2007). The Wzc proteins of Gram-negative bacteria typically have a cytoplasmic region that contains two ATP binding motifs (Walker A and B)(Whitfield and Paiment, 2003). HfsA does not have either motif; however, HfsB is a predicted 25-kDa protein that contains putative Walker A and Walker B motifs and has some amino acid similarity to Wzc protein homologs in both Gram-positive and Gram-negative bacteria (Smith et al., 2003)(J. Javens and Y. Brun, unpublished). Interestingly, in Gram-positive bacteria, Wzc homologs involved in capsule translocation are comprised of two proteins: the periplasmic membrane translocator, similar to HfsA, and a separate cytoplasmic protein with an ATP binding motif, similar to HfsB (Cozzone et al., 2004; Paulsen et al., 1997). Therefore, it is possible that the C. crescentus Wzc homologues, HfsA and HfsB, function similarly to those of Gram-positive bacteria.
Figure 7.
Model of holdfast biosynthesis and attachment in C. crescentus. HfsE transfers N-acetylglucosamine (NAG) from UDP-NAG to undecaprenol (black oval). HfsG transfers NAG to the first sugar on the undecaprenol. HfsH deacetylates some of the NAG on the growing polysaccharide chain to glucosamine. HfsF is a flippase and transfers the small saccharide repeat unit linked to undecaprenol across the cytoplasmic membrane. HfsC and HfsI are polymerases that link the saccharide repeat units together to create the holdfast polysaccharide. HfsA, HfsB and HfsD translocate the holdfast polysaccharide across the outer membrane so it can be anchored to the cell surface via the holdfast attachment proteins. An HfsA tetramer and HfsD octomer create the translocator and HfsB is an ATPase important for phosphorylation and signaling. For the holdfast attachment proteins, HfaA and HfaD are translocated via the Sec system across the cytoplasmic membrane to the periplasm where the signal sequence is cleaved by signal peptidase (SP). After processing, HfaA and HfaD interact with HfaB, the outer membrane secretin. HfaB translocates HfaA and HfaB across the outer membrane where they create the holdfast anchor and associate with the holdfast polysaccharide (only drawn for HfaA) by an unknown mechanism. Homologs to each of the Hfs and Hfa proteins are shown in parentheses adjacent to each protein. UDP, uridine diphosphate; P, phosphate; dark gray hexagon, NAG; light gray hexagon, glucosamine; IM, inner membrane; OM, outer membrane; CW, cell wall; SS, signal sequence.
The holdfast biosynthesis genes are comprised of hfsC hfsE, hfsF, hfsG, and hfsH (CC2425-29) (Fig. 6B; E. Toh, H. Kurtz and Y. Brun, in preparation). HfsC is a predicted 46-kDa protein with similarity to ExoQ from Sinorhizobium meliloti, a member of the Wzy protein family of polysaccharide polymerases (Smith et al., 2003) (Collins et al., 2007). However, mutational analysis indicates that an hfsC deletion mutant is not deficient in holdfast synthesis or surface adhesion (Smith et al., 2003). Recently, a paralog of hfsC, hfsI, was identified in C. crescentusindicating that HfsC and HfsI may work in concert to polymerize the holdfast polysaccharide (E. Toh, H. Kurtz, and Y. Brun, in preparation). This situation would be analogous to lipopolysaccharide biosynthesis in Pseudomonas aeruginosa, which involves two O-antigen polymerases, Wzyα and Wzyβ {Kaluzny, 2007 #13310}.
HfsE, a predicted integral membrane protein, exhibits significant homology to initiating glycosyltransferases that catalyze the first step in polysaccharide biosynthesis. HfsF has similarity to polysaccharide flippases. HfsG has similarity to the group 2 family of glycosyltransferases and HfsH has similarity to polysaccharide deacetylases. Interestingly, the NAG of the PGA and PIA adhesins of E. coli and Staphylococcus are deacetylated to varying degrees (Mack et al., 1996; Wang et al., 2004). The exact polysaccharide content and structure of the C. crescentus holdfast and the role of deacetylation in adherence have yet to be determined.
One possible model for polysaccharide biosynthesis based on the above gene function similarities is shown in Fig. 7 (E. Toh, H. Kurtz and Y. Brun, in preparation). HfsE transfers NAG or possibly other sugars onto the undecaprenol phosphate lipid carrier in the inner membrane. HfsG transfers additional sugars to the lipid intermediate undecaprenol phosphate and serves to lengthen the repeat unit. HfsH removes acetyl groups from some of the sugars in the polysaccharide repeat unit attached to undecaprenol phosphate. HfsF, the putative flippase, translocates the lipid-linked polysaccharide units across the inner membrane of the cell. HfsI and HfsC assemble the polysaccharide chain from the repeat units transferred by HfsF. The polysaccharide oligomers are then translocated outside the cell via the holdfast export proteins: HfsD, HfsA, and HfsB as described above.
3.4.3b Holdfast attachment locus
The class III holdfast mutants, which produce holdfasts that do not remain cell associated, are known as holdfast shedding mutants. The mutations responsible for the holdfast shedding phenotype have been mapped to three genes in the holdfast attachment (hfa) locus: hfaA (CC2628), hfaB (CC2629), and hfaD (CC2630) (Fig. 6A) (Cole et al., 2003; Kurtz and Smit, 1992; Kurtz and Smit, 1994; Smith et al., 2003). HfaA is a 12-kDa protein with limited amino acid similarity (28%) to the E. coli curlin monomer CsgA and other fimbrial family adhesins (Cole et al., 2003) (G. Hardy and Y. Brun, in preparation). CsgA is a bacterial amyloid-like protein that is the structural component of the curli fibrils involved in E. coli attachment to surfaces (Olsén et al., 1989; Prigent-Combaret et al., 2000). HfaB is a 36-kDa lipoprotein with amino acid similarity (41%) to the E. coli curli secretin CsgG, which is a lipoprotein required for the secretion of the curli monomer CsgA and the nucleator protein CsgB (Robinson et al., 2006). HfaD is a 41-kD membrane protein with low-level amino acid similarity (20% to 40%) to other adhesins and collagen triple helix proteins. The hfaC gene (CC2631) is found downstream of hfaD and encodes a predicted 66-kDa protein with similarity to ABC transporters. hfaC was initially thought to be involved in holdfast attachment based on the erroneous mapping of an hfaD transposon insertion to hfaC (Kurtz and Smit, 1994); however, mutations in hfaC do not affect holdfast anchoring or cell adhesion (Cole et al., 2003). In addition, none of the other prosthecate genomes examined in this review have homologs of HfaC associated with putative holdfast attachment loci (Fig. 6A). Taken together, this suggests that HfaC is not involved in anchoring the holdfast.
Insertions within hfaA, hfaB or hfaD result in different holdfast shedding phenotypes, reduced adherence and decreased WGA lectin binding. Insertions within hfaB are the most severe resulting in very low levels of lectin binding (5%, as compared to 75% for the parental strain CB15), low levels of adherence, no rosette formation, and high levels of holdfast shedding. hfaA insertions result in moderate levels of lectin binding (30%) and adherence, reduced levels of rosette formation, and some holdfast shedding. hfaD insertions result in moderate levels of lectin binding (43%) and adherence, rosette formation is comparable to wild-type cells, and there is a high level of holdfast shedding (Cole et al., 2003).
HfaA, HfaB, and HfaD have a predicted signal sequence suggesting that they are translocated across the cytoplasmic membrane. HfaB and HfaD are associated with the membrane fraction of C. crescentus and examination of isolated stalks and cell bodies indicates that HfaB and HfaD reside in the stalk of the cell (Cole et al., 2003). One possible model for an Hfa-mediated holdfast anchor is that HfaB is an outer membrane secretin that translocates HfaA and HfaD across the outer membrane where they form a complex that interacts with the holdfast polysaccharide to anchor it to the cell (Fig. 7). If HfaB translocates HfaA and HfaD, then deletion of hfaB would prevent their translocation to the cell surface, possibly explaining why an hfaB mutant has the most severe phenotype. Deletion of either hfaA or hfaD result in moderate phenotypes, so HfaA and HfaD may contribute equally to anchoring the holdfast. HfaA and HfaD could associate with each other in several combinations: 1) HfaD could be a nucleator protein and HfaA could be a fibrillar adhesin that mediates most of the association with the polysaccharide, 2) HfaA and HfaD could form independent adhesin structures in close proximity on the cell surface or 3) HfaD could act as the main adhesin and HfaA could be a tip adhesin.
3.4.4 Regulation of holdfast synthesis
The holdfast is synthesized during specific times in the developmental cycle. The holdfast is synthesized in the mid to late swarmer stage at the flagellar pole and throughout swarmer cell differentiation, and then remains at the tip of the stalk for the rest of the cell cycle (Levi and Jenal, 2006). Of the three holdfast loci that have been identified, only the cell cycle regulation of the hfa locus has been examined directly (Janakiraman and Brun, 1999). Transcription of the hfa operon begins in the swarmer compartment of the predivisional cell and then decreases during swarmer to stalk cell differentiation (Janakiraman and Brun, 1999). The promoter region of the hfa operon contains both a putative σ54 promoter and CtrA binding sites. While σ54 is not required for transcription of the hfa operon, deletion of rpoN, the gene encoding σ54, results in increased transcription of the hfa operon suggesting that either σ54 or a σ54 regulated gene product negatively regulates the hfa operon (Janakiraman and Brun, 1999). Transcription of the hfa operon is decreased in a ctrA mutant suggesting that CtrA acts as a transcriptional activator for this operon (Laub et al., 2002).
The fourth class of holdfast mutants is pleiotropic resulting in a variety of phenotypes in addition to the loss of holdfast (Merker and Smit, 1988; Smith et al., 2003; Yun et al., 1994). The class IV mutations were mapped to podJ and pleC (Smith et al., 2003). Mutation of podJ and pleC (see sections 2.4.1c and b) both result in loss of holdfast production and severely reduced binding to surfaces (Hinz et al., 2003; Smith et al., 2003). Functional domain analysis of PodJ indicates that the cytoplasmic region between amino acids 589 and 639 is important for holdfast biosynthesis (Lawler et al., 2006). PleD (see section 2.4.1b) has been implicated in the regulation of holdfast synthesis. Deletion of pleD results in a 70% reduction in C. crescentus adherence to polystyrene (Levi and Jenal, 2006); however, adherence levels and holdfast production are only affected during the swarmer to stalk cell differentiation. The reduced adherence of a pleD mutant during the swarmer cell differentiation results from a delay in holdfast biosynthesis. This result suggests that PleD is necessary for the appropriate timing of holdfast biosynthesis during cell cycle development and possibly in response to environmental signals (Levi and Jenal, 2006). pleD mutants also remain motile after differentiation of swarmer cells (Sommer and Newton, 1989) indicating that synthesis and degradation of c-di-GMP by PleD probably regulates the transition between motility and cell adhesion.
Motility and attachment are divergently regulated using c-di-GMP in Salmonella and E. coli. In Salmonella enterica serovar Typhimurium and E. coli, GGDEF domain proteins, such as AdrA, increase cellulose biosynthesis, an extracellular polysaccharide (1-4-β-glucose polymer) important in biofilm formation and curli biosynthesis (Kader et al., 2006; Simm et al., 2004; Zogaj et al., 2001). Increased levels of c-di-GMP also result in shedding of the flagella, and expression of adhesins (Jenal and Malone, 2006; Romling, 2005; Simm et al., 2004; Tamayo et al., 2007). In contrast, YhjH, an EAL phosphodiesterase domain protein, results in decreased levels of c-di-GMP and inactivation of flagella (Simm et al., 2004). Similarly, in C. crescentus, the phosphodiesterase activity associated with the TipF EAL domain decreases the level of c-di-GMP and results in flagellar biogenesis (Huitema et al., 2006), while increased diguanylate cyclase activity mediated by PleD results in flagellar shedding and activation of holdfast biosynthesis (Jenal and Malone, 2006; Levi and Jenal, 2006; Paul et al., 2004). Together, PleD and TipF regulate timing of expression of polar structures and holdfast elaboration to mediate permanent attachment (Fig. 4). The signals for the switch between motile and sessile lifestyles have not been identified, but may be mediated by the initial association with surfaces (Jenal and Malone, 2006). C. crescentus has numerous GGDEF and EAL containing proteins that could be involved in the regulation of the transition between a planktonic lifestyle and a sessile state associated with biofilms, which could be regulated by a variety of cellular and extracellular signals. In addition, a c-di-GMP receptor protein, DgrA (CC1599), as well as its paralog, DgrB (CC3165), are involved in the control of motility (Christen et al., 2007). Both DgrA and DgrB or other c-di-GMP receptor proteins in C. crescentus may be important for the control of holdfast production through signal transduction based on the ability to sense the concentrations of c-di-GMP and affect holdfast production by repressing or activating holdfast related genes.
C. crescentus attachment is also regulated by a photosensory two-component system, LovK and LovR (Purcell et al., 2007). These proteins contain an LOV (light, oxygen or voltage) domain that regulates blue-light dependent processes and many bacterial proteins containing these domains have been identified in bacteria (Crosson et al., 2003). LovK is a histidine-protein kinase and LovR is a single-domain response regulator (Purcell et al., 2007). Deletion of lovK (CC0285) or lovR (CC0284) results in decreased adherence in C. crescentus, and overexpression of LovK and LovR results in increased rosette formation indicating that LovK and LovR are involved in regulation of adherence associated genes. Maximal transcription of lovK and lovR occurs during swarmer cell differentiation (Purcell et al., 2007), which corresponds to holdfast synthesis and loss of pili and flagella in C. crescentus.
3.4.5 Holdfast in other Caulobacter species and prosthecate bacteria
Other prosthecate bacteria including Hyphomonas, Hyphomicrobium, Asticaccaulis, and a variety of marine bacteria and other freshwater Caulobacter, have adhesive holdfasts (MacRae and Smit, 1991; Moore and Marshall, 1981; Quintero and Weiner, 1995; Umbreit and Pate, 1978; Yun et al., 1994). Although these holdfasts are not as well characterized as the C. crescentus holdfast, some insights have been gained from the basic characterizations that have been performed.
The freshwater bacterium Asticcacaulis biprosthecum undergoes a dimorphic lifecycle resulting in a motile swarmer cell and a sessile stalked cell. The stalked cell of A. biprosthecum has two stalks that extend laterally from each side of the cell. The holdfast of A. biprosthecum is localizes to the pole of the cell, is not associated with either side stalk, and is composed of acidic polysaccharide based on Ruthenium red binding (Umbreit and Pate, 1978). A. biprosthecum holdfast mutants were isolated by chemical mutagenesis, many of which had multiple phenotypes suggesting that either there were multiple mutations or that some of the mutations isolated are in genes necessary for polar development as has been seen for C. crescentus, where mutations in pleC and podJ affect holdfast synthesis and are pleiotropic (Hinz et al., 2003; Smith et al., 2003; Umbreit and Pate, 1978).
The genus Hyphomonas includes species of marine bacteria that have a biphasic lifestyle, which results in a motile swarmer cell and a sessile prosthecate cell; however, the daughter cell is produced by budding from the tip of the stalk. Hyphomonas adherens has an adhesive polysaccharide capsule that is localized around the entire surface of the mother cell, but is not associated with the stalk or daughter cell. Based on lectin and Calcofluor binding studies, H. adherens polysaccharide capsule is composed of galactose and N-acetylgalactoscosamine possibly in a β1-4 linkage (Quintero and Weiner, 1995). Similar to holdfasts, the H. adherens capsule binds gold and polycationic ferritin, which may be due to the positive charge of the gold ions and the negative charge of the acidic polysaccharides of the capsule (Quintero and Weiner, 1995). In contrast, Hyphomonas strain VP-6, which is now classified as Hyphomonas rosenbergii (Weiner et al., 2000), has holdfast at the pole of the mother cell that binds coral tree lectin suggesting that the holdfast contains galactose-β-1-4-N-acetylglucosamine linkages. The H. rosenbergii holdfast is expressed in the newly released swarmer cell similar to C. crescentus (Langille and Weiner, 1998; Levi and Jenal, 2006). In addition to the holdfast, H. rosenbergii also has an extracellular polysaccharide capsule comprised of N-acetylated sugars that surrounds the entire cell and contributes to adherence (Langille and Weiner, 1998). While all Hyphomonas sp. studied thus far have some type of polysaccharide adhesin, Hyphomonas neptunium has not been examined for holdfast or capsular polysaccharide adhesin; however, the genome sequence for H. neptunium has been completed (Badger et al., 2006). H. neptunium does not have any orthologs to the holdfast attachment proteins indicating that H. neptunium may use an alternative mechanism to anchor holdfast polysaccharide to the cell or may not have a polar holdfast. There is some limited amino acid homology between a few of the holdfast biosynthesis and secretion proteins of C. crescentus and H. neptunium, HfsA (HNE_2241), HfsB (HNE_2240) and HfsE (HNE_2651) and HfsC (HNE_1196), but the genes are spread out around the genome and there are no homologs for the other known C. crescentus genes involved in holdfast biosynthesis, suggesting that H. neptunium synthesizes and secretes adhesive polysaccharide in a different manner than C. crescentus perhaps as a polysaccharide capsule adhesin.
Hyphomicrobium sp. can be found in soil, fresh and salt water and are noted for their ability to utilize one carbon compounds for energy (Sperl and Hoare, 1971). Hyphomicrobium sp. are similar to Hyphomonas sp. in that they produce a daughter cell by budding from the end of the mother cell stalk. In Hyphomicrobium vulgare strain ZV580, which has been renamed Hyphomicrobium zavarzinii (Hirsch, 1989), the holdfast is cell-associated but is not present at the tip of the stalk. As a result, H. zavarzinii forms rosettes with the cell body (Moore and Marshall, 1981). Inhibition of rosette formation by D-mannose and D-galactose, as well as Concanavalin-A, which binds α-linked mannose residues, suggests that the holdfast is comprised of mannose and galactose bound by α-linkages. Protein, in addition to the polysaccharide, was found to be important for H. zavarzinii attachment.
Like C. crescentus, most marine Caulobacter sp. and Maricaulis sp. examined have holdfasts comprised of NAG (Abraham et al., 1999; Merker and Smit, 1988) and bind strongly to gold particles. Conversely, lysozyme and chitinase do not affect holdfast in a variety of Maricaulis sp. suggesting that the structure of the holdfast is different from the holdfast of fresh water Caulobacter species (Merker and Smit, 1988). Holdfast mutants in Maricaulis washingtonensis MCS 6 generated similar classes of holdfast mutants that were seen in C. crescentus (Yun et al., 1994): I) those that make no holdfast II) those with less holdfast and III) those with normal holdfast but with altered adhesion. Five different clusters of genes were identified by Southern blot and complementation, but none of these loci have been further characterized (Yun et al., 1994).
Recently, the sequence of the Maricaulis maris MCS10 and Oceanicaulis alexandrii HTCC2633 genomes was completed and are available at the Joint Genome Institute website (http://genome.jgi-psf.org/mic_home.html). O. alexandrii is a member of the Hyphomonadacea family and is closely related to M. maris (Lee et al., 2005). Based on examination of their DNA sequence, both M. maris and O. alexandrii have holdfast biosynthesis, secretion and attachment loci (Fig. 6). O. alexandrii and M. maris have homologs of hfsD, hfsA, hfsB, hfsC, hfsH, hfsG, hfsF, hfaA, hfaB and hfaD, but the genes are organized in a slightly different manner than those of C. crescentus (J. Javens, E. Toh, G. Hardy and Y. Brun, unpublished). Interestingly, the hfsE homolog for M. maris is in the same region of the genome as the holdfast biosynthesis genes, but the hfsE homolog of O. alexandrii is not contained within the holdfast biosynthesis gene cluster. Both M. maris and O. alexandrii have additional genes within the holdfast biosynthesis gene cluster that appear to be involved in polysaccharide biosynthesis. There is an additional predicted glycosyltransferase gene between hfsH and hfsC. Two additional genes adjacent to hfsF are transcribed in the opposite direction. (Fig. 6B and Table 1). In both M. maris and O. alexandrii, the orf adjacent to hfsF encodes proteins with amino acid similarity to CelD in Agrobacterium tumefaciens, which is involved in cellulose biosynthesis in A. tumefaciens (Matthysse et al., 1995a; Matthysse et al., 1995b). The second orf of M. maris adjacent to hfsF encodes a protein with amino acid similarity to glycosyl hydrolases and the second orf of O. alexandrii is predicted to encode a protein with amino acid similarity to mannosyl transferases (Fig. 6B and Table 1). Both O. alexandrii and M. maris have homologs of the holdfast attachment genes that are organized in a similar manner to those found in C. crescentus (Fig. 6A). Directly upstream of the hfa promoter and divergently transcribed, there is an orf (CC2627) that encodes a protein with amino acid similarity to rhomboid proteases, which are intramembrane serine proteases (Ben-Shem et al., 2007; Koonin et al., 2003). CC2627 is conserved among C. crescentus CB15, M. maris and O. alexandrii perhaps indicating that this protease may play a role in regulation of adhesion. Interestingly, M. maris has two different hfa loci (Fig. 6A and Table 1; G. Hardy and Y. Brun, unpublished). The function of the additional hfa locus is unknown. The differences in organization among the genes predicted to be involved in holdfast biosynthesis, secretion, and attachment suggest that the holdfast structure of O. alexandrii and M. maris is likely somewhat different than that of C. crescentus.
TABLE 1.
Comparison of the Holdfast Biosynthesis, Secretion and Attachment Proteins.
| C. crescentus CB15 | Caulobacter sp. K31 | O. alexandrii HTCC2633 | M. maris MCS10 | ||||
|---|---|---|---|---|---|---|---|
| Holdfast gene | Putative Genea Function | homolog | % Ident./Sim.b | homolog | % Identi./Sim.b | homolog | % Ident./Sim.b |
| HfsD | OMP accessory (Wza) | 2177 | 75/86 | 01484 | 38/57 | 1691 | 43/61 |
| HfsA | MPA (Wzc) | 2176 | 76/87 | 01489 | 27/47 | 1690 | 26/43 |
| HfsB | ATPase (Wzc) | 2175 | 73/83 | 01494 | 23/40 | 1689 | 29/45 |
| HfsC | Polysaccharide Polymerase (Wzy) | 2174 | 68/81 | 01499 | 37/56 | 1688 | 39/58 |
| CC2277c | Glycosyl transferase | 2347c | 43/55 (45/60)d | 01504 | 43/58 (100)d | 1687 | 42/56 (50/63)d |
| HfsH | Deacetylase | 2545 | 83/90 | 01509 | 37/51 | 1686 | 41/54 |
| HfsG | Glycosyl transferase | 2170 | 76/85 | 01514 | 38/55 | 1685 | 38/55 |
| HfsF | Flippase | 2169 | 71/80 | 01519 | 39/55 | 1684 | 40/58 |
| CC3689c | Cellulose biosynthesis (CelD)e | 3380c | 28/47 | 01524 | 29/44 | 1683 | 27/46 |
| No homolog | Glycosyl hydrolase | No homolog | NAf | No homolog | NAf | 1682 | NAf (100)g |
| CC0469c | Dolichol-phosphate Mannosly transferase | 0476c | 86/92 (41/58)d | 01529 | 40/59 (100)d | 0011c | 43/60 (49/66)d |
| HfsE | Glycosyl transferase | 2168 | 66/76 | 07474 | 37/49 | 1681 | 49/63 |
| HfaA | Holdfast attachment (CsgA) | 0606 | 75/90 | 15215 | 34/55 | 2747 1083 | 37/54 35/51 |
| HfaB | Hfa protein secretin (CsgG) | 0605 | 81/89 | 15210 | 53/66 | 2748 1082 | 37/55 54/68 |
| HfaD | Holdfast attachment | 0604 | 58/73 | 15205 | 27/39 | 2749 1081 | 23/38 29/48 |
E. coli homolog is shown in parentheses.
Percent similarity and identity was determined using BLAST and are a comparison to the gene in CB15
These genes are not associated with the hfa and hfs holdfast gene clusters and are located elsewhere in the genome.
Percent identity and similarity is in comparison to O. alexandrii HTCC2633.
A. tumifaciens homolog is shown in parentheses.
NA, there was no homolog so percent identity and similarity is “not applicable”.
Percent identity is in comparison to M. maris MCS6.
Finally, the holdfasts of other freshwater Caulobacter sp. have been characterized and in contrast to Maricaulis sp., they contain a variety of polysaccharides (MacRae and Smit, 1991; Merker and Smit, 1988). The holdfasts of C. henricii and C. vibroides have α-linked N-acetylgalactosamine and NAG. C. subvibroides holdfast contains α-linked mannose, α-linked fucose and N-acetylgalactosamine. C. leidyia, a member of the Sphingomonadaceae that is closely related to A. biprosthecum (Abraham et al., 1999), contains α- or β-linked N-acetylgalactosamine or galactose and α-linked mannose. As with C. crescentus and the other prosthecates described here, all the fresh water Caulobacter species examined bind gold, a cationic metal, suggesting that the polysaccharides of the holdfast are negatively charged (Merker and Smit, 1988). The variation in sugar composition and holdfast structure may allow freshwater Caulobacter sp. to attach to surfaces with different chemistries depending on the specific environment they colonize. No examination of the genes involved in holdfast synthesis has been performed in the other freshwater Caulobacter sp. The draft genome sequence of Caulobacter sp. strain K31, available at the Joint Genome Institute website (http://genome.jgipsf.org/mic_home.html), indicates that Caulobacter sp. K31 has the same organization of genes for holdfast biosynthesis, secretion and attachment to that of C. crescentus CB15 (Fig. 6 and Table 1).
Future studies will be needed to identify the specific functions of the holdfast biosynthesis and attachment proteins as well as the biochemistry of holdfast polysaccharide transport, biosynthesis and physical attachment. As additional genomes of Caulobacter and other prosthecates are sequenced, comparisons of their holdfast loci may provide insights into the functions of the holdfast genes, their regulation and the reason for the variation in holdfast composition.
4. Chromosome Replication and Segregation, Cell Division, and Cell Shape
In order for C. crescentus to produce two different cell types after cell division, changes in cell shape must be coupled with chromosome replication and segregation and cell division. During the swarmer to stalked cell differentiation, chromosome replication and cell division are initiated. Each compartment of the predivisional cell must contain a complete chromosome prior to the completion of cell division. The complex developmental steps that are required for changes in chromosome replication and segregation, cell division, and cell shape are examined in this section.
4.1 Chromosome Replication Occurs Once and Only Once per Cell Division Cycle
Chromosome replication is subject to extensive regulation during the C. crescentus cell cycle. A combination of mechanisms including the control of CtrA and DnaA abundance and activity and the expression of DNA replication proteins ensures that chromosome replication occurs once and only once per cell division cycle. In this manner, C. crescentus differs from E. coli and many other bacteria that are able to initiate chromosome replication more than once per cell cycle, depending on growth rate.
The pattern of DNA methylation was used to precisely assess the frequency of chromosome replication initiation per cell division cycle (Marczynski, 1999). The CcrM DNA methyltransferase is subject to extremely tight control during the cell cycle (Marczynski and Shapiro, 2002). The ccrM gene is transcribed in the predivisional cell (Zweiger et al., 1994) under the positive transcriptional control of CtrA (Reisenauer et al., 1999; Stephens et al., 1995). CtrA binds to the ccrM promoter relatively weakly, ensuring that ccrM transcription is only activated late in the predivisional cell (Reisenauer et al., 1999). This late expression of ccrM results in a sudden increase in CcrM concentration and subsequent methylation of the chromosomes (Stephens et al., 1996; Zweiger et al., 1994). CcrM is then degraded by the Lon protease just prior to cell division (Wright et al., 1996) and the two daughter cells are released with fully methylated chromosomes and no CcrM. Chromosome replication results in hemimethylated DNA and the DNA remains hemimethylated until late in the predivisional cell when CcrM is synthesized. Fully methylated DNA can be distinguished from hemi-methylated DNA by restriction digestion using enzymes that are sensitive to methylation state and whose recognition sequence overlaps with the site of CcrM methylation. Therefore, the appearance of hemi-methylated DNA during the cell cycle was used as a measure of the frequency of chromosome replication initiation per cell division cycle (Marczynski, 1999). Using this strategy, hemi-methylated DNA could not be detected above background levels, indicating that less than one cell in 1000 reinitiates chromosome replication. Since plasmids can replicate in all cell types (Marczynski et al., 1990), mechanisms which restrict chromosome replication to the predivisional cell must exist. It is interesting to note that plasmids have a much higher rate of replication in stalked cells as compared to swarmer cells (Marczynski et al., 1990). This is likely to be due in part to cell cycle regulation of the expression of DNA replication genes (Laub et al., 2000). In contrast, the control of chromosome replication is regulated by the state of the origin of chromosome replication as described in the next section.
4.1.1. The origin of replication
The precise location of the C. crescentus origin of replication (Cori) was identified using three different methods. The initial discovery of the Cori region came from experiments in which chromosomal DNA from a synchronized swarmer cell population was radioactively labeled throughout the cell cycle and analyzed by pulse-field gel electrophoresis and autoradiography (Dingwall and Shapiro, 1989). Refinement of this method using a temperature sensitive DNA replication mutant narrowed the Cori region to a smaller DNA fragment (Marczynski and Shapiro, 1992). DNA fragments within this region were tested for their ability to support autonomous plasmid replication and the minimal Cori region was narrowed down to ∼500 bp (Marczynski et al., 1995; Marczynski and Shapiro, 1992). The localization of Cori was confirmed using the two-dimensional DNA neutral/neutral agarose gel method that resolves non-linear DNA fragments caused by the passage of a replication fork (Brassinga and Marczynski, 2001). All these methods determined that Cori is located between the hemE gene and the RP001 gene, both of which are transcribed divergently from Cori (Fig. 8). This gene organization is conserved in at least some Alphaproteobacteria and may be useful in the identification of the origin of chromosome replication in these bacteria. DNA binding studies with CtrA, its homolog in Rickettsia prowazekii CzcR, and IHF indicate that the binding sites for these proteins are conserved in C. crescentus Cori and in the R. prowazekii origin of replication, suggesting conserved regulatory mechanisms for the initiation of chromosome replication (Brassinga et al., 2002).
A number of regulatory sequences have been identified in the minimal Cori sequence including five CtrA binding sites (sites a-e starting with the hemE-proximal site), one DnaA box, an IHF binding site, and some conserved motifs of unidentified function (Fig. 8;(Marczynski and Shapiro, 2002)). CtrA represses the initiation of chromosome replication, DnaA stimulates it, and IHF is thought to also act positively on initiation.
A weaker promoter, PW, and a stronger promoter, PS, are located upstream of the hemE gene (Fig. 8;(Marczynski et al., 1995). PW is responsible for most of the hemE expression while PS transcription is uncoupled from HemE synthesis. The RNA transcribed from the PS promoter lacks a good ribosome binding site and the 5' untranslated region has the potential to fold because of the presence of four conserved 8-mer motifs found between PS and PW. The 8-mer motifs are essential elements of Cori function since small deletions in this region are not tolerated in the chromosome or in Cori plasmids (Marczynski et al., 1995). It may be that folding of the RNA in the 8-mer region transcript plays a role analogous to RNAs that regulate plasmid replication and copy number, but this has yet to be determined.
Another indication that transcription from PS is likely to play a role in the control of replication initiation lies in the fact that PS is regulated by CtrA and is only transcribed in stalked cells (Marczynski et al., 1995). Two of the five CtrA binding sites in Cori, sites a and b, are adjacent and lie within a 40 bp AT-rich region that also contains PS (Fig. 8). Upstream of the AT-rich region is a 40 bp region rich in purines that is essential for Cori function, although the function of this purine rich region is not known (Marczynski and Shapiro, 2002). Next is CtrA binding site c, which overlaps with the IHF binding site. IHF binding reduces CtrA binding to site c and also to CtrA binding site d (Siam et al., 2003). Deletion of CtrA site c and the IHF site is tolerated on the chromosome but cells grow slowly and are intolerant to a CtrA phosphomimetic mutant (Siam et al., 2003). Since IHF expression increases during the swarmer to stalked cell differentiation (Gober and Shapiro, 1990; Gober and Shapiro, 1992), at the same time as CtrA concentration decreases, it is possible that IHF displaces CtrA and bends Cori DNA to promote the initiation of chromosome replication (Siam et al., 2003). Finally, CtrA binding site e is adjacent to a DnaA box. DnaA is essential in C. crescentus and depletion of DnaA using a xylose inducible promoter prevents the initiation of chromosome replication, but not ongoing replication (Gorbatyuk and Marczynski, 2001). The function and regulation of DnaA is described in detail in section 2.1.3.
4.1.2. Regulation of chromosome replication by tmRNA
Another regulator of chromosome replication is the hybrid tRNA-mRNA molecule called tmRNA, which is encoded by the ssrA gene (reviewed in (Keiler, 2007)). One domain of tmRNA folds into an alanyl-tRNA-like structure and is charged by alanyl-tRNA synthetase. tmRNA lacks an anticodon stem-loop and its 3' end contains an open reading frame that encodes the degradation tag that is added to the C-terminal end of incomplete proteins. tmRNA is one of the most abundant RNAs in bacterial cells and it plays important roles in general physiology (Keiler, 2007). The main function of tmRNA is to release ribosomes that are stalled due to the presence of mRNAs without a stop codon, which result from partial degradation at the 3' end. tmRNA adds the SsrA degradation tag at the C-terminal end of the incomplete proteins. The degradation of incomplete proteins eliminates the potential for dominant-negative effects associated with some incomplete proteins. A small protein called SmpB assists tmRNA function by increasing its stability and interaction with the ribosome.
Deletion of the tmRNA gene (ssrA) in C. crescentus results in slow growth rate that is caused by a stage-specific reduction of growth rate (Keiler and Shapiro, 2003b). Synchronized ssrA swarmer cells take 195 min to complete the cell cycle as compared to 150 min for wild-type cells. This difference in the length of the cell cycle is caused by a marked increase in the amount of time ssrA cells remain in the swarmer stage, from 30 min in wild-type cells to 75 min in ssrA mutant cells. Flow cytometry experiments showed that the prolonged swarmer stage in ssrA cells correlates with a delay in the initiation of chromosome replication (Keiler and Shapiro, 2003b). CtrA degradation normally occurs just prior to chromosome initiation. In an ssrA mutant, CtrA degradation occurs at the proper time; however, there is a 40 min delay before the initiation of chromosome replication. Therefore, tmRNA is required for the proper control of replication initiation through a CtrA-independent process. These results also imply that the degradation of CtrA is not sufficient for the initiation of chromosome replication.
Is the requirement for tmRNA for the timing of replication initiation due to its role in protein degradation? Expression of tmRNA which encodes a proteolysis-resistant SsrA tag fails to complement an ssrA deletion mutant and produces a phenotype that is more severe than that of the ssrA deletion mutant (Keiler and Shapiro, 2003b). Furthermore, introduction of the mutation encoding the stable SsrA tag into wild-type cells causes a delay in replication initiation. These results indicate that the tagging of some protein(s) with a wild-type SsrA peptide is required for the timing of replication initiation.
As suggested by its involvement in the regulation of replication initiation timing, tmRNA abundance is subject to cell cycle regulation (Keiler and Shapiro, 2003a). tmRNA is transcribed and produces a stable molecule in swarmer and predivisional cells. A sharp increase in transcription from the ssrA promoter leads to high levels of tmRNA during the swarmer to stalked cell transition; however tmRNA is rapidly degraded in stalked cells. The cell cycle regulation of tmRNA stability is controlled by the opposing actions of the 3′ to 5′ exoribonuclease RNase R and the SmpB protein (Hong et al., 2005). RNase R can degrade tmRNA in vitro and is required for the cell cycle dependent degradation of tmRNA in stalked cells. The level of RNase R is relatively constant during the cell cycle, indicating that the cell cycle variation in tmRNA is not due to the regulation of RNase R expression (Keiler and Shapiro, 2003a). Deletion of the gene encoding the tmRNA binding protein SmpB causes the same delay in replication initiation as the deletion of ssrA (Keiler and Shapiro, 2003b). SmpB has high affinity for tmRNA in vitro and selectively protects it from degradation by RNase R (Keiler and Shapiro, 2003a). SmpB is also required for tmRNA stability in vivo (Keiler and Shapiro, 2003b). SmpB abundance dramatically increases during the swarmer to stalked cell differentiation when tmRNA accumulates, and SmpB is rapidly degraded in stalked cells when tmRNA is degraded (Keiler and Shapiro, 2003a). Therefore, the cell cycle dependent fluctuation in tmRNA stability is mediated by the protective action of SmpB against RNase R degradation.
A proteomic identification of tmRNA substrates identified 73 proteins that are tagged by tmRNA (Hong et al., 2007). Proteins involved in DNA replication, recombination, and repair are overrepresented in the subset of proteins identified as tmRNA substrates. This observation is consistent with the model that tmRNA regulates the timing of chromosome replication by controlling the amount or activity of protein complexes involved in the initiation of chromosome replication, chromosome replication, and DNA repair.
4.2 Coordination of Chromosome Segregation and Cell Division
Prior to cell division newly replicated chromosomes must be partitioned into the newly formed daughter cells. This important process ensures that each daughter cell contains one copy of the chromosome that is not prematurely truncated by the closing of the division site. This section describes the mechanisms that contribute to chromosome segregation in C. crescentus and its coordination with cell division.
4.2.1. Replisome movement and chromosome organization
In swarmer cells, the origin of replication is located at the flagellar pole and the terminus is located at the opposite pole (Fig. 9). Following the swarmer to stalked cell differentiation, chromosome replication is initiated and one copy of the origin rapidly migrates to the opposite pole, resulting in bipolar localization of the origins, and the terminus migrates to the mid-cell (Jensen and Shapiro, 1999). The position of the replisome during the cell cycle was monitored using GFP fusions to the DnaB, HolB, and HolC proteins, which are known components of the replisome, in strains where the gene fusions are the only copies of these genes (Jensen et al., 2001). The replication proteins are distributed throughout the swarmer cell prior to the initiation of chromosome replication. At the time of replication initiation, the replication proteins localize to the flagellar pole in a DNA replication-dependent fashion. As replication proceeds towards the terminus, the replisome gradually moves to the center of the cell and is disassembled once DNA replication is complete (Jensen et al., 2001). Movement of the replisome depends on DNA replication and it has been hypothesized that accumulation of newly replicated DNA near the stalked pole passively displaces the replisome towards the midcell (Jensen et al., 2001).
Figure 9.
Localization of replication origin and terminus and chromosome segregation proteins in C. crescentus. In each cell cycle, the light gray oval represents a non-replicating chromosome and the tetha structures represent a replicating chromosome. The origin of replication and the terminus (square and triangle in the middle cell cycle, respectively) are at opposite poles in swarmer cells, with the origin at the flagellated pole. A copy of the origin is rapidly moved to the opposite pole early in chromosome replication. The terminus progressively moves to the midcell and the two copies of the terminus are decatenated prior to cell division. The various proteins involved in chromosome segregation are represented by shaded shapes. FtsK localizes to the pole opposite the flagellum and the stalk in swarmer and stalked cells. During constriction, FtsK localizes to the mid-cell where it remains until the cell divides to stabilize the Z-ring and help complete chromosome segregation. ParB co-localizes with and tracks the origin of replication. The SMC protein has a random or spiral-like localization localization throughout the cytoplasm; however during cell division the SMC proteins additionally form discreet foci at the poles. MreB forms a spiral in swarmer cells that coalesces into a band at the mid-cell during division. MreB then returns to a spiral near the end of cell division.
Analysis of the position of over 100 loci on the C. crescentus chromosome revealed that the chromosome is highly organized (Viollier et al., 2004). Each locus has a precise location in the cell, with loci organized in a linear order along the long axis of the cell that recapitulates their physical location on the chromosome. Loci closest to the origin of replication are closest to the pole where the origin is localized and those close to the terminus are closest to the opposite pole. During DNA replication, as each locus is replicated, it rapidly moves to its proper cellular address. Therefore, the chromosome is not randomly compacted within the cell but is organized with impressive precision. Whether this organization has functional or regulatory consequences is not yet known.
Part of the mechanism responsible for the precise organization of the chromosome lies in the fact that the origin and terminus are localized to opposite poles of the cell (Fig. 9). This polar localization is also relevant to chromosome segregation and to its coordination with cell division, as is discussed in the next sections. Much of this coordination occurs by regulating the localization of the cell division initiation protein FtsZ.
4.2.2 Formation and structure of the Z-ring
FtsZ, a tubulin-like GTPase, is the most conserved protein involved in bacterial cell division and is required for the initiation of cell division (reviewed in (Lutkenhaus, 2007)). Prior to the initiation of cell division, FtsZ localizes around the circumference of the mid-cell, forming a structure called the Z-ring, and recruits other cell division proteins to this site (Fig. 10). In C. crescentus, the timing of Z-ring formation is dependent on cell cycle progression. FtsZ concentration is low in swarmer cells and increases sharply after the swarmer to stalked cell differentiation and in predivisional cells and FtsZ is then degraded in late predivisional cells (Kelly et al., 1998; Quardokus et al., 1996; Quardokus et al., 2001). Midcell Z-rings begin to form shortly after the swarmer to stalked cell differentiation (Kelly et al., 1998; Quardokus et al., 2001), coincident with the increase in FtsZ concentration.
Figure 10.
Localization and regulation of cell division proteins in C. crescentus: (A) In each cell, the light gray oval represents a non-replicating chromosome and the tetha structures represent a replicating chromosome. FtsZ (circle) initially localizes to the pole opposite the flagellum along with FtsK (star) in the swarmer cell. MipZ (diffuse shaded circle) resides at the flagellar pole. As the cell cycle is initiated, MipZ tracks with the newly replicated origins and rapidly localizes to both poles. Arrival of MipZ to the pole opposite the stalk displaces FtsZ, which migrates to localize at the midcell (Z-ring, shaded band at the mid-cell). As the cell elongates, FtsA and FtsQ initially localize in a spiral-like pattern (not shown) and FtsK localizes to the midcell. In late predivisional cells, a preponderance of FtsQ (square) and FtsA (hexagon) join the Z-ring. (B) Timing of transcription of ftsZ, ftsQ, and ftsA and variation in abundance of the transcriptional regulators CtrA and DnaA during the cell cycle. The solid lines indicate the proteins levels of the global regulators CtrA and DnaA. The dashed lines represent the transcription pattern of ftsZ and ftsQA as the cell proceeds through the cell cycle. (C) The diagram depicts how multiple short filaments of FtsZ form the Z-ring at the mid-cell. The cell is represented by the cylinder and the FtsZ filaments are represented by the gray curved lines.
Fluorescence microscopy analysis of FtsZ in many bacteria has led to the popular model that FtsZ forms a continuous ring at the site of cell division; however, the resolution of light microscopy is not sufficient to determine if FtsZ molecules are polymerized into a continuous ring around the cell circumference. Experiments in which the fluorescence recovery of FtsZ-GFP after photobleaching is observed indicate that FtsZ molecules rapidly exchange between the Z-ring and the soluble cytoplasmic pool challenging the validity of the continuous Z-ring model (Anderson et al., 2004). Electron cryotomography (ECT) of C. crescentus cells was used to probe the structure of the Z-ring (Li et al., 2007). ECT represents a major technological advance in microscopy that is revolutionizing our understanding of cellular ultrastructure (Jensen and Briegel, 2007). ECT is particularly good at revealing the three-dimensional ultrastructure of small cells in a life-like frozen-hydrated state generated by rapid freezing (Jensen and Briegel, 2007). By tilting the specimen in an electron cryomicroscope, a 3D reconstruction of the sample at “molecular” resolution can be obtained (Jensen and Briegel, 2007). C. crescentus cells are ideal specimens for ECT because their small size relative to many bacteria improves the resolution.
ECT analysis demonstrated that during cell division in C. crescentus, FtsZ forms multiple short arc-like filaments at the mid-cell. The filaments average ∼100 nm in length and are found nearly perpendicular to the long axis of the cell approximately 16 nm inside the inner membrane (Fig. 10) (Li et al., 2007). Notably, no complete Z-rings were observed in this study. Only a few filaments were present in a given cell and analysis of cells at different stages of cell division revealed that there is no obvious pattern in the number, position, or configuration of FtsZ filaments during cell division (Li et al., 2007). Filaments are ∼5 nm in diameter, suggesting that they consist of single or double protofilaments. The FtsZ filaments appear in straight and curved conformations and often appear to extend into and through the inner membrane, possibly interacting with the peptidoglycan (Li et al., 2007). The portion of the filaments extending through the membrane is probably composed of other cell division proteins. in vitro, FtsZ filaments bound to GTP are straight and become curved when GTP is hydrolyzed to guanosine diphosphate GDP (Lu and Erickson, 1999; Lu et al., 2000). An iterative pinching model has been proposed to explain how the short FtsZ filaments could drive constriction (Li et al., 2007). GTP bound FtsZ polymerizes into straight filaments that attach to the inner membrane through anchor proteins. As GTP hydrolysis occurs, the filaments bend, drawing the membrane inward in the region of the filament. While having only a small local effect for each individual filament, the repetition of this process thousands of times would be sufficient to drive cell constriction, as long as some process, presumably local peptidoglycan synthesis, stabilizes the small constriction prior to FtsZ depolymerization.
For simplicity and despite the fact that a continuous Z-ring does not appear to exist, we will continue to refer to the Z-ring in the remainder of the text to describe the ring-like localization of FtsZ as seen by fluorescence microscopy.
4.2.3 Coordination of Z-ring formation with development and chromosome replication
In addition to the importance of coordinating cell division with chromosome replication and segregation, C. crescentus imposes an additional regulatory layer that coordinates cell division with the developmental program, ensuring that cell division occurs once the asymmetry of the predivisional cell has been established. In this section, we describe how the timing of cell division is coordinated with development and with chromosome replication and segregation.
Z-ring formation is not solely driven by an increase in FtsZ concentration. Ectopic expression of FtsZ in swarmer cells does not cause premature Z-ring formation, nor does it cause earlier initiation of cell division (Quardokus et al., 2001). Overexpression of FtsZ in predivisional cells causes the formation of additional constrictions near the midcell and a delay in cell separation (Din et al., 1998; Quardokus et al., 2001). In these cells, Z-ring localization is constrained to the sites of constriction, suggesting the existence of a mechanism to constrain Z-ring formation at the midcell. These results suggested that a cell cycle or developmental cue was required for midcell Z-ring formation.
Midcell Z-ring formation requires the initiation of chromosome replication; however DNA replication per se is not required for the formation of Z-rings (Quardokus and Brun, 2002). When replication initiation is blocked, Z-rings still form but at subpolar regions of the cell, and cells constrict in an extended area mostly away from, but sometimes over the nucleoid (Quardokus and Brun, 2002). These observations suggest that early stages of chromosome replication are a major determinant for positioning of Z-rings in the cell, but not for Z-ring assembly (Quardokus and Brun, 2002). Identification of the terminus of replication (Jensen, 2006) allowed an examination of the timing of separation of the replicated termini during cell constriction. Careful fluorescence microscopy with a synchronized population of C. crescentus determined that the invagination of the inner membrane clearly occurs before separation of the termini (Jensen, 2006). In fact, the replicated termini remain associated with the deeply constricted division site for an extended period after the completion of DNA replication and termini separation occurs just before the final cell separation step (Jensen, 2006). This indicates that nucleoid occlusion does not occur in C. crescentus since cell constriction occurs in the presence of non-segregated DNA (Jensen, 2006). However the Z-ring appears to form preferentially at sites where there is less DNA present (Quardokus and Brun, 2002).
The results described above indicate that Z-ring formation is influenced by chromosome replication. Whether this influence is due to a mechanism directly monitoring chromosome replication is not known. However, it is becoming clear that the state of chromosome segregation affects Z-ring formation through the concerted action of ParA, ParB, and MipZ, as described in the next section. It is therefore possible that the effect of perturbations in chromosome replication on Z-ring formation occur indirectly through their effect on segregation.
4.2.4 Coordination of Z-ring formation with chromosome segregation
Homologs of the plasmid partition genes parA and parB are found in most prokaryotic genomes. Type I ParA and ParB are involved in plasmid partitioning, while type II ParA and ParB are involved in chromosome segregation (Gerdes et al., 2000; Mohl and Gober, 1997; Thanbichler and Shapiro, 2006b). The Type II chromosome segregation genes parA, parB, and the ParB binding site parS of C. crescentus all reside in a single operon near Cori (Mohl and Gober, 1997). ParA and ParB localize at the poles where ParB binds to the parS cis-acting region on the chromosome (Fig. 9) (Mohl and Gober, 1997; Thanbichler and Shapiro, 2006b). ParB consists of three domains: an N-terminal domain that interacts with ParA, a central DNA binding helix-turn-helix domain, and a C-terminal dimerization domain (Figge et al., 2003). The N-terminal domain of ParB regulates the ATPase activity of ParA by facilitating nucleotide exchange (Easter and Gober, 2002; Figge et al., 2003). ParA-ATP displaces ParB from parS (Easter and Gober, 2002).
ParA and ParB are essential for viability; they are both required for cell division and for chromosome segregation (Mohl et al., 2001; Mohl and Gober, 1997). Perturbation of ParA and ParB protein levels prevents Z-ring formation and arrests cell division without affecting the expression of ftsZ, ftsQ, or ftsA (Mohl et al., 2001) (see section 4.3.2). Bipolar ParB foci appear prior to Z-ring formation, suggesting that polar localization of a chromosome segregation complex provides a checkpoint that coordinates cell division and chromosome segregation (Figge et al., 2003; Mohl et al., 2001). Identification of a member of the ParA superfamily of P loop ATPases, MipZ, has provided the link between ParB function and Z-ring formation (Thanbichler and Shapiro, 2006b).
MipZ is highly conserved in alphaproteobacteria; it is essential for viability and regulates Z-ring formation in C. crescentus (Thanbichler and Shapiro, 2006b). Depletion of MipZ causes cell filamentation and random localization of FtsZ in structures that are unable to form a functional divisome. Overexpression of MipZ causes dispersion of FtsZ in the cytoplasm and its localization to both poles. In vitro, MipZ interacts directly with FtsZ and interferes with its polymerization (Thanbichler and Shapiro, 2006b). By itself, FtsZ polymerizes into long straight polymers, whereas it forms short and highly curved polymers in the presence of MipZ. Careful observations from fluorescence microscopy suggest that FtsZ and MipZ cannot co-exist in the same area of the cell, indicating that MipZ controls FtsZ positioning (Thanbichler and Shapiro, 2006b). FtsZ is localized to the pole opposite the flagellum in swarmer cells and then migrates to the mid-cell during the swarmer to stalked cell transition. The localization of MipZ has the same pattern of localization as the origin of replication and ParB during the cell cycle (Thanbichler and Shapiro, 2006b). Indeed, MipZ binds to ParB in vitro and the two proteins form a ternary complex with a parS DNA fragment, with ParB providing the parS binding activity (Thanbichler and Shapiro, 2006b). By tracking the chromosome origins with ParB, MipZ coordinates Z-ring formation with chromosome movement (Thanbichler and Shapiro, 2006b). MipZ colocalizes with the origin of replication by binding to ParB, itself bound to parS sites located close to the origin, at the flagellar pole of swarmer cells. After the initiation of chrmosome replication, one origin-ParB-MipZ complex remains at the same pole, while a MipZ-ParB complex associates with the second copy of the origin. This second origin-ParB-MipZ complex migrates to the opposite pole, where MipZ displaces FtsZ. Therefore, MipZ localization to both poles prevents FtsZ localization to those sites. In addition, the interaction of MipZ with ParB is highly dynamic and is regulated by the ATPase activity of MipZ, resulting in the formation of a gradient of MipZ with its highest concentration at the poles (Thanbichler and Shapiro, 2006b). This MipZ gradient results in a low concentration of MipZ at the midcell, where FtsZ can polymerize productively.
Interestingly, an earlier study using immunogold transmission electron microscopy showed that FtsZ can localize to the stalked pole (Quardokus et al., 2001). This localization is consistent with the requirement of FtsZ for crossband synthesis at the stalked pole (Divakaruni et al., 2007). Since MipZ is always present at the stalked pole where it should inhibit FtsZ polymerization, a mechanism has to exist to allow polymerization of a small amount of FtsZ for crossband synthesis. Alternatively, polymerization of FtsZ is not required at the pole for crossband synthesis.
4.2.5 Role of MreB in Chromosome Segregation
The actin-like cytoskeleton protein MreB that is required for cell shape determination (see section 4.4 for a discussion of the role of MreB in cell shape determination) has been implicated in chromosome segregation in a number of bacteria (Thanbichler and Shapiro, 2006a). When synchronized C. crescentus cells are treated with the MreB inhibitor A22 (S-(3,4-dichlorobenzyl)isothiourea) prior to chromosome replication initiation, replication still occurs, but origin-proximal regions fail to segregate (Gitai et al., 2005). This inhibition of segregation is rapidly reversed when A22 is removed, and does not occur when A22 is administered to a mreB mutant that is resistant to A22. When chromosome segregation is instead allowed to begin before wild-type cells are treated with A22, the origin-proximal regions segregate normally. The inhibition of chromosome segregation is specific to origin-proximal regions, since origin-distal regions still segregate normally when cells are treated with A22. Chromatin immunoprecipitation indicates that MreB associates, most likely indirectly, with origin-proximal regions but not with origin-distal regions (Gitai et al., 2005). These results indicate that MreB only controls the separation of the genes near Cori and likely only affects the initial steps of chromosome separation.
Whether MreB is directly involved in chromosome segregation or whether the effect of MreB disruption on segregation is due to effects on cell shape remains controversial. For example, deletion of mreB in a cyanobacterium, Anabaena sp. PCC 7120, or of mreBCD in E. coli causes no defect in chromosome segregation (Hu et al., 2007; Karczmarek et al., 2007). To determine if cell shape perturbation causes defects in chromosome segregation, the effects of A22 treatment and mecillinam (amdinocillin) treatment, which targets PBP2, were compared in E. coli (Karczmarek et al., 2007). Mecillinam treatment results in the formation of round cells without disrupting MreB. Altered chromosome segregation was observed following either mecillinam or A22 treatment. This observation suggests that altered cell shape, rather than a specific function of MreB, is responsible for the chromosome segregation defect (Karczmarek et al., 2007).
4.2.6 Other proteins involved in chromosome segregation
Chromosome segregation is also controlled by the structural maintenance of chromosome (SMC) proteins (Graumann, 2001; Jensen and Shapiro, 1999). The SMC proteins consist of N- and C-terminal domains with characteristic Walker A and B boxes, respectively, that are separated by a coiled coil domain with a non-coiled “hinge” patch in the middle (Löwe et al., 2001; Melby et al., 1998). SMC dimerizes in an anti-parallel fashion with the opposite adjoining terminal domains interacting with one another (Melby et al., 1998). A null smc mutant in C. crescentus causes aberrant nucleoid localization, similar to what is seen in E. coli and B. subtilis (Jensen and Shapiro, 1999). Interestingly, SMC displays punctate localization throughout much of the cell cycle and roughly 35% of predivisional cells have brighter foci of SMC at the poles, with fainter foci throughout the cell (Fig. 9) (Jensen and Shapiro, 2003). The brighter foci may stem from the increase in smc transcription from the swarmer to stalk cell transition (Jensen and Shapiro, 2003). There are roughly between 1,500 and 2,000 SMC molecules per cell, enough for one protein to 6,000 - 8,000 bp of chromosomal DNA (Jensen and Shapiro, 2003). While it is unclear if all the SMC molecules are present in the foci, multiple SMC proteins must bind the chromosome since they likely play a role in condensing replicated DNA (Jensen and Shapiro, 2003; Thanbichler and Shapiro, 2006a).
The coordination of chromosome segregation and cell division also involves FtsK (Bigot et al., 2007) (Wang et al., 2006). FtsK is a member of the FtsK/SpoIIIE/Tra family of proteins. FtsK consists of a N-terminal domain required to localize FtsK to the mid-cell and for its function in cell division and a cytoplasmic C-terminal ATPase domain required for chromosome translocation (Bigot et al., 2007). FtsK interacts with topoisomerase IV, a protein complex involved in chromosome decantenation, and the XerCD site-specific recombinase that resolves chromosome dimers (Bigot et al., 2007). Finally, FtsK translocates the separated chromosomes to their respective side of the septum (Bigot et al., 2007). In C. crescentus, FtsK localizes to the site of cell division early in the division process and is required for the assembly or maintenance of the Z-ring (Wang et al., 2006). FtsK is essential for viability and its C-terminus is required for proper segregation of the chromosome terminus. One of the roles of FtsK in chromosome segregation involves the ParC subunit of topoisomerase IV (Wang et al., 2006). The parC and parE genes encoding the topoisomerase IV subunits are essential for viability and required for proper chromosome segregation and the polar localization of the origin of replication in C. crescentus (Wang and Shapiro, 2004; Ward and Newton, 1997). ParC co-localizes with the replisome and, even though ParE is dispersed throughout the cell, ParE is required for ParC localization to the replisome (Wang et al., 2006). The FtsK C terminus is also required for the localization of ParC, which explains, at least in part, the chromosome segregation defect of ftsK mutants (Wang et al., 2006).
4.3 Regulation of Cell Division Protein Expression
In addition to the regulation of FtsZ polymerization and localization, coordination of cell division with other cell cycle and developmental events is achieved by regulating the synthesis of cell division proteins. Like other developmental regulators described in earlier sections, a combination of cell cycle dependent transcription and proteolysis is used to ensure that cell division proteins are present at the time they are required.
4.3.1 Transcriptional regulation of cell division genes
In C. crescentus, ftsZ is part of a large cell division gene cluster whose overall organization is conserved in many bacteria. The regulation of the last three genes in the cluster, ftzQ-ftsA-ftsZ, has been studies in some detail (Kelly et al., 1998; Martin et al., 2004; Sackett et al., 1998; Wortinger et al., 2000). The bitopic inner membrane protein FtsQ is essential for cell division, but it precise role remains unknown, although it may be involved in peptidoglycan synthesis (Chen et al., 2002; Martin et al., 2004). FtsQ is part of a group of cell division proteins that localizes to the mid-cell late in cell division (Fig. 10) (Buddelmeijer et al., 1998; Chen et al., 1999; Martin et al., 2004). FtsA, a peripheral membrane protein with ATPase activity, is a member of the actin family and, like FtsZ, is a highly conserved cell division protein (Bork et al., 1992; Rothfield et al., 1999; van den Ent and Löwe, 2000). In E. coli, FtsA and ZipA, an inner membrane protein that binds FtsZ, help stabilize the Z-ring and anchor it to the membrane (Addinall and Holland, 2002; Feucht et al., 2001; Pichoff and Lutkenhaus, 2002; Pichoff and Lutkenhaus, 2005; Sánchez et al., 1994).
Three promoters, PQA, PA, PZ, drive transcription of ftsQ, ftsA, and ftsZ (Kelly et al., 1998; Sackett et al., 1998). The PQA promoter drives most of ftsQ and ftsA transcription (Sackett et al., 1998). PA is located between ftsQ and ftsA and is ∼10 times weaker than PQA. While ftsQ and ftsA are cotranscribed, a strong terminator is found between ftsA and ftsZ and prevents transcription from the ftsQ and ftsA genes from extending to ftsZ. ftsZ is transcribed from a strong promoter, PZ, which is located between ftsA and ftsZ (Kelly et al., 1998; Sackett et al., 1998). CtrA directly activates PQA transcription (Wortinger et al., 2000) and represses PZ transcription (Kelly et al., 1998), while DnaA activates the expression of ftsZ (Hottes et al., 2005). Transcription from all three promoters is low in swarmer cells. During the swarmer to stalked cell transition, the level of CtrA decreases and the level of DnaA increases, leading to the transcription of ftsZ concurrently with the start of DNA replication (Hottes et al., 2005; Kelly et al., 1998; Sackett et al., 1998). In the predivisional cell, CtrA is synthesized and activates the transcription of ftsQ and ftsA and represses the transcription of ftsZ (Kelly et al., 1998; Sackett et al., 1998; Wortinger et al., 2000). Following the completion of cell division, transcription of ftsQ and ftsA ceases through an unknown mechanism (Sackett et al., 1998).
4.3.2 The proteolysis of cell division proteins helps establish a DNA replication checkpoint
Coordination between cell division and DNA replication is ensured in part by a checkpoint that targets the expression of late cell division proteins. When DNA replication is blocked in C. crescentus, the transcription of ctrA from the P2 promoter is inhibited (Wortinger et al., 2000). In the absence of high levels of CtrA, ftsA and ftsQ are not transcribed (Wortinger et al., 2000). ctrA transcription from the P2 promoter depends on the presence of a low level of phosphorylated CtrA (see section 2.1.1a). ctrA transcription from the P1 promoter is activated by GcrA and supplies the initial CtrA which is then phosphorylated (Holtzendorff et al., 2004). Therefore, the DNA replication checkpoint probably targets CtrA phosphorylation or GcrA activity, or both.
One requirement for this checkpoint is that FtsA or FtsQ should be limiting for division in the next cell cycle. Indeed, the number of FtsA and FtsQ molecules fluctuates such that their concentration is low in swarmer and stalked cells, peaks in pre-divisional cells, and then dramatically decreases after cell division (Martin et al., 2004). Even when an inducible promoter drives ftzA and ftsQ transcription, FtsA and FtsQ levels vary during the cell cycle. The half-life of FtsA increases from 13 minutes in swarmer cells to 55 minutes in stalked cells, suggesting that FtsA is specifically degraded in certain cell types (Martin et al., 2004). Similarly, the half-life of FtsZ decreases from ∼80 min in stalked and early predivisional cells to ∼10 min in late predivisional cells (Kelly et al., 1998). Following cell division, degradation of FtsA and FtsQ reduces their concentration to 1% and 10% of their maximal level, respectively (Martin et al., 2004). These results strongly suggest that de novo synthesis of cell division proteins is required for each division cycle and sets the stage for checkpoint control through the regulation of their expression.
FtsA and FtsQ are dynamically localized throughout the cell cycle (Fig. 10). After their synthesis in stalked cells, FtsA and FtsQ are distributed throughout cells in a spiral-like pattern. Both proteins are recruited to the mid-cell in late pre-divisional cells, consistent with their maximum level of expression and the requirement for FtsA in late stages of C. crescentus cell division (Martin et al., 2004).
4.4 Cell Shape
Bacteria are characterized by a myriad of different shapes and sizes, including the large viviparous Epulopiscium fishelsoni (Angert et al., 1993; Fishelson et al., 1985), the star shaped bacterium, Stella strain IFAM1312 (Young, 2006), and the small, vibrioid, prosthecate bacterium, C. crescentus (Poindexter, 1964). Why do bacteria have different shapes and sizes? The specific shape of a bacterium is generally thought to confer a selective advantage to the bacterium in a certain environment (Young, 2006). For example, different bacterial shapes have varying abilities to take up nutrients or to protect the cell from predation (Young, 2007). The vibrioid shape of C. crescentus may contribute to its ability to disperse (Ausmees et al., 2003) and the stalk functions in nutrient uptake (Wagner et al., 2006). In addition, the shape of surviving C. crescentus cells changes dramatically during late stationary phase, indicating that changes in morphology are likely to confer additional advantages (Wortinger et al., 1998). What mechanisms govern the formation and maintenance of cell shapes? In C. crescentus, changes in cell shape occur throughout out the cell cycle and are dictated by cell growth, cell division, and stalk elongation. This section will address how the shape of C. crescentus is generated, maintained, and changed throughout the cell cycle.
4.4.1 MreB, MreC, RodA, and PBP2
The mre operon (murein cluster e) contains the mreB, mreC, mreD, rodA (mrdB), and mrdA (PBP2) genes, which are involved in maintaining cell shape, are conserved in nearly all of the rod shaped bacteria (Wachi et al., 1987). In C. crescentus and other rod-shaped bacteria, the proteins encoded by these genes determine and maintain bacterial cell shape by functioning as cytoskeletal proteins and by controlling peptidoglycan synthesis (Dye et al., 2005; Wachi et al., 1987; Wagner et al., 2005). MreB is an essential actin-like cell shape protein required for the maintenance of cell shape during growth. MreB depletion or treatment with A22, a drug that inhibits MreB polymerization, results in the formation of lemon-shaped cells (Fig. 11) (Figge et al., 2004; Gitai et al., 2004; Wagner et al., 2005). In C. crescentus, MreB also plays a role in stalk elongation (see section 3.3.2) and chromosome segregation (see section 4.2.5) (Gitai et al., 2004; Wagner et al., 2005).
Figure 11.
Cell shape proteins in C. crescentus. (A) Localization of cell shape proteins during the cell cycle exhibits. The proteins are represented in gray. MreB localizes as a spiral in swarmer cells, coalesces to a ring in the stalked and predivisional cells, and returns to a spiral at the end of cell division. MreC and PBP2 localize in a similar spiral-like pattern throughout the cell cycle. CreS aligns along the inner curve of the cell throughout the cell cycle. (B) Cell shape protein localization in swarmer cells. MreC and PBP2 co-align while the MreB spiral is offset from the MreC/PBP2 spirals. CreS lies along the inner curve of the bacterium. (C) Cell shapes resulting from the absence or inhibition of cytoskeletal proteins. Gene deletions and protein depletions are indicated by (-). A22 targets MreB and cells grown in its presence have an identical phenotype to cells depleted for MreB. PBP2 is inhibited by the addition of mecillinam.
The transcript and protein levels of MreB remain constant throughout the cell cycle; however MreB is dynamically localized during the cell cycle. MreB is primarily found as a cytoplasmic spiral that condenses to a ring at the mid-cell in a FtsZ and TipN-dependent manner during cell division (Fig. 11)(Figge et al., 2004; Lam et al., 2006). Single MreB proteins have been shown to undergo treadmilling within the spiral structure in vivo (Kim et al., 2006). Treadmilling allows a protein polymer to remain stationary, despite the movement of individual proteins as new monomers are polymerized at one end of the polymer and old monomers are depolymerized from the other end (Kim et al., 2006). Addition of A22 results in a nearly immediate delocalization of MreB from the cytoplasmic spiral or mid-cell localization (Gitai et al., 2005).
RodA, MreC, and PBP2 contribute to the maintenance of cell shape and control of cell width. Depletion of either MreC or RodA results in severe cell shape defects similar to that seen following MreB depletion (Fig. 11) (Divakaruni et al., 2007; Divakaruni et al., 2005; Wagner et al., 2005). Treatment of C. crescentus with mecillinam, an antibiotic that targets PBP2 in E. coli, generates wide filamentous cells (Fig. 11) (Koyasu et al., 1983; Seitz and Brun, 1998; Wagner et al., 2005). Throughout the entire cell cycle, MreC and PBP2 form longitudinal spirals, which encompass the whole cell (Fig. 11) (Divakaruni et al., 2005; Dye et al., 2005; Figge et al., 2004; Gitai et al., 2004). The cytoplasmic spirals of MreC and PBP2 co-localize in vivo and affinity chromatography experiments have shown that MreC and PBP2 interact in vitro (Divakaruni et al., 2005; Dye et al., 2005; Figge et al., 2004). Notably, the spirals formed by MreC and PBP2 are offset from the spirals formed by MreB (Fig. 11). While the localization of MreB and MreC is independent of one another, proper localization of PBP2 requires the presence of both MreB and MreC (Divakaruni et al., 2005; Dye et al., 2005; Figge et al., 2004). In both E. coli and C. crescentus, cell elongation depends on RodA, which controls the transpeptidase activity of PBP2 (Shih and Rothfield, 2006; Wagner et al., 2005). Depleting either MreC or RodA disrupts peptidoglycan synthesis (Divakaruni et al., 2007; Ishino et al., 1986) but not the localization of PBP2. RodA and PBP2 are also required for stalk formation, indicating that stalk biosynthesis is a specialized form of cell elongation (Seitz and Brun, 1998; Wagner et al., 2005). The localization of RodA remains unknown due to the technical challenges associated with tracking the localization of inner membrane proteins. The function of MreD in maintaining the cell shape of C. crescentus has not been determined; however in E. coli, MreD is essential and its depletion results in spherical cells (Kruse et al., 2005).
4.4.2 CreS
C. crescentus cells undergo morphological changes throughout the cell cycle, but the cells always remain curved. While depletion or disruption of mreB, mreC, rodA, or mrdA causes defects in cell morphology, they are not responsible for the characteristic curved shape of C. crescentus. A transposon mutagenesis screen for cell shape defects identified two straight mutants with mutations that mapped to the creS gene, which encodes the protein crescentin (Fig. 11) (Ausmees et al., 2003). The straight phenotype was confirmed in deletion mutants of creS, suggesting that crescentin is responsible for imparting the curve shape to C. crescentus cells. Crescentin localizes along the concave side of the cell along the cytoplasmic side of the inner membrane (Fig. 11) (Ausmees et al., 2003); however CreS-GFP is nonfunctional and requires wild-type CreS for localization (Ausmees et al., 2003). A creS mutant that contains creS-gfp as its only copy of crescentin is straight and CreS-GFP localizes randomly in the cytoplasm as a curved filament (Ausmees et al., 2003). Crescentin is a curved polymeric protein that is ∼25% identical and 40% similar and to cytoskeletal intermediate filaments, which play a role in controlling eukaryotic cell shape (Ausmees et al., 2003). The existence of intermediate filaments in bacterial cells completes the set of eukaryotic cytoskeletal homologs found within bacteria including the actin homologs, MreB/ParM/FtsA, and the tubulin homolog, FtsZ (Ayako et al., 1992; Bork et al., 1992; Erickson, 1995; Jensen and Gerdes, 1997; Lutkenhaus, 1993; Margolin, 2004; Sanchez et al., 1994; van den Ent et al., 2001).
Crescentin is also required for the maintenance of cell shape during stationary phase. During late stationary phase, a small percentage of cells survive and adopt an elongated helical morphology (Wortinger et al., 1998). The elongated helical cells have a decreased level of FtsZ, explaining why the cells become elongated, but not why the cells become coiled. Crescentin is essential for the helical nature of stationary phase cells and is localized along the inner coil of the cell (Ausmees et al., 2003). This observation indicates that crescentin follows a helical pitch that is revealed only in elongated stationary cells. In exponential cells, the short filaments of crescentin appear only slight curved (Ausmees et al., 2003).
4.4.3 Peptidoglycan synthesis
While MreB, MreC, PBP2, and RodA maintain the proper cell shape throughout the cell cycle, these proteins are not directly responsible for mediating cell shape changes. How are changes in cell shape changes mediated? The generation and maintenance of bacterial cell shape requires temporal and spatial control of cell growth, cell division, and stalk elongation. Cell growth, cell division, and stalk elongation are thought to require an expansion of the peptidoglycan cell wall. During cell growth, elongation is thought to occur as new peptidoglycan is inserted into the existing peptidoglycan cell wall along each side of the cells (De Pedro et al., 1997). During cell division and stalk elongation, peptidoglycan precursor synthesis and peptidoglycan insertion is thought to be localized to the specific sites of elongation (Aaron et al., 2007; Divakaruni et al., 2007). In this section, the role of proteins involved in peptidoglycan synthesis and insertion at the sites of cell division and stalk synthesis is discussed.
4.4.3a FtsZ, MurG, and PBP2
Peptidoglycan synthesis at the site of cell division requires the presence of MurG, PBP2, and FtsZ. MurG, a glycosyltransferase involved in peptidoglycan precursor synthesis, contributes to the coordination of peptidoglycan synthesis along the length of the cell and PBPs are responsible for the insertion of new peptidoglycan to facilitate changes in cell shape (Aaron et al., 2007; Divakaruni et al., 2007). Experiments conducted in C. crescentus using D-cysteine or fluorescently labeled vancomycin, which incorporate into newly made peptidoglycan, determined that peptidoglycan synthesis occurs along the cell body and at the mid-cell (Aaron et al., 2007; De Pedro et al., 1997; Divakaruni et al., 2007). In cells depleted of FtsZ, no new synthesis was observed at the mid-cell, indicating that FtsZ directs peptidoglycan synthesis at that site (Aaron et al., 2007; Divakaruni et al., 2007).
What is the role of FtsZ in directing peptidoglycan synthesis to the mid-cell? The transpeptidase protein, PBP2, which is likely involved in the insertion of new peptidoglycan into the cell wall, is mislocalized in the absence of FtsZ (Dye et al., 2005). When MreB is depleted, PBP2 mislocalizes from a spiral shape to the mid-cell; however, in cells depleted of FtsZ and treated with the MreB inhibitor A22, PBP2 remains localized as a spiral. This indicates that FtsZ plays an indirect role in PBP2 localization. While other bacteria form septa during cell division, C. crescentus must add cell wall material as it pinches and invaginates.
To further elucidate the role of FtsZ in transverse peptidoglycan synthesis in C. crescentus, peptidoglycan synthesis was tracked using the localization of MurG, an enzyme that functions in a late step to produce a peptidoglycan precursor (Aaron et al., 2007; Divakaruni et al., 2007). Despite using similar strategies to localize MurG, conflicting data was obtained by the two research groups. Aaron et al. (2007) showed that MurG-GFP had a dynamic behavior in that it displayed diffuse localization in swarmer cells, mostly migrated to the mid-cell during cell division, and released from the mid-cell to diffuse localization again prior to completion of cell division. They determined that MurG-GFP localizes to the mid-cell in an FtsZ-dependent and MreB-independent manner (Aaron et al., 2007). This was demonstrated using a functional murG-mgfp fusion that provided the cell with its only copy of murG (Aaron et al., 2007). These observations are consistent with immunolocalization studies in E. coli, where MurG has also been shown to be localized to the mid-cell in an MreB-independent manner (Mohammadi et al., 2007).
Conversly, Divakaruni et al. (2007) showed that MurG-mCherry displayed an MreB dependent banded or punctate pattern along the cell with concentrations of localization at the poles. When the cells were treated with A22, MurG-mCherry became localized as single foci at the mid-cell or the poles, or both (Divakaruni et al., 2007). These observations were confirmed using immunolocalization (Divakaruni et al., 2007). How can the differences in MurG localization be reconciled? Both groups used C-terminal fluorescent protein fusions; however murG-mCherry was expressed in the presence of wild-type murG in one case (Divakaruni et al., 2007), while murG-mgfp was the only copy of murG in the cell in the other case (Aaron et al., 2007). Thus, the differences in MurG localization observed by the two groups may be due to the differences in their respective MurG constructs. Despite the differences in the construction of the strains used to localize MurG, it is clear that mid-cell localization occurs and likely depends on FtsZ; however it remains unclear if mid-cell localization of MurG depends on MreB. FtsZ is also required for transverse peptidoglycan synthesis to form the characteristic stalk crossbands (Divakaruni et al., 2007). The role of FtsZ in peptidoglycan synthesis of C. crescentus is consistent with results in E. coli, which show that FtsZ is required for peptidoglycan synthesis near the poles, indicating the FtsZ function is not restricted to the mid-cell (Varma et al., 2007).
4.4.3b MreC, MltA, and MipA
In order for new peptidoglycan to be inserted into the cell wall at specific locations, localized cell wall degradation must occur. MltA, a lytic transglycoslase, and its interacting partner, MipA, are localized in a band-like pattern that is similar to that of PBP2, indicating that cell wall degradation and synthesis may be coupled (Divakaruni et al., 2007; Figge and Gober, 2003). In addition, both MltA and MipA are localized at the stalked cell pole where peptidoglycan synthesis is likely to occur (Divakaruni et al., 2007). Localization of MltA and MipA depends on the presence of MreC, but not MreB. In C. crescentus, MreC has been shown to interact with complexes of PBPs (Divakaruni et al., 2005). Taken together, these results suggest that MreC coordinates the localization of multiprotein complexes containing proteins for the degradation and synthesis of peptidoglycan. Efficient peptidoglycan synthesis leading to changes in cell shape likely requires the coordination of a number of cell shape determining proteins, cell division proteins, and biosynthetic enzymes.
5. Concluding Remarks
C. crescentus is a powerful model system for the study of cell cycle progression at the molecular level. Multiple levels of regulation, including both temporal and spatial regulation, are required to coordinate polar morphogenesis, chromosome replication, and cell division throughout the cell cycle. Cell cycle progression is mediated by primarily by the activities of three global regulators, CtrA, DnaA, and GcrA, which are expressed in a specific temporal sequence. The actions of these three transcriptional regulators directly and indirectly impact the expression of genes encoding both regulatory and structural proteins, at each phase of the cell cycle. The temporal expression of additional regulatory proteins, particularly location-specific two-component signal proteins, is necessary to fine-tune the processes that occur during each phase of the cell cycle. The ability of the cell to monitor progress in each phase of the cell cycle to direct the activation (or repression) of the regulators for subsequent steps in cell cycle progression has proven to be a critical means of feedback control. Regulators not only direct cell-cycle progression, but are also subject to activation (or deactivation) based on the success of previous steps in the cell-cycle. These regulatory feedback loops ensure that cell-cycle progression does not continue until the previous phase is successfully completed and are critical for defining the physiology of the two distinct progeny cells.
Despite significant progress in identifying and understanding the regulatory elements that direct cell-cycle progression, a number of questions remain. Is cell cycle progression in C. crescentus truly hardwired? What signals, if any, are responsible for controlling cell cycle progression? Are multiple signals integrated to control global regulators? Recent studies indicate that cyclic-di-GMP may act as an important signal in mediating cell-cycle progression (for review see Jenal and Malone, 2006). Gain of motility, through flagellum assembly in the swarmer compartment of the predivisonal cell and flagellum activation in the new swarmer cell, requires a reduction in cyclic-di-GMP levels. Conversely, loss of motility, holdfast formation, and stalk biogenesis during the swarmer to stalked cell transition requires an increase in cyclic-di-GMP levels. The signal responsible for initiating production of cyclic-di-GMP levels remains unknown. Given the large number of two-component signal transduction proteins involved in mediating cell-cycle progression it seems likely that additional signals remain to be discovered which impact cell-cycle progression. Irrespective of the physiological and/or environmental signals which trigger the production, activation, and destruction of regulatory proteins, it is clear that successful progression through the cell cycle of C. crescentus requires multiple levels of regulation which must be precisely orchestrated in both time and space.
Acknowledgments
Acknowledgements
We wish to thank members of the Brun laboratory for critical reading of this manuscript and providing unpublished data. Research in our laboratory is supported by grants GM077648 and GM51986 from the National Institutes of Health, MCB0731950 from the National Science Foundation, and by the Indiana METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. P.J.B.B. is supported by a postdoctoral National Institutes of Health National Research Service Award number F32AI072992 from the National Institute of Allergy and Infectious Diseases.
Abbreviations
- A22
S-(3,4-dichlorobenzyl)isothiourea
- AFM
atomic force microscopy
- bps
basepairs
- c-di-GMP
cyclic-diguanosine monophosphate
- Cori
C. crescentus origin of replication
- ECT
electron cryotomography
- Flp
fimbrial low-weight molecular protein
- GDP
guanosine diphosphate
- GFP
green fluorescent protein
- GTP
guanosine triphosphate
- Hpt
histidine phosphotransferase
- IHF
integration host factor
- NAG
N-acetyl glucosamine
- ∼P
phosphorylated
- PBP2
penicillin binding protein 2
- WGA
wheat germ agglutinin
- YFP
yellow fluorescent protein
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- Abraham WR, Strompl C, Meyer H, Lindholst S, Moore ER, Christ R, Vancanneyt M, Tindall BJ, Bennasar A, Smit J, Tesar M. Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Int. J. Syst. Bacteriol. 1999;49(Pt 3):1053–73. doi: 10.1099/00207713-49-3-1053. [DOI] [PubMed] [Google Scholar]
- Addinall SG, Holland B. The tubulin ancester, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J. Mol. Biol. 2002;318:219–236. doi: 10.1016/S0022-2836(02)00024-4. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Hughes KT. Regulation of flagellar assembly. Currents Opinion in Microbiology. 2002;5:160–5. doi: 10.1016/s1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Jenal U. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 1999;32:379–392. doi: 10.1046/j.1365-2958.1999.01358.x. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 2003;47 doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- Alipour-Assiabi E, Li G, Powers TR, Tang JX. Fluctuation analysis of Caulobacter crescentus adhesion. Biophys. J. 2006;90:2206–2212. doi: 10.1529/biophysj.105.064592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Martinez CE, Baldini RL, Gomes SL. A Caulobacter crescentus extracytoplasmic function sigma factor mediating the response to oxidative stress in stationary phase. J. Bacteriol. 2006;188:1835–46. doi: 10.1128/JB.188.5.1835-1846.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Martinez CE, Lourenco RF, Baldini RL, Laub MT, Gomes SL. The ECF sigma factor sigma(T) is involved in osmotic and oxidative stress responses in Caulobacter crescentus. Mol. Microbiol. 2007;66:1240–55. doi: 10.1111/j.1365-2958.2007.06005.x. [DOI] [PubMed] [Google Scholar]
- Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 2004;186:5775–81. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DK, Ohta N, Wu J, Newton A. Regulation of the Caulobacter crescentus rpoN gene and function of the purified σ54 in flagellar gene transcription. Mol. Gen. Genet. 1995;246:697–706. doi: 10.1007/BF00290715. [DOI] [PubMed] [Google Scholar]
- Anderson PE, Gober JW. FlbT, the post-transcriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5′ untranslated region of flagellin mRNA. Mol. Microbiol. 2000;38:41–52. doi: 10.1046/j.1365-2958.2000.02108.x. [DOI] [PubMed] [Google Scholar]
- Angert ER, Clements KD, Pace NR. The largest bacterium. Nature. 1993;362:239–241. doi: 10.1038/362239a0. [DOI] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- Ayako N, Naterstad K, Kristensen T, Lopez R, Bork P, KolstØ A-B. Cloning of a gene from Bacillus cereus with homology to the mreB gene from Escherichia coli. Gene. 1992;122:181–185. doi: 10.1016/0378-1119(92)90047-s. [DOI] [PubMed] [Google Scholar]
- Badger JH, Hoover TR, Brun YV, Weiner RM, Laub MT, Alexandre G, Mrazek J, Ren Q, Paulsen IT, Nelson KE, Khouri HM, Radune D, Sosa J, Dodson RJ, Sullivan SA, Rosovitz MJ, Madupu R, Brinkac LM, Durkin AS, Daugherty SC, Kothari SP, Giglio MG, Zhou L, Haft DH, Selengut JD, Davidsen TM, Yang Q, Zafar N, Ward NL. Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus. J. Bacteriol. 2006;188:6841–50. doi: 10.1128/JB.00111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc. Natl. Acad. Sci. U. S. A. 2007;104:462–6. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Sivanathan V, Possoz C, Barre F-X, Cornet F. FtsK, a literate chromosome segregation machine. Mol. Microbiol. 2007;64:1434–1441. doi: 10.1111/j.1365-2958.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006a;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Biondi EG, Skerker JM, Arif M, Prasol MS, Perchuk BS, Laub MT. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol. Microbiol. 2006b;59:386–401. doi: 10.1111/j.1365-2958.2005.04970.x. [DOI] [PubMed] [Google Scholar]
- Bodenmiller D, Toh E, Brun YV. Development of surface adhesion in Caulobacter crescentus. J. Bacteriol. 2004;186:1438–47. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordes P, Wigneshweraraj SR, Zhang X, Buck M. Sigma54-dependent transcription activator phage shock protein F of Escherichia coli: a fragmentation approach to identify sequences that contribute to self-association. Biochem. J. 2004;378:735–44. doi: 10.1042/BJ20031464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassinga AK, Marczynski GT. Replication intermediate analysis confirms that chromosomal replication origin initiates from an unusual intergenic region in Caulobacter crescentus. Nucleic Acids Res. 2001;29:4441–51. doi: 10.1093/nar/29.21.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassinga AK, Siam R, McSween W, Winkler H, Wood D, Marczynski GT. Conserved response regulator CtrA and IHF binding sites in the alpha-proteobacteria Caulobacter crescentus and Rickettsia prowazekii chromosomal replication origins. J. Bacteriol. 2002;184:5789–99. doi: 10.1128/JB.184.20.5789-5799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun YV, Janakiraman R. In: Prokaryotic Development. Brun YV, Shimkets LJ, editors. American Society for Microbiology; Washington, D.C.: 2000. pp. 297–317. [Google Scholar]
- Brun YV, Shapiro L. A temporally controlled sigma factor is required for cell-cycle dependent polar morphogenesis in Caulobacter. Genes and Development. 1992;6:2395–2408. doi: 10.1101/gad.6.12a.2395. [DOI] [PubMed] [Google Scholar]
- Buddelmeijer N, Aarsman MEG, Kolk AHJ, Vicente M, Nanninga N. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J. Bacteriol. 1998;180:6107–6116. doi: 10.1128/jb.180.23.6107-6116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17084–9. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Hottes AK, McAdams HH, McGrath PT, Viollier PH, Shapiro L. Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. EMBO Journal. 2006;25:377–86. doi: 10.1038/sj.emboj.7600935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Minev M, Beckwith J. Analysis of ftsQ mutant alleles in Escherichia coli: complementation, septal localization, and recruitment of downstream cell division proteins. J. Bacteriol. 2002;184:695–705. doi: 10.1128/JB.184.3.695-705.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- Chen JC, Weiss DS, Ghigo JM, Beckwith J. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J. Bacteriol. 1999;181:521–30. doi: 10.1128/jb.181.2.521-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6590–5. doi: 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Allan MG, Folcher M, Jeno P, Grzesiek S, Jenal U. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4112–7. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J, Hardy GG, Bodenmiller D, Toh E, Hinz A, Brun YV. The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk. Mol. Microbiol. 2003;49:1671–1683. doi: 10.1046/j.1365-2958.2003.03664.x. [DOI] [PubMed] [Google Scholar]
- Collier J, Murray SR, Shapiro L. DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO Journal. 2006;25:346–56. doi: 10.1038/sj.emboj.7600927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RF, Beis K, Dong C, Botting CH, McDonnell C, Ford RC, Clarke BR, Whitfield C, Naismith JH. The 3D structure of a periplasm-spanning platform required for assembly of group 1 capsular polysaccharides in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2390–2395. doi: 10.1073/pnas.0607763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 1987;41:435–64. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Cozzone AJ, Grangeasse C, Doublet P, Duclos B. Protein phosphorylation on tyrosine in bacteria. Arch. Microbiol. 2004;181:171–181. doi: 10.1007/s00203-003-0640-6. [DOI] [PubMed] [Google Scholar]
- Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson S, McAdams H, Shapiro L. A Genetic Oscillator and the Regulation of Cell Cycle Progression in Caulobacter crescentus. Cell Cycle. 2004;3 doi: 10.4161/cc.3.10.1181. [DOI] [PubMed] [Google Scholar]
- Crosson S, Ragagopal S, Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry (Mosc) 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- Crymes WB, Jr., Zhang D, Ely B. Regulation of podJ expression during the Caulobacter crescentus cell cycle. J. Bacteriol. 1999;181:3967–73. doi: 10.1128/jb.181.13.3967-3973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva AC, Simao RC, Susin MF, Baldini RL, Avedissian M, Gomes SL. Downregulation of the heat shock response is independent of DnaK and sigma32 levels in Caulobacter crescentus. Mol. Microbiol. 2003;49:541–53. doi: 10.1046/j.1365-2958.2003.03581.x. [DOI] [PubMed] [Google Scholar]
- De Pedro M, Quintela JC, Höltje J-V, Schwartz H. Murein Segregation in Escherichia coli. J. Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnen ST, Newton A. Chromosome replication during development in Caulobacter crescentus. J. Mol. Biol. 1972;64:671–80. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- Din N, Quardokus EM, Sackett MJ, Brun YV. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol. Microbiol. 1998;27:1051–1064. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- Dingwall A, Shapiro L. Rate, origin, and bidirectionality of Caulobacter chromosome replication as determined by pulsed-field gel electrophoresis. Proc. Natl. Acad. Sci. U. S. A. 1989;86:119–123. doi: 10.1073/pnas.86.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AV, Baida C, White C, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol. Microbiol. 2007;66:174–188. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- Divakaruni AV, Loo RRO, Xie Y, Loo JA, Gober JW. The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18602–7. doi: 10.1073/pnas.0507937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Reisenauer A, Shapiro L. Feedback control of a master bacterial cell-cycle regulator. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Drummelsmith J, Whitfield C. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO Journal. 2000;19:57–66. doi: 10.1093/emboj/19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton RJ, Xu Z, Gober JW. Linking structural assembly to gene expression: a novel mechanism for regulating the activity of a sigma54 transcription factor. Mol. Microbiol. 2005;58:743–57. doi: 10.1111/j.1365-2958.2005.04857.x. [DOI] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z. Two independent spiral structures control cell shape in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18608–18613. doi: 10.1073/pnas.0507708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easter J, Jr., Gober JW. ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol. Cell. 2002;10:427–34. doi: 10.1016/s1097-2765(02)00594-4. [DOI] [PubMed] [Google Scholar]
- England JC, Gober JW. Cell cycle control of cell morphogenesis in Caulobacter. Currents Opinion in Microbiology. 2001;4:674–80. doi: 10.1016/s1369-5274(01)00268-5. [DOI] [PubMed] [Google Scholar]
- Entcheva-Dimitrov P, Spormann AM. Dynamics and control of biofilms of the oligotrophic bacterium Caulobacter crescentus. J. Bacteriol. 2004;186:8254–66. doi: 10.1128/JB.186.24.8254-8266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. FtsZ, a prokaryotic homolog of tubulin? Cell. 1995;80:367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- Feucht A, Lucet I, Yudkin MD, Errington J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 2001;40:115–125. doi: 10.1046/j.1365-2958.2001.02356.x. [DOI] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- Figge RM, Easter J, Gober JW. Productive interaction between the chromosome partitioning proteins, ParA and ParB, is required for the progression of the cell cycle in Caulobacter crescentus. Mol. Microbiol. 2003;47:1225–1237. doi: 10.1046/j.1365-2958.2003.03367.x. [DOI] [PubMed] [Google Scholar]
- Figge RM, Gober JW. Cell shape, division and development: the 2002 American Society for Microbiology (ASM) conference on prokaryotic development. Mol. Microbiol. 2003;47:1475–1483. doi: 10.1046/j.1365-2958.2003.03397.x. [DOI] [PubMed] [Google Scholar]
- Fischer B, Rummel G, Aldridge P, Jenal U. The FtsH protease is involved in development, stress response and heat shock control in Caulobacter crescentus. Mol. Microbiol. 2002;44:461–78. doi: 10.1046/j.1365-2958.2002.02887.x. [DOI] [PubMed] [Google Scholar]
- Fishelson L, Montgomery WL, Myrberg AA. A unique symbiosis in the gut of tropical herbivorous surgeonfish (Acanthuridae: Teleostei) from the Red Sea. Science. 1985;229:49–51. doi: 10.1126/science.229.4708.49. [DOI] [PubMed] [Google Scholar]
- Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Gerdes K, MØ;ller-Jensen J, Jensen RB. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Gober JW, Boyd CH, Jarvis M, Mangan EK, Rizzo MF, Wingrove JA. Temporal and spatial regulation of fliP, an early flagellar gene of Caulobacter crescentus that is required for motility and normal cell division. J. Bacteriol. 1995;177:3656–3667. doi: 10.1128/jb.177.13.3656-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gober JW, England JC. In: Prokaryotic Development. Brun YV, Shimkets LJ, editors. American Society for Microbiology; Washington, D.C.: 2000. pp. 319–339. [Google Scholar]
- Gober JW, Shapiro L. Integration Host Factor is Required for the Activation of developmentally Regulated Genes in Caulobacter. Genes Dev. 1990;4:1494–1504. doi: 10.1101/gad.4.9.1494. [DOI] [PubMed] [Google Scholar]
- Gober JW, Shapiro L. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol. Biol. Cell. 1992;3:913–926. doi: 10.1091/mbc.3.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes SL, Juliani MH, Maia JCC, Silva AM. Heat Shock Protein Synthesis During Development in Caulobacter crescentus. J. Bacteriol. 1986;168:923–930. doi: 10.1128/jb.168.2.923-930.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonin M, Quardokus EM, O'Donnol D, Maddock J, Brun YV. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J. Bacteriol. 2000;182:337–47. doi: 10.1128/jb.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol. Microbiol. 2001;40:485–97. doi: 10.1046/j.1365-2958.2001.02404.x. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol. Microbiol. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- Götz F. Staphylococcal biofilms. Mol. Microbiol. 2002;43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- Graumann PL. SMC proteins in bacteria: Condensation motors for chromosome segregation? Biochimie. 2001;83:53–59. doi: 10.1016/s0300-9084(00)01218-9. [DOI] [PubMed] [Google Scholar]
- Grunenfelder B, Gehrig S, Jenal U. Role of the Cytoplasmic C Terminus of the FliF Motor Protein in Flagellar Assembly and Rotation. J. Bacteriol. 2003;185:1624–33. doi: 10.1128/JB.185.5.1624-1633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunenfelder B, Tawfilis S, Gehrig S, Eglin D, Jenal U. Identification of the protease and the turnover signal responsible for cell cycle-dependent degradation of the Caulobacter FliF motor protein. J. Bacteriol. 2004;186:4960–71. doi: 10.1128/JB.186.15.4960-4971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht GB, Lane T, Ohta N, Newton A. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO Journal. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht GB, Newton A. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 1995;177:6223–6229. doi: 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrici AT, Johnson DE. Studies of freshwater bacteria: stalked bacteria, a new order of Schizomycetes. J. Bacteriol. 1935;30:61–93. doi: 10.1128/jb.30.1.61-93.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol. Microbiol. 2003;47:929–41. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- Hirsch P. In: Bergey's Manual of Systematic Bacteriology. 9th edn. Staley JT, Bryant MP, Pfenning N, Holt JG, editors. vol. 3. Williams and Wilkins; Baltimore: 1989. pp. 1897–1904. [Google Scholar]
- Holtzendorff J, Hung D, Brende P, Reisenauer A, Viollier PH, McAdams HH, Shapiro L. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science. 2004;304:983–987. doi: 10.1126/science.1095191. [DOI] [PubMed] [Google Scholar]
- Holtzendorff J, Reinhardt J, Viollier PH. Cell cycle control by oscillating regulatory proteins in Caulobacter crescentus. Bioessays. 2006;28:355–61. doi: 10.1002/bies.20384. [DOI] [PubMed] [Google Scholar]
- Hong SJ, Lessner FH, Mahen EM, Keiler KC. Proteomic identification of tmRNA substrates. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17128–33. doi: 10.1073/pnas.0707671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SJ, Tran QA, Keiler KC. Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB. Mol. Microbiol. 2005;57:565–75. doi: 10.1111/j.1365-2958.2005.04709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover TR, Santero E, Porter S, Kustu S. The Integration Host Factor Stimulates Interaction of RNA Polymerase with NIFA, the Transcriptional Activator for Nitrogen Fixation Operons. Cell. 1990;63:11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Hottes AK, Meewan M, Yang D, Arana N, Romero P, McAdams HH, Stephens C. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 2004;186:1448–61. doi: 10.1128/JB.186.5.1448-1461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottes AK, Shapiro L, McAdams HH. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 2005;58:1340–1353. doi: 10.1111/j.1365-2958.2005.04912.x. [DOI] [PubMed] [Google Scholar]
- Hu B, Yang G, Zhao W, Zhang Y, Zhao J. MreB is important for cell shape but not for chromosome segregation of the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 2007;63:1640–1652. doi: 10.1111/j.1365-2958.2007.05618.x. [DOI] [PubMed] [Google Scholar]
- Hu P, Brodie EL, Suzuki Y, McAdams HH, Andersen GL. Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J. Bacteriol. 2005;187:8437–49. doi: 10.1128/JB.187.24.8437-8449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Hung DY, Shapiro L. A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13160–5. doi: 10.1073/pnas.202495099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10935–40. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland MM, Karty JA, Quardokus EM, Reilly JP, Brun YV. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol. Microbiol. 2002;45:1029–41. doi: 10.1046/j.1365-2958.2002.03071.x. [DOI] [PubMed] [Google Scholar]
- Ishino F, Park W, Tomioka S, Tamaki S, Takase I, Kunugita K, Matsuzawa H, Asoh S, Ohta T, Spratt B, Matsuhashi M. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and RodA protein. J. Biol. Chem. 1986;261:7024–7031. [PubMed] [Google Scholar]
- Jacobs C, Ausmees N, Cordwell SJ, Shapiro L, Laub MT. Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol. Microbiol. 2003;47:1279–1290. doi: 10.1046/j.1365-2958.2003.03379.x. [DOI] [PubMed] [Google Scholar]
- Jacobs C, Domian IJ, Maddock JR, Shapiro L. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell. 1999;97:111–20. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- Jacobs C, Hung D, Shapiro L. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4095–100. doi: 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janakiraman RS, Brun YV. Cell cycle control of a holdfast attachment gene in Caulobacter crescentus. J. Bacteriol. 1999;181:1118–1125. doi: 10.1128/jb.181.4.1118-1125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson KK. What drives bacteria to produce a biofilm. FEMS Microbiol. Lett. 2004;236:163–173. doi: 10.1016/j.femsle.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Jenal U. Signal transduction mechanisms in Caulobacter crescentus development and cell cycle control. FEMS Microbiology Rev. 2000;24:177–91. doi: 10.1016/S0168-6445(99)00035-2. [DOI] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. An Essential Protease Involved in Bacterial Cell cycle control. EMBO Journal. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Jenal U, White J, Shapiro L. Caulobacter Flagellar Function, but Not Assembly, Requires FliL, a Non-Polarly Localized Membrane Protein Present in All Cell Types. J. Mol. Biol. 1994;243:227–244. doi: 10.1006/jmbi.1994.1650. [DOI] [PubMed] [Google Scholar]
- Jensen GJ, Briegel A. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr. Opin. Struct. Biol. 2007;17:260–7. doi: 10.1016/j.sbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Jensen RB. Analysis of the terminus region of the Caulobacter crescentus chromosome and identification of the dif site. J. Bacteriol. 2006;188:6016–9. doi: 10.1128/JB.00330-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Gerdes K. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parC complex. J. Mol. Biol. 1997;269:505–513. doi: 10.1006/jmbi.1997.1061. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Shapiro L. Cell-cycle-regulated expressoin and subcellular localization of the Caulobacter crescentus SMC chromosome structural protein. J. Bacteriol. 2003;185:3068–75. doi: 10.1128/JB.185.10.3068-3075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Wang SC, Shapiro L. A moving DNA replication factory in Caulobacter crescentus. EMBO Journal. 2001;20:4952–4963. doi: 10.1093/emboj/20.17.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HC, Schmidt JM. Ultrastructural Study of Crossbands Occurring in the Stalks of Caulobacter crescentus. J. Bacteriol. 1973;116:466–470. doi: 10.1128/jb.116.1.466-470.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Ferguson NL, Alley MR. New members of the ctrA regulon: the major chemotaxis operon in Caulobacter is CtrA dependent. Microbiology. 2001;147:949–58. doi: 10.1099/00221287-147-4-949. [DOI] [PubMed] [Google Scholar]
- Judd EM, Ryan KR, Moerner WE, Shapiro L, McAdams HH. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8235–40. doi: 10.1073/pnas.1433105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader A, Simm R, Gerstel U, Morr M, Romling U. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar typhimurium. Mol. Microbiol. 2006;60:602–616. doi: 10.1111/j.1365-2958.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- Kanbe M, Shibata S, Umino Y, Jenal U, Aizawa SI. Protease susceptibility of the Caulobacter crescentus flagellar hook-basal body: a possible mechanism of flagellar ejection during cell differentiation. Microbiology. 2005;151:433–8. doi: 10.1099/mic.0.27386-0. [DOI] [PubMed] [Google Scholar]
- Karczmarek A, Baselga RM-A, Alexeeva S, Hansen FG, Vicente M, Nanninga N, Blaauwen T. DNA and origin region segregation are not affected by the transition from rod to sphere after inhibition of Escherichia coli MreB by A22. Mol. Microbiol. 2007;65:51–63. doi: 10.1111/j.1365-2958.2007.05777.x. [DOI] [PubMed] [Google Scholar]
- Keiler KC. Physiology of tmRNA: what gets tagged and why? Curr. Opin. Microbiol. 2007;10:169–75. doi: 10.1016/j.mib.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Shapiro L. tmRNA in Caulobacter crescentus is cell cycle regulated by temporally controlled transcription and RNA degradation. J. Bacteriol. 2003a;185:1825–30. doi: 10.1128/JB.185.6.1825-1830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Shapiro L. tmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J. Bacteriol. 2003b;185:573–80. doi: 10.1128/JB.185.2.573-580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AJ, Sackett MJ, Din N, Quardokus EM, Brun YV. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biology. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyasu S, Fukuda A, Okada Y, Poindexter JS. Penicillin-binding Proteins of the Stalk of Caulobacter crescentus. J. Gen. Microbiol. 1983;129:2789–2799. [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- Kurtz HD, Jr., Smit J. Analysis of a Caulobacter crescentus gene cluster involved in attachment of the holdfast to the cell. J. Bacteriol. 1992;174:687–694. doi: 10.1128/jb.174.3.687-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz HD, Jr., Smit J. The Caulobacter crescentus holdfast: Identification of holdfast attachment complex genes. FEMS Microbiol. Lett. 1994;116:175–182. doi: 10.1111/j.1574-6968.1994.tb06697.x. [DOI] [PubMed] [Google Scholar]
- Lagenaur C, Agabian N. Caulobacter crescentus pili: structure and stage specific expression. J. Bacteriol. 1977;131:340–346. doi: 10.1128/jb.131.1.340-346.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Matroule JY, Jacobs-Wagner C. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev. Cell. 2003;5:149–59. doi: 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Langille SE, Weiner RM. Spatial and temporal deposition of Hyphomonas strain VP-6 capsules involved in biofilm formation. Appl. Environ. Microbiol. 1998;64:2906–2913. doi: 10.1128/aem.64.8.2906-2913.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson RJ, Pate JL. Glucose Transport in Isolated Prosthecae of Asticcacaulis biprosthecum. J. Bacteriol. 1976;126:282–293. doi: 10.1128/jb.126.1.282-293.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro L, McAdams HH. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–8. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- Lawler ML, Larson DE, Hinz AJ, Klein D, Brun YV. Dissection of functional domains of the polar localization factor PodJ in Caulobacter crescentus. Mol. Microbiol. 2006;59:301–16. doi: 10.1111/j.1365-2958.2005.04935.x. [DOI] [PubMed] [Google Scholar]
- Leclerc G, Wang SP, Ely B. A new class of Caulobacter crescentus flagellar genes. J. Bacteriol. 1998;180:5010–9. doi: 10.1128/jb.180.19.5010-5019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-B, Liu C-T, Anzai Y, Kim H, Aono T, Oyaizu H. The hierarchical system of the ‘Alphaproteobacteria’: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2005;55:1907–1919. doi: 10.1099/ijs.0.63663-0. [DOI] [PubMed] [Google Scholar]
- Levi A, Jenal U. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J. Bacteriol. 2006;188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Smith CS, Brun YV, Tang JX. The elastic properties of the Caulobacter crescentus adhesive holdfast are dependent on oligomers of Nacetylglucosamine. J. Bacteriol. 2005;187:257–65. doi: 10.1128/JB.187.1.257-265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Tang JX. Low flagellar motor torque and high swimming efficiency of Caulobacter crescentus swarmer cells. Biophys. J. 2006;91:2726–34. doi: 10.1529/biophysj.106.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO Journal. 2007 doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn M, Dutton RJ, Easter J, O'Donnol D, Gober JW. The conserved flaF gene has a critical role in coupling flagellin translation and assembly in Caulobacter crescentus. Mol. Microbiol. 2005;57:1127–42. doi: 10.1111/j.1365-2958.2005.04745.x. [DOI] [PubMed] [Google Scholar]
- Löwe J, Cordell SC, van den Ent F. Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled-coil inserted. J. Mol. Biol. 2001;306:25–35. doi: 10.1006/jmbi.2000.4379. [DOI] [PubMed] [Google Scholar]
- Lu C, Erickson HP. The straight and curved conformation of FtsZ protofilamentsevidence for rapid exchange of GTP into the curved protofilament. Cell Struct. Funct. 1999;24:285–290. doi: 10.1247/csf.24.285. [DOI] [PubMed] [Google Scholar]
- Lu C, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 2000;182:164–170. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. FtsZ ring in bacterial cytokinesis. Mol. Microbiol. 1993;9:403–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 2007;76:539–62. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Mack D, Nedelmann M, Krokotsch A, Schwarzkopf A. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 1996;178:175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae JD, Smit J. Characterization of Caulobacters isolated from wastewater treatment systems. Appl. Environ. Microbiol. 1991;57:751–758. doi: 10.1128/aem.57.3.751-758.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakooti J, Ely B. Principal Sigma Subunit of the Caulobacter crescentus RNA Polymerase. J. Bacteriol. 1995;177:6854–6860. doi: 10.1128/jb.177.23.6854-6860.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakooti J, Wang SP, Ely B. A Consensus Promoter Sequence for Caulobacter crescentus Genes Involved in Biosynthetic and Housekeeping Functions. J. Bacteriol. 1995;177:4372–4376. doi: 10.1128/jb.177.15.4372-4376.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan EK, Malakooti J, Caballero A, Anderson P, Ely B, Gober JW. FlbT couples flagellum assembly to gene expression in Caulobacter crescentus. J. Bacteriol. 1999;181:6160–70. doi: 10.1128/jb.181.19.6160-6170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczynski GT. Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J. Bacteriol. 1999;181:1984–93. doi: 10.1128/jb.181.7.1984-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczynski GT, Dingwall A, Shapiro L. Plasmid and chromosomal DNA replication and partitioning during the Caulobacter crescentus cell cycle. J. Mol. Biol. 1990;212:709–722. doi: 10.1016/0022-2836(90)90232-B. [DOI] [PubMed] [Google Scholar]
- Marczynski GT, Lentine K, Shapiro L. A developmentally regulated chromosomal origin of replication uses essential transcription elements. Genes Dev. 1995;9:1543–57. doi: 10.1101/gad.9.12.1543. [DOI] [PubMed] [Google Scholar]
- Marczynski GT, Shapiro L. Cell cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J. Mol. Biol. 1992;226:959–977. doi: 10.1016/0022-2836(92)91045-q. [DOI] [PubMed] [Google Scholar]
- Marczynski GT, Shapiro L. Control of Chromosome Replication in Caulobacter crescentus. Annu. Rev. Microbiol. 2002 doi: 10.1146/annurev.micro.56.012302.161103. [DOI] [PubMed] [Google Scholar]
- Margolin W. Bacterial shape: concave coiled coils curve Caulobacter. Curr. Biol. 2004;14:R242–R244. doi: 10.1016/j.cub.2004.02.057. [DOI] [PubMed] [Google Scholar]
- Marques MV, Gomes SL, Gober JW. A Gene Coding for a Putative Sigma 54 Activator Is Developmentally Regulated in Caulobacter crescentus. J. Bacteriol. 1997;179:5502–5510. doi: 10.1128/jb.179.17.5502-5510.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall KC. In: Proceedings of the Third International Congress on Marine Corrosion and Fouling. Acker RF, Brown BF, DePalma JR, Iverson WP, editors. Northwestern Universtiy Press; Gaithersburg, MD: 1972. pp. 625–632. [Google Scholar]
- Martin ME, Trimble MJ, Brun YV. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol. Microbiol. 2004;54:60–74. doi: 10.1111/j.1365-2958.2004.04251.x. [DOI] [PubMed] [Google Scholar]
- Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell. 2004;118:579–90. doi: 10.1016/j.cell.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Matthysse AG, Thomas DL, White AR. Mechanism of cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 1995a;177:1076–81. doi: 10.1128/jb.177.4.1076-1081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse AG, White S, Lightfoot R. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 1995b;177:1069–75. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–47. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, Tan MH, Hillson NJ, Hu P, Shapiro L, McAdams HH. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat. Biotechnol. 2007;25:584–92. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The Symmetrical Structure of Structural Maintenance of Chromosomes (SMC) and MukB Proteins: Long, Antiparallel Coiled Coils, Folded at a Flexible Hinge. J. Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker RI, Smit J. Characterization of the adhesive holdfast of marine and freshwater Caulobacters. Journal of Applied and Environmental Microbiology. 1988;54:2078–2085. doi: 10.1128/aem.54.8.2078-2085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiology Rev. 2002;26:355–74. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- Mitchell D, Smit J. Identification of genes affecting production of the adhesion organelle of Caulobacter crescentus CB2. J. Bacteriol. 1990;172:5425–5431. doi: 10.1128/jb.172.9.5425-5431.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, Karczmarek A, Crouvoisier M, Bouhss A, Mengin-Lecreulx D, den Blaauwen T. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol. Microbiol. 2007;65:1106–1121. doi: 10.1111/j.1365-2958.2007.05851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl DA, Easter J, Gober JW. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol. Microbiol. 2001;42:741–755. doi: 10.1046/j.1365-2958.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Gober JW. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- Moore RL, Marshall KC. Attachment and rosette formation by Hyhomicrobia. Appl. Environ. Microbiol. 1981;42:751–757. doi: 10.1128/aem.42.5.751-757.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir RE, Easter J, Gober JW. The trans-acting flagellar regulatory proteins, FliX and FlbD, play a central role in linking flagellar biogenesis and cytokinesis in Caulobacter crescentus. Microbiology. 2005;151:3699–711. doi: 10.1099/mic.0.28174-0. [DOI] [PubMed] [Google Scholar]
- Muir RE, Gober JW. Regulation of late flagellar gene transcription and cell division by flagellum assembly in Caulobacter crescentus. Mol. Microbiol. 2001;41:117–30. doi: 10.1046/j.1365-2958.2001.02506.x. [DOI] [PubMed] [Google Scholar]
- Muir RE, Gober JW. Mutations in FlbD that relieve the dependency on flagellum assembly alter the temporal and spatial pattern of developmental transcription in Caulobacter crescentus. Mol. Microbiol. 2002;43:597–615. doi: 10.1046/j.1365-2958.2002.02728.x. [DOI] [PubMed] [Google Scholar]
- Muir RE, Gober JW. Regulation of FlbD activity by flagellum assembly is accomplished through direct interaction with the trans-acting factor, FliX. Mol. Microbiol. 2004;54:715–30. doi: 10.1111/j.1365-2958.2004.04298.x. [DOI] [PubMed] [Google Scholar]
- Muir RE, Gober JW. Role of integration host factor in the transcriptional activation of flagellar gene expression in Caulobacter crescentus. J. Bacteriol. 2005;187:949–60. doi: 10.1128/JB.187.3.949-960.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir RE, O'Brien TM, Gober JW. The Caulobacter crescentus flagellar gene, fliX, encodes a novel trans- acting factor that couples flagellar assembly to transcription. Mol. Microbiol. 2001;39:1623–37. doi: 10.1046/j.1365-2958.2001.02351.x. [DOI] [PubMed] [Google Scholar]
- Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen JA, Heidelberg JF, Alley MR, Ohta N, Maddock JR, Potocka I, Nelson WC, Newton A, Stephens C, Phadke ND, Ely B, DeBoy RT, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Kolonay JF, Smit J, Craven MB, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg SL, Venter JC, Shapiro L, Fraser CM. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4136–41. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N, Newton A. The core dimerization domains of histidine kinases contain recognition specificity for the cognate response regulator. J. Bacteriol. 2003;185:4424–31. doi: 10.1128/JB.185.15.4424-4431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N, Ninfa AJ, Allaire A, Kulick L, Newton A. Identification, characterization, and chromosomal organization of cell division cycle genes in Caulobacter crescentus. J. Bacteriol. 1997;179:2169–2180. doi: 10.1128/jb.179.7.2169-2180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsén A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- Ong CJ, Wong MY, Smit J. Attachment of the adhesive holdfast organelle to the cellular stalk of Caulobacter crescentus. J. Bacteriol. 1990;172:1448–1456. doi: 10.1128/jb.172.3.1448-1456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteras M, Stotz A, Schmid Nuoffer S, Jenal U. Identification and transcriptional control of the genes encoding the Caulobacter crescentus ClpXP protease. J. Bacteriol. 1999;181:3039–50. doi: 10.1128/jb.181.10.3039-3050.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet MC, Marczynski GT. Analysis of a cell-cycle promoter bound by a response regulator. J. Mol. Biol. 2000;302:761–75. doi: 10.1006/jmbi.2000.4500. [DOI] [PubMed] [Google Scholar]
- Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. Activation of the diguanylate cyclase pleD by phosphorylation-mediated dimerization. J. Biol. Chem. 2007 doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel diguanylate cyclase output domain. Genes and Development. 2004;18:715–27. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Beness AM, Saier MH., Jr. Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology. 1997;143:2685–2699. doi: 10.1099/00221287-143-8-2685. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in spetal ring assembly in Escherichia coli. EMBO Journal. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- Pierce DL, O'Donnol DS, Allen RC, Javens JW, Quardokus EM, Brun YV. Mutations in DivL and CckA rescue a divJ null mutant of Caulobacter crescentus by reducing the activity of CtrA. J. Bacteriol. 2006;188:2473–82. doi: 10.1128/JB.188.7.2473-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter JLS, Bazire GC. The fine structure of stalked bacteria belonging to the family Caulobacteraceae. J. Cell Biol. 1964;23:587–607. doi: 10.1083/jcb.23.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter JS. Biological properties and classification of the Caulobacter group. Microbiol. Mol. Biol. Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter JS, Staley JT. Caulobacter and Asticcacaulis Stalk Bands as Indicators of Stalk Age. J. Bacteriol. 1996;178:3939–3948. doi: 10.1128/jb.178.13.3939-3948.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiology. 2000;2:450–64. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quardokus E, Din N, Brun YV. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6314–6319. doi: 10.1073/pnas.93.13.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quardokus E, Din N, Brun YV. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol. Microbiol. 2001;39:949–59. doi: 10.1046/j.1365-2958.2001.02287.x. [DOI] [PubMed] [Google Scholar]
- Quardokus EM, Brun YV. DNA replication initiation is required for mid-cell positioning of FtsZ rings in Caulobacter crescentus. Mol. Microbiol. 2002;45:605–16. doi: 10.1046/j.1365-2958.2002.03040.x. [DOI] [PubMed] [Google Scholar]
- Quardokus EM, Brun YV. Cell cycle timing and developmental checkpoints in Caulobacter crescentus. Currents Opinion in Microbiology. 2003;6:541–9. doi: 10.1016/j.mib.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Quintero EJ, Weiner RM. Evidence for the adhesive function of the expopolysaccharide of Hyphomonas strain MHS-2 in its attachment to surfaces. Appl. Environ. Microbiol. 1995;61:1897–1903. doi: 10.1128/aem.61.5.1897-1903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. U. S. A. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan G, Newton A. FlbD of Caulobacter crescentus is a Homologue of the NtrC (NRI) Protein and Activates σ54-Dependent Flagellar Gene Promoters. Proc. Natl. Acad. Sci. U. S. A. 1990;87:2369–2373. doi: 10.1073/pnas.87.6.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisenauer A, Mohr CD, Shapiro L. Regulation of a heat shock sigma32 homolog in Caulobacter crescentus. J. Bacteriol. 1996;178:1919–27. doi: 10.1128/jb.178.7.1919-1927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisenauer A, Quon K, Shapiro L. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J. Bacteriol. 1999;181:2430–2439. doi: 10.1128/jb.181.8.2430-2439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisenauer A, Shapiro L. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO Journal. 2002;21:4969–77. doi: 10.1093/emboj/cdf490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisinger SJ, Huntwork S, Viollier PH, Ryan KR. DivL performs critical cell cycle functions in Caulobacter crescentus independent of kinase activity. J. Bacteriol. 2007;189:8308–20. doi: 10.1128/JB.00868-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 2006;59:870–81. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobaceriaceae. Cell. Mol. Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L, Justice S, Garcia-Lara J. Bacterial Cell Division. Annu. Rev. Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- Ryan KR, Huntwork S, Shapiro L. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7415–20. doi: 10.1073/pnas.0402153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KR, Judd EM, Shapiro L. The CtrA response regulator essential for Caulobacter crescentus cell-cycle progression requires a bipartite degradation signal for temporally controlled proteolysis. J. Mol. Biol. 2002;324:443–55. doi: 10.1016/s0022-2836(02)01042-2. [DOI] [PubMed] [Google Scholar]
- Sackett MJ, Kelly AJ, Brun YV. Ordered expression of ftsQA and ftsZ during the Caulobacter crescentus cell cycle. Mol. Microbiol. 1998;28:421–434. doi: 10.1046/j.1365-2958.1998.00753.x. [DOI] [PubMed] [Google Scholar]
- Sanchez M, Valencia A, Ferrandiz M-J, Sander C, Vicente M. Correlation between the Structure and Biochemical Activities of FtsA, an Essential Cell Division Protein of the Actin Family. EMBO Journal. 1994;13:4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez M, Valencia A, Ferrándiz M-J, Sander C, Vicente M. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO Journal. 1994;13:4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Stanier RY. The Development of Cellular Stalks in Bacteria. J. Cell Biol. 1966;28:423–436. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciochetti SA, Ohta N, Newton A. The role of polar localization in the function of an essential Caulobacter crescentus tyrosine kinase. Mol. Microbiol. 2005;56:1467–80. doi: 10.1111/j.1365-2958.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- Seitz LC, Brun YV. Genetic anaylsis of mecillinam-resistant mutants of Caulobacter crescentus deficient in stalk biosynthesis. J. Bacteriol. 1998;180:5235–5239. doi: 10.1128/jb.180.19.5235-5239.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y-L, Rothfield L. The bacterial cytoskeleton. Microbiol. Mol. Biol. Rev. 2006;70:729–754. doi: 10.1128/MMBR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siam R, Brassinga AK, Marczynski GT. A dual binding site for integration host factor and the response regulator CtrA inside the Caulobacter crescentus replication origin. J. Bacteriol. 2003;185:5563–72. doi: 10.1128/JB.185.18.5563-5572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siam R, Marczynski GT. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO Journal. 2000;19:1138–47. doi: 10.1093/emboj/19.5.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO Journal. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J, Agabian N. Cell Surface Patterning an Morphogenesis: Biogenesis of a Periodic Surface Array During Caulobacter Development. J. Cell Biol. 1982;95:41–49. doi: 10.1083/jcb.95.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J, Sherwood CS, Turner RF. Characterization of high density monolayers of the biofilm bacterium Caulobacter crescentus: evaluating prospects for developing immobilized cell bioreactors. Can. J. Microbiol. 2000;46:339–49. [PubMed] [Google Scholar]
- Smith CS, Hinz A, Bodenmiller D, Larson DE, Brun YV. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J. Bacteriol. 2003;185:1432–42. doi: 10.1128/JB.185.4.1432-1442.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer JM, Newton A. Turning off flagellum rotation requires the pleiotropic gene pleD: pleA, pleC, and pleD define two morphogenic pathways in Caulobacter crescentus. J. Bacteriol. 1989;171:392–401. doi: 10.1128/jb.171.1.392-401.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperl GT, Hoare DS. Denitrification with methanol: A selective enrichment for Hyphomicrobium species. J. Bacteriol. 1971;108:733–736. doi: 10.1128/jb.108.2.733-736.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C, Zweiger G, Shapiro L. Cooridinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J. Bacteriol. 1995;177:1662–1669. doi: 10.1128/jb.177.7.1662-1669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies D, Costerton J. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Tam E, Pate JL. Amino Acid Transport by Prosthecae of Asticcacaulis biprosthecum: Evidence for a Broad-Range Transport System. J. Gen. Microbiol. 1985;131:2687–2699. doi: 10.1099/00221287-131-10-2687. [DOI] [PubMed] [Google Scholar]
- Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. Chromosome organization and segregation in bacteria. J. Struct. Biol. 2006a;156:292–303. doi: 10.1016/j.jsb.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006b;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nature Reviews in Microbiology. 2007;5:363–75. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- Tsang PH, Li G, Brun YV, Freund LB, Tang JX. Adhesion of single bacterial cells in the micronewton range. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5764–8. doi: 10.1073/pnas.0601705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit TH, Pate JL. Characterization of the holdfast region of wild-type cells and holdfast mutants of Asticcacaulis biprosthecum. Arch. Microbiol. 1978;118:157–168. [Google Scholar]
- van den Ent F, Amos L, Lowe J. Bacterial ancestry of actin and tubulin. Currents Opinion in Microbiology. 2001;4:634–8. doi: 10.1016/s1369-5274(01)00262-4. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Löwe J. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO Journal. 2000;19:5300–5307. doi: 10.1093/emboj/19.20.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, de Pedro MA, Young KD. FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 2007;189:5692–5704. doi: 10.1128/JB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Shapiro L. A lytic transglycosylase homologue, PleA, is required for the assembly of pili and the flagellum at the Caulobacter crescentus cell pole. Mol. Microbiol. 2003;49:331–45. doi: 10.1046/j.1365-2958.2003.03576.x. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO Journal. 2002a;21:4420–8. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc. Natl. Acad. Sci. U. S. A. 2002b;99:13831–6. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9257–62. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, Otto M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 2004;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol. 1987;169:4935–4940. doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JK, Brun YV. Out on a limb: how the Caulobacter stalk can boost the study of bacterial cell shape. Mol. Microbiol. 2007;64:28–33. doi: 10.1111/j.1365-2958.2007.05633.x. [DOI] [PubMed] [Google Scholar]
- Wagner JK, Galvani CD, Brun YV. Caulobacter crescentus requires RodA and MreB for stalk synthesis and prevention of ectopic pole formation. J. Bacteriol. 2005;187:544–553. doi: 10.1128/JB.187.2.544-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JK, Setayeshgar S, Sharon LA, Reilly JP, Brun YV. A nutrient uptake role for bacterial cell envelope extensions. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11772–7. doi: 10.1073/pnas.0602047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Shapiro L. The topoisomerase IV ParC subunit colocalizes with the Caulobacter replisome and is required for polar localization of replication origins. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9251–9256. doi: 10.1073/pnas.0402567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SCE, West L, Shapiro L. The bifunctional FtsK protein mediates chromosome partitioning and cell division in Caulobacter. J. Bacteriol. 2006;188:1497–1508. doi: 10.1128/JB.188.4.1497-1508.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SP, Sharma PL, Schoenlein PV, Ely B. A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 1993;90:630–634. doi: 10.1073/pnas.90.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Preston JF, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 2004;186:2724–34. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner BL. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt FC, editor. ASM Press; Washington, D. C.: 1996. pp. 1357–1381. [Google Scholar]
- Ward D, Newton A. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol. Microbiol. 1997;26:897–910. doi: 10.1046/j.1365-2958.1997.6242005.x. [DOI] [PubMed] [Google Scholar]
- Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. Structure of BeF3- -modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure. 2007;15:915–27. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Weiner RM, Melick M, O'Neill K, Quintero E. Hyphomonas adhaerens sp. nov., Hyphomonas johnsonii sp. nov. and Hyphomonas rosenbergii sp. nov., marine budding and prosthecate bacteria. Int J Syst Evol Microbiology. 2000;50(Pt 2):459–69. doi: 10.1099/00207713-50-2-459. [DOI] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell. 1999;4:683–94. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherchia coli. Annu. Rev. Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Whitfield C, Paiment A. Biosynthesis and assembly of group 1 capsular polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr. Res. 2003;338:2491–2502. doi: 10.1016/j.carres.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Wingrove JA, Mangan EK, Gober JW. Spatial and Temporal Phosphorylation of a Transcriptional Activator Regulates Pole-Specific Gene Expression in Caulobacter. Genes Dev. 1993;7:1979–1992. doi: 10.1101/gad.7.10.1979. [DOI] [PubMed] [Google Scholar]
- Wortinger M, Sackett MJ, Brun YV. CtrA mediates a DNA replication checkpoint that prevents cell division in Caulobacter crescentus. EMBO Journal. 2000;19:4503–4512. doi: 10.1093/emboj/19.17.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortinger MA, Quardokus EM, Brun YV. Morphological adaptation and inhibition of cell division during stationary phase in Caulobacter crescentus. Mol. Microbiol. 1998;29:963–973. doi: 10.1046/j.1365-2958.1998.00959.x. [DOI] [PubMed] [Google Scholar]
- Wright R, Stephens C, Zweiger G, Shapiro L, Alley MRK. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- Wu J, Newton A. Isolation, identification, and transcriptional specificity of the heat shock sigma factor sigma32 from Caulobacter crescentus. J. Bacteriol. 1996;178:2094–2101. doi: 10.1128/jb.178.7.2094-2101.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Newton A. The Caulobacter heat shock sigma factor gene rpoH is positively autoregulated from a sigma32-dependent promoter. J. Bacteriol. 1997;179:514–21. doi: 10.1128/jb.179.2.514-521.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Ohta N, Benson AK, Ninfa AJ, Newton A. Purification, Characterization, and Reconstitution of DNA-dependent RNA Polymerases from Caulobacter crescentus. Journal of Biological Chemistry. 1997;272:21558–21564. doi: 10.1074/jbc.272.34.21558. [DOI] [PubMed] [Google Scholar]
- Wu J, Ohta N, Newton A. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Ohta N, Zhao JL, Newton A. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13068–73. doi: 10.1073/pnas.96.23.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. Bacterial morphology: why have different shapes? Curr. Opin. Microbiol. 2007;10:596–600. doi: 10.1016/j.mib.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Shapiro L. Early C. crecentus switch genes fliL and fliM are required for flagellar gene expression and normal cell division. J. Bacteriol. 1992;174:3327–3338. doi: 10.1128/jb.174.10.3327-3338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C, Ely B, Smit J. Identification of genes affecting production of the adhesive holdfast of a marine Caulobacter. J. Bacteriol. 1994;176:796–803. doi: 10.1128/jb.176.3.796-803.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang WY, Shapiro L. Caulobacter FliQ and FliR Membrane Proteins, Required for Flagellar Biogenesis and Cell Division, Belong to a Family of Virulence Factor Export Proteins. J. Bacteriol. 1995;177:343–356. doi: 10.1128/jb.177.2.343-356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZoBell CE. The influence of solid surface upon the physiological activities of bacteria in sea water. J. Bacteriol. 1937;33:86. [Google Scholar]
- ZoBell CE. The effect of solid surfaces upon bacterial activity. J. Bacteriol. 1943;46:39–46. doi: 10.1128/jb.46.1.39-56.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- Zweiger G, Marczynski G, Shapiro L. A Caulobacter methyltransferase that functions only in the predivisional cell. J. Mol. Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
- Zweiger G, Shapiro L. Expression of Caulobacter dnaA as a function of the cell cycle. J. Bacteriol. 1994;176:401–408. doi: 10.1128/jb.176.2.401-408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]