Abstract
The PI 3-phosphatase PTEN (phosphatase and tensin homologue deleted on chromosome 10), one of the most important tumor suppressors, must associate with the plasma membrane to maintain appropriate steady-state levels of phosphatidylinositol 3,4,5-triphosphate. Yet the mechanism of membrane binding has received little attention and the key determinants that regulate localization, a phosphatidylinositol 4,5-bisphosphate (PIP2) binding motif and a cluster of phosphorylated C-terminal residues, were not included in the crystal structure. We report that membrane binding requires PIP2 and show that phosphorylation regulates an intramolecular interaction. A truncated version of the enzyme, PTEN1–351, bound strongly to the membrane, an effect that was reversed by co-expression of the remainder of the molecule, PTEN352–403. The separate fragments associated in vitro, an interaction dependent on phosphorylation of the C-terminal cluster, a portion of the PIP2 binding motif, integrity of the phosphatase domain, and the CBR3 loop. Our investigation provides direct evidence for a model in which PTEN switches between open and closed states and phosphorylation favors the closed conformation, thereby regulating localization and function. Small molecules targeting these interactions could potentially serve as therapeutic agents in antagonizing Ras or PI3K-driven tumors. The study also stresses the importance of determining the structure of the native enzyme.
Keywords: iRAP, phosphatase, PI3K, PIP2, PIP3
PTEN (phosphatase and tensin homologue deleted on chromosome 10) is a lipid phosphatase that dephosphorylates phosphatidylinositol-3,4,5-triphosphate (PIP3) and opposes PI3K signaling (1). This important tumor suppressor is frequently mutated in endometrium, prostate, and brain cancer, and alterations of PTEN activity leads to Cowden and Bannayan-Zonana syndromes in which patients develop benign hamartomas (2–4). Mice homozygous for a deletion of PTEN die in early embryogenesis whereas heterozygotes have a propensity to develop widespread neoplasms (5). Prostate-directed conditional deletions of PTEN result in metastatic prostate cancer (6–8). Neural and astrocyte-specific deletions lead to greatly enlarged brain size and abnormalities in astrocytes and neurons (9). Cultured mammalian cells lacking PTEN proliferate faster, resist apoptosis, and migrate aberrantly (10–12). Dictyostelium discoideum cells lacking PTEN have elevated PIP3 and are severely defective in chemotaxis (13).
Regulation of PTEN activity, localization, and function are controlled by a variety of mechanisms. NOTCH1, by acting through transcription factors CBF-1 and MYC or HES-1, respectively, has been reported to either increase or decrease PTEN expression (14–16). Another level of regulation is post-translational modification. Acetylation by p300/CBP-associated factor blocks PTEN activity (17). PTEN catalytic activity is negatively regulated by reactive oxygen species under oxidative stress (18–20) by disulfide linkage of catalytic cysteine 124 with cysteine 71 (18). E3 ubiquitin ligase NEDD4–1 can mono- and poly-ubiquitinate PTEN (21). The mono-ubiquitinated protein displays increased translocation to the nucleus and the poly-ubiquitinated form undergoes rapid degradation. Finally, phosphorylation of the C-terminal residues regulate protein stability and function in cells (22). In addition, PTEN is phosphorylated by a series of kinases, including RhoA-associated kinase (23), casein kinase 2, and glycogen synthase 3b (24–26).
Although the main substrate of PTEN is at the plasma membrane, the enzyme is mainly found in cytosol and the nucleus, but a small fraction is dynamically associated with the inner face of the plasma membrane (27). This interaction is critical, as mutations that do not affect catalytic activity against soluble substrates but impair membrane binding, such as deletion of an N-terminal “PIP2 binding” motif, lead to a null phenotype in cells (27–29). Consistently, addition of PIP2 to PIP3-containing vesicles makes them more effective substrates (30). Furthermore, there are tumor-derived mutations that do not reduce catalytic activity in vitro but effectively inactivate PTEN by preventing membrane association (31). In addition to the PIP2 binding motif, a globular phosphatase domain, and a C2 domain that binds lipid vesicles, human PTEN has a 51-aa C terminus that contains a cluster of phosphorylation sites. This cluster, thought to be the target of casein kinase 2, is an important regulatory region, as a version of PTEN with alanine substitutions of these phosphorylation sites, designated PTENA4, displays greatly increased membrane association (32). The PIP2 binding motif and the entire C terminus were in the crystallized protein (33). Thus, the structures and conformations of the enzyme including the regions most critical for membrane association and function in cells are unknown.
Single-molecule imaging studies have shown that the increased steady-state levels of PTENA4 on the membrane results from an increased association rate whereas the lifetimes of PTEN and PTENA4 on the membrane were identical. This behavior is consistent with a model wherein PTEN exists in “closed” and “open” conformations, regulated by phosphorylation, and that the open conformation has a more favorable interaction with the membrane. Here we demonstrate that the C terminus of PTEN directly interacts with the remainder of the molecule in a phosphorylation-dependent manner, as well as plasma membrane binding and activity. Furthermore, we demonstrate that depletion of PIP2 from the plasma membrane causes a concomitant loss of PTEN binding sites.
Results
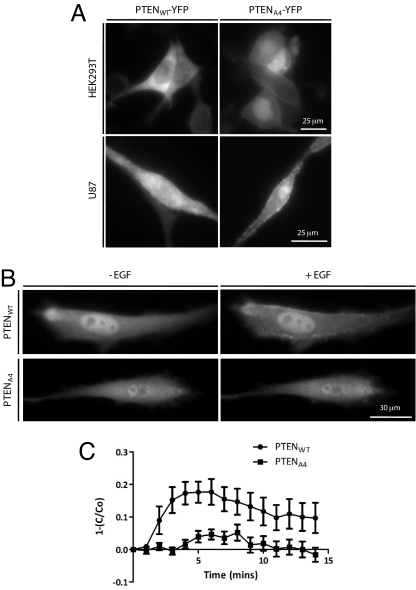
As previously shown, the localization of PTENWT-YFP and PTENA4-YFP are dramatically different. In HEK293T cells, membrane association of PTENWT-YFP was not discernible by epifluorescence, whereas PTENA4-YFP with alanine substitutions S380A, T382A, T383A, and S385A showed significant localization to the membrane (Fig. 1A). This effect did not require catalytic activity, as PTENC124S,A4-YFP, which lacks lipid and protein phosphatase activity, showed even greater association with the membrane (data not shown). Quantitative analysis of multiple cells expressing PTENWT-YFP or PTENA4-YFP indicated a membrane to cytosol ratio of 0.02 ± 0.03 (n = 7) versus 0.24 ± 0.07 (n = 8), respectively. Levels of PTENA4-YFP in the cytosol were lower and a significant pool was localized to the nucleus. A similar difference was also found when PTENWT-YFP and PTENA4-YFP were expressed in U87 cells. The membrane to cytosol ratios were 0.01 ± 0.02 (n = 7) and 0.22 ± 0.07 (n = 5) for PTENWT-YFP and PTENA4-YFP, respectively [supporting information (SI) Fig. S1].
Fig. 1.
PTEN cellular activity is proportional to the extent of membrane association. (A) HEK293T (Upper) and PTEN-null U87 cells (Lower) were transiently transfected with either PTENWT-YFP or PTENA4-YFP. Localization of the respective proteins was assessed. (B) U87 cells expressing PHAKT-GFP were co-transfected with either PTENWT or PTENA4. PHAKT-GFP localization was monitored after addition of 20 ng/mL EGF to serum starved cells. Examples are representative of 20 respective cells. Whereas two cells co-expressing PTENWT and PHAKT-GFP failed to respond to EGF stimulation, all PTENA4 co-transfected cells failed to respond. (C) Depletion of the cytosolic GFP signal was measured. Cytoplasmic GFP intensity was compared with the initial value (C/C0) and subtracted from 1. Error bars represent SEM.
Because the PTEN substrate PIP3 is localized at the membrane, we reasoned that, in a cellular context, PTENA4 would display greater activity than PTENWT. We examined the relative ability of PTENWT and PTENA4 to reduce PIP3 levels by monitoring EGF-induced translocation of the PIP3 probe PHAKT-GFP to the membrane in individual cells. U87 cells transfected with PHAKT-GFP alone showed high levels of membrane association (Fig. S2). Following co-transfection with PTENWT or PTENA4, these levels were no longer detectable (Fig. 1B Left). When cells co-expressing PTENWT were stimulated with 20 ng/mL EGF, PHAKT-GFP translocated to the plasma membrane coincident with cytosolic depletion of the probe. Membrane levels remained elevated for 15 min (Fig. 1 B and C and Movie S1). In contrast, stimulation of cells co-expressing PTENA4 elicited only a slight transient decrease in cytoplasmic levels of PHAKT-GFP, whereas no observable membrane translocation was observed (Movie S2). The extent of inhibition up to 15 min was 87%. Based on these inferred PIP3 levels, we estimated PTENA4 was at least sevenfold more active than PTENWT. Furthermore, as PTENA4 is typically expressed at slightly lower levels then PTENWT, the relative activity of PTENA4 is likely to be higher. PTENA4 was also effective at lowering steady-state levels of phosphorylated AKT1 (data not shown and ref. 35). These observations suggest that activity is dependent on extent of membrane binding and that phosphorylation of the C-terminal cluster has an inhibitory effect on membrane association and activity.
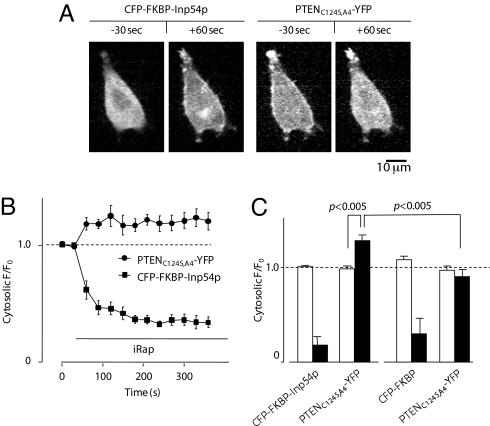
The relatively higher levels of PTENA4 binding to the membrane allowed us to ask whether PIP2 was required for the interaction. Although its PIP2 binding motif is required for membrane localization of PTEN and addition of PIP2 to PIP3-containing vesicles make them more effective substrates, a requirement of PIP2 for PTEN binding to the membrane in cells has not been demonstrated to our knowledge. To assess the need for PIP2, we expressed PTENC124S,A4-YFP in HeLa cells equipped with an engineered system previously shown to conditionally deplete PIP2 from the inner leaflet of the plasma membrane (34). The cellular distribution of PTENC124S,A4-YFP was continuously monitored whereas PIP2 was rapidly depleted by forced translocation of a PIP2-specific phosphatase, inositol polyphosphate-5-phosphatase (Inp54p), to the plasma membrane. As the probe was recruited to the membrane, PTENC124S,A4-YFP rapidly re-localized to the cytosol, indicating that membrane-associated binding sites for PTEN had disappeared (Fig. 2 A and B). PTENC124S,A4-YFP re-localization was not observed when a dimerization probe lacking a phosphatase domain was used (Fig. 2C). These studies strongly suggest that PIP2 is required for high-affinity binding of PTENC124S,A4-YFP to the membrane.
Fig. 2.
PTEN membrane binding requires PIP2. (A) HeLa cells were transiently co-transfected with CFP-FKBP-Inp54p and PTENC124S,A4-YFP. Localization of respective proteins was observed 60 seconds after iRap treatment. (B) Change in cytosolic fluorescence intensity of CFP-FKBP-Inp54p and PTENC124S,A4-YFP was analyzed upon iRap addition. (C) Effect of Inp54p on PTENC124S,A4-YFP localization was assessed by transient co-transfection of probe lacking PIP2 phosphatase domain (i.e., CFP-FKBP). Open and closed bars represent cytosolic levels (F/F0) before and after addition of iRap, respectively. Error bars represent SEM.
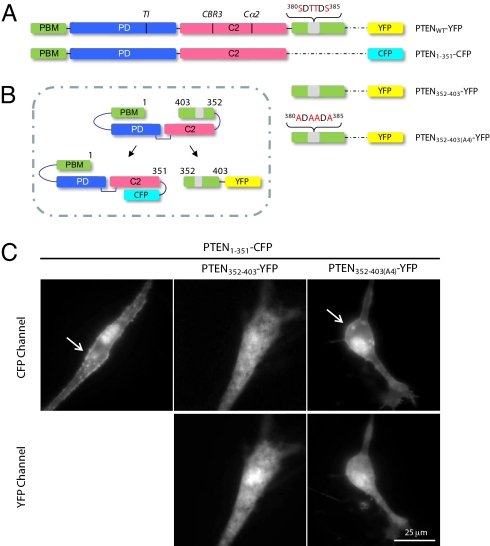
We hypothesized that the mechanism of inhibition depends on a direct association of the C-terminal region with the rest of the protein that prevents membrane binding. To test this hypothesis, we removed residues 352 through 403, the flexible region beyond the C2 domain, and fused the remainder of the protein to CFP (Fig. 3 A and B). Strikingly, when expressed in U87 cells, the association of PTEN1–351-CFP with the plasma membrane was greatly increased, with a distribution similar to that observed for PTENA4-YFP (compare Figs. 1A and 3C Upper). Similar results were obtained in HEK293T cells (data not shown). To further demonstrate that the C-terminal region inhibited the interaction of PTENWT with the membrane, we co-expressed PTEN352–403-YFP and found that it reversed the enhanced membrane binding of PTEN1–351-CFP (Fig. 3C and Fig. S7). Importantly, the C-terminal portion of PTENA4, PTEN352–403(A4)-YFP, did not reduce the increased PTEN1–351-CFP membrane binding, indicating that phosphorylation of S380, T382, T383, and/or S385 are critical for the presumed intramolecular interaction. Whether co-expressed with PTEN1–351-CFP or independently expressed, neither PTEN352–403(WT)-YFP nor PTEN352–403(A4)-YFP showed significant membrane association (Fig. 3C Lower). These observations suggest that a phosphorylation-dependent intramolecular interaction modulates PTEN membrane association and activity.
Fig. 3.
PTEN1–351 binds strongly to the plasma membrane and co-expression of the phosphorylated C-terminal domain, PTEN352–403, blocks this interaction. (A) Schematic of PTENWT-YFP, PTEN1–351-CFP, PTEN352–403-YFP, and PTEN352–403(A4)-YFP. (B) Depiction of the strategy used to assess the effects of PTEN352–403 on membrane binding of PTEN1–351. (C) U87MG cells were transiently transfected with PTEN1–351-CFP alone or co-transfected with PTEN352–403-YFP or PTEN352–403(A4)-YFP for 48 h. Images of cells expressing both constructs were taken using CFP and YFP filter sets, and localization of the respective constructs was assessed.
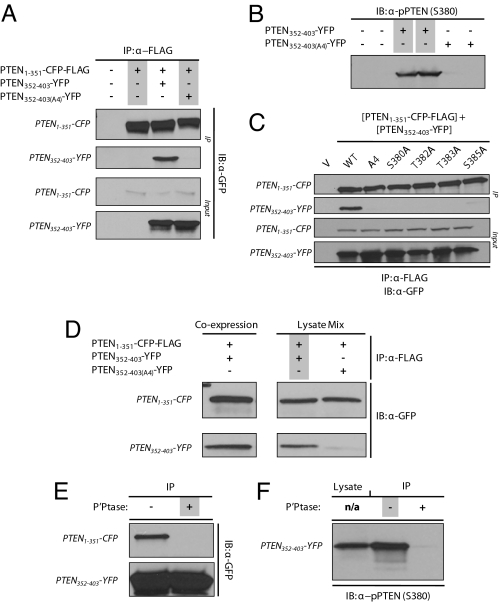
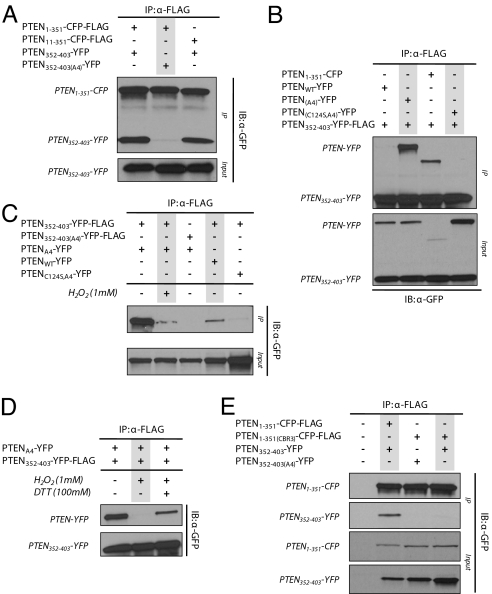
To directly test the phosphorylation-dependent interaction of the C- and N-terminal portions of the protein, we carried out a series of co-expression and co-immunoprecipitation experiments. We observed that PTEN1–351-CFP-FLAG was able to effectively co-immunoprecipitate PTEN352–403-YFP (Fig. 4A). However, it was not able to appreciably co-immunoprecipitate PTEN352–403(A4)-YFP. The C-terminal fragment was invariably expressed at much higher levels, but approximately stoichiometric amounts of the N- and C-terminal fragments were bound in the immunoprecipitates (ratio of C-terminal to N-terminal, 0.79 ± 0.14; n = 5). To verify that PTEN352–403-YFP was phosphorylated, we expressed it in HEK293T cells and performed immunoblotting with antibody specific for S380. As shown in Fig. 4B, PTEN352–403-YFP was phosphorylated, whereas, as predicted, PTEN352–403(A4)-YFP was not. These observations confirm a phosphorylation-dependent interaction between the N- and C-terminal regions of the protein. We then examined whether alanine substitutions of single phosphorylation sites would disrupt the interaction. PTEN1–351-CFP-FLAG was co-expressed with PTEN352–403-YFP containing S380A, T382A, T383A, or S385A substitution, and immunoprecipitated using anti-FLAG affinity beads. It was observed that substitution of any single phosphorylated residue in the cluster resulted in a loss of affinity for PTEN1–351-CFP (Fig. 4C), suggesting all of the sites are required for optimal interaction.
Fig. 4.
The phosphorylated C-terminal domain, PTEN352–403, interacts specifically with PTEN1–351. (A) HEK293T cells were transiently co-transfected with PTEN1–351-CFP-FLAG and either PTEN352–403-YFP or PTEN352–403(A4)-YFP. Cell lysates were immunoprecipitated and probed with anti-GFP antibody. (B) Lysates from HEK293T cells expressing PTEN352–403-YFP or PTEN352–403(A4)-YFP were analyzed by Western blot using anti-phosphorylated PTEN (i.e., S380) antibody. (C) Interaction of PTEN1–351-CFP-FLAG with single alanine substituted PTEN352–403-YFP (S380A, T382A, T383A, or S385A) was assessed. (D) Lysates from HEK293T cells expressing either PTEN1–351-CFP-FLAG or PTEN352–403-YFP were mixed in a 1:1 ratio. The mixture was subjected to previously described immunoprecipitation and Western blot analysis using anti-GFP antibody. The data were compared with co-expression experiments. (E) Cell lysate from HEK293T cells transfected with PTEN352–403-YFP-FLAG were divided and immunoprecipitated. The immunoprecipitates were treated with and without protein phosphatase, washed, and mixed with cell lysates from HEK293T cells expressing PTEN1–351-CFP. The mixture was immunoprecipitated and probed using anti-GFP antibody. (F) The cell lysate before first immunoprecipitation was analyzed by Western blot using anti-phospho-PTEN (i.e., S380) antibody and compared with immunoprecipitated fractions with and without phosphatase treatment.
We next tested whether the interaction of PTEN1–351 with PTEN352–403 required co-translation or whether the mature proteins could associate. We performed a series of lysate mixing experiments to test the interaction and to determine whether it is phosphorylation-dependent. PTEN1–351-CFP-FLAG, PTEN352–403-YFP, or PTEN352–403(A4)-YFP were separately expressed in HEK293T cells. The appropriate lysates were mixed and the samples were then immunoprecipitated using anti-FLAG affinity beads. Fig. 4D shows that PTEN1–351-CFP interacted selectively with PTEN352–403 but not with PTEN352–403(A4). Furthermore, the ratios of PTEN352–403YFP co-immunoprecipitated with PTEN1–351-CFP-FLAG were similar whether they were co-expressed or mixed after lysis. Thus, the interaction does not require co-translation and can occur at dilutions of more than 500 fold compared with the cellular volume, indicating a specific, high-affinity binding reaction.
This approach allowed us to directly show that the intramolecular interaction of PTEN1–351 and PTEN352–403 is phosphorylation-dependent. In a similar lysate mixing experiment, we separately expressed PTEN352–403-YFP-FLAG and PTEN1–351-CFP in HEK293T cells. The PTEN352–403-FLAG was immunoprecipitated from the lysate and treated with λ-protein phosphatase, while a parallel sample was similarly treated in the absence of protein phosphatase. The samples were washed, lysate containing PTEN1–351-CFP was added, the reaction was allowed to proceed, and the beads were collected again. As shown in Fig. 4E, the phosphatase treatment resulted in a nearly complete loss of the interaction. Consistently, Western blot analysis of PTEN352–403-YFP-FLAG treated with λ-phosphatase verified the elimination of S380 phosphorylation (Fig. 4F). These observations confirm the findings from the alanine substitutions and definitively show that the interaction can be regulated by reversible phosphorylation.
To show that there is a phosphorylation-dependent intramolecular interaction within the native PTEN molecule, we designed a series of experiments illustrated in Fig. S3A. We envision that phosphorylation of the C-terminal region promotes a “closed” conformation that would be expected to prevent interaction of an exogenous phosphorylated C-terminal fragment. Conversely, PTENA4 is expected to be in an “open” conformation in which the endogenous C-terminal portion would not interfere with its exogenous counterpart (Fig. S3A). These possibilities were assessed by comparing the interaction of PTEN352–403-YFP-FLAG with either PTENWT-YFP or PTENA4-YFP. As shown in Fig. S3B, PTENA4-YFP showed significantly greater association with PTEN352–403-YFP-FLAG than did PTENWT-YFP. Also, PTENA4-YFP association with PTEN352–403-YFP was comparable to that of PTEN1–351-CFP. All of these observations are consistent with a phosphorylation-dependent intramolecular interaction with the native PTEN molecule.
We next investigated the determinants involved in the binding of PTEN352–403 to PTEN1–351. As shown earlier, PTENC124S,A4 binding to the membrane requires PIP2, and previous studies have indicated that binding is mediated by the N-terminal PIP2 binding motif. We set out to determine whether the C-terminal region interacted with this critical amino-terminal region. We removed the first 10 N-terminal amino acids from PTEN1–351, which are required for membrane binding, and assessed the ability of PTEN11–351-CFP-FLAG to effectively immunoprecipitate PTEN352–403-YFP, as previously described. As shown in Fig. 5A, the N-terminal truncation had no effect on the ability of PTEN11–351-CFP-FLAG to interact with PTEN352–403-YFP. However, in separate experiments, deletion of the first 15 aa, the triple mutation K13R, R14A, and R15A, or the single substitution K13A, prevented the interaction of FLAG-PTEN1–353 with PTEN354–403-GFP or FLAG-PTENA4 with PTEN354–403-GFP (Fig. S4 A and B). Thus, the entire N-terminal PIP2-binding motif is necessary for membrane association, whereas only the most C-terminal-charged residues are needed for the intramolecular interaction.
Fig. 5.
Evaluation of binding determinants of PTEN1–351 and PTEN352–403. (A) Interaction of the C-terminal domain of PTEN with its N-terminal tail was assessed by co-expression and co-immunoprecipitation of PTEN11–351-CFP-FLAG with PTEN352–403-YFP. (B) The interaction of catalytically inactive PTEN with its C-terminal domain was assessed by co-expression of PTENC124S,A4-YFP with PTEN352–403-YFP-FLAG. Lysates were immunoprecipitated and analyzed by Western blot. PTENC124S,A4-YFP association with PTEN352–403-YFP-FLAG was compared with PTEN352–403-YFP-FLAG affinity for PTENWT-YFP, PTENA4-YFP, and PTEN1–351-CFP. (C) HEK293T cells were co-transfected with the indicated constructs. Cells expressing PTEN352–403-YFP-FLAG and PTENA4-YFP were pretreated with 1 mM H2O2 for 5 min and immunoprecipitated. Relative expression of full-length PTEN variants were also analyzed by Western blot using anti-GFP antibody. (D) HEK293T cells co-expressing PTENA4-YFP and PTEN352–403-YFP-FLAG were treated with 1 mM H2O2 for 5 min before lysis. Equal volumes of the lysate were subjected to immunoprecipitation with anti-FLAG affinity beads with and without the presence of 100 mM DTT. (E) To evaluate the possible interaction of PTEN352–403-YFP with the CBR3 loop of the C2 domain, lysines K260, K263, K266, K267, and K269 were alanine substituted to generate PTEN1–351(CBR3)-CFP-FLAG, then co-expressed with PTEN352–403-YFP in HEK293T cells and analyzed by Western blot using anti-GFP antibody. Results were compared with PTEN1–351-CFP-FLAG co-expression with PTEN352–403-YFP and PTEN1–351(CBR3)-CFP-FLAG co-expression with PTEN352–403(A4)-YFP.
We then explored whether the phosphorylated cluster might interact with the rest of the protein by docking near the catalytic pocket of the enzyme. Initial evidence of this possibility arose serendipitously when we used the catalytically dead version of PTEN, PTENC124S,A4. We observed that PTENC124S,A4-YFP did not interact with PTEN352–403-YFP-FLAG (Fig. 5B). To further explore this phenomenon, we exploited the observations of Lee et al. (18, 20). In studies investigating the effects of radical oxygen species on PTEN activity, they discovered that H2O2 can reversibly inhibit PTEN activity by covalently linking Cys-71 to Cys-124. Cells co-expressing PTEN352–403-CFP-FLAG and PTENA4-YFP were treated with 1 mM H2O2. We found that, following H2O2 pre-treatment, PTENA4 displayed notably lower binding to the exogenous C-terminal region (Fig. 5C). This inhibition could be partially reversed by inclusion of 100 mM DTT in the immunoprecipitation buffer (Fig. 5D). Furthermore, in separate experiments, mutation of charged residues at the start of the TI loop, R161A, K163A, and K164A prevented the interaction of FLAG-PTEN1–353 with PTEN354–403-GFP (Fig. S4A). These observations indicate that the phosphatase domain, particularly access to the catalytic pocket, is required for the interaction with the C-terminal region.
We next examined the possible role of the CBR3 loop and the Cα2 region and its adjacent loop of the C2 domain in binding of PTEN1–351 to PTEN352–403. We reasoned that, as the CBR3 is required for PTEN membrane association, the C-terminal region might regulate CBR3 membrane accessibility. We disrupted the CBR3 loop by alanine substitution of lysine residues K260, K263, K266, K267, and K269. PTEN1–351(CBR3)-CFP-FLAG was co-expressed with PTEN352–403-YFP and immunoprecipitated with anti-FLAG affinity beads. Following Western blot analysis, we observed that these mutations of the CBR3 loop completely disrupted the association of the C-terminal fragment with the rest of the protein (Fig. 5E). In a similar experiment, substitutions of CBR3 loop residues K263A, M264A, L265G, K266A, K267A, and K269A also disrupted the association. Conversely, alanine substitution of Cα2 and its adjacent loop had no effect (Fig. S4A). The observations suggest that the interaction of the C-terminal region with the remainder of the protein is in part dependent on intramolecular interactions with its C2 domain.
Discussion
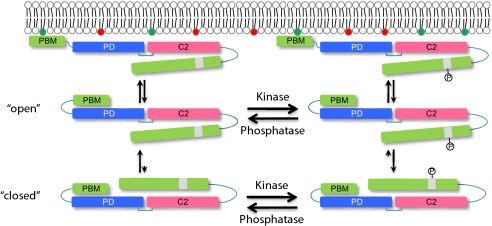
The interaction of PTEN with the membrane requires a short N-terminal PIP2 binding motif and is negatively regulated by phosphorylation of a cluster of residues in the C-terminal region of the protein. In this report we showed that indeed PIP2 is part of the binding site and that PTEN activity in cells is correlated with the level of membrane association. Furthermore, we have found that the negative regulation is caused by an intramolecular interaction between the phosphorylated C-terminal region and the phosphatase and the C2 domains. Our observations are consistent with a model in which PTEN switches between open and closed states and phosphorylation of the C-terminal cluster favors the closed conformation (Fig. 6). First, we demonstrated that removal of the C-terminal region enhances PTEN membrane localization in a variety of cell types, and that co-expression of a phosphorylatable C-terminal fragment blocks membrane binding. Second, we confirmed the suggested intramolecular interaction between the N- and C-terminal regions by showing that two separate fragments, PTEN1–351 and PTEN352–403, respectively, could be co-immunoprecipitated whether they are co-expressed or expressed separately and mixed. Eliminating phosphorylation by alanine substitution or phosphatase treatment of the C-terminal fragment prevented the association. Furthermore, perturbations of the catalytic pocket or mutation of the CBR3 loop in the C2 domain prevented the interaction of the two fragments. Removal of the N-terminal PIP2 binding motif did not. Third, substitution of the phosphorylated cluster, in context of the full-length protein, greatly increased access for exogenous phosphorylated C-terminal fragment.
Fig. 6.
Phosphorylation-dependent switch between open and closed conformations regulates rate of association of PTEN with the membrane. PTEN molecules can exist in open or closed conformations, and open molecules bind to the membrane. Membrane shows scattered molecules of PIP2 (green), which are required for binding, and PIP3 (red), which are PTEN substrates. Phosphorylation of the C-terminal cluster tilts the equilibrium toward the closed conformation, whereas most dephosphorylated molecules are in the open conformation. Alanine substitution of the C-terminal cluster or removal of the C-terminal region locks all of the molecules in an open conformation, promoting membrane association.
We propose that there is a steady-state distribution between open and closed conformations of PTEN (Fig. 6). Phosphorylated molecules are found predominantly in the closed conformation, whereas un-phosphorylated molecules are found predominantly in the open conformation. Open molecules encountering the membrane bind transiently, whereas the closed molecules fail to associate. Therefore, only a small fraction of phosphorylated WT PTEN is associated with membrane at steady state. Substitution of the phosphorylated cluster would be expected to shift the distribution toward the open conformation and thereby increase the rate of association with the membrane. Consistently, previous studies have shown that non-phosphorylatable PTEN has a more rapid rate of association with the membrane but its lifetime on the membrane is identical to WT (27). Similarly, removal of the C-terminal region would be expected to render PTEN in the open conformation. As we have shown here, this manipulation greatly increases association with the membrane and co-expression of the phosphorylated C-terminal region closes the molecule and reverses this effect. We think it is unlikely that the phosphorylation/dephosphorylation events are directly linked to each cycle of binding. Both PTENWT and PTENA4 dissociate at identical rates, yet clearly PTENA4 cannot receive a phosphate as it dissociates. We assume that a similar situation holds for PTEN1–351, although we have not directly measured its lifetime. Nevertheless, future work is needed to determine the rates of phosphate turnover and compare them with binding lifetime.
Our studies extend previous work showing that the conserved PIP2 binding motif on the N terminal of PTEN is required for membrane association and that, in mixed micelles, inclusion of PIP2 enhances phosphatase activity against PIP3 (30). The latter effect was abolished by the tumor-derived mutation K13E within the PIP2 binding motif. We show here that rapid depletion of PIP2 from the plasma membrane triggered by forced recruitment of Inp54p causes PTENC124S,A4 to immediately re-localize to the cytosol. This shows that PIP2 comprises one element of the binding site for PTEN. Taken together, these observations suggest an association of the PIP2 binding motif with PIP2. Interestingly, only a portion of the PIP2 binding motif was required for the phosphorylation-dependent intramolecular interaction between the C-terminal region and the remainder of the protein.
The transition between open and closed states favored by phosphorylation depends on direct association of C-terminal region with the phosphatase and C2 domains. Within the phosphatase domain, substitution of the catalytic cysteine by serine (i.e., C124S) completely prevented the interaction. Cross-linking of the C124 to C71 by oxidation with H2O2 also blocked the intramolecular interaction, and this effect was reversed by reduction with DTT. Based on our model (Fig. 6), these perturbations would be expected to open the molecule and increase membrane binding. Indeed, both human and Dictyostelium versions of PTENC124S show greater membrane association than their WT counterparts. In the human version, this effect appears to be partially additive, as the combined mutation PTENC124S,A4 displays higher membrane association compared with PTENC124S or PTENA4. Further studies are needed to determine if the perturbations of the catalytic pocket inhibit C-terminal interaction by conformational changes within the phosphatase domain or loss of protein phosphatase activity. In addition to the phosphatase domain, the integrity of the C2 domain is required as mutations of the CBR3 loop also prevented the intramolecular interaction. Our studies open the way for determination of the exact docking regions by biophysical methods.
During the course of our studies, Odriozola et al. presented evidence for an intramolecular interaction within the PTEN molecule (36). They show that a fragment of PTEN consisting of a portion of the C2 domain and the C-terminal region, PTEN335–403, interacts with a partially overlapping fragment consisting of the entire C2 domain and the C-terminal region, PTEN185–403. N-terminal truncations into the C2 domain of PTEN185–403 did not interfere with this interaction until removal of portions beyond the CBR3 loop. From this, the authors concluded that the CBR3 loop is critical for the interaction, yet, when they mutated all charged residues in the CBR3 loop, leaving only membrane anchoring residues M264 and L265 (33, 37), the interaction remained intact. They also show that the C-terminal domain, PTEN335–403, does not interact with the phosphatase domain, PTEN1–185. Paradoxically, phospho-mimetic peptides spanning the C-terminal cluster at high concentrations inhibit PTEN phosphatase activity, suggesting a weak interaction between the C-terminal domain and the catalytic pocket of the phosphatase domain. From these data, they derive a model whereby the C-terminal region mediates an auto-inhibitory interaction that inhibits membrane binding and catalytic activity.
Despite the similarity of our models, our data disagree in a number of ways. First, all of their immunoprecipitation studies use a portion of PTEN lacking the phosphatase domain (36). In contrast, we find that subtle perturbations of the phosphatase domain, such as the PTENC124S mutation, reversible H2O2 treatment, or mutation of the TI loop, greatly reduced interactions with the C terminus. Second, in their experiments, mutations of the charged residues in the CBR3 loop did not interfere with PTEN185–403 interaction with PTEN335–403, whereas in our experiments, similar mutations disrupted the interaction of PTEN1–351 with PTEN352–403. Third, in our study, phosphorylated closed PTEN bound significantly less C-terminal domain, presumably because of intramolecular competition. Yet in all of their work, overlapping regions of the C2 domain were present in both interacting fragments (36). These details and discrepancies, which may result in part from major differences in assays used, will be important in future strategies to manipulate PTEN localization and function.
Our findings have important implications for the function of PTEN. High levels of PIP3 promote uncontrolled cell growth and migration, and must be restricted by proper regulation. Because PIP3 is on the membrane, the localization of PTEN is expected to effectively control catalytic activity in cells. We show that phosphorylation of the C terminus controls an intramolecular interaction that determines the fraction of PTEN associated with the membrane and thereby the set point for cellular PIP3. In fact, there is some evidence that phosphorylation of PTEN can be regulated (35). It is likely that the set point is regulated in different tissues by controlling the level of phosphorylation of the cluster. Furthermore, the conformational switch in PTEN provides an attractive target. For instance, with further knowledge of the molecular interaction, peptides or small molecules can be designed to inhibit the intramolecular interaction of the C-terminal tail and increase membrane association and activity. Alternatively, opening of the molecule could be achieved by inhibiting C-terminal phosphorylation. Finally, open versions of PTEN could be used for gene therapy.
Materials and Methods
Detailed description of the materials and methods are provided in SI Text. Plasmids were prepared by standard PCR and cloning procedures. HEK293 and U87 cells were cultured using ATCC media and procedures. Fluorescence microscopy was carried out using an Olympus IX71 inverted microscope with appropriate CFP/YFP filter sets. For immunoprecipitation and Western blotting, HEK293T cells expressing described constructs were lysed in buffer containing 10 mM Tris-HCl (pH 7.5), 140 mM NaCl, 5 mM EDTA, and 0.1% Nonidet P-40 and supplemented with 1 mM DTT, as well as phosphatase and protease inhibitor mixture.
Inducible PIP2 depletion using CFP-FKBP-Inp54p, and dual color (CFP/YFP) confocal imaging of cells have been described previously (34). Briefly, HeLa cells were transfected with dimerization probes, CFP-FKBP-Inp54p, and Lyn11-FRB together with YFP-labeled PTENC124S,A4. Cells were treated with iRap, which triggered a rapid translocation of CFP-FKBP-Inp54p to the plasma membrane. Fluorescent signals from CFP and YFP in HeLa cells were alternately collected every 30 seconds on a spinning-disk confocal microscope.
Supplementary Material
Acknowledgments.
The authors wish to acknowledge members of the Devreotes lab, particularly Yoichiro Kamimura, Jonathan Franca-Koh, and Stacey Willard, for helpful discussion and advice. We also wish to thank Moira McMahon for critical reading of this manuscript and helpful advice. This work was supported by National Institutes of Health Grants GM 28007, GM 34933, and GM 71920.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811212106/DCSupplemental.
References
- 1.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 2.Waite KA, Eng C. Protean PTEN: form and function. Am J Hum Genet. 2002;70:829–844. doi: 10.1086/340026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Podsypanina K, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki A, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 6.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 7.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 8.Trotman LC, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser MM, et al. Pten loss causes hypertrophy and increased proliferation of astrocytes in vivo. Cancer Res. 2004;64:7773–7779. doi: 10.1158/0008-5472.CAN-04-2487. [DOI] [PubMed] [Google Scholar]
- 10.Ramaswamy S, et al. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA. 1999;96:2110–2115. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun H, et al. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc Natl Acad Sci USA. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liliental J, et al. Genetic deletion of the pten tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr Biol. 2000;10:401–404. doi: 10.1016/s0960-9822(00)00417-6. [DOI] [PubMed] [Google Scholar]
- 13.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 14.Chappell WH, et al. Increased protein expression of the PTEN tumor suppressor in the presence of constitutively active notch-1. Cell Cycle. 2005;4:1389–1395. doi: 10.4161/cc.4.10.2028. [DOI] [PubMed] [Google Scholar]
- 15.Whelan JT, Forbes SL, Bertrand FE. CBF-1 (RBP-J kappa) binds to the PTEN promoter and regulates PTEN gene expression. Cell Cycle. 2007;6:80–84. doi: 10.4161/cc.6.1.3648. [DOI] [PubMed] [Google Scholar]
- 16.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okumura K, et al. PCAF modulates PTEN activity. J Biol Chem. 2006;281:26562–26568. doi: 10.1074/jbc.M605391200. [DOI] [PubMed] [Google Scholar]
- 18.Lee SR, et al. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 19.Kwon J, et al. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci USA. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leslie NR, et al. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 2003;22:5501–5510. doi: 10.1093/emboj/cdg513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, et al. NEDD4–1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, et al. Regulation of PTEN by rho small GTPases. Nat Cell Biol. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 24.Al-Khouri AM, Ma Y, Togo SH, Williams S, Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. J Biol Chem. 2005;280:35195–35202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- 25.Miller SJ, Lou DY, Seldin DC, Lane WS, Neel BG. Direct identification of PTEN phosphorylation sites. FEBS Lett. 2002;528:145–153. doi: 10.1016/s0014-5793(02)03274-x. [DOI] [PubMed] [Google Scholar]
- 26.Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. implications for PTEN stability to proteasome-mediated degradation. J Biol Chem. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 27.Vazquez F, et al. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc Natl Acad Sci USA. 2006;103:3633–3638. doi: 10.1073/pnas.0510570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem. 2004;279:16606–16613. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- 29.Denning G, Jean-Joseph B, Prince C, Durden DL, Vogt PK. A short N-terminal sequence of PTEN controls cytoplasmic localization and is required for suppression of cell growth. Oncogene. 2007;26:3930–3940. doi: 10.1038/sj.onc.1210175. [DOI] [PubMed] [Google Scholar]
- 30.Campbell RB, Liu F, Ross AH. Allosteric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2003;278:33617–33620. doi: 10.1074/jbc.C300296200. [DOI] [PubMed] [Google Scholar]
- 31.Walker SM, Leslie NR, Perera NM, Batty IH, Downes CP. The tumour-suppressor function of PTEN requires an N-terminal lipid-binding motif. Biochem J. 2004;379:301–307. doi: 10.1042/BJ20031839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vazquez F, et al. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J Biol Chem. 2001;276:48627–48630. doi: 10.1074/jbc.C100556200. [DOI] [PubMed] [Google Scholar]
- 33.Lee JO, et al. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 34.Suh BC, Inoue T, Meyer T, Hille B. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ning K, et al. A novel leptin signalling pathway via PTEN inhibition in hypothalamic cell lines and pancreatic beta-cells. EMBO J. 2006;25:2377–2387. doi: 10.1038/sj.emboj.7601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odriozola L, Singh G, Hoang T, Chan AM. Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J Biol Chem. 2007;282:23306–23315. doi: 10.1074/jbc.M611240200. [DOI] [PubMed] [Google Scholar]
- 37.Georgescu MM, et al. Stabilization and productive positioning roles of the C2 domain of PTEN tumor suppressor. Cancer Res. 2000;60:7033–7038. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.