Abstract
Telomerase is a ribonucleoprotein reverse transcriptase responsible for the maintenance of one strand of the telomere terminal repeats. The catalytic protein subunit of the telomerase complex, known as TERT, possesses a reverse transcriptase (RT) domain that mediates nucleotide addition. The RT domain of TERT is distinguishable from retroviral and retrotransposon RTs in having a sizable insertion between conserved motifs A and B′, within the so-called fingers domain. Sequence analysis revealed the existence of conserved residues in this region, named IFD (insertion in fingers domain). Mutations of some of the conserved residues in Saccharomyces cerevisiae TERT (Est2p) abolished telomerase function in vivo, testifying to their importance. Significant effects of the mutations on telomerase activity in vitro were observed, with most of the mutants exhibiting a uniform reduction in activity regardless of primer sequence. Remarkably, one mutant manifested a primer-specific defect, being selectively impaired in extending primers that form short hybrids with telomerase RNA. This mutant also accumulated products that correspond to one complete round of repeat synthesis, implying an inability to effect the repositioning of the DNA product relative to the RNA template that is necessary for multiple repeat addition. Our results suggest that the ability to stabilize short RNA-DNA hybrids is crucial for telomerase function in vivo and that this ability is mediated in part by a more elaborate fingers domain structure.
Telomerase is a ribonucleoprotein (RNP) that is responsible for maintaining the terminal repeats of telomeres in most organisms (21). It acts as an unusual reverse transcriptase (RT) that was a small segment of an integral RNA component as template for the synthesis of the dG-rich strand of telomeres (22).
Telomerase activity has been characterized from a wide range of organisms, and genes encoding both the RNA and protein components of the enzyme complex have been identified (for reviews, see references 4, 23, 29, and 43). Telomerase RNAs found in ciliated protozoa, in addition to having a short templating region, share a common secondary structure. Telomerase RNAs from Saccharomyces cerevisiae and mammals are considerably larger, and within each group conserved structural elements can be identified based on phylogenetic and mutational analysis (9, 50). The catalytic reverse transcriptase protein subunit (TERT), first purified from Euplotes aediculatus as p123, was found to be homologous to Est2p, a protein from Saccharomyces cerevisiae required for telomere maintenance (31-33). Both proteins possess RT-like motifs, alterations in which render telomerase inactive both in vitro and in vivo. Subsequently, homologs of TERT were identified in a phylogenetically diverse group of organisms (7, 12, 14, 18, 30, 40, 41, 44). Because coexpression of TERT and telomerase RNA in rabbit reticulocyte lysates suffices to reconstitute enzyme activity (2, 51), these two subunits probably constitute the core of the enzyme complex. Quite a few telomerase-associated polypeptides have been identified with either biochemical or genetic tools. Studies from several laboratories suggest that these factors may participate in telomerase assembly, catalytic function, or regulation (13, 17, 24, 25, 31, 42, 46).
An unusual property of telomerase is its ability to mediate realignment of the DNA product relative to the RNA template. This was inferred from the capacity of the enzyme to add more repeats onto a starting primer than are present in the RNA template (20). The addition of multiple repeats depends on two types of movements: simultaneous translocation of the RNA-DNA duplex away from the active site after each nucleotide addition, and translocation of the RNA template relative to the DNA product after each cycle of copying the template, so that the 3′ end of the DNA realigns with the 3′ end of the RNA template. These movements have been referred to as type I and type II translocation, and the propensity to carry out the movements has been referred to as nucleotide addition and repeat addition processivity, respectively (11, 34, 45). Whereas nucleotide addition processivity is a common property of all RTs, repeat addition processivity is unique to telomerase. Analysis of mouse and human telomerase RNAs has revealed a feature of the RNA template region that modulates repeat addition processivity (8).
The RT-like domain of cloned TERTs, located near the C terminus of the protein, contains seven motifs (named 1, 2, A, B′, C, D, and E) that are conserved in all families of RTs. In the three-dimensional structural model of human immunodeficiency virus type 1 (HIV-1) RT, these motifs are located in the fingers and palm subdomain and mediate specific interactions with the template, primer, or nucleotides (27). Extensive mutational analysis indicates that these motifs perform nearly identical functions in TERTs. In particular, the same conserved residues appear to control nucleotide addition processivity in both HIV-1 RT and TERT (5, 6, 26, 45). However, none of the mutations in conserved RT motifs have been shown to specifically affect type II translocation (data not shown). This is perhaps not surprising, given the inability of conventional RTs to carry out type II translocation.
Interestingly, there is one feature of the TERT RT domain that distinguishes it from conventional RTs: a sizable insertion between conserved motifs A and B′ within the fingers subdomain (33, 41). In this report, we demonstrate by sequence analysis the existence of conserved TERT-specific residues in this region, named IFD (insertion in fingers domain). To address the functional significance of these residues, we generated substitution mutations and examined their effects on telomerase function. Our results indicate that the IFD of TERTs is required for normal telomere maintenance in vivo and for maximal telomerase activity in vitro. Various effects of the mutations on activity were observed, with most of the mutants exhibiting a uniform reduction in activity regardless of primer sequence. Remarkably, one mutant manifested a primer-specific defect, being selectively impaired in extending primers that form short hybrids with telomerase RNA. This mutant also accumulated products that correspond to one complete round of repeat synthesis, implying an inability to perform type II translocation. Our results suggest that telomerase has evolved a more elaborate fingers domain structure to optimize substrate interaction and to mediate the synthesis of multiple copies of telomere repeat. Thus, a TERT-specific motif within the catalytic domain is required for the unique enzymatic functions of telomerase.
MATERIALS AND METHODS
Yeast strains and plasmids.
The construction of an est2-Δ strain harboring the pSE-Est2-C874 plasmid (containing a protein A-tagged EST2 gene) has been described (52). This fully functional Est2p is designated wild-type telomerase throughout the text. All substitution mutations in IFD of EST2 were generated with the Quick-Change protocol (Stratagene), appropriate primer oligonucleotides, and pSE-Est2-C874 as the template. All point mutations were confirmed by sequencing. The oligodeoxynucleotide primers used for mutagenesis were purchased from Sigma-Genosis and purified by denaturing gel electrophoresis prior to use.
For overexpression of TERT mutants, a vector containing the triose phosphate isomerase promoter (pYX232; Ingenious Inc.) was utilized. The NcoI site within the polylinker of pYX232 was converted to an NdeI site, and the NdeI-SalI fragments from the pSE-Est2-C874 series of plasmids were inserted between the NdeI and SalI sites of the resulting vector (52).
Comparative sequence analysis.
All sequences used in comparative analysis were obtained from the NCBI website at http://www.ncbi.nlm.nih.gov. The final alignment was generated from a hidden Markov model trained with a combination of automated and semiautomated model-building procedures. The UCSC SAM software suite was used for all hidden Markov model work (http://www.cse.ucsc.edu/compbio/sam.html).
Analysis of telomere length.
The est2-Δ and W303a strains were transformed with the pSE and pYX series of plasmids, respectively. Independent clones were passaged on plates or in liquid cultures. Chromosomal DNAs were then isolated with the smash and grab protocol, digested with PstI, and electrophoretically separated on a 0.9% agarose gel. Following capillary transfer to nylon membranes, telomere-containing fragments were detected by hybridization with a 32P-labeled poly(dG-dT) probe (52).
Purification of and assay for yeast telomerase.
Whole-cell extracts and IgG-Sepharose-purified telomerase were prepared as previously described (10, 36, 45, 52). Each primer extension assay was carried out with 20 μl of IgG-Sepharose pretreated with 4 mg of protein extract and was initiated by the addition of a 20-μl cocktail containing 100 mM Tris-HCl, pH 8.0, 4 mM magnesium chloride, 2 mM dithiothreitol, 2 mM spermidine, primer oligodeoxynucleotides, and various combinations of labeled and unlabeled dGTP and dTTP. Primer extension products were processed and analyzed by gel electrophoresis as previously described (36, 37). The oligodeoxynucleotide primers used for telomerase assays were purchased from Sigma-Genosis and purified by denaturing gel electrophoresis prior to use. The primers have the following sequences: TEL10, GGGTGTGGTG; TEL15, TGTGTGGTGTGTGGG; OXYT1, GTTTTGGGGTTTTGGG; TEL15(m5), TGTGTGGTGTCTGGG; and TEL19, TGTGTGGTGTGTGGGTGTG.
For determination of the processivity of substitution mutants, assays were performed with OXYT1 or TEL15(m5) as the primer. The signal for each product was determined by PhosphorImager (Molecular Dynamics) and normalized to the amount of transcript by dividing against the number of labeled residues. The OXYT1 primer ends in 3 G's and can align to only one site along the yeast RNA template, supporting the addition of a defined sequence (TGTGGTG) up to the 5′ boundary of the template. For nucleotides beyond the +7 position, we assume a sequence of TGTG…. Calculations were made assuming other compatible sequences, and the conclusions were not altered:
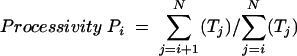 |
where Ti denotes the amount of transcript calculated for the primer + i position and N is the highest number at which a visible signal can be discerned in the PhosphorImager file for the primer + N product.
Kinetic analysis was performed with a range of primer concentrations and a time point within the linear range of product accumulation. The amounts of products were quantified by comparing the “volume” derived from scanning of PhosphorImager plates to those from known amounts of radioactivity. In control scanning experiments, we spotted known quantities of radioactive label on filter paper, exposed a PhosphorImager plate to the paper for 16 h, and scanned the plate to obtain the volume derived from the radioactive spots. From such experiments, we estimate that 1 fmol of label (3,000 Ci/mmol) gives rise to a volume of 650,000 in 16 h. Thus, we were able to calculate the molar amount of the product from the scan. The reaction rates were then calculated (by dividing the product against the duration of the reaction), plotted against primer concentrations, and the data points were fitted to the Michaelis-Menton equation with a nonlinear regression algorithm (Prism, GraphPad Software, Inc., San Diego, Calif.).
Protein and RNA analysis.
The levels of protein A-tagged yeast TERT in cell extracts were determined by enrichment on IgG-Sepharose and subsequent Western blotting as previously described (52). RNase protection studies were carried out as follows. A PCR fragment that spans nucleotides 1 to 1301 of the TLC1 gene (48) was generated and cloned between the BamHI and EcoRV sites of pBluescript II KS+ to give pBS-TLC1. For the synthesis of labeled antisense probe, pBS-TLC1 was linearized by digestion with HinfI and transcribed with T3 RNA polymerase in the presence of 12 μM [α-33P]GTP as described (35). For the protection assays, total RNAs from IgG-Sepharose beads were combined with the probe (100,000 cpm), and the mixtures were precipitated with ethanol. The RNAs were then hybridized in 80% formamide, digested successively with RNase T1, RNase A, and proteinase K, and analyzed by gel electrophoresis as described (39).
RESULTS
Conserved TERT-specific residues are required for telomere maintenance.
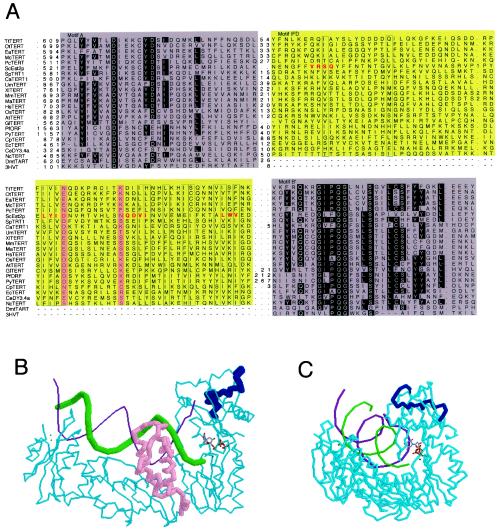
TERTs are distinguishable from conventional RTs in having sizable insertions in the region between conserved motifs A and B′ (Fig. 1A ) (33, 41); whereas the typical distance between the two motifs is 20 residues in retroviral RTs, it ranges from about 70 to 120 residues in TERTs. In the context of the three-dimensional structural model of HIV-1 RT, this region is located in the fingers domain, in close proximity to the nucleotide triphosphates, the 5′ end of the template, and the 3′ end of the primer (Fig. 1B and 1C). We therefore named the extra segments in TERTs IFD for insertion in fingers domain. A hidden Markov model-based alignment of the IFDs of TERTs revealed moderately conserved residues (Fig. 1A).
FIG. 1.
Hidden Markov model-generated multiple sequence alignment for the IFD region of known and predicted telomerase reverse transcriptases (TERT), and putative location of IFD in the context of HIV-1 RT. (A) Motifs A and B′ are shaded in gray, and highly conserved residues are shown as white letters on a black background. The IFD region is shaded in yellow, and well-conserved hydrophobic positions are boxed. Two conserved charged/hydrophilic positions are shown with a pink background. Red letters indicate the locations of the S. cerevisiae mutations examined in this work. Numbers indicate the number of residues not depicted explicitly. The sequences and their database codes are as follows: TtTERT, Tetrahymena thermophila TERT [TERT_TETTH]; OtTERT, Oxytricha trifallax TERT [TERT_OXYTR]; EaTERT, Euplotes aediculatus TERT [TERT_EUPAE]; McTERT, Moneuplotes crassus TERT [AAM95622]; PcTERT, Paramecium caudatum TERT [Q9GRC5]; ScEst2p, Saccharomyces cerevisiae TERT [TERT_YEAST]; SpTRT1, Schizosaccharomyces pombe TERT [TERT_SCHPO]; CaTERT1, Candida albicans TERT 1 [Q9P8T3]; UmTERT, Ustilago maydis TERT; XlTERT, Xenopus laevis TERT [Q9DE32]; MmTERT, Mus musculus TERT [TERT_MOUSE]; MaTERT, Mesocricetus auratus TERT [Q9QXZ4]; HsTERT, Homo sapiens TERT [TERT_HUMAN]; OsTERT, Oryza sativa TERT [Q9AU13]; AtTERT, Arabidopsis thaliana TERT [Q9SE99]; GlTERT, Giardia lamblia TERT [Q9NCP5]; PfORF, Plasmodium falciparum 3D7 hypothetical protein PF13_0080 [NP_705050]; PyTERT, Plasmodium yoelii TERT [EAA16235]; CpTERT, Cryptosporidium parvum TERT [AAK60396]; EcTERT, Encephalitozoon cuniculi TERT [NP_596954], CeDY3.4a, Caenorhabditis elegans TERT; NcTERT, Neurospora crassaTERT; DmtTART, Drosophila melanogaster TART retrotransposon RT; and 3HVT, HIV-1 RT. (B) A structural model of HIV-1 RT P66 in a complex with DNA and a nucleotide triphosphate is shown. The protein backbone, the template strand, and the primer strand are shown in cyan, purple, and green, respectively. The thumb domain of the protein is shown in pink. Residues 120 to 140 of HIV-1 RT, which correspond to the region between motifs A and B′, are shown in dark blue. The nucleotide substrate is depicted by sticks. The enzyme is trapped in the closed transcribing conformation. (C) The same structural model of the complex is shown in a different orientation. Color codes are the same as in B except that the thumb domain is shown in the same color as the rest of the protein.
To assess the functional importance of IFD, we created a series of substitution mutants in Est2p (Saccharomyces cerevisiae TERT), named VRSQ559, LYID589, QDVI602, and LWV617. Each mutant contains three to four alanine substitutions and is designated by the identity of the altered residues and the position of the first altered residue. Each mutant was tagged at the C terminus with tandem copies of the IgG-binding domain of protein A and then introduced into a yeast strain whose chromosomal EST2 had been disrupted. A similarly tagged wild-type EST2 gene (named C874) was analyzed in parallel as the control.
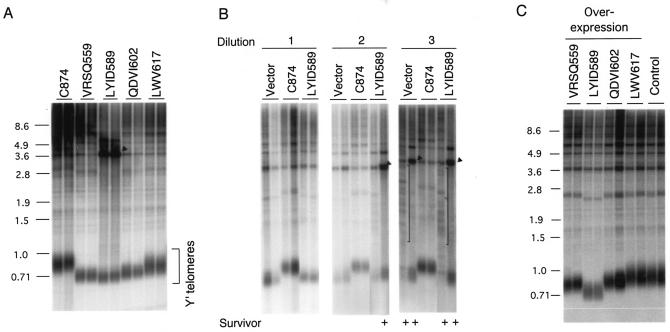
All strains were first tested for a telomere maintenance defect. Chromosomal DNAs were prepared from cells approximately 50 generations posttransformation and subjected to Southern analysis of terminal restriction fragments. As shown in Fig. 2A, all four mutants exhibited various degrees of telomere length defect. The relative severities of the defect for the mutants were as follows: LYID589 > QDVI602 ≈ VRSQ559 > LWV716. Both isolates of the LYID589 mutant showed amplification of the subtelomeric Y′ fragment (indicated by a triangle, Fig. 2A), consistent with the presence of type I survivors. To determine if the LYID589 mutant was completely nonfunctional, we compared the telomere pattern of individual transformants that had been passaged in liquid culture by serial dilution and that contained either an empty vector, wild-type EST2 (C874), or LYID589. As shown in Fig. 2B, the clones containing the LYID589 mutant gave rise to survivors with kinetics similar to those containing the empty vector, consistent with a complete loss of function for this mutant. Thus, some residues in IFD appear to be essential for telomere maintenance in vivo.
FIG. 2.
Mutations in the IFD region impair telomere maintenance. (A) Telomere lengths were determined for strains bearing wild-type or mutated TERT after the strains had been restreaked on the plate once. The identities of the mutants are indicated at the top of the panel. The location of Y′-type telomeres is indicated by brackets. The amplified Y′ fragments that are often observed in type I survivors are marked by a solid triangle. (B) Telomere lengths were determined for strains bearing wild-type EST2 (C874), the LYID589 mutant, or an empty vector after the strains were passaged in liquid culture by serial dilution (1:500 for each dilution). The amplified Y′ fragments that are often observed in type I survivors are marked by solid triangles. The heterogeneous telomere fragments observed in type II survivors are marked by vertical brackets. Samples that exhibit features of survivors are marked with + at the bottom. (C) Telomere lengths were determined for W303a strains with plasmids that overexpressed individual IFD mutant proteins. DNAs were isolated from clones that had been restreaked twice on plates after initial transformation. A strain that does not contain any overexpression plasmid (Control) was tested in parallel.
To assess the basis for the loss of function exhibited by the IFD mutants, we investigated the ability of individual mutant proteins to induce telomere shortening when overexpressed in a wild-type strain. If the mutant proteins are defective in stability or assembly, they should not compete with endogenous Est2p for association with telomerase RNA and should not act in a dominant-negative fashion even when overexpressed. Earlier analysis indicated that overexpression of wild-type Est2p does not alter telomere lengths (52). In contrast, as shown in Fig. 2C, all of the mutants except LWV617 caused various degrees of telomere shortening in the overexpression assay. Moreover, the extent of telomere shortening provoked by overexpression of the mutants correlated with those observed in the original telomere maintenance assay, with the LYID589 mutation causing the greatest shortening in both assays. Taken together, our observations imply that the telomere maintenance defects of at least three of the IFD mutants are due to a loss of function rather than stability or assembly.
Various effects of the mutations on protein and RNP levels.
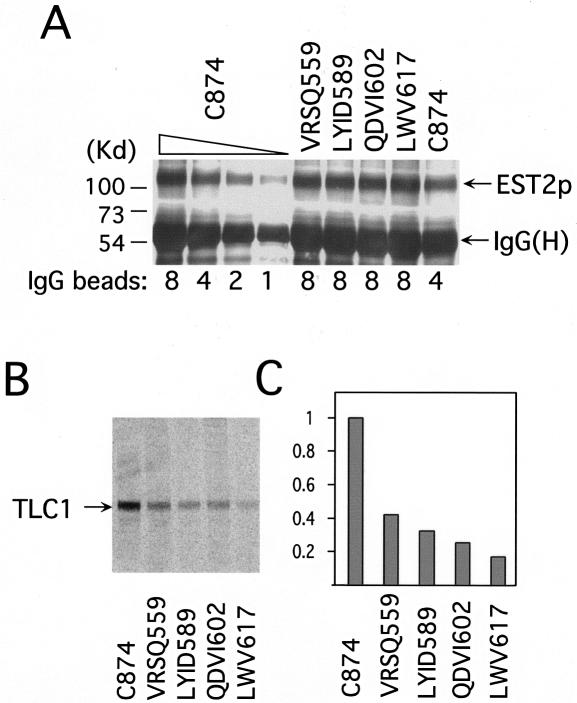
To directly test the effects of the IFD mutations on TERT protein stability, we measured the levels of the mutant proteins by enrichment on IgG-Sepharose and subsequent Western analysis with antibodies directed against the protein A tag. As shown in Fig. 3A, all of the mutant proteins were present at levels comparable to that of the wild-type protein, suggesting that the mutants are not grossly destabilized. We also determined the amount of TERT-associated TLC1 RNA by precipitating the RNP with IgG-Sepharose and then performing RNase protection assays on the resulting precipitates. As shown in Fig. 3B, the levels of TLC1 RNA were reduced by two- to threefold for the VRSQ559, LYID589, and QDVI602 mutants and by ≈5-fold for the LWV617 mutant. Together with the overexpression study, these results suggest that of the four IFD mutants, the LWV617 mutant may have the most significant defect in RNP assembly, whereas the other mutants (VRSQ559, LYID589, and QDVI602) are likely to have functional defects.
FIG. 3.
Mutations in the IFD region lead to reductions in the level of TERT-associated RNA. (A) Levels of protein A-tagged TERT in the mutant extracts were determined by enrichment on IgG-Sepharose and subsequent Western blotting with antibodies directed against protein A and compared to that of the wild-type extract. The identities of the mutants are indicated at the top, and the relative amounts of beads loaded are indicated at the bottom. The position of the protein A-tagged Est2p and the IgG heavy chain are marked by arrows on the right. (B) The levels of TERT-associated TLC1 RNA in the wild-type and mutant strains were determined by RNase protection assays. The identities of the mutants are indicated at the bottom. The position of the protected TLC1 fragment is indicated by an arrow on the left. (C) The levels of TERT-associated TLC1 RNA in the mutant strains relative to the wild-type strain were determined by assays such as those shown in B, and the average values from two experiments were plotted.
Various effects of IFD mutations on telomerase activity in vitro.
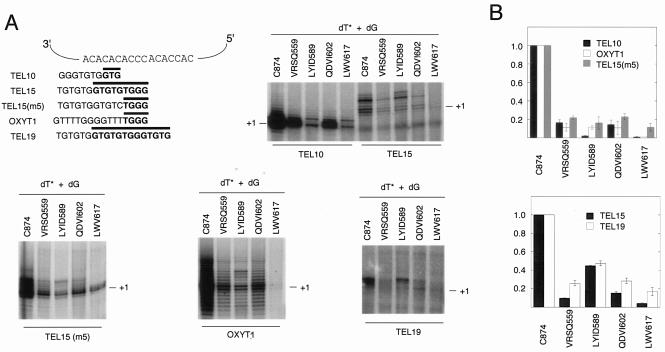
To assess potential alterations in enzyme activity, we precipitated protein A-tagged telomerase from the wild-type and mutant strains by adsorption to IgG-Sepharose and subjected the beads to primer extension analysis. The sequence and length of the DNA primer are known to affect the polymerization properties of yeast telomerase (36, 47). To detect such sequence- or length-specific defects, we examined the behavior of the wild-type and mutant enzymes with five different primers (Fig. 4A). As shown in Fig. 4A and 4B, all of the mutants exhibited substantial activity loss beyond those predicted by the moderate reduction in RNP levels. For example, the VRSQ559 and QDVI602 extracts consistently manifested a ≈10-fold reduction in total activity relatively to the wild-type enzyme despite having RNP levels that are about one half to one third of the wild-type extract. Likewise, the LWV617 extract can exhibit ≈1/100 of the wild-type activity despite having ca. one fifth of the wild-type RNP level. These results indicate that all of the IFD mutants have reduced specific activities and are in fact functionally defective.
FIG. 4.
Mutations in the IFD region result in the loss of telomerase activity. (A) Telomerase from the wild-type and various mutant strains was isolated by IgG affinity chromatography and tested in primer extension assays with several different oligonucleotides (5 μM) as the primer and [32P]dTTP (0.2 μM) and dGTP (50 μM) as the nucleotides. The sequences of the primers and how they can align with the template region of telomerase RNA are illustrated in the upper left portion of the figure. The portion of each primer that can form an uninterrupted hybrid with telomerase RNA is represented by a thick horizontal bar. Primer TEL10 can align to multiple positions along the template, with each alignment position supporting the formation of a 3- or 4-bp hybrid. For illustrative purposes, we show just one such potential alignment position. Representative assays are shown in five different panels. The identities of the mutations are indicated at the top and the location of the primer +1 (+1) product indicated by horizontal lines at the right. (B) Total DNA synthesis mediated by each mutant enzyme was determined in assays such as those presented in part A, normalized against that mediated by the wild-type enzyme, and the results were plotted. The averages of two independent assays and the deviations are shown in the plots.
Defects of the LYID589 mutants: changes in primer utilization and altered elongation properties.
Remarkably, the defect of the LYID589 mutant was primer dependent (Fig. 4A and 4B). With either TEL15 or TEL19 as the primer, the mutant activity was about half of the wild-type level. With TEL15(m5), OXYT1, or TEL10 as the primer, the mutant activity ranged from 1 to 10% of the wild-type level. The difference between the two groups of primers appears to be their potential for forming RNA-DNA duplexes with telomerase RNA, with the first group forming hybrids of 9 to 13 bp and the second group forming hybrids of 3 to 4 bp. This result raises the interesting possibility that the LYID residues replaced in the mutant are especially important for telomerase function when the RNA-DNA hybrid is short.
Close inspection of the reaction products revealed a second interesting feature of the LYID589 mutant: altered elongation. The alteration was most evident in reactions utilizing OXYT1. In particular, the LYID589 mutant appears to preferentially accumulate product at the primer +7 position (Fig. 5B, lane 3, marked by a triangle). This visual impression was confirmed by quantitative analysis of the distribution of extension products. In particular, the relative intensities of products derived from the LYID589 mutant enzyme that are longer or shorter than primer +7 were all reduced relative to the wild-type enzyme (Fig. 5C). The primer +7 product is generated upon one complete round of template copying, when the active site of the enzyme reaches the 5′ boundary of the template (Fig. 5A). Accumulation of this product can be explained by a defect in type II translocation or more efficient type I translocation (from the primer +1 to primer +6 positions). To distinguish between these alternatives, we assessed the processivity of wild-type and LYID589 telomerase at each position along the template. Interestingly, as shown in Fig. 5D, both alternatives appeared to contribute to changes in the elongation pattern of the mutant. For example, the processivity of the mutant enzyme at the primer +7 position was reduced by ≈60%, while that at the primer +3, primer +4, and primer +5 positions were increased by ≈15 to 25%. Similar results were obtained in experiments performed with the TEL15(m5) primer (data not shown).
FIG. 5.
Defective type II translocation for the LYID589 mutant. (A) The sequence of the OXYT1 primer, how it aligns with the telomerase RNA template, the expected nucleotide addition, and a type II translocation reaction are illustrated. (B) Telomerase from the wild-type and various mutant strains was isolated by IgG affinity chromatography and tested in primer extension assays with OXYT1 as the primer, and [32P]dTTP (0.2 μM) and dGTP (50 μM) as the nucleotides. The identities of the mutations are shown at the top, and the locations of the primer +1 (+1) and primer +7 (+7) products are indicated by horizontal lines to the right. The preferential accumulation of the primer +7 product by the LYID589 mutant is highlighted by a solid triangle. To allow easier visual comparison of the banding pattern, a shorter exposure (1X) of the lane containing wild-type products is shown in the rightmost lane. (C) The relative intensities of the extension products for the C874 and LYID589 telomerases from the primer +1 to +11 positions are plotted. The scales for the two reactions are set so that the primer +7 bars are of equal height. This results in better visual appreciation of the fact that the mutant enzyme preferentially accumulates products at this position. (D) The processivity of the C874 and LYID589 telomerases from the primer +1 to +7 positions was determined in duplicate assays, and the results were plotted.
In contrast to the LYID589 mutant, the banding patterns of the VRSQ559 and QDVI602 mutant were basically indistinguishable from the wild-type enzyme (Fig. 5B, compare lanes 2, 4, and 5). Quantitative analysis of the distributions of products also failed to reveal any significant changes in the elongation properties of these two mutants (data not shown). The elongation property of the LWV617 mutant was not analyzed because of weak product intensities.
LYID589 mutant exhibits reduced binding to DNA.
To test the notion that the LYID589 mutant is selectively impaired in binding primers that form short hybrids with telomerase RNA, we attempted to measure binding of the RNP to the OXYT1 primer with a previously described gel mobility shift assay (37). However, no complex could be identified even with the use of the wild-type enzyme, possibly because of the low stability of such a complex (data not shown). Attempts to determine the off-rate of the complex by primer challenge were also unsuccessful, probably for the same reason (data not shown).
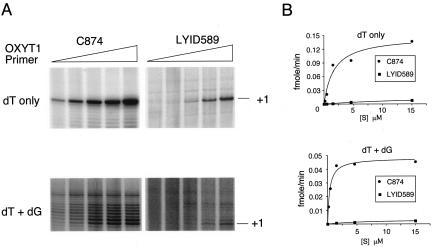
As an alternative method of assessing telomerase-primer binding, we determined the Kms of the wild-type and mutant enzymes for OXYT1 with various concentrations of the primer in primer extension assays. Because of low activity levels of yeast telomerase, it was necessary to use nonsaturating concentrations of the labeled nucleotide in these assays. To rule out the possibility that nucleotide concentrations may affect the results, we conducted three sets of analysis with different combinations and concentrations of the nucleotide substrates. As shown in Fig. 6 and Table 1, the LYID589 telomerase consistently displayed higher apparent Kms for OXYT1 than the wild-type enzyme. For example, when dTTP alone (at 0.5 μM) was used for the assays, the LYID589 mutant exhibited a 9-fold higher Km. When 1 μM dTTP and 50 μM dGTP were included in the assays, the difference in Km between the wild-type and the mutant enzyme was more than 15-fold. Assuming Michaelis-Menton kinetics, these results suggest that the binding of the LYID589 mutant to OXYT1 (a primer that forms a short hybrid with telomerase RNA) is indeed substantially weaker than that of wild-type telomerase.
FIG. 6.
Reduced primer binding for the LYID589 mutant. (A) Telomerases derived from strains carrying the C874 or LYID589 plasmid were tested for primer extension activity in vitro with increasing concentrations of the OXYT1 primer. The assays shown in the top panels were performed in the presence of 0.5 μM dTTP (dT only), whereas those shown in the bottom panel were done in the presence of 1 μM dTTP and 50 μM dGTP (dT+dG). For each set of assays, the concentrations of the primer were 0.15, 0.46, 1.5, 4.6, and 15 μM (left to right). (B) The signals from the assays shown in part A were quantified, converted to reaction rates (see Materials and Methods for a more detailed explanation), and plotted against primer concentration at the bottom. The data points were fitted to the Michaelis-Menton equation, and the resulting curves are shown.
TABLE 1.
Apparent Kms for the OXYT1 primer in reactions containing different nucleotide combinations and concentrations
| Nucleotide combination |
Km (μM)
|
|
|---|---|---|
| C874 | LYID589 | |
| 1 μM dTTP + 50 μM dGTP | 0.36 | 6.48 |
| 0.2 μM dTTP + 50 μM dGTP | 0.91 | 3.95 |
| 0.5 μM dTTP | 1.70 | 15.2 |
DISCUSSION
Mechanisms of IFD.
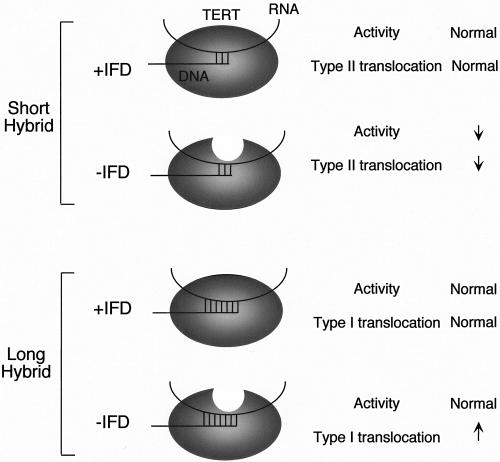
We showed in this report that a telomerase-specific region within the catalytic domain of TERT, named IFD, is required for telomerase function both in vivo and in vitro. Most remarkably, a four-amino-acid substitution in this region (resulting in the LYID589 mutant) caused complex changes in the primer utilization and elongation properties of telomerase. These changes include (i) a severe defect in extending primers that form short hybrids with telomerase RNA, (ii) reduced type II translocation efficiency, and (iii) enhanced type I translocation efficiency. At first glance, one might be tempted to attribute such a complex phenotype to direct and indirect effects of the mutations on diverse aspects of telomerase structure and function. However, we believe that a more parsimonious explanation can be proposed that attributes all of the enzymatic changes to reduced protein-DNA interactions (as suggested by the kinetic experiments) afforded by the mutant (see Fig. 7 for a graphic illustration of the following discussion.)
FIG. 7.
Interplay between RNA-DNA hybrid and IFD on telomerase activity and translocation property. The discussion concerning the effects of loss of IFD function on telomerase activity and processivity is summarized. Loss of IFD is illustrated by an oval (TERT) missing a top portion. See the Discussion section for a more detailed explanation.
We surmise that the overall affinity of the telomerase enzyme for DNA is a function of both RNA-DNA and protein-DNA interactions and that a threshold level of affinity is required for efficient binding and polymerization. In the presence of a short RNA-DNA hybrid, the protein-DNA interaction disrupted by the LYID589 mutation may be critical for this threshold level of affinity, accounting for the primer-specific impairment of activity. Because type II translocation and repeat addition processivity also entail the formation of a short hybrid (Fig. 5A), an impact of the LYID589 mutation can be readily explained by the same hypothesis. Interestingly, an N-terminal deletion mutant of human TERT simultaneously manifests reduced processivity and altered primer utilization, again suggesting that these properties can be coupled (1). The notion that the affinity of telomerase for DNA after type II translocation is important for repeat addition processivity is supported by the recent finding that the length of the RNA-DNA hybrids after type II translocation is an important determinant of mouse and human telomerase processivity in vitro (8).
Can a reduction in binding affinity stimulate more efficient type I translocation? Though less intuitively obvious, this idea has precedence in an earlier analysis of Kluyveromyces lactis telomerase (16). Specifically, the nucleotide addition processivity of K. lactis telomerase was shown to be impaired by increasing the length of complementarity between the DNA primer and RNA template. This observation suggests that excessively tight binding between the enzyme and DNA may hinder elongation. Perhaps the energy barrier for conformational transition that is necessary for nucleotide addition becomes higher with a stabler telomerase-DNA complex. In light of this idea, it is interesting that the stimulatory effect of the LYID589 mutation on type I translocation was evident only at or beyond the primer +3 position (for the OXYT1 primer) as the RNA-DNA hybrid became longer than 6 nucleotides. We surmise that with longer hybrids, the additional binding contributed by the LYID589 residues may result in an excessively stable telomerase-DNA complex that cannot elongate efficiently. Thus, the same protein-DNA interaction can have diametrically opposite effects on type I and II translocation, consistent with the different energetic requirements for these two types of translocation reactions (34, 45).
How might the insertion in the fingers domain of TERT optimize DNA binding? In comparison with other DNA polymerases, conventional RTs such as HIV-1 RT appear to have rather “stubby” fingers. Thus, even in the closed, transcribing conformation illustrated in Fig. 1B and 1C, one face of the RNA-DNA hybrid is open to solution (27). We imagine that the longer and more elaborate fingers of TERT may make additional contact with the RNA-DNA duplex on this open face to enhance stability. In fact, it is possible that the tip of the fingers may extend sufficiently to make contact with the putative thumb domain of TERT, thus fully encircling the substrate. This type of full closure may in turn augment enzyme-substrate stability.
The notion that IFD is an important determinant of primer binding is supported by recent findings in Euplotes crassus (28). Three different TERT genes can be identified in this ciliated protozoan. The expression of one of the TERT genes (named EcTERT-2) is specifically upregulated during macronuclear development, a period of de novo telomere formation. Telomerase isolated from this developmental stage exhibits a remarkably distinct primer recognition property: an ability to elongate nontelomeric primers (3, 19). Interestingly, EcTERT-2 differs from the other TERT genes in having a very divergent amino acid sequence in the IFD region, suggesting that this region is responsible for the altered primer recognition property. On the other hand, the residues altered in the LYID589 mutants are well conserved between the E. crassus TERTs, arguing that these particular residues per se do not mediate the de novo telomere formation activity of EcTERT-2 (data not shown).
It should be stressed that IFD may have functions other than primer recognition. In contrast to the LYID589 mutant, the other three mutants that we analyzed did not exhibit primer-specific defects in vitro, but rather a consistent loss of activity on all primers. One plausible explanation is that these mutants may be defective in nucleotide or template binding, which has been shown to be mediated by the fingers domain of RTs. Impairment of nucleotide or template binding may cause consistent loss of enzyme activity, as observed. Further studies will be necessary to address these possibilities.
Implications for telomere extension in vivo and telomerase evolution.
It is not immediately obvious why the LYID589 mutant should behave as a null mutant in vivo. The rate of telomere loss in the absence of telomerase is on average only about 3 to 4 bp per generation, and the LYID589 mutant appears fully capable of adding this number of nucleotides, especially given the right substrate. However, others have shown that telomerase preferentially mediates the extension of short telomeres (15, 38), which presumably entails the addition of multiple repeats, possibly through processive repeat addition. In this scenario, the LYID589 mutant would be unable to maintain telomeres because it is defective in type II translocation. Our results thus provide the first mutational evidence that the repeat addition processivity of telomerase may be essential in vivo. However, it is also possible that the synthesis of long telomeric DNA in vivo is due to multiple cycles of binding, polymerization, and dissociation (i.e., without processive repeat addition). In this scenario, the LYID589 mutant may be nonfunctional because it is unable to rebind the telomere ends with the 3′ end of the template after one round of synthesis to mediate the addition of more nucleotides. Better understanding of the in vivo dynamics of telomere addition would be required to distinguish between these alternatives. Finally, it is worth noting that telomerase has been proposed to have a protective function at telomeres (47, 49, 53), which may also be compromised by the LYID589 mutation.
The opposite effects of the LYID589 mutation on type I and II translocation raise the interesting possibility that the activity of IFD represents an evolutionary compromise between the competing demands of these two types of telomerase translocation. An IFD that binds DNA too weakly might abolish type II translocation, permitting only one round of synthesis. An IFD that binds too tightly might impair type I translocation sufficiently to prevent even a complete round of DNA synthesis. The negative effect of IFD on type I translocation may be especially significant for telomerases with long RNA templates (such as the S. cerevisiae and K. lactis enzymes), which can allow the accumulation of long RNA-DNA hybrids during telomere synthesis. Investigating the effect of comparable IFD mutations on the enzymatic properties of telomerases with short templates should be informative.
Acknowledgments
We thank Dorothy Shippen for communicating results prior to publication. We also thank an anonymous reviewer for insightful comments on processivity of the LYID589 mutant.
This work was supported by an R01 from the National Institutes of Health (N. Lue) and by the California Breast Cancer Research Program (I. S. Mian). The Department of Microbiology and Immunology at Weill Cornell Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation.
REFERENCES
- 1.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 2000. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell 11:3329-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 1998. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8:177-180. [DOI] [PubMed] [Google Scholar]
- 3.Bednenko, J., M. Melek, E. C. Greene, and D. E. Shippen. 1997. Developmentally regulated initiation of DNA synthesis by telomerase: evidence for factor-assisted de novo telomere formation. EMBO J. 16:2507-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackburn, E. H. 1992. Telomerases. Annu. Rev. Biochem. 61:113-129. [DOI] [PubMed] [Google Scholar]
- 5.Bosoy, D., and N. Lue. 2001. Functional analysis of conserved residues in the putative “finger” domain of telomerase reverse transcriptase. J. Biol. Chem. 276:46305-46312. [DOI] [PubMed] [Google Scholar]
- 6.Bryan, T. M., K. J. Goodrich, and T. R. Cech. 2000. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem. 275:24199-24207. [DOI] [PubMed] [Google Scholar]
- 7.Bryan, T. M., J. M. Sperger, K. B. Chapman, and T. R. Cech. 1998. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytrica trifallax. Proc. Natl. Acad. Sci. 95:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J., and C. Greider. 2003. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 22:304-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J. L., M. A. Blasco, and C. W. Greider. 2000. Secondary structure of vertebrate telomerase RNA. Cell 100:503-514. [DOI] [PubMed] [Google Scholar]
- 10.Cohn, M., and E. H. Blackburn. 1995. Telomerase in yeast. Science 269:396-400. [DOI] [PubMed] [Google Scholar]
- 11.Collins, K. 1999. Ciliate telomerase biochemistry. Annu. Rev. Biochem. 68:187-218. [DOI] [PubMed] [Google Scholar]
- 12.Collins, K., and L. Gandhi. 1998. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl. Acad. Sci. 95:8485-8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins, K., R. Kobayashi, and C. W. Greider. 1995. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell 81:677-686. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald, M. S., K. Riha, F. Gao, S. Ren, T. D. McKnight, and D. E. Shippen. 1999. Disruption of the catalytic telomerase subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. 96:14813-14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forstemann, K., M. Hoss, and J. Lingner. 2000. Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res. 28:2690-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fulton, T. B., and E. H. Blackburn. 1998. Identification of Kluyveromyces lactis telomerase: discontinuous synthesis along the 30-nucleotide-long templating domain. Mol. Cell. Biol. 18:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandhi, L., and K. Collins. 1998. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes Dev. 12:721-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg, R. A., R. C. Allsopp, L. Chin, G. B. Morin, and R. A. DePinho. 1998. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene 16:1723-1730. [DOI] [PubMed] [Google Scholar]
- 19.Greene, E. C., and D. E. Shippen. 1998. Developmentally programmed assembly of higher order telomerase complexes with distinct biochemical and structural properties. Genes Dev. 12:2921-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greider, C. 1991. Telomerase is processive. Mol. Cell. Biol. 11:4572-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 22.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 23.Harrington, L. 2003. Biochemical aspects of telomerase function. Cancer Lett. 194:139-154. [DOI] [PubMed] [Google Scholar]
- 24.Harrington, L., T. McPhail, V. Mar, W. Zhou, R. Oulton, A. E. program, M. B. Bass, I. Arruda, and M. O. Robinson. 1997. A mammalian telomerase-associated protein. Science 275:973-977. [DOI] [PubMed] [Google Scholar]
- 25.Holt, S. E., D. L. Aisner, J. Baur, V. M. Tesmer, M. Dy, M. Ouellette, J. B. Trager, G. B. Morin, D. O. Toft, J. W. Shay, W. E. Wright, and M. A. White. 1999. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 13:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hossain, S., S. Singh, and N. Lue. 2002. Functional analysis of the C-terminal extension of telomerase reverse transcriptase: a putative “thumb” domain. J. Biol. Chem. 277:36174-36180. [DOI] [PubMed] [Google Scholar]
- 27.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 28.Karamysheva, Z., L. Wang, T. Shrode, J. Bednenko, L. Hurley, and D. Shippen. 2003. Developmentally programmed gene elimination in euplotes crassus facilitates a switch in the telomerase catalytic subunit. Cell 113:565-576. [DOI] [PubMed] [Google Scholar]
- 29.Kelleher, C., M. Teixeira, K. Forstemann, and J. Lingner. 2002. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 27:572-579. [DOI] [PubMed] [Google Scholar]
- 30.Kilian, A., D. D. Bowtell, H. E. Abud, G. R. Hime, D. J. Venter, P. K. Keese, E. R. Duncan, R. R. Reddel, and R. A. Jefferson. 1997. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet. 6:2011-2019. [DOI] [PubMed] [Google Scholar]
- 31.Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian, and V. Lundblad. 1996. Senescence mutants of saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lingner, J., and T. R. Cech. 1996. Purification of telomerase from Euplotes aediculatis: requirement of a primer 3′ overhang. Proc. Natl. Acad. Sci. 93:10712-10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 34.Lue, N., 2002. Yeast telomerases: structure, mechanisms and regulation. p. 239-258. In G. Krupp and R. Parwaresh (ed.), Telomerases, telomeres and cancer. Landes Biosciences, Georgetown, Tex.
- 35.Lue, N. F., and R. D. Kornberg. 1987. Accurate initiation at RNA polymerase II promoters in extracts from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 84:8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lue, N. F., and Y. Peng. 1998. Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 26:1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lue, N. F., and J. Xia. 1998. Species-specific and sequence-specific recognition of the dG-rich strand of telomeres by yeast telomerase. Nucleic Acids Res. 26:1495-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcand, S., V. Brevet, and E. Gilson. 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12:7035-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyerson, M., C. M. Counter, E. N. Eaton, L. W. Ellisen, P. Steiner, S. D. Caddle, L. Ziaugra, R. L. Beijersbergen, M. J. Davidoof, and Q. e. a. Liu. 1997. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90:785-795. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955-959. [DOI] [PubMed] [Google Scholar]
- 42.Nakayama, J.-I., M. Saito, H. Nakamura, A. Matsuura, and F. Ishikawa. 1997. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell 88:875-884. [DOI] [PubMed] [Google Scholar]
- 43.Nugent, C. I., and V. Lundblad. 1998. The telomerase reverse transcriptase: components and regulation. Genes Dev. 12:1073-1085. [DOI] [PubMed] [Google Scholar]
- 44.Oguchi, K., H. Liu, K. Tamura, and H. Takahashi. 1999. Molecular cloning and characterization of AtTERT, a telomerase reverse transcriptase homolog in Arabidopsis thaliana. FEBS Lett. 457:465-469. [DOI] [PubMed] [Google Scholar]
- 45.Peng, Y., I. S. Mian, and N. F. Lue. 2001. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell 7:1201-1211. [DOI] [PubMed] [Google Scholar]
- 46.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 47.Prescott, J., and E. H. Blackburn. 1997. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal non-processivity in vivo and in vitro. Genes Dev. 11:528-540. [DOI] [PubMed] [Google Scholar]
- 48.Singer, M. S., and D. E. Gottschling. 1994. TLC1:template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 49.Singh, S., O. Steinberg-Neifach, I. Mian, and N. Lue. 2002. Analysis of telomerase in Candida albicans: potential role in telomere end protection. Eukaryot. Cell 1:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tzfati, Y., T. B. Fulton, J. Roy, and E. H. Blackburn. 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288:863-867. [DOI] [PubMed] [Google Scholar]
- 51.Weinrich, S. L., R. Pruzan, L. Ma, M. Ouellette, V. M. Tesmer, S. E. Holt, A. G. Bodnar, S. Lichtsteiner, N. W. Kim, J. B. Trager, R. D. Taylor, R. Carlos, W. H. Andrews, W. E. Wright, J. W. Shay, C. B. Harley, and G. B. Morin. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17:198-502. [DOI] [PubMed] [Google Scholar]
- 52.Xia, J., Y. Peng, I. S. Mian, and N. F. Lue. 2000. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase Mol. Cell. Biol. 20:5196-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, J., H. Wang, J. M. Bishop, and E. H. Blackburn. 1999. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc. Natl. Acad. Sci. 96:3723-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]