Abstract
Defects in human DNA mismatch repair have been reported to underlie a variety of hereditary and sporadic cancer cases. We characterized the structure of the MSH6 promoter region to examine the mechanisms of transcriptional regulation of the MSH6 gene. The 5′-flanking region of the MSH6 gene was found to contain seven functional Sp1 transcription factor binding sites that each bind Sp1 and Sp3 and contribute to promoter activity. Transcription did not appear to require a TATA box and resulted in multiple start sites, including two major start sites and at least nine minor start sites. Three common polymorphisms were identified in the promoter region (−557 T→G, −448 G→A, and −159 C→T): the latter two were always associated, and each of these functionally inactivated a different Sp1 site. The polymorphic allele −448 A −159 T was demonstrated to be a common Caucasian polymorphism found in 16% of Caucasians and resulted in a five-Sp1-site promoter that had 50% less promoter activity and was more sensitive to inactivation by DNA methylation than the more common seven Sp1 site promoter allele, which was only partially inactivated by DNA methylation. In cell lines, this five-Sp1-site polymorphism resulted in reduced MSH6 expression at both the mRNA and protein level. An additional 2% of Caucasians contained another polymorphism, −210 C→T, which inactivated a single Sp1 site that also contributes to promoter activity.
The human DNA mismatch repair (MMR) system functions to repair mispaired bases in DNA that result from DNA replication errors and thereby prevents the accumulation of mutations due to such replication errors. Biochemical and genetic studies have identified a number of mismatch repair proteins involved in this system, including those encoded by the MSH2, MSH3, MSH6, MLH1, MLH3, PMS2, and EXO1 genes, as well as the replication proteins PCNA, RFC, RPA, and DNA polymerase delta (for reviews, see references 24 and 33). Loss of MMR function is associated with both inherited cancer susceptibility and the development of sporadic tumors. Inherited mutations in MSH2 and MLH1 are the most prevalent cause of hereditary nonpolyposis colorectal carcinoma (HNPCC) (for a review, see reference 52), and epigenetic silencing of MLH1 has been found to underlie most MMR defective sporadic cancer cases (14, 25, 31, 46, 47). Inherited mutations in MSH6 have been found in a small proportion (0 to 3%) of HNPCC families and appear to underlie a higher proportion for familial colorectal cancer cases that show later onset and a less pronounced family history than HNPCC (6, 34, 45, 65, 67, 68). Mutations in PMS2 have been found in patients with Turcots syndrome but are only rarely found in patients with HNPCC (11, 39, 64, 66). Whether or not mutations in MLH3 or EXO1 underlie a significant proportion of HNPCC is unclear (2, 29, 38, 69, 70).
In the human MMR system, two heterodimeric complexes—MSH2-MSH6 (MutSα) MSH2-MSH3 (MutSβ)—function to recognize mispaired bases in DNA (1, 19, 21, 49, 61). MutSα appears to function in the repair of base-base and insertion/deletion mispairs, whereas hMutSβ only appears to function in the repair of insertion/deletion mispairs. In addition, MutSα appears to be relatively more important for the repair of smaller insertion/deletion mispairs, whereas MutSβ is relatively more important for the repair of larger insertion/deletion mispairs (1, 21, 54). As a consequence of this partial redundancy, defects in MSH2, MSH3, and MSH6 each have different effects: mutations in MSH2 cause complete loss of MMR, mutations in MSH6 cause increased accumulation of base substitution mutations, and in some cases single base insertion/deletion mutations and mutations in MSH3 cause increased accumulation of larger insertion/deletion mutations (42, 57). Several studies have also shown that alteration in expression levels of the MSH proteins can perturb MMR. Overexpression of human MSH3 inhibits the formation of the MutSα complex, resulting in increased accumulation of mutations (18, 41, 63) and in Saccharomyces cerevisiae overexpression of MSH6 results in a mutator phenotype (8, 15), although the basis for this is unclear.
Although it is known that alteration in the expression of MSH6 can perturb MMR, only a limited analysis of the structure of the human MSH6 promoter has been performed and no possible regulatory mutations that affect the expression of MSH6 have been reported. The 5′ untranslated region (UTR) of the human MSH6 gene has been cloned and sequenced (1, 60). The promoter region has a high GC content, and there appear to be multiple start sites for transcription. It is known that human MLH1, PMS1, PMS2, and MSH2 gene promoters share structural characteristic of TATA-less promoters, including multiple start sites and the sporadic presence of CAAT-boxes and GC-boxes, which may indicate a housekeeping function for these genes (27, 28, 48, 71). The transcription of a number of mammalian genes is believed to be regulated by GC box target sites for Sp family transcription factors (58), in the absence of a TATA-box close to the transcriptional start sites (3, 4, 72, 73). The limited information about the MSH6 promoter region suggests that it may be a member of this class of promoters. To better understand the regulation of the MSH6 gene, we identified seven putative Sp1 binding sites in the MSH6 promoter region and analyzed their role in the regulation of MSH6 expression. We found that the MSH6 promoter is regulated by Sp1 and that all seven Sp1 sites functionally interacted with Sp1. In addition, we identified polymorphic variants of the MSH6 promoter in which different Sp1 sites were inactivated by single-nucleotide polymorphisms (SNPs) resulting in altered promoter activity.
MATERIALS AND METHODS
Cell culture.
Chinese hamster ovary (CHO) cells were obtained from the American Type Culture Collection (Manassas, Va.). Human cervical carcinoma (HeLa) cells were provided by Giuseppina Bonizzi (UCSD School of Medicine, La Jolla, Calif.), a human skin fibroblast (BJ) cell line was provided by Jean Wang (UCSD Department of Biology), and human glioblastoma and glioma cell lines were provided by Webster Cavenee (Ludwig Institute, La Jolla, Calif.). Cells were cultured in Dulbecco modified Eagle medium (Gibco-Invitrogen) supplemented with 10% fetal bovine serum (Sigma) and 1% l-glutamine, penicillin, and streptomycin sulfate (Irvine Scientific) at 37°C in a humidified chamber with 5% CO2. Drosophila Schneider SL2 cells were from the American Type Culture Collection and were grown at 26°C under normal atmospheric conditions in Schneider's medium (Gibco-Invitrogen) supplemented with 10% fetal bovine serum.
DNA sequence analysis.
For analysis of genomic DNA samples, a 672-bp region of the human MSH6 gene containing the 5′ UTR and part of the coding region was amplified by PCR with primers 166UF (5′-GTGCCTACTCTATACAAATCTTGAG) and 817LR (5′-GTGCCTACTCTATACAAATCTTGAG) by using an Advantage GC2 PCR kit (Clontech). PCR was carried out under the following conditions: initial denaturation for 4 min at 94°C; followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 68°C for 30 s; followed by a single final cycle extension at 68°C for 4 min. The resulting PCR products were then purified by digestion with ExoI and shrimp alkaline phosphatase and sequenced by using a Perkin-Elmer ABI 3700 DNA sequencer (Applied Biosystems) (34). Plasmid DNAs were similarly sequenced with plasmid DNA templates purified by using Qiagen Plasmid Midi kits (Qiagen) and primers designed by using the predicted sequence of individual plasmids.
Sequence chromatograms were analyzed to identify base changes, including heterozygous base changes by using Sequencher 3.1 software (Gene Codes Corp.), and sequence homologies were examined by using the BLAST 2.0 and FASTA programs available on the web server of the National Center for Biotechnology Information (Bethesda, Md.) (51). The 5′-flanking sequence was analyzed for the presence of putative transcription factor binding sites by using MacVector (Oxford Molecular, Ltd.) and TESS (www.cbil.upenn.edu/cgi-bin/tess) software. The sequence of the 5′ UTR of the MSH6 gene initially used in these studies was obtained from GenBank and included previous submissions from this and other laboratories (GenBank sequences U73732 and AF334668).
To detect polymorphisms in the 5′ UTR of the MSH6 gene, normal control DNAs were obtained from Coriell Laboratories (Camden, N.J.) and sequenced as described above. The first 100 samples of the complete human diversity set (catalog no. MPDR450) were sequenced. Subsequently, all 100 DNAs from the 100 Caucasian DNA set (catalog no. HD100CAU) were sequenced.
Genomic DNA and mRNA from the LN299, LN340, and LN443 glioblastoma and LNZ308 glioma cell lines were extracted by using a Puregene DNA isolation kit (Gentra Systems) and the FastTrack mRNA isolation kit (Invitrogen, San Diego, Calif.), respectively. The 5′ UTR and all exons with flanking region of hMHS6 gene were amplified and sequenced by utilizing the primers and conditions described previously (34). cDNA was generated by using a First-Strand cDNA synthesis kit (Invitrogen), reverse transcription-PCR was performed with the previously described NS6X1UF and CS6X4LAr.5 primers (34), and the resulting PCR fragment including exons 1 and 4 was sequenced. To analyze the linkage between different nucleotide changes, the cDNA amplification product and genomic DNA amplified with primers 166UF and CS6X1L2 (32) were cloned into a Topo TA cloning pCR2.1 vector (Invitrogen) and then sequenced by using M13 forward and reverse primers.
Primer extension and RACE (rapid amplification of cDNA ends) assay.
Primer extension was performed by using total human adult normal colon RNA (ResGen; Invitrogen Corp.) and a reverse primer 817LR that was complementary to nucleotides 17 to 39 from ATG of MSH6. The primer was end labeled with [γ-32P]ATP by using T4 polynucleotide kinase, and free nucleotides were removed by using a Microspin G25 column (Amersham-Pharmacia). Primer extension assays were performed in parallel by using Superscript II RNase H− reverse transcriptase and ThermoScript RNase H− reverse transcriptase (Invitrogen/Life Technologies). Reactions containing either 5 or 10 μg of total human adult normal colon RNA, 5× buffer (provided by the manufacturer), 10 mM dithiothreitol (DTT), 10 mM deoxynucleoside triphosphates, and reverse primer were denatured at 70°C for 2 min and then incubated at 20 min at 60°C so that the primer could anneal to the RNA template, followed by cooling the reactions on ice for 10 min. Then, GC Melt (final concentration, 1 M), RNasin (final concentration, 0.5 U/μl), and either 48 U of Superscript II RNase H− reverse transcriptase or 200 U of ThermoScript RNase H− reverse transcriptase were added so that the final volume was 20 μl; the samples were then incubated at 42°C for 90 min. The reactions were stopped by precipitation with 0.5 M ammonium acetate and 100% ethanol. These samples and molecular weight markers (HinfI digest of φX174 DNA; Promega) were electrophoresed in parallel on a 7 M urea-6% polyacrylamide gel run in 0.5× TBE buffer. The radioactive DNA species were then detected by autoradiography, as well as by using a phosphorimager.
5′ RACE was carried out by using a Human Colon Marathon-Ready cDNA amplification kit (Clontech). The primary and nested PCR were performed with the adaptor oligonucleotides AP1 and AP2 (provided in the kit) and a specific reverse primer 817LR or a fluorescent derivative of 817LR. PCR amplification was performed by using an Advantage 2 PCR kit (Clontech) according to the manufacturer's instructions for the Ready Marathon cDNA amplification kit. The fluorescently labeled PCR products were electrophoresed on a Perkin-Elmer ABI 377 DNA sequencer and analyzed by using ABI GeneScan software. In addition, the unlabeled RACE products were cloned into the TA cloning vector pCR2.1 (Invitrogen) by using a TA cloning kit, and 100 independent insert containing clones were isolated and sequenced by using standard M13 and T7 vector primers.
Construction of hMSH6 reporter vectors.
To construct a wild-type MSH6 promoter-luciferase reporter plasmid, a 624-bp fragment of MSH6, including 5′-flanking sequences from positions −633 to −9 relative to the ATG was amplified by PCR by using an Advantage GC2 PCR kit (Clontech) as described above except that only 20 cycles of PCR were used. The forward primer 166KUF (5′-GTGCCTACTCTATACAAATCT) contained a KpnI site (5′-GGGGTACC) at its 5′ end and the reverse primer 9BLR (5′-CGGCAAGGCCCAACCGTTC) contained a BglII site (5′-GAAGATTC) at its 5′ end. The PCR product was digested by with KpnI and BglII and cloned between the KpnI and BglII sites of the pGL3 enhancer-luciferase reporter plasmid (Promega), and a fully wild-type clone was identified by DNA sequencing. The full-length of the promoter construct pGL3 −633/−9WT was used to construct all deletion and site-directed mutant derivatives.
A series of 5′ MSH6 promoter deletions (pGL3-490/-9, pGL3-318/-9, pGL3-248/-9, pGL3-248Mut/-9, pGL3-221/-9, pGL3-120/-9) was constructed by amplifying the MSH6 promoter region with different 5′ primers complementary to different regions of MSH6 and containing the 5′ KpnI site indicated above in combination with the reverse primer 9BLR, and these PCR products were inserted between the KpnI and BglII sites of the pGL3 enhancer-luciferase reporter plasmid. Similarly, 3′ deletion constructs (pGL3-633/-252 and pGL3-633/-193) were generated by using the 166KUF primer and different reverse primers complementary to different regions of MSH6 and containing the above indicated 5′ BglII site. The first number in the designation of each mutant construct indicates the most N-terminal MSH6 nucleotide present, and the second number indicates the most C-terminal MSH6 nucleotide present.
Plasmids containing individual nucleotide changes were constructed by using different site-directed mutagenesis methods. To construct the pGL3-633/-9+TATA box mutation, a derivative of the forward primer 166TKUF was synthesized to contain a 2-bp difference in the core of the TATA box sequence (TATA→AAAA), and then the plasmid was constructed as described above for the wild-type plasmid. The full-length promoter constructs containing mutations that created some of the promoter SNPs, including pGL3-633/-9 SNP1 (−557G), pGL3-633/-9 SNP2-3 (−448A −159T), and pGL3-633/-9 SNP1-2-3 (−557G −448A −159T), were constructed by amplifying these alleles from genomic DNAs in which these alleles were present and then cloning the resulting PCR product as described for the wild-type plasmid. A single round of overlap extension PCR (62) with appropriate mutant primers and pGL3-633/-9WT DNA as a template was used to create other single-mutant plasmids. These single-mutant plasmids included the two promoter SNP-containing plasmids pGL3-633/-9 SNP2 (−448A) and pGL3-633/-9 SNP3 (−159T) and a series of plasmids in which a 2-bp substitution was made to eliminate each individual Sp1 consensus sequence, including pGL3-Sp1-7Mut (CC→AA), pGL3-Sp1-6Mut (GG→TT), pGL3-Sp1-5Mut (CC→AA), pGL3-Sp1-4Mut (CC→AA), pGL3-Sp1-3Mut (GG→TT), and pGL3-Sp1-1/2Mut (CC→AA and GG→TT). (The exact nucleotides changed are indicated in Fig. 1A. The Sp1 sites are numbers from 1 to 7 as indicated in Fig. 1B, with 1 indicating the most N-terminal Sp1 site.) Sequential rounds of overlap extension PCR were then used to generate a series of plasmids containing different combinations of the above-described mutations, including pGL3-Sp1-7/6Mut, pGL3-Sp1-7/6/5Mut, pGL3-Sp1-7/6/5/4Mut, pGL3-Sp1-7/6/5/4/3Mut, pGL3-Sp1-7/6/5/4/3/2/1Mut, and pGL3-Sp1-7/6/5/4/3/2/1Mut+ TATA box mutation. All plasmids were sequenced to verify that they only contained the desired base changes. Note that the oligonucleotide sequences that were the basis for the 2-bp substitution mutations were also used in the many of the competitor DNAs for the electrophoretic mobility shift assay (EMSA) experiments.
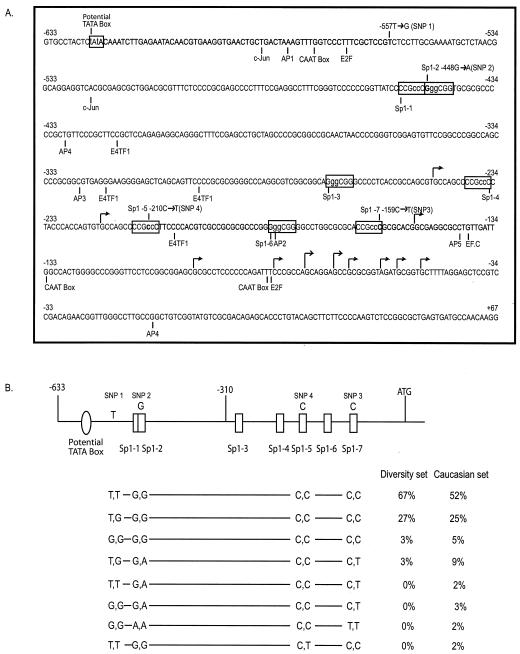
FIG. 1.
Identification of cis-acting elements and SNPs in the MSH6 promoter. (A) The sequence of nucleotides −633 to +167 relative to the ATG of the MSH6 gene is shown. The Sp1 sites and the potential TATA box analyzed in the present study are indicated in boxes. Other potential transcription factor binding sites are indicated below the sequence. The major and minor transcription start sites are indicated by the tall and short arrows, respectively. The nucleotides altered by the 2-bp substitution mutations used in the analysis of the Sp1 sites are indicated in lowercase, and the four SNPs identified are indicated above the sequence. (B) Schematic representation of the MSH6 promoter region, with the most prevalent nucleotide at each polymorphic site indicated above the promoter diagram. Below the promoter diagram are indicated the eight different genotypes of the MSH6 promoter region identified by analyzing the SNPs present at, from left to right, nucleotides −557 (SNP 1), −448 (SNP 2), −210 (SNP 4), and −159 (SNP 3), respectively. Also indicated are the percentages of the samples from the human diversity set of DNAs (first 100 DNAs) and the Caucasian DNAs found to have each genotype.
Transient-transfection and luciferase reporter gene expression analyses.
HeLa, CHO, and BJ cells (2 × 105) were plated into individual wells of a six-well plate 12 h before transfection, and the medium was changed immediately before transfection. A 1-μg DNA mixture was prepared in 100 μl of OptiMEM I medium (Invitrogen) that contained 0.1 μg of β-galactosidase expression plasmid (pβgal-control; BD Biosciences) as an internal control or 0.4 μg (HeLa cells) or 0.1 μg (CHO/BJ cells) of test plasmid and pUC18 carrier DNA and was then mixed with Fugene in a 1:3 ratio. This mixture was immediately added to one well of cells; after 24 h the cells were washed twice with phosphate-buffered saline, and cell extracts were prepared by resuspending the cells in lysis buffer provided in a luciferase assay kit (Promega). The luciferase activity was then analyzed by mixing 25 μl of cell extract and 100 μl of the luciferase assay substrate (Promega) and measuring the resulting luminescence 10 s in a luminometer (EG&G Berthold Microlumat LB 96P). The luciferase activity was then normalized to the level of β-galactosidase activity present in the same extracts measured by using a β-galactosidase activity kit (Invitrogen) according to the manufacturer's instructions. Each extract was analyzed in duplicate, and at least two independent experiments were performed. The observed luciferase activity reported in each figure is expressed as the percentage of the activity obtained with the most active construct used in each figure. The DNAs and cell lines transfected are as indicated in individual experiments. In some experiments, the plasmids were methylated in vitro with SssI methylase (New England Biolabs) prior to transfection. In these cases, complete methylation at the Sp1 and CpG sites was verified by measuring the extent of protection from digestion with the restriction enzymes AciI (CCGC) and HpaI (CCGG).
For transfection of Drosophila SL2 cells, cells were seeded at 2 × 106 cells per well of a six-well plate 24 h before transfection, and transfections were performed and analyzed essentially as described above except that CellFectin reagent (Invitrogen) was used instead of Fugene. The pPac-Vector, pPac-Sp1, and pPac-Sp3 expression plasmids used in the Drosophila cell transfection experiments were provided by K. Okumura, (Ludwig Institute for Cancer Research, La Jolla, Calif.). In initial optimization experiments, cotransfection was carried out with different amounts of pPAC-Sp1 or pPAC-Sp3 (0.1, 0.25, and 0.5 μg), along with either the pGL3-Basic vector or pGL3-633/-9WT, and maximal promoter activity was obtained with 0.25 μg of pPAC-Sp1 and pPAC-Sp3. In subsequent experiments, the DNA transfection mixture contained 0.1 μg of β-galactosidase expression plasmid, 0.4 μg of each experimental plasmid, and 0.25 μg of pPac vector, pPAC-Sp1, or pPAC-Sp3 or a mixture of pPAC-Sp1 and pPAC-Sp3, along with sufficient pUC18 DNA carrier, so that each mixture contained a total of 1 μg of DNA.
Nuclear extracts, EMSAs, and protein analysis.
HeLa cells (107) were harvested by centrifugation at 6,000 rpm for 5 min, washed twice in ice-cold phosphate-buffered saline, and lysed by resuspending them in 200 μl of ice-cold buffer A (10 mM HEPES [pH 8.0], 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.2% NP-40) and incubating the suspension for 10 min on ice. All solutions contained freshly added protease inhibitor cocktail (1× recommended concentration; Roche) and phosphatase inhibitors (final concentrations of 1 mM orthovanadate and 1 mM sodium fluoride), as well as final concentrations of 1 mM PMSF and 1 mM DTT. After centrifugation at 6,000 rpm at 4°C for 10 min to harvest the nuclei, the nuclei were washed with 200 μl of ice-cold buffer B (10 mM HEPES [pH 8], 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM PMSF) and then resuspended with ice-cold buffer C (20 mM HEPES [pH 8.0], 0.63 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 1 mM PMSF, 25% glycerol) by rotating the suspension for 20 min at 4°C. The resulting lysate was centrifuged at 12,000 rpm for 10 min (7, 17), and the protein concentration was determined by using Bio-Rad protein assay reagent.
To construct oligonucleotide duplexes, 500 ng each of sense and antisense oligonucleotides were annealed in buffer N (1 M NaCl, 25 mM Tris-HCl, 1 mM EDTA) by heating the mixtures to 94°C for 10 min and then cooling them to 65°C for 30 min, followed by cooling from 65 to 25°C over a 90-min period, followed by incubation at 25°C for 20 min. The oligonucleotide duplexes were then purified by high-pressure liquid chromatography as previously described, precipitated with ethanol from a solution containing 0.5 M ammonium acetate, and resuspended in 10 μl of TE buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA). A portion (1 μl) of oligonucleotide duplex was then 5′end labeled with [γ-32P]ATP by using T4 polynucleotide kinase, and free nucleotides were removed by using Microspin G25 columns (Amersham-Pharmacia). In some cases, the oligonucleotide duplexes were methylated in vitro with SssI methylase, and then the methylation status was verified by digesting the DNAs with the restriction enzyme AciI.
EMSAs was performed by using the gel shift assay system (Promega), with minor modifications. Nuclear extracts (8 μg in final reaction volumes of 20 μl) from HeLa cells were incubated for 20 min at room temperature in the presence or absence of a 50-fold molar excess (relative to the radioactive substrate) of oligonucleotide duplex competitors. Then 20,000 cpm of Sp1 or AP2 consensus oligonucleotide duplexes were added to the reaction and, after 20 min of incubation at room temperature, the samples were analyzed by electrophoresis through a 6% polyacrylamide gel run in 0.5× Tris-borate-EDTA for 2 h at 120 V, followed by detection of the radioactive species by autoradiography or by using a phosphorimager. For the supershift assays, the labeled oligonucleotide duplexes were incubated with the nuclear extract on ice for 20 min as described above; anti-Sp1 (PEP2) or anti-Sp3 (D20) antibody (Santa Cruz Biotechnology) was then added to the reactions on ice for 1 h, and the reaction products were analyzed as described above.
For Western blot analysis, nuclear and cytoplasm extracts from LN229, LN340, and LN443 glioblastoma cell lines were prepared as described above. The nuclear and cytoplasmic extracts from each batch of cells were mixed, and the protein concentration was determined by using a Bio-Rad protein assay kit. A total of 7.5 or 15 μg of protein/sample was electrophoresed through a 4 to 15% sodium dodecyl sulfate-polyacrylamide gel gradient and then transferred to a Hybond-P polyvinylidene difluoride membrane (Amersham Pharmacia Biotech, Inc.). The membrane was probed with monoclonal anti-hMSH6 and anti-hMSH2 (BD Biosciences) antibodies, followed by reaction with horseradish peroxidase-conjugated secondary antibody, and then visualized by using the ECL Plus Light System (Amersham Biosciences). The same membrane was then similarly analyzed by using anti-α tubulin (Sigma) antibody.
RESULTS
Structure of the MSH6 promoter region.
To characterize the promoter region of the MSH6 gene, a fragment of genomic DNA that spans positions −633 to +39 bp relative to the A of the ATG was amplified by PCR and sequenced to confirm the available sequence of this region (Fig. 1A). Analysis of the sequence by using MacVector and TESS software revealed putative binding sites for AP family c-JUN, E2F, E4TF1, and EF.C transcription factors, as well as three potential CAAT boxes and a potential TATA box, identified as a TATAWAW site (44) at position −623. No potential Inr elements were found. Also observed were seven potential GC-box/Sp1 transcription factor-binding elements. These included three consensus sequences (10, 30) (GGGCGG) at positions −448, −271, and −183 and four inverted complement sequences (CCGCCC) at positions −449, −235, −208, and −159. Comparison of the sequence of the human MSH6 upstream region with that of other species suggested that the Sp1 site at −159 was conserved in the MSH6 gene from Mus musculas, Cavia porcellus, and Mycobacterium tuberculosis, whereas the Sp1 site at −271 was conserved in the MSH6 gene from Bovine herpesvirus typ.
To identify naturally occurring sequence variations within the promoter region, the region from positions −633 to +39 was amplified and sequenced from the first 100 DNAs of the human DNA diversity set. This analysis revealed three novel SNPs at positions −557 (T→G; SNP 1), −448 (G→A; SNP 2), and −159 (C→T; SNP 3). The frequency of each genotype observed is shown in Fig. 1B. The SNP at −557 was more common than the other two polymorphisms, which were found at the same frequency. Significantly, the −448A and −159T alleles were always associated with each other. The two SNPs at −448A (SNP 2) and −159T (SNP 3) were within the consensus sequences (GGGCGG/CCCGCC) for the potential Sp1 binding sites Sp1-2 and Sp1-7, respectively, and eliminated these two potential Sp1 binding sites. Subsequent analysis of 100 normal Caucasian DNAs revealed the same three SNPs, except that the frequency of the −448 (G→A; SNP 2) and −159 (C→T; SNP 3) SNPs was much higher in these samples and homozygous −557G −448A −159T and −557T −448A −159T alleles were observed. These results suggest the existence of four haplotypes (−557T −448G −159C; −557G −448G −159C; −557T −448A −159T; and −557G −448A −159T) and further suggest that the −557G −448A −159T and −557T −448A −159T alleles represent common Caucasian polymorphisms that alter the number of Sp1 binding sites present in the MSH6 promoter. In addition, the −210 C→T change (SNP 4) was observed in 2% of the Caucasian samples in association with a −557T −448G −159C haplotype. The −210 C→T change (SNP 4) eliminates the consensus sequence for the putative Sp1-5 site.
Mapping of the MSH6 transcriptional start sites.
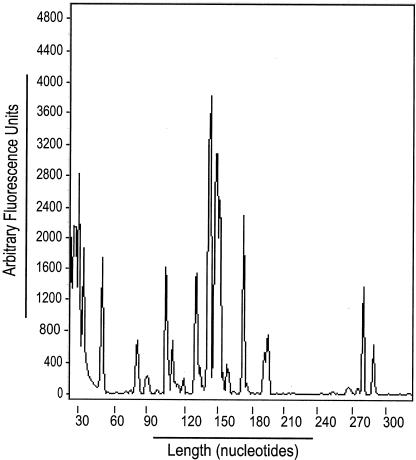
To map the transcriptional start site(s) of the MSH6 gene, primer extension analysis of total human normal colon RNA was first performed. This analysis revealed multiple potential 5′ mRNA ends mapping between positions −247 and −51 (data not shown). Because the extremely GC-rich nature of the MSH6 promoter region might cause premature termination of the primer extension products due to secondary structure, 5′-RACE analysis was performed to define the positions of the potential multiple transcription start sites. An anchor primer (AP1) and a specific antisense oligonucleotide were used to amplify Human Colon Marathon-Ready cDNA. Nested PCR was then carried out with internal adaptor primer and fluorescently labeled antisense primer, and the sizes of these PCR products were determined by using an ABI 377 sequencer. Multiple species of different lengths were observed, and the positions of the potential start sites relative to the position of the antisense primer sequence were determined by subtracting the length of the adapter sequence from the length of each PCR product (Fig. 2). These positions fell within the range of positions identified by primer extension. Finally, PCR products were generated by using an unlabeled antisense primer, and these were cloned and sequenced to determine the exact position of the start sites. Analysis of 100 clones revealed two major start sites at positions −76 and −70 (found in 49% of the clones), as well as at least nine minor start sites, all of which are indicated in Fig. 1A.
FIG. 2.
Transcription of the MSH6 gene initiates from multiple start sites. Genescan analysis of 5′-RACE-PCR products was performed as described in Materials and Methods. Below the chromatogram is indicated a molecular size scale in base pairs determined by using appropriate markers (TAMRA).
EMSA analysis of the putative Sp1 binding sites.
EMSAs were performed to characterize the seven potential Sp1 binding sites identified by computer analysis and to investigate the functional consequences of the SNPs found at −448 (SNP 2) and −159 (SNP 3). Gel shifts were carried out with HeLa cell nuclear extracts as a source of protein and mixtures of one labeled double-stranded oligonucleotide containing the consensus-binding sites for either Sp1 or AP2 and a series of unlabeled double stranded oligonucleotide competitors, including the seven Sp1 sites found between positions −454 and −159, the Sp1 sites containing the −448A and −159T SNPs, and the Sp1 and AP2 consensus sequences (Fig. 3A).
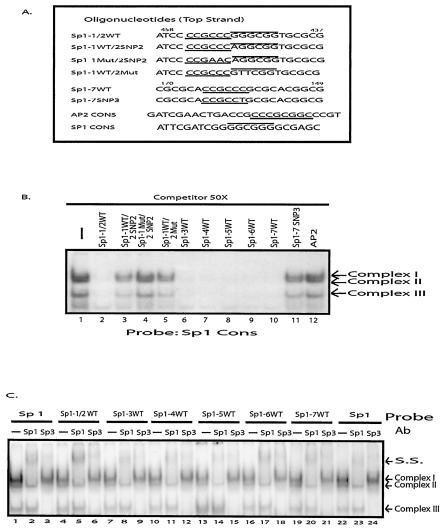
FIG. 3.
EMSA of the seven Sp1 sites found in the MSH6 promoter region. (A) Sequences of the competitor and probe DNAs used in gel mobility shift assays. Only the sequence of the top strand of the oligonucleotide duplexes is indicated and, where relevant, the MSH6 sequence coordinates are also indicated. The Sp1 binding consensus sequences (GGGCGG) and inverted complement sequences (CCGCCC) are indicated by the overlining and underlining, respectively. The numbering system for each of the Sp1 sites is as defined in Fig. 1B. Each Sp1 sequence is wild type except that Sp1-1WT/2SNP 2 refers to the sequence containing the −448A SNP 2, Sp1-1Mut/2SNP 2 refers to the sequence containing the 2-bp substitution mutation in the Sp1-1 site and the −448A SNP 2, Sp1-1WT/2Mut refers to the sequence containing the 2-bp substitution mutation in the Sp1-2 site, and Sp1-7SNP 3 refers to the sequence containing the −159T SNP 3. See Fig. 1A for the sequences that are changed by these nucleotide substitutions. (B) Gel mobility shift assays performed with HeLa cell extract, radioactively labeled Sp1 consensus sequence as a probe, and a 50-fold molar excess of the indicated competitor DNAs. The arrows indicate the three specific protein-DNA complexes formed. (C) Effect of anti-Sp1 and anti-Sp3 antibodies on gel shift assays. Assays were performed with HeLa cell extract, and the radioactive substrate is indicated above each set of three lanes. After complex formation, the antibody indicated above each individual lane was added. The arrows indicate the three specific protein-DNA complexes formed and the position of the supershifted (S.S.) species formed after the addition of antibodies.
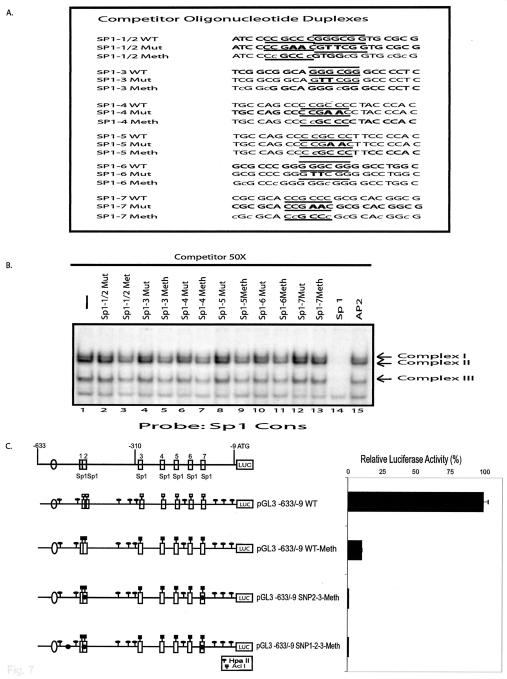
When binding to the labeled Sp1 consensus sequence was examined in the absence of competitor, three different protein-DNA complexes were observed (Fig. 3B). A 50-fold excess of competitor DNAs comprising the first two Sp1 sites (Sp1-1/2) or each of the individual Sp1 sites Sp1-3 through Sp1-7 completely eliminated the formation of the three protein DNA complexes (lanes 2 and 6 to 10), whereas the AP2 competitor had no effect on complex formation (lane 12). An Sp1 consensus sequence competitor also eliminated the formation of the three complexes (data not shown). These results suggest that all seven putative Sp1 sites can bind Sp1. The −159T SNP 3 completely eliminated the ability of the Sp1-7 oligonucleotide to act as a competitor of complex formation (lane 11), indicating that this SNP eliminates the Sp1-7 site. The −448A SNP 2 partially eliminated the ability of the Sp1-1/2 oligonucleotide to act as a competitor, and a similar effect was seen with a GG→TT that eliminated the second Sp1 binding site (lanes 3 and 5). However, combining CC→AA mutation eliminating the first Sp1 site with the −448A SNP 2 completely eliminated the ability of the oligonucleotide containing the first two Sp1 sites to act as a competitor of complex formation (lane 4). These latter results suggest that both the Sp1-1 and Sp1-2 sites can bind Sp1 and indicate that the −448A SNP 2 eliminates the Sp1-2 site. As a control, the same unlabeled oligonucleotides were used as competitors in binding reactions containing the labeled AP2 oligonucleotide (data not shown), and competition was only observed with the AP2 competitor and with the Sp1-6 competitor that contains an overlapping AP2 site.
To better characterize the seven Sp1 transcription factor-binding sites, supershift experiments were performed with anti-Sp1 and anti-Sp3 antibodies (Fig. 3C). When the Sp1 consensus sequence and the Sp1-1/2, Sp1-3, Sp1-4, Sp1-5, Sp1-6, or Sp1-7 oligonucleotides were used as labeled substrates in gel shift experiments, the same three protein-DNA complexes were observed (lanes 1, 4, 7, 10, 13, 16, 19, and 22). The addition of an anti-Sp1 antibody to the reactions caused a supershift of only the upper band (complex 1) (lanes 2, 5, 8, 11, 14, 17, 20, and 23), whereas the addition of anti-Sp3 antibody caused a supershift of both of the lower bands (complexes 2 and 3) (lanes 3, 6, 9, 12, 15, 18, 21, and 24). This pattern of supershifting has been observed during the analysis of other promoters whose expression involves Sp1 and Sp3 proteins (4, 37) (36, 58). These results indicate that complex 1 results from Sp1 binding, that complexes 2 and 3 result from Sp3 binding, and that the Sp1 sites Sp1-1/2, Sp1-3, Sp1-4, Sp1-5, Sp1-6, and Sp1-7 can all bind both Sp1 and Sp3.
Regulation of the MSH6 promoter by Sp1 binding sites.
To examine the role of the seven Sp1 binding sites on the activity of the MSH6 promoter, a DNA fragment comprising nucleotides −633 to −9 of MSH6 containing the core promoter was cloned into pGL3 enhancer vector to construct a luciferase reporter vector. When this construct was transiently transfected into either HeLa or CHO cells, strong luciferase expression was observed compared to transfection of the vector alone (Fig. 4). This MSH6 promoter reporter vector was then used in a series of experiments in which mutations were tested for their effect on expression after transfection into both HeLa and CHO cells; all reported experiments were performed with both cell lines; however, since the same results were obtained with both cell lines, data from only one cell line is presented for each experiment. A 5′ deletion that removed the potential TATA box partially reduced expression; however, a two-base substitution mutation (TATA→AAAA) that eliminated the TATA box had no effect on expression, suggesting the potential TATA box was not important for expression (Fig. 4A). A series of 5′ deletions that progressively eliminated the Sp1-1/2 and Sp1-3 sites resulted in progressive, but not complete, loss of expression. Mutations or deletion of the Sp1-4 site resulted in further reduced expression, and finally deletion of the remaining three Sp1 sites reduced expression to the level seen with the pGL3 vector alone. An internal deletion that eliminated the Sp1-6 and Sp1-7 sites eliminated ca. 80% of the promoter activity, and a deletion that eliminated the Sp1-4, Sp1-5, Sp1-6, and Sp1-7 sites eliminated ca. 90% of the promoter activity. These results suggest that all seven Sp1 sites contribute to the expression of MSH6 but that the four Sp1 sites closest to the transcriptional start sites are most important.
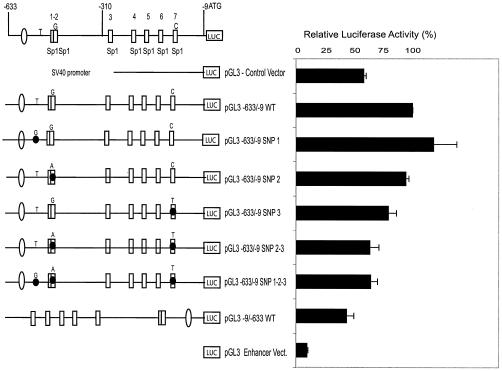
FIG. 4.
Effects of deletion and two-base substitution mutations on MSH6 promoter activity in transient-transfection assays. The wild-type MSH6 promoter and different mutant derivatives were inserted into the pGL3 enhancer luciferase reporter vector, and the promoter activity of each construct was assayed after transfection into either HeLa or CHO cells. (A) Analysis of deletion mutant derivatives by transfection into HeLa cells. (B) Analysis of individual 2-bp substitution mutations after transfection into HeLa cells. (C) Analysis of combinations of different 2-bp substitution mutations after transfection into CHO cells. The boxes and circles indicate the Sp1 sites and a potential TATA box as indicated in Fig. 1, respectively, with the open boxes and circles indicating the wild-type sequences and the solid boxes and circles indicating the presence of 2-bp substitution mutations: M TATA is a 2-bp substitution in the TATA box, M 1-2 indicates 2-bp substitutions in both the Sp1-1 and Sp1-2 sites, and M 3 through M 7 indicate 2-bp substitutions in the Sp1-3 through Sp1-7 sites, respectively (see Materials and Methods and Fig. 1A for details). The DNA present in the deletion mutants is indicated by the horizontal line and structural features (boxes and circles) present in the diagram: pGL3 WT contains MSH6 nucleotides −633 to −9, pGL3 Δ1 contains MSH6 nucleotides −490 to −9, pGL3 Δ2 contains MSH6 nucleotides −318 to −9, pGL3 Δ3 contains MSH6 nucleotides −248 to −9, pGL3 Δ4 contains MSH6 nucleotides −221 to −9, pGL3 Δ5 contains MSH6 nucleotides −120 to −9, pGL3 Δ6 contains MSH6 nucleotides −633 to −252, pGL3 Δ7 contains MSH6 nucleotides −633 to −193, and pGL3 Δ8 does not contain any MSH6 sequences.
The deletion mutations discussed above likely eliminate more than just the seven Sp1 sites. To determine the functional significance of the seven individual Sp1 sites, a series of two-base substitution mutations (GGGCGG→GTTCGG or CCGCCC→CCGAAC) eliminating the core Sp1 binding site for each Sp1 site were constructed and tested. The data (Fig. 4B) show that mutation of the individual Sp1-4, Sp1-5, Sp1-6, and Sp1-7 sites significantly reduced MSH6 promoter activity. In this analysis, the Sp1-4 and Sp1-5 mutations appeared to have a somewhat greater effect than the Sp1-6 and Sp1-7 mutations. In contrast, mutations of the Sp1-1/2 and Sp1-3 sites only modestly reduced the promoter activity whereas mutation of the TATA box had little or no effect on promoter activity. A series of successive 2-bp substitution mutations inactivating the seven Sp1 binding sites were then tested to characterize the contribution of all seven Sp1 sites to MSH6 promoter activity (Fig. 4C). Mutation of the Sp1-7 site decreased the expression of luciferase and successive mutation of the Sp1-6 site resulted in a slight increase in the expression of luciferase. This suggests that the presence of the overlapping Ap2 binding site combined with mutation of the Sp1-6 site might result in increased transcription. In contrast, successive mutation of the Sp1-5, Sp1-4, and Sp1-3 resulted in progressive loss of luciferase expression. Mutation of the Sp1-1/2 sites and the TATA box resulted in little if any further loss of luciferase expression; however, this mutant construct still directed luciferase expression at levels which were above that of the vector alone. Clearly, when all of the Sp1 sites were mutated, the TATA box did not drive promoter activity. Overall, these results suggest that (i) all seven Sp1 binding sites contribute to MSH6 promoter activity, (ii) that the Sp1-4, Sp1-5, Sp1-6, and Sp1-7 sites are the most important, and (iii) that the promoter does not contain a functional TATA box.
Functional analysis of the three MSH6 promoter SNPs.
Two of the three most common SNPs present in the MSH6 promoter region eliminate Sp1 sites and could have an effect on the promoter activity. To investigate this possibility, we generated a series of luciferase reporter constructs containing different combinations of the three SNPs and measured their luciferase expression activity in transient-transfection assays with CHO cells (Fig. 5). The individual −557G (SNP 1) and −448A (SNP 2) changes did not significantly affect luciferase expression, whereas the single −159T change (SNP 3) reproducibly reduced luciferase expression. These observations are consistent with the above results, indicating that the individual Sp1-7 site is more important for promoter activity than the Sp1-2 site and that the −557 change does not alter a potential transcription factor binding site. The −448A −159T double SNP (SNP 2-3) construct and the −557G −448A −159T triple SNP construct (SNP 1-2-3), the latter of which likely corresponds to the major naturally occurring polymorphic allele, each reproducibly yielded less luciferase expression than the wild-type construct, although expression was higher than that seen for either the promoterless vector or a construct in which the MSH6 promoter was present in reverse orientation. Virtually identical results were obtained when the same constructs were transfected into the human BJ cell line (human skin fibroblasts), whereas the SNPs caused somewhat less reduction of luciferase expression than when the constructs were transfected into HeLa cells.
FIG. 5.
Effect of SNPs on the activity of the MSH6 promoter. The pGL3 wild-type MSH6 promoter reporter vector containing MSH6 nucleotides −633 to −9 and derivatives containing single base substitutions corresponding to the −557G (SNP 1), −448A (SNP 2), and −159T (SNP 3) SNPs or combinations of these substitutions were analyzed for promoter activity after transfection into CHO cells. The presence of base substitutions of interest is indicated by the nucleotides indicated above each promoter diagram and by the black circle on the relevant structural feature of the promoter diagram when an SNP is present. The pGL3 control vector contains the simian immunodeficiency virus 40 promoter and the vector pGL3-9/-633WT contains the MSH6 promoter region in reverse orientation.
Transactivation of MSH6 promoter activity by Sp1 and Sp3.
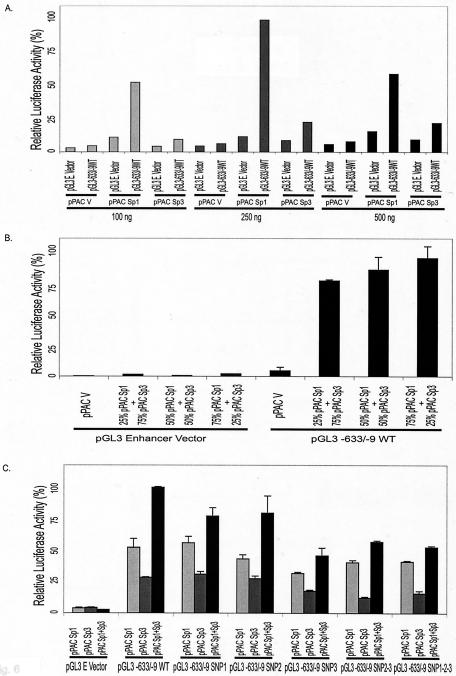
To investigate whether Sp1 and Sp3 transfection factors could directly regulate MSH6 promoter activity, Drosophila SL2 cells, which are deficient in Sp-related proteins (59), were cotransfected with different combinations of Sp1 and Sp3 expression vectors along with different wild-type or mutant MSH6-luciferase reporter constructs (Fig. 6). When the full-length MSH6-luciferase construct or the empty vector (pGL3-Enhancer vector) was cotransfected into SL2 cells, along with increasing amounts of the pPAC-Sp1 and pPAC-Sp3 expression vectors, luciferase expression by the full-length construct was strongly and specifically stimulated by Sp1 expression, whereas Sp3 expression resulted in a lower level of specific expression (Fig. 6A). Consistent with this observation, when different ratios of the Sp1 and Sp3 expression vectors were cotransfected into Drosophila SL2 cells, along with the full-length MSH6-luciferase construct or the empty vector, strong MSH6 promoter-dependent luciferase expression was observed with the extent of expression increasing as the ratio of Sp1 to Sp3 increased (Fig. 6B). Finally, the five different SNP containing reporter constructs described above were cotransfected into Drosophila SL2 cells, along with either the Sp1 expression construct, the Sp3 expression construct, or a 2:2 ratio of the Sp1 and Sp3 constructs (Fig. 6C). Under the three different Sp transcription factor expression conditions, the −557T (SNP 1) and −448A (SNP 2) single SNPs did not appear to significantly reduce luciferase expression, whereas the −159T SNP (SNP 3), the −448A −159T double SNP (SNP 2-3), and the −557G −448A −159T triple SNP (SNP 1-2-3) combinations all reproducibly reduced luciferase expression. These results support the view that the MSH6 promoter is regulated by the Sp transcription factors and that the naturally occurring polymorphic promoter allele containing five Sp1 sites has less promoter activity than the wild-type promoter, which contains seven Sp1 sites.
FIG. 6.
Activation of the MSH6 promoter by Sp1 and Sp3 proteins in SL2 cells. The effect of Sp1 and Sp3 transcription factors on the expression of luciferase driven by the MSH6 promoter was analyzed by cotransfection of the pGL3 vector or the pGL3-633/-9 vector containing the wild-type MSH6 promoter and either Sp1 or Sp3 expression plasmids into Drosophila SL2 cells which lack endogenous Sp1 and Sp3. (A) Drosophila SL2 cells were cotransfected with the indicated amounts of the pPAC-Sp1, pPAC-Sp3, or pPAC (empty) expression vectors and 0.4 μg of either the promoterless reporter construct pGL3-Basic vector or the pGL3-633/-9 vector containing the wild-type MSH6 promoter. (B) Drosophila SL2 cells were cotransfected with 0.4 μg of either the promoterless pGL3-Enhancer vector or the pGL3-633/-9 MSH6 promoter vector and 0.25 μg of either the pPAC empty vector or the indicated mixture of the pPAC-Sp1 and pPAC-Sp3 expression vectors. (C) Drosophila SL2 cells were cotransfected with 0.4 μg of either the promoterless pGL3-Enhancer vector, the pGL3-633/-9 MSH6 promoter vector, or derivatives of pGL3-633/-9 containing the indicated SNPs (see Fig. 5 for details about the SNP containing plasmids) and 0.25 μg of either the pPAC-Sp1, pPAC-Sp3, or an equimolar mixture of the pPAC-Sp1 and pPAC-Sp3 expression vectors.
DNA methylation inhibits Sp1 binding and MSH6 promoter activity.
It has previously been reported that DNA methylation can be associated with silencing of MSH6 expression (5). Because the Sp1 binding site consensus sequence is a potential substrate for CpG methylation, such methylation-dependent effects could be mediated by methylation of Sp1 sites. To investigate this possibility, the oligonucleotide duplexes containing the Sp1-1/2, Sp1-3, Sp1-4, Sp1-5, Sp1-6, and Sp1-7 sites (Fig. 7A) were methylated in vitro with SssI methylase and tested for their ability to compete with the unmethylated Sp1 consensus site for Sp1/Sp3 binding in EMSA assays (Fig. 7B). Methylation of all six Sp1 site DNAs significantly reduced their ability to compete with the labeled Sp1 consensus oligonucleotide for binding by Sp1 and Sp3 (lanes 3, 5, 7, 9, 11, and 13), although possibly not to the same extent as two-base mutations (GGGCGG→GTTCGG and CCGCCC→CCGAAC) that eliminated the Sp1 recognition sequence in each of the MSH6 Sp1 site oligonucleotides (lanes 2, 4, 6, 8, 10, and 12). In comparison, the Sp1 consensus competitor completely eliminated binding. Some previous studies have shown that methylation does not inhibit Sp1 binding (26), whereas other studies have found that methylation inhibits Sp1 binding depending on the exact sequence of the Sp1 binding site (12); thus, our results suggest that the sequence context of the MSH6 Sp1 sites allows DNA methylation to inhibit Sp1 binding. To further analyze the role of DNA methylation in regulation of the MSH6 promoter, the full-length MSH6 promoter luciferase reporter construct and the two versions containing either the −448A −159T double SNP (SNP 2-3) or the −557G −448A −159T triple SNP (SNP 1-2-3) combinations were treated with SssI methylase. The DNAs were then transfected into HeLa cells (Fig. 7C) or CHO cells (data not shown) to analyze the effect of DNA methylation on luciferase expression. Methylation of the wild-type construct reduced luciferase expression by 90%, whereas methylation of the two SNP-containing constructs reduced luciferase expression to <1% of that seen with the unmethylated wild-type construct. These results suggest that the MSH6 promoter could be regulated by methylation, including methylation of its Sp1 binding sites, and that the naturally occurring triple SNP five Sp1 site promoter allele is more sensitive to this type of regulation than the major seven Sp1 site promoter allele.
FIG. 7.
Effect of DNA methylation on the MSH6 promoter Sp1 sites and promoter activity. (A) Sequences of the competitor DNAs used in gel mobility shift assays. Only the sequence of the top strand of the oligonucleotide duplexes is indicated (see Fig. 3 for more details). The Sp1 binding consensus sequences (GGGCGG) and inverted complement sequences (CCGCCC) are indicated by the overlining and underlining, respectively. The numbering system for each of the Sp1 sites is as defined in Fig. 1B. The wild-type sequences are indicated (WT). The mutant sequence Sp1-1/2Mut refers to the sequence containing the indicated 2-bp substitution mutations in both the Sp1-1 and Sp1-2 sites, whereas otherwise “Mut” refers to the presence of the indicated 2-bp substitution mutations in Sp1 sites Sp1-3 through Sp1-7. See Fig. 1A for sequences that are changed by these nucleotide substitutions. “Meth” refers to the wild-type sequences that have been methylated in vitro at the Cs designated in lowercase. (B) Gel mobility shift assays performed with HeLa cell extract, radioactively labeled Sp1 consensus sequence as probe (see Fig. 3A), and a 50-fold molar excess of the indicated competitor DNAs. The arrows indicate the three specific protein-DNA complexes formed. (C) The pGL3 wild-type MSH6 promoter reporter vector containing MSH6 nucleotides −633 to −9 and methylated (Meth) versions of this plasmid or derivatives containing single base substitutions corresponding to the −448A (SNP 2) and −159T (SNP 3) SNPs or the −557G (SNP 1), −448A (SNP 2), and −159T (SNP 3) SNPs were analyzed for promoter activity after transfection into HeLa cells. The positions of the HpaII and AciI sites that are substrates for DNA methylation are indicated by the symbols above the line diagram of each promoter, and the presence and position of the SNPs are indicated by the black circles on each line diagram.
Analysis of MSH6 expression in tumor cell lines containing MSH6 polymorphisms.
To begin to assess the possible in vivo significance of the five Sp1 site promoter, a number of tumor cell lines were screened to identify lines containing different MSH6 polymorphic variants. To do this, the genomic MSH6 locus was sequenced and analyzed for mutations and polymorphisms (34). In some cases, fragments of the MSH6 genomic locus and cDNA were amplified by PCR, cloned, and sequenced to determine the linkage between different nucleotide changes present in the promoter, exon 1, and exon 4. Three glioblastoma cell lines were studied. LN229 was found to be homozygous for the seven Sp1 site promoter allele −557T −448G −159C 186C (exon 1) 642C (exon 4) and appears to have at least two copies of MSH6, as evidenced by the presence of heterozygous nucleotides at the site of common silent intronic polymorphisms (34). LN340 was found to be heterozygous for the five and seven Sp1 site alleles and for two silent coding sequence polymorphisms; one allele was −557G −448G −159C 186A (exon 1) 642C (exon 4), and the other allele was −557G −448A −159T 186C (exon 1) 642T (exon 4). LN443 only contained the five Sp1 site allele −557G −448A −159T 186C (exon 1) 642T (exon 4). This cell line was probably hemizygous because no heterozygous nucleotides were observed at the site of common silent intronic polymorphisms. None of the cell lines contained a loss-of-function MSH6 mutation or amino-acid-changing variant.
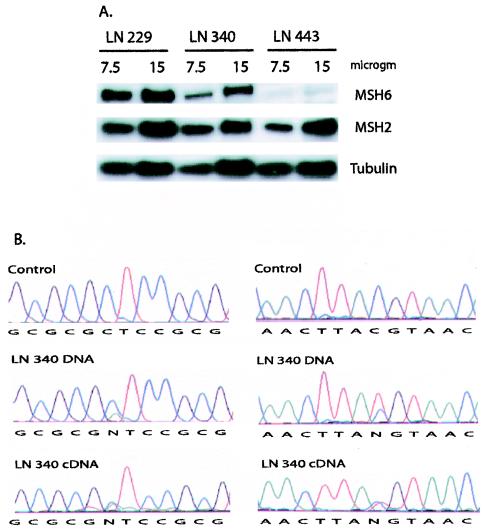
The cell lines LN229, LN340, and LN443 were first analyzed by Western blotting with antibodies to detect MSH6, MSH2, and tubulin as a loading control (Fig. 8A). LN340, the cell line that is heterozygous for the five and seven Sp1 site promoters, had reduced MSH6 levels compared to that of LN229, the cell line that was homozygous for the seven Sp1 site promoter. LN443, the cell line that was homozygous for the five Sp1 site promoter, had further reduced very low, but still detectable, levels of MSH6. All three cell lines expressed similar levels of MSH2 and tubulin. Treatment of LN340 and LN443 cells with 5-aza-2′-deoxycytidine did not increase the MSH6 protein levels, suggesting that the reduced MSH6 expression was not due to DNA methylation associated silencing in these cell lines. These results are consistent with the idea that the five Sp1 site promoter is less active than the seven Sp1 site promoter in vivo. To further analyze the activity of the five Sp1 site promoter, mRNA was isolated from LN340 cells and used as a template for synthesis of cDNA. Then the region around nucleotides 186 and 642 was amplified by PCR from cDNA and genomic DNA and sequenced to determine the relative amounts of the two polymorphic nucleotides present at each site in each sample (Fig. 8B). Compared to the genomic DNA control, analysis of the relative peak heights in the cDNA indicated that the 186C and 642T variants that are linked to the five Sp1 site promoter were reduced by ca. 60% relative to the 186A and 642C variants linked to the seven Sp1 site promoter. In a control experiment with the LNZ308 cell line that is homozygous for the seven Sp1 site allele and heterozygous for the 186C/A (exon 1) polymorphism, there was no difference between the relative amounts of the 186C and 186A alleles when genomic DNA and cDNA were similarly analyzed (data not shown). Similar to the Western blotting analysis, these results support the view that the five Sp1 site promoter is less active than the seven Sp1 site promoter, a finding consistent with the conclusion from the transfection analysis that the −448A and −159T polymorphisms each inactivate an Sp1 site that is important for the full activity of the promoter. We are currently performing a detailed molecular analysis of the consequences of reduced MSH6 expression in these cell lines.
FIG. 8.
Analysis of MSH6 expression in glioblastoma cell lines. (A) Western blot analysis of MSH6, MSH2, and tubulin expression. Cell extracts were prepared from the LN229, LN340, and LN443 and analyzed by Western blotting as described in Materials and Methods. Portions (7.5 and 15 μg) of each extract were analyzed as indicated above the individual lanes. The MSH6, MSH2, and tubulin bands are indicated on the right side of the gel. (B) Sequence analysis of MSH6 mRNA expression from the five and seven Sp1 site promoters. Genomic DNA and cDNA from LN340 cells were prepared and sequenced as described in Materials and Methods. LN340 cells are heterozygous for two different MSH6 alleles: one allele is the seven Sp1 site promoter allele −557G −448G −159C 186A (exon 1) 642C (exon 4), and the other allele is the five Sp1 site promoter allele −557G −448A −159T 186C (exon 1) 642T (exon 4). The sequences on the left cover the region around nucleotide 186 from a control (186C) DNA and LN340 genomic and cDNA as indicated. The sequences on the right cover the region around nucleotide 642 from a control (642C) DNA and LN340 genomic and cDNA as indicated. Reduction of the relative levels of the 186C and 642T nucleotides linked to the five Sp1 site promoter compared to the levels of the 186A and 642C nucleotides linked to the seven Sp1 site promoter is seen in the cDNA relative to the genomic DNA.
DISCUSSION
In the present study, we investigated the structure of the human MSH6 core promoter located within a DNA fragment containing nucleotides −633 to +1 relative to the MSH6 translational start site. Computer analysis indicated the presence of seven GC boxes potentially capable of binding Sp1 and Sp3 transcription factors located between positions −454 and −159, as well as other potential transcription factor binding sequences, including AP family, c-JUN, E2F, E4TF1, EF.C, and CAAT box sequences. All seven of the GC boxes partially contributed to promoter activity and were able to bind Sp1 and Sp3. MSH6 transcription appeared to initiate from multiple start sites located between nucleotides −250 to −50 and, consistent with this, deletion analysis showed that a potential TATA box at −623 was not required for transcription. These features are similar to those reported for other human MMR and MMR-related genes such as MSH2 and MLH1 (27, 28), where multiple start sites and the absence of a functional TATA box have been reported, and for PMS1 and PMS2, where the promoter regions have been reported to contain CAAT boxes, CpG islands, and Sp1 binding sites (48, 71). TATA-less promoters are often activated by Sp family proteins. Similar to the results reported here for MSH6, Sp1-dependent promoters contain multiple GC boxes located within several hundred base pairs upstream of multiple transcription start sites, and these GC boxes are recognized by the Sp1 protein (20), which actives mRNA synthesis by RNA polymerase II (13). In this respect, MMR genes, including MSH6, appear to be housekeeping genes. As part of the present study, we identified two different polymorphic alleles of MSH6 that had either five or six functional Sp1 sites and provided evidence that the five Sp1 site promoter is less active in vivo than the seven Sp1 site promoter, further supporting the view that MSH6 is regulated by Sp1. This suggests that the human population contains individuals with different levels of MSH6 promoter activity.
To characterize the transcriptional regulation of the MSH6 gene, we performed a functional analysis of the 5′-flanking region of the gene. The mapping of the 5′ ends of the MSH6 mRNA by 5′-RACE and primer extension assays revealed multiple start sites, including two major start sites that mapped between −76 and −70 bp from ATG. This finding confirmed the previous mapping of MSH6 start sites by using an RNase protection assay (60), although we observed a greater number of potential start sites than previously reported and mapped them to the nucleotide level. Seven potential Sp1 binding sites were observed in the proximity of the multiple transcription start sites, which suggests that the MSH6 promoter might be regulated by Sp1 and Sp3 transcription factors. To investigate this possibility, a series of deletion and two-base substitution mutations altering these Sp1 sites were tested for their effect on expression of a reporter construct in transient-transfection assays. This analysis demonstrated that all seven Sp1 sites contributed to maximal expression and also suggested that the first four Sp1 sites upstream of the ATG were more important for expression than the three most upstream Sp1 sites. This type of regulation by multiple Sp1 sites has been observed for numerous other genes (23, 43, 50) for which synergistic activation due to binding of multiple Sp1 proteins is important for transcription and regulation of gene expression. Consistent with the view that MSH6 is an Sp1-regulated promoter, all seven Sp1 sites were found to bind both Sp1 and Sp3 and expression of an MSH6 core promoter construct in Drosophila SL2 cells that lack Sp1 and Sp3 proteins was absolutely dependent on cotransfection with either Sp1 or Sp3 expression constructs. In addition, the one potential TATA box identified in the MSH6 promoter region was not required for expression of MSH6, a finding consistent with that observed with other Sp1-regulated genes. Overall, the MSH6 promoter region resembles that of other MMR genes and related genes, such as MSH2, MLH1, PMS1, and PMS2, and is consistent with the idea that these genes are typical housekeeping genes that are transcribed by RNA polymerase II.
The presence of multiple Sp1 sites in the MSH6 promoter has a number of implications for the regulation of MSH6 expression. It is known that Sp1 and Sp3 can act as transcriptional activators or repressors, depending on the specific promoter involved and also on the form of Sp3 (short or long) present in the cell (3, 16, 23, 53). In addition, the hypothetical model of Kwon et al. (35) has suggested that the number of sites occupied by Sp1 or Sp3, which compete for the same binding site, are determined by levels and the relative proportion of Sp1 and Sp3 in the cell and affect both transcription initiation and promoter activation. The cotransfection analysis with the Drosophila SL2 cells presented here indicates that MSH6 is more strongly activated by Sp1 than by Sp3. This suggests that MSH6 expression could be affected by the relative levels of these two transcription factors present in different cell types.
Analysis of the promoter of other MMR genes and of related genes such as MLH1, MSH2, and PMS1 has identified polymorphisms in the 5′ upstream of these genes that did not appear to affect the transcription of the genes (27, 28) (71). Recently, there has been a report of germ line mutations in the MHS2 promoter that may have pathogenic effects in some suspected HNPCC and sporadic colorectal cancer patients (56). To investigate the presence of polymorphisms in the MSH6 promoter region, nucleotides −633 to +39 were sequenced from 100 samples of the human diversity set of normal control DNAs and from 100 normal Caucasian control DNAs. Compared to the wild-type sequence (−557T −448G −159C), the most common polymorphic allele was −557G −448G −159C, which was heterozygous or homozygous in 30% of the diversity and Caucasian sample sets. The −557G (SNP 1) change does not appear to alter any sequence required for transcription factor binding and, consistent with this, the −557G −448G −159C allele did not cause altered promoter activity in transfection experiments. Interestingly, 16% of the Caucasian DNAs were heterozygous or homozygous for the −557G −448A −159T or −557T −448A −159T alleles, which were present in the diversity set at a lower frequency, a finding consistent with the proportion of Caucasian DNAs in the diversity set. The −448A (SNP 2) and −159T (SNP 3) changes eliminated the Sp1-2 and Sp1-7 consensus sites and, a finding consistent with this, the mutant sites were no longer bound by Sp1 or Sp3. In transfection experiments, the −557G −448A −159T allele showed a 50% reduction in promoter activity, and this allele was significantly more sensitive to inactivation by DNA methylation than the wild-type allele. Analysis of MSH6 expression in cell lines containing the −557G −448A −159T allele showed that the five Sp1 site promoter was less active than the seven Sp1 site promoter, resulting in reduced MSH6 expression at both the mRNA and protein level; this result provides further evidence that MSH6 is a Sp1 regulated gene in vivo. We also observed that 2% of the Caucasian DNAs were heterozygous for the −210T change (SNP 4); these data and two additional examples (not shown) are consistent with the polymorphic allele being −557T −448G −210T −159C. The −210T change (SNP 4) is predicted to inactivate the Sp1-5 site, which is particularly important for promoter activity, although this polymorphic variant was not analyzed directly for its effect on promoter activity. These results suggest that Caucasians contain two relatively common polymorphic variants that result in reduced MSH6 promoter activity. However, at present, we have no evidence that either variant is associated with increased cancer susceptibility. Finally, we observed five other SNPs at low frequencies in the diversity set but not the Caucasian set, none of which appear to alter sequences predicted to be important for promoter activity.
The MSH6 promoter region has a high GC content, a finding consistent with the presence of seven Sp1 sites in the promoter region. This raises the possibility that the MSH6 promoter region could be a target of DNA methylation at CpG sites (12, 22). No studies have yet examined MSH6 expression and methylation in human tumors; however, a human tumor cell line has been reported in which MSH6 expression was absent but could be reactivated by treatment with 5-azacytidine, indicating that MSH6 expression can be eliminated by DNA methylation associated gene silencing (5). Consistent with this, we have observed that methylation of the MSH6 promoter region at CpG sites by SssI methylase significantly reduces promoter activity. In addition, consistent with some published reports (12), methylation of oligonucleotides containing the different MSH6 Sp1 sites reduced the ability of each site to bind Sp1 and Sp3, although not as much as a mutation eliminating each Sp1 site. This suggests that methylation of individual Sp1 sites may result in reduced promoter activity by reducing Sp1 binding. In addition, it is thought (12) that binding of Sp1 transcription factors to their GC-rich binding sites helps maintain CpG islands in an unmethylated state. As a consequence, mutation or deletion of Sp1 sites could reduce binding of Sp1 transcription factors, allowing de novo methylation of CpG islands (9, 32, 40). This has been reported for the Rb gene, where mutations resulted in promoter silencing (55). This finding suggests that, because the two polymorphisms (−448A, SNP 2; −159T, SNP 3) decrease the amount of Sp1 bound to the MSH6 promoter and hence increase the sensitivity of the MSH6 promoter to silencing by DNA methylation in vitro, this polymorphic allele could be a preferential target of de novo methylation, resulting in transcriptional silencing of the gene compared to the more prevalent seven Sp1 site promoter. It is also possible that reduced transcription due to methylation of Sp1 sites could then facilitate the binding of other factors to methylated DNA, resulting in more complete silencing of MSH6. However, to our knowledge, there have been no reports of epigenetic silencing of MSH6 in tumors, although there have been reports of loss of expression of MSH6 in tumors.
Acknowledgments
We are grateful to K. Arden, G. Bonizzi, W. Cavenee, A. C. Li, and J. Wang for providing cell lines and for technical support. We thank J. Weger and J. Mueller for performing DNA sequencing and A. Rojas and T. Sternsdorf for help with sequence analysis software. Finally, we thank C. Glass, B. Guerra, H. Hoffman, A. R. Houweling, and K. Okumura for helpful discussions and comments on the manuscript.
This work was supported by NIH grant ES11040.
REFERENCES
- 1.Acharya, S., T. Wilson, S. Gradia, M. F. Kane, S. Guerrette, G. T. Marsischky, R. Kolodner, and R. Fishel. 1996. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. USA 93:13629-13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiyama, Y., H. Nagasaki, T. Nakajima, H. Sakai, T. Nomizu, and Y. Yuasa. 2001. Infrequent frameshift mutations in the simple repeat sequences of hMLH3 in hereditary nonpolyposis colorectal cancers. Jpn. J. Clin. Oncol. 31:61-64. [DOI] [PubMed] [Google Scholar]
- 3.Andrew, S. D., P. J. Delhanty, L. M. Mulligan, and B. G. Robinson. 2000. Sp1 and Sp3 transactivate the RET proto-oncogene promoter. Gene 256:283-291. [DOI] [PubMed] [Google Scholar]
- 4.Aslam, F., L. Palumbo, L. H. Augenlicht, and A. Velcich. 2001. The Sp family of transcription factors in the regulation of the human and mouse MUC2 gene promoters. Cancer Res. 61:570-576. [PubMed] [Google Scholar]
- 5.Bearzatto, A., M. Szadkowski, P. Macpherson, J. Jiricny, and P. Karran. 2000. Epigenetic regulation of the MGMT and hMSH6 DNA repair genes in cells resistant to methylating agents. Cancer Res. 60:3262-3270. [PubMed] [Google Scholar]
- 6.Berends, M. J., Y. Wu, R. H. Sijmons, R. G. Mensink, T. van der Sluis, J. M. Hordijk-Hos, E. G. de Vries, H. Hollema, A. Karrenbeld, C. H. Buys, A. G. van der Zee, R. M. Hofstra, and J. H. Kleibeuker. 2002. Molecular and clinical characteristics of MSH6 variants: an analysis of 25 index carriers of a germline variant. Am. J. Hum. Genet. 70:26-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonizzi, G., J. Piette, S. Schoonbroodt, M. P. Merville, and V. Bours. 1999. Role of the protein kinase C lambda/iota isoform in nuclear factor-kappaB activation by interleukin-1beta or tumor necrosis factor-alpha: cell type specificities. Biochem. Pharmacol. 57:713-720. [DOI] [PubMed] [Google Scholar]
- 8.Bowers, J., T. Sokolsky, T. Quach, and E. Alani. 1999. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2-MSH6 complex disrupts mismatch recognition. J. Biol. Chem. 274:16115-16125. [DOI] [PubMed] [Google Scholar]
- 9.Brandeis, M., D. Frank, I. Keshet, Z. Siegfried, M. Mendelsohn, A. Nemes, V. Temper, A. Razin, and H. Cedar. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371:435-438. [DOI] [PubMed] [Google Scholar]
- 10.Briggs, M. R., J. T. Kadonaga, S. P. Bell, and R. Tjian. 1986. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 234:47-52. [DOI] [PubMed] [Google Scholar]
- 11.Chadwick, R. B., J. E. Meek, T. W. Prior, P. Peltomaki, and A. de La Chapelle. 2000. Polymorphisms in a pseudogene highly homologous to PMS2. Hum. Mutat. 16:530. [DOI] [PubMed] [Google Scholar]
- 12.Clark, S. J., J. Harrison, and P. L. Molloy. 1997. Sp1 binding is inhibited by (m)Cp(m)CpG methylation. Gene 195:67-71. [DOI] [PubMed] [Google Scholar]
- 13.Courey, A. J., and R. Tjian. 1988. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55:887-898. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham, J. M., E. R. Christensen, D. J. Tester, C. Y. Kim, P. C. Roche, L. J. Burgart, and S. N. Thibodeau. 1998. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 58:3455-3460. [PubMed] [Google Scholar]
- 15.Das Gupta, R., and R. D. Kolodner. 2000. Novel dominant mutations in Saccharomyces cerevisiae MSH6. Nat. Genet. 24:53-56. [DOI] [PubMed] [Google Scholar]
- 16.Dennig, J., M. Beato, and G. Suske. 1996. An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J. 15:5659-5667. [PMC free article] [PubMed] [Google Scholar]
- 17.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drummond, J. T., J. Genschel, E. Wolf, and P. Modrich. 1997. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSα/hMutSβ ratio and reduces the efficiency of base-base mismatch repair. Proc. Natl. Acad. Sci. USA 94:10144-10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drummond, J. T., G. M. Li, M. J. Longley, and P. Modrich. 1995. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science 268:1909-1912. [DOI] [PubMed] [Google Scholar]
- 20.Dynan, W. S., and R. Tjian. 1985. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. Nature 316:774-778. [DOI] [PubMed] [Google Scholar]
- 21.Genschel, J., S. J. Littman, J. T. Drummond, and P. Modrich. 1998. Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J. Biol. Chem. 273:19895-19901. [DOI] [PubMed] [Google Scholar]
- 22.Graff, J. R., J. G. Herman, S. Myohanen, S. B. Baylin, and P. M. Vertino. 1997. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J. Biol. Chem. 272:22322-22329. [DOI] [PubMed] [Google Scholar]
- 23.Hagen, G., S. Muller, M. Beato, and G. Suske. 1994. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 13:3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harfe, B. D., and S. Jinks-Robertson. 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34:359-399. [DOI] [PubMed] [Google Scholar]
- 25.Herman, J. G., A. Umar, K. Polyak, J. R. Graff, N. Ahuja, J. P. Issa, S. Markowitz, J. K. Willson, S. R. Hamilton, K. W. Kinzler, M. F. Kane, R. D. Kolodner, B. Vogelstein, T. A. Kunkel, and S. B. Baylin. 1998. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl. Acad. Sci. USA 95:6870-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holler, M., G. Westin, J. Jiricny, and W. Schaffner. 1988. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 2:1127-1135. [DOI] [PubMed] [Google Scholar]
- 27.Ito, E., Y. Yanagisawa, Y. Iwahashi, Y. Suzuki, H. Nagasaki, Y. Akiyama, S. Sugano, Y. Yuasa, and K. Maruyama. 1999. A core promoter and a frequent single-nucleotide polymorphism of the mismatch repair gene hMLH1. Biochem. Biophys. Res. Commun. 256:488-494. [DOI] [PubMed] [Google Scholar]
- 28.Iwahashi, Y., E. Ito, Y. Yanagisawa, Y. Akiyama, Y. Yuasa, T. Onodera, and K. Maruyama. 1998. Promoter analysis of the human mismatch repair gene hMSH2. Gene 213:141-147. [DOI] [PubMed] [Google Scholar]
- 29.Jagmohan-Changur, S., T. Poikonen, S. Vilkki, V. Launonen, F. Wikman, T. F. Orntoft, P. Moller, H. Vasen, C. Tops, R. D. Kolodner, J. P. Mecklin, H. Jarvinen, S. Bevan, R. S. Houlston, L. A. Aaltonen, R. Fodde, J. Wijnen, and A. Karhu. 2003. EXO1 variants occur commonly in normal population: evidence against a role in hereditary nonpolyposis colorectal cancer. Cancer Res. 63:154-158. [PubMed] [Google Scholar]
- 30.Kadonaga, J. T., K. R. Carner, F. R. Masiarz, and R. Tjian. 1987. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51:1079-1090. [DOI] [PubMed] [Google Scholar]
- 31.Kane, M. F., M. Loda, G. M. Gaida, J. Lipman, R. Mishra, H. Goldman, J. M. Jessup, and R. Kolodner. 1997. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 57:808-811. [PubMed] [Google Scholar]
- 32.Knight, S. W., T. J. Vulliamy, B. Morgan, K. Devriendt, P. J. Mason, and I. Dokal. 2001. Identification of novel DKC1 mutations in patients with dyskeratosis congenita: implications for pathophysiology and diagnosis. Hum. Genet. 108:299-303. [DOI] [PubMed] [Google Scholar]
- 33.Kolodner, R. D., and G. T. Marsischky. 1999. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9:89-96. [DOI] [PubMed] [Google Scholar]
- 34.Kolodner, R. D., J. D. Tytell, J. L. Schmeits, M. F. Kane, R. D. Gupta, J. Weger, S. Wahlberg, E. A. Fox, D. Peel, A. Ziogas, J. E. Garber, S. Syngal, H. Anton-Culver, and F. P. Li. 1999. Germ-line msh6 mutations in colorectal cancer families. Cancer Res. 59:5068-5074. [PubMed] [Google Scholar]
- 35.Kwon, H. S., M. S. Kim, H. J. Edenberg, and M. W. Hur. 1999. Sp3 and Sp4 can repress transcription by competing with Sp1 for the core cis-elements on the human ADH5/FDH minimal promoter. J. Biol. Chem. 274:20-28. [DOI] [PubMed] [Google Scholar]
- 36.Lania, L., B. Majello, and P. De Luca. 1997. Transcriptional regulation by the Sp family proteins. Int. J. Biochem. Cell Biol. 29:1313-1323. [DOI] [PubMed] [Google Scholar]
- 37.LeVan, T. D., J. W. Bloom, T. J. Bailey, C. L. Karp, M. Halonen, F. D. Martinez, and D. Vercelli. 2001. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J. Immunol. 167:5838-5844. [DOI] [PubMed] [Google Scholar]
- 38.Lipkin, S. M., V. Wang, D. L. Stoler, G. R. Anderson, I. Kirsch, D. Hadley, H. T. Lynch, and F. S. Collins. 2001. Germline and somatic mutation analyses in the DNA mismatch repair gene MLH3: evidence for somatic mutation in colorectal cancers. Hum. Mutat. 17:389-396. [DOI] [PubMed] [Google Scholar]
- 39.Liu, T., H. Yan, S. Kuismanen, A. Percesepe, M. L. Bisgaard, M. Pedroni, P. Benatti, K. W. Kinzler, B. Vogelstein, M. Ponz de Leon, P. Peltomaki, and A. Lindblom. 2001. The role of hPMS1 and hPMS2 in predisposing to colorectal cancer. Cancer Res. 61:7798-7802. [PubMed] [Google Scholar]
- 40.Macleod, D., J. Charlton, J. Mullins, and A. P. Bird. 1994. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 8:2282-2292. [DOI] [PubMed] [Google Scholar]
- 41.Marra, G., I. Iaccarino, T. Lettieri, G. Roscilli, P. Delmastro, and J. Jiricny. 1998. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc. Natl. Acad. Sci. USA 95:8568-8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marsischky, G. T., N. Filosi, M. F. Kane, and R. Kolodner. 1996. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10:407-420. [DOI] [PubMed] [Google Scholar]
- 43.Mastrangelo, I. A., A. J. Courey, J. S. Wall, S. P. Jackson, and P. V. Hough. 1991. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc. Natl. Acad. Sci. USA 88:5670-5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKnight, S. L., and R. Kingsbury. 1982. Transcriptional control signals of a eukaryotic protein-coding gene. Science 217:316-324. [DOI] [PubMed] [Google Scholar]
- 45.Miyaki, M., M. Konishi, K. Tanaka, R. Kikuchi-Yanoshita, M. Muraoka, M. Yasuno, T. Igari, M. Koike, M. Chiba, and T. Mori. 1997. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat. Genet. 17:271-272. [DOI] [PubMed] [Google Scholar]
- 46.Miyakura, Y., K. Sugano, F. Konishi, N. Fukayama, S. Igarashi, K. Kotake, T. Matsui, Y. Koyama, M. Maekawa, and H. Nagai. 2003. Methylation profile of the MLH1 promoter region and their relationship to colorectal carcinogenesis. Genes Chromosomes Cancer. 36:17-25. [DOI] [PubMed] [Google Scholar]
- 47.Miyakura, Y., K. Sugano, F. Konishi, A. Ichikawa, M. Maekawa, K. Shitoh, S. Igarashi, K. Kotake, Y. Koyama, and H. Nagai. 2001. Extensive methylation of hMLH1 promoter region predominates in proximal colon cancer with microsatellite instability. Gastroenterology 121:1300-1309. [DOI] [PubMed] [Google Scholar]
- 48.Nicolaides, N. C., K. W. Kinzler, and B. Vogelstein. 1995. Analysis of the 5′ region of PMS2 reveals heterogeneous transcripts and a novel overlapping gene. Genomics 29:329-334. [DOI] [PubMed] [Google Scholar]
- 49.Palombo, F., I. Iaccarino, E. Nakajima, M. Ikejima, T. Shimada, and J. Jiricny. 1996. hMutSβ, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 6:1181-1184. [DOI] [PubMed] [Google Scholar]
- 50.Pascal, E., and R. Tjian. 1991. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev. 5:1646-1656. [DOI] [PubMed] [Google Scholar]
- 51.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peltomaki, P., H. F. Vasen, et al. 1997. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. Gastroenterology 113:1146-1158. [DOI] [PubMed] [Google Scholar]
- 53.Rajakumar, R. A., S. Thamotharan, R. K. Menon, and S. U. Devaskar. 1998. Sp1 and Sp3 regulate transcriptional activity of the facilitative glucose transporter isoform-3 gene in mammalian neuroblasts and trophoblasts. J. Biol. Chem. 273:27474-27483. [DOI] [PubMed] [Google Scholar]
- 54.Risinger, J. I., A. Umar, J. Boyd, A. Berchuck, T. A. Kunkel, and J. C. Barrett. 1996. Mutation of MSH3 in endometrial cancer and evidence for its functional role in heteroduplex repair. Nat. Genet. 14:102-105. [DOI] [PubMed] [Google Scholar]
- 55.Sakai, T., N. Ohtani, T. L. McGee, P. D. Robbins, and T. P. Dryja. 1991. Oncogenic germ-line mutations in Sp1 and ATF sites in the human retinoblastoma gene. Nature 353:83-86. [DOI] [PubMed] [Google Scholar]
- 56.Shin, K. H., J. H. Shin, J. H. Kim, and J. G. Park. 2002. Mutational analysis of promoters of mismatch repair genes hMSH2 and hMLH1 in hereditary nonpolyposis colorectal cancer and early onset colorectal cancer patients: identification of three novel germ-line mutations in promoter of the hMSH2 gene. Cancer Res. 62:38-42. [PubMed] [Google Scholar]
- 57.Sia, E. A., R. J. Kokoska, M. Dominska, P. Greenwell, and T. D. Petes. 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17:2851-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suske, G. 1999. The Sp-family of transcription factors. Gene 238:291-300. [DOI] [PubMed] [Google Scholar]
- 59.Suske, G. 2000. Transient transfection of Schneider cells in the study of transcription factors. Methods Mol. Biol. 130:175-187. [DOI] [PubMed] [Google Scholar]
- 60.Szadkowski, M., and J. Jiricny. 2002. Identification and functional characterization of the promoter region of the human MSH6 gene. Genes Chromosomes Cancer 33:36-46. [DOI] [PubMed] [Google Scholar]
- 61.Umar, A., J. I. Risinger, W. E. Glaab, K. R. Tindall, J. C. Barrett, and T. A. Kunkel. 1998. Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics 148:1637-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Urban, A., S. Neukirchen, and K. E. Jaeger. 1997. A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res. 25:2227-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaisman, A., M. Varchenko, A. Umar, T. A. Kunkel, J. I. Risinger, J. C. Barrett, T. C. Hamilton, and S. G. Chaney. 1998. The role of hMLH1, hMSH3, and hMSH6 defects in cisplatin and oxaliplatin resistance: correlation with replicative bypass of platinum-DNA adducts. Cancer Res. 58:3579-3585. [PubMed] [Google Scholar]
- 64.Viel, A., E. Novella, M. Genuardi, E. Capozzi, M. Fornasarig, M. Pedroni, M. Santarosa, M. P. De Leon, L. Della Puppa, M. Anti, and M. Boiocchi. 1998. Lack of PMS2 gene-truncating mutations in patients with hereditary colorectal cancer. Int. J. Oncol. 13:565-569. [DOI] [PubMed] [Google Scholar]
- 65.Wagner, A., Y. Hendriks, E. J. Meijers-Heijboer, W. J. de Leeuw, H. Morreau, R. Hofstra, C. Tops, E. Bik, A. H. Brocker-Vriends, C. van Der Meer, D. Lindhout, H. F. Vasen, M. H. Breuning, C. J. Cornelisse, C. van Krimpen, M. F. Niermeijer, A. H. Zwinderman, J. Wijnen, and R. Fodde. 2001. Atypical HNPCC owing to MSH6 germline mutations: analysis of a large Dutch pedigree. J. Med. Genet. 38:318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, Q., C. Lasset, F. Desseigne, J. C. Saurin, C. Maugard, C. Navarro, E. Ruano, L. Descos, V. Trillet-Lenoir, J. F. Bosset, and A. Puisieux. 1999. Prevalence of germline mutations of hMLH1, hMSH2, hPMS1, hPMS2, and hMSH6 genes in 75 French kindreds with nonpolyposis colorectal cancer. Hum. Genet. 105:79-85. [DOI] [PubMed] [Google Scholar]
- 67.Wijnen, J., W. de Leeuw, H. Vasen, H. van der Klift, P. Moller, A. Stormorken, H. Meijers-Heijboer, D. Lindhout, F. Menko, S. Vossen, G. Moslein, C. Tops, A. Brocker-Vriends, Y. Wu, R. Hofstra, R. Sijmons, C. Cornelisse, H. Morreau, and R. Fodde. 1999. Familial endometrial cancer in female carriers of MSH6 germline mutations. Nat. Genet. 23:142-144. [DOI] [PubMed] [Google Scholar]
- 68.Wu, Y., M. J. Berends, R. G. Mensink, C. Kempinga, R. H. Sijmons, A. G. van Der Zee, H. Hollema, J. H. Kleibeuker, C. H. Buys, and R. M. Hofstra. 1999. Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am. J. Hum. Genet. 65:1291-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu, Y., M. J. Berends, J. G. Post, R. G. Mensink, E. Verlind, T. Van Der Sluis, C. Kempinga, R. H. Sijmons, A. G. van der Zee, H. Hollema, J. H. Kleibeuker, C. H. Buys, and R. M. Hofstra. 2001. Germline mutations of EXO1 gene in patients with hereditary nonpolyposis colorectal cancer (HNPCC) and atypical HNPCC forms. Gastroenterology 120:1580-1587. [DOI] [PubMed] [Google Scholar]
- 70.Wu, Y., M. J. Berends, R. H. Sijmons, R. G. Mensink, E. Verlind, K. A. Kooi, T. van der Sluis, C. Kempinga, A. G. van der Zee, H. Hollema, C. H. Buys, J. H. Kleibeuker, and R. M. Hofstra. 2001. A role for MLH3 in hereditary nonpolyposis colorectal cancer. Nat. Genet. 29:137-138. [DOI] [PubMed] [Google Scholar]
- 71.Yanagisawa, Y., E. Ito, Y. Iwahashi, Y. Akiyama, Y. Yuasa, and K. Maruyama. 1998. Isolation and characterization of the 5′ region of the human mismatch repair gene hPMS1. Biochem. Biophys. Res. Commun. 243:738-743. [DOI] [PubMed] [Google Scholar]
- 72.Zaveri, N. T., C. J. Green, W. E. Polgar, N. Huynh, and L. Toll. 2002. Regulation of transcription of the human prepronociceptin gene by Sp1. Gene 290:45-52. [DOI] [PubMed] [Google Scholar]
- 73.Zhao, L., and L. S. Chang. 1997. The human POLD1 gene: identification of an upstream activator sequence, activation by Sp1 and Sp3, and cell cycle regulation. J. Biol. Chem. 272:4869-4882. [PubMed] [Google Scholar]