Abstract
FrCasE is a mouse retrovirus that causes a fatal noninflammatory spongiform neurodegenerative disease with pathological features strikingly similar to those induced by transmissible spongiform encephalopathy (TSE) agents. Neurovirulence is determined by the sequence of the viral envelope protein, though the specific role of this protein in disease pathogenesis is not known. In the present study, we compared host gene expression in the brain stems of mice infected with either FrCasE or the avirulent virus F43, differing from FrCasE in the sequence of the envelope gene. Four of the 12 disease-specific transcripts up-regulated during the preclinical period represent responses linked to the accumulation of unfolded proteins in the endoplasmic reticulum (ER). Among these genes was CHOP/GADD153, which is induced in response to conditions that perturb endoplasmic reticulum function. In vitro studies with NIH 3T3 cells revealed up-regulation of CHOP as well as BiP, calreticulin, and Grp58/ERp57 in cells infected with FrCasE but not with F43. Immunoblot analysis of infected NIH 3T3 cells demonstrated the accumulation of uncleaved envelope precursor protein in FrCasE- but not F43-infected cells, consistent with ER retention. These results suggest that retrovirus-induced spongiform neurodegeneration represents a protein-folding disease and thus may provide a useful tool for exploring the causal link between protein misfolding and the cytopathology that it causes.
The transmissible spongiform encephalopathy (TSE) agents or prions induce an unusual form of neurodegeneration characterized primarily by the appearance of vacuoles in neurons and the neuropil and are associated with neuronal dropout and astrocytosis. These are chronic diseases with incubation periods ranging from several months in rodents to several years in larger animals and humans. The chronicity of these diseases has made it difficult to identify the proximal events in the pathogenesis of this unusual form of neurodegeneration. The only other infectious agents known to cause spongiform neurodegeneration are a group of murine retroviruses that induce neuropathology essentially indistinguishable from that induced by the TSE agents (2, 52) but without the accumulation of prion protein in the brain. Through genome manipulation, the tempo of disease can be dramatically shortened, which has facilitated the identification of molecular and cellular events associated with the induction of spongiform degeneration (reviewed in reference 43).
The first of these retroviruses to be identified was CasBrE, a murine virus originally isolated from wild mice (14). CasBrE causes a paralytic disease with variable incidence and a long incubation period of several months to more than a year (13). The determinants of neurovirulence of CasBrE have been localized to the viral envelope gene (12), though the role of the envelope protein in disease pathogenesis remains unknown. We constructed a chimeric virus FrCasE, which contains the envelope gene of CasBrE inserted into the genome of a strain of Friend murine leukemia virus (MLV) FB29 (44). When inoculated intraperitoneally into neonatal mice, FrCasE causes an acute fatal spongiform neurodegenerative disease with a clinical course beginning at 14 days postinoculation (dpi) and lasting approximately 1 week. Early clinical signs are characterized by tremor and muscle weakness that increase in severity with time in all infected mice. Mice reach a terminal stage that includes wasting, hind limb and forelimb paralysis, and seizures by 18 to 21 dpi. Small foci of spongiform lesions are first noted at 10 dpi and become widely distributed by 17 dpi (11). This timeline has been found to be highly predictable (11).
We have previously shown that the neurons and astrocytes that exhibit cytopathology are not infected by FrCasE, indicating an indirect mechanism of virus-induced neuropathology (43). Instead, it is infection of microglial cells that appears to be necessary for the induction of spongiosis (35, 37). Interestingly, in light of the extensive cytopathology and pronounced clinical manifestations caused by FrCasE, we have not observed activation of the infected microglial cells until late in the disease (34), and there is little evidence that up-regulation of genes encoding proinflammatory cytokines is a determinant of virulence (4).
Given the lack of a clear understanding of the molecular pathways involved in retrovirus-induced spongiform neurodegeneration, we conducted a study with high-density oligonucleotide microarrays to investigate transcriptional profiles in FrCasE-infected brain stem. To discriminate host responses to retrovirus infection per se from those responses associated specifically with spongiform neurodegeneration, we compared gene expression in the brain stem of mice infected with FrCasE with that induced by a nonpathogenic retrovirus (F43). While FrCasE contains the envelope gene of CasBrE, F43 contains the envelope gene of a nonneurovirulent mouse retrovirus Friend MLV57 (5). FrCasE and F43 utilize the mouse cationic amino acid transporter as a receptor for virus entry (1) and infect the same spectrum of cells in the brain, yet F43 is nonpathogenic and thus serves as a useful control in this study. Transcriptional profiles for both early (10 dpi) and late (17 dpi) time points in the disease process were generated to follow the progression of gene expression changes.
We show here both in vivo and in vitro that induction of a program of endoplasmic reticulum (ER) stress responses in infected cells is a correlate of retrovirus-induced spongiform neurodegeneration. The ER stress was found to be associated with the accumulation of envelope protein in an unprocessed form consistent with ER retention. By analogy with a growing number of human degenerative diseases associated with the misfolding of host proteins, this disease appears to be associated with the misfolding of a viral protein.
MATERIALS AND METHODS
Mice and virus inoculations.
Inbred Rocky Mountain White mice were bred and raised at the Rocky Mountain Laboratories and were handled according to polices of the Rocky Mountain Laboratories Animal Care and Use Committee. Mice were inoculated with virus stocks prepared in Mus dunni cells as described previously (44) or for mock infections, with tissue culture media alone. Mice were inoculated intraperitoneally at 24 to 48 h after birth with 30 μl of virus stock containing between 5 × 106 and 1 × 107 focus-forming units of infectivity per ml.
Total RNA preparation.
Mice were sacrificed under deep isoflurane anesthesia by cardiac perfusion with ice-cold phosphate-buffered saline (PBS) at 10 (early) and 17 (late) dpi. RNA was isolated, by using an RNeasy mini kit (Qiagen), from the brain stem (diagram in Fig. 1A), consisting of medulla oblongata, pons, and midbrain. Prior to array and RT-PCR experiments, the integrity of RNA was evaluated with an RNA 6000 nano assay kit and Bioanalyzer 2100 (Agilent) to visualize and compare 18S and 28S rRNA bands.
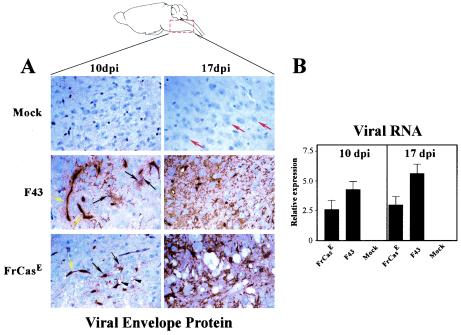
FIG. 1.
The region of the brain analyzed in this study (brain stem) is illustrated above. Panel A shows sections of brain stem from mice either mock inoculated or inoculated as neonates intraperitoneally with F43 or FrCasE. Sections are stained with a polyclonal antiserum to MLV surface glycoprotein (SU). Immunoreactivity was detected with the substrate AEC (red), and the sections were counterstained with hematoxylin (blue). Both viruses infected cells associated with the microvasculature (yellow arrows), as well as cells in the parenchyma consisting of primarily microglial cells (black arrows). Microglial cells are distinguished by highly arborized processes and small ovoid nuclei. Neurons in these sections (red arrows) are identified by the largeness of their nuclei and their prominent nucleoli. At 10 dpi both viruses have spread beyond the microvasculature and infected parenchymal microglial cells. At 17 dpi infection of microglial cells by both viruses is extensive. Focal spongiosis can be detected at 10 dpi only in the FrCasE-infected brain stem (black arrowheads), and by 17 dpi the spongiosis is extensive. No neuropathology is detectable in the F43-infected brain stem at either time point. Magnification before reduction, ×125. Panel B shows viral RNA levels in the brain stem as measured by real-time RT-PCR with a probe specific for sequences within the gag gene shared by both viruses (n = 8). The difference between FrCasE and F43 was not statistically significant at 10 dpi (P > 0.05) but was significant at 17 dpi (P < 0.001). Note the inverse relationship between viral RNA levels and neurovirulence. Data were normalized with GAPDH and are shown as mean ± standard error of the mean. In panel A there appears to be a dramatic increase in the level of staining of the SU protein from 10 to 17 dpi, which is not apparent in respective levels of viral RNA shown in panel B. RNA levels correlate with previous quantitative Western blot analyses for FrCasE that indicate no difference in viral protein levels between 10 and 17 dpi (11). Immunohistochemistry is not a quantitative technique and is subject to substantial sampling error, so it is not used here to indicate levels of virus infection.
Affymetrix arrays.
Generation of cDNA and cRNA for Affymetrix arrays and hybridization conditions were set up according to the manufacturer's protocol (Affymetrix). Each treatment group contained five mice, except for the group of mock-inoculated mice sacrificed at 17 dpi, which contained four mice. Gene expression profiles for 10 and 17 dpi were analyzed separately and were then compared for similarities. Microarray suite 4.0 (Affymetrix) was used to generate cell intensity files and presence/absence calls. These data were imported into the program dChip, where data were normalized with an invariant set normalization method to a common baseline array with the median overall brightness (30), and gene expression values were calculated with a model-based approach (31).
Several criteria were used to determine significant gene expression changes by applying dChip-calculated expression values. First, significant differences in transcript levels were determined with the program Statistical Analysis of Microarrays (SAM) (56). SAM calculates gene-specific significance values and provides an estimate of the number of genes incorrectly called significant, also known as the false discovery rate (FDR). False positives are particularly a problem when multiple statistical comparisons are made for large numbers of transcripts. For any set of potentially significant differences between groups, an FDR is determined based on the number of significant genes in randomized data and in the original data set (reviewed in reference 10). For each group of genes identified as significant in this study, an FDR is indicated (footnotes in Tables 1, 2, and 3). In addition to a statistical criterion, an arbitrary threshold of average 1.4-fold change was used to identify differentially expressed transcripts. Finally, a P value was calculated for each gene with analysis of variance (ANOVA) to complement SAM results. Genes were classified with the Gene Ontology Consortium's (GO) classifications (3), or, in cases where no GO classification was available, classifications were determined from relevant literature.
TABLE 1.
Transcripts up-regulated by both viruses at 10 and 17 dpi
| Functiona and GenBankb | Gene name | 10-dpi change (n-fold)
|
17-dpi change (n-fold)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cas:F43c | Cas:Md | F43:M | Pe | Cas:F43 | Cas:M | F43:M | P | ||
| Immune response/ interferon induced | |||||||||
| X01838 | Beta-2 microglobulin | 1.1 | 2.2 | 2.1 | 2.3E-07 | 1.4 | 2.9 | 2.1 | 1.2E-06 |
| Y00629 | Histocompatibility 2, T region locus 23 | 1.1 | 2.9 | 2.7 | 1.0E-07 | 1.9 | 3.6 | 1.9 | 1.5E-06 |
| X52490 | Histocompatibility 2, D region | 1.3 | 1.7 | 1.3 | 2.1E-03 | 1.2 | 1.9 | 1.6 | 1.9E-04 |
| X00246 | Histocompatibility 2, D region locus 1 | 1.3 | 2.5 | 2.0 | 3.7E-04 | 1.6 | 4.2 | 2.6 | 2.5E-04 |
| V00746 | Histocompatibility 2, K region | 1.3 | 2.0 | 1.6 | 1.1E-04 | 1.5 | 2.6 | 1.8 | 1.2E-05 |
| X16202 | Mouse Q4 MHC class 1 gene (exon 5) | 1.2 | 1.9 | 1.6 | 4.9E-05 | 1.3 | 2.1 | 1.6 | 3.9E-06 |
| U43084 | Interferon-induced protein with tetratricopeptide repeats 1 | −1.2 | 10.1 | 10.8 | 2.8E-08 | 1.4 | 15.8 | 11.4 | 8.0E-05 |
| U43085 | Interferon-induced protein with tetratricopeptide repeats 2 | −1.2 | 1.5 | 1.8 | 6.8E-04 | 1.1 | 1.9 | 1.7 | 8.6E-06 |
| X56602 | Interferon-stimulated protein (15 kDa) | 1.0 | 2.0 | 2.1 | 5.9E-08 | −1.2 | 1.3 | 1.6 | 2.8E-04 |
| L38444 | T-cell-specific GTPase | 2.2 | 7.0 | 3.1 | 9.9E-06 | 1.0 | 2.2 | 2.3 | 2.7E-02 |
| U51992 | Interferon-dependent positive-acting transcription factor 3 gamma | 1.1 | 1.7 | 1.6 | 4.8E-07 | 1.1 | 1.7 | 1.5 | 5.9E-05 |
| AV152244 | Interferon-stimulated protein (15 kDa) | 1.0 | 3.2 | 3.1 | 1.3E-06 | −1.1 | 2.3 | 2.5 | 4.6E-05 |
| U43086 | Interferon-induced protein with tetratricopeptide repeats 3 | −1.1 | 5.7 | 6.4 | 3.9E-06 | 1.2 | 3.7 | 3.1 | 2.4E-05 |
| U19119 | Interferon-inducible protein 1 | 1.1 | 1.7 | 1.6 | 1.7E-06 | 1.1 | 1.7 | 1.6 | 4.5E-05 |
| X04653 | Lymphocyte antigen 6 complex | 1.3 | 2.1 | 1.6 | 3.7E-05 | 1.1 | 1.3 | 1.3 | 8.3E-03 |
| Ion transport/potassium transport | |||||||||
| AV367240 | Potassium voltage-gated channel, subfamily Q, member 1 | −1.6 | 3.0 | 4.8 | 1.3E-04 | −1.2 | 5.9 | 7.0 | 2.9E-07 |
| GTPase | |||||||||
| AW047476 | Guanylate nucleotide binding protein 3 | 1.1 | 2.4 | 2.2 | 1.3E-08 | 0.9 | 1.5 | 1.6 | 4.9E-05 |
| Transcription regulation, signal transduction | |||||||||
| U06924 | Signal transducer and activator of transcription 1 (STAT1) | 1.4 | 3.8 | 2.8 | 1.1E-07 | 0.8 | 1.4 | 1.9 | 4.9E-02 |
| Unknown function | |||||||||
| AA816121 | ESTs, weakly similar to B chain B, crystal structuref | 1.1 | 2.1 | 2.0 | 2.0E-09 | 0.7 | 1.4 | 1.8 | 3.5E-04 |
| AW047653 | EST AW047653 similar to ubiquitin-specific protease 18 | 0.9 | 3.7 | 3.9 | 2.9E-07 | 0.9 | 1.3 | 1.4 | 5.7E-04 |
Function based on GO annotation (3) or deduced from relevant literature.
GenBank accession numbers.
Changes (n-fold) in pathogenic versus nonpathogenic viruses.
Mock inoculated.
P was determined with ANOVA. The FDR was <10%.
EST, expressed sequence tag.
TABLE 2.
Disease-specific transcripts up-regulated at 10 dpi
| Functiona and GenBankb | Gene name | Change (n-fold) for:
|
|
|---|---|---|---|
| Case:F43 | Pc | ||
| Protein disulfide isomerase | |||
| *M73329d | Glucose-regulated protein 58 kDa (Grp58/ERp57) | 1.4 | 5.9E-03 |
| Protein amino acid dephosphorylation | |||
| U92437 | Phosphatase and tensin homolog | 1.6 | 4.8E-03 |
| Cell adhesion, signal transduction | |||
| L25274 | Activated leukocyte cell adhesion molecule | 1.4 | 2.3E-04 |
| Guanyl-nucleotide release factor | |||
| *AF061529 | rjs-containing HECT domain | 1.7 | 4.1E-04 |
| Transcription regulation | |||
| L35032 | SRY box containing gene 18 | 1.4 | 3.2E-03 |
| *X67083 | CHOP/GADD153 | 1.4 | 5.2E-03 |
| Ligase, protein modification | |||
| AI837415 | cDNA similar to ubiquitin carrier protein | 1.7 | 4.4E-03 |
| Unknown function | |||
| AI837830 | Similar to MOP-3 | 1.8 | 3.1E-03 |
| AI852838 | Expressed sequence R75394 | 1.8 | 6.4E-04 |
| AB028921 | Neighbor of A-kinase anchoring protein 95 | 1.4 | 1.5E-04 |
| AI849035 | RIKEN cDNA 2700043121 | 1.5 | 6.9E-04 |
| AW123907 | RIKEN cDNA 2310022K15 | 1.4 | 7.1E-03 |
Function based on GO annotation (3) or deduced from literature.
GenBank accession numbers.
Determined using ANOVA. FDR was <20%.
*, gene involved in ER stress.
TABLE 3.
Disease-specific transcripts differentially expressed at 17 dpi
| Functiona and GenBankb | Gene name | Change (n-fold) for:
|
|||
|---|---|---|---|---|---|
| Cas:F43c | Cas:Md | F43:M | Pe | ||
| Acute phase | |||||
| X58861 | Complement component 1, q a | 1.6 | 2.1 | 1.3 | 4.5E-06 |
| M22531 | Complement component 1, q b | 1.8 | 2.7 | 1.5 | 6.8E-06 |
| X66295 | Complement component 1, q c | 1.7 | 2.1 | 1.0 | 2.6E-05 |
| X81627 | Lipocalin 2 | 8.6 | 19.6 | 2.3 | 5.5E-09 |
| AF045887 | Angiotensinogen | 1.7 | 1.7 | 1.0 | 2.6E-09 |
| Apoptosis | |||||
| D83698 | BH3 interacting (with BCL2 family) domain (DP5) | 2.2 | 2.9 | 1.3 | 5.1E-04 |
| AV138783 | Gadd 45 | 1.4 | 1.3 | 0.9 | 8.8E-05 |
| U88909 | Baculoviral IAP repeat-containing 3 (IAP2) | −2.3 | −1.6 | 1.4 | 1.8E-07 |
| Calcium binding | |||||
| X66449 | Calcium binding protein A6 (calcyclin) | 1.6 | 1.8 | 1.1 | 8.7E-04 |
| X59382 | Parvalbumin | 1.7 | 1.4 | 0.8 | 6.5E-03 |
| AB006758 | Protocadherin 7 | 2.1 | 2.9 | 1.4 | 5.7E-05 |
| D13003 | Reticulocalbin | −1.6 | −1.7 | 1.0 | 1.5E-04 |
| X14432 | Thrombomodulin | −1.9 | −2.1 | 0.9 | 1.8E-04 |
| Cell adhesion | |||||
| M70642 | Connective tissue growth factor | 2.9 | 2.4 | 0.9 | 1.1E-04 |
| AA592182 | Nephronectin | 1.5 | 1.6 | 1.1 | 7.3E-03 |
| U47323 | Stromal interaction molecule 1 | 1.5 | 1.4 | 0.9 | 7.4E-06 |
| U83903 | Tumor necrosis factor-induced protein 6 | 2.6 | 1.6 | −1.7 | 8.9E-04 |
| U12884 | Vascular cell adhesion molecule 1 | 2.3 | 3.0 | 1.3 | 3.9E-06 |
| X52046 | Procollagen, type III, alpha 1 | −2.1 | −2.0 | 1.0 | 8.0E-05 |
| M77123 | Vitronectin | −1.4 | −1.5 | 1.0 | 1.3E-04 |
| D50086 | Neuropilin | −1.6 | −1.5 | 1.1 | 1.8E-03 |
| U03419 | Procollagen, type I, alpha 1 | −1.4 | −1.4 | 1.0 | 1.3E-02 |
| L02918 | Procollagen, type V, alpha 2 | −1.5 | −1.5 | 1.0 | 7.3E-03 |
| U16162 | Procollagen-proline 2-oxoglutarate 4-dioxygenase | −1.8 | −2.0 | 0.9 | 3.2E-07 |
| M31811 | Myelin-associated glycoprotein | −1.6 | −1.6 | 1.0 | 7.6E-04 |
| AI132491 | Bystin-like | −1.6 | −1.5 | 1.1 | 3.4E-04 |
| A1853217 | VE-cadherin gene Cdh5 | −1.5 | −1.5 | 1.0 | 1.2E-04 |
| L04678 | Integrin beta 4 | −1.5 | −2.0 | 0.7 | 2.1E-05 |
| Cell cycle | |||||
| U83902 | Mad211 | 1.4 | 1.2 | 0.8 | 5.5E-03 |
| AW048937 | Cyclin-dependent kinase inhibitor 1A (p21) | 5.4 | 4.7 | 0.9 | 6.9E-11 |
| *AI849615f | Growth arrest specific 5 (Gas5) | 1.5 | 1.5 | 1.0 | 1.7E-04 |
| AW121294 | Wnt1 responsive Cdc42 homolog | 1.4 | 1.2 | 0.9 | 2.8E-03 |
| Z67745 | Protein phosphatase 2a, catalytic subunit | 1.5 | 1.6 | 1.1 | 6.7E-04 |
| AW049584 | Protein phosphatase 1, regulatory (inhibitor) 2 | 1.9 | 2.5 | 1.3 | 1.6E-04 |
| D83745 | B-cell translocation gene 3 | 1.4 | 1.8 | 1.3 | 7.1E-04 |
| AI843106 | p53-regulated PA26 nuclear protein | 1.6 | 1.3 | 0.8 | 4.2E-04 |
| *AI849928 | Cyclin D1 | −1.5 | −1.4 | 1.1 | 2.0E-06 |
| M34094 | Midkine | −1.4 | −1.4 | 1.1 | 2.5E-05 |
| Cell growth | |||||
| AI842277 | Insulin-like growth factor binding protein 3 | 2.2 | 2.4 | 1.1 | 7.3E-04 |
| Endocytosis | |||||
| D86066 | Rabaptin 5 | −1.5 | −1.5 | 1.0 | 2.4E-04 |
| AI747654 | Caveolin, caveolar protein, 22 kDa | −1.7 | −1.6 | 1.1 | 1.4E-05 |
| Cytoskeleton | |||||
| L29468 | Cofilin 2, muscle | 1.5 | 1.4 | 0.9 | 4.1E-03 |
| X57377 | Myosin Va | 1.9 | 1.9 | 1.0 | 1.1E-04 |
| AV170878 | Rhotekin | 1.6 | 1.4 | 0.9 | 9.6E-06 |
| J04953 | Gelsolin | −2.2 | −2.0 | 1.1 | 3.3E-09 |
| AA657164 | Rab6, kinesin-like | −1.7 | −1.7 | 1.0 | 2.0E-03 |
| Creatine biosynthesis | |||||
| AI844626 | Glycine amidinotransferase | −1.6 | −1.6 | 1.0 | 5.6E-08 |
| AF010499 | Guanidinoacetate methyltransferase | −1.5 | −1.4 | 1.1 | 2.9E-04 |
| Glycolysis/glucose metabolism | |||||
| Z11911 | Glucose-6-phosphate dehydrogenase X-linked | 1.6 | 1.3 | 0.8 | 6.5E-03 |
| J03928 | Phosphofructokinase, liver, B-type | −1.4 | −1.5 | 0.9 | 3.9E-06 |
| X13586 | 2,3-Bisphosphoglycerate mutase | −1.4 | −1.3 | 1.1 | 6.9E-05 |
| GTP binding | |||||
| AF009246 | RAS, dexamethasone-induced 1 | 1.5 | 1.9 | 1.3 | 1.6E-03 |
| AW061337 | Adenylate kinase 4 | −1.4 | −1.7 | 0.9 | 9.1E-08 |
| ER | |||||
| AA615951 | Dolichyl-phosphate beta-glucosyltransferase | 1.5 | 1.5 | 1.0 | 6.8E-04 |
| AI642389 | Similar to KDEL. ER protein retention receptor 3 | 1.6 | 1.9 | 1.2 | 6.6E-06 |
| AI851230 | Homolog to ER lumen protein retaining receptor 1 | −1.5 | −1.2 | 1.2 | 7.4E-04/PICK> |
| Heat shock response/protein folding | |||||
| D85904 | Heat shock protein, 110 kDa | 1.4 | 1.6 | 1.1 | 6.9E-05 |
| U16959 | FK506 binding protein 5 (51 kDa) | 1.6 | 1.6 | 1.0 | 1.0E-04 |
| AI852161 | Tubulin-specific chaperone | 1.6 | 1.2 | 0.8 | 1.7E-04 |
| *X60676 | Heat shock protein 47, member 1 (Hsp47) | −2.6 | −2.6 | 1.0 | 1.8E-06 |
| L40406 | Heat shock protein, 105 kDa | −1.6 | −1.9 | 0.8 | 1.5E-05 |
| *AA615831 | Rat ischemia responsive 94-kDa protein | −1.6 | −1.6 | 1.0 | 4.0E-06 |
| AW122022 | Peptidylprolyl isomerase D (cyclophilin D) | −1.5 | −1.7 | 0.9 | 1.3E-06 |
| Immune response | |||||
| AV370035 | Chemokine (C-C) receptor 5 | −2.8 | −1.7 | 1.6 | 4.5E-04 |
| AV139913 | Stromal cell-derived factor 1 | −3.1 | −2.9 | 1.1 | 4.8E-05 |
| AF081789 | Lymphocyte antigen 68 | −1.4 | −1.3 | 1.0 | 1.1E-06 |
| Lipid/cholesterol metabolism | |||||
| Z19521 | Low-density lipoprotein receptor | −1.4 | −1.5 | 0.9 | 2.4E-05 |
| AB016248 | Sterol-C5-desaturase homolog (Saccharomyces cerevisiae) | −1.8 | −1.7 | 1.1 | 2.8E-03 |
| AW106745 | NAD(P)-dependent steroid dehydrogenase-like | −1.7 | −1.7 | 1.0 | 6.2E-09 |
| AW122653 | Mevalonate kinase | −1.4 | −1.4 | 1.0 | 7.8E-03 |
| D29016 | Farnesyl diphosphate farnesyl transferase 1 | −1.5 | −1.5 | 1.0 | 1.5E-06 |
| AW122260 | Cytochrome P450, 51 | −1.7 | −1.9 | 0.9 | 7.8E-05 |
| Al848668 | Sterol-C4-methyl oxidase-like | −1.9 | −2.1 | 0.9 | 1.0E-05 |
| Al846851 | Farnesyl pyrophosphate synthase | −1.8 | −1.7 | 1.1 | 1.1E-05 |
| AF057368 | 7′-Dehydrocholesterol reductase | −1.8 | −1.7 | 1.0 | 1.9E-04 |
| D42048 | Squalene epoxidase | −1.7 | −1.7 | 1.0 | 7.0E-07 |
| AW122523 | Long-chain fatty-acyl elongase | −2.1 | −2.0 | 1.1 | 2.5E-07 |
| AW060927 | 2,3-Oxidosqualene: lanosterol cyclase | −2.3 | −2.2 | 1.0 | 1.1E-06 |
| U48896 | UDP-glucoronosyltransferase 8 | −2.1 | −2.2 | 1.0 | 1.1E-06 |
| U28960 | Phospholipid transfer protein | −1.8 | −1.8 | 1.0 | 1.0E-06 |
| X95279 | Thyroid hormone responsive SPOT14 homolog | −1.5 | −1.7 | 0.9 | 2.4E-05 |
| M72394 | Phospholipase A2, group IV | −3.0 | −2.9 | 1.0 | 1.5E-07 |
| AW212131 | Choline phosphotransferase 1 | 1.5 | 1.8 | 1.2 | 7.2E-03 |
| U44389 | Hydroxyprostaglandin dehydrogenase 15 (NAD) | 1.9 | 2.4 | 1.3 | 1.5E-03 |
| Lysosome | |||||
| M32017 | Lysomal membrane glycoprotein 2 | 1.4 | 1.4 | 1.0 | 6.8E-06 |
| AV356071 | Lysosome-associated protein transmembrane 5 | 1.4 | 1.6 | 1.1 | 1.7E-03 |
| U87240 | Mannosidase 2, alpha B1 | 1.5 | 1.5 | 1.0 | 2.2E-05 |
| Nucleosome assembly | |||||
| X13605 | H3 histone, family 3B | 1.4 | 1.4 | 1.0 | 1.9E-02 |
| X05862 | H2B and H2A histone genes | −1.5 | −1.4 | 1.1 | 2.5E-06 |
| X61449 | Nucleosome assembly protein 1-like 1 | −1.5 | −1.6 | 1.0 | 1.2E-06 |
| Neurogenesis/survival | |||||
| U81317 | Myelin-associated oligodendrocytic basic protein | −2.7 | −2.3 | 1.2 | 3.0E-04 |
| U04827 | Fatty acid binding protein 7, brain | −1.7 | −1.2 | 1.4 | 2.5E-05 |
| AB017270 | Tmeff2 | −2.1 | −2.2 | 1.0 | 1.1E-07 |
| Polyamine biosynthesis | |||||
| L10244 | Spermidine/spermine N1-acetyl transferase | 2.2 | 2.1 | 1.0 | 3.8E-07 |
| Proteinase inhibitor | |||||
| X93037 | Extracellular proteinase inhibitor | 5.0 | 6.7 | 1.4 | 3.9E-11 |
| X70393 | Inter-alpha trypsin inhibitor, heavy chain 3 | 1.5 | 1.4 | 0.9 | 1.4E-04 |
| M64086 | Serine protease inhibitor 2-2 | 1.7 | 1.7 | 1.0 | 1.1E-07 |
| U26437 | Tissue inhibitor of metalloproteinases-3 | 1.5 | 1.3 | 0.9 | 1.2E-04 |
| Protein biosynthesis | |||||
| U10118 | Endothelial monocyte-activating polypeptide 2 | 1.4 | 1.3 | 0.9 | 1.0E-03 |
| AW125491 | Eukaryotic translation initiation factor 2, subunit 2 | 1.7 | 1.7 | 1.0 | 2.9E-03 |
| AI132207 | Eukaryotic translation initiation factor 1A | 1.6 | 1.7 | 1.0 | 2.3E-03 |
| AI839632 | Eukaryotic translation elongation factor 1 delta | 1.4 | 1.4 | 0.9 | 2.4E-03 |
| *Z50159 | Suil | 1.5 | 1.3 | 0.9 | 7.7E-05 |
| AI835709 | Ribosomal protein S25 | 2.0 | 1.5 | 0.8 | 9.1E-03 |
| AW048899 | Ribosomal protein S19 | 1.4 | 1.4 | 1.0 | 3.1E-03 |
| AW125120 | Mitochondrial ribosomal protein S25 | −1.5 | −1.3 | 1.1 | 2.3E-03 |
| C77227 | Mitochondrial ribosomal protein S25 | −1.4 | −1.4 | 1.0 | 7.8E-04 |
| Protein kinase/phosphatase | |||||
| AW046181 | Serum/glucocorticoid-regulated kinase | 3.8 | 2.9 | 0.8 | 2.1E-08 |
| AF039574 | Serine/threonine kinase 2 | 1.5 | 1.6 | 1.0 | 4.2E-05 |
| D45859 | Protein phosphatase 1B | 3.0 | 3.0 | 1.0 | 1.3E-04 |
| AI849109 | Dual specificity protein phosphatase 9 | −1.5 | −1.7 | 0.9 | 5.6E-05 |
| AF062076 | Serine/threonine kinase 16 | −1.5 | −1.6 | 0.9 | 9.4E-06 |
| M95408 | PTK2 protein tyrosine kinase 2 | −1.6 | −1.6 | 1.0 | 1.6E-05 |
| AV235418 | Tyrosine kinase receptor 1 | −1.7 | −1.5 | 1.1 | 3.3E-05 |
| X71426 | Endothelium-specific receptor tyrosine kinase | −1.5 | −1.6 | 1.0 | 4.5E-03 |
| AF077659 | Homeodomain interacting protein kinase 2 | −1.6 | −1.4 | 1.1 | 2.6E-04 |
| Protein degradation/proteolysis | |||||
| AI242663 | Cathepsin Z | 2.6 | 2.5 | 1.0 | 2.2E-06 |
| AI851255 | Cathepsin B | 1.7 | 1.7 | 1.0 | 2.4E-05 |
| AI848382 | Ubiquitin-specific protease 1 | 1.4 | 1.3 | 1.0 | 5.0E-03 |
| AI846522 | Ubiquitin-specific protease 2 | 1.7 | 1.5 | 0.9 | 1.1E-06 |
| AW228316 | Hypothetical serine protease | 1.5 | 1.4 | 0.9 | 6.1E-03 |
| AI746365 | Beta-transducin repeat containing protein | −1.6 | −1.5 | 1.0 | 4.4E-04 |
| AA726223 | a disintegrin and metalloproteinase domain 19 | −1.7 | −1.7 | 1.0 | 1.0E-05 |
| RNA binding/processing | |||||
| AB016424 | RNA binding motif protein 3 | 1.9 | 2.3 | 1.2 | 5.3E-06 |
| AI226368 | Heterogenous nuclear ribonucleoprotein D-like | 1.8 | 1.8 | 1.0 | 1.2E-04 |
| U11274 | Heterogenous nuclear ribonucleoprotein D | 1.6 | 2.0 | 1.3 | 4.1E-04 |
| AI183202 | Heterogenous nuclear ribonucleoprotein A1 | 2.2 | 2.0 | 0.9 | 1.4E-03 |
| AI840643 | Heterogenous nuclear ribonucleoprotein K | 1.4 | 1.3 | 0.9 | 2.8E-03 |
| *AA684508 | RNA, U22 small nucleolar (Rnu22) | 1.5 | 1.5 | 1.0 | 3.0E-06 |
| AW227345 | U2 small nuclear ribonucleoprotein polypeptide A | 1.7 | 1.7 | 1.0 | 1.2E-02 |
| AF073993 | Heterogenous nuclear ribonucleoprotein A2/B1 | −1.5 | −1.5 | 1.0 | 9.1E-06 |
| AI846595 | Homolog to splicing factor, arginine/serine-rich 6 | −1.9 | −1.5 | 1.3 | 7.8E-05 |
| U65735 | Elavll | −1.4 | −1.3 | 1.1 | 2.9E-05 |
| Scrapie | |||||
| AJ223206 | Scrapie-responsive gene 1 | 1.9 | 2.0 | 1.1 | 1.5E-09 |
| Cell signaling | |||||
| AJ001418 | Pyruvate dehydrogenase kinase 4 | 1.5 | 1.5 | 1.0 | 9.0E-03 |
| AB015978 | Oncostatin receptor | 2.0 | 1.9 | 1.0 | 8.2E-08 |
| AJ010045 | Neuroepithelial cell transforming gene 1 | 1.8 | 1.4 | 0.8 | 1.2E-03 |
| AW123650 | Gap junction membrane channel protein beta 6 | 1.8 | 1.5 | 0.9 | 4.5E-05 |
| X84215 | Gap junction membrane channel protein beta 1 | −1.5 | −1.4 | 1.1 | 4.3E-04 |
| AF105222 | Leukemia-associated gene-like | −1.6 | −1.6 | 1.0 | 2.2E-05 |
| Transcription regulation | |||||
| *U19118 | Activating transcription factor 3 (ATF3) | 2.4 | 2.6 | 1.1 | 5.4E-08 |
| X61800 | CCAAT/enhancer binding protein (C/EBP), delta | 2.1 | 2.1 | 1.0 | 1.7E-05 |
| M61007 | CCAAT/enhancer binding protein (C/EBP), beta | 3.0 | 3.2 | 1.1 | 4.9E-03 |
| AW045443 | Myocyte enhancer factor 2A | 1.5 | 1.6 | 1.1 | 7.6E-04 |
| AI848050 | Kruppel-like factor 9 | 1.4 | 1.4 | 1.0 | 2.2E-03 |
| M28845 | Early growth response 1 | 1.6 | 1.3 | 1.0 | 7.7E-05 |
| X63866 | Cellular nucleic acid binding protein | 1.4 | 1.3 | 1.0 | 2.3E-03 |
| *U20344 | Kruppel-like factor 4 (gut) (GKLF) | 1.6 | 1.5 | 0.9 | 9.1E-03 |
| U60453 | Enhancer of zeste homolog 1 (Drosophila) | 1.4 | 1.3 | 0.9 | 7.7E-05 |
| *AI843895 | Ethanol-induced 6 (Srebpl) | 1.4 | 1.4 | 1.0 | 9.2E-09 |
| AF038848 | Transcriptional regulator, S1N3B (yeast) | 1.5 | 1.4 | 1.0 | 4.6E-06 |
| *A67083 | CHOP/GADD153 | 3.6 | 3.5 | 1.0 | 4.6E-05 |
| U79748 | MAD homolog 4 (Drosophila) | 1.5 | 1.3 | 0.9 | 1.2E-02 |
| AF022992 | Period homolog (Drosophila) | 3.8 | 2.3 | −1.7 | 2.3E-06 |
| Y18505 | Mus musculus mRNA for XAP-5 protein | 1.5 | 1.5 | 1.0 | 9.0E-05 |
| U183148 | NFIL3/E4BP4 | 1.8 | 1.7 | 1.0 | 4.8E-06 |
| AB023485 | TATA box binding protein, Taf2s | 1.6 | 1.5 | 0.9 | 2.2E-04 |
| AI842969 | TAF9 RNA polymerase II | 1.5 | 1.7 | 1.1 | 1.4E-03 |
| AI553024 | Zinc finger gene PLZF | 2.9 | 2.0 | 0.7 | 1.6E-08 |
| U57524 | I kappa B alpha gene | 1.8 | 1.6 | 0.9 | 5.1E-04 |
| AW047554 | Zinc finger protein 96 | 2.2 | 1.7 | 0.8 | 8.1E-07 |
| AF017085 | General transcription factor II 1 | −1.5 | −1.5 | 1.0 | 8.2E-07 |
| U31566 | NK2 transcription factor related, locus 2 | −1.7 | −1.5 | 1.2 | 2.1E-05 |
| AF047389 | SRY box containing gene 10 | −1.4 | −1.3 | 1.1 | 6.8E-05 |
| L38607 | Forkhead box D1 | −1.6 | −1.5 | 1.0 | 4.9E-04 |
| AF043220 | General transcription factor II 1 | −1.5 | −1.3 | 1.2 | 1.9E-03 |
| L35032 | SRY-box containing gene 18 | −1.7 | −1.6 | 1.0 | 1.2E-02 |
| AW124932 | Pre B-cell leukemia transcription factor 1 | −1.9 | −1.9 | 1.0 | 3.2E-08 |
| AF015260 | MAD homolog 7 (Drosophila) | −1.6 | −1.5 | 1.1 | 8.0E-05 |
| AW046227 | WD40 protein Ciao 1 | −1.4 | −1.7 | 0.9 | 3.2E-04 |
| Transport | |||||
| *M22998 | Solute carrier family 2 member 1 (Glut 1) | 2.0 | 1.6 | 0.8 | 1.9E-07 |
| AI845514 | ATP-binding cassette, subfamily A (ABC1) | 2.3 | 2.2 | 1.0 | 1.9E-05 |
| AI853978 | Similar to solute carrier family 21 member 11 | −1.6 | −1.8 | 0.9 | 9.4E-08 |
| Miscellaneous | |||||
| AI854154 | 89% identical to Human ACP33 | 1.6 | 1.4 | 0.9 | 1.6E-04 |
| AW125865 | Histocompatibility antigen | 1.5 | 1.6 | 1.0 | 1.5E-03 |
| AW046124 | Cytochrome b-245, alpha-polypeptide | 1.5 | 1.7 | 1.1 | 1.7E-04 |
| U883327 | Cytokine-inducible SH2-containing protein 2 | 1.5 | 1.6 | 1.1 | 9.5E-03 |
| AI854020 | Cytosolic cysteine dioxygenase 1 | 1.4 | 1.4 | 1.0 | 4.0E-04 |
| X15591 | Cytotoxic-T-lymphocyte-associated protein 2 alpha | 1.6 | 1.9 | 1.2 | 2.1E-03 |
| AI326963 | Fibrinogen/angiopoietin-related protein | 1.6 | 1.6 | 1.0 | 9.7E-04 |
| AI844178 | HLA-B-associated transcript 3 | 2.0 | 1.3 | −1.5 | 8.7E-03 |
| V00756 | Interferon-related developmental regulator 1 | 1.4 | 1.6 | 1.1 | 3.0E-03 |
| C85523 | MLV VLeco LTRg | 1.7 | 1.6 | 0.9 | 2.8E-04 |
| AW125442 | Protein kinase inhibitor, alpha | 1.5 | 1.6 | 1.1 | 1.2E-03 |
| *AF063095 | Sell (suppressor of lin-12) 1 homolog (C. elegans) | 1.7 | 1.6 | 0.9 | 2.7E-04 |
| AI853531 | Similar to rat gene 33 polypeptide (GO) | 1.4 | 1.2 | 0.8 | 1.4E-04 |
| AF077527 | Syndecan binding protein | 1.7 | 1.4 | 0.8 | 5.4E-04 |
| D29797 | Syntaxin 3 | 1.4 | 1.5 | 1.1 | 7.8E-04 |
| X52102 | Synaptotagmin interacting protein | 1.4 | 1.4 | 0.9 | 2.3E-03 |
| Z70023 | Gap junction membrane channel protein beta 6 | 1.5 | 1.5 | 1.0 | 5.1E-03 |
| AW212859 | Axotrophin | 1.4 | 1.5 | 1.1 | 1.7E-02 |
| AA790056 | Cysteine- and histidine-rich protein | 1.8 | 1.9 | 1.0 | 1.7E-03 |
| AA674143 | Rattus norvegicus synaptojanin 2 binding protein | −1.5 | −1.6 | 0.9 | 3.9E-06 |
| AW061161 | Asparaginase-like sperum autoantigen | −1.7 | −1.7 | 1.0 | 4.7E-06 |
| L16992 | Branched chain ketoacid dehydrogenase E1, beta | −1.4 | −1.4 | 1.1 | 2.3E-03 |
| X99641 | Chromobox homolog 5 (Drosophila HP1a) | −1.4 | −1.3 | 1.1 | 8.7E-05 |
| U82758 | Claudin 5 | −1.7 | −1.4 | 1.2 | 1.5E-03 |
| *D16333 | Coproporphyrinogen oxidase (Cpo) | −1.7 | −1.8 | 0.9 | 1.1E-07 |
| AW122453 | Cys- and His-rich domain (CHORD)-containing | −1.8 | −2.1 | 0.9 | 3.3E-07 |
| D88792 | Cysteine-rich protein 2 | −1.6 | −2.0 | 0.8 | 4.2E-05 |
| AI842825 | Glycolipid transfer protein | −1.6 | −1.5 | 1.1 | 5.8E-06 |
| U18773 | GPI-anchored membrane protein 1 | −1.6 | −1.4 | 1.1 | 3.4E-05 |
| AI839882 | Homolog to oligo-RNase | −1.8 | −1.7 | 1.1 | 1.3E-06 |
| AI465543 | Homolog to protein ad-004 (protein cgi-137) | −1.7 | −2.0 | 0.9 | 1.9E-04 |
| AI843232 | Succinyl-coa: 3-ketoacid-coenzyme a transferase | −1.7 | −1.9 | 0.9 | 2.6E-05 |
| AF079528 | Immediate early response 5 | −1.4 | −1.5 | 1.0 | 6.0E-04 |
| AI850297 | Junction cell adhesion molecule 2 | −2.0 | −2.6 | 0.8 | 1.1E-05 |
| D16141 | Lethal giant larval homolog | −1.5 | −1.4 | 1.1 | 3.2E-07 |
| AW124555 | Methylmalonyl CoA epimerase (MCEE), mRNA | −1.4 | −1.5 | 1.0 | 4.8E-05 |
| AF060883 | Mucin 14, endothelial | −1.4 | −1.5 | 1.0 | 3.2E-04 |
| AI850965 | NADPH oxidase homolog | −1.4 | −1.4 | 1.0 | 1.5E-05 |
| AA986258 | Phosphorylated adaptor for RNA export | −1.4 | −1.4 | 1.0 | 3.1E-04 |
| AV319920 | Protein kinase WNK1 (WNK1) mRNA | −1.7 | −2.0 | 0.9 | 9.4E-03 |
| U35374 | Purine-nucleoside phosphorylase | −1.5 | −1.4 | 1.1 | 6.0E-04 |
| AI592541 | Ret finger protein 2 | −1.4 | −1.4 | 1.0 | 8.6E-04 |
| AA716963 | Similar to isopentenyl-diphosphate delta | −2.7 | −2.6 | 1.0 | 1.3E-07 |
| AI854285 | Similar to kelch family protein Ndl-L | −1.5 | −1.6 | 0.9 | 2.5E-04 |
| *U44088 | T-cell death associated gene (Tdag) | −1.5 | −1.4 | 1.1 | 2.5E-05 |
| X70956 | Topoisomerase (DNA) I | −1.5 | −1.5 | 1.0 | 1.3E-04 |
| U67611 | Transaldolase 1 | −1.6 | −1.5 | 1.1 | 1.1E-05 |
| AF061516 | Vesicle-associated membrane protein 4 | −1.7 | −1.9 | 0.9 | 1.9E-05 |
Function based on GO annotation (3) or deduced from relevant literature.
GenBank accession numbers.
Changes (n-fold) in pathogenic versus nonpathogenic viruses.
Mock inoculated.
Calculated with ANOVA FDR < 5%.
*, gene involved in ER stress.
LTR, long terminal repeat.
Real-time quantitative RT-PCR.
Quantitative reverse transcriptase PCR (RT-PCR) was performed to verify some changes in gene expression determined by array analysis by using an ABI PRISM 7900 Sequence Detection System (Applied Biosystems). Total RNA samples used in microarray analysis were analyzed with real-time RT-PCR with the addition of mice to each treatment group for a total sample size of eight mice. After DNase treatment, each sample was reverse transcribed and was PCR amplified in triplicate by using TaqMan one-step RT-PCR master mix (Applied Biosystems) in a total reaction volume of 10 μl. Reactions were performed with approximately 10 ng of total RNA, 500 nM each primer, and 250 nM TaqMan probe. Relative quantification was performed by constructing a standard curve of 10-fold serial dilutions by using RNA from brain extracted through protocols described above. This standard curve was used to determine a relative, arbitrary quantity based on the threshold cycle for each amplified sample with probes specific for the target. The relative quantity of target genes was normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each sample by amplifying GAPDH simultaneously with target genes but in separate triplicate-reaction tubes. Probe and primers for GAPDH were obtained from the Rodent GAPDH Control Reagent kit (Applied Biosystems). GAPDH was a suitable gene for normalization because it showed little variation between treatments, as determined in Affymetrix array experiments reported here (10 dpi, FrCasE versus F43 = 0.97-fold change and FrCasE versus mock = 0.95-fold change; and 17 dpi, FrCasE versus F43 = 0.99-fold change and FrCasE versus mock = 0.98-fold change). Normalized average relative gene expression values for each treatment were compared to determine relative change. Data were analyzed with ANOVA with Tukey's multiple-comparison test and a level of statistical significance of ≤0.05. Primer and probe sequences used for real-time RT-PCR are listed 5′ to 3′. F is forward strand and R is reverse. Virus RNA gag: F-AAACCAATGTGGCCATGTCATT, R-AAATCTTCTAACCGCTCTAACTTTCG, probe -ATCTGGCAGTCCGCCCCGG; CHOP: F - GTCCCTAGCTTGGCTGACAGA, R-TGGAGAGCGAGGGCTTTG, probe-CAGGGCCAACAGAGGTCACACGC; Grp58/ERp57: F-TCAAGGGTTTTCCTACCATCTACTTC, R-TTAATTCACGGCCACCTTCAT, probe-CACCAGCCAACAAGAAGCTAACTCCAAAGA; BiP: F-TCATCGGACGCACTTGGAA, R-CAACCACCTTGAATGGCAAGA, probe-ACCCTTCGGTGCAGCAGGACATCA, calreticulin: F-TTACGCACTGTCCGCCAAA, R-GCTCATGCTTCACCGTGAACT, probe-CGAACCCTTCAGCAATAAGGGCCAG; and Perk: F-AAGTAGATGACTGCAATTACGCTATCAA, R-TTTAACTTCCCGCATTACCTTCTC, probe-ATCCGGCTCCCCAACAGGGAGTT.
Immunohistochemistry.
Mice were anesthetized with isofluorane and were perfused with ice-cold PBS. Brains were removed and were fixed in 3.7% formaldehyde in PBS for 24 h at room temperature. Coronal blocks were subjected to processing for dehydration and were embedded in paraffin. Six-micrometer sections were deparaffinized, subjected to antigen retrieval with heated citrate buffer, and stained with goat antiserum to viral surface glycoprotein (SU) as described previously (5).
Cell culture studies.
NIH 3T3 cells (ATCC CRL 1658) were grown in Dulbecco's modified Eagle's medium supplemented with 10% calf serum at 37°C in a 5% CO2 humidified incubator. Cells were infected as described previously (44) in triplicate in the presence of 8 μg of Polybrene/ml. Mock-infected cells were exposed to Polybrene alone. After 48 h, RNA was extracted with an RNeasy mini kit (Qiagen) by lysing cells directly on plates. Cells were also infected and passed once at 72 h, and RNA was extracted at 96 h after infection. To determine the magnitude of expression changes caused by a known ER stress-inducing agent that blocks glycosylation, we treated NIH 3T3 cells with 2.0 μg of tunicamycin/ml for 6 h. This treatment has been used in previous studies where ER stress gene expression profiles were examined (19). NIH 3T3 cells were plated at a density of 5 × 104 cells/well in 12-well plates (Corning 3513). Tunicamycin (Sigma) dissolved in dimethyl sulfoxide (DMSO) or DMSO alone was added 18 h later when cells were 50 to 60% confluent. After 6 h, RNA was extracted and was subjected to real-time RT-PCR with the probes (TaqMan) described above. All real-time RT-PCR data were normalized with GAPDH as described above, though in one experiment we normalized with 18S rRNA (Applied Biosystems) with similar results. Statistical analysis was performed by using ANOVA for virus infection and unpaired t tests for tunicamycin experiments.
Immunoblot analysis.
Western blot analysis was performed on FrCasE-, F43-, and mock-infected NIH 3T3 cells at 48 h postinfection. Cells were washed with ice-cold PBS and were then lysed with 0.5% NP-40 in 0.01 M Tris-HCl, pH 7.4, 0.15 M NaCl, and 0.001 M EDTA and a protease inhibitor cocktail (Sigma) on ice for 10 min. Lysates were briefly centrifuged in a Capsule Tomy HF-120 centrifuge for 1 min to pellet nuclei. The supernatant was diluted in 5× sodium dodecyl sulfate (SDS) sample buffer containing 2-mercaptoethanol and was boiled for 5 min prior to loading on an SDS-10% polyacrylamide gel. The gel was then electroblotted onto an Immobilon P membrane (Millipore) and was probed with rabbit anti-SU protein kindly provided by G. Hunsmann, Goettingen, Germany, that recognizes both the precursor pr85env and the cleaved product, SU. A mouse anti-GAPDH antibody (Biodesign) was used to control for loading. Anti-SU was used at a 1:1,000 dilution, and anti-GAPDH was used at a 1:5,000 dilution. Membranes were developed with Attophos substrate (Promega), and bands were quantified with ImageQuant 5.2 (Molecular Dynamics).
RESULTS
Characterizing the model used for transcriptional profiling.
In this study we focused on the brain stem, a region of the nervous system where spongiform neurodegeneration is consistently observed after infection with FrCasE (11). Both FrCasE and F43 have been shown previously to infect the same spectrum of cell types in the nervous system. In the brain stem these include elements of the microvasculature (yellow arrows in Fig. 1A) as well as microglial cells (black arrows in Fig. 1A) (5). Astrocytes and neurons in the brain stem are not infected (34). Neuropathology in the form of small foci of spongiosis was first observed in the brain stem at 10 dpi of FrCasE (arrowheads in Fig. 1A), approximately 4 days prior to the development of mild tremor, which is the first sign of clinical disease (11). Lesions continued to spread in parallel with the progression of clinical disease, which reached the preterminal stage 17 dpi (Fig. 1A). In contrast, no neuropathology was seen in the F43-infected brain stem at either time point, despite extensive infection of microglial cells (Fig. 1A). Measurement of the relative levels of viral RNA in extracts of brain stem from FrCasE- and F43-inoculated mice revealed that at both time points F43 was expressed at higher levels than was FrCasE. These results support previous studies that showed higher levels of viral protein in the brains of F43- than in those of FrCasE -infected mice (5), indicating a curious inverse relationship between virus load and neurovirulence.
Genes up-regulated by both FrCasE and F43.
The importance of discriminating disease-specific responses from those responses to virus infection per se was readily apparent when expression profiles of FrCasE-, F43-, and mock-infected mice were examined. At both 10 and 17 dpi, 20 genes were up-regulated by infection with either retrovirus compared to mock-infected mice (Table 1). Interestingly, up-regulated transcripts of STAT1 as well as a variety of interferon-responsive genes, including major histocompatibility complex (MHC) class I genes and β2 microglobulin, suggested that both viruses induced brisk interferon responses. Since neither MHC class II nor the class II transactivator genes were induced (not shown), this profile appeared to represent an alpha/beta interferon response. Because these genes were up-regulated by both the virulent and avirulent viruses, they were omitted from further analysis.
Disease-specific transcriptional profile early in the disease is suggestive of an unfolded protein response.
It is widely recognized that perturbations of ER homeostasis such as the accumulation of misfolded proteins in the ER activate a stereotypical “unfolded protein response” (UPR) (15, 29). This response is controlled by stress-induced signaling that results in transcriptional up-regulation of ER chaperones or glucose-regulated proteins (Grps), which help alleviate stress by promoting proper protein folding. In addition to increased chaperone expression, increased proteasome degradation of misfolded proteins and decreased translation initiation allow cellular adaptation to the increased protein folding demands placed on the organelle. Ultimately, perturbations in the ER may lead to the transcriptional activation of the ER stress-regulated, proapoptotic gene CHOP/GADD153 and cell death (58, 61).
At 10 dpi, a time coincident with the onset of lesions in vivo, but 4 days prior to the onset of clinical disease, there was a group of 12 up-regulated genes that were observed only in mice inoculated with FrCasE (Table 2). Two genes in this group encode proteins that may be involved in protein ubiquitination. One gene encodes the protein rjs, which has a HECT domain that may have common function with E3 ubiquitin-protein ligases and has recently been shown to be induced by tunicamycin, a known inducer of ER stress (19). The other gene is unnamed but appears to be related to a ubiquitin carrier protein. In addition, the up-regulation of glucose-regulated protein 58 (Grp58/ERp57) as well as of CHOP supports the notion that FrCasE infection may be associated with the induction of an ER stress response. Grp58/ERp57 is a thiol oxidoreductase involved in ER protein folding (32). Thus, 4 out of the 12 up-regulated disease-specific genes detected at this early time point appear to represent a response to the accumulation of unfolded proteins in the ER.
Disease-specific transcriptional profile detected in advanced disease.
At 17 dpi there were 263 transcripts that were differentially expressed in FrCasE-inoculated mice relative to F43- and mock-inoculated mice (Table 3). Of these, 41 transcripts were from genes of unknown function and are not shown in Table 3. Consistent with the expression profile at 10 dpi suggesting perturbation of ER function, we found differential expression of several genes that are known to be involved in the response to ER stress, including CHOP, ATF3, Sui1, GADD45, Tdag, Cyclin D1, Sel-1, Srebp-1, ischemia-responsive 94kDa protein, Hsp47, Gas5, Rnu22, NFIL3/E4BP4, GKLF, Cpo, and Glut1 (8, 19, 20, 27, 46, 47, 49, 57, 59) (marked by asterisks in Table 3). CHOP, ATF3, GADD45, and cyclin D1 also are implicated in cell cycle control and/or apoptosis. Additional genes differentially expressed late in disease and involved in cell cycle/apoptosis include inhibitor of apoptosis protein 2 (IAP2) (down-regulated), the cell death-promoting gene DP5 (up-regulated), which interacts with BCL2 family members, and the cyclin-dependent kinase inhibitor p21 (up-regulated).
Strikingly, of these 263 differentially expressed genes, only one gene, CHOP, was scored as up-regulated at both the early and late time points in FrCasE-inoculated mice. The quantitative differences in CHOP transcripts between FrCasE- and F43-inoculated mice increased from 1.4- at 10 dpi to 3.6-fold at 17 dpi. No change in levels of CHOP transcripts was observed in the F43- versus mock-inoculated mice at either time point.
Does FrCasE induce ER stress?
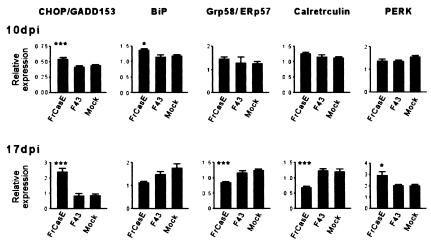
In view of the progressive increase in CHOP expression as well as other signs of ER stress detected at 10 dpi, we used real-time RT-PCR to determine transcript levels for several important players in ER stress responses (Fig. 2). Some of these transcripts were not detected with our initial filtering criteria or were not present on the arrays used. Genes investigated included CHOP, three ER chaperones, i.e., BiP (Grp78), Grp58/ERp57, and calreticulin, and PERK, a protein kinase involved in the attenuation of protein translation during ER stress. PERK activation occurs posttranslationally (18), and transcriptional activation has not, to our knowledge, been reported. Nevertheless, we included this gene because previous studies in our lab with non-Affymetrix arrays suggested up-regulation of PERK in FrCasE-infected mice. When the sample size was increased from five mice in microarrays to eight mice used in real-time RT-PCR, we found that BiP was elevated at 10 dpi in the FrCasE-inoculated mice (Fig. 2). Although the change (n-fold) relative to the F43-inoculated mice was small, it was significant. At 10 dpi, Grp58/ERp57 was up-regulated as determined by Affymetrix arrays (Table 2). Real-time RT-PCR confirmed this small increase in Grp58/ERp57 expression in FrCasE-inoculated mice (Fig. 2), although the difference was not significant. At 17 dpi PERK was indeed up-regulated in the FrCasE-inoculated mice, but curiously transcripts for calreticulin and Grp58/ERp57, appeared to be depressed in these mice (Fig. 2). BiP expression reflected this trend, although the difference in BiP expression was not significant. Consistent with the late down-regulation of ER chaperones, heat shock protein 47 (Hsp47) and ischemia-responsive element 94 RNAs were also depressed in FrCasE-inoculated mice at 17 dpi as seen in Affymetrix array analysis (see asterisks under “Heat shock response/protein folding” in Table 3). These results, thus, provided support for the microarray data, which suggested early transcriptional activation of a subset of genes involved in ER stress.
FIG. 2.
Quantification of mRNA levels of select ER stress genes in brain stems of FrCasE-infected mice. Real-time RT-PCR was used to quantify transcript levels in groups of eight mice sacrificed at 10 and 17 dpi. At 10 dpi there was a small increase in FrCasE-infected brain stem in CHOP, BiP, Grp58/ERp57, and calreticulin, though statistical significance was reached only for CHOP and BiP. At 17 dpi CHOP was up-regulated, but BiP, GrP58/ERp57, and calreticulin each appeared to be down-regulated. PERK was up-regulated as well at 17 dpi (Fig. 3). Data are shown as mean ± standard error of the mean. P for FrCasE versus: F43: *, <0.05; **, <0.01; and ***, <0.001.
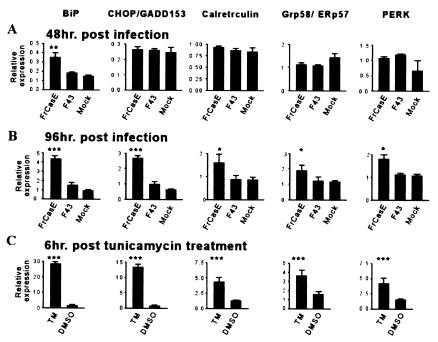
In order to address more directly whether infection of cells with FrCasE was associated with the induction of an ER stress response, we carried out in vitro experiments with NIH 3T3 cells that are highly permissive for both FrCasE and F43. Cells were infected, and RNA was extracted at either 48 h or 96 h postinfection. Quantitative RT-PCR was performed with probes for BiP, calreticulin, Grp58/ERp57, CHOP, and PERK (Fig. 3A and B). After 48 h BiP was significantly up-regulated only in the FrCasE-infected cultures (Fig. 3A), a result that has been repeated in three independent experiments. In contrast, at this time point expression of the other ER stress-associated genes appeared not to be affected. At 96 h postinfection, however, all of these genes were up-regulated in the FrCasE-infected cultures (Fig. 3B). In contrast, none of these genes was up-regulated in the F43-infected cultures at either time point.
FIG. 3.
Expression of ER stress genes in FrCasE-infected and tunicamycin-treated NIH 3T3 cells. NIH 3T3 cells were infected with either FrCasE or F43 or were mock infected. RNA was extracted at 48 (A) or 96 (B) h after infection and was analyzed by real-time RT-PCR with probes for BiP, CHOP/GADD153, calreticulin, Grp58/ERp57, and PERK. Each data set represents infections in triplicate. (C) NIH 3T3 cells were treated with 2.0 μg of tunicamycin or control (DMSO) per ml for 6 h and with RNA subject to real-time RT-PCR with the probes listed above (n = 4). Virus infection data were analyzed with ANOVA with Tukey's multiple-comparison test and are shown as average relative expression values normalized to GAPDH. Data from tunicamycin-treated cells were analyzed with unpaired t tests. For comparisons of FrCasE, F43, or tunicamycin with DMSO, *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Data are shown as mean ± standard error of the mean.
To place these data in perspective, we exposed NIH 3T3 cells to tunicamycin, a known inducer of ER stress. RNA was extracted after 6 h of exposure and was analyzed with the same set of probes (Fig. 3C). The results indicated that the responses of BiP and CHOP to tunicamycin were more robust than those to FrCasE. However, the up-regulation of calreticulin, Grp58/ERp57, and PERK was quantitatively similar for the two treatments, implying that FrCasE was a relatively potent inducer of ER stress. The up-regulation of PERK mRNA by tunicamycin was unexpected but was consistent with its up-regulation by FrCasE. This suggests that PERK activation under conditions of ER stress may occur both posttranslationally and at the transcriptional level, an observation that merits further investigation.
These in vitro results therefore support the in vivo data suggesting the early activation of an ER stress response in the brain stems of FrCasE-infected mice and imply that even small changes discovered with microarray technology in complex tissues can represent larger changes when a single affected cell type is examined. Taken together, these results suggest that the induction of an ER stress response in the brain stems of FrCasE-inoculated mice likely represents a proximal event originating from virus-infected cells.
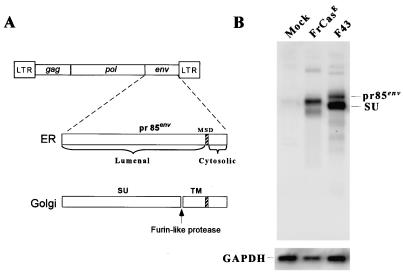
Since FrCasE and F43 differ in the sequence of their envelope genes, it is reasonable to suggest that it is the misfolding of the viral envelope protein of FrCasE that is responsible for the induction of ER stress. The viral envelope protein is synthesized as a precursor polyprotein called pr85env (Fig. 4A) (51) consisting of the N-terminal SU glycoprotein and the C-terminal transmembrane protein (TM) (Fig. 4A). The FrCasE SU protein contains seven and the F43 SU protein eight potential N-linked glycosylation sites (36). Furthermore, the FrCasE SU protein contains two deletions of 4 and 7 residues within the proline-rich domain when compared to F43. These differences account for differences in molecular sizes of the respective proteins in SDS-polyacrylamide gel electrophoresis (Fig. 4B). pr85env is cleaved by a furin-like protease in the Golgi, yielding a membrane-anchored TM protein and an SU protein associated with TM by noncovalent and disulfide bonds. Thus, at steady state, the ratio of pr85env to SU can be used as a relative measure of the partitioning of the respective proteins within the ER and Golgi. Immunoblot analysis of whole-cell lysates of NIH 3T3 cells infected with FrCasE and F43 revealed a reproducible difference in the pr85env-to-SU ratio (Fig. 4B). For F43 the ratio was 1:2.5; for FrCasE the ratio was 2:1, suggesting that relative to F43, the FrCasE Env precursor protein was retained in the ER.
FIG. 4.
Steady-state levels of viral envelope protein in infected NIH 3T3 cells. Panel A schematically shows the proteolytic processing of the envelope protein of MLV (see text). The viral genome is shown at the top. MSD, membrane-spanning domain. Panel B shows an immunoblot of lysates of FrCasE-, F43-, and mock-infected cells probed with a polyclonal antiserum to MLV SU protein. This antiserum recognizes pr85env as well as SU protein but does not react with the TM protein (p15E). The blot was stripped and was reprobed with anti-GAPDH to control for loading differences. Note that the ratio of pr85env to SU is different for these two viruses, indicating that in FrCasE-infected cells there is accumulation of the envelope precursor protein, suggestive of ER retention.
DISCUSSION
The neurologic disease caused by the wild mouse virus CasBrE and its derivatives appears to have more in common with a degenerative process than with a typical inflammatory viral encephalitis. Most striking is the complete absence of inflammatory cellular infiltrates in the brain, despite progressive neuronal dropout and gliosis (2). Indeed, by using a highly sensitive RNase protection assay, we found essentially no evidence for up-regulation of inflammatory cytokines in the central nervous system until very late in the course of the disease (4). These results were confirmed by microarray analysis in this study. Thus, it appears unlikely that this disease has an inflammatory etiology.
Spongiform neurodegeneration is a fairly rare type of pathology that is generally considered to be diagnostic of the prion diseases, though similar lesions have also been observed in SOD2 (40) and attractin knockout (16) mice. In none of these conditions is the molecular pathogenesis of the neurodegeneration understood. Here we have utilized oligonucleotide microarray technology to approach, at the transcriptional level, the early events in the genesis of spongiform lesions induced by the mouse retrovirus FrCasE, a highly neuropathogenic derivative of CasBrE.
The number of disease-specific genes up-regulated early was small, and curiously only one of these genes, CHOP, was also up-regulated at the late time point. In recent years it has become apparent that CHOP is up-regulated in response to conditions that perturb ER function, such as the accumulation of unfolded proteins (58). Indeed, evidence for up-regulation of genes encoding proteins involved in both the sensing and resolution of ER stress was detected at the early time point (10 dpi) and only in mice inoculated with the pathogenic virus FrCasE. These genes included BiP, an ER resident chaperone, and Grp58/ERp57, an oxidoreductase known to complex with calreticulin in the ER and to be involved in protein folding. Finally, there was disease-specific up-regulation of two genes encoding molecules that appear to be involved in the ubiquitin/proteosome system; one of these (rjs) has recently been implicated in the response to ER stress (19). Cumulatively, these results provide evidence for engagement of a host response to perturbation of ER homeostasis early in the course of the disease.
Although the ER stress response was observed very early in the course of the neurodegenerative disease caused by FrCasE, there was already evidence for small but clear-cut foci of spongiform neurodegeneration. It was possible that these ER stress-associated transcripts were expressed in the uninfected neuronal and glial cells exhibiting the degenerative changes. However, we found clear evidence that infection of fibroblasts in vitro with FrCasE but not F43 induced a similar spectrum of ER stress responses as was observed in vivo. Thus, it appears likelier that these responses were derived from the infected cells in the brain stem. Finally, since FrCasE and F43 differ in the sequence of their respective envelope genes, it seems likely that it is misfolding of the envelope protein in the ER of FrCasE-infected cells that is responsible for the induction of ER stress both in vivo and in vitro. In support of this notion was the finding of the accumulation of unprocessed envelope precursor protein (pr85env) in FrCasE- compared to F43-infected NIH 3T3 cells. Since proteolytic processing of pr85env occurs in the Golgi, this observation suggests that the FrCasE envelope protein is retained in the ER. Interestingly, infection of microglial cells with FrCasE as well as another retrovirus that induces spongiform neurodegeneration, NT40, results in defective processing of the viral envelope protein (17, 33). These results are similar to processing defects described here that were encountered in the use of infected NIH 3T3 cells, suggesting that infected microglia are also undergoing ER stress. To further test our model, it will be important to fully investigate ER stress in microglia both in vivo and in vitro.
It has long been recognized through genetic studies that the envelope genes of the neuropathogenic MLVs carry the determinants of their neurovirulence, but the virus-host interactions underlying this neurotoxicity have not been resolved. The notion that the neuropathogenicity of these viruses may be related to protein misfolding in the ER has been proposed previously, though the evidence was derived exclusively from in vitro studies (33, 48). The most compelling evidence supporting a role for protein misfolding in this disease has come from a series of genetic studies on a temperature-sensitive mutant of Moloney MLV (ts1) that induces a disease indistinguishable pathologically from that caused by FrCasE. The temperature sensitivity of this virus in vitro involved the improper assembly of envelope protein oligomers in the ER at the nonpermissive temperature, resulting in ER retention (26). Interestingly, the neurovirulence of ts1 in vivo was found to map to the same sequence in the N terminus of the envelope protein, as did the temperature sensitivity in vitro (53). Nevertheless, the present study is the first to provide evidence suggesting that protein misfolding is a determinant of MLV-induced neurodegeneration in vivo.
Although more than 250 disease-specific transcripts were differentially expressed in the brain stems of mice with advanced disease, it is difficult to distinguish those responses specifically involved in disease pathogenesis from those induced as a consequence of secondary effects of the poor clinical condition of mice at a preterminal phase of the disease. These mice not only exhibited severe tremor and paralysis but were also wasted, suggesting nutritional deficiency. Thus, some of the late responses observed in the brain stems of FrCasE-inoculated mice were undoubtedly a consequence of secondary metabolic effects.
CHOP was up-regulated at both the early and late time points in the disease. In contrast, at 17 dpi some ER stress-associated transcripts analyzed here, including calreticulin and Grp58/ERp57, were actually down-regulated in FrCasE-inoculated mice. The explanation for these fluxes in gene expression is not readily apparent, though they may also represent secondary effects of the poor clinical status of these mice with advanced neurologic disease. Alternatively, this response could indicate that infected cells were no longer able to adapt to the stress of increased misfolded proteins and may have been in the process of undergoing cell death. This is consistent with the progressive increase in expression of the proapoptotic gene CHOP between 10 and 17 dpi.
Two types of ER-stress responses have been associated with virus infection. One is termed the ER overload response (EOR) and appears to be induced by the accumulation of normally folded viral glycoproteins in the ER (42). This response can apparently be induced by increased expression of any ER-targeted protein and results in the activation of NF-κB and the consequent up-regulation of a variety of genes involved in inflammatory responses as well as interferon-responsive elements. The early up-regulation of interferon-responsive genes by both FrCasE and F43 (Table 1) may represent an EOR but clearly is not a determinant of neurovirulence. The second type of ER stress is the unfolded protein response (UPR) described above, which is induced by the accumulation specifically of misfolded proteins in the ER. In vitro studies have shown that infection with several different viruses can result in the induction of the UPR. These include at least three members of the Flaviviridae (25, 50, 54), measles virus (7), and respiratory syncytial virus (6). As yet, however, there is no evidence that the UPR is involved in the pathogenesis of the diseases caused by these viruses. The observation that the induction of an unfolded protein response is a determinant of neurovirulence of FrCasE suggests the possibility that this neurodegenerative disease may represent a protein folding or conformational disease induced by a virus.
The induction of ER stress by FrCasE could explain the unusual inverse relationship between the level of viral RNA (this study) and viral protein (5) in the brain stem. We have consistently observed that F43 replicates to higher levels in the brain than does FrCasE, an observation that has, until now, remained counterintuitive. However, retention of the FrCasE envelope protein by the ER quality control system could influence the amount of protein available for virus assembly and therefore might impact the level of virus spread in the brain.
What remains unclear is the connection between the UPR induced in FrCasE-infected cells (primarily microglial and endothelial cells) and the ultimate expression of cytotoxicity that is exhibited by uninfected neurons and glial elements in the vicinity of the infected cells. One possibility is that the UPR induced in infected microglial cells leads ultimately to their demise and perhaps a loss of trophic factors secreted by these cells. This hypothesis is supported by the up-regulation of proapoptotic genes, such as CHOP, DP5, p21, and GADD45, and the down-regulation of IAP2, an antiapoptotic gene (Table 3). Alternatively, it is also possible that the neuropathology induced by FrCasE is a consequence of a gain-of-function in the infected cells. It has been reported that protein misfolding as well as overexpression of CHOP is associated with the depletion of reducing equivalents such as glutathione in the cell (19, 39). Since glutathione is a major scavenger of reactive oxygen species, this could conceivably tie unfolded proteins and ER stress to the accumulation of ROS in the vicinity of the infected cells. Indeed, the spongiform lesions induced by another neurovirulent MLV, PVC211, appear to be associated with evidence of local oxidative damage, and feeding mice vitamin E measurably lengthened the incubation period of this disease (60).
Whether apoptosis or the generation of ROS represents the link between the UPR and the neurotoxicity, it is likely that understanding the events downstream of this virus-induced UPR will shed light on the pathogenesis of human neurodegenerative disorders associated with misfolded host proteins. Recent studies suggest that ER stress may play a role in the pathogenesis of Alzheimer's (28, 55), Huntington's (41), and Parkinson's (22, 45) diseases. Interestingly, the activation of ER stress responses appears to be induced both by the accumulation of misfolded proteins in the ER as well as the accumulation of protein aggregates in the cytosol. The latter effect may be mediated through inhibition of ubiquitin/proteasome-mediated protein degradative pathways (9, 41). In this regard, it is of interest that a spongiform neurodegenerative disease seen in mice with the mahaganoid coat color was recently linked to a mutation in a gene that resembles a ubiquitin ligase (21). Finally, several studies suggest that misfolding and retention of PrP in the ER (23, 24) and perhaps retrograde transport of misfolded PrP to the cytosol (38) may participate in the pathogenesis of familial TSE diseases. It remains to be determined whether the spongiform neurodegeneration induced by these mutant host proteins involves the induction of ER stress responses related to those observed here in this retroviral disease.
Acknowledgments
We thank Gary Hettrick and Anita Mora for assistance with figures and Byron Caughey, Bruce Chesebro, Kim Hasenkrug, and Ina Vorberg for critical reading of the manuscript.
REFERENCES
- 1.Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659-666. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, J. M., and M. B. Gardner. 1974. Lower motor neuron degeneration associated with type C RNA virus infection in mice: neuropathological features. J. Neuropathol. Exp. Neurol. 33:285-307. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, and G. Sherlock. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Askovic, S., C. Favara, F. J. McAtee, and J. L. Portis. 2001. Increased expression of MIP-1α and MIP-1β mRNAs in the brain correlates spatially and temporally with the spongiform neurodegeneration induced by a murine oncornavirus. J. Virol. 75:2665-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Askovic, S., F. J. McAtee, C. Favara, and J. L. Portis. 2000. Brain infection by neuroinvasive but avirulent murine oncornaviruses. J. Virol. 74:465-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bitko, V., and S. Barik. 2001. An endoplasmic reticulum-specific stress-activated caspase (caspase-12) is implicated in the apoptosis of A549 epithelial cells by respiratory syncytial virus. J. Cell. Biochem. 80:441-454. [DOI] [PubMed] [Google Scholar]
- 7.Bolt, G., K. Berg, and M. Blixenkrone-Moller. 2002. Measles virus-induced modulation of host-cell gene expression. J. Gen. Virol. 83:1157-1165. [DOI] [PubMed] [Google Scholar]
- 8.Brewer, J. W., L. M. Hendershot, C. J. Sherr, and J. A. Diehl. 1999. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl. Acad. Sci. USA 96:8505-8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush, K. T., A. L. Goldberg, and S. K. Nigam. 1997. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J. Biol. Chem. 272:9086-9092. [DOI] [PubMed] [Google Scholar]
- 10.Clement, K., N. Viguerie, M. Diehn, A. Alizadeh, P. Barbe, C. Thalamas, J. D. Storey, P. O. Brown, G. S. Barsh, and D. Langin. 2002. In vivo regulation of human skeletal muscle gene expression by thyroid hormone. Genome Res. 12:281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czub, S., W. P. Lynch, M. Czub, and J. L. Portis. 1994. Kinetic analysis of spongiform neurodegenerative disease induced by a highly virulent murine retrovirus. Lab. Investig. 70:711-723. [PubMed] [Google Scholar]
- 12.DesGroseillers, L., M. Barrette, and P. Jolicoeur. 1984. Physical mapping of the paralysis-inducing determinant of a wild mouse ecotropic neurotropic retrovirus. J. Virol. 52:356-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner, M. B., B. E. Henderson, J. D. Estes, R. W. Rongey, J. Casagrande, M. Pike, and R. J. Huebner. 1976. The epidemiology and virology of C-type virus-associated hematological cancers and related diseases in wild mice. Cancer Res. 36:574-581. [PubMed] [Google Scholar]
- 14.Gardner, M. B., B. E. Henderson, J. E. Officer, R. W. Rongey, J. C. Parker, C. Oliver, J. D. Estes, and R. J. Huebner. 1973. A spontaneous lower motor neuron disease apparently caused by indigenous type-C RNA virus in wild mice. J. Natl. Cancer Inst. 51:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gething, M. J., and J. Sambrook. 1992. Protein folding in the cell. Nature 355:33-45. [DOI] [PubMed] [Google Scholar]
- 16.Gunn, T. M., T. Inui, K. Kitada, S. Ito, K. Wakamatsu, L. He, D. M. Bouley, T. Serikawa, and G. S. Barsh. 2001. Molecular and phenotypic analysis of Attractin mutant mice. Genetics 158:1683-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen, R., S. Czub, E. Werder, J. Herold, G. Gosztonyi, H. Gelderblom, S. Schimmer, S. Mazgareanu, V. ter Meulen, and M. Czub. 2000. Abundant defective viral particles budding from microglia in the course of retroviral spongiform encephalopathy. J. Virol. 74:1775-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 19.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto, Y., C. Zhang, J. Kawauchi, I. Imoto, M. T. Adachi, J. Inazawa, T. Amagasa, T. Hai, and S. Kitajima. 2002. An alternatively spliced isoform of transcriptional repressor ATF3 and its induction by stress stimuli. Nucleic Acids Res. 30:2398-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, L., X. Y. Lu, A. F. Jolly, A. G. Eldridge, S. J. Watson, P. K. Jackson, G. S. Barsh, and T. M. Gunn. 2003. Spongiform degeneration in mahoganoid mutant mice. Science 299:710-712. [DOI] [PubMed] [Google Scholar]
- 22.Imai, Y., M. Soda, H. Inoue, N. Hattori, Y. Mizuno, and R. Takahashi. 2001. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell 105:891-902. [DOI] [PubMed] [Google Scholar]
- 23.Ivanova, L., S. Barmada, T. Kummer, and D. A. Harris. 2001. Mutant prion proteins are partially retained in the endoplasmic reticulum. J. Biol. Chem. 276:42409-42421. [DOI] [PubMed] [Google Scholar]
- 24.Jin, T., Y. Gu, G. Zanusso, M. Sy, A. Kumar, M. Cohen, P. Gambetti, and N. Singh. 2000. The chaperone protein BiP binds to a mutant prion protein and mediates its degradation by the proteasome. J. Biol. Chem. 275:38699-38704. [DOI] [PubMed] [Google Scholar]
- 25.Jordan, R., L. Wang, T. M. Graczyk, T. M. Block, and P. R. Romano. 2002. Replication of a cytopathic strain of bovine viral diarrhea virus activates PERK and induces endoplasmic reticulum stress-mediated apoptosis of MDBK cells. J. Virol. 76:9588-9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamps, C. A., Y. C. Lin, and P. K. Wong. 1991. Oligomerization and transport of the envelope protein of Moloney murine leukemia virus-TB and of ts1, a neurovirulent temperature-sensitive mutant of MoMuLV-TB. Virology 184:687-694. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko, M., M. Ishiguro, Y. Niinuma, M. Uesugi, and Y. Nomura. 2002. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 532:147-152. [DOI] [PubMed] [Google Scholar]
- 28.Katayama, T., K. Imaizumi, N. Sato, K. Miyoshi, T. Kudo, J. Hitomi, T. Morihara, T. Yoneda, F. Gomi, Y. Mori, Y. Nakano, J. Takeda, T. Tsuda, Y. Itoyama, O. Murayama, A. Takashima, P. George-Hyslop, M. Takeda, and M. Tohyama. 1999. Presenilin-1 mutations downregulate the signalling pathway of the unfolded-protein response. Nat. Cell Biol. 1:479-485. [DOI] [PubMed] [Google Scholar]
- 29.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 30.Li, C., and W. W. Hung. 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:1-11. [DOI] [PMC free article] [PubMed]
- 31.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindquist, J. A., O. N. Jensen, M. Mann, and G. J. Hammerling. 1998. ER-60, a chaperone with thiol-dependent reductase activity involved in MHC class I assembly. EMBO J. 17:2186-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lynch, W. P., W. J. Brown, G. J. Spangrude, and J. L. Portis. 1994. Microglial infection by a neurovirulent murine retrovirus results in defective processing of envelope protein and intracellular budding of virus particles. J. Virol. 68:3401-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch, W. P., S. Czub, F. J. McAtee, S. F. Hayes, and J. L. Portis. 1991. Murine retrovirus-induced spongiform encephalopathy: productive infection of microglia and cerebellar neurons in accelerated CNS disease. Neuron 7:365-379. [DOI] [PubMed] [Google Scholar]
- 35.Lynch, W. P., S. J. Robertson, and J. L. Portis. 1995. Induction of focal spongiform neurodegeneration in developmentally resistant mice by implantation of murine retrovirus-infected microglia. J. Virol. 69:1408-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynch, W. P., and A. H. Sharpe. 2000. Differential glycosylation of the Cas-Br-E Env protein is associated with retrovirus-induced spongiform neurodegeneration. J. Virol. 74:1558-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lynch, W. P., E. Y. Snyder, L. Qualtiere, J. L. Portis, and A. H. Sharpe. 1996. Late virus replication events in microglia are required for neurovirulent retrovirus-induced spongiform neurodegeneration: evidence from neural progenitor-derived chimeric mouse brains. J. Virol. 70:8896-8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma, J., R. Wollmann, and S. Lindquist. 2002. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science 298:1781-1785. [DOI] [PubMed] [Google Scholar]
- 39.McCullough, K. D., J. L. Martindale, L. O. Klotz, T. Y. Aw, and N. J. Holbrook. 2001. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol. Cell. Biol. 21:1249-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melov, S., J. A. Schneider, B. J. Day, D. Hinerfeld, P. Coskun, S. S. Mirra, J. D. Crapo, and D. C. Wallace. 1998. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat. Genet. 18:159-163. [DOI] [PubMed] [Google Scholar]
- 41.Nishitoh, H., A. Matsuzawa, K. Tobiume, K. Saegusa, K. Takeda, K. Inoue, S. Hori, A. Kakizuka, and H. Ichijo. 2002. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 16:1345-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pahl, H. L., and P. A. Baeuerle. 1997. The ER-overload response: activation of NF-kappa B. Trends Biochem. Sci. 22:63-67. [DOI] [PubMed] [Google Scholar]
- 43.Portis, J. L. 2001. Genetic determinants of neurovirulence of murine oncornaviruses. Adv. Virus Res. 56:3-38. [DOI] [PubMed] [Google Scholar]
- 44.Portis, J. L., S. Czub, C. F. Garon, and F. J. McAtee. 1990. Neurodegenerative disease induced by the wild mouse ecotropic retrovirus is markedly accelerated by long terminal repeat and gag-pol sequences from nondefective Friend murine leukemia virus. J. Virol. 64:1648-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryu, E. J., H. P. Harding, J. M. Angelastro, O. V. Vitolo, D. Ron, and L. A. Greene. 2002. Endoplasmic reticulum stress and the unfolded protein response in cellular models of Parkinson's disease. J. Neurosci. 22:10690-10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 47.Sheikh, M. S., E. Fernandez-Salas, M. Yu, A. Hussain, J. D. Dinman, S. W. Peltz, Y. Huang, and A. J. Fornace, Jr. 1999. Cloning and characterization of a human genotoxic and endoplasmic reticulum stress-inducible cDNA that encodes translation initiation factor 1(eIF1(A121/SUI1)). J. Biol. Chem. 274:16487-16493. [DOI] [PubMed] [Google Scholar]
- 48.Shikova, E., Y.-C. Lin, K. Saha, B. R. Brooks, and P. K. Y. Wong. 1993. Correlation of specific virus-astrocyte interactions and cytopathic effects induced by ts1, a neurovirulent mutant of Moloney murine leukemia virus. J. Virol. 67:1137-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Southwood, C. M., J. Garbern, W. Jiang, and A. Gow. 2002. The unfolded protein response modulates disease severity in Pelizaeus-Merzbacher disease. Neuron 36:585-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su, H. L., C. L. Liao, and Y. L. Lin. 2002. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J. Virol. 76:4162-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 52.Swarz, J. R., B. R. Brooks, and R. T. Johnson. 1981. Spongiform polioencephalomyelopathy caused by a murine retrovirus. II. Ultrastructural localization of virus replication and spongiform changes in the central nervous system. Neuropathol. Appl. Neurobiol. 7:365-380. [DOI] [PubMed] [Google Scholar]
- 53.Szurek, P. F., E. Floyd, P. H. Yuen, and P. K. Y. Wong. 1990. Site-directed mutagenesis of the codon for Ile-25 in gPr80env alters the neurovirulence of ts1, a mutant of Moloney murine leukemia virus TB. J. Virol. 64:5241-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tardif, K. D., K. Mori, and A. Siddiqui. 2002. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 76:7453-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terro, F., C. Czech, F. Esclaire, W. Elyaman, C. Yardin, M. C. Baclet, N. Touchet, G. Tremp, L. Pradier, and J. Hugon. 2002. Neurons overexpressing mutant presenilin-1 are more sensitive to apoptosis induced by endoplasmic reticulum-Golgi stress. J. Neurosci. Res. 69:530-539. [DOI] [PubMed] [Google Scholar]
- 56.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urano, F., M. Calfon, T. Yoneda, C. Yun, M. Kiraly, S. G. Clark, and D. Ron. 2002. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J. Cell Biol. 158:639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, X. Z., B. Lawson, J. W. Brewer, H. Zinszner, A. Sanjay, L. J. Mi, R. Boorstein, G. Kreibich, L. M. Hendershot, and D. Ron. 1996. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153). Mol. Cell. Biol. 16:4273-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Werstuck, G. H., S. R. Lentz, S. Dayal, G. S. Hossain, S. K. Sood, Y. Y. Shi, J. Zhou, N. Maeda, S. K. Krisans, M. R. Malinow, and R. C. Austin. 2001. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J. Clin. Investig. 107:1263-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilt, S. G., N. V. Dugger, N. D. Hitt, and P. M. Hoffman. 2000. Evidence for oxidative damage in a murine leukemia virus-induced neurodegeneration. J. Neurosci. Res. 62:440-450. [DOI] [PubMed] [Google Scholar]
- 61.Zinszner, H., M. Kuroda, X. Wang, N. Batchvarova, R. T. Lightfoot, H. Remotti, J. L. Stevens, and D. Ron. 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12:982-995. [DOI] [PMC free article] [PubMed] [Google Scholar]