Abstract
Human immunodeficiency virus type 1 (HIV-1) exonic splicing silencers (ESSs) inhibit production of certain spliced viral RNAs by repressing alternative splicing of the viral precursor RNA. Several HIV-1 ESSs interfere with spliceosome assembly by binding cellular hnRNP A/B proteins. Here, we have further characterized the mechanism of splicing repression using a representative HIV-1 hnRNP A/B-dependent ESS, ESSV, which regulates splicing at the vpr 3′ splice site. We show that hnRNP A/B proteins bound to ESSV are necessary to inhibit E complex assembly by competing with the binding of U2AF65 to the polypyrimidine tracts of repressed 3′ splice sites. We further show evidence suggesting that U1 snRNP binds the 5′ splice site despite an almost complete block of splicing by ESSV. Possible splicing-independent functions of U1 snRNP-5′ splice site interactions during virus replication are discussed.
The single human immunodeficiency virus type 1 (HIV-1) precursor RNA species transcribed from proviral DNA integrated in the host cell genome contains all the translated viral open reading frames (20). Production of a viral mRNA pool that expresses all open reading frames requires inefficient splicing of the precursor RNA (37). Unspliced precursor viral RNA serves as mRNA expressing the virion structural components Gag and Pol and as genomic RNA, which is packaged in progeny virions (11, 14). Spliced mRNAs express the viral regulatory and accessory genes, as well as the virion structural component Env, and are produced by alternative splicing of the viral precursor RNA in reactions catalyzed by the cellular splicing machinery (4, 40, 41). The spliced mRNA species in infected cells are, from highest to lowest concentration, the viral genes are env/vpu, nef, rev, tat, vpr, and vif (37). This pattern of steady-state mRNA production is essentially conserved among all HIV-1 isolates, suggesting that regulation of alternative splicing of the viral precursor RNA is essential for efficient viral replication (37).
Each spliced mRNA species is defined by an alternative 3′ splice site utilized during the splicing reactions. For example, production of vpr and tat mRNAs requires splicing to the viral 3′ splice sites A2 and A3, respectively (20, 22, 25). Therefore, selection of 3′ splice sites by the cellular splicing machinery is one of the defining steps in the production of HIV-1 regulatory, accessory, and Env proteins. The mechanism of 3′ splice site selection by the major spliceosome can be deduced from the sequence characteristics of the polypyrimidine tract (PPT) (38). Short PPTs, less than 14 nucleotides (nt), inefficiently bind the 65-kDa subunit of the essential heterodimeric splicing factor U2AF (U2AF65) (39). The binding of the 35-kDa subunit of U2AF (U2AF35) to the region surrounding the 3′ splice junction stabilizes the binding of U2AF65 to these weak PPTs, resulting in an increase in the splicing efficiency of the 3′ splice site (26, 35, 49, 52). Therefore, stable binding of U2AF65 to a PPT is a critical event in selection of a 3′ splice site by the splicing machinery.
The observed splicing efficiencies of HIV-1 3′ splice sites do not strictly correlate with predicted PPT length or PPT content (36, 37). This lack of correlation results from the presence of cis-acting splicing regulatory elements that lie outside the 3′ splice sites (2, 3, 8, 28, 42, 43, 45-47). Efficient use of 3′ splice sites A2, A3, and A7 is repressed by the activity of viral exonic splicing silencers (ESSs) ESSV, ESS2, and ESS3, respectively (Fig. 1A) (2, 3, 8, 45). ESSV, ESS2, and ESS3 share several characteristics. All members of the hnRNP A/B protein family bind to each of these ESS sequences, and reduced binding affinity correlates with functionally inactive mutant ESS sequences (8, 12, 51). Depletion of hnRNP A/B proteins from HeLa cell nuclear extracts (HNEs) by ESSV RNA alleviates splicing inhibition mediated by ESS2 and ESS3 (6). Finally, the HIV-1 hnRNP A/B-dependent ESSs are functionally redundant because replacement of ESS2 with ESS3 or ESSV results in repression of splicing at the viral 3′ splice site A3 (8, 43). Because of these common binding and functional requirements, the three HIV-1 ESSs can be classified as HIV-1 hnRNP A/B-dependent ESSs.
FIG. 1.
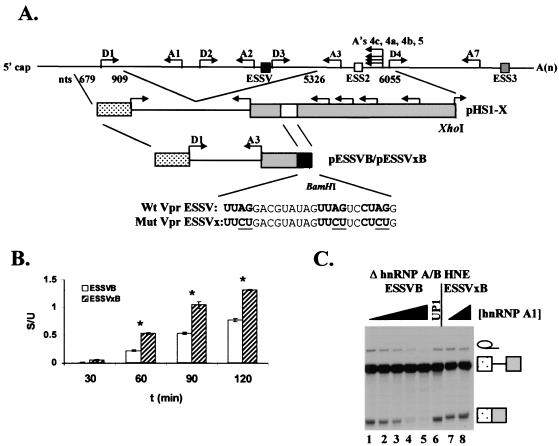
Effect of hnRNP A1 on ESSV activity in substrates ESSVB and ESSVxB. (A) Schematic diagram of the in vitro splicing constructs pESSVB and pESSVxB. A minigene containing major 5′ splice site D1 and 3′ splice site A3 was constructed by insertion of a BamHI site immediately downstream of ESSV or ESSVx in pHS1-ESSV or pHS1-ESSVx, respectively. Endogenous locations of ESSV, ESS2, and ESS3 are shown as at the top. Sequences of the wild-type (Wt) ESSV and mutant (Mut) ESSVx are shown below the schematic diagrams. (B) Kinetics of splicing of ESSVB and ESSVxB in hnRNP A/B-depleted HNE. Radiolabeled, capped substrates were transcribed from BamHI-linearized plasmids. Substrates were spliced in hnRNP A/B-depleted HNE. RNAs were electrophoresed on denaturing polyacrylamide gels, band intensity was quantified, and amounts (femtomoles) of unspliced and spliced RNAs were calculated. The ratio of spliced (S) to unspliced (U) RNA is shown. Asterisks, samples with calculated (Student's t test) P values <0.05 compared to results at the 30-min time point. (C) Purified hnRNP A1 or its N-terminal fragment UP1 (0, 0.3, 0.5, 0.7. 0.9, 0.9, and 0, 0.9 μM in lanes 1 to 8, respectively) was added back to hnRNP A/B-depleted extract. Capped, radiolabeled ESSVB or ESSVxB was spliced, and RNAs were electrophoresed on denaturing polyacrylamide gels. The positions of the unspliced RNA, spliced RNA, and intron products are shown on the right.
Previously, we have shown that ESS2 and ESS3 repress splicing by interfering with early events in the spliceosome assembly pathway (3, 43). In this report, we have further investigated the mechanism by which the HIV-1 hnRNP A/B-dependent ESS element ESSV represses splicing. Our results indicate that cellular hnRNP A/B proteins bound to ESSV inhibit the binding of U2AF65 to the PPT of the repressed 3′ splice site, resulting in inhibition of 3′ splice site splicing efficiency. These data suggest that the inhibition of splicing occurs prior to formation of an E complex.
MATERIALS AND METHODS
Plasmids.
Plasmids pHS1-X, pHS1-ESSV, and pHS1-ESSVx were previously described (2, 8). Plasmids pESSVB and pESSVxB were generated by introducing BamHI sites with the QuikChange mutagenesis kit (Stratagene) downstream of ESSV in pHS1-ESSV with the mutagenic primers WVPR5857s (5′CGTATAGTTAGTCCTAGGGGATCCATCCAGGAAGTCAGCC3′) and WVPR5857a (5′GGCTGACTTCCTGGATGGATCCCCTAGGACTAACTATACG3′) and downstream of ESSVx in pHS1-ESSVx with the mutagenic primers MVPR5857s (5′CGTATAGTTCTTCCTCTGGGATCCATCCAGGAAGTCAGCC3′) and MVPR5857a (5′GGCTGACTTCCTGGATGGATCCCAGAGGAAGAACTATACG3′). Plasmids pESSVB3′ and pESSVxB3′ were generated by introducing an XhoI site 40 bp upstream of 3′ splice site A3 in pESSVB and pESSVxB, respectively, with the QuikChange mutagenesis kit (Stratagene) with the mutagenic primers Xho sense (5′GGGCAAGCAGGGAGCTCGAGATATCGAATTCTGC3′) and Xho antisense (5′GCAGAATTCGATATCTCGAGCTCCCTGCTTGCCC3′). The fragment from the XhoI site in the polylinker of the Bluescript SK(+) (Stratagene) backbone to the engineered XhoI site in the HIV-1 insert was then excised. Plasmid pESSVxBΔ3′ was generated by excising the fragment from the XhoI to HincII sites in the HIV-1 insert in plasmid pESSVxB3′. pAd81 was supplied by R. Reed (Harvard Medical School). Plasmids pESSVA2-3′ and pESSVxA2-3′ were constructed by first generating PCR products spanning nt 5332 to 5661 from pHS3-ESSV and pHS3-ESSVx with sense primer 5′GCGCGAGCTCGTAGAGACCCTGACCTAG3′ and antisense primer 5′CGCGTCTAGATGTTATGTCCTGCTTG3′ (8). The PCR primers were engineered to incorporate a SacI site in the sense primer and an XbaI site in the antisense primer. The PCR product was then cleaved with SacI and XbaI and inserted into Bluescript SK(+), which was cleaved with SacI and XbaI.
RNA substrate synthesis.
In vitro transcription template plasmids pHS1-ESSV and pHS1-ESSVx were linearized with XhoI. All other in vitro transcription template plasmids were linearized with BamHI, and the linearized templates were gel purified. Capped and radiolabeled HS1-ESSV, HS1-ESSVx, ESSVB, ESSVxB, and Ad81 pre-mRNA splicing substrates were synthesized by runoff transcription using either T3 or T7 RNA polymerase (RNAP) in the presence of [α-32P]UTP (specific activity: 16 mCi/mmol). Uncapped, radiolabeled RNAs used for UV cross-linking experiments were synthesized by runoff transcription using T7 RNAP (for ESSVB3′, ESSVxB3′, and ESSVxBΔ3′ RNAs) or T3 RNAP (for ESSVA2-3′ and ESSVxA2-3′ RNAs) in the presence of [α-32P]UTP (specific activity: 80 mCi/mmol). RNAs were extracted from the synthesis reactions with phenol-chloroform and purified on Sephadex G-50 (Sigma) spin columns.
In vitro splicing reactions.
In vitro splicing reactions were carried out as previously described (31). Briefly, 6.0 fmol of radiolabeled pre-mRNA was incubated at 30°C in 60% (vol/vol) HNE in Dignam's buffer D, 20 mM creatine phosphate, 3.0 mM MgCl2, 0.5 mM ATP, and 2.6% (wt/vol) polyvinyl alcohol in a final volume of 25 μl (19). Reactions were carried out for times up to 2.5 h. RNAs were extracted from the reaction mixture with phenol-chloroform, precipitated, and electrophoresed on denaturing polyacrylamide gels.
Western blots.
Proteins were electrophoretically transferred at 15 V for 1 h from sodium dodecyl sulfate (SDS)-10% polyacrylamide gels onto Hybond ECL nitrocellulose membranes (Amersham) in 50 mM Tris-glycine-20% (vol/vol) methanol transfer buffer. Membranes were then treated with 10% (wt/vol) nonfat dry milk-Tris buffered saline (TBS)-0.1% (vol/vol) Tween 20 for 1 h at 37°C and probed with either anti-U2AF65 mouse immunoglobulin G (IgG) monoclonal antibody MC3 (supplied by J. Valcarcel, European Molecular Biology Laboratory) or an anti-U2AF35 rabbit IgG polyclonal antibody (supplied by R. Reed, Harvard Medical School) at dilutions of 1:100 and 1:1,000, respectively, in TBS-0.1% (vol/vol) Tween 20 for 1 h at 37°C. Washed membranes were then probed for 1 h at 37°C with an anti-mouse IgG-conjugated horseradish peroxidase secondary antibody (Amersham) or an anti-rabbit IgG-conjugated alkaline phosphatase secondary antibody (Vector Labs) at dilutions of 1:100,000 and 1:5,000, respectively, in TBS-0.1% (vol/vol) Tween 20. Blots were developed with the ECL Plus detection system (Amersham) or the alkaline phosphatase detection system (Vector Labs).
U2AF depletion.
Depletion of U2AF from HNE was performed as previously described, except that 2.5 ml of HNE in Dignam's buffer D was applied to a 5.0-ml poly(U) Sepharose column (Amersham Pharmacia) and eluted at a flow rate of ∼0.4 ml/min (19, 50). One-milliliter fractions were collected, and peak protein fractions were determined by the Bradford protein concentration assay (Pierce). Peak fractions were pooled, dialyzed against buffer D, aliquoted, and stored at −70°C.
Protein purification.
Recombinant hnRNP A/B proteins were purified as previously described (33, 34). Baculovirus expressing a histidine-tagged U2AF heterodimer (HisU2AF) was supplied by B. Graveley (University of Connecticut Health Center). For purification of HisU2AF, a shaker culture of Sf9 cells (150 ml at 2.0 × 106 cells/ml) was infected with 9 ml of freshly amplified virus (multiplicity of infection = ∼3.0). Cells were pelleted at 2 days postinfection and washed with 10 ml of cold 1× phosphate-buffered saline. The pellet was resuspended in 20 ml of lysis buffer (50 mM Tris [pH 8.0], 1.0% NP-40, 1.0 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml) and sonicated (setting 4; VirSonic 475) for 20 s on ice. After the debris was pelleted, the supernatant was incubated for 2 h at 4°C with 1 ml of Ni-nitrilotriacetic acid beads preequilibrated in lysis buffer. Beads were washed with twice with 10 ml of 50 mM Tris (pH 8.0)-500 mM NaCl and then five times with 10 ml of 50 mM Tris (pH 8.0)-500 mM NaCl-5.0 mM imidazole. Proteins were eluted with 2 ml of 50 mM Tris (pH 8.0)-500 mM NaCl-500 mM imidazole and dialyzed against Dignam's buffer D overnight at 4°C (19). The final concentration of U2AF was estimated to be 0.5 pmol/μl. Aliquots were stored at −70°C.
hnRNP A/B depletions.
HNE was depleted of hnRNP A/B proteins as previously described (6). Briefly, in vitro-transcribed biotinylated ESSV RNA bait was immobilized on streptavidin-coated paramagnetic beads (Dynal) and incubated in HNE for 15 min at 30°C. Beads were then separated from the HNE by application of a magnetic microconcentrator.
Spliceosome assembly analyses.
For A complex analysis, capped, radiolabeled ESSVB or ESSVxB RNA was incubated in 60% (vol/vol) HNE under in vitro splicing conditions for various times indicated in Fig. 2 (30). Heparin was then added to a 0.5-mg/ml final concentration, and reactions were stopped on ice for 5 min. A 6.0-μl aliquot of each sample was loaded onto 4.0% native polyacrylamide gels and electrophoresed for 6 h at 300 V in 50 mM Tris-glycine running buffer. For E complex analysis, endogenous ATP was depleted from 7.5 μl of HNE by incubation at 25°C for 20 min (16). Capped, radiolabeled ESSVB or ESSVxB RNA was then added, and the final reaction volume was brought to 12.5 μl with diethyl pyrocarbonate-treated H2O. Reaction mixtures were incubated at 30°C for various times and then stopped by incubation on ice for 5 min. Heparin was added to a final concentration of 0.5 mg/ml as indicated in Fig. 2. Prior to electrophoresis, 2.5 μl of 5× loading dye (20% [vol/vol] glycerol, 1× TBE, 0.25% [wt/vol] bromophenol blue, 0.25% [wt/vol] xylene cyanol) was added to each sample. A 7.2-μl aliquot of each sample was loaded onto 1.0% low-melting-point agarose gels-0.5× TBE and electrophoresed at 16 mA for 3.5 h at 4°C. All equipment, buffers, and gels were cooled to 4°C prior to use. The number of counts per band was quantitated on an Instant Imager (Packard).
FIG. 2.
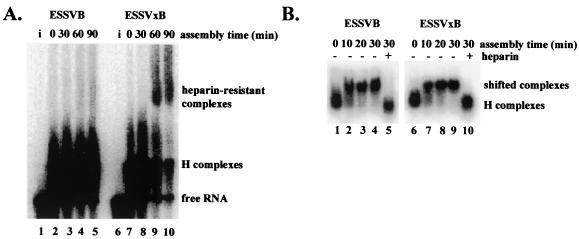
ESSV inhibits spliceosome assembly prior to A complex formation. (A) Spliceosomes were assembled with [α-32P]UTP-labeled ESSVB or ESSVxB substrates in HNE in the presence of ATP. Aliquots of reaction mixtures were sampled at the times indicated and electrophoresed on native 4.0% polyacrylamide gels. Lanes i, RNA substrates loaded directly onto gel. The location of the ATP-dependent, heparin-resistant complex is shown on the right. (B) Spliceosomes were assembled as in panel A, except that ATP was depleted from the HNE prior to assembly. Lanes +, addition of heparin to samples at the 30-min time point prior to electrophoresis. Aliquots of reaction mixtures were sampled at the times indicated and electrophoresed on native 1.0% low-melting point agarose gels. The locations of H complexes and heparin-sensitive complexes are noted on the right.
Oligonucleotide-directed RNase H digestions.
Digestion of U1 snRNAs in HNE was performed as previously described (9). Briefly, DNA oligonucleotides complementary to the first 16 nt of U1 snRNA were added to HNE, and the RNA portion of the RNA-DNA hybrid was digested by exogenous RNase H. Reaction products were analyzed by extracting total RNA from the HNE. RNAs were then precipitated and electrophoresed on denaturing 10% polyacrylamide-7 M urea gels. RNA was visualized with ethidium bromide stain. HNE was aliquoted and stored at −70°C.
Protein binding analysis.
For UV cross-linking studies, 10 fmol of radiolabeled ESSVB3′, ESSVxB3′, ESSVxBΔ3′, ESSVA2-3′, or ESSVxA2-3′ RNA was incubated in 20% (vol/vol) HNE for 30 min at 30°C. Reaction mixtures were placed on ice 2.0 cm from a UV source and irradiated for 15 min at 3 mW/cm2/s (Stratagene; UV Stratalinker 1800). Samples were then digested with RNase A and separated on SDS-10% polyacrylamide gels. For immunoprecipitations, cross-linking reactions were scaled up fourfold. Reaction mixtures were incubated for 15 min at 30°C with 1 mg of RNase A/ml. After digestion, 8.0 μl of anti-U2AF65 mouse IgG monoclonal antibody MC3 or a nonspecific mouse IgG antibody was added and the samples were incubated on ice for 2 h. IgG was then precipitated with anti-mouse IgG-conjugated agarose beads for 2 h at 4°C. The beads were then extensively washed, and the remaining bound proteins were eluted by denaturation. Samples were then electrophoresed on SDS-10% polyacrylamide gels.
RESULTS
ESSV is functional in the context of a two-exon splicing substrate.
We have previously shown that the hnRNP A/B-dependent ESSs ESS2 and ESS3 inhibit splicing by interfering with early steps in the spliceosome assembly pathway (3, 43). Because ESS2, ESS3, and ESSV share many characteristics, including the ability to repress splicing at the viral 3′ splice site A3, we hypothesized that ESSV also inhibits splicing by interfering with early steps in the spliceosome assembly pathway.
To maximize the observable difference between HIV-1 hnRNP A/B-dependent ESS-repressed and nonrepressed splicing conditions, we had previously created the splicing substrate HS1-ESSV, where a 24-nt sequence necessary and sufficient for ESSV silencer activity was substituted for a 21-nt sequence necessary and sufficient for ESS2 silencer activity (Fig. 1A) (8). This substitution resulted in an almost complete inhibition of splicing at 3′ splice site A3, indicating that ESSV is a stronger silencer of A3 than ESS2 (8). While this substrate was used to assess silencer activity of ESSV by in vitro splicing, the substrate cannot be used to study assembly of spliceosome intermediates because of the presence of five alternative 3′ splice sites in the second exon. To create simplified splicing substrates which would prevent the formation of heterogeneous mixtures of intermediates assembled at different 3′ splice sites, we derived two-exon splicing constructs containing either wild-type ESSV (ESSVB) or mutant ESSVx (ESSVxB) sequences from the alternative splicing construct (Fig. 1A). The portion of tat exon 2 downstream of the ESS substitution was deleted such that only one 5′ splice site and one 3′ splice site are present in the splicing substrates.
We first determined if the new substrates were capable of splicing. To inactivate the silencer in the ESSVB substrate, hnRNP A/B proteins were depleted from HNE as previously described (8). As expected, spliced RNAs were detected for both ESSVB and ESSVxB substrates (Fig. 1B). To assess the functionality of ESSV, recombinant hnRNP A1 was added to the hnRNP A/B-depleted HNE. This treatment resulted in a dose-dependent decrease in spliced RNA produced from ESSVB, with maximal silencer activity occurring at 0.9 μM hnRNP A1 (Fig. 1C, lanes 1 to 5). The same dose had no effect on the splicing efficiency of the ESSVxB substrate, showing that the silencer effect was specific for the wild-type ESSV (Fig. 1C, lanes 7 and 8). Addition of 0.9 μM UP1, which contains the RNA binding domains of hnRNP A1 but which lacks the C-terminal glycine-rich domain, did not restore ESSV activity to the hnRNP A/B-depleted HNE, indicating that the C-terminal domain is required for silencing (Fig. 1C, lane 6). These results are consistent with the previously described characteristics of ESSV, and we conclude that ESSV is functional in the context of the simplified two-exon splicing substrate (8).
ESSV does not affect the rate of formation of a heparin-sensitive complex that has characteristics similar to those of E complexes.
Assembly of a catalytically active metazoan spliceosome sequentially proceeds through a series of intermediate complexes termed H, E, A, B, and C, which can be distinguished by gel electrophoresis under nondenaturing conditions (4, 30). Formation of A, B, and C complexes in vitro requires addition of ATP to the assembly reaction mixtures (30). Once formed, these three complexes can be resolved on nondenaturing polyacrylamide gels and are resistant to heparin-mediated dissociation to complexes that migrate with efficiencies similar to that of H complexes (30). We assembled ATP-dependent splicing complexes using radiolabeled ESSVB or ESSVxB splicing substrates and analyzed the rates of formation by electrophoretic mobility shift assay (EMSA). The ESSVB substrate did not form detectable heparin-resistant complexes over the time course of the experiment (Fig. 2A, lanes 2 to 5). In contrast, the ESSVxB substrate formed a heparin-resistant complex that was first detected at the 60-min time point (Fig. 2A, lanes 7 to 10). Formation of a shifted, ATP-dependent, heparin-resistant complex indicated that A complexes can form on the splicing substrates when ESSV is mutated. We conclude that ESSV represses splicing by inhibiting spliceosome assembly prior to A complex formation.
We could not assess the rate of E complex formation using the same analytical method as that described above because E and H complexes do not resolve on the polyacrylamide gels. We therefore analyzed the effect of ESSV on spliceosome assembly using a modified EMSA developed to detect E complexes (16). Capped and radiolabeled ESSVB or ESSVxB splicing substrates were incubated at 30°C in HNE depleted of ATP, and aliquots of the reaction mixtures were electrophoresed on native low-melting-point agarose gels. Incubation of reaction mixtures at 30°C resulted in formation of a shifted complex on both ESSVB and ESSVxB substrates that represents nearly 100% of the population upon a 30-min incubation at 30°C (Fig. 2B, lanes 2 to 4 and 7 to 9). The shifted complexes appear to be stable because, once formed, they can be resolved on the agarose gels despite incubation at 4°C for up to 6 h prior to electrophoresis (data not shown). Addition of heparin prior to electrophoresis resulted in dissociation of the shifted complex to a faster-migrating species (Fig. 2B, lanes 5 and 10), which has a migration rate similar to that of the complex which forms without the requirement for incubation at 30°C (Fig. 2B, lanes 1 and 6). Based on these characteristics, the faster-migrating species appeared to be H complexes, whereas the shifted species migrated similarly to E complexes. These data suggest that a complex similar to E complexes assembles on in vitro splicing substrates containing ESSV.
hnRNP A/B proteins bound to ESSV inhibit binding of U2AF65 to the PPT of the repressed 3′ splice site.
We further characterized the shifted complexes detected in Fig. 2B to determine if they were authentic E complexes. We first identified factors which associated with the 3′ splice site under the same conditions as those for the E complex EMSAs. We identified HNE factors that cross-link to the 3′ halves of ESSVB and ESSVxB RNA substrates under the same conditions as in Fig. 2B (Fig. 3A). The cross-linking of an ∼65-kDa protein to the RNA substrate was detected when ESSV was mutated (Fig. 3B, lane 2). When wild-type ESSV was present in the RNA substrate, cross-linking to the ∼65-kDa band was below background (Fig. 3B, lane 1). To further characterize the ∼65-kDa cross-linked protein, we performed immunoprecipitation analyses using the anti-U2AF65 monoclonal antibody MC3 (23). A cross-linked protein of ∼65 kDa was precipitated from the ESSVxB3′ reactions (Fig. 3C, lane 2). An isotype control antibody failed to precipitate the ∼65-kDa protein cross-linked to ESSVxB3′ (Fig. 3C, lane 4). These results indicate that the ∼65-kDa protein has an epitope recognized by the anti-U2AF65 monoclonal antibody. Precipitation of the ∼65-kDa protein cross-linked to ESSVB3′ was not detected (Fig. 3C, lane 1). These results indicate that the ∼65-kDa protein is U2AF65.
FIG. 3.
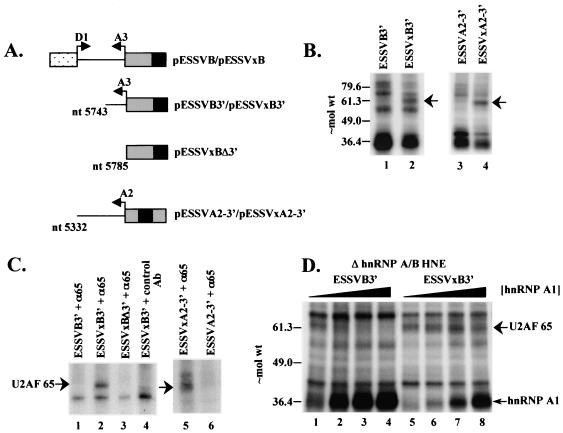
Inhibition of U2AF65 cross-linking to ESSV-containing RNAs is dependent on hnRNP A/B proteins. (A) Schematic diagram of the 3′ half RNA probes used in the binding studies. Arrows, splice sites; black boxes, ESSV and ESSVx. (B) UV cross-linking of HNE proteins to [α-32P]UTP-labeled ESSVB3′ (lane 1), ESSVxB3′ (lane 2), ESSVA2-3′ (lane 3), and ESSVxA2-3 (lane 4) RNAs. Proteins cross-linked to RNase A-digested samples were electrophoresed on an SDS-10% polyacrylamide gel. Arrow, ∼65-kDa band. Approximate locations of molecular weight (in thousands) markers are shown on the left. (C) Immunoprecipitation of proteins cross-linked to [α-32P]UTP-labeled RNAs. Anti-U2AF65 monoclonal antibody MC3 (α65) and nonspecific murine IgG control antibody (control Ab) used for immunoprecipitations are noted above the lanes. Precipitated proteins were electrophoresed on SDS-10% polyacrylamide gels. The location of U2AF65 is noted on the left. (D) Purified hnRNP A1 (0, 0.3, 0.5, and 0.7 μM in lanes 1 to 4 and 5 to 8, respectively) was added back to hnRNP A/B-depleted HNEs. Cross-linking was performed as in panel B. Locations of U2AF65 and hnRNP A1 are noted on the right.
To confirm that the precipitated ∼65-kDa band is U2AF65, we deleted the PPT from ESSVxB3′ to create ESSVxBΔ3′ (Fig. 3A). ESSVxBΔ3′ failed to cross-link to the ∼65-kDa protein (Fig. 3C, lane 3), showing that the cross-linking of the ∼65-kDa band requires the predicted binding site for U2AF65. In addition, this result indicates that the ∼65-kDa protein does not cross-link to the altered sequences created by the ESSVx mutations (Fig. 1A). We conclude that ESSV inhibits the binding of U2AF65 to the A3 PPT.
To confirm that the binding of U2AF65 to the PPT was also inhibited when ESSV was present in its native position downstream of 3′ splice site A2, we performed UV cross-linking experiments using radiolabeled 3′ half RNAs corresponding to wild-type and mutant A2 region sequences (ESSVA2-3′ and ESSVxA2-3′, respectively). The results given in Fig. 3B (lanes 3 and 4) indicated that the mutant RNA bound U2AF65 more efficiently than the wild-type RNA. The major band labeled by the ESSV mutant substrate was recognized by the anti-U2AF65 monoclonal antibody as shown in Fig. 3C (lane 5 and 6). The results strongly suggest that ESSV inhibits splicing by similar mechanisms in its native context downstream of A2 and in the heterologous context downstream of A3.
The above results led us to hypothesize that hnRNP A/B proteins bound to ESSV inhibit the binding of U2AF65 to the PPT of the repressed 3′ splice site. This model predicts that depletion of hnRNP A/B proteins from HNE would result in an increased efficiency of the cross-linking of U2AF65 to the PPT on ESSVB3′. In support of this hypothesis, the cross-linking of U2AF65 to ESSVB3′ in hnRNP A/B-depleted HNE was enhanced (Fig. 3D, lane 1, versus B, lane 1). Addition of purified hnRNP A1 to the reaction mixtures resulted in a dose-dependent decrease in the efficiency of the cross-linking of U2AF65 to ESSVB3′, with nearly complete inhibition of cross-linking occurring at 0.7 μM hnRNP A1 (Fig. 3D, lanes 1 to 4). In contrast, the cross-linking of U2AF65 to ESSVxB3′ was not inhibited by up to 0.7 μM hnRNP A1 (Fig. 3D, lanes 5 to 8), indicating that hnRNP A1-mediated inhibition of U2AF65 cross-linking requires ESSV. We conclude that hnRNP A1 bound to ESSV is necessary for inhibition of the binding of U2AF65 to the PPT.
In contrast to the differences in the amounts of U2AF65 bound, the amount of bound hnRNP A1 when high levels of the protein were added to the mutant substrate ESSVxB3′ was not very different from the amount bound to the wild-type substrate ESSV3′ at a lower concentration of protein (Fig. 3D, compare lanes 3 and 8). hnRNP A/B proteins bind both specifically to high-affinity sites present in ESS elements and nonspecifically at lower affinities to all RNA sequences (1, 10). Our results suggest that, at high concentrations of hnRNP A1, most of the radioactive signal results from the nonspecific binding of hnRNP A1, which does not lead to disruption of U2AF65 recruitment to the PPT of the mutant substrate.
Purified U2AF stimulates splicing of ESSVB in in vitro splicing assays.
If hnRNP A/B proteins bound to ESSV inhibit the binding of U2AF to the regulated 3′ splice site, then increasing the dose of U2AF in an in vitro splicing reaction would be expected to increase the splicing efficiency of the ESSVB splicing substrate. To test this prediction, we expressed a histidine-tagged U2AF heterodimer (HisU2AF) utilizing a baculovirus expression system and purified the heterodimer based on affinity for Ni+-nitrilotriacetic acid beads (24). Coomassie staining of an SDS-polyacrylamide gel showed two bands of approximately 65 and 35 kDa present in the eluate (Fig. 4A, left), and immunoblotting showed single distinct bands detected by antibodies against U2AF65 (Fig. 4A, center) and U2AF35 (Fig. 4A, right) comigrating with the stained bands. To determine if the purified HisU2AF was functional, we performed in vitro splicing assays on radiolabeled adenovirus-derived splicing substrate in HNE or HNE depleted of U2AF based on affinity for poly(U)-Sepharose 4B (50). No spliced RNA was detected from reactions using the U2AF-depleted HNE (ΔU2AF) since U2AF is an essential splicing factor (Fig. 4B, lane 2). Addition of HisU2AF restored splicing activity to ΔU2AF HNE (Fig. 4B, lanes 1 and 3), indicating that the purified HisU2AF was functional in in vitro splicing assays.
FIG. 4.
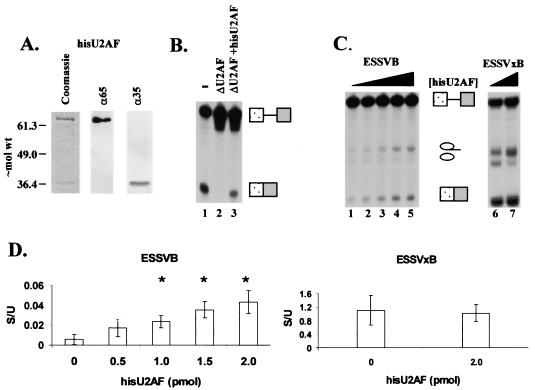
Recombinant U2AF stimulates the splicing of a 3′ splice site regulated by ESSV. (A) Characterization of baculovirus-expressed HisU2AF. Left, Coomassie blue stain of purified protein; middle and right, Western blot analysis for purified U2AF65 and U2AF35. Molecular weights are in thousands. (B) Functional activity of purified HisU2AF. U2AF was depleted from HNE based on affinity for poly(U)-Sepharose 4B. −, undepleted HNE; ΔU2AF, depleted HNE. Purified HisU2AF (2 pmol) was added to ΔU2AF HNE (ΔU2AF+hisU2AF). The α-32P-labeled Ad81 substrate was spliced for 2 h in the various HNEs. RNAs were electrophoresed on 4% denaturing polyacrylamide gels. Locations of unspliced and spliced RNA are on the right. (C) In vitro splicing competition assays. hnRNP A/B-depleted HNE was reconstituted with 0.7 μM hnRNP A1. Increasing amounts of HisU2AF were added to the splicing reaction mixtures for ESSVB (left; 0, 0.5, 1, 1.5, and 2 pmol) and ESSVxB (right; 0 and 2 pmol) substrates. Locations of unspliced and spliced RNA species are indicated between gels. (D) Ratios of spliced to unspliced species for ESSVB (left) and ESSVxB (right) substrates. Quantitations are from three independent experiments. Error bars indicate 1 standard deviation. Asterisks, samples with calculated P values <0.05 compared to lane 1 of panel C, as determined by Student's t test.
Next, we performed in vitro splicing assays using ESSVB or ESSVxB substrates in hnRNP A/B-depleted HNE reconstituted with hnRNP A1 (0.7 μM). We found that the partial depletion of hnRNP A/B proteins was necessary to reduce the large excess of endogenous hnRNP A/B proteins present in our extract preparations (data not shown) (8). Addition of HisU2AF resulted in a dose-dependent increase of spliced RNA produced from the ESSVB substrate (Fig. 4C, lanes 1 to 5). Quantitations from three independent trials indicated that addition of HisU2AF resulted in an approximately fourfold increase in the ratio of spliced to unspliced RNA (Fig. 4D, left). Addition of HisU2AF to splicing reaction mixtures containing ESSVxB substrate resulted in an increase in both unspliced- and spliced-RNA levels (Fig. 4C, lanes 6 and 7). However, the ratio of spliced to unspliced RNA from reaction mixtures containing ESSVxB was not significantly affected by HisU2AF (Fig. 4D, right), indicating that HisU2AF did not cause a generalized increase in the splicing efficiency of the HNE. The results from Fig. 3 and 4 are consistent with the hypothesis that hnRNP A/B proteins bound to ESSV repress splicing by inhibiting the binding of U2AF to the regulated 3′ splice site.
Formation of heparin-sensitive complexes detected by EMSA requires intact U1 snRNA.
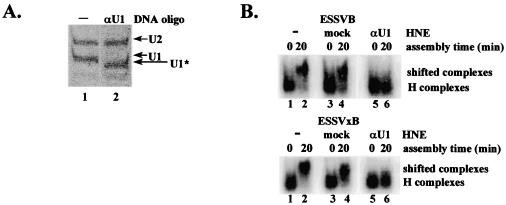
Because U2AF65 is a component of purified E complexes, our data in Fig. 3 and 4 suggested that the shifted complexes detected using the ESSVB substrate in Fig. 2B do not contain U2AF65 and therefore are not authentic E complexes (17). To further characterize the shifted complexes detected in Fig. 2B, we attempted to detect another component of purified E complexes, U1 snRNP, by transferring the complexes directly from the agarose gels and performing Northern blotting for U1 snRNA. However, we found that transfer to the nitrocellulose membranes was inefficient due to the large size of the complexes. Therefore, we utilized an alternative approach in which a portion of U1 snRNA necessary for interaction of U1 snRNP with a 5′ splice site was specifically cleaved by antisense DNA-directed RNase H digestion (Fig. 5A, lane 1 versus 2). E complex EMSAs showed that the shifted complexes did not form on radiolabeled in vitro splicing substrates ESSVB and ESSVxB in the U1-digested HNE (Fig. 5B, top and bottom, lane 6). The shifted complexes were detected in HNE treated with RNase H alone, showing that the lack of shifting seen in lane 6 was not due to inactivation of the HNE by the addition of exongenous RNase H (Fig. 5B, lane 4, top and bottom). We conclude that intact U1 snRNA is required for formation of the shifted complexes using ESSVB and ESSVxB substrates and that U1 snRNP is stably associated with the 5′ splice site.
FIG. 5.
Intact U1 snRNA is required for formation of an ATP-independent, heparin-sensitive complex. (A) U1 snRNA in HNE was partially digested with RNase H as described in Materials and Methods. Products of the digestions were electrophoresed on the same 10% polyacrylamide-7 M urea gel and visualized by ethidium bromide staining. Locations of intact U1 and U2 snRNAs (U1 and U2) and partially digested U1 snRNA (U1*) are shown on the right. (B) Analysis of E complex assembly in untreated (−), mock-digested, and U1 snRNA-digested (αU1) HNEs. Locations of H complexes and heparin-sensitive complexes are noted on the right.
ESSV does not affect function of the 5′ splice site.
The results shown in Fig. 5B suggested that ESSV does not affect the binding of U1 snRNP to the 5′ splice site present in the HIV-1 ESSVB splicing substrate. However, this assay did not assess whether ESSV inhibited the function of the 5′ splice site. We reasoned that, if ESSV did not inhibit the function of U1 snRNP bound at the 5′ splice site, then the 5′ splice site should be free to pair with nonrepressed alternative 3′ splice sites present in the splicing substrate.
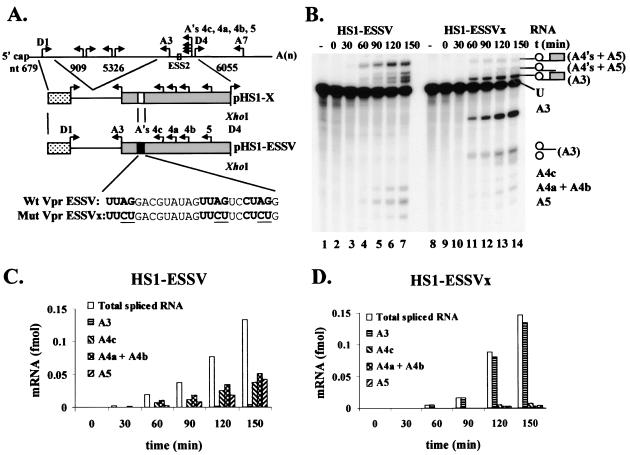
To determine if 5′ splice site D1 was inactivated by ESSV, we performed in vitro splicing assays using the alternative splicing substrates HS1-ESSV and HS1-ESSVx (Fig. 6A). These substrates contain the portion of tat exon 2 that was deleted from the ESSVB and ESSVxB substrates. Splicing to the alternative 3′ splice sites downstream of ESSV predominated (Fig. 6B, lanes 1 to 7). Mutation of ESSV alleviated repression of splicing to A3, concomitant with a decrease in splicing to the downstream alternative 3′ splice sites A4c, A4a, A4b, and A5 (Fig. 6B, lanes 8 to 14). Despite the change in the splicing patterns upon ESSV mutation, the total molar amounts of the spliced products produced at each time point were similar for both splicing substrates, as calculated from the quantitated counts per minute per band and the specific activity of the [γ-32P]UTP RNAs (Fig. 6C versus D). We conclude that ESSV does not affect the overall rate of splicing of the HS1-ESSV and HS1-ESSVx substrates, but rather regulates a balance of splicing between 3′ splice site A3 and the downstream 3′ splice sites. These results suggest that the U1 snRNP molecule bound to 5′ splice site D1 is not inactivated by ESSV and that repression is specific to the 3′ splice site.
FIG. 6.
ESSV does not inhibit 5′ splice site splicing efficiency. (A) Schematic diagram of the in vitro splicing constructs pHS1-ESSV and pHS1-ESSVx. A minigene containing 5′ splice site D1 and 3′ splice sites A3, A4c, A4a, A4b, and A5 was generated as previously described. ESS2 was replaced by ESSV or ESSVx sequences as shown. (B) Capped and radiolabeled substrates were transcribed from linearized plasmids and spliced in HNE. RNAs were electrophoresed on denaturing polyacrylamide gels. The identities of the unspliced and spliced RNAs are shown on the right. Note that RNAs spliced at A4a and A4b comigrate on these gels. The positions of introns and exon lariats are in parentheses. (C and D) Splicing kinetics of substrates HS1-ESSV (C) and HS1-ESSVx (D). The amounts of radioactivity in the spliced RNA bands was quantitated on Instant Imager (Packard). The amounts (femtomoles) of the spliced RNA species and the total amount of spliced RNA from each reaction were calculated based on the specific activity of the [α-32P]UTP precursor and the UTP content of each RNA species.
DISCUSSION
Previous studies have shown that increasing the pyrimidine content of the PPT of HIV-1 hnRNP A/B-dependent repressed 3′ splice sites reduces ESS silencing activity (8, 42, 43). These results suggested that reduced binding of the essential splicing factor U2AF65 to the PPT may be the rate-limiting step in hnRNP A/B-dependent ESS inhibition (3). Our studies have directly tested this hypothesis and showed that, although 5′ splice site selection was not inhibited by interference with U2AF65 binding to the PPT of either 3′ splice site A2 or A3, 3′ splice site selection was inhibited. Negative regulation of alternative splicing by interference with U2AF65 binding is a common feature of numerous alternative splicing pathways. The Drosophila melanogaster sex-specific splicing regulatory factor Sxl competes with U2AF65 for binding to a region containing the PPT of the male-specific default 3′ splice site within tra pre-mRNA (48). The splicing of msl-2 pre-mRNA is also inhibited by competition between Sxl and U2AF65 for the PPT of the repressed 3′ splice site (35). The human PPT binding protein represses splicing of an adenovirus-derived in vitro splicing substrate by competing with U2AF65 for binding to the precursor RNA (44). In each of the above cases, inhibition of U2AF65 binding to the PPT results from the binding of a negative regulatory factor to an adjacent or overlapping region of the PPT within the intron of the pre-mRNA. In contrast, we have shown that inhibition of U2AF binding to the PPT was caused by the activity of an exonic negative regulatory element positioned downstream of the 3′ splice site.
How does the ESS element exert its effects on the binding of U2AF65 from a position approximately 75 nt downstream of the PPT? Our previous data have suggested that splicing repression by ESSV does not occur as a result of a simple binding of one trans-acting factor to one cis element, since an N-terminal-truncated hnRNP A1 protein containing the RNA-binding domain (UP1) does not restore ESSV activity to hnRNP A/B-depleted HNE (8) (Fig. 1C). Similar results have been reported for inhibition of splicing by HIV-1 ESS2 and ESS3, the ESS in the K-SAM alternative exon within the human fibroblast growth factor receptor 2 pre-mRNA, and the ESS in the cytoskeletal 4.1R protein (12, 18, 27, 51). These results suggest that the binding of hnRNP A/B proteins to an ESS via the RNA binding domain is not sufficient to inhibit splicing and that the C-terminal region of hnRNP A1, which may mediate multimerization of the protein, is also necessary for splicing inhibition (13). In addition, we have shown above that ESSV activity apparently proceeds in a unidirectional manner. Within the context of substrate HS1-ESSVB (Fig. 6A), ESSV repressed splicing only to 3′ splice site A3 approximately 75 nt upstream and not to the 3′ splice sites (A4c, A4a, A4b, and A5) located in a region approximately 75 to 125 nt downstream. Together, these results are consistent with a recently described model proposing that ESSs serve as high-affinity binding sites for hnRNP A/B proteins and as a nucleation points for multimerization of hnRNP A/B proteins to regions upstream of the ESS (51). In further support of this model, footprinting analyses of tat exon 3 RNA, which contains an HIV-1 hnRNP A/B-dependent ESS (ESS3), have shown that purified hnRNP A1 protects several regions within the RNA upstream of the silencer (15, 32). This multimerization of hnRNP A/B proteins may not be sufficient to explain inhibition of U2AF65 binding since we were unable to show inhibition of binding of purified HisU2AF to ESSVB3′ by addition of excess purified hnRNP A1 in the absence of HNE (data not shown). This result suggests that there may be an additional factor(s) necessary to transmit inhibition of U2AF65 binding from hnRNP A/B proteins bound at the ESS. Together, these results are consistent with the model shown in Fig. 7, where inhibition of U2AF65 binding to an upstream PPT requires a higher-order RNA-protein structure relying on protein-protein interactions between hnRNP A/B proteins and possibly other proteins.
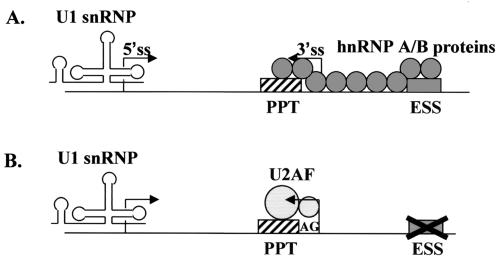
FIG. 7.
Working model of the ATP-independent complexes formed in vitro. (A) Assembly of splicing components under conditions of repression. hnRNP A/B proteins (circles) bound to ESSV (gray box) nucleate binding of additional hnRNP A/B proteins to upstream cis sites, resulting in the blocking of the PPT (striped box). Note that, in the absence of splicing, U1 snRNP is bound to the 5′ splice site (5′ss). Only contacts between adjacent hnRNP A/B proteins are shown, although long-range protein-protein interactions may also occur. (B) Assembly of splicing components under nonrepressing conditions. Mutation of ESSV prevents stimulation of binding of additional hnRNP A/B proteins to upstream sites, allowing U2AF65 (large circle) to bind the PPT and U2AF35 (small circle) to bind a region including the 3′ splice site AG. Progression through the spliceosome assembly pathway then results in the splicing of the substrate.
Note that the HIV-1 3′ splice site A3 used throughout this study contains a short PPT interspersed with purines, and therefore splicing of the intron is predicted to be AG dependent (39). The binding of U2AF65 to short PPTs, which are characteristic of AG-dependent introns, is stimulated by the binding of U2AF35 to a region that includes the 3′ splice junction (26, 35, 49, 50). Therefore, it is possible that ESSV indirectly inhibits the binding of U2AF65 to the A3 PPT by interfering with stabilization mediated by the binding of U2AF35 to the 3′ splice junction. It is also possible that ESSV interferes with the binding of the branch point binding protein (mBBP), which is also a component of E complexes and which acts to stabilize the binding of U2AF65 (7). Further characterization of the ATP-independent, heparin-sensitive complex observed in the E complex EMSAs presented here will be necessary clarify these issues.
We do not yet know if the other hnRNP A/B-dependent ESS elements inhibit U2AF65 binding by the same mechanism as does ESSV. Based on the previously described shared characteristics of the hnRNP A/B-dependent silencers, this would be an expected result. However, Tange et al. (47) have reported that the hnRNP A/B proteins in HNE supplemented with the SR protein SF2/ASF do not inhibit U2AF65 binding to the PPT of HIV 3′ splice site A7. In its native context A7 is negatively regulated by ESS3 and an intronic splicing enhancer. The binding of U2 snRNP to the branch point sequence in this case appears to be inhibited (47). Thus, it is possible that, in different sequence contexts, different rate-limiting steps of spliceosome assembly may be affected by hnRNP A/B-dependent ESS elements.
We have shown that ESSV did not affect the rate of formation of an ATP-independent, heparin-sensitive complex that requires intact U1 snRNP. Our results indicate that this complex, containing U1 snRNP, presumably bound at the 5′ splice site, is stable, even though splicing of the substrate is almost completely inhibited by ESSV. It is not yet clear whether this complex is a functional precursor to the E complex or is an aberrant splicing complex. As evidence for the former, we have found that the 5′ splice site is capable of pairing with nonrepressed alternative 3′ splice sites despite the presence of an ESSV-repressed 3′ splice site in the substrate. However, we cannot exclude the possibility that the selected alternative 3′ splice sites influence the activity of the 5′ splice site.
Our results suggest that U1 snRNP is stably associated with 5′ splice site D1 independent of splicing, as intact U1 snRNP is required for formation of a stable complex that forms on a splicing substrate which is almost completely inhibited from splicing by ESSV. The tethering of U1 snRNP to HIV-1 5′ splice sites independent of splicing may serve several functions during virus replication. Hybridization of U1 snRNA to 5′ splice site D4 has been shown to be necessary for stabilization of both env mRNA and unspliced RNA, suggesting that U1 snRNP bound to 5′ splice site D4 has a function independent of splicing (29). In addition, it has been proposed that the tethering of U1 snRNP to HIV-1 5′ splice site D1 is primarily responsible for prevention of premature cleavage and polyadenylation at a 3′ processing signal located approximately 70 nt downstream of the cap site in the primary viral transcript (5, 6, 21). According to this model, synthesis of all viral transcripts would require the binding of U1 snRNP to 5′ splice site D1 independent of splicing, because approximately 50% of the viral precursor RNA remains unspliced in an infected cell. A splicing-independent function of U1 snRNP in the context of HIV-1 infection remains to be established.
Acknowledgments
We thank R. Reed for the U2AF35 antibody and the adenovirus-derived splicing construct, B. Graveley for the U2AF-expressing baculovirus, and J. Valcarcel for the U2AF 65 antibody. We also thank S. Perlman and D. Price for critical reviews of the manuscript.
HeLa cells were obtained from the National Cell Culture Center, which is sponsored by the National Center for Research Resources of the NIH. This work was supported by PHS grant AI36073 from the NIAID to C.M.S., by an institutional research grant (IRG-98-277-04) from ACS and Florida Biomedical Research Program grant (BM031) from FDH to A.M., and by PHS grant CA13106 from the NCI to A.R.K. A.M. is a research member of the Sylvester Comprehensive Cancer Center.
REFERENCES
- 1.Abdul-Manan, N., and K. R. Williams. 1996. hnRNP A1 binds promiscuously to oligoribonucleotides: utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 24:4063-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amendt, B. A., D. Hesslein, L.-J. Chang, and C. M. Stoltzfus. 1994. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol. Cell Biol. 14:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amendt, B. A., Z.-H. Si, and C. M. Stoltzfus. 1995. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell Biol. 15:4606-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrigo, S. J., S. Weitsman, J. A. Zack, and I. S. Chen. 1990. Characterization and expression of novel singly spliced RNA species of human immunodeficiency virus type 1. J. Virol. 64:4585-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashe, M. P., A. Furger, and N. J. Proudfoot. 2000. Stem-loop 1 of the U1 snRNP plays a critical role in the suppression of HIV-1 polyadenylation. RNA 6:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashe, M. P., L. H. Pearson, and N. J. Proudfoot. 1997. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 16:5752-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berglund J. A., N. Abovich, and M. Rosbash. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilodeau, P. S., J. K. Domsic, A. Mayeda, A. R. Krainer, and C. M. Stoltzfus. 2001. RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol. 75:8487-8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black, D. L., B. Chabot, and J. A. Steitz. 1985. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell 42:737-750. [DOI] [PubMed] [Google Scholar]
- 10.Burd, C. G., and G. Dreyfuss. 1994. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 13:1197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butsch, M., and K. Boris-Lawrie. 2000. Translation is not required to generate virion precursor RNA in human immunodeficiency virus type 1-infected T cells. J. Virol. 74:11531-11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartegni, L., M. Maconi, E. Morandi, F. Cobianchi, S. Riva, and G. Biamonti. 1996. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 259:337-348. [DOI] [PubMed] [Google Scholar]
- 14.Cullen, B. R. 1998. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology 249:203-210. [DOI] [PubMed] [Google Scholar]
- 15.Damgaard, C. K., T. O. Tange, and J. Kjems. 2002. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA 8:1401-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, R., and R. Reed. 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5:1504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das, R., Z. Zhou, and R. Reed. 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell 5:779-787. [DOI] [PubMed] [Google Scholar]
- 18.Del Gatto, F., M. C. Gesnel, and R. Breathnach. 1996. The exon sequence TAGG can inhibit splicing. Nucleic Acids Res. 24:2017-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freed, E. O. 2001. HIV-1 replication. Somat. Cell Mol. Genet. 26:13-33. [DOI] [PubMed] [Google Scholar]
- 21.Furger, A., J. Monks, and N. J. Proudfoot. 2001. The retroviruses human immunodeficiency virus type 1 and Moloney murine leukemia virus adopt radically different strategies to regulate promoter-proximal polyadenylation. J. Virol. 75:11735-11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furtado, M. R., R. Balachandran, P. Gupta, and S. M. Wolinsky. 1991. Analysis of alternatively spliced human immunodeficiency virus type-1 mRNA species, one of which encodes a novel tat-env fusion protein. Virology 185:258-270. [DOI] [PubMed] [Google Scholar]
- 23.Gama-Carvalho, M., R. D. Krauss, L. Chiang, J. Valcarcel, M. R. Green, and M. Carmo-Fonseca. 1997. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 137:975-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graveley, B. R., K. J. Hertel, and T. Maniatis. 2001. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA 7:806-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guatelli, J. C., T. R. Gingeras, and D. D. Richman. 1990. Alternative splice acceptor utilization during human immunodeficiency virus type 1 infection of cultured cells. J. Virol. 64:4093-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guth, S., T. O. Tange, E. Kellenberger, and J. Valcarcel. 2001. Dual function for U2AF(35) in AG-dependent pre-mRNA splicing. Mol. Cell. Biol. 21:7673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou, V. C., R. Lersch, S. L. Gee, J. L. Ponthier, A. J. Lo, M. Wu, C. W. Turck, M. Koury, A. R. Krainer, A. Mayeda, and J. G. Conboy. 2002. Decrease in hnRNP A/B expression during erythropoiesis mediates a pre-mRNA splicing switch. EMBO J. 21:6195-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacquenet, S., D. Ropers, P. S. Bilodeau, L. Damier, A. Mougin, C. M. Stoltzfus, and C. Branlant. 2001. Conserved stem-loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res. 29:464-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kammler, S., C. Leurs, M. Freund, J. Krummheuer, K. Seidel, T. O. Tange, M. K. Lund, J. Kjems, A. Scheid, and H. Schaal. 2001. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA 7:421-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konarska, M. M. 1989. Analysis of splicing complexes and small nuclear ribonucleoprotein particles by native gel electrophoresis. Methods Enzymol. 180:442-453. [DOI] [PubMed] [Google Scholar]
- 31.Krainer, A. R., T. Maniatis, B. Ruskin, and M. R. Green. 1984. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell 36:993-1005. [DOI] [PubMed] [Google Scholar]
- 32.Marchand, V., A. Mereau, S. Jacquenet, D. Thomas, A. Mougin, R. Gattoni, J. Stevenin, and C. Branlant. 2002. A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J. Mol. Biol. 323:629-652. [DOI] [PubMed] [Google Scholar]
- 33.Mayeda, A., and A. R. Krainer. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68:365-375. [DOI] [PubMed] [Google Scholar]
- 34.Mayeda, A., S. H. Munroe, J. F. Caceres, and A. R. Krainer. 1994. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 13:5483-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merendino, L., S. Guth, D. Bilbao, C. Martinez, and J. Valcarcel. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402:838-841. [DOI] [PubMed] [Google Scholar]
- 36.O'Reilly, M. M., M. T. McNally, and K. L. Beemon. 1995. Two strong 5′ splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology 213:373-385. [DOI] [PubMed] [Google Scholar]
- 37.Purcell, D. F., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67:6365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12:340-345. [DOI] [PubMed] [Google Scholar]
- 39.Reed, R. 1989. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 3:2113-2123. [DOI] [PubMed] [Google Scholar]
- 40.Robert-Guroff, M., M. Popovic, S. Gartner, P. Markham, R. C. Gallo, and M. S. Reitz. 1990. Structure and expression of tat-, rev-, and nef-specific transcripts of human immunodeficiency virus type 1 in infected lymphocytes and macrophages. J. Virol. 64:3391-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz, S., B. K. Felber, D. M. Benko, E. M. Fenyo, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Si, Z., B. A. Amendt, and C. M. Stoltzfus. 1997. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 25:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Si, Z.-H., D. Rauch, and C. M. Stoltzfus. 1998. The exon splicing silencer in human immunodeficiency virus type 1 Tat exon 3 is bipartite and acts early in spliceosome assembly. Mol. Cell. Biol. 18:5404-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh, R., J. Valcarcel, and M. R. Green. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268:1173-1176. [DOI] [PubMed] [Google Scholar]
- 45.Staffa, A., and A. Cochrane. 1995. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell Biol. 15:4597-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staffa, A., and A. Cochrane. 1994. The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3′ splice site. J. Virol. 68:3071-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tange, T. O., C. K. Damgaard, S. Guth, J. Valcarcel, and J. Kjems. 2001. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 20:5748-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valcarcel, J., R. Singh, P. D. Zamore, and M. R. Green. 1993. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362:171-175. [DOI] [PubMed] [Google Scholar]
- 49.Wu, S., C. M. Romfo, T. W. Nilsen, and M. R. Green. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832-835. [DOI] [PubMed] [Google Scholar]
- 50.Zamore, P. D., and M. R. Green. 1991. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 10:207-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu, J., A. Mayeda, and A. R. Krainer. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8:1351-1361. [DOI] [PubMed] [Google Scholar]
- 52.Zorio, D. A., and T. Blumenthal. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402:835-838. [DOI] [PubMed] [Google Scholar]