Abstract
The Mre11/Rad50/Xrs2 complex initiates IR repair by binding to the end of a double-strand break, resulting in 5′ to 3′ exonuclease degradation creating a single-stranded 3′ overhang competent for strand invasion into the unbroken chromosome. The nuclease(s) involved are not well understood. Mre11 encodes a nuclease, but it has 3′ to 5′, rather than 5′ to 3′ activity. Furthermore, mutations that inactivate only the nuclease activity of Mre11 but not its other repair functions, mre11-D56N and mre11-H125N, are resistant to IR. This suggests that another nuclease can catalyze 5′ to 3′ degradation. One candidate nuclease that has not been tested to date because it is encoded by an essential gene is the Dna2 helicase/nuclease. We recently reported the ability to suppress the lethality of a dna2Δ with a pif1Δ. The dna2Δ pif1Δ mutant is IR-resistant. We have determined that dna2Δ pif1Δ mre11-D56N and dna2Δ pif1Δ mre11-H125N strains are equally as sensitive to IR as mre11Δ strains, suggesting that in the absence of Dna2, Mre11 nuclease carries out repair. The dna2Δ pif1Δ mre11-D56N triple mutant is complemented by plasmids expressing Mre11, Dna2 or dna2K1080E, a mutant with defective helicase and functional nuclease, demonstrating that the nuclease of Dna2 compensates for the absence of Mre11 nuclease in IR repair, presumably in 5′ to 3′ degradation at DSB ends. We further show that sgs1Δ mre11-H125N, but not sgs1Δ, is very sensitive to IR, implicating the Sgs1 helicase in the Dna2-mediated pathway.
Introduction
Double-strand breaks (DSBs) are a major result of endogenous DNA replication errors and of exogenous DNA damaging agents, and failure to repair such breaks leads to the gross chromosomal rearrangements characteristic of tumor cells. In yeast, homologous recombination (HR), carried out by proteins encoded by the RAD52 epistasis group, is used to repair DSBs induced by ionizing radiation (IR). The time course of IR repair in vivo has been assayed by observing the post-irradiation appearance of foci of the various HR repair proteins tagged with green fluorescent protein [1]. Mre11, Rad50, and Xrs2 form a complex (MRX) and are the first arrivals after DSB formation. The MRX complex regulates the processing of the end of the DSB, resulting in resection of the 5′ end and leaving a 3′ single-stranded overhang [2]. RPA binds the single-stranded DNA, followed by Rad52 and Ddc2/Mec1 binding. Ddc2/Mec1 is the apical checkpoint-activating protein kinase. Rad52 can mediate the exchange of RPA for the Rad51 strand exchange protein, generating the 3′ terminal filament that initiates invasion into the intact sister or homologous chromosome [3]. Rad51 association is followed by the binding of Rad54 and additional strand exchange proteins, Rad55 and Rad57.
Mechanistically, the initial steps are poorly understood. Our studies address the function of the MRX complex, which has ATP binding, ATPase, adenylate kinase, nuclease, and DNA end-bridging activity [4], [5]. Mre11 houses the nuclease activity in a domain containing five phosphodiesterase motifs homologous to the E. coli sbcD exonuclease. Mre11 has 3′ to 5′ exonuclease activity on double-stranded DNA and single-strand endonuclease activity on DNA hairpins [6]–[8]. Rad50 is an ATP binding protein, with the Walker A motif in the N terminus and Walker B motif in the C terminus separated by a long heptad repeat domain [9]. A signature (arginine finger) region in the C terminus is also part of the ATP binding motif [10]. Mutations in the ATP binding motif result in strains with meiotic recombination null phenotypes [9], [11]. The Xrs2 protein (known as Nbs1 in mammals) contains a forkhead domain at the N terminus and a binding domain for Tel1 at the C-terminus. Tel1 is a kinase involved in telomere maintenance and checkpoint signaling. Mre11/Rad50/Nbs1 has weak ATP-stimulated DNA unwinding activity [12].
In bacterial recombinational repair, the strand invasion intermediate is generated by the RecBCD helicase/nuclease in a complicated reaction requiring 5′ to 3′ helicase, 3′ to 5′ helicase, 5′ to 3′ nuclease, 3′ to 5′ nuclease, and a specific DNA sequence called chi. A counterpart to this complex has not been identified in eukaryotic cells. The most highly radioresistant organism known, Micrococcus radiodurans, does not have a RecBCD helicase/nuclease, but does possess a complete RecF pathway [13]. A major question has been what pathway is found in eukaryotes. MRE11 is a nuclease involved in DSB repair and mre11Δ strains are extremely sensitive to irradiation. After 17 krads the survival of an mre11Δ strain is about 0.06%. If one assumes 2 krads results in 1 DSB per haploid cell [14], then a survival of about 0.02% would be expected in the absence of repair. Therefore, repair is nearly absent in a mre11Δ strain. mre11-D65N and mre11-H125N are more sensitive to IR than MRE11 strains but much more resistant than mre11Δ strains. At a high dose of 70 krads about 75 DSBs are induced, which would predict a survival value of e−35 or 10−13% in the complete absence of repair. The survival of mre11-D56N and mre11-H125N mutants at 70 krads is about 5 to 10 fold less than MRE11 strains (3% survival), which is much higher than the predicted survival in the absence of DSB repair [15], [16]. In addition, paradoxically, Mre11 has 3′ to 5′ exonuclease activity rather than the 5′ to 3′ nuclease expected for 5′ resection [6]–[8]. Furthermore, resection is not reduced in mre11 mutants defective in the nuclease active site but proficient in the other functions of Mre11 [17], [18]. This has led to the proposal that other nucleases are required and that they can compensate for the absence of the Mre11 nuclease. Sae2 is another nuclease that appears to be associated with the MRX complex, but also has 3′ to 5′ activity [19]. Exo1, a 5′ to 3′ nuclease of the Fen1 family involved in mismatch repair, may play a role. Overproduction of Exo1 indeed increases the IR resistance of an mre11Δ strain [20], [21]. However, exo1Δ mre11Δ and exoΔ mre11-nuclease deficient double mutants are viable, are no more sensitive to IR than mre11Δ or mre11-nuclease deficient mutants, respectively, and can carry out gene conversion as efficiently in vivo as EXO1 MRE11 strains [20]. This suggests that yet another nuclease must compensate for Mre11 nuclease deficiency.
Dna2 is a candidate for this role. Dna2 is a 5′ to 3′ helicase, 5′ to 3′ exo/endonuclease, 3′ to 5′ exo/endonuclease, and has single strand DNA annealing and strand exchange activity [22]–[28]. Dna2 plays a role in Okazaki fragment processing (OFP), assisting FEN1 in RNA primer removal [26], [29]. dna2 mutants, however, are extremely sensitive to bleomycin and IR, consistent with an additional role in DSB repair [30]–[32]. Also, we showed many years ago that dna2-2 rad50-5 mutants are synergistically defective in IR repair [31]. Finally, Dna2 forms a hub in a genetic network of 322 proteins that preserve genome stability [33]. Since the DNA2 gene is essential, however, it has been difficult to test the hypothesis that Dna2 nuclease participates in generation of the 3′ overhang during DSB repair. Recently, we found that the inviability of dna2Δ mutants is efficiently suppressed by deletion of Pif1, a helicase orthologous to E. coli RecD helicase, that functions in DNA replication and telomere homeostasis [34], [35]. Using a dna2Δ pif1Δ strain and mre11 nuclease defective mutants, we show that the nucleases of Dna2 and Mre11 are required in repair of IR-induced damage, presumably in 5′ resection, and that they can compensate for each other's loss. In addition, we show that the helicase of Dna2 is dispensable but that the Sgs1 helicase is required for Dna2 nuclease to be able to compensate for lack of Mre11 nuclease.
Results
Dna2 nuclease can compensate for the loss of Mre11 nuclease in DSB repair and vice versa
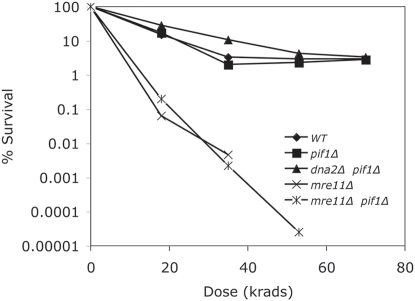
The relative importance of repair proteins can be measured by determining the survival of mutants defective in the respective proteins after increasing doses of a DNA damaging agent. We have used assays that measure survival after an acute dose of IR followed by growth in the absence of the DNA damaging agent. We were surprised that the dna2Δ pif1Δ mutant was resistant to X-rays (Figure 1, the sensitivity of mre11Δ and mre11Δ pif1Δ are shown as controls), since several dna2 hypomorphic mutants are sensitive to IR [31], [36]. This suggested that in the absence of Pif1, another protein might carry out the role of Dna2 in repair. Both hypomorphic dna2 mutants and the dna2Δ pif1Δ strain are synthetically lethal with mutants affecting three nuclease mutants defective in DSB repair, mre11Δ, sae2Δ, and exo1Δ [31], [33], [34]. We reasoned that the Mre11 nuclease might be compensating for the loss of Dna2 during X-ray repair in the dna2Δ pif1Δ strain.
Figure 1. X-ray resistance of the dna2Δ pif1Δ strain compared to sensitivity of mre11Δ pif1Δ.
The strains MB120-5A (WT), MB121- pif1Δ, MB161B- dna2Δ pif1Δ, MB122-17C- mre11Δ, MB124-2D- mre11Δ pif1Δ were harvested in mid-log phase, resuspended in water, irradiated, serially diluted (1∶10), and plated.
To test the hypothesis that Mre11 and Dna2 had compensatory functions, we wished to compare the X-ray sensitivity of dna2Δ pif1Δ and dna2Δ pif1Δ mre11-nuclease defective strains. We first created dna2Δ pif1Δ mre11Δ strains carrying MRE11, mre11-D56N or mre11-H125N on centromeric plasmids. The D56N and H125N mutations lie in the conserved phosphodiesterase motif of Mre11, and the mutant proteins have no in vitro endonuclease activity [18], [37]; however, they are able to form a complex with Rad50 and Xrs2. As shown in Figure 2, both wild-type and nuclease-defective alleles of MRE11 suppress the lethality of dna2Δ pif1Δ mre11Δ strains. As predicted, however, the strains deficient in both Dna2 and the Mre11 nuclease are X-ray sensitive, while strains expressing wild-type Mre11 are resistant to IR. Thus, in the absence of Dna2, Mre11 nuclease is required for X-ray repair. By contrast, the X-ray sensitivity of pif1Δ mre11Δ, in which there is a functional Dna2 nuclease, is complemented not only by wild-type MRE11 but also by nuclease defective mre11-D56N or mre11-H125N. These results suggest that Dna2 nuclease can function in X-ray repair in the absence of Mre11 nuclease, but not in the complete absence of Mre11. It also says that Mre11 nuclease can function in X-ray repair in the absence of Dna2.
Figure 2. dna2Δ pif1Δ mre11Δ (pRS414mre11-D56N) and dna2Δ pif1Δ mre11Δ (pRS414mre11-H125N) strains exhibit similar sensitivity to X-rays as mre11Δ pif1Δ.
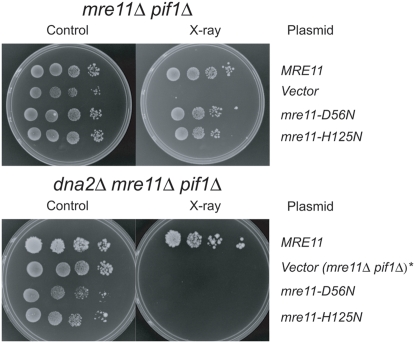
These strains were created by dissecting tetrads from the following crosses: MATa dna2Δ pif1Δ trp1Δ bar1Δ/ MATα mre11Δ trp1Δ (pRS414MRE11), MATa dna2Δ pif1Δ trp1Δ bar1Δ/ MATα mre11Δ trp1Δ (pRS414mre11-D56N) and MATa dna2Δ pif1Δ trp1Δ bar1Δ/ MATα mre11Δ trp1Δ (pRS414mre11-H125N) diploids. Trp+ spores were replica plated, and irradiated. Control strains mre11Δ pif1Δ (pRS414MRE11) and dna2Δ pif1Δ mre11Δ (pRS414MRE11) were identified from the respective cross and were X-ray resistant. dna2Δ pif1Δ mre11Δ (pRS414mre11D56N) or dna2Δ pif1Δ mre11Δ (pRS414mre11H125N), from the two respective crosses were radiation sensitive. To compare the radiation sensitivity of the dna2Δ pif1Δ mre11Δ (pRS414mre11-D56N) and the dna2Δ pif1Δ mre11Δ (pRS414mre11-H125N) (bottom panel) to mre11Δ pif1Δ (pRS414mre11-D56N) and to mre11Δ pif1Δ (pRS414mre11-H125N) strains (top panel), serially diluted cells were irradiated with 26 krads on a plate and allowed to grow for three days. Note that a dna2Δ pif1Δ mre11Δ strain carrying only the pRS vector is inviable, so an mre11Δ pif1Δ strain carrying the pRS vector alone is shown as the control, as indicated by the * in the bottom panel.
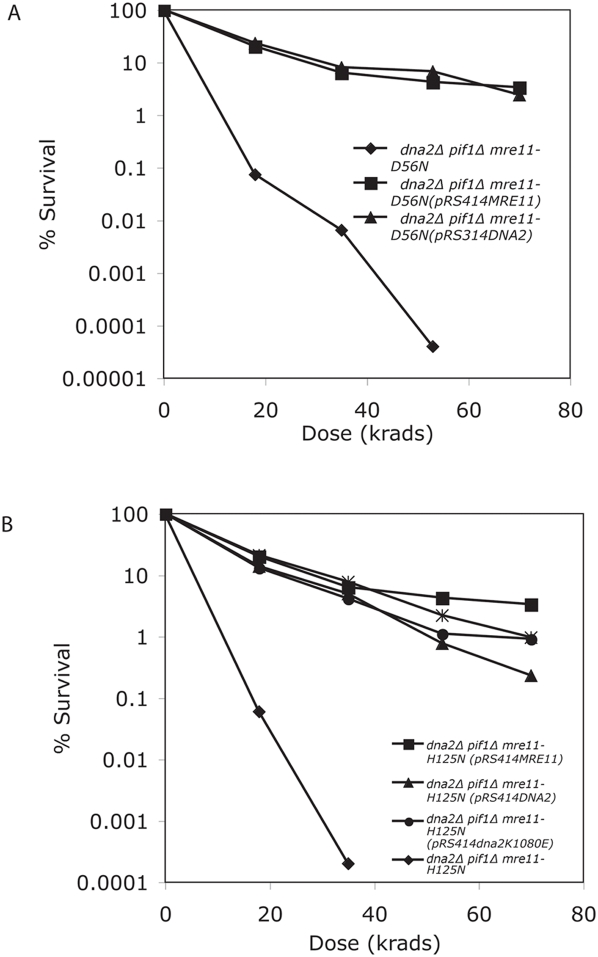
To insure that the X-ray sensitivities of the nuclease-dead mre11 mutants were not the result of inefficient expression due to employing extrachromosomal plasmids to express the mre11 alleles, the mre11 nuclease defective genes were integrated into the chromosome by two-step gene replacement. The dna2Δ pif1Δ mre11-D56N and dna2Δ pif1Δ mre11-H125N clones were as X-ray sensitive as the mre11Δ pif1Δ strain (compare Figure 3A and Figure 3B to Figure 1). Importantly, in both dna2Δ pif1Δ mre11-D56N and dna2Δ pif1Δ mre11-H125N X-ray sensitive clones, X-ray resistance is restored by introduction of a plasmid expressing either DNA2 or MRE11 (Figure 3A,B). The helicase activity of Dna2 was not essential for complementation, since the X-ray resistance of the dna2Δ pif1Δ mre11-H125N clone was restored by plasmids expressing Dna2 with either of two mutations in the helicase domain: pRS314dna2K1080E plasmid, which has a mutation in the conserved Walker P loop but has a functional nuclease, and pRS314dna2R1253Q (dna2-2) (Figure 3B). We conclude that it is the nucleolytic activities of Mre11 and Dna2 that are complementary.
Figure 3.
(A) X-ray sensitivity of a dna2Δ pif1Δ mre11-D56N strain and complementation by CEN plasmids containing Mre11 or Dna2. DNA fragments containing mre11-D56N and mre11-H125N mutant genes with 800 bp 5′ and 140 bp 3′ to the gene were cloned into pRS306URA3, cleaved in the mre11 promoter and transformed into a dna2Δ pif1Δ MRE11 strain to allow integration at the MRE11 locus. The resulting transformants were propagated on YPD media then grown on 5-FOA containing media to excise the MRE11 allele. Approximately 15% of the pRS306mre11-D56N and pRS306mre11-H125N Ura− pop outs were X-ray sensitive. Strains MB161B-56- dna2Δ pif1Δ mre11D56N, MB161B-56 (pRS414MRE11), MB161B (pRS314DNA2) were then compared for X-ray sensitivity as in Figure 1. (B) X-ray sensitivity of a dna2Δ pif1Δ mre11-H125N strain and complementation by plasmids containing MRE11 or nuclease proficient DNA2 genes. Strain MB161B-125 – dna2Δ pif1Δ mre11H125N, MB161B-125 (pRS414MRE11), MB161B-125 (pR314DNA2), MB161B (pRS314dna2K1080E), MB161B-125 (pRS314dna2-2) were compared for X-ray sensitivity as described in Figure 1.
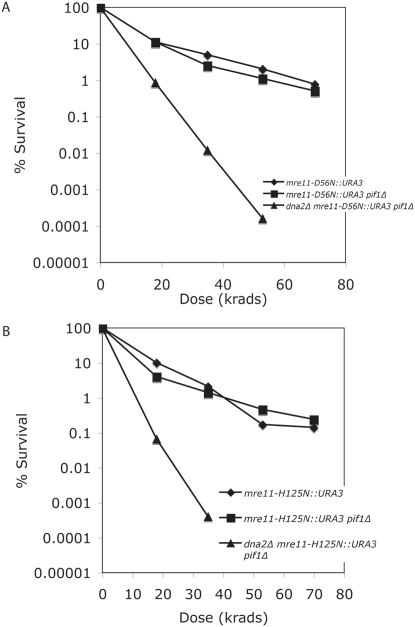
mre11-D56N and mre11-H125N mutants are not sensitive enough to DNA damaging agents to allow direct scoring in genetic crosses. Therefore, to compare the DNA2 PIF1 mre11 nuclease-minus with the dna2Δ pif1Δ mre11 nuclease-minus mutants, triple mutants were also constructed by introduction of a genetically tagged copy of the mre11 nuclease mutant genes into the chromosome, as described in the legend to Figure 4. dna2Δ pif1Δ MRE11 were X-ray resistant (see Figure 1). As before, dna2Δ pif1Δ mre11Δ::NatR::306 mre11-D56N::URA3 or dna2Δ pif1Δ mre11Δ::NatR::306 mre11-H125N::URA3 were highly sensitive to IR compared to the mre11Δ::NatR::306 mre11D56N::URA3 and the mre11Δ::NatR::306 mre11H125N::URA3 strains (Figure 4 A,B). Clearly, in the absence of Dna2 nuclease, Mre11 nuclease is essential for repair of IR-induced damage.
Figure 4.
(A) Comparison of the X-ray sensitivity of a mre11-D56N pif1Δ strain with a dna2Δ mre11-D56N pif1Δ strain. The integrating plasmids pRS306mre11-D56N::URA3 and pRS306mre11-H125N::URA3 were cut with Sph1 which cleaves about 300 bp 5′ to the ATG of MRE11 and transformed into a MATa mre11Δ::natR strain and insertion at the MRE11 locus was selected for on G418 plates lacking uracil. These strains were mated with MB161B-MATa dna2Δ pif1Δ MRE11, and the diploids were sporulated and dissected. When the resulting spores were scored, the NatR gene always segregated with the URA3 gene, demonstrating that the pRS306mre11nuclease minus plasmids were integrated at the MRE11 loci in the respective strains. Strains MB126-mre11Δ::natR::mre11D56N::URA3, MB128- mre11Δ::natR::mre11-D56N::URA3 pif1Δ, and MB133 - dna2Δ pif1Δ mre11Δ::natR::mre11-D56N::URA3 were treated as in Figure 1 to determine IR sensitivity. (B) Comparison of the X-ray sensitivity of a mre11-H125N pif1Δ strain with a dna2Δ mre11-H125N pif1Δ strain. Strains were constructed as in Figure 4A. Strains MB127-mre11Δ::natR::mre11-H125N::URA3, MB129-mre11Δ::natR::mre11-H125N::URA3 pif1Δ, and MB134 - dna2Δ mre11Δ::natR::mre11-H125N::URA3 pif1Δ, were treated as in Figure 1.
Sgs1 is Required for the Dna2-mediated Repair Pathway
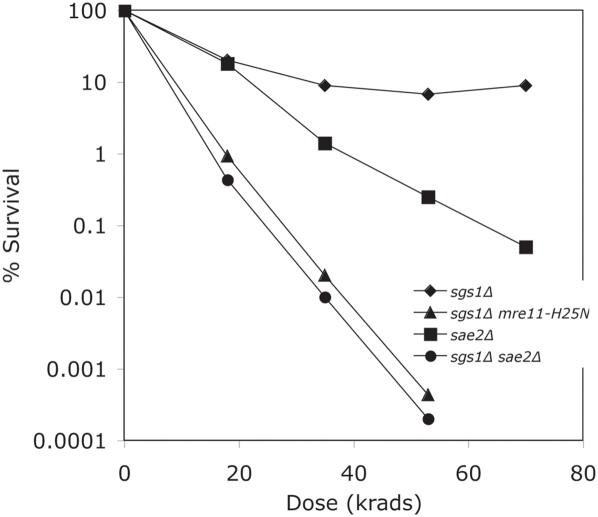
Based on the above results, we hypothesized that Dna2 nuclease might play a role in resection of the 5′ end at IR-induced breaks. Since the Dna2 helicase was not required, we asked what helicase might participate to convert the DSB into a substrate recognized by Dna2, which recognizes only single-stranded termini. Sgs1, which encodes a helicase of the RecQ family, was a likely candidate, since helicase defective dna2-2 mutants are defective in X-ray repair but the double sgs1Δ dna2-2 mutant is significantly more sensitive to IR than dna2-2, even though sgs1Δ itself is X-ray resistant [31], [38]. Furthermore, we also recently demonstrated that human BLM, a RecQ family helicase, can suppress the MMS and bleomycin sensitivity of dna2-2 mutants [39], [40]. We therefore investigated whether yeast Sgs1 might be involved in the Dna2 repair pathway. We found, as have others, that sgs1Δ mutants are as resistant to IR as wildtype (Figure 5). We then constructed an sgs1Δ mre11-H125N double mutant. As we predicted, the double mutant was as sensitive to IR as the dna2Δ mre11-H125N mutant (compare Figures 3 and 4 with Figure 5). We conclude that Sgs1 is required in order for Dna2 to compensate for the loss of Mre11 nuclease during repair of IR-induced damage.
Figure 5. Comparison of the X-ray sensitivity of strain sgs1Δ with sgs1Δ mre11-H125N and sgs1Δ sae2Δ.
Strains MB137 sgs1Δ, MB138 sgs1Δ mre11-H125N and MB135 sae2Δ and MB136 sgs1Δ sae2Δ were constructed in this study and treated as in Figure 1.
Sae2 is also a nuclease that functions in the MRX complex and can function as a nuclease on its own [19]. Thus, we wanted to test whether sae2Δ sgs1Δ strains have the same survival as sgs1Δ mre11-H125N, dna2Δ mre11-H125N, and mre11Δ strains. As illustrated in Figure 5, sae2Δ sgs1Δ strains are indeed as sensitive to IR as sgs1Δ mre11-H125N strains. These results put Mre11 nuclease and Sae2 nuclease in one pathway and Dna2 nuclease and Sgs1 helicase in another pathway in the initiation of exonucleolytic degradation after DSB formation.
Dna2 Helicase Activity produces the Requirement for Dna2 Nuclease Activity
Separation of function of MRX ATP binding and nuclease functions has been investigated extensively so we attempted a similar analysis of Dna2. The nuclease of Dna2 maps to a region N-terminal to the helicase, between amino acids 649–744 in a domain containing conserved E. coli RecB nuclease motifs. Two mutants of Dna2, D657A and E675A, specifically affecting RecB active site motifs, result in Dna2 proteins with greatly reduced or undetectable nuclease activity but with intact helicase and ATPase activity [25], [41]. The plasmid, pGAL::DNA2, is a high copy vector containing a GAL10 inducible Dna2 but expresses sufficient Dna2 without induction to complement dna2-1 mutants at 37°C on glucose plates [25]. The plasmids pGAL::dna2-K1080E, pGAL::dna2-D657A pGAL::dna2-E675A do not complement a dna2-1 mutant on glucose plates at 37°C [25]. Also, we found here, that the dna2-K1080E, dna2-D657A, and dna2-E675A genes expressed from centromeric plasmids fail to complement the dna2Δ PIF1 mutant (data not shown). This indicates that the ATP binding and nuclease functions of Dna2 are essential.
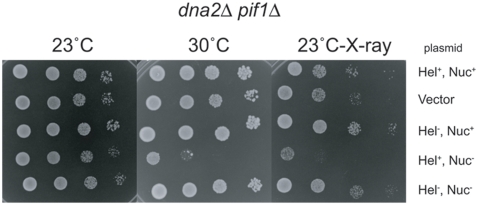
To verify that an active Dna2 nuclease was necessary for IR repair in the absence of Mre11 nuclease, we compared the growth and X-ray sensitivity of dna2Δ pif1Δ strains transformed with centromeric plasmids carrying DNA2, dna2K1080E, and dna2E675A, or dna2K1080E,E675A. The dna2K1080E,E675A is doubly deficient in both helicase and nuclease. When the dna2Δ pif1Δ transformants were incubated at 23°C on tryptophan deficient plates all five transformants grew, as illustrated in Figure 6. At 30°C, the transformants containing the plasmids pRS314, pRS314DNA2, pRS314dna2K1080E (helicase minus, nuclease plus) formed viable colonies. Thus, pif1Δ suppresses the inviability of a dna2Δ PIF1 (pRS314dna2-K1080E) strain, and in fact the dna2Δ pif1Δ (RS314dna2-K1080E) strain even grows at 37°C, unlike the dna2Δ pif1Δ strain [34]. Interestingly, the pRS314dna2E675A transformant (helicase plus, nuclease minus) did not form colonies at 30°C (Figure 6). However, the pRS314dna2K1080E,E675A (helicase minus, nuclease minus) did support viability at 30°C. We conclude that at this temperature the absence of the nuclease was deleterious to normal DNA replication, and furthermore that it is the Dna2 helicase activity that generates a need for the Dna2 nuclease.
Figure 6. Expression of nuclease deficient Dna2 is toxic in a dna2Δ pif1Δ strain.
Strain MB21-dna2Δ pif1Δ carrying either pRS314DNA2, Hel+, Nuc+; pRS314, Vector; pRS314dna2K1080E, Hel−Nuc+; pRS314dna2E675A, Hel+Nuc− or pRS314dna2K1080E,dna2E675A, Hel−Nuc− plasmids was grown, serially diluted 10 fold and plated on trptophan deficient plates with or without 26 krads IR, and allowed to grow for four days at the temperatures indicated.
Next, the dna2Δ pif1Δ transformants were irradiated after plating and incubated at 23°C. The pRS314, pRS314DNA2, pRS314dna2K1080E, and pRS314dna2K1080E,E675A transformants were resistant to IR, whereas the transformant containing the pRS314dna2E675A plasmid was sensitive to IR. Thus, as with growth at the restrictive temperature of 30°C, after IR, the Dna2 nuclease minus strain is sensitive to damage, but only in the presence of an active Dna2 helicase. We conclude that the activities of the Dna2 helicase and nuclease must be properly coordinated for both DNA replication and repair.
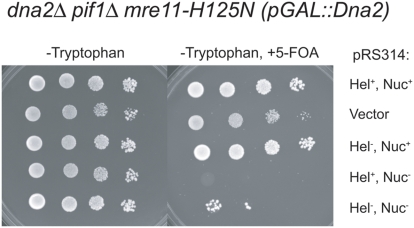
The requirement for coordination of the Dna2 helicase and nuclease was further examined in the dna2Δ pif1Δ mre11-H125N mutant. The dna2Δ pif1Δ mre11-H125N (pGAL::DNA2) was transformed with the same set of Dna2 mutant plasmids used in Figure 6, to yield double transformants, dna2Δ pif1Δ mre11-H125N (pGAL::DNA2) (pRS414-dna2 mutant). Each double transformant was grown in tryptophan deficient liquid media to select for the pRS314dna2 mutant and to allow loss of the URA3 containing the pGAL::DNA2 plasmid. The resulting cultures were spotted on tryptophan-deficient or tryptophan-deficient media containing 5-FOA and grown at 23°C to determine the ability of the dna2 mutants to support growth. dna2Δ pif1Δ mre11-H125N containing the plasmids pRS314, pRS314DNA2, pRS314dna2K1080E grew, but strains containing the plasmids pRS314dna2E675A (nuclease minus) did not grow on 5-FOA-containing plates (Figure 7). In this Mre11-nuclease deficient background, there is also reduced viability of the transformant containing the pRS314dna2K1080E,E675A (nuclease minus, helicase minus). We conclude that the viability of the Dna2-nuclease defective dna2Δ pif1Δ (pRS314dna2E675A) at 23°C shown above (Figure 6) depends on a functional Mre11 nuclease, suggesting their complementarity not only during IR repair but also during DNA replication or repair of replication errors. This is consistent with a model in which Dna2 helicase makes a potentially toxic structure during DNA replication that is preferentially and coordinately processed by the Dna2 nuclease, but that can also be processed by Mre11 nuclease. Inactivation of Dna2 helicase is sufficient to remove the requirement for nucleolytic processing in the absence Dna2 alone but is insufficient in the absence of both Dna2 and Mre11 nuclease. In the dna2Δ pif1Δ mre11-H12N mutant, the residual MRX helicase may now contribute to generating a structure needing nuclease processing.
Figure 7. Inactivation of the Dna2 nuclease is lethal in a pif1Δ mre11-H125N strain.
A dna2Δ pif1Δ mre11-H125N trp1Δ (pGAL::DNA2URA3) strain was transformed with the Trp+ plasmids pRS314DNA2, Hel+, Nuc+; pRS314, Vector; pRS314dna2K1080E, Hel−Nuc+; pRS314dna2E675A, Hel+Nuc− or pRS314dna2K1080E,dna2E675A, Hel−Nuc−. The Trp+Ura+ transformants were grown in tryptophan deficient media and spotted onto tryptophan minus (left) or tryptophan minus, 5-FOA containing (right) media at 23°C and photographed after 4 days at 23°C.
Discussion
We have shown that the Dna2 5′ to 3′ nuclease can function in IR repair in the absence of an active Mre11 nuclease. The key result is that the dna2Δ pif1Δ strain is resistant to X-rays but the dna2Δ pif1Δ mre11-H125N mutant is as sensitive to X-rays as an mre11Δ strain, showing that in the absence of both the Mre11 nuclease and Dna2 nuclease, cells are nearly blocked in repair. Thus, strikingly, either nuclease can support repair efficiently (Figure 3). The most direct mechanistic inference is that Dna2 and Mre11 are performing at least partially overlapping nucleolytic functions in processing the ends of DSBs. Since Mre11 has 3′ to 5′ nuclease rather than 5′ to 3′ activity in vitro, the question arises as to how it can perform 5′ to 3′ nucleolytic degradation in the absence of Dna2. Mre11 may achieve this in the absence of Dna2 by cleaving hairpin structures in ends that are unwound by an unknown helicase [8]. Another interpretation is that Mre11 nuclease and Dna2 function sequentially and that a second nuclease, exonuclease 1, can substitute for Dna2 nuclease, when Mre11 is present. In the absence of Mre11 nuclease, however, Dna2 but not Exo1, can overcome the requirement for the Mre11 nucleolytic step. We favor the latter interpretation (see below).
While our studies show that the Dna2 nuclease can compensate entirely, in terms of repair and survival, for the absence of Mre11 nuclease, Dna2 cannot compensate for the absence of Mre11. Mre11 is clearly required to form the MRX end processing complex.
The interaction between Mre11 and Dna2 at DSB damage is paralleled by the fact that Dna2, like Mre11, is found at telomeres, the linear ends of chromosomes that resemble DSBs [32]. The Mre11 nuclease is required during the de novo synthesis of telomeres at a DSB with a telomere seed sequence, though an mre11 nuclease deficient mutant does not have short telomeres [42]. Dna2 also appears to be required for extending a short telomere, and its nucleolytic function may be at play [32], [43], [44]. Interestingly, dna2Δ pif1Δ mutants have short telomeres compared to pif1Δ mutants [34].
Unexpectedly, our study has also supplied genetic evidence that supports biochemical studies suggesting that the Dna2 helicase and nuclease activities are coupled. dna2Δ pif1Δ expressing a Dna2-E675A (helicase plus, nuclease minus) protein have a reduced maximum permissive growth temperature and they are sensitive to IR. However, dna2Δ pif1Δ expressing a helicase minus, nuclease minus double Dna2 mutant protein has no growth or repair defect, i.e. inactivation of the helicase removes the requirement for the nuclease (Figure 6). This suppression occurs only if Mre11 nuclease is functional (Figure 7). Therefore, we propose that the Dna2 helicase creates a structure during DNA replication and DNA repair that is potentially toxic unless coordinately processed by the nuclease of either Dna2 or, in its absence, Mre11. Further, the observed lethality at all temperatures and in the absence of DNA damage in mre11-nuclease minus dna2Δ pif1Δ expressing nuclease defective Dna2 leads to the hypothesis that Mre11 nuclease may play a role in Okazaki fragment processing or in repair of lethal endogenous DNA damage. A role for Mre11 in OFP is consistent with previous results showing that mre11-H125N rad27 mutants are inviable [18].
Also worth emphasizing is the fact that our findings suggest an additional role for Dna2, besides 5′ resection, in X-ray repair. dna2Δ pif1Δ strains are as resistant to IR as wildtype (this work), whereas dna2Δ PIF1 overexpressing FEN1 to maintain viability is as X-ray sensitive as rad52Δ [31]. Therefore, during IR repair, Pif1 may contribute to formation of a structure that is lethal unless processed by Dna2, and this function is not complemented by MRE11, EXO1, or SGS1. Furthermore, a dna2-2 PIF1 mutant (nuclease plus, helicase defective) is 100 times more sensitive to IR than DNA2 PIF1 (at 75 krads) [31]. However, the dna2-2 gene complements the X-ray sensitivity of the dna2Δ pif1Δ mre11-H125N strain (Figure 3B). These latter two results appear at odds but could be explained if Dna2-2 protein can perform resection but not some other function necessary for X-ray resistance in the presence of PIF1, either during resection or during strand exchange.
Mechanistic studies that were published after our work was completed show that dna2 mutants as well as sgs1 mutants exhibit significantly delayed long-range resection in a Rad51-independent single-strand annealing (SSA) repair reaction at an enzymatically-induced DSB [45], [46]. Based on these studies and the enzymatic activities of Dna2 and Sgs1, we propose that our results imply an interaction between Mre11 nuclease and Dna2/Sgs1 during resection in Rad51-dependent homologous recombination. At the same time, however, our new results also raise a conundrum. The reduced exonuclease resection in the dna2Δ pif1m2 mutant observed in the SSA studies does not appear to cause a defect in subsequent Rad51-dependent repair [46], since we show that the dna2Δ pif1Δ strain is as resistant to IR as a wild-type strain after 70 krads IR (35 DSB or 2 per 17 chromosomes). Similarly, sgs1Δ strains, though they show reduced SSA, are as resistant to IR as wild-type strains, even after 75 krads. Thus, the mechanisms at work during the long-range resection in SSA may not be entirely interchangeable with the resection pathway leading to strand exchange. In this regard it is interesting that Exo1, another 5′ to 3′ nuclease, appears able to compensate for Dna2 or Sgs1 deficiency in SSA [45], [46], but does not appear to be able to do so in IR repair, since dna2 mre11-H125N and sgs1 mre11-H125N mutants are as sensitive to IR as mre11Δ, even though Exo1 is present at normal levels and since mre11 nuclease-defective exo1 double mutants are not significantly more sensitive to IR than mre11 nuclease deficient mutants. Previous studies showed that only 100–300 bp of homology are required for Rad51-dependent gene conversion [47], [48], which differs from SSA in that gene conversion requires strand invasion of a homologous sister chromatid or homolog. We propose that the Dna2/Sgs1 pathway is the major pathway of resection in homologous recombination.
The observed IR sensitivity of the sgs1 mre11-H125N mutant we report here, combined with the defect in resection in sgs1Δ demonstrated by others, may help explain both some of our previous and our current results. We have shown that helicase defective dna2-2 mutants are very defective in X-ray repair, but the double sgs1Δ dna2-2 mutant is significantly more sensitive to IR than dna2-2, even though sgs1Δ itself is X-ray resistant [31]. Thus, Dna2 may compensate for the absence of Sgs1 helicase, but Sgs1 does not fully compensate for a defective Dna2 helicase. We have previously shown that BLM, a human counterpart of Sgs1 helicase, complements the MMS sensitivity of dna2-2 mutants as well as the lethality of dna2-1 mutants [39], and this suppression can now be attributed to suppression of defects in DSB processing. We expect that overexpression of BLM should also suppress the IR-sensitivity of dna2-2 mutants. Genetic evidence suggests Pif1 helicase is an inhibitor of the Sgs1 helicase [49]. Another possible role for Dna2 may be to counteract Pif1 inhibition of Sgs1 during the 5′ to 3′ processing step.
What do our results tell us about the role of Dna2 in endogenous damage repair? We have previously identified the DNA replication fork pause at the replication fork barrier (RFB) in the ribosomal DNA as a specific site of endogenous DNA damage requiring the combined activities of Dna2 and Sgs1 for repair. In the presence of Fob1, DNA replication forks pause at the RFB and a DSB arises, the occurrence of which is elevated in dna2-2 and sgs1Δ mutants [40], [50]. Interestingly, Pif1 is also required for normal levels of pausing at the RFB and subsequent chromosomal breakage [51]. These endogenous DSBs and the inability to repair them contribute to the synthetic sickness/lethality in the dna2-2 sgs1Δ strain, since the sickness/lethality is suppressed by the fob1Δ mutation [40], [50]. A similar situation occurs with Ctf4, a replication/cohesion protein, in which the dna2-2 ctf4Δ mutant is lethal, but the dna2-2 ctf4Δ fob1Δ is viable and slow growing. The dna2-2 ctf4Δ fob1Δ mutant is significantly more sensitive to IR than either dna2-2 fob1Δ or ctf4 fob1Δ mutants [33]. Presumably dna2-2 ctf4Δ inviability also results from failure to repair endogenous DSBs at the RFB. Thus, Dna2 plays a role in repairing endogenous breaks in collaboration with Sgs1 and Ctf4 and this is likely a reflection of the role of Dna2 and Sgs1 in resection. It is probably this role of Dna2 that accounts for the remarkable observation that increasing the gene dosage of Dna2 by one copy leads to partial genome stabilization and is absolutely required for viability of a mec1-21 strain [52].
In conclusion, while our results and those of Zhu et al. [46] clarify the multiple events occurring early in repair at DSBs, especially the previously unappreciated centrality of Dna2, they also highlight several fundamental questions for the future: First, how much resection is sufficient for repair and what is the rate-limiting step in repair? Second, what is the mechanism of resection per se? How do Mre11 and Dna2, which may form a link between DNA replication and repair, coordinate repair of endogenous DSBs occurring due to replication fork failure?
Materials and Methods
Strains and Plasmids
The plasmids pRS314DNA2, pRS314dna2K1080E, pRS314dna2E675A, and pRS314dna2K1080E,E675A have the 6 kb EcoRI fragment containing Dna2 or Dna2 with the indicated mutations cloned into pRS314CEN TRP at the EcoR1 site. The construction of the Dna2 containing fragment and further construction of the mutants was described previously [25]. The plasmid pSEY18GALDNA2, referred to herein as pGAL::DNA2, was described previously [25], except that in this work the DNA2 gene carried an additional FLAG tag, MDYKDDDK, at the N terminus of the protein.
The plasmids pRS414MRE11, pRS414mre11-D56N, and pRSmre11-H125N (gift of K. Lewis, Texas State University, San Marcos, TX) contain the MRE11 genes with about 800 bp 5′ to the ATG and 136 bp 3′ to the TAG contained on a Kpn1 Not1fragment. The KpnI NotI fragments containing mre11-D56N and mre11-H125N were cloned into the KpnI NotI site of the integrating plasmid, pRS306. The pRS306mre11-D56N and pRS306mre11-H125N plasmids were cut with SphI and transformed into yeast.
All MRE11 and DNA2 alleles used in this study are expressed under the control of their native promoters.
Strains used are described in Table 1.
Table 1. Strains.
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| BY4742 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| MB120-5A | 4742 MATα bar1Δ::kanMX trp1Δ |
| MB121 | 4741 MATa pif1Δ::HIS3 bar1Δ::kanMX |
| MB122-17C | 4741 MATa mre11Δ::natR bar1Δ::kanMX trp1Δ |
| MB123-1D | 4742 MATα mre11Δ::natR trp1Δ |
| MB124-2D | 4741MATa mre11Δ::natR pif1Δ::HIS3 bar1Δ::kanMX trp1Δ |
| MB126 | 4742 MATα mre11Δ::natR mre11-D56N::URA3 trp1Δ |
| MB127 | 4741 MATa mre11Δ::natR mre11-H125N::URA3 trp1Δ |
| MB128 | 4741MATa mre11Δ::natR mre11-D56N::URA3 pif1Δ::HIS3 bar1Δ::kanMX |
| MB129 | 4741 MATa mre11Δ::natR mre11-H125N::URA3 pif1Δ::HIS3 bar1Δ::kanMX |
| MB203 | 4741 MATa dna2Δ::kanMX pif1Δ::HIS3 trp1Δ |
| MB213 | 4741 MATa dna2Δ::natR pif1Δ::HIS3 trp1Δ |
| MB161B | 4741 MATa dna2Δ::kanMX pif1Δ::HIS3 trp1Δ bar1Δ::kanMX |
| MB161B-56 | 4741 MATa dna2Δ::kanMX pif1Δ::HIS3 trp1Δ bar1Δ::kanMX mre11-D56N |
| MB161B-56 | (pRS414MRE11) |
| MB161B-56 | (pRS314DNA2) |
| MB161B-125 | 4741 MATa dna2Δ::kanMX pif1Δ::HIS3 trp1Δ bar1Δ::kanMX mre11-H125N |
| MB161B-125 | (pRS414MRE11) |
| MB161B-125 | (pRS314DNA2) |
| MB161B-125 | (pRS314dna2-K1080E) |
| MB161B-125 | (pRS314dna2-E675A) |
| MB161B-125 | (pRS314dna2-K1080E,K673A) |
| MB161B-125 | (pRS314dna2-2) |
| MB130 | 4741 MATa dna2Δ::kanMX pif1Δ::HIS3 trp1Δ bar1Δ::kanMX mre11Δ::natR (pRS414MRE11) |
| MB131 | 4741 MATa dna2Δ::kanMX pif1Δ::HIS3 trp1Δ bar1Δ::kanMX mre11Δ::natR (pRS414mre11-D56N) |
| MB132 | 4741 MATa dna2Δ::kanMX pif1Δ::HIS3 trp1Δ bar1Δ::kanMX mre11Δ::natR (pRS414mre11-H125N) |
| MB133 | 4741 MATα dna2Δ::kanMX pif1Δ::HIS3 trp1Δ bar1Δ::kanMX mre11Δ::natR mre11-D56N::URA3 |
| MB134 | 4741 MATα dna2Δ::kanMX pif1Δ::HIS3 trp1Δ bar1Δ::kanMX mre11Δ::natR mre11-H125N::URA3 |
| MB135 | 4742 MATα sgs1Δ::kanMX sae2Δ::natR |
| MB136 | 4742 MATα sae2Δ::natR |
| MB137 | 4742 MATα sgs1Δ::kanMX |
| MB138 | 4741 MATα sgs1Δ::kanMX mre11Δ::natR mre11-H125N::URA3 |
X-ray Treatment
Cells were treated with X-rays as previously described using a Pantak MKII X-ray machine operated at 20 mA and 70 kev [31]. The dosimetry was determined using a Radcaλ; 0.6 cubic centimeter ion chamber (model 10X5-0.6, serial no. 9352) connected to a Radiation Monitor Controller (model 9010; serial no. 90-2910).
Acknowledgments
We thank Kevin Lewis for the cloned wild-type and mutant MRE11 genes.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by NIH GM087666, the Philip Morris External Research Program, and the Ellison Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 2.Ivanov EJ, Sugawara N, White CI, Fabre F, Haber JE. Mutants of XRS2 and RAD50 delay, but do not prevent, mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 4.D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 5.D'Amours D, Jackson SP. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 2001;15:2238–2249. doi: 10.1101/gad.208701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 7.Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 8.Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- 9.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 10.Moncalian G, Lengsfeld B, Bhaskara V, Hopfner KP, Karcher A, et al. The rad50 signature motif: essential to ATP binding and biological function. J Mol Biol. 2004;335:937–951. doi: 10.1016/j.jmb.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 11.Alani E, Subbiah S, Kleckner N. The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics. 1989;122:47–57. doi: 10.1093/genetics/122.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Resnick MA, Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976;143:119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 15.Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis LK, Storici F, Van Komen S, Calero S, Sung P, et al. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics. 2004;166:1701–1713. doi: 10.1534/genetics.166.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llorente B, Symington LS. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol. 2004;24:9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis LK, Karthikeyan G, Westmoreland JW, Resnick MA. Differential suppression of DNA repair deficiencies of Yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics. 2002;160:49–62. doi: 10.1093/genetics/160.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bae S-H, Seo Y-S. Characterization of the enzymatic properties of the yeast Dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J Biol Chem. 2000;275:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 23.Bae S-H, Choi E, Lee K, Park J, Lee S, et al. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J Biol Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 24.Masuda-Sasa T, Polaczek P, Campbell JL. Single strand annealing and ATP-independent strand exchange activities of yeast and human DNA2: possible role in Okazaki fragment maturation. J Biol Chem. 2006;281:38555–38564. doi: 10.1074/jbc.M604925200. [DOI] [PubMed] [Google Scholar]
- 25.Budd ME, Choe W-C, Campbell JL. The nuclease activity of the yeast Dna2 protein, which is related to the RecB-like nucleases, is essential in vivo. J Biol Chem. 2000;275:16518–16529. doi: 10.1074/jbc.M909511199. [DOI] [PubMed] [Google Scholar]
- 26.Budd ME, Campbell JL. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budd ME, Choe W-C, Campbell JL. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J Biol Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 28.Budd ME, Campbell JL. A new yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc Natl Acad Sci USA. 1995;92:7642–7646. doi: 10.1073/pnas.92.17.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bae SH, Bae K-H, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 30.Formosa T, Nitiss T. Dna2 mutants reveal interactions with DNA polymerase alpha and Ctf4, a Pol α accessory factor, and show that full DNA2 helicase activity is not essential for growth. Genetics. 1999;151:1459–1470. doi: 10.1093/genetics/151.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Budd ME, Campbell JL. The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat Res. 2000;459:173–186. doi: 10.1016/s0921-8777(99)00072-5. [DOI] [PubMed] [Google Scholar]
- 32.Choe W, Budd M, Imamura O, Hoopes L, Campbell JL. Dynamic localization of an Okazaki fragment processing protein suggests a novel role in telomere replication. Mol Cell Biol. 2002;22:2002–2017. doi: 10.1128/MCB.22.12.4202-4217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budd ME, Tong AH, Polaczek P, Peng X, Boone C, et al. A network of multi-Tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 2005;1:634–650. doi: 10.1371/journal.pgen.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bessler JB, Torre JZ, Zakian VA. The Pif1p subfamily of helicases: region-specific DNA helicases? Trends in Cell Biology. 2001;11:60. doi: 10.1016/s0962-8924(00)01877-8. [DOI] [PubMed] [Google Scholar]
- 36.Fiorentino DF, Crabtree GR. Characterization of Saccharomyces cerevisiae dna2 mutants suggests a role for the helicase late in S phase. Mol Biol Cell. 1997;8:2519–2537. doi: 10.1091/mbc.8.12.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, et al. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 38.Gangloff S, de Massy B, Arthur L, Rothstein R, Fabre F. The essential role of yeast topoisomerase III in meiosis depends on recombination. Embo J. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imamura O, Campbell JL. The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc Natl Acad Sci USA. 2003;100:8193–8198. doi: 10.1073/pnas.1431624100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weitao T, Budd M, Campbell JL. Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutation Res. 2003;532:157–172. doi: 10.1016/j.mrfmmm.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 41.Lee K-H, Kim DW, Bae S-H, Kim J-A, Ryu G-H, et al. The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucl Acids Res. 2000;28:2873–2881. doi: 10.1093/nar/28.15.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank CJ, Hyde M, Greider CW. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell. 2006;24:423–432. doi: 10.1016/j.molcel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 43.Tomita K, Kibe T, Kang H-Y, Seo Y-S, Uritani M, et al. Fission yeast Dna2 is required for generation of the telomeric single-strand overhang. Mol Cell Biol. 2004;24:9557–9567. doi: 10.1128/MCB.24.21.9557-9567.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parenteau J, Wellinger RJ. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol Cell Biol. 1999;19:4143–4152. doi: 10.1128/mcb.19.6.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Z, Chung W-H, Shi EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jinks-Robertson S, Michelitch M, Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ira G, Haber JE. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol Cell Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner M, Price G, Rothstein R. The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics. 2006;174:555–573. doi: 10.1534/genetics.104.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weitao T, Budd M, Mays Hoopes LL, Campbell JL. Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J Biol Chem. 2003;278:22513–22522. doi: 10.1074/jbc.M301610200. [DOI] [PubMed] [Google Scholar]
- 51.Ivessa AS, Zhou J-Q, Zakian V. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have oppostie effects on replication fork progression in the ribsomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 52.Vernon M, Lobachev K, Petes TD. High rates of “unselected” aneuploidy and chromosome rearrangements in tel1 mec1 haploid yeast strains. Genetics. 2008;179:237–247. doi: 10.1534/genetics.107.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]