Abstract
The Rex protein of human T-cell leukemia virus (HTLV) acts posttranscriptionally to induce the cytoplasmic expression of the unspliced and incompletely spliced viral RNAs encoding the viral structural and enzymatic proteins and is therefore essential for efficient viral replication. Rex function requires nuclear import, RNA binding, multimerization, and nuclear export. In addition, it has been demonstrated that the phosphorylation status of HTLV-2 Rex (Rex-2) correlates with RNA binding and inhibition of splicing in vitro. Recent mutational analyses of Rex-2 revealed that the phosphorylation of serine residues 151 and 153 within a novel carboxy-terminal domain is critical for function in vivo. To further define the functional domain structure of Rex-2, we evaluated a panel of Rex-2 mutants for subcellular localization, RNA binding capacity, multimerization and trans-dominant properties, and the ability to shuttle between the nucleus and the cytoplasm. Rex-2 mutant S151A,S153A, which is defective in phosphorylation and function, showed diffuse cytoplasmic staining, whereas mutant S151D,S153D, previously shown to be functional and in a conformation corresponding to constitutive phosphorylation, displayed increased intense speckled staining in the nucleoli. In vivo RNA binding analyses indicated that mutant S151A,S153A failed to efficiently bind target RNA, while its phosphomimetic counterpart, S151D,S153D, bound twofold more RNA than wild-type Rex-2. Taken together, these findings provide direct evidence that the phosphorylation status of Rex-2 is linked to cellular trafficking and RNA binding capacity. Mutants with substitutions in either of the two putative multimerization domains or in the putative activation domain-nuclear export signal displayed a dominant negative phenotype as well as defects in multimerization and nucleocytoplasmic shuttling. Several carboxy-terminal mutants that displayed wild-type levels of phosphorylation and localized to the nucleolus were also partially impaired in shuttling. This is consistent with the hypothesis that the carboxy terminus of Rex-2 contains a novel domain that is required for efficient shuttling. This work thus provides a more detailed functional domain map of Rex-2 and further insight into its regulation of HTLV replication.
Human T-cell leukemia virus types 1 and 2 (HTLV-1 and HTLV-2) are closely related complex retroviruses that have been causally associated with a variety of human diseases. HTLV-1 is associated with adult T-cell leukemia, an aggressive CD4+ T-cell malignancy, and a chronic neurodegenerative disorder termed HTLV-associated myelopathy/tropical spastic paraparesis (HAM/TSP) (24, 41, 69). HTLV-2 is less clearly associated with disease, with only a few cases of leukemia or neurological disease reported (34, 55, 56). The genetic basis for the difference in pathobiology of HTLV-1 and HTLV-2 is not yet clear but likely resides in the activities of the regulatory and/or accessory proteins, thus highlighting the importance of comparative structure and function studies of the viral gene products.
In addition to the structural and enzymatic genes, gag, pol, and env, HTLV encodes the Tax and Rex trans-regulatory gene products essential for viral replication as well as several accessory gene products shown to be important for viral persistence in vivo (17, 18). The role of the Tax trans-activator protein in HTLV replication and cellular transformation is quite well established (3, 42, 57, 61). Tax increases the rate of transcription from the viral long terminal repeat (LTR) and modulates the transcription or activity of numerous cellular genes involved in cell growth and differentiation, cell cycle control, and DNA repair (4, 5, 37, 51, 66, 68). Strong evidence indicates that these pleiotropic effects of Tax on cellular processes are critical in HTLV-mediated oncogenesis (20, 29, 46, 53, 57).
HTLV Rex is a trans-acting regulatory protein involved in preferential binding, stabilization, and selective export of unspliced and incompletely spliced viral RNA transcripts from the nucleus to the cytoplasm (10, 39). Rex function is mediated by a cis-acting Rex response element (RxRE) located in the R region of the viral LTR (8, 48, 65, 71). Mutational analyses of HTLV-1 Rex (Rex-1) have defined several domains within the protein that are critical for function (11, 13, 32, 36, 49, 52, 60, 70). These include the arginine-rich N-terminal RNA binding domain (RBD) that overlaps with a nuclear localization signal (NLS), a leucine-rich activation domain encompassing the nuclear export signal (NES), and two regions flanking the NES that are important for Rex-Rex multimerization. Rex-1 and Rex-2 and their RNA response elements are structurally similar and functionally interchangeable (38), suggesting a similar domain structure. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of HTLV-2-infected cells revealed two major forms of Rex-2 migrating at about 24 and 26 kDa (54, 59) whose size differences are due to a phosphorylation-induced conformational change rather than to differences in amino acid sequence (25-27). Studies have demonstrated that phosphorylation of Rex-2 is required for efficient RNA binding and inhibition of splicing in vitro and may be linked to subcellular compartmentalization (9, 16, 28, 71). Recent mutational analyses of Rex-2, in conjunction with amino acid alignment and sequence comparison with Rex-1, revealed a similar functional domain structure between the two proteins but also identified potential differences, including a novel carboxy-terminal domain in Rex-2 important for function (45). Phosphorylation of Rex-2 on two serine residues (S151 and S153) within this carboxy-terminal domain is critical for the ability of the protein to augment expression of RxRE-containing mRNA (45). While Rex-1 (p27rex-1) is known to be phosphorylated on three major sites, a conclusive functional role for phosphorylation has not been directly established for this protein (1, 2).
The goal of the present study was to further define the functional domain structure of Rex-2. We evaluated a panel of Rex-2 mutants (45) for various biochemical properties, including subcellular localization, RNA binding capacity, multimerization and trans-dominant properties, and the ability to shuttle between the nucleus and the cytoplasm. Carboxy-terminal mutant S151A,S153A showed diffuse cytoplasmic staining and was defective for in vivo binding to RxRE-containing RNA. On the contrary, mutant S151D,S153D, previously shown to mimic a constitutively phosphorylated active conformation, displayed increased accumulation in the nucleoli and in nucleolar speckles and was complexed with twofold more target RNA than wild-type Rex-2 (wtRex-2). Two mutants with substitutions in the putative multimerization domains and a mutation in the putative NES exhibited dominant negative activity, were defective for multimerization, and were partially impaired in shuttling. Two carboxy-terminal mutants that displayed partial functional activity also showed a shuttling defect. A more detailed understanding of the regions of Rex-2 required for biological activity is provided by this study and will facilitate future analyses aimed at determining how its function modulates viral gene expression and ultimately the pathobiology of HTLV.
MATERIALS AND METHODS
Cells.
293T and HeLa-Tat (58) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM glutamine.
Plasmids.
The Rex expression vector BC20.2, containing the HTLV-2 tax-rex cDNA expressed from the cytomegalovirus (CMV) immediate-early gene promoter, has been described previously (15, 27). Various rex mutants were generated by site-directed mutagenesis (Quickchange; Stratagene) by using BC20.2 as the template. Mutations were confirmed by dideoxy DNA sequencing. The human immunodeficiency virus type 1 (HIV-1) Tat expression vector, pctat, contains the HIV-1 tat cDNA cloned downstream of the CMV promoter. pCgagRxRE-II contains the HIV-1 LTR promoter and gag gene linked to a 445-bp fragment of HTLV-2 spanning the RxRE (nucleotides 316 to 760 of the R-U5 region) (16). CMV-luciferase plasmid was used to control for transfection efficiency in each experiment (Luciferase Assay System; Promega).
Indirect immunofluorescence.
To determine the subcellular localization of HTLV-2 Rex mutants, 7 × 104 HeLa-Tat cells were plated overnight and subsequently transfected with 2 μg of BC20.2 expressing wtRex-2 or various Rex-2 mutants or with control plasmid (BC12). At 48 h posttransfection, cells were washed and fixed for 30 min at room temperature with 4% paraformaldehyde in phosphate-buffered saline. After incubation with anti-Rex-2 primary antibody (1:250 dilution in phosphate-buffered saline) for 1 h at room temperature, cells were washed and incubated with a 1:200 dilution of Alexa 488-conjugated goat anti-rabbit secondary antibody (Molecular Probes) for 1 h at room temperature. Cells were washed with phosphate-buffered saline and photographed with a Nikon TE 300 quantum fluorescence microscope. For analysis of the subnuclear distribution of Rex-2 by laser scanning microscopy, transfected HeLa-Tat cells were triple-stained with rabbit anti-Rex-2 serum, goat anti-B23 serum (Santa Cruz), and mouse anti-Nup62 monoclonal antibody (Transduction Laboratories) followed by Alexa 488-conjugated chicken anti-rabbit (Molecular Probes), Alexa 546-conjugated donkey anti-goat (Molecular Probes), and CY5-conjugated donkey anti-mouse antibodies (Jackson ImmunoResearch). Images were obtained by using a Zeiss LSM 510 laser scanning microscope with an Apochromat ×63/1.4 oil immersion objective and ×2 zoom. Scans were made in the Multitracker mode with argon (488-nm emission), helium-neon (543-nm emission), and helium-neon (633-nm emission) lasers; the optical slice was set at <0.8 μm for the Rex-2 and B23 signals and <0.9 μm for the Nup62 signal.
Measurement of Rex-2 functional activity.
We transfected 293T cells (2 × 105) by using calcium phosphate with 1 μg of pctat, 3 μg of pCgagRxRE-II, 1 μg of CMV-luciferase, 5 μg of wtRex-2, and increasing concentrations (0, 0.2, 0.5, 2, and 5 μg) of various Rex-2 mutant expression plasmids. Total DNA per transfection was kept constant by using the empty rex-2 backbone vector BC12. At 48 h posttransfection, cell lysates were made as described previously (45), and luciferase activity in each sample was determined to control for transfection efficiency. HIV-1 p24gag levels in cell lysates were determined by using a p24gag enzyme-linked immunosorbent assay (p24 HIV antigen assay kit; Beckman Coulter). p24gag calibration curves were generated by using HIV-1 p24 antigen standards as described by the kit manufacturer (detection sensitivity was 1 pg/ml). All the experiments were performed in triplicate and normalized for transfection efficiency; the statistical significance of resulting data was calculated by using the analysis of variance with the MINITAB statistical software package (version 13.31).
In vivo multimerization assay.
The ability of wtRex-2 and Rex-2 mutants to multimerize in vivo was determined by using a mammalian matchmaker assay (11). The assay is based on chimeric expression plasmids producing Rex fused to the DNA binding domain of the yeast Gal4 transcription factor and to the activation domain of the herpes simplex virus VP16 transcription factor. For the Gal4 chimeras, the coding sequences of wtRex-2 or various Rex-2 mutants were amplified by PCR to generate a BamHI-SacI fragment. The PCR product was ligated in frame downstream of the first 147 codons of the Gal4 DNA binding domain contained within the pSG424 plasmid (a kind gift from H. Bogerd, Duke University) to produce the various Gal4-Rex-2 expression vectors. For the VP16 chimera, the coding sequence of wtRex-2, except for the stop codon, was amplified by PCR to generate a NcoI-BglII fragment. The PCR product was ligated upstream of the 78-residue acidic activation domain (amino acids [aa] 413 to 490) of the VP16 gene supplemented with a C-terminal stop codon to produce the Rex2-VP16 expression vector. The integrity of all plasmids was confirmed by dideoxy sequencing. The 293T cells were cotransfected by the Lipofectamine method (Gibco) with 0.4 μg of Gal4 chimeric expression vector, 0.4 μg of VP16 chimeric expression vector, and 0.2 μg of the luciferase reporter plasmid pG5Luc, containing five binding sites for Gal4, in addition to pCMV-β-galactosidase to control for transfection efficiency. As needed, BC12 plasmid DNA was added to maintain a constant DNA concentration. At 48 h posttransfection, cells were lysed in passive lysis buffer (Promega). To test for luciferase activity, 20 μl of cell lysate (adjusted for transfection efficiency by β-galactosidase activity) was added to 100 μl of luciferase assay buffer (Promega). The average luciferase values for three independent experiments are presented as the change in induction (n-fold) over Gal4Rex-2 alone (set at 1).
Isolation of RNA from Rex-2 immune complexes and RT-PCR.
Detection of RNA bound to a specific protein in vivo was performed as described previously (6, 7) with modifications. Briefly, 10 μg of various Rex-2 expression vectors or negative control (BC12) along with 5 μg of pCgagRxRE-II, 2 μg of pctat, and 2 μg of CMV-luciferase were electroporated into 107 293T cells at 250 V and 975 mF. At 48 h posttransfection, cell lysates were prepared in NP-40 lysis buffer (150 mM NaCl, 10 mM Tris [pH 8.0], 1.5 mM MgCl2, 0.65% NP-40) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, and 1 μg of pepstatin per ml) and 100 U of RNasin. Equivalent amounts of cell lysate based on transfection efficiency were immunoprecipitated by using anti-Rex-2 antiserum, and the RNA bound to Rex-2 immune complex was extracted with TRI reagent (Molecular Research Center, Inc.). Yeast tRNA (50 μg; Sigma) was added to the RNA extraction protocol to facilitate recovery of RNA bound to Rex-2. Equal amounts of total RNA (1 μg) were subjected to a coupled primer extension 25-cycle PCR containing HIV-1 gag-specific oligonucleotide primer pairs (6, 7, 39). The 50-μl volume-coupled primer extension PCR contained RNA, 0.25 mM deoxynucleoside triphosphates, 50 mM KCl, 10 mM Tris (pH 8.0), 1.5 mM MgCl2, 0.01% gelatin, 100 ng of 3′ (antisense) oligonucleotide, 50 ng of 5′ (sense) oligonucleotide that was 32P end labeled with T4 DNA kinase to a specific activity of approximately 2 × 108 cpm/μg, and 2.5 U of Taq DNA polymerase (Promega) in the presence or absence of 5 U of murine leukemia virus reverse transcriptase (Amersham). The reaction was performed in a Perkin-Elmer model 9600 thermal cycler as follows: 65°C for 10 min, 50°C for 8 min, and 95°C for 5 min followed by 25 cycles of 95°C for 1 min, 55°C for 2 min, and 72°C for 2 min. The reverse transcriptase PCR (RT-PCR) primers M667 (5′ GGCTAACTAGGGAACCCACTG 3′) and M668 (5′ CAGGTCCCTGTTCGGGCGCC 3′) were slightly modified from the previously published sequence to accommodate the detection of HIV-1 gag from HIV-1 HBX2 strain and to amplify a 161-bp fragment from pCgagRxRE-II mRNA. PCR-amplified products were separated on a 6% polyacrylamide gel and visualized and quantitated by phosphorimager analysis. Intensities of the 161-bp fragment were quantified over three independent experiments and expressed as the percentage of the signal obtained for wtRex-2; significance relative to wtRex-2 was calculated with the Student's t test (MINITAB software).
Rex shuttling assay.
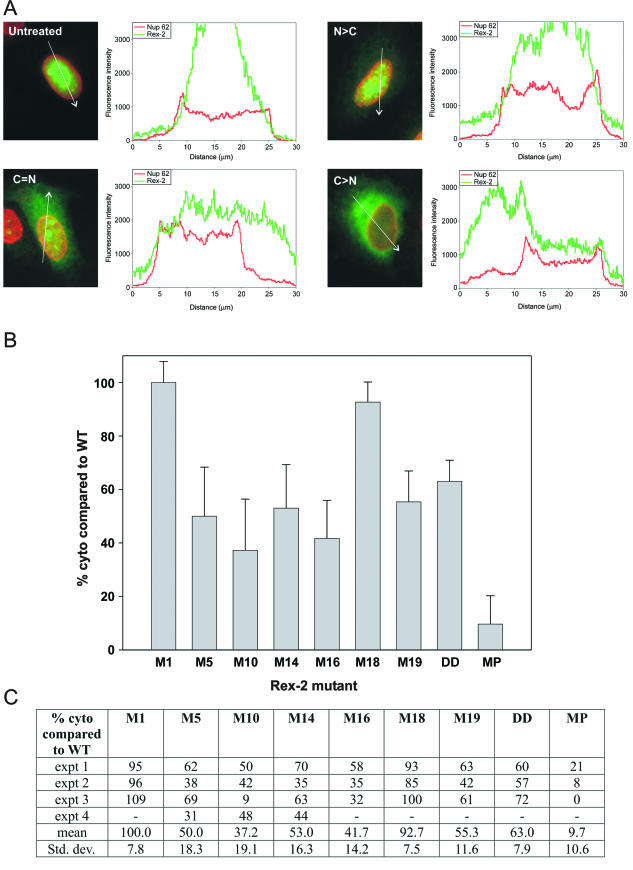
HeLa-Tat cells (2 × 105) were seeded onto 35-mm tissue culture plates. The next day, the cells were transfected with Rex-2 plasmids using Fugene 6 (Roche). Twenty-four hours later, the cultures were pretreated for 30 min with 50 μg of cycloheximide per ml and then incubated for 3 h in the presence of both 50 μg of cycloheximide per ml and 5 μg of actinomycin D (Sigma) per ml prepared in ethanol; control cultures were incubated with cycloheximide and an equivalent amount of ethanol. The cells were then rinsed twice with phosphate-buffered saline, fixed for 20 min with 3.7% formaldehyde-phosphate-buffered saline, and permeabilized for 10 min with 0.05% NP-40-phosphate-buffered saline. Indirect immunofluorescence was carried out by using a mixture of rabbit anti-Rex and mouse anti-Nup62 antibodies (used to identify the nucleus; Transduction Laboratories) followed by Alexa 488-conjugated anti-rabbit and Alexa 546-conjugated anti-mouse secondary antibodies (Molecular Probes). The stained cells were examined with an Olympus IX70 fluorescence microscope and photographed. Rex-expressing cells were divided into two categories of Rex localization: nuclear > cytoplasmic and cytoplasmic ≥ nuclear. To verify the validity of the visual scoring method, the distribution of Rex in representative cells showing these different patterns was quantified by laser scanning microscopy with a Zeiss LSM 510 microscope with a ×63/1.4 oil immersion objective. Scans were obtained with the pinhole fully opened using argon and helium-neon laser sources to detect the Rex and Nup62 signals, respectively; the distributions of the two signals in the nucleus and cytoplasm were quantified with the Profile software tool (see Fig. 5A).
FIG.5.
Response of Rex-2 mutants to actinomycin D treatment. HeLa-Tat cells were transfected with plasmids expressing either wt or mutant Rex-2 and then treated with actinomycin D, analyzed by indirect immunofluorescence, and separated into nuclear > cytoplasmic (N > C) and cytoplasmic ≥ nuclear (C ≥ N) categories, all as described in Materials and Methods. (A) Examples of untreated and actinomycin D-treated cells expressing wtRex-2 that were quantitated using the Zeiss profile software tool to compare the intensities of the Rex-2 (green) and Nup62 (red) signals along a line indicated by the white arrow. Results are presented as a graph showing the relative intensities of the two signals (y axis) against the distance (in micrometers) from the origin of the arrow (x axis), with the Nup62 signal plateaus corresponding to the nuclear compartment; cells in images labeled C = N and C > N were classified in the C ≥ N category. (B) Bars represent the average extent of cytoplasmic accumulation (i.e., percentage of cells in C ≥ N category) relative to that observed for wtRex-2 tested in at least three independent experiments, as summarized in the table shown in panel C. Control mutant MP, containing a Ser-to-Pro substitution at residue 92 within the NES, was generated by site-directed mutagenesis (Quickchange; Stratagene) of plasmid pLsRex2-AU1 (16).
RESULTS
Rex-2 functional domains.
In a recent study, we mutated serine and threonine residues throughout the 170-amino-acid Rex-2 protein to identify regions or domains important for function, with specific emphasis on identifying phosphorylation mutants. We identified a panel of Rex-2 mutants that were significantly defective in function (Table 1). Based on previous reports and on alignment with Rex-1, we predicted that the defect in several of these mutants (M1, M5, M10, and M14) would be in RNA binding and nuclear localization (aa 1 to 19; M1), activation and nuclear export (aa 81 to 94; M10), or multimerization (aa 57 to 66 and 106 to 124; M5 and M14, respectively). Interestingly, several nonfunctional Rex-2 mutants (M16, M17, M18, and M19) mapped to a novel region or domain at the carboxy terminus (aa 144 to 164) (45). Furthermore, phosphorylation of Rex-2 on two serine residues (S151 and S153) within this carboxy-terminal domain was shown to be critical for function (45). In the present study, we further evaluated Rex-2 mutants for key biochemical properties, including subcellular localization, RNA binding capacity, multimerization and trans-dominant properties, and the ability to shuttle from the nucleus to the cytoplasm to better define Rex-2 functional domain structure.
TABLE 1.
Biological activity of Rex-2 proteins
| Rex-2 proteins | Positions (aa)a | Mutationsb | % Biological activity relative to wtRexc |
|---|---|---|---|
| wtRex-2 | 100 | ||
| Rex-2 M1 | 4, 9 | T, T → A, A | 54 |
| Rex-2 M5 | 65, 71 | S, S → A, A | 20 |
| Rex-2 M10 | 92, 95 | S, S → A, A | 2 |
| Rex-2 M14 | 125, 131, 132 | S, S, T → A, A, A | 30 |
| Rex-2 M16 | 144, 147, 148 | S, S, S → A, A, A | 35 |
| Rex-2 M17 | 151, 153 | S, S → A, A | 25 |
| Rex-2 DD | 151, 153 | S, S → D, D | 200 |
| Rex-2 M18 | 158 | S → A | 45 |
| Rex-2 M19 | 162, 163, 164 | T, S, T → A, A, A | 30 |
Positions mutated within the 170-aa HTLV-2 Rex.
S, serine; T, threonine; A, alanine; D, aspartic acid.
Biological activity relative to wtRex-2 (set at 100%) and determined to be statistically significant by Student's t test. Data were taken from Narayan et al. (45).
Subcellular localization of Rex-2 mutants.
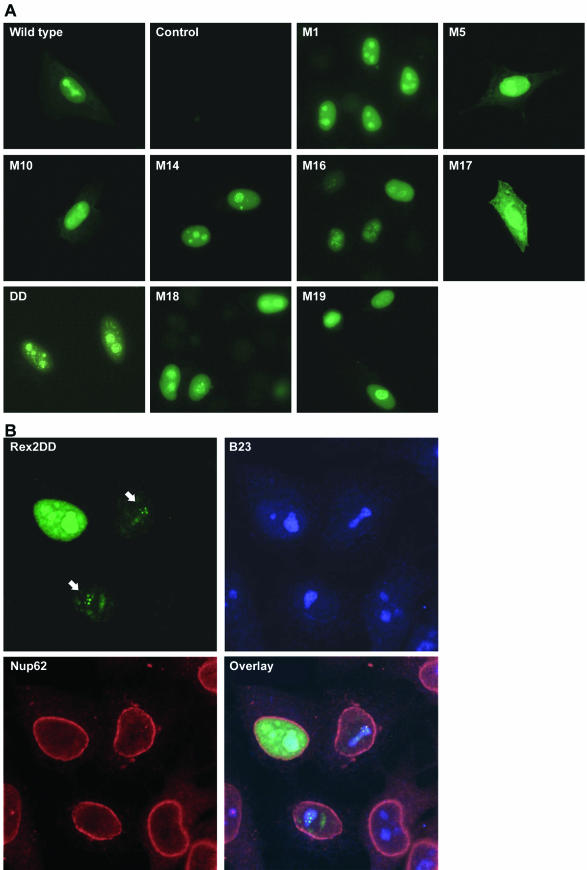
Previous studies have indicated that Rex and its HIV-1 homologue Rev localize primarily to the nucleus and nucleolus. We and others have presented data indicating that a phosphorylated form of Rex-2 found primarily in the nucleus is the active form of the protein (9, 16, 28, 71). Indirect immunofluorescence was used to determine whether the functional defects of the Rex-2 mutants could be attributed to improper subcellular localization. HeLa-Tat cells were transfected with wtRex-2 or various Rex-2 mutants, and Rex subcellular localization was evaluated. As expected, wtRex-2 exhibited predominant nuclear and nucleolar staining (Fig. 1A), with additional weak cytoplasmic staining occurring in about 20% of the cells (data not shown). In contrast, no fluorescence was detected in mock-transfected cells or cells stained with preimmune serum only (Fig. 1A). The majority of the functionally defective Rex-2 mutants tested displayed a subcellular distribution similar to that of wtRex-2. In contrast, M17 (S151A,S153A) showed diffuse cytoplasmic accumulation with some nuclear localization. We previously reported that serine residues 151 and 153 are critical phosphorylation sites and that replacement of both serines with phosphomimetic aspartic acid residues (S151D,S153D) locks the protein in a phosphorylated conformation with greater functional activity than wtRex-2 (45). Interestingly, mutant S151D,S153D (DD) exhibited increased accumulation in nucleoli and nucleolar speckles compared to wtRex-2. These data are consistent with the hypothesis that a Rex-2 conformation favored under normal circumstances by phosphorylation or artificially induced by phosphomimetic amino acid substitution (aspartic acid) is linked to nucleolar localization. To examine the speckle pattern of mutant DD in greater detail, we analyzed DD-transfected cells by confocal microscopy after staining them with the anti-Rex-2 antibody, an antibody against the nuclear envelope component Nup62 (added to delineate the nucleus), and an antibody recognizing B23, a multifunctional nucleolar protein that is proposed to act as a chaperone for nuclear import, nucleolar accumulation, and functional activity of Rev and Rex (21, 44, 62-64). The resulting images shown in Fig. 1B illustrate accumulation of mutant DD (green signal) in the nucleus and nucleoli, which were identified by the red Nup62 and blue B23 signals, respectively, as well as in intensely staining speckles within the nucleoli (indicated by arrows). The DD and B23 signals showed clear overlap, as indicated by the pale blue-green and white colors in the overlay panel. Interestingly, a similar pattern of accumulation in nucleolar speckles was previously reported for Rex of HTLV-1 in experiments carried out using Rex-1-transfected cells and cell lines chronically infected with HTLV-1 (47).
FIG.1.
Subcellular localization of Rex-2 mutants. The top set of panels (A) shows anti-Rex-2 immunofluorescence assays of HeLa-Tat cells that were transfected with wild-type rex or various rex mutant expression vectors. The bottom set of panels (B) shows laser scanning microscopy images of HeLa-Tat cells that were transfected with mutant DD and subjected to indirect immunofluorescence staining using antibodies against Rex-2, B23, and Nup62; the arrows point to DD-containing nucleolar speckles, and the pale blue-green and white areas in the overlay image indicate colocalization of DD with B23. Immunofluorescence assays were carried out as described in Materials and Methods.
Rex phosphorylation status is important for RNA binding.
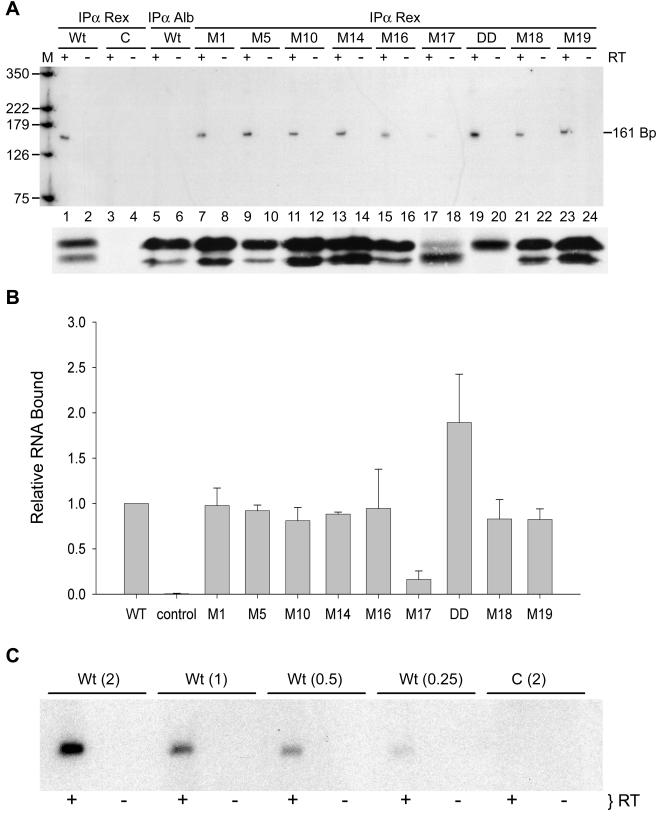
We next evaluated our panel of Rex-2 mutants for their ability to bind RxRE-containing RNA in living cells. The RNA binding capacity of each mutant protein was evaluated with a previously described quantitative functional bioassay based on the Rex-dependent reporter plasmid, pCgagRxRE-II, in conjunction with immunoprecipitation and RT-PCR. pCgagRxRE-II contains the HIV-1 LTR and gag gene linked to the RxRE of HTLV-2 and the simian virus 40 polyadenylation signal site; its efficient expression of Gag is dependent on Tat-mediated transcription and functional Rex binding to the RxRE (16). 293T cells were cotransfected with pCgagRxRE-II, pctat, the transfection standard CMV-luciferase, and either wtRex-2, Rex-2 mutants, or the corresponding empty vector. Cell lysates were assayed for luciferase activity, and equivalent amounts based on transfection efficiency were immunoprecipitated by using anti-Rex-2 serum or anti-albumin control serum; RNA was then isolated from the immune complexes and subjected to RT-PCR. In addition, we subjected equivalent amounts of cell lysate based on transfection efficiency to immunoprecipitation, SDS-PAGE, and Western blot analysis to confirm our previous results that the Rex-2-specific antiserum used in this study immunoprecipitates wtRex-2 and each of the Rex-2 mutant proteins with similar efficiency (45). Data from a representative experiment are presented in Fig. 2A. The expected 161-bp HIV-1 gag-specific product was amplified from wtRex-2-specific immune complexes (Fig. 2A, lane 1) but not from anti-albumin immune complexes (Fig. 2A, lane 5). In addition, we did not detect a Gag-specific product in cells transfected with control vector (Fig. 2A, lane 3). Furthermore, the 161-bp fragment was amplified only in the presence of RT, indicating that the template for PCR amplification was RNA and not contaminating plasmid DNA. Figure 2B summarizes the in vivo RNA binding data obtained for various Rex-2 mutants, with results expressed as the mutant/wtRex-2 ratios averaged over three independent experiments. Mutants M17 (S151A,S153A) and DD (S151D,S153D) displayed significant differences in RxRE binding compared to wtRex-2 (Student's t test, P < 0.001). M17 had a binding capacity of 10% of that of wtRex-2, whereas mutant DD consistently bound twofold more RNA than wtRex-2. To determine whether our RT-PCR approach was quantitative, sequential twofold dilutions of RNA isolated from the wtRex-2 immune complexes were subjected to RT-PCR (Fig. 2C). The signals produced were quantitative across a fourfold range (0.25 to 2.0 μg of RNA), revealing increasing signal intensities from increasing amounts of RNA. In addition, the low concentration of RNA used in this assay was within the linear range. These results provide direct in vivo evidence that the phosphorylation status of Rex-2 is important for RNA binding and that the amount of RNA bound is consistent with the subcellular localization (Fig. 1) and previously described functional capacity (45) of these mutants. It is important to note that mutant M1, containing amino acid substitutions in the amino-terminal RNA binding-NLS domain, did not display significant alterations in RNA binding capacity or subcellular localization.
FIG. 2.
In vivo RNA binding assay reveals that phosphorylation-induced Rex-2 conformation is important for RNA binding. (A) Gag RNA bound to Rex (α Rex) or Albumin (α Alb) was detected by RT-PCR as described in Materials and Methods. Equal amounts of RNA were subjected to coupled-primer extension 25-cycle PCR in the presence (+) or absence (−) of reverse transcriptase. The primer pairs were designed to amplify a 161-bp fragment of HIV-1 gagRxRE-containing reporter RNA. The PCR product was separated on a 6% polyacrylamide gel and visualized byautoradiography or with a phosphorimager. M, size markers (positions are indicated in base pairs). Levels of wtRex-2 and mutant Rex-2 (shown below the RT-PCR panel) were measured in equivalent amounts of cell lysate based on transfection efficiency following immunoprecipitation, SDS-PAGE, and Western blot analysis. (B) Quantitation of the 161-bp fragment. The numbers, which represent relative phosphorimager-quantified values of the 161-bp PCR product for three independent experiments, are normalized to the value obtained for wtRex-2 (set to 1). Error bars indicate standard deviations. Significance relative to wtRex-2 was determined by the Student's t test; *, values that are statistically different. (C) Quantitation of RT-PCR. Sequential twofold dilutions of RNA (0.25 to 2.0 μg) isolated from the wtRex-2 immune complexes were subjected to RT-PCR and analyzed as described for panel A.
Rex-2 dominant negative mutants fail to multimerize.
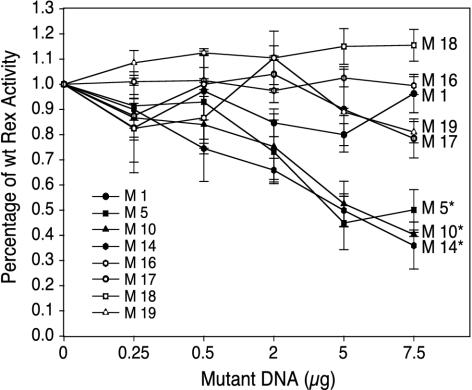
The Rex-2 mutants were next tested for their capacity to block the biological action of wtRex-2 by using the pCgagRxRE-II reporter assay described above. 293T cells were cotransfected with pCgagRxRE-II, pctat, wtRex-2, and increasing concentrations of Rex-2 mutants (up to a 1.5 M excess). As shown in Fig. 3, mutants M5, M10, and M14 significantly inhibited the activity of wtRex-2 (P < 0.01) and thus displayed a dominant negative phenotype. In contrast, M1, M16, M17, M18, and M19 displayed a recessive negative phenotype, as the action of wtRex-2 was not significantly altered in their presence.
FIG. 3.
Rex-2 functional assay reveals that mutants M5, M10, and M14 inhibit the function of wtRex-2. 293T cells were transfected with 1 μg of pctat, 3 μg of pCgagRxRE, 1 μg of CMV-luciferase, 5 μg of wtRex-2 plasmid, and increasing concentrations of various Rex-2 mutants. Cell lysates were prepared 48 h posttransfection, and p24 Gag levels were determined by HIV-1 p24 Gag enzyme-linked immunosorbent assay. *, statistically significant curves determined by analysis of variance.
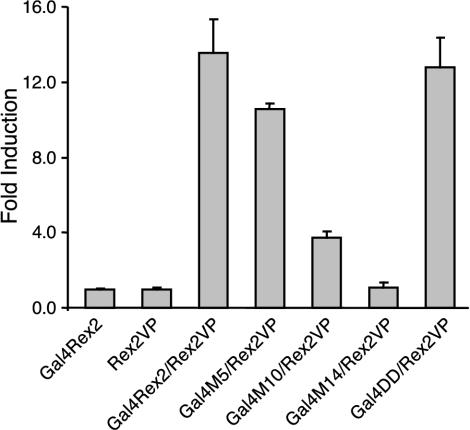
Several laboratories have reported that Rex-1 and HIV-1 Rev dominant negative mutants map to either multimerization or activation-NES domains, both of which are important for protein-protein interactions (11, 14, 35, 40, 67). Based on amino acid sequence alignment of Rex-2 and Rex-1, we predicted that the M5 and M14 mutations would disrupt the protein's multimerization domain and that the M10 mutation would disrupt the activation-NES domain. To confirm this hypothesis, we next evaluated whether these mutants were defective in multimerization in vivo by using a modified version of a previously described transcriptional activation assay (11). This assay screens for multimerization based on the ability of separate Gal4 DNA binding domain chimeras and VP16 activation domain chimeras to activate transcription when tethered together via multimerizing fusion partners (11). Gal4-wtRex-2, wtRex-2-VP16, and mutant Rex-2-VP16 gene fusion constructs were generated and cotransfected into 293T cells along with pG5Luc reporter plasmid. In this assay, expression of the pG5Luc reporter plasmid is expected only when the Gal4 and VP16 functional domains are juxtaposed through multimerization of the Rex-2 portion of the fusion proteins. As shown in Fig. 4, coexpression of Gal4-wtRex-2 and wtRex-2-VP16 induced approximately 14-fold activation of luciferase activity compared with the negative controls. Screening of each of the Rex-2 mutants in the context of the Gal4 expression vector revealed that the three dominant negative mutants (M5, M10, and M14) displayed markedly reduced capacity to interact with wtRex2 in vivo (Fig. 4), whereas other biologically inactive and active mutants displayed wtRex-2 multimerization activity (Fig. 4). Using this same approach, we further showed that mutants M5, M10, and M14 were also defective for self-interaction (data not shown).
FIG. 4.
Mammalian cell-based two-hybrid reporter assay reveals that mutants M5, M10, and M14 fail to efficiently interact or multimerize in vivo. 293T cells were cotransfected with 0.4 μg of Gal4 chimeric expression vector, 0.4 μg of VP16 chimeric expression vector, and 0.2 μg of the luciferase reporter plasmid pG5Luc. At 48 h posttransfection, cells were lysed and tested for luciferase activity. The average luciferase values for three independent experiments are presented as the change in induction (n-fold) over Gal4Rex-2 alone (set at 1).
Identification of mutations that affect shuttling.
We next tested the nucleocytoplasmic shuttling capacity of our panel of Rex-2 mutants in actinomycin D assays. Upon incubation with this drug, shuttling proteins that normally accumulate in the nucleus (e.g., Rex and Rev) are redistributed to the cytoplasm, whereas nuclear proteins that are either naturally nonshuttling or that are export defective because of mutations in their nuclear export signal are retained in the nucleus (43, 49). Initial examination of the treated cultures gave the impression that the panel of mutants responded to actinomycin D to various degrees, with none of them displaying complete nuclear retention in all cells. Therefore, we compared the response of each mutant to that of wtRex-2 in at least three independent experiments by counting cells exhibiting nuclear > cytoplasmic or cytoplasmic ≥ nuclear localization patterns (see Materials and Methods and Fig. 5A). For comparative purposes, we also tested the shuttling properties of Rex-2 mutant MP, which contained a Ser-to-Pro substitution at residue 92; this mutation corresponded to a previously described substitution made in HTLV-1 Rex that rendered the protein inactive and defective for shuttling (mutant S91P in reference 49). M17 was not tested, as the assay requires that the protein of interest efficiently localize to the nucleus prior to treatment. The graph and table shown in Fig. 5B and C summarize the results obtained, expressed as percent cytoplasmic accumulation compared to wtRex-2. M1 and M18 showed efficient accumulation in the cytoplasm, indicating that their functional defect is not due to impaired shuttling. In contrast, MP was almost completely defective for cytoplasmic accumulation, thus verifying that the NES of Rex-2 is structurally similar to that of Rex-1. All of the remaining mutants showed some degree of reduced accumulation in the cytoplasm compared to wtRex2, and could be ordered as follows: M10 < M16 < M5 < M14 < M19 < DD < wt.
DISCUSSION
In a recent study, we characterized a panel of Rex-2 mutants in which potential serine/threonine phosphorylation sites scattered throughout the protein were replaced with alanines, and we identified several mutations that substantially impaired functional activity. The present study focused on a subset of functionally relevant mutations which, based on sequence alignment with Rex-1, are predicted to fall within either the amino-terminal RBD-NLS (M1), the activation-NES domain (M10), or a bipartite multimerization domain flanking the NES (M5 and M14); additional mutations defined a functionally important carboxy-terminal domain not previously described for Rex-1 (M16, M17, M18, M19, and DD). The latter included two mutants in Ser151 and Ser153 that displayed opposite effects on Rex-2 activity: functionally defective M17, containing alanine substitutions, and hyperactive DD, containing aspartic acid substitutions. We took advantage of HTLV-1 Rex and HIV-1 Rev studies indicating that function is dependent on their ability to localize to the nucleus and nucleolus, specifically bind viral RNA, multimerize, and interact with cellular proteins facilitating export from the nucleus to the cytoplasm. Our results, summarized in Table 2, indicate that Rex-1 and Rex-2 have a similar RBD-NLS, activation-NES, and bipartite multimerization domain structures, and our results also provided further information regarding the novel multifunctional carboxy-terminal domain in Rex-2 (Fig. 6).
TABLE 2.
Summary of Rex-2 mutant proteins
| Rex-2 proteins | Localizationa | RNA bindingb | Multimerization/trans dominantc | Shuttlingd |
|---|---|---|---|---|
| wtRex-2 | Nuc | + | +/− | + |
| Rex-2 M1 | Nuc | + | +/− | + |
| Rex-2 M5 | Nuc | + | −/+ | ± |
| Rex-2 M10 | Nuc | + | −/+ | ± |
| Rex-2 M14 | Nuc | + | −/+ | ± |
| Rex-2 M16 | Nuc | + | +/− | ± |
| Rex-2 M17 | Cyt | ± | NDe/− | NDe |
| Rex-2 DD | Nuc | ++ | +/NDe | ± |
| Rex-2 M18 | Nuc | + | +/− | + |
| Rex-2 M19 | Nuc | + | +/− | ± |
Predominant subcellular localization. Nuc, nucleus and nucleolus; Cyt, cytoplasmic.
Positive (+), impaired (±), or enhanced (++) Rex binding to RxRE-containing RNA as determined by in vivo binding experiments.
Positive (+) or negative (−) for multimerization and trans-dominant capacity.
Positive (+) or impaired (±) for nucleocytoplasmic shuttling as determined by actinomycin D assays.
ND, not determined.
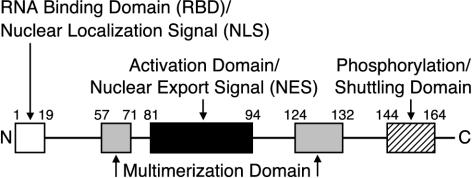
FIG. 6.
Domain structure of the HTLV-2 Rex protein. The functional domains or regions of the 170-amino-acid Rex protein based on this study and alignment with HTLV-1 Rex are indicated. RNA binding domain-NLS, aa 1 to 19. Core activation effector-NES domain, aa 81 to 94. Multimerization domains, aa 57 to 71 and 124 to 132. In Rex-2, the region spanning residues 144 to 164 is important for efficient function and includes key phosphorylation sites (Ser151 and -153). Mutations in this region also impair nuclear-to-cytoplasmic cycling.
Our subcellular localization analyses revealed that wtRex-2 and the majority of our panel of Rex-2 mutants localized primarily to the nucleus and nucleolus, with a subpopulation of nucleoli containing intensely staining speckles (Fig. 1), a pattern previously reported for Rex-1 (47). Interestingly, the Rex-2 mutants containing amino acid substitutions of serine residues 151 and 153 displayed subcellular distributions distinct from wtRex-2: functionally defective M17 (S151A,S153A) showed marked cytoplasmic accumulation, while hyperactive mutant DD (S151D,S153D) showed a more striking pattern of nucleolar speckles than did wtRex-2. Therefore, Ser151 and Ser153 appear to be particularly important for directing appropriate subcellular targeting of Rex-2. We previously showed that M17 corresponds to the 24-kDa hypophosphorylated form of the 26- and 24-kDa wtRex-2 doublet, whereas mutant DD, containing phosphomimetic aspartic acids, is locked in the fully phosphorylated conformation migrating at 26 kDa and exhibits greater functional activity than wtRex-2 (45). Together, these data indicate that a Rex-2 conformation normally conferred by full phosphorylation or artificially induced by phosphomimetic amino acid substitution is required for or tightly linked to nuclear and nucleolar localization and might favor accumulation of the protein in nucleolar speckles. A speculative explanation for this observation is that a phosphorylation-induced conformational change in Rex-2 stabilizes the protein or uncovers a domain, allowing it to efficiently interact with cellular components that direct import into the nucleus and/or favor nucleolar accumulation. Future characterization of M17 and DD will thus include assessment of their ability to bind to cellular proteins involved in these processes, such as importin β and B23.
The results of actinomycin D assays (Fig. 5) identified several Rex-2 mutants that were impaired in nuclear-to-cytoplasmic shuttling. Consistent with previous Rex-1 and HIV-1 Rev mutational studies, mutant M10, containing two Ser-to-Ala substitutions within the core activation-NES domain, showed the greatest defect in nuclear shuttling. This observation indicates that defective shuttling due to a disrupted NES contributes to M10's functional impairment, although substitution of Ser92 and -95 with alanines is apparently better tolerated than the single proline substitution at Ser92 in control mutant MP. Detailed studies of both Rev and Rex-1 showed that mutations in the NES interfere with the ability of the proteins to associate with CRM1, a cellular protein belonging to the importin β family that functions as a nuclear export receptor for NES-containing proteins and ribonucleoprotein complexes containing U snRNAs and 5S rRNA (reviewed in reference 22), some cellular mRNAs (23), the Rev- and Rex-dependent viral mRNAs encoded by complex retroviruses and lentiviruses (reviewed in reference 19), and mRNAs of several other viruses (50). It is therefore very likely that the M10 mutation interferes with the Rex-2-CRM1 interaction.
The partial shuttling defect observed for M16, M5, M14, M19 and DD was unexpected and could not be directly attributed to NES disruption, as the mutations in question are distant from this domain. One explanation is that these mutations introduce global conformational changes that affect the ability of the NES to interact with cellular factors. It is noteworthy that, like M10, M5 and M14 were also defective for multimerization (Fig. 4). Although this observation suggests that multimer formation might positively influence NES activity, such a hypothesis is not supported by results of studies of Rex-1, which showed that mutants lacking either of the two multimerization domains accumulated in the cytoplasm upon treatment with actinomycin D (33). It will therefore be interesting to directly compare the extent of actinomycin D-induced cytoplasmic accumulation of our Rex-2 multimerization domain mutants with the corresponding Rex-1 mutants and to determine whether they are all able to interact with CRM1.
The increased nuclear retention of M16, M19, and DD, the mutations in which are clustered in the extreme carboxy terminus of Rex-2, suggests that this region might constitute a domain that influences shuttling in conjunction with the NES. To date, there is no evidence suggesting a similar functional importance of the carboxy terminus of Rex-1. However, a recent study that examined sequences in HIV-1 Rev lying carboxy terminal to the protein's NES revealed that this region is also required for efficient binding to CRM1, thus facilitating the nuclear export leg, and augments formation of Rev-Rev multimers; the authors proposed that these carboxy-terminal sequences are important for Rev to express its full functional activity when present in low quantities within the cell—for example, during the initial phase of viral infection (31). The overall functional conservation between Rex and Rev suggests such a role for the carboxy terminus of Rex. It is tempting to propose that phosphorylation of the residues mutated in M16 and M19 (i.e., Ser144, Ser147, and Ser148 in M16; Thr162, Ser163, and Thr164 in M19) might have a direct impact on this aspect of Rex function. However, our initial characterization of M16 and M19 indicated that they display wt phosphorylation levels (45), suggesting that the residues in question are not phosphorylated. Nevertheless, we cannot rule out the possibility that they undergo transient phosphorylation events that are below the detection level of our analysis. In the case of DD, the mean 40% reduction in cytoplasmic accumulation observed suggests either that this level of impairment is not sufficient to impinge on functional activity or that it is counteracted by other properties of this mutant, e.g., its increased nucleolar localization and association with RxRE-containing RNA.
Our in vivo RxRE binding assays revealed that, among all the mutants tested, only M17 and DD differed substantially from wtRex-2. It is interesting that the amount of RNA bound correlated with altered subcellular localization. M17, found primarily in the cytoplasm, consistently displayed 10% of the binding capacity of wtRex-2, whereas DD, detected almost exclusively in the nucleolus and nucleolar speckles, bound approximately twofold more RNA than wtRex-2. We were surprised that mutant M1 (T4A,T9A), containing amino acid substitutions in the amino-terminal RBD-NLS domain, was not significantly impaired in RNA binding capacity or subcellular localization, despite the fact that its biological activity was approximately 50 to 60% of that of wtRex-2. One possible explanation for this result is that the amino-terminal domain of Rex-2 has other undefined properties important for full function, such as interacting with key cellular proteins.
Using a mammalian cell-based two-hybrid reporter assay, we showed that mutants M5, M10, and M14 all failed to efficiently interact or multimerize with wtRex-2 in vivo, with M14 being completely negative for multimerization (Fig. 4). Mutants M5 and M14 align with the two multimerization domains identified for Rex-1 (12, 33). Therefore, our data identify a similar bipartite multimerization domain structure in Rex-2. The multimerization-defective phenotype observed for M10 is consistent with observations made for an NES mutant of Rex-1 (RexM90), which was shown to be both shuttling defective and unable to efficiently multimerize due to a defect in CRM1 binding (30). Such a coupled defect in shuttling and multimerization has also been described for HIV-1 Rev activation-NES domain mutants (11, 40) and stresses the crucial role for a fully productive export receptor interaction for Rex-Rev functional activity.
We further showed that M5, M10, and M14 behave as dominant negative inhibitors of wtRex-2 function. Therefore, all Rex-2 mutants displaying dominant negative activity map to either the multimerization domains or the activation-NES domain. Our RNA binding data for multimerization domain mutants M5 and M14 indicate that they bind RxRE with efficiency similar to that of wtRex-2. This finding, taken together with data accumulated from studies of Rex-1 (30), suggests that although M5 and M14 are able to bind to the RxRE and interact with CRM1, their reduced ability to form large-scale multimers ultimately compromises the formation and export of functional ribonucleoprotein complexes containing sufficient amounts of CRM1 and other critical factors. The dominant negative activity of these mutants is in turn likely to reflect competition with wtRex-2 for RxRE molecules. The observation that M10 is competent for RxRE binding but defective for both shuttling and multimerization (likely due to the inability to associate with CRM1) suggests that the dominant negative activity of M10 results mainly from its binding of RxRE molecules that remain trapped in the nucleus due to the lack of a CRM1 interaction.
Taken together, our studies provide a more detailed map of the HTLV-2 Rex domain structure, underscore the structural similarities between Rex-2 and Rex-1, and provide further evidence that the carboxy terminus contains a novel multifunctional domain that regulates subcellular localization. Further studies are required to elucidate the precise role of this domain in Rex-2 function to ultimately understand the pathogenesis of HTLV-2.
Acknowledgments
We thank Vincenzo Ciminale for critical comments and Eliana Scarponi for technical assistance. We also thank Tim Vojt for figure preparation.
This work was supported by grants from the National Institutes of Health (grant numbers CA93584, CA100730, and TWO6053), the Istituto Superiore di Sanità, Progetto AIDS (grant number 40D.28), and the Associazione Italiana per la Ricerca sul Cancro.
REFERENCES
- 1.Adachi, Y., T. D. Copeland, C. Takahashi, T. Nosaka, A. Ahmed, S. Oroszlan, and M. Hatanaka. 1992. Phosphorylation of the Rex protein of human T-cell leukemia virus type I. J. Biol. Chem. 267:21977-21981. [PubMed] [Google Scholar]
- 2.Adachi, Y., T. Nosaka, and M. Hatanaka. 1990. Protein kinase inhibitor H-7 blocks accumulation of unspliced mRNA of human T-cell leukemia virus type I (HTLV-I) Biochem. Biophys. Res. Commun. 169:469-475. [DOI] [PubMed] [Google Scholar]
- 3.Akagi, T., H. Ono, H. Nyunoya, and K. Shimotohno. 1997. Characterization of peripheral blood T-lymphocytes transduced with HTLV-I Tax mutants with different trans-activating phenotypes. Oncogene 14:2071-2078. [DOI] [PubMed] [Google Scholar]
- 4.Akagi, T., H. Ono, and K. Shimotohno. 1996. Expression of cell-cycle regulatory genes in HTLV-I infected T-cell lines: possible involvement of Tax1 in the altered expression of cyclin D2, p18Ink4, and p21Waf1/Cip1/Sdi1. Oncogene 12:1645-1652. [PubMed] [Google Scholar]
- 5.Armstrong, A. P., A. A. Franklin, M. N. Henbogaard, H. A. Giebler, and J. K. Nyborg. 1993. Pleiotropic effect of the human T-cell leukemia virus Tax protein on the DNA binding activity of eukaryotic transcription factors. Proc. Natl. Acad. Sci. USA 90:7303-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrigo, S. J., S. Weitsman, J. D. Rosenblatt, and I. S. Y. Chen. 1989. Analysis of rev gene function on human immunodeficiency virus type 1 replication in lymphoid cells by using quantitative polymerase chain reaction method. J. Virol. 63:4875-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arrigo, S. J., S. Weitsman, J. A. Zack, and I. S. Y. Chen. 1990. Characterization and expression of novel singly spliced RNA species of human immunodeficiency virus type 1 (HIV-1). J. Virol. 64:4585-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Askjaer, P., and J. Kjems. 1998. Mapping of multiple RNA binding sites of the human T-cell lymphotropic virus type 1 Rex protein with 5′- and 3′-Rex response elements. J. Biol. Chem. 273:11463-11471. [DOI] [PubMed] [Google Scholar]
- 9.Bakker, A. L. X., C. T. Ruland, D. W. Stephens, A. C. Black, and J. D. Rosenblatt. 1996. Human T-cell leukemia virus type 2 Rex inhibits pre-mRNA splicing in vitro at an early stage of spliceosome formation. J. Virol. 70:5511-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballaun, C., G. R. Farrington, M. Dobrovnik, J. Rusche, J. Hauber, and E. Bohnlein. 1991. Functional analysis of human T-cell leukemia virus type I Rex-response element: direct RNA binding of Rex protein correlates with in vivo binding activity. J. Virol. 65:4408-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogerd, H., and W. C. Greene. 1993. Dominant negative mutants of human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev fail to multimerize in vivo. J. Virol. 67:2496-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogerd, H. P., R. A. Fridell, S. Madore, and B. R. Cullen. 1995. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell 82:485-494. [DOI] [PubMed] [Google Scholar]
- 13.Bogerd, H. P., G. L. Huckaby, Y. F. Ahmed, S. M. Hanly, and W. C. Greene. 1991. The type 1 human T-cell leukemia virus (HTLV-I) Rex trans-activator binds directly to the HTLV-I Rex and the type 1 human immunodeficiency virus Rev RNA response elements. Proc. Natl. Acad. Sci. USA 88:5704-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohnlein, S., F. P. Pirker, L. Hofer, K. Zimmermann, H. Bachmayer, E. Bohnlein, and J. Hauber. 1991. Transdominant repressors for human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev function. J. Virol. 65:81-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cann, A. J., Y. Koyanagi, and I. S. Y. Chen. 1988. High efficiency transfection of primary human lymphocytes and studies of gene expression. Oncogene 3:123-128. [Google Scholar]
- 16.Ciminale, V., L. Zotti, D. M. D'agostino, and L. Chieco-Bianchi. 1997. Inhibition of human T-cell leukemia virus type 2 Rex function by truncated forms of Rex encoded in alternately spliced mRNAs. J. Virol. 71:2810-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cockerell, G. L., J. Rovank, P. L. Green, and I. S. Y. Chen. 1996. A deletion in the proximal untranslated pX region of human T-cell leukemia virus type II decreases viral replication but not infectivity in vivo. Blood 87:1030-1035. [PubMed] [Google Scholar]
- 18.Collins, N. D., G. C. Newbound, B. Albrecht, J. Beard, L. Ratner, and M. D. Lairmore. 1998. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood 91:4701-4707. [PubMed] [Google Scholar]
- 19.Cullen, B. R. 1998. Retroviruses as model systems for the study of RNA export pathways. Virology 249:203-210. [DOI] [PubMed] [Google Scholar]
- 20.Endo, K., A. Hirata, K. Iwai, M. Sakurai, M. Fukushi, M. Oie, M. Higuchi, W. W. Hall, F. Gejyo, and M. Fujii. 2002. Human T-cell leukemia virus type 2 (HTLV-2) Tax protein transforms a rat fibroblast cell line but less efficiently than HTLV-1 Tax. J. Virol. 76:2648-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fankhauser, C., E. Izaurralde, Y. Adachi, P. Wingfield, and U. K. Laemmli. 1991. Specific complex of human immunodeficiency virus type 1 Rev and nucleolar B23 proteins: dissociation by the Rev response element. Mol. Cell. Biol. 11:2567-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fornerod, M., and M. Ohno. 2002. Exportin-mediated nuclear export of proteins and ribonucleoproteins. Results Probl. Cell Differ. 35:67-91. [DOI] [PubMed] [Google Scholar]
- 23.Gallouzi, I.-E., and J. A. Steitz. 2002. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294:1895-1901. [DOI] [PubMed] [Google Scholar]
- 24.Grant, C., K. Barmak, T. Alefantis, J. Yao, S. Jacobson, and B. Wigdahl. 2002. Human T cell leukemia virus type I and neurologic disease: events in bone marrow, peripheral blood, and central nervous system during normal immune surveillance and neuroinflammation. J. Cell. Physiol. 190:133-159. [DOI] [PubMed] [Google Scholar]
- 25.Green, P. L., and I. S. Y. Chen. 1994. Molecular features of the human T-cell leukemia virus: mechanisms of transformation and leukemogenicity, p. 227-311. In J. A. Levy (ed.), The Retroviridae, vol. 3. Plenum Press, New York, N.Y.
- 26.Green, P. L., and I. S. Y. Chen. 2001. Human T-cell leukemia virus types 1 and 2, p. 1941-1969. In D. M. Knipe, P. M. Howley, D. Griffin, R. Lamb, M. Martin, and S. Straus (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 27.Green, P. L., Y. Xie, and I. S. Y. Chen. 1991. The Rex proteins of human T-cell leukemia virus type II differ by serine phosphorylation. J. Virol. 65:546-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green, P. L., M. T. Yip, Y. Xie, and I. S. Y. Chen. 1992. Phosphorylation regulates RNA binding by the human T-cell leukemia virus Rex protein. J. Virol. 66:4325-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grossman, W. J., J. T. Kimata, F. H. Wong, M. Zutter, T. J. Ley, and L. Ratner. 1995. Development of leukemia in mice transgenic for the tax gene of human T-cell leukemia virus type I. Proc. Natl. Acad. Sci. USA 92:1057-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hakata, Y., T. Umemoto, S. Matsushita, and H. Shida. 1998. Involvement of human CRM1 (exportin 1) in the export and multimerization of the Rex protein of human T-cell leukemia virus type 1. J. Virol. 72:6602-6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hakata, Y., M. Yamada, N. Mabuchi, and H. Shida. 2002. The carboxy-terminal region of the human immunodeficiency virus type 1 protein has multiple roles in mediating CRM1-related Rev function. J. Virol. 76:8079-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammes, S. R., and W. C. Green. 1993. Multiple arginine residues within the basic domain of HTLV-1 Rex are required for specific RNA binding and function. Virology 193:41-49. [DOI] [PubMed] [Google Scholar]
- 33.Heger, P., O. Rosorius, C. Koch, G. Casari, R. Grassmann, and J. Hauber. 1998. Multimer formation is not essential for nuclear export of human T-cell leukemia virus type 1 rex trans-activator protein. J. Virol. 72:8659-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hjelle, B., O. Appenzeller, R. Mills, S. Alexander, N. Torrez-Martinez, R. Jahnke, and G. Ross. 1992. Chronic neurodegenerative disease associated with HTLV-II infection. Lancet 339:645-646. [DOI] [PubMed] [Google Scholar]
- 35.Hofer, L., I. Weichselbraun, S. Quick, G. K. Farrington, E. Bohnlein, and J. Hauber. 1991. Mutational analysis of the human T-cell leukemia virus type I trans-acting rex gene product. J. Virol. 65:3379-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hope, T. J., B. L. Bond, B. McDonald, N. P. Klein, and T. G. Parslow. 1991. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J. Virol. 65:6001-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeang, K. T., S. G. Widen, O. J. Semmes, and S. H. Wilson. 1990. HTLV-I transactivator protein, Tax, is a transrepressor of human B-polymerase gene. Science 247:1082-1084. [DOI] [PubMed] [Google Scholar]
- 38.Kim, J. H., P. A. Kaufman, S. M. Hanly, L. T. Rimsky, and W. C. Greene. 1991. Rex transregulation of human T-cell leukemia virus type II gene expression. J. Virol. 65:405-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kusuhara, K., M. Anderson, S. M. Pettiford, and P. L. Green. 1999. Human T-cell leukemia virus type 2 Rex protein increases stability and promotes nuclear to cytoplasmic transport of gag/pol and env RNAs. J. Virol. 73:8112-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malim, M. H., S. Bohnlein, J. Hauber, and B. R. Cullen. 1989. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell 58:205-214. [DOI] [PubMed] [Google Scholar]
- 41.Manns, A., M. Hisada, and L. La Grenade. 1999. Human T-lymphotropic virus type I infection. Lancet 353:1951-1958. [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto, K., H. Shibata, J.-I. Fujisawa, H. Inoue, A. Hakura, T. Tsukahara, and M. Fujji. 1997. Human T-cell leukemia virus type 1 Tax protein transforms rat fibroblasts via two distinct pathways. J. Virol. 71:4445-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer, B. E., and M. H. Malim. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538-1547. [DOI] [PubMed] [Google Scholar]
- 44.Miyazaki, Y., T. Nosaka, and M. Hatanaka. 1996. The post-transcriptional regulator Rev of HIV: implications for its interaction with the nucleolar protein B23. Biochimie 78:1081-1086. [DOI] [PubMed] [Google Scholar]
- 45.Narayan, M., K. Kusuhara, and P. L. Green. 2001. Phosphorylation of two serine residues regulates human T-cell leukemia virus type 2 Rex function. J. Virol. 75:8440-8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nerenberg, M., S. M. Hinrichs, R. K. Reynolds, G. Khoury, and G. Jay. 1987. The tat gene of human T-lymphotropic virus type I induces mesenchymal tumors in transgenic mice. Science 237:1324-1329. [DOI] [PubMed] [Google Scholar]
- 47.Nosaka, T., Y. Miyazaki, T. Takamatsu, K. Sano, M. Nakai, S. Fujita, T. E. Martin, and M. Hatanaka. 1995. The post-transcriptional regulator Rex of the human T-cell leukemia virus type I is present as nucleolar speckles in infected cells. Exp. Cell Res. 219:122-129. [DOI] [PubMed] [Google Scholar]
- 48.Ohta, M., H. Nyunoya, H. Tanako, T. Okamoto, T. Akagi, and K. Shimotohno. 1988. Identification of a cis-regulatory element involved in accumulation of human T-cell leukemia virus type II genomic mRNA. J. Virol. 62:4445-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmeri, D., and M. H. Malim. 1996. The human T-cell leukemia virus type 1 posttranscriptional trans-activator Rex contains a nuclear export signal. J. Virol. 70:6442-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Popa, I., M. E. Harris, J. E. Donello, and T. J. Hope. 2002. CRM1-dependent function of a cis-acting RNA export element. Mol. Cell. Biol. 22:2057-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ressler, S., G. F. Morris, and S. J. Marriott. 1997. Human T-cell leukemia virus type 1 Tax transactivates the human proliferating cell nuclear antigen promoter. J. Virol. 71:1181-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rimsky, L., M. Duc Dodon, E. P. Dixon, and W. C. Greene. 1989. Trans-dominant inactivation of HTLV-I and HIV-1 gene expression by mutation of the HTLV-I Rex transactivator. Nature 341:453-456. [DOI] [PubMed] [Google Scholar]
- 53.Robek, M. D., and L. Ratner. 1999. Immortalization of CD4+ and CD8+ T lymphocytes by human T-cell leukemia virus type 1 Tax mutants expressed in a functional molecular clone. J. Virol. 73:4856-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenblatt, J. D., A. J. Cann, D. J. Slamon, I. S. Smalberg, N. P. Shah, J. Fujii, W. Wachsman, and I. S. Y. Chen. 1988. HTLV-II trans-activation is regulated by two overlapping nonstructural genes. Science 240:916-919. [DOI] [PubMed] [Google Scholar]
- 55.Rosenblatt, J. D., J. C. Gasson, J. Glaspy, S. Bhuta, M. Aboud, I. S. Chen, and D. W. Golde. 1987. Relationship between HTLV-II and atypical hairy-cell leukemia: a serologic study of hairy-cell leukemia patients. Leukemia 1:397-401. [PubMed] [Google Scholar]
- 56.Rosenblatt, J. D., D. W. Golde, W. Wachsman, A. Jacobs, G. Schmidt, S. Quan, J. C. Gasson, and I. S. Y. Chen. 1986. A second HTLV-II isolate associated with atypical hairy-cell leukemia. N. Engl. J. Med. 315:372-377. [DOI] [PubMed] [Google Scholar]
- 57.Ross, T. M., S. M. Pettiford, and P. L. Green. 1996. The tax gene of human T-cell leukemia virus type 2 is essential for transformation of human T lymphocytes. J. Virol. 70:5194-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz, S., B. K. Felber, D. M. Benko, E. M. Fenyo, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shima, H., M. Takano, K. Shimotohno, and M. Miwa. 1986. Identification of p26xb and p24xb of human T-cell leukemia virus type II. FEBS Lett. 209:289-294. [DOI] [PubMed] [Google Scholar]
- 60.Siomi, H., H. Shida, S. H. Nam, T. Nosaka, M. Maki, and M. Hatanaka. 1988. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell 55:197-209. [DOI] [PubMed] [Google Scholar]
- 61.Smith, M. R., and W. C. Greene. 1991. Type I human T-cell leukemia virus Tax protein transforms rat fibroblasts through the cyclic adenosine monophosphate response element binding protein/activating transcription factor pathway. J. Clin. Investig. 88:1038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stauber, R. H., and G. N. Pavlakis. 1998. Intracellular trafficking and interactions of the HIV-1 Tat protein. Virology 252:126-136. [DOI] [PubMed] [Google Scholar]
- 63.Szebeni, A., J. E. Herrera, and M. O. Olson. 1995. Interaction of nucleolar protein B23 with peptides related to nuclear localization signals. Biochemistry 34:8037-8042. [DOI] [PubMed] [Google Scholar]
- 64.Szebeni, A., B. Mehrotra, A. Baumann, S. A. Adam, P. T. Wingfield, and M. O. Olson. 1997. Nucleolar protein B23 stimulates nuclear import of the HIV-1 Rev protein and NLS-conjugated albumin. Biochemistry 36:3941-3949. [DOI] [PubMed] [Google Scholar]
- 65.Toyoshima, H., M. Itoh, J.-I. Inoue, M. Seiki, F. Takaku, and M. Yoshida. 1990. Secondary structure of the human T-cell leukemia virus type I Rex-responsive element is essential for Rex regulation of RNA processing and transport of unspliced RNAs. J. Virol. 64:2825-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trejo, S. R., W. E. Fah, and L. Ratner. 1996. c-sis/PDGF-B promoter transactivator by the Tax protein of the human T-cell leukemia virus type 1. J. Biol. Chem. 271:14584-14590. [DOI] [PubMed] [Google Scholar]
- 67.Venkatesh, L. K., and G. Chinnadurai. 1990. Mutants in a conserved region near the carboxy-terminus of HIV-1 Rev identify functionally important residues and exhibit a dominant negative phenotype. Virology 178:327-330. [DOI] [PubMed] [Google Scholar]
- 68.Wano, Y., M. Feinberg, J. B. Hosking, H. Bogerd, and W. C. Greene. 1988. Stable expression of the HTLV-I Tax protein in human T-cells activates specific cellular genes involved in growth. Proc. Natl. Acad. Sci. USA 85:9733-9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watanabe, T. 1997. HTLV-1-associated diseases. Int. J. Hematol. 66:257-278. [DOI] [PubMed] [Google Scholar]
- 70.Weichselbraun, I., J. Berger, M. Dobrovnik, H. Bogerd, R. Grassmann, W. C. Greene, J. Hauber, and E. Bohnlein. 1992. Dominant-negative mutants are clustered in a domain of the human T-cell leukemia virus type I Rex protein: implications for trans dominance. J. Virol. 66:4540-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yip, M. T., W. S. Dynan, P. L. Green, A. C. Black, S. J. Arrigo, A. Torbati, S. Heaphy, C. Ruland, J. D. Rosenblatt, and I. S. Y. Chen. 1991. HTLV-II Rex protein binds specifically to RNA sequences of the HTLV LTR but poorly to the HIV-1 RRE. J. Virol. 65:2261-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]