Abstract
The Saccharomyces cerevisiae DBR1 gene encodes a 2′-5′ phosphodiesterase that debranches intron RNA lariats following splicing. Yeast dbr1 mutants accumulate intron lariats and are also defective for mobility of the retrotransposons Ty1 and Ty3. We used a mutagenic PCR method to generate a collection of dbr1 mutant alleles to explore the relationship between the roles of DBR1 in transposition and debranching. Eight mutants defective for Ty1 transposition contained single amino acid changes in Dbr1p. Two mutations, G84A and N85D, are in a conserved phosphoesterase motif that is believed to be part of the active site of the enzyme, supporting a connection between enzymatic activity and Ty1 transposition. Two other mutations, Y68F and Y68D, occur at a potential phosphorylation site, and we have shown that Dbr1p is phosphorylated on tyrosine. We have developed an RNase protection assay to quantitate intron RNA accumulation in cells. The assay uses RNA probes that hybridize to ACT1 intron RNA. Protection patterns confirm that sequences from the 5′ end of the intron to the lariat branch point accumulate in dbr1 mutants in a branched (lariat) conformation. RNase protection assays indicate that all of the newly generated dbr1 mutant alleles are also deficient for debranching, further supporting a role for 2′-5′ phosphodiesterase activity in Ty1 transposition. A Ty1 element lacking most of its internal sequences transposes independently of DBR1. The existence of Dbr1p-dependent Ty1 sequences raises the possibility that Dbr1p acts on Ty1 RNA.
Saccharomyces cerevisiae Ty1 is a long terminal repeat (LTR) retroelement with a life cycle similar to that of retroviruses (2). Like retroviruses, Ty1 replicates via reverse transcription of an RNA intermediate, subsequently integrating element cDNA into the host genome. Retroviruses and LTR retrotransposons rely on their hosts to provide most of the factors necessary for propagation. We identified dbr1 mutants in a screen to identify host cell genes required for Ty1 transposition (15). The dbr1 mutant had been identified previously in a similar screen by others (6). Although dbr1 cells produce wild-type levels of Ty1 proteins, they accumulate Ty1 cDNA at a slower rate than do wild-type cells, suggesting that Dbr1p plays a role in Ty1 reverse transcription or cDNA stability (15).
In its cellular role, Dbr1p acts at the end of the mRNA splicing process, cleaving the 2′-5′ linkage at the branch point of intron RNA lariats released after exon ligation, converting them to linear RNAs that are rapidly degraded (6, 23). In a dbr1 mutant, intron RNAs accumulate as lariats (6). The uncleaved lariat branch point blocks the progression of 3′ exonucleases, and there is no 5′ end available on which a 5′ exonuclease can act. DBR1 is highly conserved, and homologues have been cloned from Schizosaccharomyces pombe, S. cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, and Homo sapiens (6, 17, 18, 24). Deletion of DBR1 has no significant effect on the growth rate of S. cerevisiae, but S. pombe dbr1 null mutants display severe growth defects and abnormal cell morphology (24). The differential effect of the loss of DBR1 function in these organisms could be related to numbers of introns that are processed by the spliceosome and released as lariats. The S. pombe genome contains ∼4,730 introns distributed among 43% of the genes (34), while the S. cerevisiae genome contains ∼255 introns distributed among 5% of the genes (11, 21). However, almost half of the introns in S. cerevisiae are in highly expressed genes encoding ribosomal proteins. Therefore, the difference between S. cerevisiae and S. pombe dbr1 mutants in the amount of intron RNA lariat accumulation is less than that indicated by the numbers of introns encoded in their genomes.
The role that Dbr1p plays in Ty1 transposition is not understood, and it is not known if Dbr1p's 2′-5′ phosphodiesterase activity, which catalyzes the debranching of intron RNA lariats, is necessary for Ty1 transposition. In work presented here, a library of dbr1 missense mutants was created and tested for both Ty1 transposition and debranching activity. The purpose of this screen was to identify amino acid residues in Dbr1p that are necessary for Ty1 transposition and to determine if dbr1 mutants can be identified that are defective for Ty1 transposition but can still debranch intron RNA lariats. The existence of such mutants could be an indication that 2′-5′ phosphodiesterase activity is not needed for Ty1 transposition. Further experiments would then be warranted to test that possibility.
Although screening is ongoing, all of the dbr1 missense mutants created in this study are defective for both Ty1 transposition and debranching intron RNA lariats. Two of the mutants have changes in the Dbr1p phosphoesterase signature motif, which is believed to form part of the enzyme active site. The transposition deficiency of these two mutants supports the hypothesis that 2′-5′ phosphodiesterase activity is necessary for Ty1 transposition. Experiments with a Ty1 deletion element lacking most of the Ty1 internal sequences suggest that Dbr1p may act on Ty1 RNA.
MATERIALS AND METHODS
Yeast and bacterial strains, plasmids, and general procedures.
The plasmids and yeast strains used in this study are described in Tables 1 and 2, respectively. For plasmid manipulations, Escherichia coli strain TOP10 was used (F-mcrA Δ[mrr-hsdRMS-mcrBC] φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 D[ara-leu]7697 galU galK rpsL [Strr] endA1 nupG). When not specifically described, general molecular techniques (1) and standard yeast media and general procedures (14) were used. DNA oligonucleotides used in the procedures described below are listed in Table 3.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pCR4-TOPO | TOPO cloning vector | Invitrogen |
| pYES2.1-TOPO | TOPO cloning shuttle vector; URA3 2μm | Invitrogen |
| pBJC35 | pGAL1-Ty1::his3AI; URA3 2μm | 9 |
| pDG784 | pGAL1-Ty1::his3AI; TRP1 2μm | 10 |
| pX3 | pGAL1-Ty1::TRP1; URA3 2μm | 35 |
| pX100 | pGAL1-Ty1(Δ620-5562)::TRP1; URA3 2μm | 36 |
| pTM235 | DBR1 ORF in pYES2/GS; URA3 2μm | Invitrogen |
| pTM263 | DBR1-V5-6xHis in pRS314 | This work |
| pTM400 | ACT1 lariat probe template | This work |
| pTM411 | ACT1 3′ intron probe template | This work |
| pTM426 | ACT1 5′ intron probe template | This work |
| pTM428 | dbr1-N8 derivative of pTM235 | This work |
| pTM431 | dbr1-N14 derivative of pTM235 | This work |
| pTM432 | dbr1-N16 derivative of pTM235 | This work |
| pTM434 | dbr1-N20 derivative of pTM235 | This work |
| pTM435 | dbr1-N29 derivative of pTM235 | This work |
| pTM436 | dbr1-C30 derivative of pTM235 | This work |
| pTM442 | dbr1-N52 derivative of pTM235 | This work |
| pTM444 | dbr1-N70 derivative of pTM235 | This work |
| pTM454 | dbr1-C86 derivative of pTM235 | This work |
TABLE 2.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| YPH499 | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | 31 |
| TMY30 | MATaura3-52 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | 15 |
| TMY43 | MATatrp1-H3 ura3-52 his3-200 ade2-101oc lys2-1 leu1-12 can1-100 bar1::hisG spt3-Δ202 GAL3+ ΔTy3 | 16 |
| TMY60 | MATadbr1::neorura3-52 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | 29 |
| TMY176 | MATarad52::hisG ura3-52 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | This work |
| TMY344 | MATa dbr1::neorrad52::hisG ura3-52 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | This work |
TABLE 3.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| 99 | CGGATCGGACTACTAGCAGCT |
| 100 | CCATTGAGGCCGCAAATCTCT |
| 101 | GCTGAGCCACGATTGGCCCAA |
| 102 | GATGCGGCCCTCTAGAAACTC |
| 146 | CTCTCCCATAACCTCCTA |
| 147 | CTCTCGAGCAATTGGGACCG |
| 219 | GGATCCTCAAACCAAGAAGAA AAAGAAA |
| 220 | TCGTGGTTATTACAGATCAGTCA |
| 221 | GGATCCGTCCCAATTGCTCGAG AGATTT |
| 222 | ATAGCAACAAAAAGAATGAAGC |
| 237 | GTAAGGGTAGACCAAGACACC AAGGT |
| 238 | TAATACGACTCACTATAGGAAG AGACCGGAAGAGTACAAGGACAA |
Transposition assays.
Ty1 transposition was measured with a replica-plating assay, a cell suspension spotting assay, and a quantitative plating assay.
Replica-plating assays have been described previously (15, 29, 35). Briefly, patches of cells containing pGAL1-Ty1 are replica plated to medium containing 2% d-(+)-galactose to induce transcription of Ty1. After the induction period, the patches are replica plated to medium that selects for Ty1 transposition events. For pGAL1-Ty1::his3AI (pBJC35 and pDG784), the induction period is 16 to 20 h at 26°C and Ty1 transposition events are indicated by His+ colonies after 48 h on SD-His selection plates (14). For pGAL1-Ty1::TRP1 (pX3), the induction period is 5 days at 22°C. After the Ty1 expression period is finished for pX3, patches are grown nonselectively for 16 to 24 h to allow passive loss of the plasmid. Ty1 transposition events are then selected by growth on plates that contain 5-fluoroorotic acid (+ FOA) and lack tryptophan (− trp). FOA selects against cells containing pX3 because of its URA3 marker (3), while the lack of tryptophan selects for the presence of transposed Ty1::TRP1 elements.
The cell suspension spotting assay is semiquantitative, providing a visual comparison of Ty1 transposition rates between strains. For this assay, cells containing pGAL1-Ty1 are grown in patches as described above or in liquid cultures (YNB/CAA medium) (16). For liquid cultures, Ty1 transcription is induced by subculturing into medium containing 2% d-(+)-galactose and growing for 16 to 18 h at 26°C. After the Ty1 expression period in either liquid medium or on plates, equivalent numbers of cells for each strain are spotted in a series of dilutions onto media to select for Ty1 transpositions and separately to show cell titers (complete medium). The starting suspension of each dilution series has an optical density at 600 nm of 0.5. For cells growing on solid medium, patches of cells are scraped off the plates after the Ty1 expression period and suspended in sterile water prior to dilution and spotting.
The quantitative plating assay provides Ty1 transposition frequencies. For this assay, cells are grown in patches or in liquid cultures as described above. After the Ty1 expression period, liquid dilutions of cells are plated for single colonies onto media to select for Ty1 transpositions and separately to show cell titers. Colony counts yield the number of cells containing Ty1 transposition events out of the total number of cells in the population (transposition frequency).
For experiments with the Ty1 deletion element, the frequency of Ty1 transposition was determined by a method described by Boeke and coworkers (20). Transformants of strain TMY43 and TMY380, carrying either pX3 or pX100, were patched onto SD-Ura (to maintain selection for the plasmids) and grown for 2 days at 30°C. Patches were replica plated to SG-Ura to induce transcription of the plasmid-borne Ty1 elements. SG-Ura plates were incubated at 22°C for 5 days, replica plated to SD-Ura for overnight growth at 30°C, and then replica plated to YPD (14) for overnight growth at 30°C again. Patches grown on YPD were scraped and suspended in water. Dilutions of the suspensions were plated onto YPD at a density of 200 to 400 colonies per plate. When colonies were grown, they were replica plated to SD-Ura and SD-Trp. For a given patch, the number of Ura-Trp+ cells relative to the total number of Ura− cells is used as the measure of Ty1 transposition. At least three independent transformants were tested for each strain-plasmid combination. The results for each of the transformants for a strain-plasmid combination were combined to generate an overall frequency.
PCR mutagenesis of DBR1.
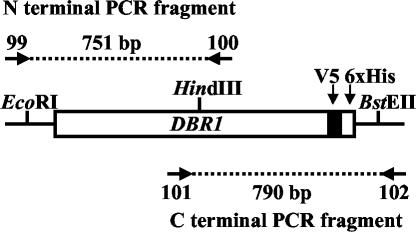
The DBR1 gene was mutagenized by error-prone PCR (4, 5) with the GeneMorph kit from Stratagene (La Jolla, Calif.). The kit contains an error-prone thermophilic DNA polymerase that produces all possible transition and transversion mutations. Mutation frequency is determined by the DNA polymerase error rate and the number of target DNA duplications. In this procedure, the initial target DNA concentration in the PCR dictates the number of target duplications. Few target duplications produces a low mutation frequency (one to three mutations per kilobase of target). The pTM235 plasmid, linearized with EcoRI, was used as the PCR template. Mutagenic PCRs were carried out separately for the 5′ and 3′ regions of the DBR1 open reading frame (ORF). Primers 99 and 100 were used for mutagenic PCR of the 5′ region of the DBR1 ORF (Fig. 1). Primer 99 corresponds to bp 453 to 474 of pYES2/GS (Invitrogen, Carlsbad, Calif.), and primer 100 corresponds to nucleotides (nt) 679 to 658 of the DBR1 ORF. Primers 101 and 102 were used for mutagenic PCR of the 3′ region of the DBR1 ORF (Fig. 1). Primer 101 corresponds to bp 528 to 549 of DBR1, and primer 102 corresponds to bp 635 to 614 of pYES2/GS. PCR conditions were as follows (amounts specified by the GeneMorph kit): 41.5 μl of H2O, 5 μl of 10× DNA polymerase buffer, 1 μl of a deoxynucleoside triphosphate mixture, 1 μl of mutazyme DNA polymerase, 1 μl (60 ng) of pTM235 template DNA, and 0.25 μl (250 ng) of each primer. Cycling conditions were as follows: 94°C, 40 s; 52°C, 40 s; 72°C, 2 min (35 cycles).
FIG. 1.
PCR mutagenesis of DBR1. The pTM235 plasmid was linearized and used for PCR with either primers 99 and 100 or primers 101 and 102 for the N- and C-terminal regions of DBR1, respectively. Sizes of PCR products are shown. Most point mutations form a fusion with V5 and a six-His tag (6xHis), facilitating Western analysis and protein purification.
Purification of Dbr1p-V5-6xHis.
Wild-type and mutant versions of Dbr1p were purified under denaturing conditions with the ProBond Purification System (Invitrogen). The ProBond system is designed for purification of proteins tagged with six tandem histidine residues (six-His tag). Whole-cell extracts were prepared from yeast cells (TMY60) containing either pTM235 (DBR1) or dbr1 mutant derivatives (pTM434 and pTM435). Cells were initially grown in YNB/CAA medium lacking uracil and containing 2% d-(+)-raffinose. Expression of Dbr1p-V5-6xHis was induced prior to extract preparation by addition of d-(+)-galactose to 2% and incubation at 26°C for 16 to 20 h. Cells were collected by centrifugation and lysed in buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 3 mM MgCl2, 0.2% Triton X-100, 0.1% β-mercaptoethanol, yeast protease inhibitors (Sigma, St. Louis, Mo.), 1 mM phenylmethylsulfonyl fluoride, 100 mM sodium orthovanadate, and 50 mM sodium fluoride. In some cases, the phosphatase inhibitors (sodium orthovanadate and NaF) were omitted. Dbr1p-V5-6xHis was purified over ProBond columns under conditions recommended by the manufacturer. Multiple fractions containing purified Dbr1p-V5-6xHis were combined and concentrated with Centricon filter devices (Millipore; Billerica, Mass.). The amount of Dbr1p-V5-6xHis recovered from individual ProBond fractions was estimated with Western blot assays done with anti-V5 antibody (1/2,500 dilution). Tyrosine phosphorylation of Dbr1p-V5-6xHis was assessed with an antiphosphotyrosine antibody (PY99; 1/1,000 dilution) from Santa Cruz Biotech (Santa Cruz, Calif.). Blots probed with the PY99 antibody were blocked in TBS-T with 1% bovine serum albumin (1). In vitro phosphatase treatment of Dbr1p-V5-6xHis was performed with λ protein phosphatase (New England Biolabs, Beverly, Mass.). Phosphatase reactions were performed at 30°C for 30 min under conditions recommended by the manufacturer.
RNase protection assays.
Plasmid pTM426 was used as a template to create the 5′ ACT1 intron probe, which spans the 5′ border of the ACT1 intron. The PCR product generated with oligonucleotides 219 and 220 was cloned into pCR4-TOPO to create pTM426. Plasmid pTM411 was used as a template to create the 3′ ACT1 intron probe, which includes the lariat branch point of the ACT1 intron. The PCR product generated with oligonucleotides 221 and 222 was cloned into pYES2.1-TOPO to create pTM411. Yeast genomic DNA was used as the template for both PCRs. Plasmid pTM400 was used as a template to create the ACT1 lariat probe, which contains sequences on either side of the 2′-5′ lariat branch linkage. The RT-PCR product generated with oligonucleotides 146 and 147 was cloned into pYES2.1-TOPO to create pTM400. The RT-PCR of total yeast RNA was performed with the One-Step kit from Qiagen (Valencia, Calif.). The PCR and RT-PCR products described above were gel purified and cloned into pCR4-TOPO or pYES2.1-TOPO, as appropriate, with TOPO cloning kits from Invitrogen.
The PCR product generated with oligonucleotides 237 and 238 was directly used as a template to create the ACT1 3′ exon probe, which hybridizes to sequences in the 3′ exon of ACT1 RNA. Oligonucleotide 238 includes the sequence of the T7 promoter at its 5′ end.
RNA probes for RNase protection assays were generated from the various templates described above by in vitro transcription reactions with T7 polymerase with the MAXIscript kit from Ambion (Austin, Tex.). Plasmid templates were cleaved immediately downstream of probe sequences prior to transcription to limit transcript length. Plasmids pTM411 and pTM426 were cleaved with BamHI; pTM400 was cleaved with Sau3AI. [α-32P] CTP (Amersham Biosciences, Piscataway, N.J.) was used in transcription reactions to radioactively label the probes, which were subsequently gel purified (1).
RNase protection assays were performed with the RPA III kit from Ambion. Hybridizations of probes with RNA samples were performed at 42°C for 16 to 24 h and were followed by digestion with RNaseT1 to remove probe sequences that did not hybridize. Probe amounts in RNase protection assays were ∼105 cpm. Yeast total RNA amounts ranged from 1 to 10 μg, depending on the experiment. RNase protection assay products were run on 6% polyacrylamide gels. RNA size markers were generated by in vitro transcription of RNA Century Marker templates from Ambion incorporating [α-32P]CTP (Amersham Biosciences). Phosphor screens exposed to the gels were read on a Storm phosphorimager (Amersham Biosciences), and the signal was quantified. Size estimates for the products of RNase protection assays were based on careful determination of mobilities, as well as established knowledge about the properties of the ACT1 RNA-processing pathway. The products of RNase protection assays were run on large DNA sequencing gels 45 cm in length. Size standards on each gel included 100-, 200-, 300-, 400-, and 500-nt species. Mobilities were carefully plotted on semilog graphs, and size estimates for the undigested probes match expectations within 5 nt. The identities and precise sizes of the protected products were assigned on the basis of the known sizes and structures of ACT1 RNA splicing products derived from 20 years of intense study by many laboratories.
RESULTS
PCR mutagenesis of DBR1.
DBR1 is required for debranching of intron RNA lariats and for full levels of transposition of the yeast LTR retrotransposon Ty1. Comparison of DBR1 genes from different organisms shows strong conservation of coding sequences. However, the importance of specific amino acid residues for Dbr1p function has not been demonstrated. Furthermore, the relationship between Dbr1p's roles in debranching and Ty1 transposition is unknown. If the 2′-5′ phosphodiesterase activity of Dbr1p is required for both processes, it would suggest that Ty1 transposition involves an RNA with this type of linkage, for which there is no evidence at present. Alternatively, Dbr1p may act differently in Ty1 transposition and debranching. In this case, dbr1 mutant alleles could be identified that affect one process but not the other.
In order to study how Dbr1p functions in Ty1 transposition, a library of dbr1 alleles was generated by mutagenic PCR. The method used (GeneMorph; Stratagene) was chosen because it can produce all possible transition and transversion mutants with minimal bias. Mutations were introduced in the DBR1 ORF present in pTM235. This plasmid contains the 1,218-nt DBR1 ORF fused at its 5′ end to the yeast GAL1 promoter and at its 3′ end to a V5 epitope tag followed by a six-His tag (1) (Invitrogen) (Fig. 1). Mutagenic PCR was performed with two different sets of primers: one set (primers 99 and 100) for the 5′ portion of the DBR1 ORF (nt 1 to 679) and the other set (primers 101 and 102) for the 3′ portion of the ORF (nt 528 to 1218). PCR conditions were adjusted to limit multiple mutations in the DBR1 gene in order to generate a collection of single-point mutations (see Materials and Methods). The PCR products were digested either with EcoRI and HindIII (5′ region of DBR1) or with HindIII and BstEII (3′ region of DBR1), and purified fragments were used to replace the corresponding portions of pTM235. Because the mutation rate in the PCR is low, many plasmids in the resulting collection of pTM235 derivatives contain a wild-type DBR1 allele. Therefore, the collection of plasmids was screened to identify those containing dbr1 mutant alleles. Yeast strain TMY60 (dbr1Δ) was cotransformed with the collection of pTM235 derivatives and pDG784 (pGAL1-Ty1::his3AI). Individual transformants were screened for the ability to support Ty1 transposition with a replica-plating assay (see Materials and Methods). In this assay, a pTM235 derivative is the only source of Dbr1p in the cell and expression of both Dbr1p and Ty1 is activated by d-(+)-galactose. Positive and negative controls were patched on each plate containing transformants being screened. The positive control (wild-type Ty1 transposition phenotype) was a TMY60 transformant containing pDG784 and the original (unmutagenized) pTM235 plasmid (pGAL1-DBR1-V5-6xHis). The negative control (dbr1 mutant Ty1 transposition phenotype) was a TMY60 transformant containing pDG784 and a vector plasmid (pYES2.1) containing no DBR1 sequences. Patches of cells unable to support wild-type levels of Ty1 transposition were single colony purified and retested.
The pTM235 derivatives from those TMY60 transformants that retained a mutant phenotype for Ty1 transposition were rescued into E. coli. Purified plasmids were subjected to restriction enzyme analysis to eliminate those with apparent deletions from further studies. Full-length plasmids were reintroduced into TMY60 and retested for Ty1 transposition. Only transformants with a Ty1 transposition defect were analyzed further.
Transformants were subjected to Western blot analysis (1) with α-V5 antibody (Invitrogen). For structure-function studies, we are interested in identifying missense mutants that express wild-type levels of full-length Dbr1p. The expression of the V5 epitope on an ∼50 kDa protein indicates Dbr1p-V5-6xHis expression because V5 is expressed as a C-terminal tag on the plasmid-encoded Dbr1p proteins. Only those mutants that express Dbr1p-V5-6xHis were analyzed further.
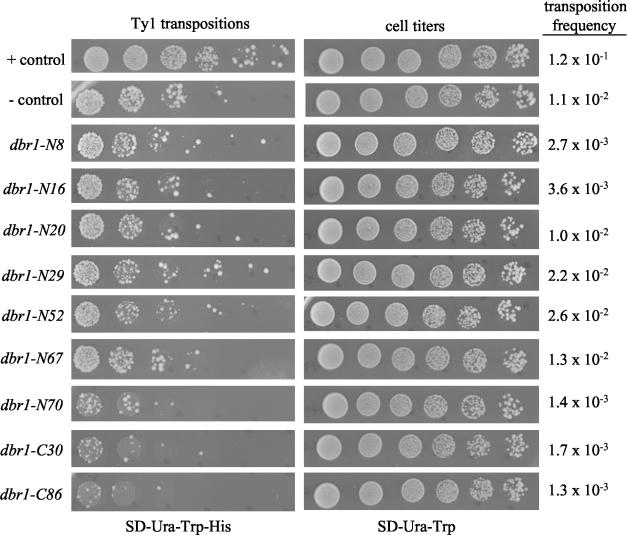
A total of 1,098 pTM235 derivatives created by replacement of the 5′ portion of the DBR1 ORF with a mutagenic PCR fragment were screened by the process described above. From this collection, 15 plasmids were identified that contain dbr1 missense alleles resulting in a Ty1 transposition deficiency and of these, 7 plasmids encoded single amino acid substitutions. A total of 955 pTM235 derivatives created by replacement of the 3′ portion of the DBR1 ORF with a mutagenic PCR fragment were screened. From this collection, seven plasmids were identified that contain dbr1 missense alleles resulting in a Ty1 transposition deficiency, and of these, one plasmid encoded a single amino acid substitution. The transposition phenotypes of the eight single amino acid substitution alleles are shown in Fig. 2 along with the phenotypes of control strains that include mutant dbr1-C30, a nonsense allele that produces a truncated form of Dbr1p.
FIG. 2.
Ty1 transposition phenotypes of dbr1 mutants. Transposition of Ty1 was measured with the pGAL1-Ty1::his3AI element as described in Materials and Methods. Individual transformants containing pDG784 and a derivative of pTM235 (with mutagenized dbr1 DNA) were grown in SD-Ura-Trp before induction of transcription of Ty1 and DBR1 with galactose for 16 to 20 h. Serial dilutions were spotted on either SD-Ura-Trp-His (to select for transpositions) or SD-Ura-Trp (for cell titers). The calculated transposition frequency of each mutant is the average of three independent experiments. The positive (+) control is pTM235 (DBR1) cotransformed with pDG784. The negative (−) control is pYES2.1 (vector) cotransformed with pDG784.
All of the mutations that encode single amino acid changes in Dbr1p were assessed for dominance or recessiveness. Yeast strain TMY30 (DBR1) was cotransformed with plasmids bearing the dbr1 point mutations and pDG784 (pGAL1-Ty1::his3AI). Individual transformants were screened for the ability to support Ty1 transposition by the replica-plating assay used in the initial mutant screen. Because the TMY30 strain expresses wild-type Dbr1p, recessive dbr1 mutations on the plasmid will not alter the wild-type phenotype of the strain, but dominant DBR1 mutations will produce a mutant phenotype. All of the mutations encoding single amino acid changes in Dbr1p are recessive (data not shown).
Mutations in DBR1 lie in conserved regions.
Amino acid changes in each of the dbr1 missense mutants are shown in Table 4. Eight of the missense mutations result in single amino acid changes in Dbr1p. The positions of these single amino acid changes are shown in Fig. 3, in which the S. cerevisiae Dbr1p sequence is shown aligned with amino acid residues conserved in at least 10 of the 12 Dbr1p sequences available in the GenBank database (Conserved in Fig. 3). Importantly, two mutations (dbr1-N16 and dbr1-N30) map to a GD/GNH phosphoesterase signature motif (19) (Phos in Fig. 3). This motif has been identified in all enzymes that catalyze phosphate bond cleavage. Two independent mutations (dbr1-N20 and dbr1-N29) were found to reside in a potential tyrosine phosphorylation site at Y68 (pattern [RK]-X[2,3]-[DE]-X[2,3]-Y) (8, 13, 26).
TABLE 4.
dbr1 missense mutants used in this study
| Mutant name | Mutated residue(s) in Dbr1p |
|---|---|
| dbr1-N1 | I26N, I37N, K135M, W148R |
| dbr1-N8 | H97Y |
| dbr1-N10 | I44T, G126W |
| dbr1-N14 | D180Y |
| dbr1-N16 | G84A |
| dbr1-N20 | Y68F |
| dbr1-N29 | Y68D |
| dbr1-N30 | N85D |
| dbr1-N31 | I66L, H92L |
| dbr1-N33 | G38V, A75V |
| dbr1-N37 | I26N, I37N, K135M |
| dbr1-N39 | S52L, G99D, H171Y |
| dbr1-N51 | I32M, L124S |
| dbr1-N52 | S212C |
| dbr1-N70 | I37T, S43C, N182Y |
| dbr1-C5 | I240S, D273N, G280N, K290E, P304S, E335G, E361K |
| dbr1-C7 | F236I, A298V, N353S |
| dbr1-C8 | M330I, E335G, E361K |
| dbr1-C9 | E284D, K290E |
| dbr1-C33 | I240S, D273N, G277D, P304S |
| dbr1-C68 | L218Q, L268S, M330I |
| dbr1-C86 | E284D |
FIG. 3.
Single-point mutations in dbr1 lie mostly in conserved residues of Dbr1p. The amino acid sequence of S. cerevisiae Dbr1p is shown on the top line of each sequence block. Positions of amino acid changes in the point mutations are indicated by asterisks above the Dbr1p sequence. The phosphoesterase domain is indicated by underlined residues in Dbr1p (amino acids 4 to 238). Residues conserved in phosphoesterases are indicated on the “Phos” line. The GD/GNH phosphoesterase signature domain is part of this conserved sequence. Residues conserved in debranching enzymes are indicated on the “Conserved” line. The 12 organisms for which Dbr1p sequences are available are S. cerevisiae, S. pombe, Neurospora crassa, Plasmodium falciparum, Arabidopsis thaliana, D. melanogaster, Anopheles gambiae, C. elegans, Xenopus laevis, Rattus norvegicus, M. musculus, and H. sapiens. Residues identical in all 12 proteins are underlined in the “Conserved” row, while residues conserved in at least 10 of the 12 proteins are shown without underlining.
Dbr1p is a phosphoprotein.
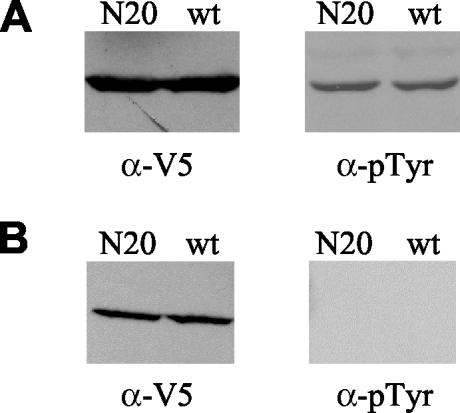
Identification of two mutations that produce changes in the Y68 residue prompted us to investigate the possibility that Dbr1p is phosphorylated. Dbr1p-V5-6xHis was purified from TMY60 cells containing DBR1 (pTM235), dbr1-N20 (pTM434), and dbr1-N29 (pTM435) as outlined in Materials and Methods. The wild-type and mutant forms of Dbr1p were purified with the ProBond system (Invitrogen) in the presence of phosphatase inhibitors. The wild-type and mutant forms of Dbr1p were detected on Western blots with anti-V5 antibody (Fig. 4A, left side) (data for N29 mutant not shown). Western analysis with antiphosphotyrosine antibody indicates that the wild-type and mutant forms of Dbr1p are phosphorylated (Fig. 4A, right side) (data for N29 mutant not shown). When purified Dbr1p proteins are treated with λ protein phosphatase, they no longer react with antiphosphotyrosine antibody (Fig. 4B). λ protein phosphatase removes phosphates from tyrosine, serine, and threonine residues. These data indicate that Dbr1p is phosphorylated on at least one tyrosine other than Y68. Although wild-type Dbr1p may also be phosphorylated on Y68, that cannot be determined from this experiment. As expected, when phosphatase inhibitors were absent during purification, antiphosphotyrosine antibody did not react with either wild-type or mutant Dbr1p (data not shown).
FIG. 4.
Dbr1p is a phosphoprotein. (A) Western blots of Dbr1p-V5-6xHis following purification of the protein in the presence of phosphatase inhibitors. The left half shows purified wild-type (wt) and N20 mutant Dbr1p probed with anti-V5 antibody (α-V5). The right half shows the same samples probed with antiphosphotyrosine antibody (α-pTyr). (B) Western blots of purified Dbr1p-V5-6xHis after treatment with λ protein phosphatase.
Intron lariat RNA accumulation within cells.
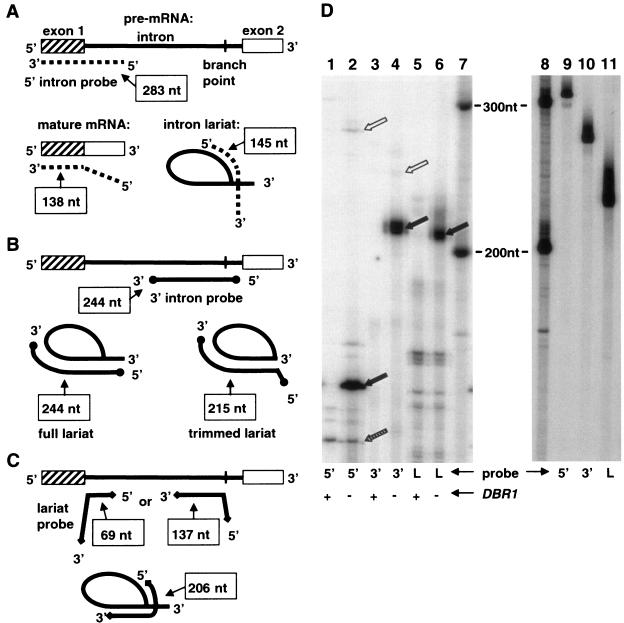
We have developed an RNase protection assay, partly based on the lariat protection assay of Vogel and coworkers (33), to measure the accumulation of intron RNA lariats within cells. The assay uses RNA probes that hybridize to ACT1 RNA. ACT1 contains an intron that accumulates in dbr1 strains after it is spliced from ACT1 pre-mRNA (6). Hybridizations of the probes described below with ACT1 RNA are shown in Fig. 5. The 5′ intron probe hybridizes to 283 nt in ACT1 RNA: 145 nt at the 5′ end of the ACT1 intron and an adjacent 138 nt in the 5′ exon. The 3′ intron probe hybridizes to 244 nt within the ACT1 intron: 30 nt downstream of the lariat branch point and 214 nt upstream of and including the lariat branch point. The lariat probe hybridizes to 206 nt within the ACT1 intron: 69 nt at the 5′ end of the intron, as well as 137 nt starting at the lariat branch point and extending upstream. As a control for the amount of protection observed by intron RNA sequences, a 3′ exon probe was used in some experiments. This probe hybridizes to 333 nt of the ACT1 3′ exon.
FIG. 5.
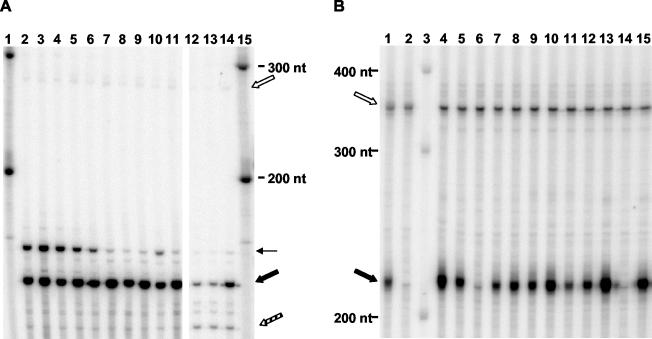
RNase protection assay. (A) Diagram of the 5′ intron probe annealing to different ACT1 RNA species. The three different ACT1 RNA species are pre-mRNA, mature mRNA, and the intron lariat. The different RNA species are not drawn to scale. The extents of probe annealing are indicated in boxes and reflect the sizes of the probe fragments protected in RNase protection assays. The relative amounts of the different RNA species are reflected by the amounts of protected probe fragments in the RNase protection assay. (B) Diagram of the 3′ intron probe annealing to different ACT1 RNA species. The ACT1 RNA species are the pre-mRNA and the intron lariat. The intron lariat is also depicted in a form in which all of the nucleotides of the 3′ tail but one have been removed by nucleases (trimmed lariat). (C) Diagram of the lariat probe annealing to different ACT1 RNA species. The ACT1 RNA species are the pre-mRNA and the intron lariat. (D) Results of the RNase protection assay for ACT1 RNA species. Lanes: 1 and 2, protection of 5′ intron probe (indicated by 5′ at bottom of gel); 3 and 4, protection of 3′ intron probe (indicated by 3′ at bottom of gel); 5 and 6, protection of lariat probe (indicated by L at bottom of gel); 7, RNA size markers. Reactions run in lanes 1, 3, and 5 used 2 μg of total RNA from DBR1 cells (wild type) (indicated by plus sign at bottom of gel). Reactions run in lanes 2, 4, and 6 used 2 μg of total RNA from dbr1 mutant cells (indicated by minus sign at bottom of gel). Solid black arrows in lanes 2 (156 nt), 4 (215 nt), and 6 (211 nt) indicate fragments protected by ACT1 intron lariat. Stippled arrow in lane 2 indicates fragment of 5′ intron probe protected by ACT1 mature mRNA (143 nt). White arrow in lane 2 indicates fragment of 5′ intron probe protected by ACT1 pre-mRNA (283 nt). White arrow in lane 4 indicates fragment of 3′ intron probe protected by ACT1 intron RNA containing its full 3′ tail (250 nt). Lanes 9, 10, and 11 are undigested probes: 5′ intron probe, 3′ intron probe, and lariat probe, respectively. Lane 8 contains RNA size markers.
With RNase protection assays, we have measured the accumulation of ACT1 RNA species in both a dbr1Δ mutant (TMY60) and the wild type (TMY30) (Fig. 5D). Cells of each strain were grown in rich medium (YPD) to mid-log phase. Total RNA was isolated from the cells and used for RNase protection assays. Extensive accumulation of particular ACT1 RNA species occurs in the dbr1 mutant compared to the wild type, evidenced by the extensive probe protection by dbr1 RNA seen in Fig. 5D, lanes 2, 4, and 6, compared to the minimal protection by wild-type RNA seen in lanes 1, 3, and 5. The sizes of the major protected fragments in lanes 2, 4, and 6 correspond to expectations for protection by ACT1 intron lariat RNA (positions of black arrows). The relative amounts of protected fragments indicate that introns accumulate in the dbr1 mutant at levels 50 to 100 times the levels in the wild type (data not shown).
For the lariat probe, the observed protection of ∼211 nt (Fig. 5D, lane 6) is expected if the 2′-5′ phosphodiester bond between the 5′ end of the intron and the lariat branch point is intact and the probe hybridization spans the lariat branch point. Also in line with expectations, enhanced protection of 138- and 74-nt fragments, predicted if intron lariats are debranched, is not observed for the dbr1 mutant. Debranched introns do not accumulate in the wild-type strain either, consistent with their rapid degradation.
As mentioned above, the ACT1 intron lariat protects 206 nt of the lariat probe. The extra nucleotides that bring the protected fragment size to 211 nt are due to the positions of the first RNase T1 cut sites on either site of the hybridizing region. Differences for other probes between the extents of probe hybridization with ACT1 RNAs and the size of protected fragments in the RNase protection assays are also due to the positions of the RNase T1 cut sites.
RNase protection assays with the 5′ intron probe indicate that a fragment of ∼156 nt is extensively protected by RNA from the dbr1 strain (Fig. 5D, lane 2). This result is expected if the 5′ portion of the probe, the part that hybridizes to the 5′ portion of the ACT1 intron, is protected because of accumulation of ACT1 intron RNA (Fig. 5A). Mature ACT1 mRNA will protect only the 3′ part of the probe, a fragment of 143 nt. Evidence of such protection by the mature ACT1 mRNA is indicated in lanes 1 and 2 of Fig. 5D by the stippled arrow. Note that this fragment is present in roughly equal amounts in the dbr1 mutant and the wild type. This result indicates that production of mature ACT1 mRNA is not grossly affected in the dbr1 mutant.
RNase protection assays with the 3′ intron probe indicate that a fragment of ∼215 nt is extensively protected by RNA from the dbr1 strain (Fig. 5D, lane 4). This result is expected if ∼30 nt at the 5′ end of the probe are not protected. The hybridizations diagrammed in Fig. 5B show both the entire intron lariat and a trimmed intron lariat in which the 3′ end is 1 base downstream of the lariat branch point. Our experimental results indicate that the 3′ tail of the intron lariat is degraded to the branch point or just downstream of it.
Accumulation of ACT1 intron lariat RNA in dbr1 mutants.
We have performed RNase protection assays to measure the accumulation of ACT1 intron lariat RNA in the newly generated dbr1 mutants. These RNase protection assays are a means by which to assess the debranching capability encoded by the various dbr1 mutant alleles. The newly generated alleles are carried on plasmids in a dbr1Δ strain (TMY60) and are under the control of the GAL1 promoter. TMY60 cells containing the various plasmids were pregrown on selective raffinose medium to early log phase. The precultures were split into two portions. d-(+)-Galactose was added to one portion to a final concentration of 2% to induce expression of the plasmid-borne dbr1 alleles. d-(+)-Glucose was added to the other portion to a final concentration of 2% to repress expression of dbr1 (negative control).
All of the dbr1 point mutant alleles identified in this study on the basis of their Ty1 transposition deficiencies are also deficient for debranching of intron lariats (Fig. 6). The extent of the debranching deficiency of each mutant is consistent with a severe loss of function relative to the wild-type allele. Figure 6 depicts RNase protection assays with both the 5′ intron probe and the 3′ intron probe. The black arrows in each panel indicate the fragments that represent accumulation of intron RNAs.
FIG. 6.
Intron lariat accumulation in dbr1 mutant strains. (A) RNase protection assay with the 5′ intron probe and 1.5 μg of total RNA from transformants of dbr1 strain TMY60 carrying different plasmids. Lanes: 2 and 3, pTM431 (N14 mutant); 4 and 5, pTM432 (N16 mutant); 6 and 7, pTM434 (N20 mutant); 8 and 9, pTM435 (N29 mutant); 10 and 11, pTM436 (C30 mutant); 12, pTM235 (pGAL1-DBR1-V5-6xHis); 13, pTM263 (DBR1-V5-6xHis); 14, pYES2.1 (vector); 1 and 15, RNA size markers. Reaction mixtures loaded into lanes 2, 4, 6, 8, 10, 12, 13, and 14 contained total RNA from cells grown in galactose for 18 h. Reaction mixtures loaded into lanes 3, 5, 7, 9, and 11 contained total RNA from cells grown in glucose. The solid black arrow indicates fragments protected by the ACT1 intron lariat (156 nt). The stippled arrow indicates the fragment of the 5′ intron probe protected by ACT1 mature mRNA (143 nt). The white arrow indicates the fragment of the 5′ intron probe protected by ACT1 pre-mRNA (283 nt). The band of variable intensity indicated by the thin horizontal arrows is most likely an incompletely digested product of protection by the ACT1 intron. (B) RNase protection assay with 3′ intron probe and 1 μg of total RNA from control strains and transformants of dbr1 mutant strain TMY60 carrying different plasmids. Lanes: 1, TMY60 (dbr1); 2, TMY30 (DBR1), 3, RNA size markers; 4, TMY60 plus pTM442 (N52 mutant); 5, TMY60 plus pTM428 (N8 mutant); 6, TMY60 plus pTM235 (pGAL1-DBR1-V5-6xHis); 7, TMY60 plus pTM444 (N70 mutant); 8, TMY60 plus pTM454 (C86 mutant); 9, TMY60 plus pTM436 (C30 mutant); 10, TMY60 plus pTM435 (N29 mutant); 11, TMY60 plus pTM434 (N20 mutant); 12, TMY60 plus pTM432 (N16 mutant); 13, TMY60 plus pTM431 (N14 mutant); 14, TMY60 plus pTM235 (pGAL1-DBR1-V5-6xHis); 15, TMY60 plus pYES2.1 (vector). All cultures were grown in galactose for 18 h prior to RNA isolation. The solid black arrow indicates the part of the ACT1 3′ intron probe protected by the ACT1 intron lariat (215 nt). The white arrow indicates the control reaction product, showing protection of the ACT1 3′ exon probe by ACT1 mature mRNA (333 nt). The C30 mutant has a nonsense mutation and does not make full-length Dbr1p.
The sensitivity of the RNase protection assay uncovers the fact that the Dbr1p-V5-6xHis fusion protein is not as efficient in debranching intron RNA lariats as native Dbr1p. Two different alleles encoding this fusion protein were tested: one allele has the GAL1 promoter driving expression of the fusion protein (pTM235), and the other allele has the native DBR1 promoter (pTM263). As shown in lanes 12 and 13 of Fig. 6A, the 156-nt fragment of the 5′ intron probe is readily detected in RNA samples from strains containing the two different DBR1-V5-6xHis fusion alleles, indicating protection by ACT1 intron sequences. The pGAL1-driven fusion needs galactose for full expression, and the result in lane 12 of Fig. 6A is for an RNA sample from cells grown for 18 h in 2% galactose. This debranching inefficiency can be contrasted with the absence of the 156-nt fragment in lane 1 of Fig. 5D, for which RNA from a strain bearing native, wild-type DBR1 was used. Interestingly, in Ty1 transposition assays, the wild-type Dbr1p-V5-6xHis fusion protein supports levels of transposition that are not readily distinguishable from levels supported by native, wild-type Dbr1p (data not shown).
Despite the inefficiency of the wild-type Dbr1p-V5-6xHis fusion protein in debranching intron RNA lariats, all of the mutants are clearly much less active. In terms of absolute levels of RNA lariat accumulation, the results for the RNase protection assays fluctuate slightly from one experiment to the next. This is most likely due to slight differences in growth state or other factors that vary from experiment to experiment. The sensitivity of the RNase protection assay then displays this variation. However, relative comparisons of the mutants to the wild type clearly show the correlation between Ty1 transposition and debranching activity.
Integration versus recombination of Ty1 cDNA in dbr1 mutant strains.
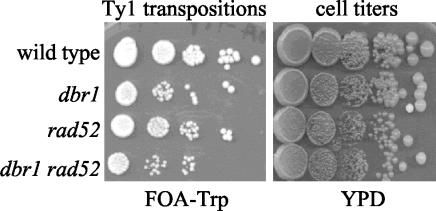
Ty1 cDNA produced by reverse transcription of Ty1 RNA can become incorporated into yeast chromosomal DNA by two independent processes. One process, integration, requires the Ty1-encoded integrase protein IN. IN binds the ends of the Ty1 cDNA, cleaves the chromosomal target sequence, and joins the Ty1 cDNA to the target DNA. Cellular repair enzymes complete the integration process (fill in gaps and ligate nicks). Interactions between homologous DNA sequences in the Ty1 cDNA and the chromosomal target are not part of the integration process. Integration sites seem to be chosen by protein-protein interactions, possibly between IN and a protein bound to target DNA. Homologous recombination is the second process for incorporation of Ty1 cDNA into host chromosomes. This process can only occur at sites where Ty1 sequences are already present. Rad52p is an essential component of the homologous recombination machinery, so rad52 mutants exhibit severe recombination defects (32), including Ty1 cDNA recombination. Previous work has shown that incorporation of Ty1 cDNA into the host chromosomes can occur in a rad52 mutant by the integration process described above (30). Likewise, incorporation of Ty1 cDNA for an IN mutant element can occur very efficiently by homologous recombination (30). However, incorporation of Ty1 cDNA for an IN mutant element does not occur in a rad52 mutant yeast strain because both integration and recombination are defective (30; data not shown). In addition, Ty1 mutants that produce alterations of the sequences at the Ty1 cDNA ends transpose only by homologous recombination. IN can no longer bind the cDNA ends in a way that promotes integration. Therefore, Ty1 mutants that produce altered cDNA ends do not transpose in a rad52 mutant.
We have compared the transposition rates of dbr1 and dbr1 rad52 strains to infer structural aspects of the Ty1 cDNA produced in a dbr1 mutant background. Strains YPH499 (wild type), TMY60 (dbr1), TMY176 (rad52), and TMY344 (dbr1 rad52) were transformed with the pGAL1-Ty1::his3AI plasmid (pBJC35) and, separately, the pGAL1-Ty1::TRP1 plasmid (pX3). Three transformants for each strain-plasmid combination were tested for Ty1 transposition. The results for transformants containing pX3 are depicted in Fig. 7. We observed the expected reduction in Ty1 transposition in the dbr1 mutant strain compared to the wild type. However, the frequency of the residual transposition events occurring in the dbr1 mutant strain is not significantly reduced any further in a dbr1 rad52 strain. If integration of the Ty1 cDNA was defective in a dbr1 mutant, transposition would be reduced at least 25-fold in the dbr1 rad52 strain compared to the dbr1 strain. This conclusion is based on the fact that Ty1 elements defective for integration transpose at wild-type levels in RAD52 yeast strains but at least 100-fold less efficiently in rad52 strains (30; data not shown). Therefore, we conclude that chromosomal incorporation of Ty1 cDNA produced in dbr1 strains in the absence of the major homologous recombination machinery must be catalyzed by the Ty1 IN protein. Because IN requires a precisely formed cDNA, these results suggest that the Ty1 cDNA produced in dbr1 strains is full length and contains the normal ends that interact with the Ty1 IN protein.
FIG. 7.
Ty1 cDNA from a dbr1 mutant can interact with IN protein. Transposition rates were measured by a dilution-spotting method. Strains YPH499 (wild type), TMY60 (dbr1), TMY176 (rad52), and TMY344 (dbr1 rad52) were spotted onto FOA-Trp (to select for transpositions) and YPD (to indicate cell titers). The dilution factor between adjacent spots is fivefold.
Loss of DBR1 dependence by Ty1 deletion elements.
We have compared the transposition frequency of a Ty1 element containing an internal deletion (pX100) (36) to a full-length Ty1 element (pX3) (35) to determine if Dbr1p acts on Ty1 RNA (Table 5). Ty1 bases 620 to 5562 are deleted in pX100, and Ty1 proteins are supplied in trans by an unmarked, transposition-defective Ty1 element carried on the same plasmid (36). Transposition was measured in wild-type and dbr1 deletion strains. As expected, the full-length element transposed at a much lower frequency in a dbr1 strain than it did in the wild-type strain (29% of the wild-type strain frequency in this experiment). However, the deletion element transposed at much more similar frequencies in wild-type and dbr1 strains (the dbr1 strain frequency was 72% of the wild-type strain frequency; Table 5). These data are consistent with the loss of DBR1-dependent sequences in the deletion element. It is notable, however, that the Ty1 deletion element transposed at an overall lower frequency in both wild-type and dbr1 mutant strains.
TABLE 5.
Transposition of a Ty1 deletion element
| Strain/plasmid | Transposition frequency (Ura− Trp+/Ura− colonies) | Relative frequency (%) |
|---|---|---|
| Wild type/pX3 | 2.88 × 10−1 (312/1,082) | 100.0 |
| dbr1/pX3 | 8.49 × 10−2 (79/931) | 29.4 |
| Wild type/pX100 | 2.98 × 10−2 (63/2,117) | 10.3 |
| dbr1/pX100 | 2.12 × 10−2 (50/2,353) | 7.4 |
DISCUSSION
The roles of Dbr1p in Ty1 transposition and intron lariat debranching are related.
Our goal is to determine the role that Dbr1p plays in Ty1 transposition. On the one hand, Dbr1p's role in Ty1 transposition may involve its 2′-5′ phosphodiesterase activity, which would suggest that an RNA with a 2′-5′ linkage is involved in transposition. Two RNAs are already known to play central roles in Ty1 transposition: Ty1 RNA acts as both an mRNA and the genome of the Ty1 virus-like particle (2), while tRNAimet acts as the primer for the initiation of Ty1 reverse transcription (7). Neither of these RNAs is known to contain a 2′-5′ linkage. On the other hand, Dbr1p's role in Ty1 transposition may not involve its 2′-5′ phosphodiesterase activity. Among the possibilities is one in which only the RNA binding capacity of Dbr1p is used in some way during transposition.
Dbr1p can be divided into different regions based on sequence conservation (Fig. 3). On the one hand, the N-terminal 242 amino acids show the strongest conservation with the Dbr1p sequences from 11 other species. The N-terminal region contains the phosphoesterase domain, comprising 20 conserved amino acids. However, the debranching enzymes exhibit additional extensive conservation. The 12 Dbr1p sequences in the GenBank database have 54 identical amino acids in their phosphoesterase domains, with an additional 84 amino acids conserved in at least 10 of the enzymes. A second region of conservation among the debranching enzymes lies outside of the phosphoesterase domain. For S. cerevisiae Dbr1p, this region encompasses approximately amino acids 294 to 340. In this region, the 12 Dbr1p sequences in the GenBank database have 6 identical amino acids, with an additional 21 amino acids conserved in at least 10 of the enzymes. The C-terminal regions of the different Dbr1p sequences vary greatly in length and do not contain significant similarities, although potential nuclear localization sequences are present in all of the enzymes.
Of the eight dbr1 mutations encoding a single amino acid change, seven lie within the phosphoesterase domain. Two dbr1 alleles (dbr1-N16 and dbr1-N30) have changes that alter the GD/GNH phosphoesterase signature motif, which is thought to form part of the active site of the enzyme (19). If this is the case and other functions of Dbr1p are intact in these mutants (e.g., RNA binding), the decrease in Ty1 transposition for these mutants would be a strong indication of the importance of phosphodiesterase activity for transposition.
Two other dbr1 alleles (dbr1-N20 and dbr1-N29) have changes that alter a potential tyrosine phosphorylation site at Y68. We have shown that Dbr1p is phosphorylated, although the specific phosphorylated residues remain to be identified. The Y68 mutations encoded by dbr1-N20 and dbr1-N29 remain phosphorylated on tyrosine, indicating that Dbr1p may have multiple phosphorylation sites. Alignment of Dbr1p sequences in Fig. 3 shows that the tyrosine represented by Y68 in S. cerevisiae is conserved among all of the Dbr1p proteins identified. A potential kinase phosphorylation site associated with this tyrosine is identified only for the S. cerevisiae, S. pombe, N. crassa, and P. falciparum proteins with Prosite software (12). Analysis of Dbr1p phosphorylation sites and identification of a Dbr1p kinase(s) will provide more information about the posttranslational regulation of Dbr1p.
Among the dbr1 mutants isolated in this study is one (dbr1-C86) that affects a region of Dbr1p that is not conserved and changes a glutamate to an aspartate. This region probably does not participate in the catalytic function of Dbr1p but may be involved in interactions that Dbr1p has with other proteins (e.g., spliceosomal proteins).
All of the dbr1 mutants defective for Ty1 transposition identified so far are also defective for RNA lariat debranching. Although we are continuing to identify additional dbr1 alleles, our results are consistent with a role for 2′-5′ phosphodiesterase activity in Ty1 transposition.
Intron RNA accumulation in dbr1 mutants.
Previous work by others, based on RNA levels and mobility patterns, suggested that intron RNAs accumulate in dbr1 mutants and maintain their lariat structure (6). RNase protection assays in our work also demonstrate the high levels to which intron RNAs accumulate in dbr1 mutants and confirm the maintenance of the lariat branch.
Another suggestion from previous work is that intron sequences accumulating in a dbr1 mutant lack most of their 3′ tails (6), presumably because of the action of exonucleases. The 3′ tail is the RNA sequence from the lariat branch point to the 3′ end of the excised intron. The protection of ∼215 nt of the 3′ intron probe in our RNase protection assay experiments demonstrates this directly. Our data do not distinguish whether the 3′ tail of the intron is degraded all the way to the branch point or to a position 1 nt or a few nucleotides downstream of the branch point. However, in RNA lariats, the 2′-5′ phosphodiester bond renders the adjacent 3′-5′ bond resistant to RNase and nuclease P1 (27). In addition, Dbr1p needs the adjacent 3′-5′ phosphodiester bond at the branch point in order to cleave the 2′-5′ phosphodiester bond (25, 28). Therefore, it is likely that the 3′ tail remaining on intron RNA lariats that accumulate in dbr1 mutant strains extends only 1 nt past the branch point.
If the entire intron lariat, including the 3′ tail, were accumulating in dbr1 strains, protection of 250 nt of the 3′ intron probe would occur. A fragment of ∼250 nt is observed accumulating in the dbr1 mutant strain (position of white arrow in Fig. 5D, lane 4), although it is present at a much lower level than the 215-nt fragment. This fragment is best observed on long exposures of RNase protection assay gels (data not shown). Enhanced accumulation of the entire intron lariat in the dbr1 mutant may indicate that other steps in the splicing reaction are slowed when the debranching reaction does not occur and intron lariats accumulate to a high level.
On the basis of what is known about mRNA splicing, debranching of intron lariats occurs after splicing is complete and is part of the process of degrading excised introns. The production of mature mRNA in the dbr1 mutant strain is expected to occur similarly to that in the wild type. Direct molecular evidence that this is the case is provided by the RNase protection assays with the 5′ intron probe described in Results. Protection of the probe by the mature ACT1 mRNA produced the 143-nt fragment indicated in lanes 1 and 2 of Fig. 5D by the stippled arrow. The levels of this fragment are very similar for dbr1 mutant and wild-type samples, especially compared to levels of the probe fragments protected by intron sequences (black arrow). However, on the basis of the accumulation of the 143-nt fragment, there is ∼30% more mature ACT1 mRNA in the wild-type strain than in the dbr1 mutant. This observation is consistent with slowing of other steps in the splicing reaction for the dbr1 mutant, such as trimming of the 3′ tail of the ACT1 intron lariat noted above.
Further support for slowing of other steps in the splicing reaction in the dbr1 mutant can be seen in the RNase protection assay that uses the 5′ intron probe. ACT1 pre-mRNA will protect a 283-nt fragment of the 5′ intron probe. In lane 2 of Fig. 5D, enhanced protection of a ∼283-nt fragment is observed for the dbr1 mutant compared to the wild type (position of white arrow). Slowing of the first transesterification step of the splicing reaction would account for the enhanced protection.
A side issue regarding the creation of the ACT1 lariat probe template (pTM400) is worth mentioning. Construction of pTM400 required the creation of an RT-PCR product that spans the ACT1 lariat branch point. One way to accomplish this is to reverse transcribe across the 2′-5′ linkage. Szostak and coworkers demonstrated that RTs can read across a lone 2′-5′ linkage in a template in which the remaining bonds are 3′-5′ linkages (22). We performed a similar type of RT reaction in our RT-PCRs. The major difference is that the intron branch point nucleotide with the 2′ linkage also has an adjacent 3′ linkage. Interestingly, in our reactions with the ACT1 branch, we found that deoxyadenosine was incorporated into the cDNA at the first position RT reads after crossing the branch point in 16 out of 17 RT-PCR clones (data not shown). Thymidine, the deoxynucleoside dictated by Watson-Crick base pairing, was present in the remaining clone. Another group found the same incorporation pattern during RT of a plant intron RNA (33). Szostak's group reported that RT stalls when RT incorporates the first nucleotide after crossing the 2′-5′ bond (22). If the Szostak template and an intron RNA lariat act similarly, the 2′-5′ linkage on the template seems to promote non-Watson-Crick base recognition during incorporation that results in altered nucleotide specificity. However, this type of base recognition appears to have only a minor effect on the rate of incorporation, as evidenced by minor stalling in cDNA synthesis at this position (22). However, once the nucleotide is incorporated, the base pairing interaction may position this nucleotide in a way that does not promote efficient incorporation of the subsequent nucleotide into the cDNA, resulting in very significant stalling in cDNA synthesis. It is not known if stalling occurs in the rare instances in which the correct nucleotide is incorporated (i.e., the nucleotide predicted by Watson-Crick pairing).
Ty1 cDNA produced in a dbr1 mutant can interact with the Ty1 IN protein.
Our previous work showed that Ty1 cDNA accumulates at a slower rate in dbr1 strains, suggesting that Dbr1p might be involved in Ty1 reverse transcription or cDNA stability. One possibility is that Ty1 RNA is a substrate for Dbr1p. However, because Ty1 reverse transcription is primed by the cellular tRNAimet, it is also possible that a 2′-5′ RNA-DNA branch remains on one end of the Ty1 cDNA. This would occur if the tRNA primes from a 2′-OH of instead of a 3′-OH, a possibility suggested previously (6). In work presented here, the Ty1 cDNA accumulating in dbr1 strains is capable of integration into the host chromosomal DNA in the absence of recombination machinery (dbr1 rad52 strain). This result indicates that the Ty1 cDNA has ends capable of interacting with the Ty1 IN protein, which rules out a 2′-5′ linkage remaining on one of the two ends. If Dbr1p plays a direct role in Ty1 transposition, these results point to Ty1 RNA as a possible substrate for the enzyme.
Identification of DBR1-dependent sequences in Ty1 RNA.
A Ty1 deletion element (pX100) missing 4,942 out of the 5,918 bases of Ty1 transposed at approximately the same frequency in wild-type and dbr1 strains. This result is consistent with the notion that Dbr1p acts on some feature that is part of the missing bases. One possibility is that an RNA branch forms in those bases or that the bases are important for the formation of a branch in the remaining Ty1 RNA sequences. Because RNA branching would not occur when the internal Ty1 sequences are missing, debranching would not be necessary and transposition would be similar in wild-type and dbr1 strains. An additional, striking feature of the Ty1 deletion element is that it transposes at an overall lower frequency than the full Ty1 element (Table 5) (36). This result indicates that the deleted sequences are important for Ty1 transposition. One possibility is that supplying Ty1 proteins in trans, as is the case for the deletion element, is inferior to supplying them in cis. Alternatively, just as removal of a branch in Ty1 RNA may be important for transposition, the formation of that branch may also be important. Accordingly, the Ty1 deletion element would not form the RNA branch and would transpose poorly as a result. However, as noted above, the transposition frequency would no longer vary between wild-type and dbr1 mutant strains.
The results presented here point to the possibility that a branch forms in Ty1 RNA during transposition. In experiments to be reported elsewhere (Z. Cheng and T. M. Menees, unpublished data), we have identified a branch in Ty1 RNA and are working to determine the functions of branch formation and removal in transposition. The relatedness of LTR retrotransposons and retroviruses raises the possibility that a debranching deficiency could similarly decrease the mobility of retroviruses. If such turns out to be the case, it could open new approaches for antiretroviral therapy.
Acknowledgments
This work was supported by grant 9983116 from the National Science Foundation.
We thank David Garfinkel, Joan Curcio, and Jef Boeke for providing plasmids; Gerald Wyckoff for help with Dbr1 protein alignments; and Antony Cooper and Chris Mattison for helpful discussions.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2003. Current protocols in molecular biology on CD-ROM. Current Protocols Inc., Brooklyn, N.Y.
- 2.Boeke, J., and J. Stoye. 1997. Retrotransposons, endogenous retroviruses and the evolution of retroelements, p. 343-435. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 3.Boeke, J. D., F. Lacroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 4.Cadwell, R. C., and G. F. Joyce. 1994. Mutagenic PCR. PCR Methods Appl. 3:S136-S140. [DOI] [PubMed] [Google Scholar]
- 5.Cadwell, R. C., and G. F. Joyce. 1992. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2:28-33. [DOI] [PubMed] [Google Scholar]
- 6.Chapman, K. B., and J. D. Boeke. 1991. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 65:483-492. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, K. B., A. S. Bystrom, and J. D. Boeke. 1992. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc. Natl. Acad. Sci. USA 89:3236-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper, J. A., F. S. Esch, S. S. Taylor, and T. Hunter. 1984. Phosphorylation sites in enolase and lactate dehydrogenase utilized by tyrosine protein kinases in vivo and in vitro. J. Biol. Chem. 259:7835-7841. [PubMed] [Google Scholar]
- 9.Curcio, M. J., and D. J. Garfinkel. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garfinkel, D. J., A. M. Hedge, S. D. Youngren, and T. D. Copeland. 1991. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J. Virol. 65:4573-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grate, L., and M. Ares, Jr. 2002. Searching yeast intron data at Ares lab web site. Methods Enzymol. 350:380-392. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database, its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter, T. 1982. Synthetic peptide substrates for a tyrosine protein kinase. J. Biol. Chem. 257:4843-4848. [PubMed] [Google Scholar]
- 14.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Karst, S. M., M.-L. Rutz, and T. M. Menees. 2000. The yeast retrotransposons Ty1 and Ty3 required the RNA lariat debranching enzyme, Dbr1p, for efficient accumulation of reverse transcripts. Biochem. Biophys. Res. Commun. 268:112-117. [DOI] [PubMed] [Google Scholar]
- 16.Karst, S. M., N. Sadeghi, and T. M. Menees. 1999. Cell cycle control of reverse transcriptase activity for the yeast retrotransposon Ty3. Biochem. Biophys. Res. Commun. 254:679-684. [DOI] [PubMed] [Google Scholar]
- 17.Kim, H. C., G. M. Kim, J. M. Yang, and J. W. Ki. 2001. Cloning, expression, and complementation test of the RNA lariat debranching enzyme cDNA from mouse. Mol. Cell 11:198-203. [PubMed] [Google Scholar]
- 18.Kim, J. W., H. C. Kim, G. M. Kim, J. M. Yang, J. D. Boeke, and K. Nam. 2000. Human RNA lariat debranching enzyme cDNA complements the phenotypes of Saccharomyces cerevisiae dbr1 and Schizosaccharomyces pombe dbr1 mutants. Nucleic Acids Res. 28:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin, E. V. 1994. Conserved sequence pattern in a wide variety of phosphoesterases. Protein Sci. 3:356-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauermann, V., M. Hermankova, and J. D. Boeke. 1997. Increased length of long terminal repeats inhibits Ty1 transposition and leads to the formation of tandem multimers. Genetics 145:911-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez, P. J., and B. Seraphin. 2000. YIDB: the yeast intron database. Nucleic Acids Res. 28:85-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorsch, J. R., D. P. Bartel, and J. W. Szostak. 1995. Reverse transcriptase reads through a 2′-5′ linkage and a 2′-thiophosphate in a template. Nucleic Acids Res. 23:2811-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nam, K., R. H. Hudson, K. B. Chapman, K. Ganeshan, M. J. Damha, and J. D. Boeke. 1994. Yeast lariat debranching enzyme. Substrate and sequence specificity. J. Biol. Chem. 269:20613-20621. [PubMed] [Google Scholar]
- 24.Nam, K., G. Lee, J. Trambley, S. E. Devine, and J. D. Boeke. 1997. Severe growth defect in a Schizosaccharomyces pombe mutant defective in intron lariat degradation. Mol. Cell. Biol. 17:809-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ooi, S. L., C. Dann III, K. Nam, D. J. Leahy, M. J. Damha, and J. D. Boeke. 2001. RNA lariat debranching enzyme. Methods Enzymol. 342:233-248. [DOI] [PubMed] [Google Scholar]
- 26.Patschinsky, T., T. Hunter, F. S. Esch, J. A. Cooper, and B. M. Sefton. 1982. Analysis of the sequence of amino acids surrounding sites of tyrosine phosphorylation. Proc. Natl. Acad. Sci. USA 79:973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruskin, B., and M. R. Green. 1990. RNA lariat debranching enzyme as tool for analyzing RNA structure. Methods Enzymol. 181:180-188. [DOI] [PubMed] [Google Scholar]
- 28.Ruskin, B., and M. R. Green. 1985. An RNA processing activity that debranches RNA lariats. Science 229:135-140. [DOI] [PubMed] [Google Scholar]
- 29.Sadeghi, N., M. L. Rutz, and T. M. Menees. 2001. Thermal blockage of viruslike particle formation for the yeast retrotransposon Ty3 reveals differences in the cellular stress response. Arch. Virol. 146:1919-1934. [DOI] [PubMed] [Google Scholar]
- 30.Sharon, G., T. J. Burkett, and D. J. Garfinkel. 1994. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol. Cell. Biol. 14:6540-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogel, J., W. R. Hess, and T. Borner. 1997. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 25:2030-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, S. Bowman, K. Brooks, D. Brown, S. Brown, T. Chillingworth, C. Churcher, M. Collins, R. Connor, A. Cronin, P. Davis, T. Feltwell, A. Fraser, S. Gentles, A. Goble, N. Hamlin, D. Harris, J. Hidalgo, G. Hodgson, S. Holroyd, T. Hornsby, S. Howarth, E. J. Huckle, S. Hunt, K. Jagels, K. James, L. Jones, M. Jones, S. Leather, S. McDonald, J. McLean, P. Mooney, S. Moule, K. Mungall, L. Murphy, D. Niblett, C. Odell, K. Oliver, S. O'Neil, D. Pearson, M. A. Quail, E. Rabbinowitsch, K. Rutherford, S. Rutter, D. Saunders, K. Seeger, S. Sharp, J. Skelton, M. Simmonds, R. Squares, S. Squares, K. Stevens, K. Taylor, R. G. Taylor, A. Tivey, S. Walsh, T. Warren, S. Whitehead, J. Woodward, G. Volckaert, R. Aert, J. Robben, B. Grymonprez, I. Weltjens, E. Vanstreels, M. Rieger, M. Schafer, S. Muller-Auer, C. Gabel, M. Fuchs, A. Dusterhoft, C. Fritzc, E. Holzer, D. Moestl, H. Hilbert, K. Borzym, I. Langer, A. Beck, H. Lehrach, R. Reinhardt, T. M. Pohl, P. Eger, W. Zimmermann, H. Wedler, R. Wambutt, B. Purnelle, A. Goffeau, E. Cadieu, S. Dreano, S. Gloux, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871-880. [DOI] [PubMed] [Google Scholar]
- 35.Xu, H., and J. D. Boeke. 1987. High-frequency deletion between homologous sequences during retrotransposition of Ty elements in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 84:8553-8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, H., and J. D. Boeke. 1990. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: analysis of mini-Ty1 elements. Mol. Cell. Biol. 10:2695-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]