Abstract
Substitution of a methionine residue at position 79 in poliovirus protein 3A with valine or threonine caused defective viral RNA synthesis, manifested as delayed onset and reduced yield of viral RNA, in HeLa cells transfected with a luciferase-containing replicon. Viruses containing these same mutations produced small or minute plaques that generated revertants upon further passage, with either wild-type 3A sequences or additional nearby compensating mutations. Translation and polyprotein processing were not affected by the mutations, and 3AB proteins containing the altered amino acids at position 79 showed no detectable loss of membrane-binding activity. Analysis of individual steps of viral RNA synthesis in HeLa cell extracts that support translation and replication of viral RNA showed that VPg uridylylation and negative-strand RNA synthesis occurred normally from mutant viral RNA; however, positive-strand RNA synthesis was specifically reduced. The data suggest that a function of viral protein 3A is required for positive-strand RNA synthesis but not for production of negative strands.
The molecular mechanism of viral RNA replication in picornavirus-infected cells has been the subject of much investigation for many years. Although the enzymatic role of 3D as an RNA-dependent RNA polymerase was assigned quite early (20, 21, 40), it was also recognized that the properties of this protein precluded its ability to initiate RNA strand synthesis by itself. It can elongate a primer bound to a template, but in the absence of a primer, it can only catalyze addition of a few nucleotides to the 3′ end of RNA (49).
Subsequent studies showed that 3D catalyzes the uridylylation of 3B (VPg) to generate a primer for initiation of RNA synthesis (55, 78) and also exhibits an RNA duplex-unwinding activity in vitro (11). Genetic analyses have shown that many other proteins are required for viral RNA synthesis, but specific biochemical roles have not been defined. The analyses are complicated by the fact that many proteins perform multiple functions during genome replication, and some intermediate products of protein processing manifest independent functional roles. For example, viral protein 2C is highly conserved among picornaviruses, and it appears to be involved in several different steps of virus replication. Analyses of mutants produced by site-directed mutagenesis and localization of mutations that confer guanidine resistance have implicated 2C sequences in virion uncoating, host cell membrane rearrangement, RNA synthesis, and encapsidation (17, 37, 43, 57, 72, 75, 79). In agreement with these proposed multiple functions of the 2C protein, biochemical studies have shown that 2C exhibits several different activities: (i) ATPase activity (42, 62) which is inhibited by 2 mM guanidine hydrochloride (GuHCl), an inhibitor of poliovirus (PV) RNA replication (56); (ii) membrane-binding activity that may induce or participate in the rearrangement of intracellular membranes (1, 12, 18, 67, 71) (membrane binding and induction of rearrangement is also a property of 2BC and 2B); and (iii) RNA binding activity (61), with reported specificity for sequences at the 3′ terminus of negative-strand PV RNA (4, 5).
Viral protein 2B also plays an important role in viral RNA replication. Several noncomplementable mutants with mutations in 2B exhibited dose-dependent dominance over the replication of wild-type virus, suggesting that 2B plays a structural role in replication complexes (31). Expression of coxsackie B3 virus protein 2B disrupted the endoplasmic reticulum (ER) membrane, and it was suggested that such alterations could facilitate plasma membrane lesions and virus release (80). For rhinoviruses RV2 and RV39, variants that could replicate in normally nonpermissive mouse cells were found to contain altered 2B proteins (38, 84).
The 2A proteinase of enteroviruses also has been shown to exhibit several different biochemical activities. It is responsible for the primary cleavage event in the enterovirus polyprotein at the 1D-2A junction, and its proteolytic activity is required for the cleavage of eukaryotic translation initiation factor 4G (32, 86). It has been shown that 2A stimulates utilization of the PV ribosome entry site (25, 60). In addition, genetic data suggest that 2Apro has an essential function in PV RNA replication, but its role remains unknown (3, 39, 45, 52, 85).
Viral protein 3A also contains sequences essential for several different steps in viral RNA replication. 3A (but not 3AB) expressed in mammalian cells in the absence of other viral proteins causes a swelling of ER membranes and inhibits protein secretion by preventing traffic from the ER to the Golgi (15, 16). This activity likely requires the membrane-binding properties of 3A conferred by its hydrophobic C terminus, but the activity is lost by deletion of amino acid residues 1 to 10 at the N terminus of the protein. It has been proposed that this inhibition of protein trafficking may allow the virus to evade the host immune response to infection (50). Expression of 3AB in the absence of other viral proteins in HeLa cells induced significant rearrangement of intracellular membranes into concentric myelin-like structures (19).
Both 3A and its putative precursor 3AB (65, 68, 69) are membrane associated in infected cells (13, 65), and cleavage of 3AB to 3A and 3B (VPg) by 3CD in vitro requires this membrane environment (34). VPg is a strongly basic peptide predicted not to bind to the membranes of the RNA replication complex; thus, it was postulated that VPg is delivered to the replication complex in the form of a precursor molecule, possibly 3AB (65). Membrane association is determined by a hydrophobic region in the C-terminal portion of 3A (76). Several mutations engineered into the hydrophobic region of 3A have lethal or severely impaired replication phenotypes (22, 35). The ∼22-amino-acid hydrophobic region of 3A is highly conserved among enterhinoviruses, and the aphthovirus foot-and-mouth disease virus 3A protein sequences also have been shown to localize to intracellular membranes and to disrupt the structure of Golgi stacks (51). It is not clear whether 3A exhibits functional activities independently of its precursor forms or is generated only as a by-product of cleavage following participation of 3AB or other precursors in replication reactions. It has been reported that a portion of both 3AB and 3A synthesized in rabbit reticulocyte lysate in vitro is glycosylated (13), but the biological significance of this observation is questionable, since inhibitors of N glycosylation do not affect PV replication in infected cells.
PV 3AB is a nonspecific RNA-binding protein (36). In addition, specific protein interactions occur between 3AB and 3D (30, 36, 41, 58, 81) as well as 3CD (44). In vitro these interactions have been linked to several different activities: (i) stimulation of the elongation activity of 3Dpol (34, 53, 58, 59, 63); (ii) stimulation of self-cleavage of 3CDpro (44); (iii) stimulation of specific complex formation with 3CDpro on the 5′ cloverleaf (26), which may be essential for genome replication (83); and (iv) binding of 3Dpol to membranes (41). Only 3AB, not 3A, manifests these activities. Precise biochemical mechanisms of these proposed “cofactor-like” roles for 3AB have not been elucidated.
In this report, we describe analysis of a 3A mutant defective in viral RNA synthesis. We show that the defect does not affect uridylylation of VPg or synthesis of negative-strand RNA but markedly reduces positive-strand synthesis.
MATERIALS AND METHODS
Recombinant DNA procedures.
Restriction endonucleases, avian myeloblastosis virus reverse transcriptase, and T7 RNA polymerase were obtained from commercial sources (New England Biolabs, Roche Molecular Biochemicals, Promega, and Ambion) and used according to standard procedures (64). DNA sequencing was performed using a BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems), and the sequencing products were resolved on an automated sequencer (ABI Prism 310; Applied Biosystems). If not indicated otherwise, PCR amplifications were done with cloned Pfu polymerase (Stratagene), using reaction conditions recommended by the manufacturer, and the primers (Invitrogen) listed in Table 1.
TABLE 1.
Oligonucleotides used for mutagenesis
| Oligonucleotide | Sequence (5′-3′)a |
|---|---|
| 3AM79V frwd | GGAGTTGTCTATGTCGTGTATAAACTGTTTGCTGG |
| 3AM79V rev | CCAGCAAACAGTTTATACACGACATAGACAACTCC |
| 3AM79T frwd | GTTGTCTATGTCACGTATAAACTGTTTGC |
| 3AM79T rev | GCAAACAGTTTATACGTGACATAGACAAC |
| 3AM79L frwd | GTTGTCTATGTCCTGTATAAACTGTTTGC |
| 3AM79L rev | GCAAACAGTTTATACAGGACATAGACAAC |
| 3AH86Y frwd | GTTTGCTGGATATCAGGGAGCATACAC |
| 3AH86Y rev | GCTCCCTGATATCCAGCAAACAG |
| 3AM79VH86Y frwd | GGAGTTGTCTATGTCGTGTATAAACTGTTTGCTGGATACCAGGGAGCATACACTGG |
| 3AM79VH86Y rev | CCAGTGTATGCTCCCTGGTATCCAGCAAACAGTTTATACACGACATAGACAACTCC |
| 3DK276L frwd | CACCTGTACCTGAATAAAACATACTGTGTCAAGGG |
| 3DK276L rev | CACAGTATGTTTTATTCAGGTACAGGTGGTGTGAG |
Substituted nucleotides are indicated in bold and underlined.
Random mutagenesis of the PV P2-P3 region.
cDNA fragments were generated and amplified by PCR with Taq polymerase (Gibco) under conditions favoring mutagenesis (7 mM MgCl2), utilizing a forward primer from nucleotide (nt) 2382 to 2404 and a backward primer from nt 4693 to 4672 in pRLuc31 cDNA. The PCR products were digested with XhoI and BglII endonucleases, inserted into plasmid pRLuc31 (2), and then used to transform Escherichia coli DH5α. Approximately 100 colonies were isolated by ampicillin selection, and plasmids were prepared from each clone. These were used individually as templates for transcription by T7 RNA polymerase, and the RNA products were used to transfect HeLa cells.
Viral cDNA clones.
Plasmid pT7PV1 (24) contains a full-length infectious cDNA of PV1; plasmid pRLuc31 (2) contains cDNA of a PV1-luciferase replicon, and plasmid pTM1FG3ABwt (76) encodes the encephalomyocarditis virus internal ribosome entry site, FLAG epitope tag (DYKDDDDK), and PV 3AB protein. pLucPV-3A-M79V, pLucPV-3A-M79T, pLucPV-3A-M79L, pLucPV-3A-H86Y, pLucPV-3A-M79VH86Y, and pLucPV-3D-K276L were engineered by overlap extension site-directed mutagenesis (29) using the primer pairs listed in Table 1. All plasmids were analyzed by restriction mapping and sequencing of the entire PCR-generated region to verify that no inadvertent mutations were present. Conventional subcloning methods were used to introduce the same mutations in full-length pT7PV1 cDNA to create pT7PV-3A-M79V and pT7PV-3A-M79T. The cis-active hammerhead ribozyme coding sequence was introduced at the 5′ end of PV cDNA by substituting the 363-bp StuI-AgeI fragment from the plasmid prib(+) XpA (27) for the corresponding fragment in pT7PV1 wild-type or mutant plasmids cut with the same enzymes to create plasmids p5′Rz-PV1, p5′Rz-PV-3A-M79V, and p5′Rz-PV-3A-M79T.
The QuickChange site-directed kit (Stratagene) was utilized to introduce mutations 3A-M79V and 3A-M79T into expression plasmid pTM1FG3AB-wt (76). PCR mutagenesis was conducted according to the manufacturer's protocol, and clones were screened by sequence analysis of the entire FLAG-3AB region.
RNA transcription.
Full-length plasmids pT7PV1 (wild type and mutants) were linearized with the restriction enzyme EcoRI, and pRLuc31 derivative plasmids (wild type and mutants) were linearized with MluI. Transcription reactions were performed with T7 RNA polymerase using a MEGAscript in vitro transcription kit (Ambion, Inc.). Transcription reactions were incubated at 37°C for 60 min, and reactions were terminated by treatment with DNase I for 15 min at 37°C. RNAs were purified by phenol-chloroform extractions and precipitated with ethanol. For in vitro translation-replication reactions, RNAs were additionally purified by passage on ChromaSpin-100 columns (Clontech). The RNA concentration was estimated by the A260.
RNA transfection.
One microgram of RNA was used to transfect HeLa cells at 80% confluency in 35-mm dishes. Transfections were performed with DEAE-dextran (molecular weight, 500,000; Pharmacia) as described previously (73). At the indicated times, cells were washed with phosphate-buffered saline and used either for luciferase assays or for isolation of total cytoplasmic RNA.
Luciferase assay.
Promega's luciferase assay system was used for quantitation of luciferase. Cells were washed once with phosphate-buffered saline and lysed directly on the dish with 600 μl of lysis buffer. Five to 10 μl of lysate was used for the assay. Luciferase activity was determined in a luminometer (Wallac's Trilux 1450 microbeta) and expressed as relative light units.
Cell-free translation-replication assays.
In vitro translation assays were performed in microccocal nuclease-treated HeLa cell extracts essentially as described previously (6, 70, 77). Reaction mixtures (60 μl) were programmed with 20 nM full-length or replicon viral RNA. For analysis of translation products, 10-μl aliquots were supplemented with 15 μCi of [35S]methionine (1,000 Ci/mmol; Amersham) and incubated at 34°C for 3.5 h. Samples of 1 μl were taken to determine methionine incorporation into trichloroacetic acid (TCA)-insoluble material, and the remainder was diluted with an equal volume of 2× sample buffer for electrophoresis in sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gels. The remaining 50-μl portion of each reaction mixture was used for analysis of RNA synthesis, VPg uridylylation, or virus production.
PV RNA synthesis was assayed using preinitiation RNA complexes formed in translation-replication reactions at 34°C for 3.5 h in the presence of 2 mM GuHCl, essentially as described elsewhere (6). The reactions were terminated by addition of buffer RLT (Qiagen), and total RNA was isolated by using an RNeasy Mini kit (Qiagen) and precipitated with ethanol. RNAs were denatured by incubation in glyoxal-dimethyl sulfoxide buffer at 60°C for 30 min and separated by electrophoresis in 1% agarose. 32P-labeled RNA was detected by autoradiography and quantitated using a PhosphorImager (Molecular Dynamics).
VPg uridylylation was assayed in preinitiation RNA replication complexes formed in 50-μl reaction mixtures containing 2 mM GuHCl as described above. Pellets containing preinitiation RNA replication complexes were resuspended in 50-μl labeling reaction mixtures similar to those used for RNA synthesis but supplemented with 100 μCi of [α-32P]UTP (3,000 Ci/mmol; NEN). The reaction mixtures were incubated for 60 min at 37°C and then centrifuged at 14,000 × g to pellet replication complexes containing uridylylated VPg. The pellets were resuspended in Tris-EDTA buffer, and proteins were solubilized by addition of an equal volume of 2× protein sample buffer. Samples were fractionated by electrophoresis in a polyacrylamide-Tris-Tricine gel system (14). After electrophoresis gels were dried without fixation, and radiolabeled products were detected by phosphorimaging.
Cell-free virus production was measured in reaction mixtures (50 μl) incubated at 34°C for 16 h. The reaction mixtures were treated with 20 μg of RNase A and used for plaque assays on HeLa cell monolayers.
Membrane association assay.
3AB proteins were synthesized in the presence or absence of canine microsomal membranes in rabbit reticulocyte lysates programmed for coupled transcription-translation, essentially as recommended by the manufacturer (Promega). A 120-μl coupled transcription-translation reaction mixture was prepared containing the following: a 0.75 volume (90 μl) of TnT Quick master mix (Promega), 1.5 to 2 μg of wild-type or mutant pTMFG3AB plasmid, 1.5 μg of pTMFG cytochrome b5, 70 μCi of [35S]methionine, and 14 μl of canine microsomal membranes (Promega) if required. Reactions were incubated for 90 min at 30°C. Each sample was then subjected to equilibrium centrifugation in high-density sucrose gradients as described by Towner et al. (76). Briefly, reaction mixtures were diluted to 400 μl with 30% (wt/wt) sucrose in RSB (10 mM Tris [pH 7.4], 10 mM NaCl, and 1.5 mM MgCl2) and layered on a discontinuous sucrose gradient formed with 400 μl of 60% (wt/wt) sucrose, 600 μl of 45% (wt/wt) sucrose, and 400 μl of 40% (wt/wt) sucrose. The sample was overlaid with 400 μl of RSB and spun in a TLS-55 rotor (Beckman) at 93,000 × g for 17 h at 4°C. Gradient fractions (120 μl) were collected, diluted with an equal volume of 2× SDS sample buffer, and boiled for 4 min. Each sample was diluted to 1.5 ml with immunoprecipitation buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 2 mM EDTA) and subjected to immunoprecipitation using the anti-FLAG M2 monoclonal antibody (Sigma) and protein A agarose (Invitrogen). The immunoprecipitation was carried out under rotation at 4°C for 16 to 18 h. Immunoprecipitated proteins were separated in an SDS-12.5% polyacrylamide gel.
RESULTS
Isolation and mapping of a replication-defective PV with a mutation in PV protein 3A.
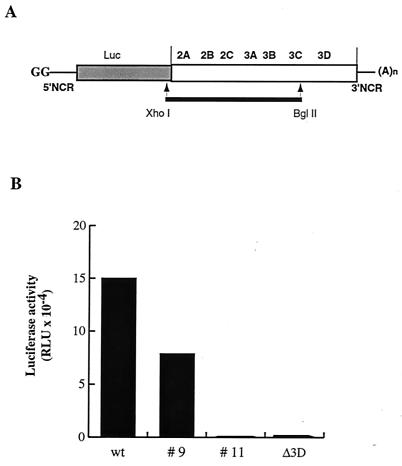
To identify defects in PV RNA replication caused by mutations in viral gene sequences encoding various nonstructural proteins, we generated random point mutations in the P2-P3 region of a PV replicon. The cDNA region representing nt 3385 to 5601 of the PV genome was amplified by PCR under conditions favoring random nucleotide misincorporation. The mutagenized fragments were used to replace the corresponding region of the luciferase-expressing PV replicon cDNA in plasmid pRLuc31 (2) (Fig. 1A), and individual clones of E. coli containing plasmid DNA were isolated after transformation. Cloned plasmids were then screened for their abilities to produce PV RNA genomes defective in RNA replication. HeLa cells were transfected with PV replicon RNAs transcribed in vitro, and luciferase activity was determined in extracts of cells harvested 7 and 11 h posttransfection. It had been shown previously (2) that the levels of luciferase activity in the transfected cells are a measure of replication of the RNA, with replicating RNAs generating exponential increases in luciferase activity and nonreplicating RNAs producing only low basal levels of luciferase activity translated from the input RNA.
FIG. 1.
Mutant PV replicon RNAs with defects in RNA synthesis. (A) PV replicon RNAs with structural genes replaced by a luciferase reporter gene are diagrammed. PV coding sequences are indicated in the white box, and the sequence coding for luciferase is shaded. XhoI-BglII fragments produced by PCR under conditions favoring random mutations are shown by a black line. The RNAs contain two nonviral guanylate residues at the 5′ end, generated by use of the T7 promoter. Luciferase is released from the PV polyprotein by 2A protease cleavage. (B) Luciferase expression was measured in HeLa cell extracts transfected with RNA transcripts produced from individual clones of mutated pLuc-PV. Luciferase activity (in relative light units [RLU]) is shown in extracts from 1.4 × 104 cells at 7 h after transfection.
The screened RNAs manifested three phenotypes, examples of which are shown in Fig. 1B. Some produced luciferase activity at the same level as wild-type Luc-PV RNA (Fig. 1B, wt). These RNAs either contained no mutations or contained mutations that had no effect on viral RNA replication. Some produced no detectable increase in luciferase activity (Fig. 1B, mutant 11). These RNAs contained one or more mutations that inhibited viral RNA replication and produced luciferase signals comparable to that induced by a control 3Dpol mutant, LucPVΔ3D, previously shown to be lacking RNA-dependent RNA polymerase activity (10). One RNA induced detectable but diminished luciferase activity (Fig. 1B, mutant 9). Among 105 RNAs screened, approximately 58% had wild-type activity and 42% demonstrated either a defective or a null RNA synthesis phenotype.
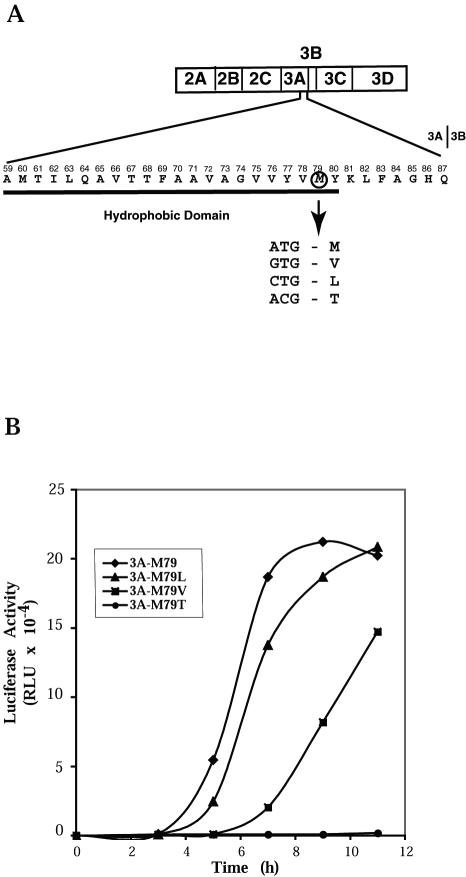
The nucleotide sequence of the entire region produced by PCR was determined for mutant 9. A single A-to-G transition at position 5345 of the PV genome was identified, which predicted an M79V change in the 3A protein sequence. The altered M residue is located at the border of the hydrophobic region near the C-terminal end of protein 3A (Fig. 2A). To confirm that this single mutation was responsible for the observed mutant phenotype, we reconstructed the mutation in the wild-type pRLuc31 background by site-directed mutagenesis to produce pLucPV-3A-M79V. We also introduced alternative single nucleotide substitutions in the codon for residue 79 of protein 3A (Fig. 2A), to generate pLucPV-3A-M79L and pLucPV-3A-M79T. The reconstituted M79V mutant RNA behaved identically to mutant 9 when HeLa cells were transfected with mutant replicon RNA: it manifested delayed onset of replication and a reduced yield of replicated RNA. Other substitutions of amino acid 79 produced 3A sequence-containing proteins that caused lesser or greater defects in RNA synthesis, as shown in Fig. 2B. RNA encoding the M79L substitution behaved similarly to the wild-type PV-Luc replicon, whereas RNA encoding threonine at 3A residue 79 failed to demonstrate detectable replication in transfected cells.
FIG. 2.
Mutation in the sequence coding for PV protein 3A causing defects in RNA replication. (A) The amino acid sequence of the C-terminal part of wild-type PV 3A protein is shown with the residue numbers in 3A indicated above it. The cleavage site between 3A and 3B is indicated after amino acid 87. The hydrophobic domain of protein 3A is underlined. Residue methionine 79 is circled. The codon for methionine and the changes introduced by site-directed mutagenesis are shown. (B) Time course of RNA replication measured by luciferase accumulation following RNA transfection of HeLa cells at 37°C.
Characterization of virus harboring mutations in protein 3A.
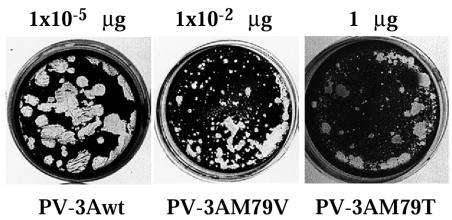
The M79V and M79T mutations were introduced in PV full-length cDNA in plasmid pT7-PV1 (24). Serial dilutions of RNA transcripts produced from these cDNAs were used to transfect HeLa cell monolayers and assayed for the ability to produce infectious progeny (Fig. 3). Wild-type RNA from pT7-PV1 manifested a specific infectivity of 4 × 10 6 PFU/μg of RNA. RNA from pT7-PV-3A-M79V showed a much lower specific infectivity of ∼1 × 10 4 PFU/μg of RNA and produced plaques that were significantly smaller in size; the mutant virus was not temperature sensitive (data not shown). RNA from pT7PV-3A-M79T showed a very low specific infectivity, ∼10 PFU/μg of RNA, and produced minute-size plaques. Among the small plaques produced upon transfection of HeLa cells with PV-3A-M79V or PV-3A-M79T RNA, some larger plaques were observed, and reversion to a large plaque phenotype occurred consistently upon subsequent passage of the mutant virus stock, so that large plaques were dominant after the second passage (data not shown). To characterize reversions that restored the wild-type plaque phenotype for vPV-3A-M79V, individual plaques were picked and virus stock was amplified once in HeLa cells and used to infect fresh HeLa cell monolayers. Total RNA isolated from infected cells was subjected to reverse transcription-PCR (RT-PCR), and the sequence corresponding to PV genome nt 5220 to 5570 was determined. Approximately half of the plaques contained revertant virus that had undergone a G-to-A transition to regenerate wild-type 3A sequence; a similar number yielded virus that had retained the M79V mutation but also had acquired an additional mutation resulting from a C-to-U transition at nt 5366, coding for substitution H86Y in viral protein 3A (Table 2). Reconstruction of the latter mutation in the pLucPV-3A-M79V construct restored wild-type kinetics of luciferase (data not shown) and RNA production (see below).
FIG. 3.
Plaque morphology of the PV-3A-79 mutants. HeLa cell monolayers were transfected with serial dilutions of wild-type or mutant RNA transcripts and overlaid with minimal essential medium containing 0.4% agarose 1 h after transfection. Plates were incubated at 37°C and stained with crystal violet 48 h after transfection. The plate transfected with the indicated amount of each RNA is shown.
TABLE 2.
Characteristics of viruses recovered after transfection of HeLa cells with RNAs bearing mutant 3A coding sequences
| RNA | 3A C-terminal sequencea | Transfection efficiency (log10 PFU/μg) | RNA synthesis | 3A C-terminal sequenceb in virus recovered after passage |
|---|---|---|---|---|
| PV1wt | GVVYVMYKLFAGHQ | 6 | Normal | GVVYVMYKLFAGHQ |
| PV-3A-M79V | GVVYVVYKLFAGHQ | 4 | Delayed kinetics and reduced yield | GVVYVMYKLFAGHQ |
| GVVYVVYKLFAGYQ | ||||
| PV-3A-M79T | GVVYVTYKLFAGHQ | 1 | Not detectable in single-step growth curve | GVVYVMYKLFAGHQ |
| GVVYVTYKLFAGYQ | ||||
| GVVHVTYKLFAGHQ |
Amino acids 74 to 87 of protein 3A. The amino acid in position 79 is shown in bold.
The amino acid in position 79 and acquired mutations are in bold and underlined.
To characterize vPV-3A-M79T, HeLa cells were transfected with the corresponding RNA transcript and incubated at 37°C for approximately 48 h. This extended incubation was required to achieve complete cytopathic effect and was significantly longer than was required for the wild-type RNA transcript (∼20 h). The virus stock obtained after transfection was used to infect fresh monolayers of HeLa cells without plaque purification. RNA was isolated from cells in four separate plates at 4.5 h after infection and was subjected to RT-PCR. Sequence analyses revealed revertant virus with wild-type 3A sequence, as well as virus that retained the M79T mutation and had acquired additional compensating mutations either at H86Y or Y77H, the latter resulting from a U-to-C transition at nt 5339 (Table 2).
Synthesis and processing of proteins in HeLa cell extracts.
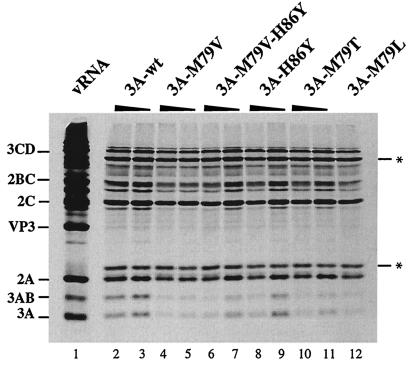
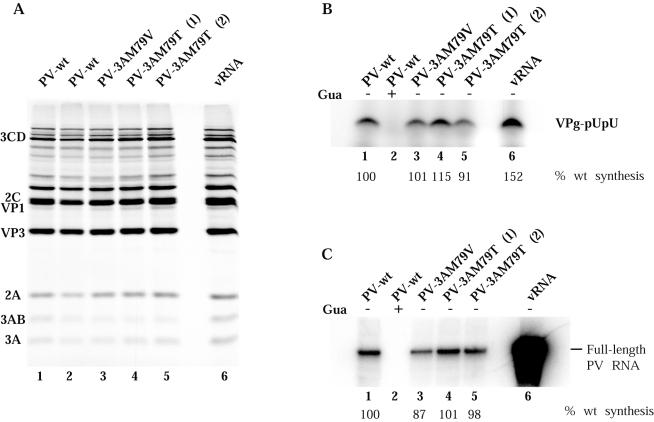
Residue 79 of protein 3A lies within 10 residues of the 3A-3B cleavage site, and both compensating mutations were located at nearby residues, either upstream or downstream of residue 79. A previous study showed that mutations in this hydrophobic region of 3A often caused aberrant proteolytic processing of the polyprotein with very poor cleavage at the 3AB-3CD cleavage site (22, 35). To determine whether the defects in RNA synthesis caused by 3A-M79 substitutions described above resulted from an effect on protein processing, transcripts from luciferase-containing PV replicon clones coding for wild-type, 3A mutant, or revertant sequences were used to program HeLa cell extracts competent for translation (46). [35S]methionine-labeled proteins were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Figure 4 shows the protein products, translated from two concentrations of each RNA. Translation of all RNAs tested occurred with equal efficiencies and produced similar patterns of processed products. Although some minor variability in overall translation activity was observed for the various RNAs in different experiments, there were no consistent differences in protein products. Thus, the M79V, M79T, M79L, and H86Y changes did not alter the proteolytic processing pattern of the polyprotein.
FIG. 4.
In vitro translation of replicon RNAs. T7 RNA transcripts (20 and 10 nM) from wild-type pRLuc-31 (3A-wt) (lanes 2 and 3), pLuc-3A-M79V (lanes 4 and 5), pLuc-3A-M79V-H86Y (lanes 6 and 7), pLuc-3A-H86Y (lanes 8 and 9), pLuc-3A-M79T (lanes 10 and 11), and pLuc-3A-M79L (lane 12; 20 nM) were used to program translation in HeLa S10 extracts supplemented with ribosomal salt wash in the presence of [35S]methionine. Lane 1 shows the products of translation programmed with 10 nM PV1 virion RNA. Reaction products were subjected to electrophoresis on a 12.5% polyacrylamide-SDS gel. The asterisk indicates Luc-related protein bands.
Membrane association of mutant 3AB proteins.
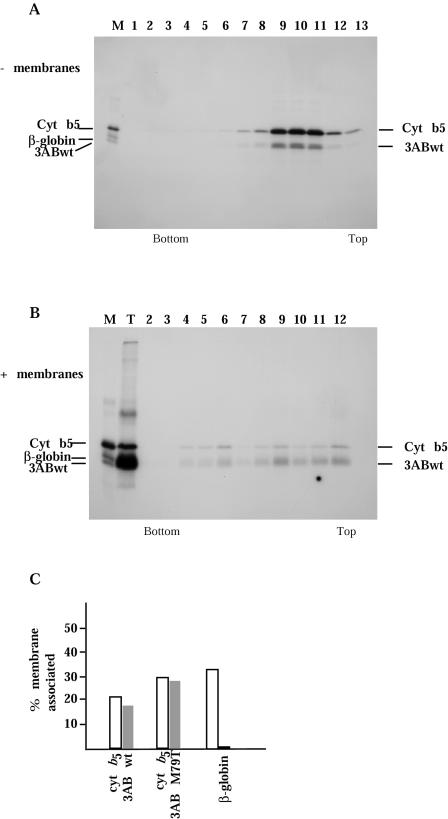
The role of PV protein 3A in the viral replication cycle is not known. It is found in infected cells exclusively associated with membranes in RNA replication complexes, generally in the form of protein 3AB. It has been proposed to serve as the membrane anchor for VPg (3B) in the replication complex by virtue of its hydrophobic membrane-binding domain. The conserved hydrophobic domain in the carboxy-terminal region of 3A was shown previously to direct the association of protein 3AB with membranes (13, 76). As residue 79 of 3A is located in this conserved hydrophobic domain, we attempted to determine whether these mutations affected the membrane association properties of 3AB proteins. To this end, we introduced the M79T and M79V mutations into a 3AB expression plasmid, pTM1-FLAG-3ABwt (76), which can be used to generate protein 3AB fused to a FLAG epitope in the absence of other PV proteins. Evidence for potential differences in membrane association of the mutant 3AB proteins was sought by examining the sedimentation profiles of FLAG-tagged radiolabeled 3AB proteins synthesized in vitro in the presence of canine pancreas membranes. Previous work has demonstrated that FLAG-tagged 3AB proteins cosediment with membranes in sucrose gradients, whereas those carrying deletions in the hydrophobic domain or multiple replacements of hydrophobic residues by charged residues within this region fail to demonstrate membrane association (76). Wild-type and mutant FLAG-tagged 3AB proteins were synthesized in vitro in the presence or absence of canine pancreas membranes. As a positive control for membrane association, we used FLAG-tagged rabbit cytochrome b5 cotranslated with wild-type or mutant 3AB. Negative controls were performed with the non-membrane-binding model protein FLAG-β-globin. Translation reaction mixtures were layered on discontinuous sucrose gradients and subjected to centrifugation for 16 h. Gradient fractions were collected, immunoprecipitated with anti-FLAG antibody, and analyzed by SDS-PAGE. When translated in the absence of membranes, both 3AB and cytochrome b5 were present in the top half of the sucrose gradient following centrifugation (Fig. 5A), along with the FLAG-β-globin model protein (data not shown).
FIG. 5.
Sucrose density gradient analysis of 3AB proteins. (A and B) Translation reactions were programmed with FLAG epitope-tagged wild-type 3AB and cytochrome b5 in the absence (A) or presence (B) of canine pancreas microsomal membranes. Products were sedimented through discontinuous sucrose gradients, and fractions were collected and immunoprecipitated with anti-FLAG antibody before analysis on SDS-PAGE gels. A mixture of individually translated FLAG epitope-tagged wild-type 3AB, cytochrome b5, and β-globin proteins served asmarkers for the gel (lane M); lane T in panel B represents a sample of the translation reaction not subjected to gradient sedimentation. (C) Radioactivity in each fraction of the sucrose gradients containing proteins from translation reactions programmed with cytochrome b5 mRNA and either wild-type 3AB, 3AB-M79T, or β-globin mRNAs, in the presence of canine pancreas microsomal membranes, was quantitated by phosphorimager. The percentage of total radioactivity cosedimenting with membranes (fractions 2 to 6) was calculated for each protein and plotted.
When wild-type 3AB and cytochrome b5 were translated in the presence of canine microsomal membranes, both proteins were also found in the bottom half of the gradient (Fig. 5B, lanes 3 to 5), indicating membrane association of these proteins. No such redistribution was observed for the negative control non-membrane-binding model protein FLAG-β-globin (data not shown). When microsomal membranes were present during in vitro translation of protein 3AB, the protein band migrating in the SDS-polyacrylamide gel was diffuse, possibly indicating posttranslational modifications, as previously reported (13). The ratio between membrane-bound and free protein fractions was similar for wild-type 3AB and cytochrome b5 and varied between 20 and 40% in different experiments. Similar analyses of 3AB-M79V and 3AB-M79T proteins translated in vitro in the presence and absence of canine microsomal membranes were performed. Both proteins showed the same distributions in sucrose gradients as wild-type 3AB and cytochrome b5. The results from a set of gradients containing cytochrome b5 and β-globin, wild-type 3AB, or 3AB-M79T, the most defective of the mutants, were quantitated by phosphorimager and are shown in Fig. 5C. The data indicated that the mutant proteins retained membrane-binding properties that were not distinguishable from those of wild-type 3AB protein by this assay.
We also expressed wild-type and mutant 3AB proteins in HeLa cells by simultaneous plasmid transfection and infection with recombinant vaccinia virus vTF7-3 to provide T7 RNA polymerase. Immunofluorescence analysis of cells probed with anti-FLAG antibodies showed a typical pattern of punctate staining resulting from association of 3AB protein with intracellular membranes (data not shown). The mutant and wild-type 3AB proteins all exhibited similar patterns. Electron microscopic examination of cells expressing the different 3AB proteins confirmed 3AB-induced membrane structural alterations; again, no differences between wild-type and mutant 3AB-transfected cells were detected (data not shown).
RNA replication in transfected cells.
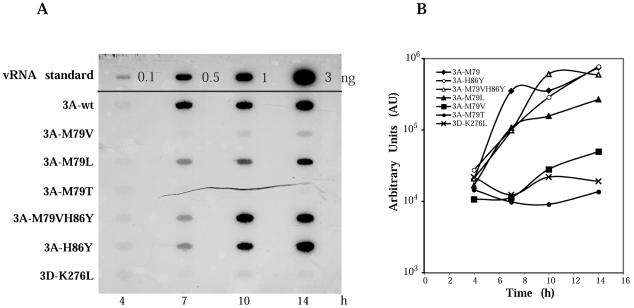
The absence of detectable defects in viral protein synthesis or processing directed by 3A-79 mutant constructs allowed us to examine viral RNA synthesis directly. Figure 6 shows the analysis of the accumulation of positive-strand virus-specific RNA in HeLa cells transfected with equal amounts of wild-type or 3A mutant replicon transcripts by slot blot hybridization of RNA. Replication of wild-type Luc-replicon (3Awt) was detectable by 7 h after transfection. Mutant replicon 3A-M79L produced RNA levels somewhat less than that produced by wild-type 3A-containing RNA (Fig. 6B). M79T failed to replicate, while M79V yielded low amounts of positive-strand RNA. As expected, addition of the second mutation, H86Y to M79V, restored the level of positive-strand RNA accumulation to the wild-type level (3A-M79VH86Y). When mutation H86Y was introduced by itself in the wild-type 3A background, it had no effect on replication (3A-H86Y). Thus, the mutations encoding 3A-79 appear to have a primary and direct effect on viral RNA synthesis.
FIG. 6.
Accumulation of PV replicon-specific RNAs in transfected cells. (A) HeLa cell monolayers were transfected with RNA transcripts in the presence of DEAE-dextran. Cells were harvested at the indicated times after transfection; total cytoplasmic RNAs isolated from approximately 104 cells were bound to a nylon membrane and hybridized to a 32P-labeled riboprobe complementary to nt 220 to 460 of PV RNA. Mutant replicon 3D-K276L bearing a mutation in 3D was used as negative control. The vRNA standards show increasing amounts of purified PV RNA (0.1 to 3 ng) hybridized in parallel. (B) Quantitative presentation of data obtained by analysis with a PhosphorImager and ImageQuant software (Molecular Dynamics).
Effect of 3A mutations on RNA synthesis in vitro.
To examine individual steps in the RNA replication process, we utilized HeLa cell extracts that support translation and replication of PV RNA (6, 46) and encapsidation into virions when capsid proteins are synthesized (46). The HeLa extract thus appears to contain all the cellular components required for PV replication, and it is generally assumed that all steps in the process of viral RNA replication mimic those that occur in an infected cell. We first examined the abilities of these extracts, containing proteins translated from wild-type or mutant RNA transcripts, to catalyze uridylylation of VPg to generate a primer for initiation of RNA strand synthesis. RNA transcripts were translated in HeLa cell extracts in the presence of 2 mM GuHCl to accumulate prereplication complexes (6) (Fig. 7A). The complexes were collected by centrifugation and resuspended in the absence of guanidine to allow VPg uridylylation and synchronous initiation of RNA synthesis. The complexes were labeled with [α-32P]UTP, and uridylylated VPg bands were resolved in SDS-Tris-Tricine-polyacrylamide gels. Figure 7B shows that proteins from all RNAs tested supported VPg uridylylation with equal efficiency.
FIG. 7.
Effect of substitutions at residue M79 in PV protein 3A on VPg uridylylation and negative-strand RNA synthesis. Translation-replication extracts were programmed with full-length RNA transcripts as indicated. (A) Synthesis and processing of viral proteins were measured in reactions carried out in the presence of [35S]methionine, and labeled viral proteins were analyzed as described in the legend to Fig. 4. (B and C) VPg uridylylation (B) and negative-strand RNA synthesis (C) were measured in reactions containing preinitiation RNA replication complexes isolated from translation reactions containing 2 mM GuHCl and the indicated RNAs after sedimentation and resuspension in the absence of guanidine (lanes 1, 3, 4, 5, and 6). GuHCl was added to the samples in lane 2. Negative-strand RNA synthesis and VPg uridylylation assays were performed as described in Materials and Methods; the data were analyzed with a PhosphorImager and are expressed as a percentage of the product observed with wild-type PV RNA transcript. The major uridylylated VPg band in panel B is thought to be diuridylylated, but it has not been rigorously characterized.
The same extracts also provide a specific assay for negative-strand RNA synthesis (9, 28, 70). When reactions are programmed with RNA transcripts containing two non-PV guanylate residues at their 5′ ends, generated by transcription from a T7 RNA polymerase promoter, the synthesis of negative-strand RNA is readily detected but further transcription to synthesize positive-strand RNA is severely inhibited (8). Figure 8A, lane 1, shows that under nondenaturing conditions, the product of this reaction is detected only as double-stranded (replicative form) RNA, indicating its hybridization to the positive-strand RNA template, as previously demonstrated (27). Direct confirmation of the RNA polarity was additionally demonstrated by one-dimensional gel analysis of oligonucleotides produced after complete RNase T1 digestion (data not shown). Therefore, product RNAs synthesized during the first 1 to 2 h in extracts programmed with T7 transcripts provided a measure of initial negative-strand synthesis. Figure 7C shows the products of such reactions programmed with transcripts harboring wild-type or 3A mutations. For comparison, lane 6 shows RNA products produced in the same extracts programmed with virion RNA, which are able to replicate progeny positive strands. Surprisingly, negative-strand synthesis occurred in all reactions with equal efficiency, despite large variations in total RNA synthesis observed in cells transfected with transcripts containing mutations in protein 3A (Fig. 2B). Although initial production of a single round of negative-strand RNA is not detectable in transfected cells due to limitations in the sensitivity of the assay (70), we assume that the requirements for negative-strand synthesis in vitro are the same as those in vivo (although this has not been rigorously proven). Thus, the defect in RNA synthesis caused by mutations near the C terminus of viral protein 3A occurs at a step after uridylylation of VPg primer and negative-strand synthesis. Apparently positive-strand RNA synthesis is specifically and differentially affected by these mutations in protein 3A.
FIG. 8.
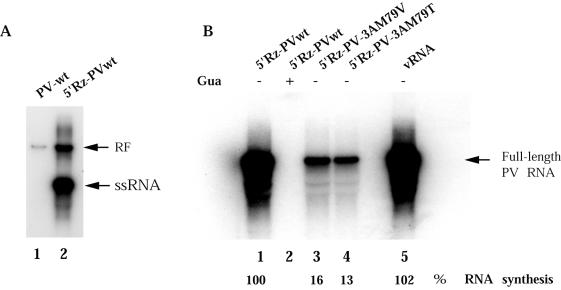
Replication of RNA transcripts with an authentic PV 5′ end. (A) Preinitiation RNA replication complexes were isolated from translation-replication reactions programmed with RNA transcripts produced from constructs that did not (lane 1) or did (lane 2) contain the cis-active hammerhead ribozyme attached to the 5′ end of the poliovirus genome sequence. Preinitiation RNA replication complexes were incubated for 60 min at 37°C in reaction mixtures containing [α-32P]CTP; RNA was phenol extracted and analyzed on nondenaturing 1% agarose gels. (B) RNA transcripts produced from constructs containing the cis-active hammerhead ribozyme (5′Rz-PV-3Awt [lanes 1 and 2], 5′Rz-PV-3A-M79V [lane 3], and 5′Rz-PV-3A-M79T [lane 4[) or vRNA (lane 5) were used to program translation-replication reactions and form preinitiation RNA replication complexes. The complexes were isolated and incubated as above in the presence (lane 2) or absence (lanes 1, 3, 4, and 5) of 2 mM GuHCl, and labeled RNAs were analyzed in denaturing gels, as for Fig. 7. The replication activity of each substrate as the percentage of synthesis observed with wild-type 5′Rz-PV RNA transcript (lane 1) is shown under each lane and was measured using a PhosphorImager.
In order to measure positive-strand viral RNA synthesis in vitro, viral cDNAs containing wild-type or mutant 3A coding sequences were engineered into plasmids containing the cis-active hammerhead ribozyme coding sequence downstream of the T7 promoter (27). These plasmids generate RNA transcripts (5′Rz-PV) that autocatalytically cleave their 5′ ends to yield RNAs with no nonviral nucleotides and thus can serve as efficient templates for subsequent synthesis of progeny positive strands. Figure 8A, lane 2, shows that the majority of the newly synthesized RNA is detected as single-stranded RNA, indicating the production of positive strands in reactions programmed with 5′Rz-PVwt. The wild-type ribozyme transcript generates equivalent amounts of labeled product RNA as are produced from virion RNA (Fig. 8B, lanes 1 and 5). The total amounts of product RNA synthesized in these reactions were significantly greater than in reactions containing PVwt RNA transcript without the ribozyme (Fig. 7C, lanes 1 and 6). Although each ribozyme-containing 3A mutant transcript was translated with equal efficiency (data not shown), total viral RNA synthesis was reduced significantly from that of M79V RNA and M79T RNA (Fig. 8B). Since these values include both negative- and positive-strand RNA, and negative-strand synthesis is not affected by the 3A mutations, the effects on positive-strand synthesis are even more severe.
Virus production in vitro.
Prolonged translation and incubation of viral RNA in HeLa cell extracts similar to those used in this study were shown previously to result in the generation of infectious virus particles (6, 46, 74). The effect of 3A-M79 mutations was examined by measuring plaque-forming activity generated in translation reactions. Translation of the T7 transcripts from wild-type and 3A mutant constructs demonstrated a large reduction in virus yield from the mutant RNAs as well as the expected reduction in plaque size (Table 3). Since these reactions were conducted under conditions where negative-strand synthesis occurred with equal efficiency from both mutant and wild-type RNA, the data were consistent with a positive-strand-specific defect in RNA synthesis resulting from the 3A mutations.
TABLE 3.
Recovery of virus from translation-replication reactions
| RNA | Virus yield in cell-free reactions (PFU/ml)a | Plaque phenotype |
|---|---|---|
| PV1wt | 105 | Large |
| PV-3A-M79V | 104 | Small |
| PV-3A-M79T | 102 | Minute |
| 5′Rz-PV1 | >108 | Large |
| 5′Rz-PV-3A-M79V | 107 | Small |
| 5′Rz-PV-3A-M79T | 105 | Minute + large |
Cell-free reactions programmed with corresponding RNA transcripts were incubated at 34°C for 16 h. Each reaction mixture was serially diluted and used to infect HeLa cell monolayers, from which virus yield was calculated.
Virus yields determined by plaque assay of the products formed from the ribozyme-containing transcripts confirmed the expected increase in virion production due to introduction of the ribozyme sequence, as well as replication defects caused by the M79 mutations (Table 3).
DISCUSSION
In this study, a point mutation causing a single amino acid change in the C-terminal portion of PV protein 3A was shown to cause a defect in viral RNA synthesis. Replacement of residue M79 with threonine abolished detectable RNA synthesis in HeLa cells transfected with replicon sequences containing this 3A mutation and produced minute plaques in cells transfected with full-length viral RNA containing the M79T mutation. Both the reduction in viral RNA synthesis and the small plaque phenotype could be spontaneously restored to wild type by a second mutation from histidine to tyrosine at 3A residue 86 or from tyrosine to histidine at 3A residue 77. Replacement of 3A residue M79 with valine caused a similar but less severe phenotype: delayed onset and reduced yield of RNA from mutant-containing replicons and small plaques after transfection with full-length viral RNA containing the M79V mutation. The same compensating second mutation, H86Y, also restored the M79V mutation to the wild-type phenotype. In a previous study of several different 3A mutations, M79 was replaced with a lysine residue. In this case, the resulting virus was not viable due to aberrant polyprotein processing (35).
The most intriguing finding of this study was the discovery that these mutations in protein 3A selectively inhibit only positive- and not negative-strand RNA synthesis. This result was demonstrated by utilizing an in vitro assay that allowed us to distinguish the two reactions. RNA transcripts bearing extra nonviral nucleotides at their 5′ ends are translated in HeLa cell extracts to produce all viral proteins required for synthesis of complementary negative strands from the input RNA; however, utilization of the product negative strands as templates for synthesis of new positive strands does not occur at detectable levels during the 1-h incubation period (7, 8, 27) (Fig. 8A). Positive-strand synthesis in vitro is accomplished when ribozyme sequences are inserted in the initial RNA template to generate a natural 5′ end with no nonviral nucleotides (27, 47, 48) (Fig. 8A). The ability to distinguish these two reactions provides a means of complementing assays of RNA synthesis in infected or transfected cells, since a single round of negative-strand RNA synthesis cannot be measured in vivo due to technical limitations in the assay (70). Utilizing the in vitro assay, we have shown that neither synthesis of uridylylated VPg primer nor synthesis of negative-strand RNA was reduced in extracts programmed by mutant 3A sequence-containing RNAs. Thus, the first steps in initiating synthesis of a new strand of RNA were independent of these 3A mutations, and our data indicate that some function of 3A is required for subsequent steps involved in synthesis of positive-strand RNA. A previously studied 3A mutation in the middle of the hydrophobic region, 3A T67I, has been reported also to selectively affect positive-strand synthesis; however, that mutant appeared to manifest reduced VPg uridylylation activity as well as decreased positive-strand synthesis (23). In that study, measurements were performed with crude replication complexes isolated from virus-infected cells, and the failure to amplify positive strands may have reduced the number of active complexes in which VPg uridylylation and negative-strand synthesis occur, thereby resulting in an apparent reduction of these activities.
Recently, two laboratories (47, 48) reported that initiation of negative-strand PV RNA synthesis does not require uridylylation of VPg directed by the cis-acting replication element (CRE) located in the 2C coding sequence (54). RNAs containing genetically disrupted CRE structures directed synthesis of negative-strand RNA but did not accumulate uridylylated forms of VPg and produced no positive-strand RNAs. A model was proposed in which VPg and 3Dpol are juxtaposed at the poly(A) tail of positive-strand template RNA, and the tyrosine hydroxyl of VPg primes negative-strand RNA synthesis without production of stable uridylylated VPg intermediates. In contrast, initiation of positive-strand RNA synthesis requires a pool of VPgpUpUOH to prime elongation by 3Dpol after base pairing with the two terminal adenylate residues at the 3′ ends of negative-strand RNA templates. This model suggests that positive- and negative-strand RNA synthesis are initiated via different mechanisms. The results described in our present study support and extend this proposal. A mutation in the 3A protein coding sequence also had no effect on negative-strand synthesis but severely affected positive-strand synthesis. However, in this case uridylylated VPg primers were produced and accumulated because the CRE-dependent reaction was not impaired. Future studies are required to elucidate the biochemical function of 3A that is affected by mutation at residue 79 and what role this function plays in initiation of positive-strand RNA synthesis, apparently at the step of primer utilization.
Although numerous biochemical activities have been demonstrated for 3A and/or 3AB proteins (see the introduction and reference 81 and references therein), the biochemical role(s) of 3A or 3AB in viral RNA replication is not known. 3AB has a propensity to dimerize and form even higher oligomers in solution in the absence of detergent, and high-affinity homointeractions of 3AB have been shown by yeast two-hybrid analysis (81). The hydrophobic domain of 3A was mapped as the region mostly responsible for these interactions (81). Recently, however, the atomic structure of a truncated 3A protein was determined by nuclear magnetic resonance spectroscopy (66). The soluble, N-terminal domain was shown to form a symmetric dimer by cross-linking and analytical ultracentrifugation studies, even in the absence of the C-terminal hydrophobic domain, where amino acid M79 resides.
Residue 79 of protein 3A lies just near the C-terminal end of a 22-amino-acid hydrophobic domain present in the C-terminal portion of 3A (residues 59 to 80). Amino acid changes in the hydrophobic domain of 3A commonly result in virus death or defective viruses with impairment in the replication of their RNAs (22, 35, 82). The domain is highly conserved among enterhinoviruses and has been described as being composed of a predicted amphipathic helix formed by residues 59 to 73 (22, 33), followed by a 7-amino-acid hydrophobic stretch. Insertion of charged residues or deletions of the major portion of the predicted amphipathic helix failed to demonstrate reductions in the mutated 3ABs' membrane association by sedimentation in sucrose gradients (76), although a similar 15-amino-acid deletion resulted in a more diffuse immunofluorescence pattern in transfected cells (13). Deletion of residues 71 to 80, on the other hand, or replacement of three valines with glutamate residues at positions 75, 76, and 78, inhibited membrane association in vitro and demonstrated that the hydrophobic amino acids in this region are crucial for 3AB membrane association (76).
We utilized two assays to assess membrane-binding properties of the M79V and M79T mutant 3AB proteins: equilibrium sedimentation in sucrose density gradients and confocal immunofluorescence patterns in cells expressing 3AB proteins. Both assays revealed no detectable differences between wild-type and mutant proteins, although neither assay likely represents a very sensitive measure of membrane affinity. Replacement of a methionine residue with valine would not be expected to result in a significant change in hydrophobicity; threonine, however, would introduce a more polar but uncharged character. Interestingly, analysis of amino acid alignments of 3A proteins in other enterhinoviruses reveals that isoleucine and valine are the most frequent amino acids occupying the equivalent position 79 in protein 3A. Our engineered M79L virus grew with properties almost indistinguishable from those of wild-type virus. Only the three PV serotypes and enterovirus C encode methionine at this position.
Two spontaneously occurring compensatory mutations that resulted in restoration of a faster-growing “wild-type” phenotype to the M79T mutant were Y77H and H86Y. The atomic structure of intact 3AB protein or even of the C-terminal portion of the 3A protein has not been determined, so the structural implications of these substitutions within a few residues up- or downstream of the original mutation cannot be predicted. Y77 is almost universally conserved among all sequenced enterhinoviruses, and it has been reported previously that Y77H mutation, by itself, was lethal (35). Position 86 is quite variable among different viruses, with H, Y, F, I, T, Q, or C being represented in different 3A proteins. H86Y by itself had no effect on virus replication. The same H86Y mutation restored the replication phenotype of the M79V mutant. No revertants were identified that contained compensating mutations in proteins other than 3A, and so no functional interactions between 3A and other proteins were implicated.
Acknowledgments
We thank Bert L. Semler and Jon Towner for plasmid pTM1-FG3ABwt and for advice on the membrane association assays. Raul Andino kindly provided pRLuc31 and prib(+)XpA. We are grateful to Xi-Yu Jia, who performed some of the initial screening and isolation of replicon mutants. We thank Vadim I. Agol, Kurt Bienz, Denise Egger, and Oliver Richards for critical reading of the manuscript.
REFERENCES
- 1.Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. [DOI] [PubMed] [Google Scholar]
- 2.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansardi, D. C., R. Pal-Ghosh, D. Porter, and C. D. Morrow. 1995. Encapsidation and serial passage of a poliovirus replicon which expresses an inactive 2A proteinase. J. Virol. 69:1359-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee, R., A. Echeverri, and A. Dasgupta. 1997. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J. Virol. 71:9570-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee, R., W. Tsai, W. Kim, and A. Dasgupta. 2001. Interaction of poliovirus-encoded 2C/2BC polypeptides with the 3′ terminus negative-strand cloverleaf requires an intact stem-loop b. Virology 280:41-51. [DOI] [PubMed] [Google Scholar]
- 6.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3Dpol, and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. [DOI] [PubMed] [Google Scholar]
- 8.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell, Y. C., B. L. Semler, and E. Ehrenfeld. 1999. Requirements for RNA replication of a poliovirus replicon by coxsackievirus B3 RNA polymerase. J. Virol. 73:9413-9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, M. W., O. C. Richards, T. M. Dmitrieva, V. Agol, and E. Ehrenfeld. 1993. RNA duplex unwinding activity of poliovirus RNA-dependent RNA polymerase 3Dpol. J. Virol. 67:3010-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 13.Datta, U., and A. Dasgupta. 1994. Expression and subcellular localization of poliovirus VPg-precursor protein 3AB in eukaryotic cells: evidence for glycosylation in vitro. J. Virol. 68:4468-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dayhuff, T. J., R. F. Gesteland, and J. F. Atkins. 1992. Electrophoresis, autoradiography and electroblotting of peptides: T4 gene 60 hopping. BioTechniques 13:500-503. [PubMed] [Google Scholar]
- 15.Doedens, J. R., T. H. Giddings, Jr., and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71:9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dove, A. W., and V. R. Racaniello. 1997. Cold-adapted poliovirus mutants bypass a postentry replication block. J. Virol. 71:4728-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Echeverri, A. C., and A. Dasgupta. 1995. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208:540-553. [DOI] [PubMed] [Google Scholar]
- 19.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flanegan, J. B., and D. Baltimore. 1979. Poliovirus polyuridylic acid polymerase and RNA replicase have the same viral polypeptide. J. Virol. 29:352-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flanegan, J. B., and D. Baltimore. 1977. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A). Proc. Natl. Acad. Sci. USA 74:3677-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giachetti, C., S.-S. Hwang, and B. L. Semler. 1992. cis-acting lesions targeted to the hydrophobic domain of a poliovirus membrane protein involved in RNA replication. J. Virol. 66:6045-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giachetti, C., and B. L. Semler. 1991. Role of a viral membrane polypeptide in strand-specific initiation of poliovirus RNA synthesis. J. Virol. 65:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haller, A. A., and B. L. Semler. 1992. Linker scanning mutagenesis of the internal ribosome entry site of poliovirus RNA. J. Virol. 66:5075-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hambridge, S. J., and P. Sarnow. 1992. Translational enhancement of the poliovirus 5′ noncoding region mediated by virus-encoded polypeptide 2A. Proc. Natl. Acad. Sci. USA 89:10272-10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris, K. S., W. Xiang, L. Alexander, W. S. Lane, A. V. Paul, and E. Wimmer. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004-27014. [PubMed] [Google Scholar]
- 27.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 30.Hope, D. A., S. E. Diamond, and K. Kirkegaard. 1997. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J. Virol. 71:9490-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, K. L., and P. Sarnow. 1991. Three poliovirus 2B mutants exhibit noncomplementable defects in viral RNA amplification and display dosage-dependent dominance over wild-type poliovirus. J. Virol. 65:4341-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuechler, E., J. Seipelt, H. D. Liebig, and W. Sommergruber. 2002. Picornavirus proteinase-mediated shutoff of host cell translation: direct cleavage of cellular initiation factor, p. 301-311. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 33.Lama, J., and L. Carrasco. 1992. Expression of poliovirus nonstructural proteins in Escherichia coli cells. Modification of membrane permeability induced by 2B and 3A. J. Biol. Chem. 267:15932-15937. [PubMed] [Google Scholar]
- 34.Lama, J., A. V. Paul, K. S. Harris, and E. Wimmer. 1994. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J. Biol. Chem. 269:66-70. [PubMed] [Google Scholar]
- 35.Lama, J., M. A. Sanz, and L. Carrasco. 1998. Genetic analysis of poliovirus protein 3A: characterization of a non-cytopathic mutant virus defective in killing Vero cells. J. Gen. Virol. 79:1911-1921. [DOI] [PubMed] [Google Scholar]
- 36.Lama, J., M. A. Sanz, and P. L. Rodriguez. 1995. A role for 3AB protein in poliovirus genome replication. J. Biol. Chem. 270:14430-14438. [DOI] [PubMed] [Google Scholar]
- 37.Li, J.-P., and D. Baltimore. 1990. An intragenic revertant of a poliovirus 2C mutant has an uncoating defect. J. Virol. 64:1102-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lomax, N. B., and F. H. Yin. 1989. Evidence for the role of the P2 protein of human rhinovirus in its host range change. J. Virol. 63:2396-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu, H.-H., X. Li, A. Cuconati, and E. Wimmer. 1995. Analysis of picornavirus 2Apro proteins: separation of proteinase from translation and replication functions. J. Virol. 69:7445-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lundquist, R. E., E. Ehrenfeld, and J. V. Maizel. 1974. Isolation of a viral polypeptide associated with poliovirus RNA polymerase. Proc. Natl. Acad. Sci. USA 71:773-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyle, J. M., A. Clewell, K. Richmond, O. C. Richards, D. A. Hope, S. C. Schultz, and K. Kirkegaard. 2002. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J. Biol. Chem. 277:16324-16331. [DOI] [PubMed] [Google Scholar]
- 42.Mirzayan, C., and E. Wimmer. 1994. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology 199:176-187. [DOI] [PubMed] [Google Scholar]
- 43.Mirzayan, C., and E. Wimmer. 1992. Genetic analysis of an NTP-binding motif in poliovirus polypeptide 2C. Virology 189:547-555. [DOI] [PubMed] [Google Scholar]
- 44.Molla, A., K. S. Harris, A. V. Paul, S. H. Shin, J. Mugavero, and E. Wimmer. 1994. Stimulation of poliovirus proteinase 3Cpro-related proteolysis by the genome-linked protein VPg and its precursor 3AB. J. Biol. Chem. 269:27015-27020. [PubMed] [Google Scholar]
- 45.Molla, A., A. V. Paul, M. Schmid, S. K. Jang, and E. Wimmer. 1993. Studies on dicistronic polioviruses implicate viral proteinase 2Apro in RNA replication. Virology 196:739-747. [DOI] [PubMed] [Google Scholar]
- 46.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 47.Morasco, B. J., N. Sharma, J. Parilla, and J. B. Flanegan. 2003. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 77:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77:4739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neufeld, K. L., J. M. Galarza, O. C. Richards, D. Summers, and E. Ehrenfeld. 1994. Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3Dpol. J. Virol. 68:5811-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neznanov, N., A. Kondratova, K. M. Chumakov, B. Angres, B. Zhumabayeva, V. I. Agol, and A. V. Gudkov. 2001. Poliovirus protein 3A inhibits tumor necrosis factor (TNF)-induced apoptosis by eliminating the TNF receptor from the cell surface. J. Virol. 75:10409-10420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Donnell, V. K., J. M. Pacheco, T. M. Henry, and P. W. Mason. 2001. Subcellular distribution of the foot-and-mouth disease virus 3A protein in cells infected with viruses encoding wild-type and bovine-attenuated forms of 3A. Virology 287:151-162. [DOI] [PubMed] [Google Scholar]
- 52.O'Neill, R. E., and V. R. Racaniello. 1989. Inhibition of translation in cells infected with a poliovirus 2Apro mutant correlates with phosphorylation of the alpha subunit of eucaryotic initiation factor 2. J. Virol. 63:5069-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paul, A. V., X. Cao, K. S. Harris, J. Lama, and E. Wimmer. 1994. Studies with poliovirus polymerase 3Dpol. Stimulation of poly(U) synthesis in vitro by purified poliovirus protein 3AB. J. Biol. Chem. 269:29173-29181. [PubMed] [Google Scholar]
- 54.Paul, A. V., E. Reider, D. W. Kim, J. H. Van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paul, A. V., J. H. van Boom, D. Fillipov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 56.Pfister, T., and E. Wimmer. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992-7001. [DOI] [PubMed] [Google Scholar]
- 57.Pincus, S. E., and E. Wimmer. 1986. Production of guanidine-resistant and -dependent poliovirus mutants from cloned cDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J. Virol. 60:793-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plotch, S. J., and O. Palant. 1995. Poliovirus protein 3AB forms a complex with and stimulates the activity of the viral RNA polymerase, 3Dpol. J. Virol. 69:7169-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richards, O. C., and E. Ehrenfeld. 1998. Effects of poliovirus 3AB protein on 3D polymerase-catalyzed reaction. J. Biol. Chem. 273:12832-12840. [DOI] [PubMed] [Google Scholar]
- 60.Roberts, L. O., R. A. Seamons, and G. J. Belsham. 1998. Recognition of picornavirus internal ribosome entry sites within cells; influence of cellular and viral proteins. RNA 4:520-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez, P. L., and L. Carrasco. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105-10112. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez, P. L., and L. Carrasco. 1993. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268:8105-8110. [PubMed] [Google Scholar]
- 63.Rodriguez-Wells, V., S. J. Plotch, and J. J. DeStefano. 2001. Primer-dependent synthesis by poliovirus RNA-dependent RNA polymerase (3Dpol). Nucleic Acids Res. 29:2715-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 65.Semler, B. L., C. W. Anderson, R. Hanecak, L. F. Dorner, and E. Wimmer. 1982. A membrane-associated precursor to poliovirus VPg identified by immunoprecipitation with antibodies directed against a synthetic heptapeptide. Cell 28:405-412. [DOI] [PubMed] [Google Scholar]
- 66.Strauss, D. M., L. W. Glustrom, and D. S. Wuttke. 2003. Towards an understanding of the poliovirus replication complex: the solution structure of the soluble domain of the poliovirus 3A protein. J. Mol. Biol. 330:225-234. [DOI] [PubMed] [Google Scholar]
- 67.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takeda, N., R. J. Kuhn, C.-F. Yang, T. Takegami, and E. Wimmer. 1986. Initiation of poliovirus plus-strand synthesis in a membrane complex of infected HeLa cells. J. Virol. 60:43-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takegami, T., R. J. Kuhn, C. W. Anderson, and E. Wimmer. 1983. Membrane-dependent uridylylation of the genome-linked protein VPg of poliovirus. Proc. Natl. Acad. Sci. USA 80:7447-7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teterina, N. L., D. Egger, K. Bienz, D. M. Brown, B. L. Semler, and E. Ehrenfeld. 2001. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 75:3841-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teterina, N. L., K. M. Kean, E. Gorbalenya, V. I. Agol, and M. Girard. 1992. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J. Gen. Virol. 73:1977-1986. [DOI] [PubMed] [Google Scholar]
- 73.Teterina, N. L., W. D. Zhou, M. W. Cho, and E. Ehrenfeld. 1995. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J. Virol. 69:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Todd, S., J. S. Towner, and B. L. Semler. 1997. Translation and replication properties of the human rhinovirus genome in vivo and in vitro. Virology 229:90-97. [DOI] [PubMed] [Google Scholar]
- 75.Tolskaya, E. A., L. I. Romanova, M. S. Kolesnikova, A. P. Gmyl, A. E. Gorbalenya, and V. I. Agol. 1994. Genetic studies on the poliovirus 2C protein, an NTPase—a plausible mechanism of guanidine effect on the 2C function and evidence for the importance of 2C oligomerization. J. Mol. Biol. 236:1310-1323. [DOI] [PubMed] [Google Scholar]
- 76.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 77.Towner, J. S., M. M. Mazanet, and B. L. Semler. 1998. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J. Virol. 72:7191-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Toyoda, H., C.-F. Yang, N. Takeda, A. Nomoto, and E. Wimmer. 1987. Analysis of RNA synthesis of type I poliovirus by using an in vitro molecular genetic approach. J. Virol. 61:2816-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vance, L. M., N. Moscufo, M. Chow, and B. A. Heinz. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71:8759-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Kuppeveld, F. G. M., J. G. J. Hoenderop, R. L. L. Smeets, P. H. G. M. Willems, H. B. P. M. Dijkman, J. M. D. Galama, and W. J. G. Melchers. 1997. Coxsackie-virus protein 2B modifies the ER membrane and plasma membrane permeability and facilitates virus release. EMBO J. 16:3519-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiang, W., A. Cuconati, D. Hope, K. Kirkegaard, and E. Wimmer. 1998. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J. Virol. 72:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiang, W., A. Cuconati, A. V. Paul, and E. Wimmer. 1995. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA 1:892-904. [PMC free article] [PubMed] [Google Scholar]
- 83.Xiang, W., K. S. Harris, L. Alexander, and E. Wimmer. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yin, F. H., and N. B. Lomax. 1983. Host range mutants of human rhinovirus in which nonstructural proteins are altered. J. Virol. 48:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu, S. F., P. Benton, M. Bovee, J. Sessions, and R. E. Lloyd. 1995. Defective RNA replication by poliovirus mutants deficient in 2A protease cleavage activity. J. Virol. 69:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zamora, M., W. E. Marrisen, and R. Lloyd. 2002. Poliovirus-mediated shutoff of host translation: an indirect effect, p. 313-320. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.