Abstract
At the G1/S phase cell cycle transition, multiple histone genes are expressed to ensure that newly synthesized DNA is immediately packaged as chromatin. Here we have purified and functionally characterized the critical transcription factor HiNF-P, which is required for E2F-independent activation of the histone H4 multigene family. Using chromatin immunoprecipitation analysis and ligation-mediated PCR-assisted genomic sequencing, we show that HiNF-P interacts with conserved H4 cell cycle regulatory sequences in vivo. Antisense inhibition of HiNF-P reduces endogenous histone H4 gene expression. Furthermore, we find that HiNF-P utilizes NPAT/p220, a substrate of the cyclin E/cyclin-dependent kinase 2 (CDK2) kinase complex, as a key coactivator to enhance histone H4 gene transcription. The biological role of HiNF-P is reflected by impeded cell cycle progression into S phase upon antisense-mediated reduction of HiNF-P levels. Our results establish that HiNF-P is the ultimate link in a linear signaling pathway that is initiated with the growth factor-dependent induction of cyclin E/CDK2 kinase activity at the restriction point and culminates in the activation of histone H4 genes through HiNF-P at the G1/S phase transition.
The G1/S phase transition represents the critical stage during the somatic cell cycle that defines cellular commitment to replicate the genome and to progress towards mitotic division. Passage beyond the G1/S boundary depends initially on the activation of a cyclin/cyclin-dependent kinase (CDK) cascade by growth factors and induction of the cyclin E/CDK2 kinase complex at the restriction (R) point (9, 14, 29). Subsequently, at the onset of S phase, de novo synthesis of histone proteins is required to package nascent DNA into chromatin immediately upon initiation of DNA synthesis (28, 35, 36). The exquisitely stringent coupling between histone biosynthesis and DNA replication necessitates the transcriptional activation of the 14 distinct human genes encoding histone H4, the most highly conserved nucleosomal protein (1, 13, 19, 28).
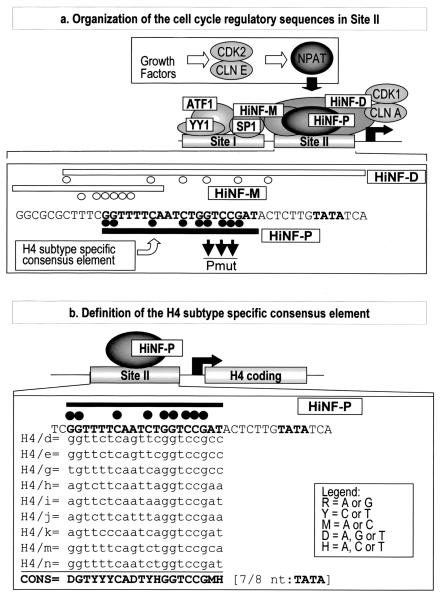
Control of histone genes provides a paradigm for gene expression that is temporally and functionally linked to DNA synthesis. It has long been postulated but not yet experimentally validated that the multiple histone H4 genes are coordinately regulated by a single histone H4 gene subtype-specific factor. However, the identity of this protein has never been established. We and others have previously demonstrated that histone H4 genes are regulated by multiple elements and cognate DNA binding activities (6, 7, 16, 30, 31, 44). The histone gene proximal promoter element site II interacts with three factors (HiNF-M, -D, and -P) and mediates cell cycle control of transcription at the onset of S phase (3, 6, 7, 16, 30, 31, 36, 43-47). Site II encompasses the H4 subtype-specific element that is phylogenetically conserved among multiple histone H4 genes in metazoan species. The cell cycle regulatory mechanisms operative at site II at the onset of S phase function independently of E2F (28, 31, 45). E2F factors control many genes, including those encoding enzymes involved in nucleotide metabolism (i.e., thymidine kinase, dihydrofolate reductase), at the R point late in G1 (9, 26). Thus, gene regulatory mechanisms controlling histone genes and E2F-dependent genes are temporally and functionally distinct.
To understand cell cycle regulation of histone H4 gene transcription within the broad context of key signaling pathways that control cell cycle progression, it is necessary to identify the rate-limiting factor that mediates histone H4 gene activation at the G1/S phase transition. Here, we have purified HiNF-P, the principal binding protein of the cell cycle regulatory element in the histone H4 gene promoter. We demonstrate that HiNF-P activates and is required for H4 gene transcription. Furthermore, we show that HiNF-P responds to the cyclin E/CDK2/NPAT signaling pathway (8, 18, 21, 51) and is necessary for efficient entry into S phase. Hence, we have defined a critical step in the cell cycle-dependent activation of histone gene expression that is stringently and functionally coupled to the onset of DNA replication.
MATERIALS AND METHODS
Biochemical purification of HiNF-P.
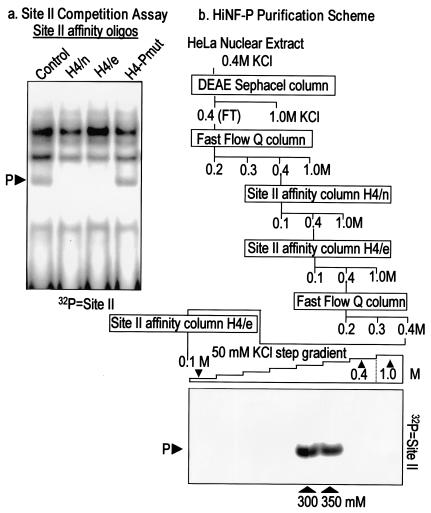
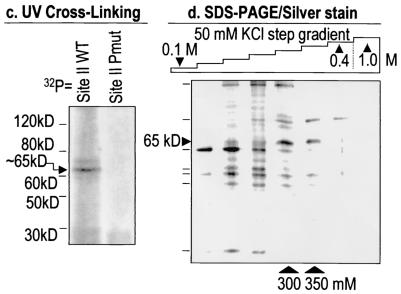
HeLa S3 cells were provided by the National Cell Culture Center. Nuclear extracts were prepared essentially as described previously (43, 44). Nuclear proteins were first passed through a DEAE Sephacel column primed with KN400 buffer (20 mM HEPES-NaOH [pH 7.5], 20% glycerol, 400 mM KCl, 0.2 mM EDTA, 1 mM EGTA [pH 8.0], and 0.01% NP-40). The flowthrough containing HiNF-P DNA binding activity was diluted with an equal volume of the same buffer without KCl (KN0) and loaded onto a Fast Q-Sepharose column. The column was washed with KN300, KN400, and KN1000. The KN400 fraction, which contains HiNF-P DNA binding activity, was mixed with three volumes of KN0 containing 6,400 μg of salmon sperm DNA, 1 mM dithiothreitol (DTT), 0.2 mM MgCl2, and 0.1 mM ZnCl2. The mixture was then passed through a DNA affinity column containing a multimerized oligonucleotide spanning site II in the H4/n gene and washed with KN400 and KN1000. The KN400 fraction was collected and diluted fourfold with KN0 containing 640 μg of salmon sperm DNA, 1 mM DTT, 0.2 mM MgCl2, and 0.1 mM ZnCl2 and loaded onto a DNA affinity column by using a multimerized oligonucleotide spanning the HiNF-P site in the H4/e gene. The H4/e site II column was washed with KN400 and KN1000, and the KN400 fraction containing HiNF-P was concentrated by passage through a Fast Q-Sepharose column (KN400 fraction collected). This fraction was reapplied to the H4/e site II affinity column in the presence of 10 μg of a site II oligonucleotide with a mutation in the HiNF-P binding site. Final fractions were eluted from the H4/e site II affinity resin with a 50 mM KCl step gradient, and all fractions were analyzed for HiNF-P DNA binding activity. Only KN300 and KN350 fractions were found to contain HiNF-P DNA binding activity. Proteins present in each of the fractions were salt exchanged analyzed on sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) gels followed by silver staining. A prominent 65-kDa band present only in the 300 and 350 mM KCl fractions was analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) (mass spectrometry) and peptide microsequencing by the University of Massachusetts Protein Microsequencing Core Facility. Peptide microsequencing indicated that the 65-kDa protein contains the amino acid sequence RYESVELTQQLLRQPQE (peptide 1).
Cloning and expression of recombinant HiNF-P.
Peptide 1 is contained within a 2.1-kb cDNA (GenBank accession no. BC017234) with an open reading frame that encodes a Zn finger protein with a predicted size of 517 amino acids. Specific primers (forward, TCA GGG ATC CGT CCG CCT CCT GGG AAA GTT CC; reverse, 5′-TCC CTC GAG CAA CCA TCT GGA TCT CTG GCT CCT C-3′) were used to amplify an ∼1.5-kb DNA segment spanning the entire open reading frame by reverse transcription-PCR with total HeLa mRNA. The reverse transcription-PCR product was cloned as a BamHI-XhoI fragment into pCDNA3.1/A (Invitrogen, Carlsbad, Calif.) for expression in eukaryotic cells. In vitro transcription and translation (IVTT) reactions were performed by incubating plasmid DNA (1.2 μg) with 25 μl of rabbit reticulocyte lysate, 30 U of RNasin, 1 μl of T7 RNA polymerase (Promega), and 1 μl of 1 mM l-methionine or 45 μCi of [35S]methionine (1,000 Ci/mmol; New England Nuclear) in a reaction mixture with a total volume of 60 μl at 30°C for 2 to 3 h. Synthesis was stopped by adding 20 μl of 80% glycerol, and an aliquot was analyzed by SDS-10% PAGE to confirm synthesis of the full-length protein.
Antibody preparation.
Antibodies were directed against peptide 1 (RYESVEKTQQLLRQPQE), which corresponds with the peptide identified by mass spectroscopy, and peptide 2 (CEKLQGIAEEPEIQMV), which is identical to the C terminus of the predicted HiNF-P protein based on the cDNA sequence (GenBank accession no. BC017234). Antisera for both peptides were generated in duplicate in rabbits (ResGen; Invitrogen Corporation, Huntsville, Ala.) and used as an antiserum, immunoglobulin G (IgG) fraction, or peptide affinity-purified fraction for Western blots, electrophoretic mobility shift assays (EMSAs), immunoprecipitations, and immunofluorescence microscopy.
EMSAs and UV cross-linking experiments.
EMSAs were performed exactly as described by van Wijnen et al. (43, 44). For competition experiments, 1 pmol of unlabeled double-stranded oligonucleotide was added per reaction. For immuno-EMSAs, antisera against peptide 1 or peptide 2 were added to the binding reaction mixture. As controls, the antisera were neutralized by incubation with excess amounts of the matching peptides before addition to the binding mixtures. UV cross-linking experiments were performed by adapting a standard protocol (2) with a radiolabeled and BrdU-substituted probe which was prepared by Klenow polymerase (New England Biolabs) of a partially double-stranded HiNF-P-specific oligonucleotide (top strand, 5′-CGC TTT CGG-3′; bottom strand, 5′-CAA GAG TAT CGG ACC AGA TTG AAA ACC GAA AGC G) as described previously (41). A Fast Flow Q fraction (7.5 μl, 0.4 M KCl) was incubated in a 50-μl reaction mixture containing 10 fmol of BrdU-substituted probe, 1 mM DTT, and 100 μM (each) Zn2+ and Mg2+. Cross-linking was carried out with an inverted UV transilluminator (305 nm). Following nuclease digestion, the products were analyzed by SDS-10% PAGE.
Transient transfection.
Transcription assays were performed with Saos-2 cells in a six-well plate with 100 ng of an H4 promoter-luciferase construct or the matching HiNF-P binding site mutant. In different experiments, cells were transiently transfected per well with expression constructs for HiNF-P (200 ng), NPAT (300 ng), cyclin E (500 ng), CDK2 (500 ng), and/or p57 (200 ng) by using FuGENE-6 (Roche Molecular Biochemical). The expression constructs for NPAT, NPATΔCDK2, cyclin E, CDK2, and p57 were described previously (21). After 24 h, cells were harvested and lysates were used for measurement of luciferase reporter activity and Western blot analysis.
Immunofluorescence microscopy.
Saos-2 cells grown on coverslips (Fisher Scientific, Springfield, N.J.) were extracted according to procedures documented previously (4, 37, 50) to obtain whole-cell and nuclear matrix intermediate filament (NMIF) preparations. Antibody staining was performed by incubating whole-cell and NMIF preparations with rabbit polyclonal antibodies against HiNF-P for 1 h at 37°C. The secondary antibody (Alexa 488 goat anti-rabbit; Molecular Probes, Eugene, Oreg.) was used at a 1:800 dilution. The cells were then stained with 4′,6-diamidino-2-phenylindole (DAPI) (5 μg/μl). Immunostaining of cell preparations was recorded by an epifluorescence microscope attached to a charge-coupled device camera, and the digital images were analyzed with the Metamorph software programs.
Genomic DNase I footprinting by LM-PCR.
Ligation-mediated PCR (LM-PCR) was carried out essentially by using the method of Mueller et al. (24) with some modifications. Initial extension and amplification/labeling was with Vent DNA polymerase (New England Biolabs). Linker ligation was carried out with 3 U of T4 DNA ligase (Promega). 5′ Primer labeling with [γ-32P]ATP was carried out by T4 polynucleotide kinase (New England Biolabs). The primers used were as follows: LM7 (nucleotides [nt] +10 to +35), 5′-GAC ATG ACC GCT GGA GCC CGA TA-3′; LM8 (nt −2 to +22), 5′-GCT GGA GCC CGA TAG ACA GCT TTC TG-3′; LM9 (nt −6 to +22), 5′-GCT GGA GCC CGA TAG ACA GCT TCT GTC A-3′. The initial extension reaction was initiated with 4 μg of DNA, but after linker ligation and precipitation, only half of this amount was used for the amplification step. Annealing temperatures used were 64°C (LM7), 66°C (LM8), and 68°C (LM9). Amplification was for 20 cycles. PCR products were labeled by primer extension with 32P-labeled nested primers for 2 cycles, extracted by phenol-chloroform, precipitated by ethanol, and analyzed on 6% denaturing polyacrylamide gels.
ChIP assay.
HL-60 human promyelocytic leukemia cells, HeLa S3 cervical carcinoma cells, Saos-2 osteosarcoma cells, and T98G glioblastoma cells were used in chromatin immunoprecipitation (ChIP) experiments, which were performed as described previously (17). Cells were incubated with 1% formaldehyde and subjected to sonication. Immune complexes were precipitated with protein A/G agarose beads. Immunocomplexes were eluted, the cross-linking reaction was reversed, and the DNA was purified by phenol-chloroform and by the DNA Clean&Concentrator purification kit (Zymo Research, Orange, Calif.). The following primer pairs were used for quantitative PCR to detect the presence of specific DNA fragments: 5′ region, F−219 (GAT CTG AAT TCT CCC GGG GAC TGT) and R+32 (GAC ATG ACC GCT GGA GCC CGA TA); 3′ region, F+644 (GAG CAC TGC TTT CTC GGC TTG CTC) and R+848 (AGG TGG GAA AGC CGG CAT CTC TAG).
Antisense inhibition experiments.
T98G cells were incubated for 4 h with each oligonucleotide (Oligo Etc., Inc.) at 400 nM with Lipofectin (Invitrogen) by following the manufacturer's instructions. The HiNF-P-related oligonucleotides used were 5′-GGG CAT TGG TCT GAT TCA CC-3′ (antisense), 5′-CCA CTT AGT CTG GTT ACG GG-3′ (reverse), and 5′-AGG CGT TGA TCT CAT TAA CC-3′ (scramble). T98G cells were assayed for the inhibition of H4 gene expression after 30 h. For cell proliferation experiments, T98G cells were treated for 4 h with antisense oligonucleotides, serum deprived for 72 h, and then released in fresh Dulbecco's minimal essential medium supplemented with 20% fetal bovine serum. Cell cycle distribution was monitored by fluorescence-activated cell sorting at different time points after release.
Northern and Western blot analyses.
Total RNA from T98G cells was prepared with Trizol reagent by following the manufacturer's instructions (GIBCO BRL). Total RNA (20 μg) was fractionated in a formaldehyde-containing 1.2% agarose gel, transferred to a Zeta-Probe GT membrane (Bio-Rad), and hybridized with DNA probes labeled by the random primer method (Redi Prime II random primer labeling system; Amersham). Whole-cell extracts were electrophoresed on an SDS-10% PAGE gel and blotted onto a polyvinylidene difluoride Immobilon-P membrane (Millipore). Immunodetection was performed with an appropriate dilution of specific antibodies and by using the Western Lightning chemiluminescence reagent plus (Perkin Elmer Life Sciences).
RESULTS
Identification of HiNF-P as a 65-kDa Zn finger protein that interacts with the histone H4 subtype-specific cell cycle element.
The cell cycle regulatory element of histone H4 genes (site II) contains a subtype-specific sequence that is highly conserved among the 14 H4 gene copies present in the human genome (Fig. 1). The conserved G residues of this sequence are protected in vivo based on genomic fingerprinting and are essential protein-DNA contacts for a DNA binding activity designated HiNF-P/H4TF-2 (5-7, 30, 43, 44). The chromatographic fractionation of H4TF-2 has previously been described, and this DNA binding activity was tentatively assigned to a 65-kDa protein band observed by UV cross-linking and in SDS-PAGE gels (7). However, this previous study did not provide genetic information (i.e., protein or DNA sequence) or immunological characterization (i.e., antibody). Therefore, it was necessary to identify an HiNF-P-specific peptide sequence, obtain a full-length cDNA, and generate protein-specific antibodies to assess the role of HiNF-P in histone H4 gene activation, involvement in cell cycle-related signaling pathways, and contribution to biological control of cell cycle progression.
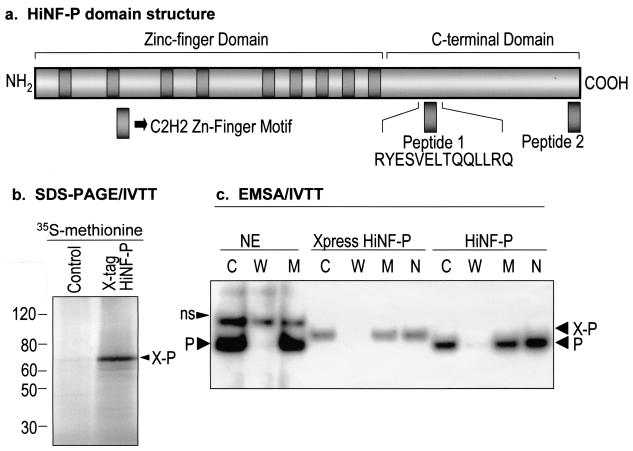
FIG. 1.
Site II-dependent cell cycle control of histone H4 gene transcription. (a) The diagram of the H4 promoter (middle) shows two genomic sites of protein-DNA interactions (site I and site II) and the cognate factors (36). The three site II proteins, HiNF-P, -D, and -M, recognize overlapping motifs (lower portion). Minimal element boundaries defined by DNase I footprinting and deletion analyses are indicated by the open or closed lines, and protein-DNA contacts established by dimethyl sulfate fingerprinting are depicted by closed or open circles. The growth factor-dependent CDK2/CLNE/NPAT pathway that functions to activate histone H4 gene transcription is also indicated (top). (b) The histone H4 subtype-specific consensus element is located upstream from the TATA box within site II and was defined based on nine representative functionally expressed H4 genes.
Here, we purified HiNF-P to homogeneity (Fig. 2) after DNA affinity chromatography. We used two distinct sequence-specific affinity resins that are based on known high affinity binding sites for HiNF-P in the H4/n and H4/e genes (Fig. 2a). Nuclear proteins were isolated from HeLa S3 cells and fractionated by anion-exchange chromatography followed by three cycles of sequence-specific DNA affinity chromatography with excess nonspecific DNA in the mobile phase (Fig. 2b). UV cross-linking of HiNF-P to a BrdU- and 32P-labeled probe spanning site II sequences reveals that HiNF-P is a 65-kDa protein (Fig. 2c). Consistent with this result, chromatographic fractions containing maximal HiNF-P activity in gel shift assays (Fig. 2b) also exhibit significant and reproducible enrichment of a 65-kDa protein in silver-stained SDS-PAGE gels (Fig. 2d).
FIG. 2.
Biochemical purification of HiNF-P. (a) Site II competition assay. We performed EMSA competitions to test binding of HiNF-P to two different oligonucleotides (H4n, 5′-GAT CTT CGG TTT TCA ATC TGG TCC GAT-3′, and H4/e, 5′-GAT CTC AGG TTC TCA GTT CGG TCC GCC-3′) that were used to make DNA affinity columns. HiNF-P in HeLa nuclear extracts binds to a 32P-labeled minimal binding sequence (control). HiNF-P binding is totally abolished when cold excess double-stranded oligo is present (lanes H4/n and H4/e), but binding is unaffected by the addition of excess HiNF-P mutant oligonucleotide (5′-GAT CTT CGG TTT TCA ATC TTC TAC GAT-3′) (lane H4-Pmut). (b) Chromatographic fractionation of HiNF-P. A diagram of the chromatographic separation procedure used in our studies is shown. Nuclear extracts from HeLa S3 cells were successively applied to the indicated chromatographic resins. Fractions from each column were analyzed by EMSA. HiNF-P activity was eluted with a 50 mM step gradient from the final site II affinity column (H4/e) and was detected by EMSA only in the 300 and 350 mM fractions (bottom right panel). (c) UV cross-linking of HiNF-P-site II complexes. Oligonucleotides spanning wild type (WT) or mutant (Pmut) Site II sequences were labeled with bromodeoxy-UTP and [32P]dCTP and incubated with a fraction (7.5 μl, 0.4 M KCl Fast Flow Q column fraction, step 2) enriched in HiNF-P binding activity. The reaction mixture was exposed to UV light (305 nm) for 60 min and then digested with DNase I and MNase. Cross-linked, labeled proteins were analyzed by SDS-10% PAGE and subjected to autoradiography. Molecular mass markers are shown at the left. The arrow indicates the 65-kDa HiNF-P-DNA complex. (d) SDS-PAGE-silver stain detection of HiNF-P. Aliquots (125 μl) from each fraction of the DNA affinity step gradient (see panel b) were mixed with an equal volume of 2× SDS-PAGE sample buffer, loaded onto SDS-10% PAGE gels, and stained with silver after electrophoresis. The two arrowheads at the bottom of the panel indicate the same 300 and 350 mM fractions exhibiting maximal HiNF-P binding activity in panel b. Both fractions contain a unique 65-kDa band (indicated by the horizontal arrowhead).
After DNA affinity chromatography, HiNF-P was purified to homogeneity by preparative SDS-PAGE and subjected to MALDI-TOF (mass spectrometry) and peptide microsequencing. A single 14-amino-acid sequence (peptide 1) (Fig. 3a) was reproducibly detected in three HiNF-P preparations of 2 × 1010 HeLa cells each. This peptide identifies a cDNA with a predicted open reading frame for a full-length protein with multiple Zn finger motifs diagnostic of sequence-specific DNA binding proteins (Fig. 3a). The protein translated from this cDNA in vitro migrates with the expected molecular mass based on SDS-PAGE (Fig. 3b). The recombinant protein with or without the epitope tag interacts specifically with Site II and exhibits oligonucleotide competition characteristics in EMSAs that are indistinguishable from endogenous HiNF-P isolated from HeLa cells (Fig. 3c and d). The DNA-bound protein without the epitope tag comigrates with the HeLa HiNF-P site II complex (Fig. 3c). Furthermore, expression of the recombinant protein in HeLa cells increases the amount of HiNF-P binding activity (Fig. 3d). Thus, we have purified a sequence-specific DNA binding protein with the biochemical properties of the histone H4 gene-specific regulatory factor HiNF-P.
FIG. 3.
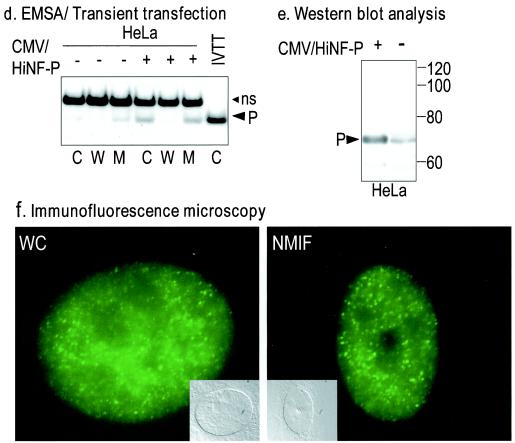
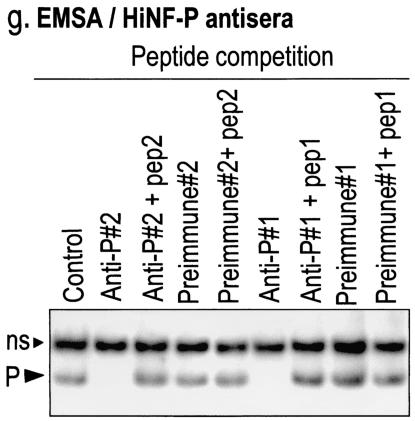
Molecular characterization of HiNF-P. (a) Schematic representation of the HiNF-P protein. The domain structure of HiNF-P is based on the conceptual translation of the open reading frame in the HiNF-P cDNA. HiNF-P contains at least nine conserved C2H2 Zn finger domains (gray shades). Peptide 1, shown below the diagram, was identified by MALDI-TOF (mass spectrometry) analysis of the purified HiNF-P protein. Both peptide 1 and peptide 2, spanning the 15 C-terminal residues, were used to raise antibodies in rabbits. (b) Analysis of recombinant HiNF-P protein synthesized by coupled IVTT. SDS-PAGE analysis of [35S]methionine-labeled proteins synthesized in a reticulocyte lysate programmed with a vector containing HiNF-P coding sequences fused to an N-terminal epitope tag (Xpress). A 5-μl aliquot of the programmed lysate or the corresponding control lysate was mixed with an equal volume of 2× SDS-PAGE sample buffer and subjected to SDS-10% PAGE. The autoradiogram reveals the synthesis of a radiolabeled recombinant protein of the expected size (indicated by the arrowhead). Molecular mass markers (in kilodaltons) are shown on the left. (c) EMSA with recombinant HiNF-P. Unlabeled recombinant HiNF-P with or without the Xpress tag was synthesized by IVTT and compared by EMSA to endogenous HiNF-P in HeLa nuclear extracts (NE; first 3 lanes). Samples were subjected to oligonucleotide competition (50 nM, 100-fold excess) with wild-type (W) or mutant (M) HiNF-P oligonucleotides or an SP1 oligonucleotide (N). The Xpress-tagged HiNF-P complex (X-P) migrates more slowly than the complexes formed with the untagged recombinant or endogenous HiNF-P (P). C, control; ns, nonspecific complex detected in nuclear extracts from HeLa cells. (d) Forced expression of HiNF-P increases HiNF-P binding activity. An EMSA was performed with nuclear extract from HeLa cells transiently transfected with HiNF-P cDNA (cytomegalovirus-HiNF-P). After 36 h, cells were harvested and nuclear proteins (10 μg) from cells transfected with expression vector containing HiNF-P (+) or vector alone (−) were used for EMSA. Expression of HiNF-P elevates HiNF-P binding. See the legend to panel c for abbreviations used on the figure. (e) HiNF-P antipeptide antibody recognizes a 65-kDa protein. HeLa cells were transfected with cytomegalovirus-HiNF-P (5 μg) or vector alone. After 36 h, nuclear extracts were prepared and 20 μg of protein was separated by SDS-10% PAGE. Western blot analysis was performed with the affinity-purified antibody directed against peptide 2 (anti-802k, 1:1,000 dilution). Both recombinant (+) and endogenous (−) HiNF-P are detected as 65-kDa proteins on the Western blot (indicated by arrow). (f) Subcellular localization of HiNF-P. Human osteosarcoma Saos-2 cells (0.4 × 106 cells/well) were grown on gelatin-coated coverslips and analyzed by in situ immunofluorescence microscopy as whole-cell (WC) and NMIF preparations. HiNF-P was detected by using the IgG-purified HiNF-P antibody (anti-802k) and a polyclonal Alexa 488 goat anti-rabbit antibody conjugated with fluorescein isothiocyanate fluorochrome. The fluorescent signals were visualized with a Zeiss Axioplan microscope equipped with a charge-coupled device camera, and MetaMorph imaging software was used for image acquisition. (g) Immunoreactivity of HiNF-P-site II complexes. Antibodies directed against HiNF-P peptide 1 (Anti-P#1) and peptide 2 (Anti-P#2) and the corresponding preimmune sera were incubated with endogenous HiNF-P from HeLa nuclear extracts complexed with a site II oligonucleotide (P). HiNF-P binding activity is blocked by both antisera unless the matching peptides (pep1 and pep2, respectively) are present. See the legend to panel c for abbreviations used on the figure.
To establish unequivocally that the recombinant protein is identical to HiNF-P, we prepared antibodies against two different peptides. We directed one antibody against the segment of the protein spanning peptide 1, which was identified by microsequencing of HiNF-P. The second antibody was raised against the predicted C-terminal sequence (peptide 2) of the protein (Fig. 3a). Western blot analysis reveals that the antibodies detect both endogenous and recombinant HiNF-P as proteins with the same molecular mass (Fig. 3e and data not shown). Immunofluorescence microscopy reveals that both the recombinant and endogenous proteins are localized in the nucleus and are stably associated with distinct subnuclear foci even upon removal of chromatin (Fig. 3f and data not shown). Most importantly, we find that both antisera disrupt formation of the endogenous HiNF-P complex in gel shift assays, but preincubation of the antisera with the corresponding antigenic peptide prevents this disruption (Fig. 3g). Because the two antibodies directed against the recombinant protein also recognize the H4 subtype-specific binding activity of HiNF-P, we conclude that they are indeed the same.
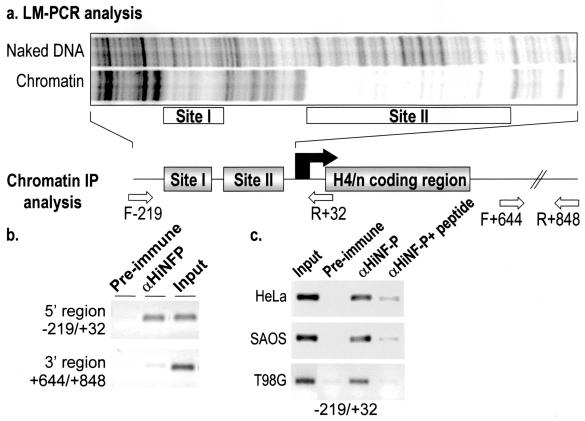
To assess directly whether HiNF-P interacts with the site II region of the H4 locus in vivo, we performed ChIP assays and genomic footprinting with LM-PCR (Fig. 4). Chromatin spanning the H4 promoter contains the expected two regions of DNase I protection that represent protein-DNA interaction domains, which we refer to as sites I and II (Fig. 4a), with site II including the HiNF-P interaction site (Fig. 1). Chromatin immunoprecipitates obtained with the HiNF-P antibody are enriched for a DNA segment that encompasses site II of the H4 promoter but not for a DNA segment derived from the 3′ noncoding region of the H4 gene that lacks an HiNF-P binding site (Fig. 4b). The in vivo interaction of HiNF-P with the histone H4 promoter is observed in all proliferating cell types examined (Fig. 4b and c). As essential controls, we show that this interaction is not observed with the preimmune serum or when the antigenic peptide is preincubated with the HiNF-P antiserum (Fig. 4b and c). We conclude that we have purified the bona fide protein, HiNF-P, which interacts in vivo with the site II cell cycle regulatory sequences of the histone H4 gene.
FIG. 4.
Genomic occupancy of HiNF-P at the site II cell cycle element. (a) Genomic occupancy of the HiNF-P binding element within Site II was established in proliferating HL-60 cells by LM-PCR-assisted DNase I footprinting with primers that amplify the proximal promoter of the H4/n gene. The locations and sizes of site I and II are similar to those previously established in HeLa cells by genomic Southern blotting (30). IP, immunoprecipitation. (b) The interaction of HiNF-P with the H4/n promoter in vivo was determined by ChIP with an HiNF-P antibody (αHiNFP). Genomic segments derived from the H4 locus in chromatin precipitates from HL-60 cells were detected by using PCR primers spanning 5′ (−219 to +32) or 3′ (+644 to +848) regions. Amplified products from the 5′ region are observed in input chromatin and in the precipitate with the HiNF-P antibody but not in precipitates obtained with preimmune serum. The 3′ primer pair amplifies DNA only from input samples and serves as a negative control. (c) ChIP was performed with preimmune serum (IgG), HiNF-P antiserum (αHiNF-P), and HiNF-P antiserum preincubated with the antigenic peptide (αHiNF-P + peptide) by using chromatin (input) isolated from different cell types (HeLa, Saos-2, and T98G). The presence of the H4 promoter in chromatin immunoprecipitates was detected with the F−219-R+32 PCR primer pair (see panel a for diagram).
HiNF-P regulates endogenous histone H4 gene expression.
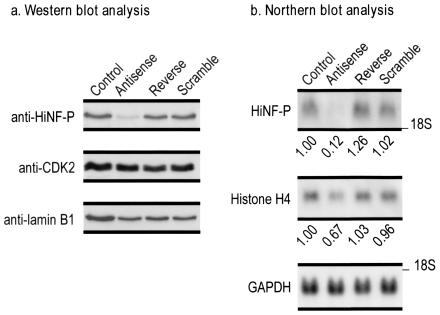
To establish that HiNF-P is rate limiting for endogenous H4 gene expression, we performed antisense inhibition experiments. Several HiNF-P antisense oligonucleotides with the corresponding negative controls (i.e., scramble and reverse) were tested for their ability to inhibit HiNF-P mRNA levels. Cells treated with the most efficacious HiNF-P antisense oligonucleotide (directed against a segment in the 3′ end of the HiNF-P mRNA) exhibit a dramatic reduction in HiNF-P mRNA and protein levels, whereas the matching scrambled and reverse oligonucleotides do not influence HiNF-P levels (Fig. 5). Significantly, treatment with the antisense oligonucleotide reduces endogenous histone H4 mRNA levels in actively proliferating cells (Fig. 5). The modest magnitude of the effect (i.e., 33% reduction of H4 mRNA) may be due in part to post-transcriptional compensatory mechanisms. The antisense-mediated reduction of H4 mRNAs indicates that HiNF-P functions to control histone H4 gene expression in vivo.
FIG. 5.
Antisense inhibition of HiNF-P reduces endogenous histone H4 mRNA levels. T98G cells treated with antisense, scrambled, or reverse oligonucleotides or mock treated (control) for 30 h were analyzed for protein (a) and RNA (b). Western (a) and Northern (b) blot analyses both reveal that HiNF-P levels are downregulated by the antisense oligonucleotide. The CDK2 kinase and the nuclear structural protein lamin B1 are shown as controls for protein loading. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is used as an internal control for changes in mRNA levels. The quantitation of H4 mRNA levels is shown below the lanes. The reduction of histone H4 mRNA upon HiNF-P antisense inhibition is clearly evidenced, even though reduced histone H4 gene transcription can be mitigated in part by posttranscriptional compensatory mechanisms (e.g., 3′ end processing and mRNA stability) (28, 36).
HiNF-P activates transcription of a DNA replication-dependent histone H4 gene.
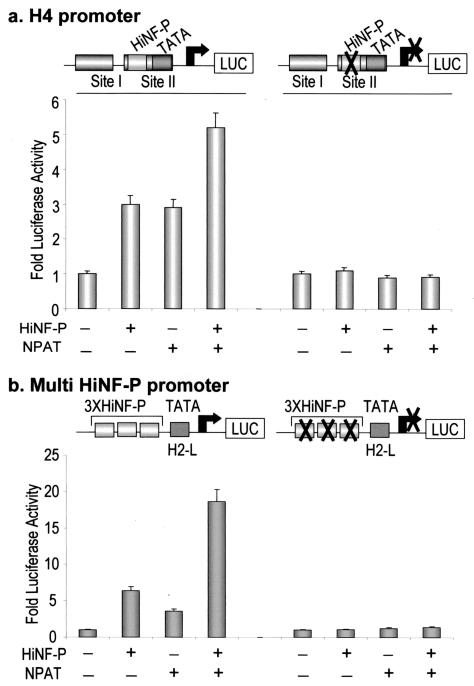
We directly addressed the functional contribution of HiNF-P to transcription of the DNA replication-dependent histone H4 gene family. The native H4/n promoter interacts with multiple transcription factors and contains a single HiNF-P binding site that corresponds to the H4 subtype-specific consensus element (Fig. 1a). Forced expression of HiNF-P results in a reproducible threefold enhancement of histone H4 gene promoter activity as measured by the luciferase (luc) reporter (Fig. 6a). This enhancement is not observed when the HiNF-P binding site in the promoter is mutated. Consistent with previous data from our laboratory (31), mutation of the HiNF-P binding site has only limited effects on basal H4 promoter activity, because other transcription factors appear to compensate. The mutant promoter mediates a basal transcription level that is no more than two- to threefold lower than the wild-type promoter and easily detectable above the background levels. We also tested the function of HiNF-P on a chimeric reporter gene containing three tandem copies of the conserved HiNF-P binding site fused to a minimal TATA box-containing promoter. In this promoter context, HiNF-P mediates a six- to sevenfold activation of transcription (Fig. 6b). The corresponding construct in which the HiNF-P sites are mutated is not responsive to forced expression of HiNF-P. Thus, HiNF-P is a potent activator of histone H4 gene transcription, which is known to be cell cycle regulated at the G1/S phase transition, and the H4 subtype-specific consensus sequence represents an autonomous element that can mediate HiNF-P-dependent transcriptional activation.
FIG. 6.
HiNF-P and NPAT-dependent activation of histone H4 gene transcription requires an intact HiNF-P binding element. Saos-2 cells were transfected with wild-type H4 promoter luciferase reporter constructs or the corresponding constructs containing mutated HiNF-P binding sites together with HiNF-P and/or NPAT expression vectors (diagrams are shown at the top of each panel). (a) Histone H4/n promoter; (b) promoter with multimerized HiNF-P elements fused to a minimal TATA box. Relative promoter activity (measured as luciferase activity) is shown as a function of the presence (+) or absence (−) of cytomegalovirus-driven HiNF-P (200 ng/well) or NPAT (300 ng/well) expression vectors. Expression of either HiNF-P or NPAT alone results in activation of histone H4 transcription from the wild-type (left) but not the HiNF-P mutant (right) promoters. However, coexpression of HiNF-P and NPAT elevates wild-type promoter activity above that of either factor alone.
NPAT is an HiNF-P-dependent transcriptional coactivator of cell cycle-controlled H4 genes.
A fundamental question is whether HiNF-P is the missing functional link between the NPAT/cyclin E/CDK2 signaling pathway and transcriptional control of histone H4 genes at the G1/S phase cell cycle transition. NPAT, a substrate of the cyclinE/CDK2 kinase, enhances histone gene transcription (21, 51). However, NPAT does not directly bind DNA, and the downstream effector through which NPAT regulates histone H4 genes has not been characterized. Transient transfection assays show that elevating NPAT levels enhance the activity of the wild type but not the activity of the HiNF-P mutant H4 promoter (Fig. 6a and b). These results clearly establish that NPAT coactivation requires recognition nucleotides to which HiNF-P binds. In addition, the remaining site II DNA binding proteins (i.e., HiNF-M/IRF2 and the HiNF-D DNA binding subunit CDP-cut) are not sufficient to support the function of NPAT on the H4 promoter. Furthermore, the multimerized HiNF-P binding site is sufficient for NPAT enhancement of a minimal heterologous promoter (Fig. 6b).
To address whether NPAT and HiNF-P cooperate to regulate H4 promoter activity, we expressed both proteins and monitored reporter gene expression. The results show that coexpression of NPAT and HiNF-P increases transcription more than expression of either factor alone (Fig. 6). The observed increase in transcription when both HiNF-P and NPAT are elevated suggests that these factors are rate-limiting components of the same linear pathway. The relationship between activation by HiNF-P and NPAT has been tested at a range of concentrations, and the extent of HiNF-P/NPAT activation is typical for the standard amounts we used in this study. In more recent experiments, in which we used lower concentrations of both NPAT and HiNF-P expression vectors, we have observed a greater level of coactivation (i.e., up to 10-fold), which is suggestive of a synergistic interaction (data not shown). Importantly, coexpression of NPAT and HiNF-P has no effect on transcription when the HiNF-P binding site is mutated in either the native H4 promoter or the heterologous promoter with the multimerized HiNF-P binding site (Fig. 6). Taken together, our results demonstrate that NPAT operates via HiNF-P and functions as a coactivator of H4 gene transcription.
Cyclin E/CDK2 signaling increases the activation potential of HiNF-P and NPAT.
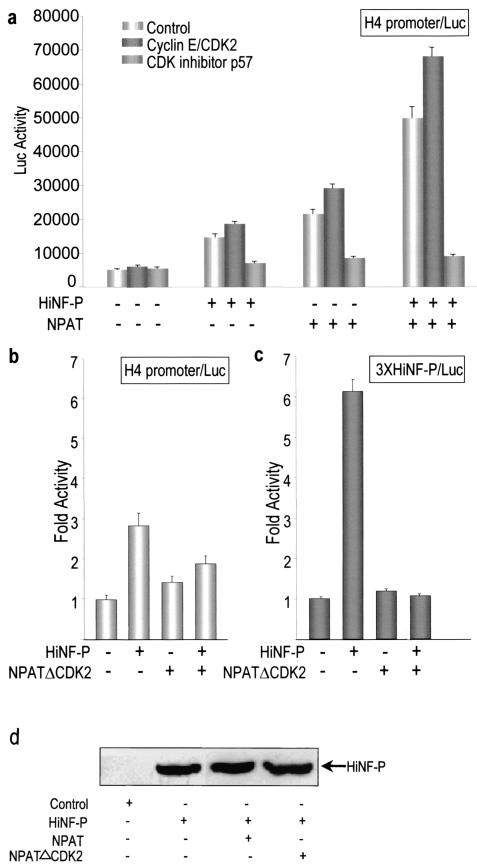
Because NPAT is phosphorylated by cyclin E/CDK2 complexes, this event may represent a critical step that links the growth factor-dependent induction of cyclin E/CDK2 complexes with the NPAT- and HiNF-P-mediated coactivation of histone H4 gene transcription. To test this hypothesis directly, we analyzed NPAT- and HiNF-P-related transcriptional effects upon elevating cyclin E/CDK2 levels. In addition, we assessed the effect of inhibiting the activity of cyclin E/CDK2 kinase complexes by expressing the CDK inhibitor p57 (Fig. 7a). Forced expression of cyclin E and CDK2 consistently results in an additional 10 to 30% increase in H4 promoter activity above levels established by HiNF-P and/or NPAT (Fig. 7a). This modest but reproducible additional increase by exogenously supplemented cyclin E/CDK2 complexes suggests that only a relatively minor fraction of HiNF-P and/or NPAT is not yet activated by endogenous cyclin E/CDK2 complexes. In agreement with the contribution of both endogenous and exogenous cyclin E/CDK2 complexes to HiNF-P activation, the cyclin E/CDK2 inhibitor p57 quantitatively blocks HiNF-P-mediated H4 transcription (Fig. 7a). These data suggest that cyclin E/CDK2 signaling supports HiNF-P and NPAT coactivation of H4 gene transcription. The effect of p57 may be direct, by interfering with NPAT phosphorylation, or indirect, by influencing cell cycle progression. However, forced expression of p57 does not influence H4 gene transcription when the HiNF-P binding site is mutated (data not shown), indicating that cyclin E/CDK2 signaling acts through the HiNF-P recognition motif.
FIG. 7.
Cyclin E/CDK2 signaling modulates the HiNF-P- and NPAT-mediated activation of H4 gene transcription. (a) Saos-2 cells were transfected with the wild-type H4 promoter-luciferase reporter gene construct as well as HiNF-P (200 ng/well) and/or NPAT (300 ng/well) expression constructs in the absence (control) or presence of expression vectors for cyclinE/CDK2 (500 ng each/well) or the CDK2 inhibitor p57 (200 ng/well). The graphs show relative luciferase activity upon expression of HiNF-P and/or NPAT as indicated by plus (present) and minus (absent) signs. H4 promoter activity, as measured by luciferase levels, is consistently increased upon elevating cyclin E/CDK2 levels and dramatically inhibited in the presence of p57. (b and c) Saos-2 cells were transfected with either the wild-type H4 promoter-luciferase construct (b) or the promoter with multimerized HiNF-P elements (c). Transfections were performed in the absence (−) or presence (+) of vectors expressing HiNF-P (200 ng/well) and/or a mutant NPAT protein (NPATΔCDK2) (300 ng/well) with mutations that abolish cyclinE/CDK2 phosphorylation sites and are known to affect NPAT function (21). Data are presented as activation (n-fold) relative to control transfections with reporter gene constructs alone (arbitrarily set at a value of 1). The data show that forced expression of NPATΔCDK2 does not enhance promoter activity and suppresses HiNF-P-dependent transactivation. (d) Levels of HiNF-P were measured by Western blot analysis in cells transfected with expression vectors for HiNF-P (200 ng/well), NPAT (300 ng/well), and/or NPATΔCDK2 (300 ng/well). +, present; −, absent.
To assess whether cyclin E/CDK2 phosphorylation of NPAT is required for activation of H4 gene transcription, we monitored reporter gene expression in the presence of a mutant NPAT protein (NPATΔCDK2) in which all known CDK2 phosphorylation sites are eliminated (i.e., Ala substitutions at S775, S779, S1100, T1270, and T1350) (21). The results show that the NPATΔCDK2 mutant does not support transcriptional enhancement of the natural H4 gene promoter (Fig. 7b) or the chimeric promoter with multimerized HiNF-P elements (Fig. 7c). Furthermore, NPATΔCDK2 inhibits the HiNF-P-dependent enhancement of H4 gene transcription (Fig. 7b and c). However, NPATΔCDK2 does not alter the levels of HiNF-P (Fig. 7d), indicating that NPATΔCDK2 acts by perturbing HiNF-P activity. These findings establish that, first, CDK2 phosphorylation sites of NPAT are essential for the HiNF-P-dependent regulation of H4 gene transcription and, second, the NPATΔCDK2 mutant is a functional inhibitor of HiNF-P activity. Taken together, our results provide strong evidence that HiNF-P is a critical component for activation of histone H4 gene transcription and is functionally linked to the cyclin E/CDK2/NPAT signaling pathway.
Inhibition of HiNF-P delays cell cycle progression into S phase.
We examined the biological coupling of HiNF-P, histone H4 gene expression and cell proliferation. The DNA binding activity of HiNF-P is detected in proliferating HL-60 cells that actively transcribe H4 mRNAs but is below the level of detection in postproliferative, differentiated HL-60 cells that have ceased histone H4 gene expression (data not shown). Western blot analysis with an HiNF-P-specific antibody shows that protein levels are also regulated with respect to the proliferative status of HL-60 cells (data not shown). Thus, HiNF-P DNA binding activity and protein levels are maximal in proliferating cells which express histone H4 genes during S phase.
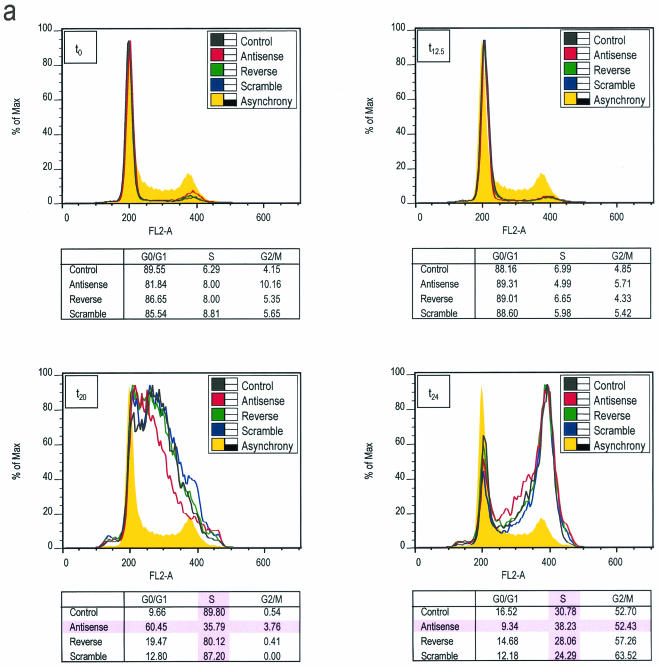
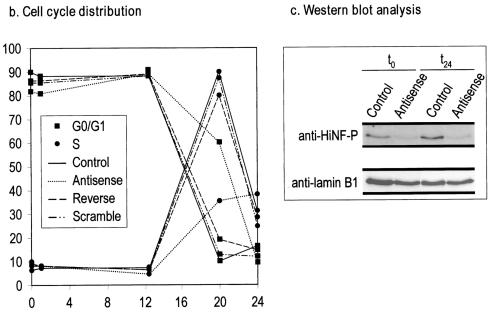
We directly assessed whether the G1/S phase-related gene regulatory role of HiNF-P reflects broader participation in cell growth control and whether HiNF-P has a rate-limiting function in cell cycle progression. We generated HiNF-P-deficient quiescent cells by treatment with antisense oligonucleotides (Fig. 8c) and monitored competency for progression into S phase following serum stimulation of proliferation. Fluorescence-activated cell sorting reveals that cells treated with the HiNF-P antisense oligonucleotide exhibit a significant delay in S phase entry relative to control cells (mock treated) or cells treated with scrambled or reverse oligonucleotides (Fig. 8a and b). Compromised cell cycle progression is directly reflected by a dramatic decrease in the percentage of antisense-treated cells in S phase at 20 h after serum stimulation (Fig. 8) and a commensurate increase in cells remaining in G1 phase. Similarly, at 24 h after stimulation, more antisense-treated cells remain in S phase (Fig. 8). The observation that antisense-treated cells progress into G2 indicates that HiNF-P deficiency does not cause an absolute cell cycle block. More importantly, our findings indicate that HiNF-P is among a limited number of transcription factors that, independent of E2F, can influence the G1/S phase cell cycle transition.
FIG. 8.
HiNF-P inhibition delays progression into S phase. (a) Analysis of cell cycle distribution by fluorescence-activated cell sorting. Fluorescence-activated cell sorting data were obtained for cells released into the cell cycle following serum stimulation. The graphs show the distribution of cells as a function of DNA content, and the tables provide a mathematical interpretation of the percentage of cells in specific cell cycle stages. (b) Graphical representation of cell cycle distribution. The fraction of cells in the G0/G1 and S phase stages as a function of time after serum addition was plotted to show an HiNF-P antisense-dependent delay in S phase progression compared to control cells. The data show that control cells reach the G1/S phase transition after 12 h. HiNF-P deficiency induced by antisense treatment decreases the S phase population with a commensurate increase in cells remaining in the G1 phase. (c) Antisense inhibition of HiNF-P. Western blot analysis of HiNF-P protein levels was performed with serum-deprived T98G cells (t0) and cells harvested 24 h after serum stimulation (t24). Lamin B1 was used as control for protein loading. Cells were treated with or without HiNF-P antisense oligonucleotide.
DISCUSSION
In this study, we have purified the histone H4 gene-specific protein HiNF-P and thereby established that HiNF-P is the previously elusive subtype-specific factor that coordinately regulates transcription of the H4 multigene family. We find that HiNF-P is a 65-kDa nuclear DNA binding protein with multiple Zn finger motifs. The molecular mass of HiNF-P is in agreement with previous UV cross-linking results obtained for a zinc-dependent 65-kDa binding activity designated H4TF-2 which was not purified to homogeneity but is most likely identical to HiNF-P (5-7, 43, 44). HiNF-P is required for H4 gene expression and is the bona fide protein that binds to the site II cell cycle sequences in vivo to regulate endogenous histone H4 genes.
Our data show that HiNF-P-dependent activation of histone H4 gene transcription is enhanced by the NPAT protein (nuclear protein mapped to the ATM locus, also known as p220) (18). Recently, it was shown that NPAT/p220, which is essential for normal mammalian development, is a direct downstream target of the cyclin E/CDK2 signaling pathway and enhances histone gene transcription (8, 21, 51). However, NPAT does not bind directly to DNA, thus invoking a requirement for a sequence-specific transcription factor that can transduce the NPAT-mediated signal in a histone gene-selective manner. We find here that HiNF-P binding to the H4 site II cell cycle element is required for NPAT-dependent enhancement of H4 gene transcription. In addition, HiNF-P and NPAT levels are each rate limiting for H4 gene transcription. This coactivation by HiNF-P and NPAT is enhanced by cyclin E/CDK2, inhibited by the CDK inhibitor p57, and critically depends on CDK2 phosphorylation sites in NPAT. Furthermore, a mutant NPAT protein lacking CDK2 phosphorylation sites can abolish HiNF-P activation of H4 gene transcription. This finding characterizes the CDK2 phosphorylation-deficient NPAT mutant as a dominant-negative inhibitor of HiNF-P function, placing both proteins in the same H4-related signaling pathway. Taken together, several lines of evidence establish that HiNF-P is the ultimate molecular link that couples growth factor stimulation and the cyclin/CDK signaling cascade with NPAT-dependent activation of H4 gene transcription at the G1/S phase transition.
The results we presented show that HiNF-P and NPAT together enhance H4 gene transcription, and this activation event is channeled through the same recognition site (i.e., for HiNF-P) in the H4 gene promoter. Furthermore, ChIP data indicate that both HiNF-P (this study) and NPAT (reference 21 and unpublished data) can interact with a representative endogenous H4 gene. Based on these results, we propose that NPAT functions as a coactivator of HiNF-P and that these proteins may act in close proximity of each other. Results from yeast two-hybrid assays, immunoprecipitations, and ChIP assays support the concept that both proteins form a complex (unpublished data). It remains to be established whether the HiNF-P and NPAT proteins are in direct contact and/or associate indirectly within a larger promoter-bound complex together with other gene regulatory factors (e.g., enhanceosome).
We also note that both HiNF-P and NPAT are tightly associated with nuclear architecture. In situ immunofluorescence microscopy data presented here indicate that endogenous HiNF-P is localized in multiple dispersed subnuclear foci while NPAT/p220 is concentrated in Cajal bodies (21). The difference in the subnuclear distribution of NPAT and HiNF-P suggests limited intersection of the functions of the two proteins. Thus, while one of the major findings of the present study is the definition of the HiNF-P/H4-site II interaction as the genomic endpoint of the cyclin E CDK2/NPAT signaling pathway, future studies should focus on the precise molecular mechanisms that mediate functional linkage between NPAT and HiNF-P in situ at the H4 promoter.
The H4 subtype-specific factor HiNF-P is known to interact with multiple histone H4 genes and recognizes conserved nucleotides of the histone H4 gene-specific consensus element (5, 7, 30, 40, 44). Significantly, all guanine residues in this sequence are known protein-DNA contacts for HiNF-P based on in vitro methylation interference and in vivo genomic fingerprinting (30, 44). Subtype-specific elements have also been described for the histone H1 and H2B genes (15, 49). H1-specific elements and analogous elements in the H2A and H3 genes are recognized by two distinct classes of CCAAT box binding proteins (i.e., NF-Y/HiNF-B/H1TF-2 and HiNF-D/CDP-cut) (10, 22, 23, 25, 39, 42), and the H2B-specific element is recognized by octamer transcription factor 1 (OTF1/OCT1/NFIII) (12). The identification of HiNF-P completes the list of the sequence-specific transcription factors that control histone gene transcription through classical, phylogenetically conserved subtype-specific regulatory elements.
Database analysis reveals that HiNF-P is identical to MIZF, a recently discovered protein that interacts with the methyl-CpG-binding protein MBD2, a subunit of the MeCP1 histone deacetylase (27, 32). Consistent with results from this and previous studies on the DNA binding activity of HiNF-P (33, 38, 43, 44), MIZF mRNA is ubiquitously present in many cell types and encodes a nuclear protein. Although MIZF and MBD2 are capable of interacting in yeast two-hybrid assays, epitope-tagged MIZF in transfected mammalian cells exhibits only limited colocalization with MBD2 in situ (32). The histone H4 gene promoter is not known to be methylated at CpG dinucleotides. Furthermore, CpG methylation of the highly conserved 5′-GGTCCG core motif within the H4 subtype-specific consensus sequence inhibits HiNF-P binding (44). Thus, it is not yet obvious whether the interaction of HiNF-P/MIZF with MBD2 is a biological component of the mechanism controlling H4 gene transcription. The previous studies used GAL4/MIZF fusion proteins and synthetic promoters to show that MIZF is a transcriptional repressor which is recruited by MBD2 to support histone deacetylation (32). Our data demonstrate that HiNF-P functions as a transcriptional activator and use NPAT as a coactivator to control the native cell cycle-regulated histone H4 gene promoter. It is possible that HiNF-P may be bifunctional and may support activation or repression depending on associated cofactors.
Biological characterization of all three principal factors, HiNF-P, -M and -D, which interact with the site II cell cycle element in histone H4 genes, has provided insight into the physiological role of these factors in E2F-independent mechanisms mediating cell growth control. Deregulated expression of HiNF-M/IRF-2 causes cell cycle defects resulting in polyploidy and apoptosis (48). Genetic inactivation of CDP-cut, the DNA binding subunit of the HiNF-D complex, causes several developmental abnormalities (e.g., in cells of the skin and the immune system) that are attributable to in vivo defects in cell growth and differentiation (11, 20, 34). We show here that antisense inhibition of HiNF-P activity impedes progression beyond the G1/S phase transition following induction of quiescent cells to enter the cell cycle. Hence, all three site II binding proteins have distinct functional roles in cell growth control.
Eukaryotic cells have developed complex mechanisms to mediate the E2F-dependent regulation of genes involved in nucleotide metabolism (e.g., thymidine kinase and dihydrofolate reductase) at the growth factor-related R point in anticipation of DNA replication. The functions of individual E2F transcription factors are partially redundant, and these factors promote either proliferation or exit from the cell cycle, depending on the biological context (26). The E2F-independent activation of DNA replication-dependent histone H4 genes at the G1/S phase transition, temporally and functionally downstream of the R point, appears to involve the intricately choreographed functions of the principal site II binding activities. Our data establish that HiNF-P, through its interaction with the highly conserved H4 subtype-specific element, coordinates transcription of the histone H4 multigene family by transducing signals from the growth factor-responsive NPAT/cyclin E/CDK2 signaling pathway. In the broader biological context of cell cycle control, we have defined a critical component of a regulatory cascade that is initiated by cyclin E/CDK2 kinase at the R point late in G1 and mediates activation of histone genes at the onset of S phase.
FIG. 8—Continued.
Acknowledgments
We thank John Leszyk (University of Massachusetts Protein Microsequencing Facility) for expert advice and assistance with MALDI-TOF analysis and peptide sequencing. We thank the National Cell Culture Center for providing HeLa cell cultures. We also thank Martin Montecino, Corey Braastad, Mai Luong, Caroline van der Meijden, Angela Miele, and Patricia Sue Vaughan for stimulating discussions and/or experimental assistance.
This study was supported by National Institutes of Health grant GM32010.
R.-L.X. and R.M. contributed equally to this work.
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Albig, W., and D. Doenecke. 1997. The human histone gene cluster at the D6S105 locus. Hum. Genet. 101:284-294. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. Greene and Wiley, New York, N.Y.
- 3.Aziz, F., A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1998. HiNF-D (CDP-cut/CDC2/cyclin A/pRB-complex) influences the timing of IRF-2 dependent cell cycle activation of human histone H4 gene transcription at the G1/S phase transition. J. Cell. Physiol. 177:453-464. [DOI] [PubMed] [Google Scholar]
- 4.Choi, J.-Y., A. J. van Wijnen, F. Aslam, J. D. Leszyk, J. L. Stein, G. S. Stein, J. B. Lian, and S. Penman. 1998. Developmental association of the β-galactoside-binding protein galectin-1 with the nuclear matrix of rat calvarial osteoblasts. J. Cell Sci. 111:3035-3043. [DOI] [PubMed] [Google Scholar]
- 5.Dailey, L., S. M. Hanly, R. G. Roeder, and N. Heintz. 1986. Distinct transcription factors bind specifically to two regions of the human histone H4 promoter. Proc. Natl. Acad. Sci. USA 83:7241-7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dailey, L., S. B. Roberts, and N. Heintz. 1987. RNA polymerase II transcription factors H4TF-1 and H4TF-2 require metal to bind specific DNA sequences. Mol. Cell. Biol. 7:4582-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dailey, L., S. B. Roberts, and N. Heintz. 1988. Purification of the human histone H4 gene-specific transcription factors H4TF-1 and H4TF-2. Genes Dev. 2:1700-1712. [DOI] [PubMed] [Google Scholar]
- 8.Di Fruscio, M., H. Weiher, B. C. Vanderhyden, T. Imai, T. Shiomi, T. A. Hori, R. Jaenisch, and D. A. Gray. 1997. Proviral inactivation of the Npat gene of Mpv 20 mice results in early embryonic arrest. Mol. Cell. Biol. 17:4080-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dou, Q. P., A. H. Levin, S. Zhao, and A. B. Pardee. 1993. Cyclin E and cyclin A as candidates for the restriction point protein. Cancer Res. 53:1493-1497. [PubMed] [Google Scholar]
- 10.El-Hodiri, H. M., and M. Perry. 1995. Interaction of the CCAAT displacement protein with shared regulatory elements required for transcription of paired histone genes. Mol. Cell. Biol. 15:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis, T., L. Gambardella, M. Horcher, S. Tschanz, J. Capol, P. Bertram, W. Jochum, Y. Barrandon, and M. Busslinger. 2001. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 15:2307-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher, C., N. Heintz, and R. G. Roeder. 1987. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell 51:773-781. [DOI] [PubMed] [Google Scholar]
- 13.Green, L., R. Van Antwerpen, J. Stein, G. Stein, P. Tripputi, B. Emanuel, J. Selden, and C. Croce. 1984. A major human histone gene cluster on the long arm of chromosome 1. Science 226:838-840. [DOI] [PubMed] [Google Scholar]
- 14.Harper, J. W., and P. D. Adams. 2001. Cyclin-dependent kinases. Chem. Rev. 101:2511-2526. [DOI] [PubMed] [Google Scholar]
- 15.Harvey, R. P., A. J. Robins, and J. R. Wells. 1982. Independently evolving chicken histone H2B genes: identification of a ubiquitous H2B-specific 5′ element. Nucleic Acids Res. 10:7851-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holthuis, J., T. A. Owen, A. J. van Wijnen, K. L. Wright, A. Ramsey-Ewing, M. B. Kennedy, R. Carter, S. C. Cosenza, K. J. Soprano, J. B. Lian, et al. 1990. Tumor cells exhibit deregulation of the cell cycle histone gene promoter factor HiNF-D. Science 247:1454-1457. [DOI] [PubMed] [Google Scholar]
- 17.Hovhannisyan, H., B. Cho, P. Mitra, M. Montecino, G. S. Stein, A. J. van Wijnen, and J. L. Stein. 2003. Maintenance of open chromatin and selective genomic occupancy at the cell-cycle-regulated histone H4 promoter during differentiation of HL-60 promyelocytic leukemia cells. Mol. Cell. Biol. 23:1460-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imai, T., T. Sugawara, A. Nishiyama, R. Shimada, R. Ohki, N. Seki, M. Sagara, H. Ito, M. Yamauchi, and T. Hori. 1997. The structure and organization of the human NPAT gene. Genomics 42:388-392. [DOI] [PubMed] [Google Scholar]
- 19.Lichtler, A. C., F. Sierra, S. Clark, J. R. Wells, J. L. Stein, and G. S. Stein. 1982. Multiple H4 histone mRNAs of HeLa cells are encoded in different genes. Nature 298:195-198. [DOI] [PubMed] [Google Scholar]
- 20.Luong, M. X., C. M. van der Meijden, D. Xing, R. Hesselton, E. S. Monuki, S. N. Jones, J. B. Lian, J. L. Stein, G. S. Stein, E. J. Neufeld, and A. J. van Wijnen. 2002. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol. Cell. Biol. 22:1424-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma, T., B. A. Van Tine, Y. Wei, M. D. Garrett, D. Nelson, P. D. Adams, J. Wang, J. Qin, L. T. Chow, and J. W. Harper. 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14:2298-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantovani, R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239:15-27. [DOI] [PubMed] [Google Scholar]
- 23.Martinelli, R., and N. Heintz. 1994. H1TF2A, the large subunit of a heterodimeric, glutamine-rich CCAAT-binding transcription factor involved in histone H1 cell cycle regulation. Mol. Cell. Biol. 14:8322-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller, P., P. Garrity, and B. Wold. 1997. Ligation-mediated PCR for genomic sequencing and footprinting, p. 15.5.1-15.5.26. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and J. A. Smith (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 25.Nepveu, A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270:1-15. [DOI] [PubMed] [Google Scholar]
- 26.Nevins, J. R. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699-703. [DOI] [PubMed] [Google Scholar]
- 27.Ng, H. H., Y. Zhang, B. Hendrich, C. A. Johnson, B. M. Turner, H. Erdjument-Bromage, P. Tempst, D. Reinberg, and A. Bird. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23:58-61. [DOI] [PubMed] [Google Scholar]
- 28.Osley, M. A. 1991. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60:827-861. [DOI] [PubMed] [Google Scholar]
- 29.Pardee, A. B. 1974. A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. USA 71:1286-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pauli, U., S. Chrysogelos, G. Stein, J. Stein, and H. Nick. 1987. Protein-DNA interactions in vivo upstream of a cell cycle-regulated human H4 histone gene. Science 236:1308-1311. [DOI] [PubMed] [Google Scholar]
- 31.Ramsey-Ewing, A., A. J. van Wijnen, G. S. Stein, and J. L. Stein. 1994. Delineation of a human histone H4 cell cycle element in vivo: the master switch for H4 gene transcription. Proc. Natl. Acad. Sci. USA 91:4475-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekimata, M., A. Takahashi, A. Murakami-Sekimata, and Y. Homma. 2001. Involvement of a novel zinc finger protein, MIZF, in transcriptional repression by interacting with a methyl-CpG-binding protein, MBD2. J. Biol. Chem. 276:42632-42638. [DOI] [PubMed] [Google Scholar]
- 33.Shakoori, A. R., A. J. van Wijnen, C. Cooper, F. Aziz, M. Birnbaum, G. P. Reddy, X. Grana, A. De Luca, A. Giordano, J. B. Lian, J. L. Stein, P. Quesenberry, and G. S. Stein. 1995. Cytokine induction of proliferation and expression of CDC2 and cyclin A in FDC-P1 myeloid hematopoietic progenitor cells: regulation of ubiquitous and cell cycle-dependent histone gene transcription factors. J. Cell. Biochem. 59:291-302. [DOI] [PubMed] [Google Scholar]
- 34.Sinclair, A. M., J. A. Lee, A. Goldstein, D. Xing, S. Liu, R. Ju, P. W. Tucker, E. J. Neufeld, and R. H. Scheuermann. 2001. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood 98:3658-3667. [DOI] [PubMed] [Google Scholar]
- 35.Stein, G., W. Park, C. Thrall, R. Mans, and J. Stein. 1975. Regulation of cell cycle stage-specific transcription of histone genes from chromatin by non-histone chromosomal proteins. Nature 257:764-767. [DOI] [PubMed] [Google Scholar]
- 36.Stein, G. S., J. L. Stein, A. J. van Wijnen, and J. B. Lian. 1996. Transcriptional control of cell cycle progression: the histone gene is a paradigm for the G1/S phase and proliferation/differentiation transitions. Cell Biol. Int. 20:41-49. [DOI] [PubMed] [Google Scholar]
- 37.Tang, L., B. Guo, A. Javed, J.-Y. Choi, S. Hiebert, J. B. Lian, A. J. van Wijnen, J. L. Stein, G. S. Stein, and G. W. Zhou. 1999. Crystal structure of the nuclear matrix targeting signal of the transcription factor AML-1/PEBP2αB/CBFα2. J. Biol. Chem. 274:33580-33586. [DOI] [PubMed] [Google Scholar]
- 38.van den Ent, F. M., A. J. van Wijnen, T. J. Last, R. Bortell, J. L. Stein, J. B. Lian, and G. S. Stein. 1993. Concerted control of multiple histone promoter factors during cell density inhibition of proliferation in osteosarcoma cells: reciprocal regulation of cell cycle-controlled and bone-related genes. Cancer Res. 53:2399-2409. [PubMed] [Google Scholar]
- 39.van den Ent, F. M., A. J. van Wijnen, J. B. Lian, J. L. Stein, and G. S. Stein. 1994. Cell cycle controlled histone H1, H3, and H4 genes share unusual arrangements of recognition motifs for HiNF-D supporting a coordinate promoter binding mechanism. J. Cell. Physiol. 159:515-530. [DOI] [PubMed] [Google Scholar]
- 40.van der Meijden, C. M. J., P. S. Vaughan, A. Staal, W. Albig, D. Doenecke, J. L. Stein, G. S. Stein, and A. J. van Wijnen. 1998. Selective expression of specific histone H4 genes reflects distinctions in transcription factor interactions with divergent H4 promoter elements. Biochim. Biophys. Acta 1442:82-100. [DOI] [PubMed] [Google Scholar]
- 41.van Wijnen, A. J., F. Aziz, X. Grana, A. De Luca, R. K. Desai, K. Jaarsveld, T. J. Last, K. Soprano, A. Giordano, J. B. Lian, et al. 1994. Transcription of histone H4, H3, and H1 cell cycle genes: promoter factor HiNF-D contains CDC2, cyclin A, and an RB-related protein. Proc. Natl. Acad. Sci. USA 91:12882-12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Wijnen, A. J., R. F. Massung, J. L. Stein, and G. S. Stein. 1988. Human H1 histone gene promoter CCAAT box binding protein HiNF-B is a mosaic factor. Biochemistry 27:6534-6541. [DOI] [PubMed] [Google Scholar]
- 43.van Wijnen, A. J., A. L. Ramsey-Ewing, R. Bortell, T. A. Owen, J. B. Lian, J. L. Stein, and G. S. Stein. 1991. Transcriptional element H4-site II of cell cycle regulated human H4 histone genes is a multipartite protein/DNA interaction site for factors HiNF-D, HiNF-M, and HiNF-P: involvement of phosphorylation. J. Cell. Biochem. 46:174-189. [DOI] [PubMed] [Google Scholar]
- 44.van Wijnen, A. J., F. M. van den Ent, J. B. Lian, J. L. Stein, and G. S. Stein. 1992. Overlapping and CpG methylation-sensitive protein-DNA interactions at the histone H4 transcriptional cell cycle domain: distinctions between two human H4 gene promoters. Mol. Cell. Biol. 12:3273-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Wijnen, A. J., M. F. van Gurp, M. C. de Ridder, C. Tufarelli, T. J. Last, M. Birnbaum, P. S. Vaughan, A. Giordano, W. Krek, E. J. Neufeld, J. L. Stein, and G. S. Stein. 1996. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc. Natl. Acad. Sci. USA 93:11516-11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaughan, P. S., F. Aziz, A. J. van Wijnen, S. Wu, H. Harada, T. Taniguchi, K. J. Soprano, J. L. Stein, and G. S. Stein. 1995. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature 377:362-365. [DOI] [PubMed] [Google Scholar]
- 47.Vaughan, P. S., C. M. J. van der Meijden, F. Aziz, H. Harada, T. Taniguchi, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1998. Cell cycle regulation of histone H4 gene transcription requires the oncogenic factor IRF-2. J. Biol. Chem. 273:194-199. [DOI] [PubMed] [Google Scholar]
- 48.Xie, R. L., A. J. van Wijnen, C. M. van der Meijden, J. L. Stein, and G. S. Stein. 2002. Forced expression of the interferon regulatory factor 2 oncoprotein causes polyploidy and cell death in FDC-P1 myeloid hematopoietic progenitor cells. Cancer Res. 62:2510-2515. [PubMed] [Google Scholar]
- 49.Younghusband, H. B., R. Sturm, and J. R. Wells. 1986. Mutagenesis of conserved 5′ elements and transcription of a chicken H1 histone gene. Nucleic Acids Res. 14:635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng, C., S. McNeil, S. Pockwinse, J. A. Nickerson, L. Shopland, J. B. Lawrence, S. Penman, S. W. Hiebert, J. B. Lian, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1998. Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc. Natl. Acad. Sci. USA 95:1585-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao, J., B. K. Kennedy, B. D. Lawrence, D. A. Barbie, A. G. Matera, J. A. Fletcher, and E. Harlow. 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14:2283-2297. [PMC free article] [PubMed] [Google Scholar]