Abstract
Salmonella enterica subspecies 1 serovar Typhimurium encodes a type III secretion system (TTSS) within Salmonella pathogenicity island 1 (SPI-1). This TTSS injects effector proteins into host cells to trigger invasion and inflammatory responses. Effector proteins are recognized by the TTSS via signals encoded in their N termini. Specific chaperones can be involved in this process. The chaperones InvB, SicA, and SicP are encoded in SPI-1 and are required for transport of SPI-1-encoded effectors. Several key effector proteins, like SopE and SopE2, are located outside of SPI-1 but are secreted in an SPI-1-dependent manner. It has not been clear how these effector proteins are recognized by the SPI-1 TTSS. Using pull-down and coimmunoprecipitation assays, we found that SopE is copurified with InvB, the known chaperone for the SPI-1-encoded effector protein Sip/SspA. We also found that InvB is required for secretion and translocation of SopE and SopE2 and for stabilization of SopE2 in the bacterial cytosol. Our data demonstrate that effector proteins encoded within and outside of SPI-1 use the same chaperone for secretion via the SPI-1 TTSS.
Type III secretion systems (TTSS) have been identified in many pathogenic and symbiotic gram-negative bacteria (34). TTSS allow the bacteria to secrete and inject bacterial toxins (effector proteins) directly into the cytosol of host cells, where the toxins induce responses which are beneficial for the bacterium. However, how the effector proteins are recognized and transported into host cells by TTSS is still poorly understood.
Due to the presence of two independent signals, analysis of effector protein recognition by TTSS has been complicated. The first signal is located at the N terminus of the effector protein. Some workers have suggested that this signal is located within the first ∼15 amino acids (aa) of the secreted polypeptide (43), while others have argued that the mRNA sequence at the 5′ end of the open reading frame (ORF) represents the secretion signal (1). This first signal does not depend on accessory proteins designated chaperones (1, 43, 63). The second signal found in effector proteins is chaperone dependent (6, 70). It represents the chaperone binding site and is generally located between aa 15 and 70 to 140 of the secreted protein (1, 42, 44, 71, 72).
The type III secretion chaperones have some common features, although they do not have sequence similarities. They are small acidic proteins with a predicted amphipathic α-helix at the C terminus. Chaperones generally bind to the N-terminal regions of secreted proteins (aa 15 to 140) in the bacterial cytoplasm, which results in protection from degradation, prevention of premature interactions, and/or mediation of recognition by the TTSS (3, 4, 7, 9, 46, 64).
Salmonella enterica subspecies I serovar Typhimurium is a gram-negative enteropathogen which is responsible for a large number of gastrointestinal infections in the human population. Among many other virulence factors, Salmonella serovar Typhimurium encodes two TTSS which are expressed at different stages of the disease (22, 26, 32). The TTSS encoded in Salmonella pathogenicity island 1 (SPI-1) is required for induction of proinflammatory responses, invasion of intestinal epithelial cells, induction of cell death in macrophages, and elicitation of diarrhea (22, 60, 69).
So far, 12 Salmonella serovar Typhimurium effector proteins which are transported via the SPI-1 TTSS have been identified (22). In contrast to the proteins of many other enteric pathogens, only some of the Salmonella effector proteins (Sip/SspA, Sip/SspB, Sip/SspC, SptP, and AvrA) are encoded in the vicinity of the TTSS apparatus. Many additional effector proteins (SopE, SopE2, SopA, SopB/SigD, SopD, SlrP, and SspH1) are encoded elsewhere in the chromosome (2, 31, 37, 48, 66, 67, 73-75). So far, there is little information about how expression and specific transport of the latter group of effector proteins via the SPI-1 TTSS are controlled.
Chaperones have been described for several SPI-1-encoded effector proteins. The effector proteins Sip/SspB and Sip/SspC and their cognate chaperone SicA (68), Sip/SspA and its chaperone InvB (5), and SptP and its chaperone SicP (21) are all encoded in SPI-1. In the case of SipB/C-SicA and SptP-SicP the proteins are even encoded in the same operon. Similarly, the effector protein SopB/SigD and its specific chaperone PipC (SigE) are encoded next to each other in SPI-5 (12, 73). However, it is not clear whether the other effector proteins, most of which are encoded outside of SPI-1, require chaperones and where the chaperones are encoded.
In the case of the effector protein SopE this was especially interesting because SopE is encoded by the temperate P2-like bacteriophage SopEφ (50). This phage frequently infects new Salmonella serovar Typhimurium strains, which are normally sopE negative, and thereby introduces SopE as an additional effector protein by lysogenic conversion (50). Interestingly, SopEφ does not have an ORF with the properties typical of a TTSS chaperone (C. Pelludat and W.-D. Hardt, unpublished data). Considering the high frequency of horizontal gene transfer of sopE between different Salmonella strains (31, 49, 50, 57), we wondered how SopE is recognized by the SPI-1 TTSS.
In a pull-down experiment we identified the SPI-1-encoded protein InvB as a SopE binding partner and analyzed the role of InvB in SopE secretion and translocation via the SPI-1 TTSS. The results are discussed below in the context of horizontal transfer of effector proteins between different Salmonella strains and their functional integration into the TTSS.
MATERIALS AND METHODS
Bacterial strains.
Salmonella serovar Typhimurium wild-type strain SL1344 (33) and its isogenic derivatives SB161 (ΔinvG) (38) and SB566 (invC::aphT) (14) (Table 1) were generously provided by J. E. Galán (Yale University, New Haven, Conn.)
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype and relevant markers | Source (reference) |
|---|---|---|
| Salmonella serovar Typhimurium SL1344 strains | ||
| SL1344 | Wild type | Hoiseth and Stocker (33) |
| SB161 | ΔinvG | Kaniga et al. (38) |
| SB566 | invC::aphT | Eichelberg et al. (14) |
| M509 | ΔsopB | Mirold et al. (49) |
| M516 | ΔsopB sopE sopE2 | Mirold et al. (49) |
| M566 | ΔsopE ΔsopE2 ΔsopB ΔsipA | This study |
| M568 | invB::aphT | This study |
| M573 | ΔsopB invB::aphT | This study |
| M574 | ΔsopE ΔsopE2 ΔsopB ΔsipA invB::aphT | This study |
| M590 | ΔinvG invB::aphT | This study |
| M608 | sopE::sopEM45 ΔsopE2 ΔsopB ΔsipA | This study |
| Salmonella serovar Typhimurium ATCC 14028 strains | ||
| CS401 | phoN2 zxx::6251 Tn10d-Cm Strr | Bronstein et al. (5) |
| PB502 | CS401 ΔinvB | Bronstein et al. (5) |
| M622 | CS401 invC::aphT | This study |
| Plasmids | ||
| pM113 | pBAD24 derivative, expression of SpE-GST fusion protein, arabinose-inducible promoter | This study |
| pM672 | pGEX-KG derivative, expression of GST-InvB fusion protein, lac promoter | This study |
| pM136 | pBAD24 derivative, expression of sopEM45, native sopE promoter | Stender et al. (66) |
| pM226 | pBAD24 derivative, expression of sopE2M45, native sopE2 promoter | Stender et al. (66) |
| pM580 | pACYC184 derivative, expression of sipAM45, sopE promoter | This study |
| pM249 | pBAD24 derivative, expression of invB, sopE promoter | This study |
| pM250 | pBAD24 derivative, expression of invB, arabinose-inducible promoter | This study |
| pM438 | pACYC184 derivative, expression of sopEM45, native sopE promoter | This study |
| pM256 | pACYC184 derivative, expression of sopE2M45, native sopE2 promoter | This study |
| pSB1130 | pACYC184 derivative, expression of sopE, native sopE promoter | Hardt et al. (31) |
| pM149 | pACYC184 derivative, expression of sopE2, native sopE2 promoter | Stender et al. (66) |
| pM416 | pBAD24 derivative, expression of sopE1-95 sopE296-240M45, sopE promoter | This study |
| pM417 | pBAD24 derivative, expression of sopE21-95, sopE96-240M45, sopE2 promoter | This study |
| pM185 | pBAD24 derivative, sopE-lacZ transcriptional fusion, sopE promoter | This study |
| pM687 | pBAD24 derivative, sopE2-lacZ transcriptional fusion, sopE2 promoter | This study |
The Salmonella serovar Typhimurium ATCC 14028s derivatives CS401 (5) and PB502 (ΔinvB) (5) were from the S. I. Miller lab (University of Washington, Seattle).
For all functional assays bacteria were grown for 12 h at 37°C in Luria-Bertani (LB) medium supplemented with 0.3 M NaCl, diluted 1:20 into fresh medium, and grown for another 4 h with mild aeration to obtain an optical density at 600 nm (OD600) of 0.8 to 0.9 (SPI-1-inducing conditions). When required, antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 30 μg/ml; and tetracycline, 12 μg/ml.
M566, carrying in-frame deletions of sopB, sipA, sopE, and sopE2, was obtained by sequential allelic exchange in the chromosome of M509 (ΔsopB) (49) by using the suicide vectors pM585, pM608, and pM586 (see below). The suicide vectors were first integrated into the chromosome by homologous recombination. Allelic exchange was completed by a second round of recombination and selection on sucrose plates (39).
To construct M568 (invB::aphT), invB was replaced by a terminatorless aphT gene cassette which conferred kanamycin resistance. The suicide vector pM670 (Tetr) (see below) was introduced into Salmonella serovar Typhimurium SL1344 by conjugation and selection on LB agar plates containing kanamycin, which yielded M568. Strains M573 (ΔsopB invB::aphT), M574 (ΔsopB ΔsipA ΔsopE ΔsopE2 invB::aphT), and M590 (ΔinvG invB::aphT) were constructed by phage P22-mediated transduction of the invB::aphT allele of M568 into M509 (49), M566, and SB161, respectively. M608, carrying chromosomal sopEM45, was constructed by P22-mediated transduction of the sopE::sopEM45 allele (Tetr) of SB875 (31) into M566 (see above).
The ATCC 14028s derivative M622 was created by P22-mediated transduction of the invC::aphT allele of SB566 (14) into CS401 (5).
The gene disruptions and deletions were confirmed by Western blot analyses by using polyclonal antisera directed against SopE (50) and by PCR by using the primers and conditions listed in Table 2.
TABLE 2.
Primers and conditions used to confirm in-frame deletions
| Deletion | Primers | Conditions |
|---|---|---|
| sopB | 5′-CGGGATCCGCGTTACGCAATCACTATC | 33 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 5 min |
| 5′-GCGGCCGCCCGTTGACATCCTCCAGAA | ||
| sopE | 5′-CGGGATCCTCTTGGCGCGTAGTCCTTC | 33 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 4.5 min |
| 5′-TTTGCGGCCGCGCACTGGATACGCTGAACGA | ||
| sopE2 | 5′-CGGGATCCGCGCAGGCGTTTAGAAGACAGTT | 33 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 5 min |
| 5′-GCGGCCGCGTTCCAGCATCAGCCACTTG | ||
| sipA | 5′-GCGGCCGCACCTGGGGTTGAGTCCTAC | 33 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 6 min |
| 5′-CCCGGGACACCAAGGCACGAG | ||
| invB | 5′-GGGCGCCAAGAGAAAAAGATGTC | 33 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1.75 min |
| 5′-CAGGGGTAAAACGCTCAACGATT |
Recombinant DNA techniques.
Cloning of DNA fragments was performed by using standard protocols (59).
Construction of suicide vectors for generating chromosomal in-frame deletions.
To construct a suicide vector for deletion of sipA, the sequences located directly upstream (primers 5′-GCGGCCGCACCTGGGGTTGAGTCCTAC and 5′-TCTAGAAGGGGGCTGAGT CCTTACAC; 33 cycles of 92°C for 30 s, 53°C for 30 s, and 68°C for 3 min) and downstream (primers 5′-TCTAGAGGCCCGGCTTACGAGTC and 5′-CCCGGGACACCAAGGCACGAG; 33 cycles of 92°C for 30 s, 55°C for 30 s, and 72°C for 3 min) of the sipA coding sequence were amplified by PCR, and the PCR products were cloned into pCR-BluntII-Topo (Invitrogen), which yielded pM582 and pM583. The insert of pM582 was cloned into the XbaI sites of pM583, which yielded pM584, and the resulting insert was subcloned into the SmaI/NotI sites of the suicide vector pSB890 (a derivative of pGP704; oriR6K Tetr sacAB) (W.-D. Hardt and J. E. Galán, unpublished data); this yielded the suicide vector pM585, which was used for deletion of sipA.
To construct a suicide vector for deletion of sopE, we amplified the sequences located directly upstream (primers 5′-CGGGATCCTCTTGGCGCGTAGTCCTTC and 5′-GCTCTAGACACGGTAATGATCCTTTTATATGT; 33 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min) and downstream (primers 5′-GCTCTAGACCCTGAACACTGAAAAACCA and 5′-TTTGCGGCCGCGCACTGGATACGCTGAACGA; 33 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min) of the ORF by PCR and cloned the PCR products into pBluescript SKII+ (Stratagene), which yielded pM593 and pM606. The insert of pM606 was cloned via XbaI/NotI into pM593, which yielded pM607, and the resulting insert was subcloned into the BamHI/NotI sites of pSB890; this yielded the suicide vector pM608.
To construct a suicide vector for deletion of sopE2, the sequences located directly upstream (primers 5′-CGGGATCCGCGCAGGCGTTTAGAAGACAGTT and 5′-GCTCTAGAAGTCACGGTAGTTCTCCTTTT; 33 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 2 min) and downstream (primers 5′-GCTCTAGAAATGCCTCCTGATGGTAGTAA and 5′-GCGGCCGCGTTCCAGCATCAGCCACTTG; 33 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 3.5 min) of the ORF were amplified by PCR, and the PCR products were cloned into pBluescript SKII+ (Stratagene), which yielded pM559 and pM560. The insert of pM560 was cloned via XbaI/NotI into pM559, which yielded pM581, and the resulting insert was subcloned into the BamHI/NotI sites of pSB890; this yielded the suicide vector pM586.
To construct a suicide vector for deletion of invB, we amplified the sequences located directly upstream (primers 5′-GCGGCCGCCGCTCTTTCGTCTGGCATTATC and 5′-GCTAGCCAAATGTTGCATAGATCTTTTCCTT; 33 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 2 min) and downstream (primers 5′-GCTAGCCCCGGGTCGCTAATGAGATGAAAACACCTC and 5′-TCTAGACGTGGCGTTATCGGTTACTTCA; 33 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min 20 s) of the ORF by PCR and cloned the PCR products into pCR2.1-TOPO (Invitrogen), which yielded pM666 and pM667. pM667 contained an additional SmaI site, which was in frame with the invB reading frame. The insert of pM666 was cloned into pM667, which yielded pM668. A modified aminoglycoside 3′-phosphotransferase (aphT) gene which lacks a transcription terminator (25) was cloned into the SmaI site of pM668, which yielded pM669, and the resulting insert was subcloned into the NotI/XbaI sites of pSB890; this yielded the suicide vector pM670.
Chromosomal DNA of Salmonella serovar Typhimurium SL1344 was used as a template for each PCR. All inserts were verified by DNA sequence analysis.
SopE-GST fusion protein expression vector pM113.
The glutathione S-transferase (GST) cassette was amplified by PCR (primers 5′-CCGGGAGCTGCATGTGTCAGAGG and 5′-GGGCTAGCAGAAGAGTGTCTTCACCGCGGCTCGAGAATGTCCCCTATACTAGGTTATTGG; template, pGEX-KG [27]) and was cloned via NheI/HindIII into pBAD24 (28), which yielded pM102. A DNA fragment containing the Shine-Dalgarno sequence, the ATG start codon, and SopE aa 1 to 78 was amplified by PCR (template, pSB1136; primers 5′-CCGCGGATCCCGGGTGCCCGGCCCTCAGAT and 5′-GTCTAGAGCTCCTGAAGGAATTCTAATGACAAAAAT) and cloned via EcoRI (Klenow treated)/SacII into BbsI (Klenow treated)/SacII sites of pM102, which yielded pM106. pSB1136 had been constructed by amplification of the SopE coding sequence (template, SL1344 chromosomal DNA; primers 5′-GGTGGAATTCTAATGACAAAAATAACTTTATCTCC and 5′-CCGATCCATGGCACCACCCCCGGGAGTGTTTTGTATATATTTAT) and cloning via EcoRI/NcoI digestion into pSB616, a pBAD24 derivative carrying the M45 epitope tag and kindly provided by J. E. Galán (8).
A fragment of pSB1136 (containing aa 56 to 240 of SopE) was cloned via BbsI/SmaI into pM106, which yielded pM113, which encoded SopE (aa 1 to 240) with a C-terminal GST fusion under control of the arabinose-inducible promoter of pBAD24.
Expression vector for the GST-InvB fusion protein.
The ORF of invB was amplified by PCR (primers 5′-GGAATTCTAATGCAACATTTGGATATCGCTGA and 5′-CCCAAGCTTACGGCGTATTTCACACAGTTCG; 33 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min 20 s; template, SL1344 chromosomal DNA) and cloned into pGEX-KG (27), which yielded pM672, which expressed GST-InvB under control of the lac promoter.
Expression vectors.
pM136 and pM226 are pBAD24 derivatives which express sopE1-240M45 and sopE21-240M45, respectively, under control of their native promoters (approximately 20 copies/cell [66]). pSB1130 and pM149 are pACYC184 derivatives which express sopE1-240 and sopE21-240, respectively, under control of their native promoters (66).
A low-copy-number expression vector for sopEM45 was constructed by subcloning the insert of pM136 (Eco47III/SalI) into the EcoRV and SalI sites of pACYC184 (approximately five copies/cell; NEB), which yielded pM438.
A low-copy-number expression vector for sopE2M45 was constructed by digesting pM226 with SpeI (Klenow treated) and HindIII and subcloning the insert into the EcoRV and HindIII sites of pACYC184 (NEB), which yielded pM256.
A DNA fragment of Salmonella serovar Typhimurium SL1344 harboring 877 bp upstream of the ORF of sopE was amplified by PCR (primers 5′-GCTGCCTGCCACCATACCCAC and 5′-GCCGCTAGCGTACATAATTCATTTATATATAGATAGC; 33 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min 20 s; template, pM136) and cloned into pSB1136, which yielded pM542, which expressed sopEM45 under control of the native promoter and the Shine-Dalgarno sequence of the arabinose-inducible promoter of pBAD24.
A chromosomal DNA fragment of Salmonella serovar Typhimurium SL1344 harboring the ORF of invB was amplified by PCR (primers 5′-GGAATTCTAATGCAACATTTGGATATCGCTGA and 5′-CCCAAGCTTTCATCTCATTAGCGACCGACTA; 33 cycles of 94°C for 30 s, 54°C for 30 s, and 68°C for 45 s) and cloned into pM542 and pSB1136, which yielded pM249 and pM250, respectively, which expressed invB under control of the sopE promoter and the arabinose-inducible promoter, respectively. The constructs were verified by DNA sequence analysis.
A chromosomal DNA fragment of Salmonella serovar Typhimurium SL1344 harboring the ORF of sipA was amplified by PCR (primers 5′-GGAATTCTAATGGTTACAAGTGTAAGGACTCAGCCC and 5′-TCCCCCCGGGACGCTGCATGTGCAAGCCATC; 33 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 3.5 min) and cloned into pM542, which yielded pM578, which expressed sipAM45 under control of the sopE promoter. The construct was verified by DNA sequence analysis. The insert was subcloned via Eco47III (partial digestion)/HindIII into the EcoRV and HindIII sites of pACYC184, which yielded pM580.
A DNA fragment of Salmonella serovar Typhimurium SL1344 harboring 870 bp upstream and aa 1 to 95 of the ORF of sopE was amplified by PCR (primers 5′-TCGATACTAGTATGCCCGTTTTCTTACCGTCG and 5′-ATGTCGATATCTATATCATTGAGCGTTTGAAGC; 33 cycles of 96°C for 30 s, 55°C for 45 s, and 72°C for 3 min; template, pM136) and cloned via SpeI/EcoRV into pM226, which yielded pM416, which expressed sopE1-95 sopE296240M45 under control of the sopE promoter.
A DNA fragment of Salmonella serovar Typhimurium SL1344 harboring aa 96 to 240 of the ORF of sopE was amplified by PCR (primers 5′-CGGGCGATATCAGAGGTAGTGCGAGTAAAGACCC and 5′-GCATGGGGTCAGGTGGG; 33 cycles of 96°C for 30 s, 55°C for 45 s, and 72°C for 3 min; template, pM136) and cloned via EcoRV/HindIII into pM226, which yielded pM417, which expressed sopE21-95 sopE96-240M45 under control of the sopE2 promoter.
lacZ transcriptional fusions.
A sopE expression vector with a lacZ transcriptional reporter cassette integrated downstream of the sopE stop codon under control of the native sopE promoter was constructed by cloning the lacZ transcriptional reporter cassette of pSB1040 (kindly provided by D. Zhou and J. E. Galán) into the XbaI site of pM136, which yielded pM185. A DNA fragment of pM185 (carrying the lacZ reporter cassette) was cloned via NcoI/HindIII into pM226, which yielded pM687, a sopE2 expression vector with a lacZ transcriptional reporter cassette under control of the native sopE2 promoter.
Protein preparation and analysis.
For analysis of secreted proteins bacteria were grown for 12 h at 37°C in LB medium supplemented with 0.3 M NaCl, diluted 1:20 into fresh medium, and grown for another 4 h with mild aeration to obtain an OD600 of 0.8 to 0.9. Culture supernatants for analysis of secretion were prepared by precipitation with trichloroacetic acid and acetone as described previously (40). Samples of whole bacterial cultures, bacterial pellets, and culture supernatants were separated on a sodium dodecyl sulfate (SDS) gel and transferred to a nitrocellulose membrane. Proteins were detected by Western blotting by using a monoclonal anti-M45 antibody (52), a polyclonal anti-SopE antiserum (50), a polyclonal anti-SopE2 antiserum (66), polyclonal anti-SipC and anti-SipA antisera (kindly provided by J. E. Galán [41]), a monoclonal anti-OmpC antibody (62), appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies, and a chemoluminescent detection kit, as recommended by the manufacturer (Amersham Pharmacia).
For analysis of protein stability, bacterial cultures were grown as described above. A 100-μl aliquot was removed after 4 h of subculturing (zero-time control). Chloramphenicol (30 μg/ml) was added to the culture, and samples were removed after 5, 10, 20, and 40 min, mixed with Laemmli sample buffer, and shock frozen. Proteins were separated on a 15% SDS gel and detected by Western blotting as described above.
Generation of an InvB-specific antiserum.
The GST-InvB expression plasmid pM672 was transformed into Escherichia coli, and cultures were grown overnight, diluted 1:50, and grown to an OD600 of 0.6. The cultures were then induced with isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h and pelleted. The GST-InvB fusion protein was purified by using glutathione (GSH)-Sepharose beads (Amersham Pharmacia), and the GST tag was removed by cleavage with thrombin. The purified InvB protein was used by Neosystem (Strasbourg, France) to raise a polyclonal rabbit antiserum.
Pull-down assay.
Strain M566(pM113) was grown for 12 h at 37°C in LB medium supplemented with 0.3 M NaCl, diluted 1:20 into 100 ml of fresh medium, and grown for another 3 h with mild aeration. Expression of the SopE-GST fusion protein was induced by addition of 0.2% arabinose and growth for another 1 h at 37°C with mild aeration. Bacteria were harvested by centrifugation (6,000 × g, 10 min, 4°C). The bacterial pellet was resuspended in 25 ml of buffer B (50 mM Tris [pH 7.6], 1 mM MgCl2, 100 mM NaCl; 4°C) supplemented with 2 mM dithiothreitol, and the cells were lysed in a French pressure cell. Cell debris was removed by centrifugation. The supernatant was passed through a 0.4-μm-pore-size filter. The SopE-GST fusion protein was bound to 200 μl of glutathione-Sepharose 4B beads (Amersham Pharmacia) at 4°C. The beads were washed extensively seven times in 10 ml of buffer B at 4°C, and Laemmli sample buffer was added. The proteins bound to 100 μl of beads were analyzed on a 15% SDS-10% SDS discontinuous gel. The gel was stained with Coomassie brilliant blue. As a control a 5-μl aliquot of beads was analyzed on a 15% SDS-10% SDS discontinuous gel, and the SopE-GST fusion protein was detected by Western blotting with monoclonal anti-GST antibody (Sigma) and polyclonal anti-SopE antiserum as the primary antibodies. A monoclonal HRP-coupled anti-rabbit antibody (Sigma) and a monoclonal HRP-coupled anti-mouse antibody (Sigma) were used as secondary antibodies. Bands of interest were excised from the Coomassie brilliant blue-stained gel. Cysteines were treated with iodoacetamide to form carbamidocysteines, and proteins were digested with trypsin. The protein fragments were analyzed by matrix-assisted laser desorption ionization mass spectrometry, and proteins were identified by a fingerprint analysis by using the PeptIdent program (http://www.expasy.ch).
To verify the specificity of the pull-down experiment, a GST-InvB fusion protein and all associated proteins were purified from the lysate of wild-type Salmonella serovar Typhimurium strain SL1344(pM672) by glutathione-Sepharose affinity chromatography as described above. Samples were removed at each step of the purification procedure, separated on a 15% SDS-polyacrylamide gel electrophoresis (PAGE) gel, analyzed by Western blotting by using an anti-SopE antibody, and later reprobed by using anti-SipC and anti-SipA antisera (kindly provided by J. E. Galán).
Coimmunoprecipitation.
Strains M566 and M608 were grown for 12 h at 37°C in LB medium containing 0.3 M NaCl, diluted 1:20 into 40 ml of fresh medium, and grown for another 4 h. Bacteria were harvested by centrifugation (6,000 × g, 10 min, 4°C). Each bacterial pellet was resuspended in 20 ml of buffer B supplemented with 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and a complete protease inhibitor mixture. The cells were lysed in a French pressure cell, and the cell debris was removed by centrifugation. The supernatant was passed through a 0.4-μm-pore-size filter. Five milliliters of the lysate was precleared by incubation with 20 μl of protein A-Sepharose CL-4B beads (Amersham Pharmacia) for 1 h at 4°C. We then transferred the lysate into a new tube without touching the beads, and the lysate was incubated with 10 μl of anti-M45 antibody (1 mg/ml) for 1 h at 4°C. The lysate was centrifuged for 10 min at 10,000 × g in a table top centrifuge to remove nonspecific aggregates. Ten microliters of protein A-Sepharose beads was added to the lysate and incubated for 1 h at 4°C. After this the beads were washed four times in 1 ml of buffer B at 4°C, and Laemmli sample buffer was added. Samples were removed at each step of the immunoprecipitation procedure, separated by 16% SDS-PAGE, and analyzed by Western blotting by using an anti-InvB antiserum. Later the blot was reprobed by using an anti-SopE antiserum. A monoclonal HRP-coupled anti-rabbit antibody (Sigma) was used as the secondary antibody.
Analysis of sopE and sopE2 transcription by lacZ expression.
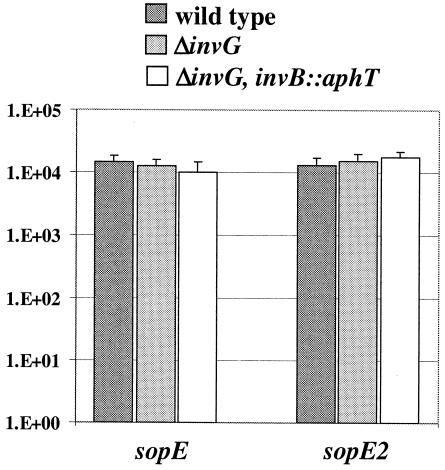
Salmonella serovar Typhimurium strains SL1344, SB161, and M590 harboring pM185 or pM687 were grown under SPI-1-inducing conditions as described above. Samples (100 μl) were removed and analyzed for β-galactosidase activity by using standard methods (59). Briefly, 100 μl of culture, 900 μl of buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol), 2 drops of CHCl3, and 1 drop of 0.1% SDS were mixed with vortexing for 10 s and equilibrated to 28°C. Then 0.2 ml of o-nitrophenyl-β-d-galactopyranoside (4 mg/ml) was added. The reaction was stopped by adding 0.5 ml of 1 M Na2CO3. The time at which a yellow color developed was recorded. The A600 of the culture and the A550 and A420 of the reaction samples were determined, and the numbers of β-galactosidase activity units were calculated.
Gentamicin protection assay.
The invasiveness of mutant Salmonella strains for COS-7 cells was analyzed as described previously (49). COS-7 tissue culture cells were grown for 2 days in Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal bovine serum (FBS) in 24-well dishes to obtain 80% confluence. The culture medium was removed, and 500 ml of Hanks' balanced salt solution (HBSS) was added 3 min before addition of the bacteria. The bacteria were grown for 12 h in LB medium supplemented with 0.3 M NaCl, diluted 1:20 into fresh medium, and grown for another 4 h with mild aeration to obtain an OD600 of 0.8 to 0.9. The actual bacterial density was determined by plating appropriate dilutions on LB agar. To start the assay, bacteria were added to COS-7 cells at a multiplicity of infection (MOI) of 15 bacteria/cell and incubated for 50 min at 37°C in the presence of 5% CO2. The cells were washed three times with HBSS and incubated in 500 ml of DMEM containing 5% FBS and 400 μg of gentamicin per ml for 2 h at 37°C in the presence of 5% CO2. The cells were washed three times with 1× phosphate-buffered saline (PBS) and lysed in 1× PBS containing 0.1% sodium deoxycholate, and the number of intracellular bacteria (CFU) was determined by plating on LB agar. Usually, about 25% of the inoculum of the wild-type strain was recovered from COS-7 cells at the end of the assay. The numbers given below were determined by performing at least five independent experiments for each strain. The statistical significance of differences in invasiveness between different strains was analyzed by using the Mann-Whitney U test.
Immunofluorescence analysis of SipC, SopEM45, and SopE2M45 translocation.
COS-7 cells were grown in 24-well dishes for 2 days in DMEM containing 5% FBS on glass coverslips to about 60% confluence. The culture medium was removed, and 500 ml of HBSS was added. For analysis of SipC translocation, COS-7 cells were infected for 50 min with bacteria grown as described above (MOI, 30 bacteria/cell). After this, extracellular bacteria were removed by washing the cells twice with HBSS, and COS-7 cells were incubated in 500 ml of DMEM containing 5% FBS and 400 μg of gentamicin per ml for 1 h at 37°C in the presence of 5% CO2. The cells were fixed with 3.7% formaldehyde in PBS and permeabilized with 0.1% Triton X-100 in PBS. The translocated SipC was detected by using an anti-SipC antiserum (kindly provided by J. E. Galán) and an anti-rabbit fluorescein isothiocyanate (FITC) conjugate (1:400 dilution in 3% bovine serum albumin-2% dry milk in PBS; Sigma). The DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma). For analysis of translocation of SopEM45 and SopE2M45, COS-7 cells were infected with SPI-1-induced Salmonella serovar Typhimurium strains harboring the sopEM45 (pM438) and sopE2M45 (pM256) expression vectors (MOI, 60 bacteria/cell), respectively. The translocated M45-tagged proteins were detected by using a monoclonal anti-M45 antibody (kindly provided by P. Hearing) and an anti-mouse FITC conjugate (1:400 dilution in 3% bovine serum albumin-2% dry milk in PBS; Sigma). Coverslips were mounted and analyzed by fluorescence microscopy. To quantify effector protein translocation, at least 100 cells that were infected (harboring more than five bacteria per cell) were scored in a blinded manner for detectable effector protein-specific staining. The proportion of infected host cells that were positive for each effector protein was determined as follows: (number of infected, effector protein-positive cells)/(total number of infected cells) × 100.
Images were recorded by using a Perkin-Elmer Ultraview confocal imaging system and a Zeiss Axiovert 200 microscope; green fluorescence was recorded confocally, and the DAPI fluorescence was recorded by epifluorescence microscopy.
RESULTS
Identification of InvB in a SopE-GST pull-down assay.
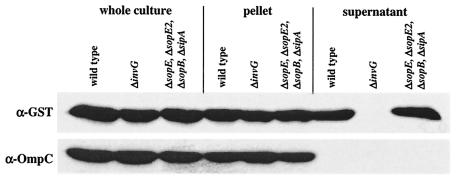
So far, no specific chaperone has been identified for the phage-encoded SPI-1 effector protein SopE. In the genome of the P2-like temperate phage SopEφ, the 723-nucleotide ORF encoding SopE is located in a 1.2-kb gene cassette (a moron [51]) which includes an autonomous promoter (11) and a putative transcriptional terminator but no ORF which might encode a chaperone. If SopE requires a chaperone for stabilization or secretion, it must be encoded elsewhere in the Salmonella serovar Typhimurium chromosome. We used an affinity purification approach to identify potential chaperones for SopE. This approach is based on the observation that type III effector proteins from Salmonella serovar Typhimurium can form stable complexes with their cognate chaperones in vitro and in vivo (5, 13, 23, 69). We constructed the pBAD24-related plasmid pM113, which allowed arabinose-inducible expression of a SopE-GST fusion protein (see Materials and Methods). pM113 was transformed into Salmonella serovar Typhimurium SL1344, the secretion-deficient mutant SB161 (ΔinvG), and M566 (ΔsopE ΔsopE2 ΔsopB ΔsipA) (Table 1), which lacked several known effector proteins that might compete for SopE binding proteins. The SopE-GST fusion protein was secreted by SL1344 and M566(pM113) but not by SB161 (Fig. 1), which indicates that the SopE-GST fusion protein is recognized and transported by the SPI-1 TTSS.
FIG. 1.
Western blot analysis of expression of the SopE-GST fusion protein and secretion via the SPI-1 TTSS. SL1344(pM113), SB161(pM113), and M566(pM113) were grown under SPI-1-inducing conditions and induced with arabinose (see Materials and Methods). One hundred microliters of a whole culture, a pellet from 100 μl of a culture, and proteins recovered from 1 ml of a culture supernatant were separated by SDS-PAGE and analyzed by Western blotting by using an anti-GST antibody (α-GST). The blot was reprobed by using an anti-OmpC (α-OmpC) antibody to verify that equal amounts were loaded.
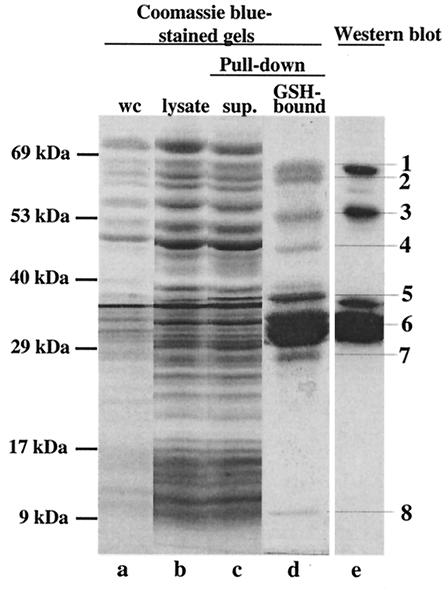
After expression of SopE-GST in M566, the cleared lysate was incubated with GSH-Sepharose beads (see Materials and Methods). Bound proteins were analyzed by discontinuous PAGE. Eight distinct protein bands were purified from the M566(pM113) lysate (Fig. 2). Western blot analysis performed with a polyclonal rabbit anti-SopE antiserum and a monoclonal anti-GST antibody suggested that bands 1, 3, and 6 correspond to the full-length SopE-GST fusion protein (55 kDa) and fragments of this protein (Fig. 2). Bands 2, 4, 5, 7, and 8 were excised from the gel, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization mass spectrometry. The fingerprint analysis revealed that bands 5 and 7 were also fragments of the SopE-GST fusion protein. Bands 2 and 4 corresponded to glycerol-3-phosphate dehydrogenase (11 matching peptides, 24.5% of the sequence covered) and the DNA polymerase III β-chain (eight matching peptides, 29.2% of the sequence covered) and were probably copurified due to nonspecific binding. Band 8 corresponded to the Salmonella serovar Typhimurium protein InvB (five matching peptides, 57.8% of the sequence covered) (14). InvB is encoded in SPI-1 (23) and has been described as a secretion chaperone for the SPI-1 effector protein Sip/SspA (5). This suggested that InvB might also be a type III secretion chaperone for SopE.
FIG. 2.
Pull-down assay to isolate putative binding partners of SopE. A SopE-GST fusion protein (pM113) was expressed in Salmonella serovar Typhimurium M566 (ΔsopB ΔsopE ΔsopE2 ΔsipA) (see Materials and Methods). The bacterial lysate (25 ml) was incubated with 200 μl of GSH-Sepharose beads. Samples from different purification steps were analyzed on a discontinuous SDS gel and were detected by staining with Coomassie brilliant blue (lanes a to d). Proteins bound to GSH-Sepharose were analyzed on a discontinuous 10% SDS-15% SDS gel and by Western blotting by using a mixture of anti-SopE and anti-GST antibodies (lane e). Lane a, 100 μl of a whole culture (wc) before harvesting of the cells; lane b, 25 μl of cleared cell lysate after lysis with a French pressure cell; lane c, 25 μl of supernatant after incubation of the cell lysate with GSH-Sepharose (sup.); lane d, SDS-PAGE analysis of proteins bound to 100 μl of GSH-Sepharose beads; lane e, Western blot analysis of 5% of the sample loaded in lane d performed with anti-SopE antiserum and an anti-GST antibody.
GST-InvB affinity purification of SopE and SipA but not SipC.
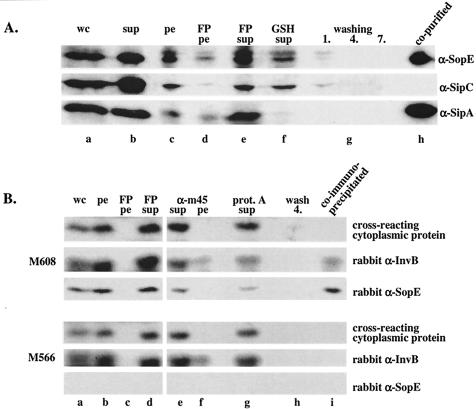
To confirm that SopE can indeed be copurified specifically with InvB, we repeated the affinity purification assay using a GST-InvB expression vector (pM672) (see Materials and Methods). Salmonella serovar Typhimurium SL1344(pM672) was grown and a lysate was prepared as described in Materials and Methods. GST-InvB and the bound bacterial proteins were purified by using GSH-Sepharose beads. A Western blot analysis demonstrated that the affinity purification procedure removed SopE and SipA but not SipC from the cleared cell lysate (Fig. 3A, compare lanes e and f). The remaining amounts of SopE and SipA detected in the depleted lysate (lane f) might be attributable to the presence of some misfolded SopE or SipA fraction or to SopE/SipA binding to the chromosomally encoded wild-type InvB. On the washed glutathione-Sepharose beads we detected large amounts of SopE and SipA which were copurified with GST-InvB (Fig. 3A, lane h) but no SipC, which is known to be secreted independently of InvB (5). These data confirmed that InvB bound directly or indirectly to SipA (5) and SopE (this study). This protein formed a specific complex with SopE and SipA but not with just any protein transported via the SPI-1 TTSS.
FIG. 3.
(A) Western blot analysis of a GST-InvB affinity purification assay. GST-InvB (pM672) was expressed in wild-type Salmonella serovar Typhimurium SL1344 (see Materials and Methods). The bacterial lysate (25 ml) was incubated with 200 μl of GSH-Sepharose beads. Samples from the purification steps were analyzed by Western blotting by using anti-SopE (α-SopE), anti-SipA (α-SipA), and anti-SipC (α-SipC) antisera. Lane a, 100 μl of a whole culture before harvesting of the cells (wc); lane b, proteins recovered from 250 μl of culture supernatant after pelleting of the cells (sup); lane c, 25 μl of a bacterial pellet resuspended in 25 ml of buffer B; lane d, 50 μl of pelleted cell debris resuspended in 25 ml of buffer B after lysis with a French pressure cell (FP pe); lane e, 50 μl of cleared French pressure cell lysate (total volume, 25 ml) (FP sup); lane f, 50 μl of cleared cell lysate after binding of GST-InvB and its associated proteins to 200 μl of GSH-Sepharose beads (GSH sup); lanes g, 100 μl of washing solution after the first, fourth, and seventh washes of the GSH-Sepharose beads with buffer B (washing 1, 4, and 7, respectively); lane h, 10 μl of GSH-Sepharose beads. (B) Western blot analysis of a coimmunoprecipitation experiment performed with the sopEM45-expressing strain M608 and an anti-M45 antibody (see Materials and Methods). The bacterial lysate (5 ml) was incubated with 10 μl of monoclonal mouse anti-M45 antibody and 10 μl of protein A-Sepharose beads. Samples from the precipitation procedure were analyzed by Western blotting by using polyclonal rabbit anti-InvB (rabbit α-InvB) and anti-SopE (rabbit α-SopE) antisera. Lane a, 100 μl of a whole culture before harvesting of the cells (wc); lane b, 100 μl of bacterial pellet resuspended in 20 ml of buffer B (pe); lane c, 100 μl of pelleted cell debris resuspended in 20 ml of buffer B after lysis with a French pressure cell (FP pe); lane d, 100 μl of cleared French pressure cell lysate (FP sup); lane e, 100 μl of cleared cell lysate after incubation with anti-M45 antibody and removal of nonspecific aggregates by centrifugation (α-m45 sup); lane f, 100 μl of pelleted nonspecific aggregates resuspended in buffer B (α-m45 pe); lane g, 100 μl of cleared cell lysate after incubation with 10 μl of protein A-Sepharose beads (prot. A sup); lane h, 100 μl of washing solution after the fourth wash of the protein A-Sepharose beads with buffer B (wash 4); lane i, 10 μl of protein A-Sepharose beads.
Coimmunoprecipitation experiments confirmed the SopE-InvB interaction.
The pull-down experiments (Fig. 2 and 3) were performed with strains overexpressing large quantities of GST fusion proteins. Therefore, altered protein concentrations might have affected complex formation. In order to eliminate this possibility, we wanted to confirm the SopE-InvB interaction under more physiological conditions. To do this, we performed a coimmunoprecipitation assay using the SL1344 derivative M608, which encoded an M45 epitope-tagged version of SopE (SopEM45) and native InvB in the chromosome (Table 1) (see Materials and Methods). This strain was grown under SPI-1-inducing conditions, and the cleared bacterial lysate was incubated with an anti-M45 monoclonal antibody and protein A-Sepharose beads. The beads were washed extensively, and the material bound to the beads was analyzed by Western blotting (Fig. 3B, top panel). Analysis with polyclonal rabbit anti-InvB and rabbit anti-SopE antisera revealed that InvB coprecipitated with SopEM45 (Fig. 3B, top panel, lane i). In contrast, an unknown 18-kDa cytosolic polypeptide that cross-reacted with the anti-InvB antiserum was not coprecipitated. The specificity of the coimmunoprecipitation was verified by a parallel control experiment in which a cleared lysate of M566 was used. This strain was identical to M608 except that it lacked SopEM45 (Table 1 and Fig. 3B, bottom panel). In this control experiment, InvB was not coprecipitated with the anti-M45 monoclonal antibody (Fig. 3B, compare anti-InvB signals in lane i from M608 and M566). In conclusion, the data confirmed that SopE bound directly or indirectly to InvB in the cytosol of Salmonella serovar Typhimurium.
Construction and initial characterization of an invB mutant.
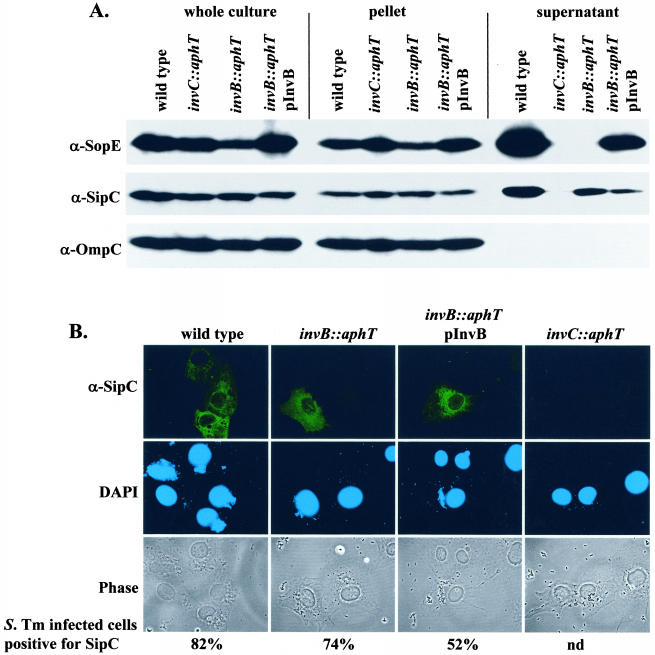
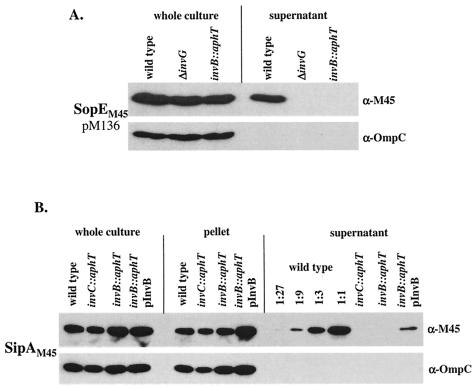
To analyze the role of InvB in SopE secretion, we constructed the isogenic mutant strain M568 (invB::aphT) carrying a terminatorless aphT cassette instead of invB (SL1344 [invB::aphT]) (see Materials and Methods). Because invB is located in an operon encoding several essential subunits of the SPI-1 type III secretion apparatus, it was crucial to verify that insertion of the aphT cassette did not interfere with the general function of the SPI-1 TTSS. To do this, we analyzed secretion and translocation of the effector protein SipC, which is transported via the SPI-1 TTSS in an InvB-independent manner (5). Western blot analysis performed with a polyclonal anti-SipC antiserum confirmed that expression and secretion of SipC in wild-type Salmonella serovar Typhimurium SL1344 were virtually identical to expression and secretion of SipC in M568 (invB::aphT). The small reduction in SipC secretion in the invB mutant M568 (Fig. 4A, compare the lanes for the wild-type and invB::aphT strains) might be attributable to a slight polar effect of the aphT cassette on downstream genes encoding essential components of the SPI-1 type III apparatus. Alternatively, InvB might have some minor regulatory function. Nevertheless, SipC secretion by a secretion-deficient Salmonella serovar Typhimurium mutant (SB566 [invC::aphT]) (Fig. 4A) was affected much more severely than SipC secretion by M568 (invB::aphT). Overall, our results confirmed that the invB::aphT mutation did not interfere with the general function of the SPI-1 TTSS.
FIG. 4.
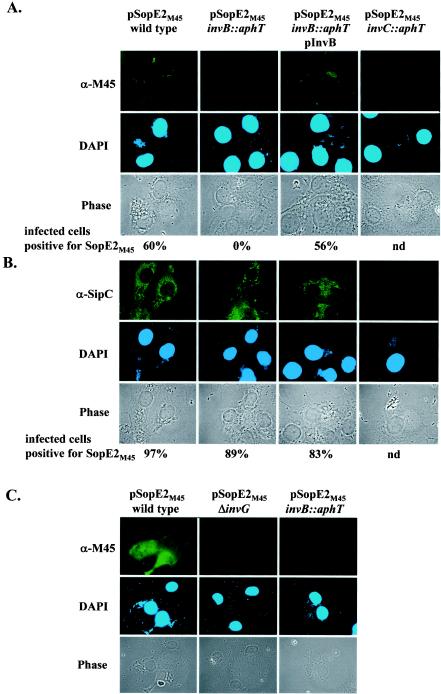
Effect of the invB::aphT mutation on expression and secretion of SopE and on expression, secretion, and translocation of SipC. (A) Western blot analysis of InvB-dependent expression and secretion of SopE and SipC. SL1344 (wild type), SB566 (invC::aphT), M568 (invB::aphT), and M568(pM249) (pM249 is an invB expression plasmid) were grown under SPI-1-inducing conditions (see Materials and Methods). One hundred microliters of a whole culture, pelleted bacteria recovered from 100 μl of a culture, and the proteins recovered from 1 ml of a culture supernatant were separated by SDS-PAGE and analyzed by Western blotting with anti-SopE (α-SopE) and anti-SipC (α-SipC). To ensure equal loading of the lanes, the blot was reprobed by using an anti-OmpC (α-OmpC) antibody. (B) Immunofluorescence analysis of SipC translocation into COS-7 cells. COS-7 cells were infected for 2 h with the strains described above (MOI, 30 bacteria/cell). The cells were fixed, permeabilized, and stained with a rabbit anti-SipC (α-SipC) antiserum, with an anti-rabbit FITC conjugate, and with DAPI. At least 100 cells infected with more than five bacteria were evaluated to quantify SipC translocation (see Materials and Methods). S. Tm, Salmonella serovar Typhimurium; nd, not determined.
These results were supported by an analysis of the translocation of SipC by M568 into host cells (Fig. 4B). For this analysis, COS-7 tissue culture cells were infected with wild-type Salmonella serovar Typhimurium, M568 (invB::aphT), the complemented strain M568(pM249), and the secretion-deficient mutant SB566 (invC::aphT [see Materials and Methods]) (MOI, 30 bacteria/cell). The cells were fixed 2 h postinfection, and the translocated SipC was stained by using an anti-SipC antibody (Fig. 4B) (41). Our results show that M568 (invB::aphT) translocates SipC into infected host cells at wild-type levels. In conclusion, the data demonstrate that disruption of invB by the aphT cassette does not compromise the general function of the SPI-1 TTSS. Therefore, this strain could be used to study the specific role of InvB in the secretion and translocation of SopE.
Role of invB in secretion of SopE.
We compared secretion of SopE into the bacterial culture supernatant by M568 (invB::aphT), the secretion-deficient mutant SB566 (invC::aphT), and the isogenic wild-type strain SL1344. This was done by reanalyzing the samples shown in Fig. 4A in a Western blot by using a polyclonal anti-SopE antibody. We found that M568 (invB::aphT) and SB566 (invC::aphT) do not secrete the chromosomally encoded SopE into the culture supernatant (Fig. 4A). SopE secretion by M568 (invB::aphT) could be partially complemented by transformation with the invB expression vector pM249 (Fig. 4A). The partial complementation might be attributable to a minor polar effect of the aphT cassette. Nevertheless, the data in Fig. 4A support the notion that InvB is required for secretion of SopE via the SPI-1 TTSS.
The anti-SopE antiserum used in the initial experiments (Fig. 4A) showed some cross-reactivity with the effector protein SopE2 (66). In order to develop a more specific assay for SopE, we transformed Salmonella serovar Typhimurium SL1344 (wild type), SB161 (ΔinvG), and M568 (invB::aphT) with pM136, an expression vector for an M45 epitope-tagged version of SopE (66). M568(pM136) and the secretion-deficient mutant SB161(pM136) were incapable of SopEM45 secretion, whereas SopEM45 was secreted by wild-type strain SL1344(pM136) (Fig. 5A).
FIG. 5.
Western blot analysis of invB-dependent expression and secretion of SopEM45 and SipAM45. (A) SL1344 (wild type), SB161 (ΔinvG), and M568 (invB::aphT) carrying the sopEM45 expression plasmid pM136 were grown under SPI-1-inducing conditions (see Materials and Methods). One hundred microliters of a whole culture and 100 μl of a culture supernatant were separated by SDS-PAGE and analyzed by Western blotting by using an anti-M45 (α-M45) antibody. α-OmpC, anti-OmpC antibody. (B) SL1344 (wild type), SB566 (invC::aphT), and M568 (invB::aphT) carrying the sipAM45 expression plasmid pM580, as well as M568 harboring invB (pM249) and sipAM45 (pM580) expression plasmids, were grown under SPI-1-inducing conditions (see Materials and Methods). Twenty-five microliters of a whole culture, pelleted bacteria from 25 μl of a culture, and proteins recovered from 300 μl of a culture supernatant (and dilutions for SL1344) were analyzed by Western blotting by using an anti-M45 (α-M45) antibody. The blots were reprobed by using an anti-OmpC (α-OmpC) antibody to control the amount of protein loaded.
The data show that SopE and SopEM45 are not secreted by M568 (invB::aphT) and mutants with a defective SPI-1 TTSS apparatus (SB566 [invC::aphT] and SB161 [ΔinvG], respectively). Therefore, all of the SopE/SopEM45 present in cultures or bacterial pellets of M568, SB566, and SB161 is attributable to intracellular protein pools (Fig. 4A and 5A). No differences were detected between the intracellular pools of SopEM45 in M568 (invB::aphT) and SB566 (invC::aphT) (Fig. 4A and 5A). Therefore, invB is required for secretion but not stabilization of intracellular SopE (see below).
Since InvB has been described as a secretion chaperone for the effector protein Sip/SspA (5), we analyzed the secretion of SipAM45 in the invB mutant M568 as a control. In line with the results of Bronstein et al. (5), we found that the invB mutant M568 secreted less than 5% of the wild-type levels of SipAM45 (Fig. 5B). The secretion defect could be partially complemented with the invB expression vector pM249. Again, incomplete complementation of M568 by pM249 might be attributable to a minor polar effect of the invB::aphT mutation. Nevertheless, our data support the results of Bronstein et al. (5) which demonstrated that InvB is a secretion chaperone for SipA.
Role of invB in the translocation of SopE.
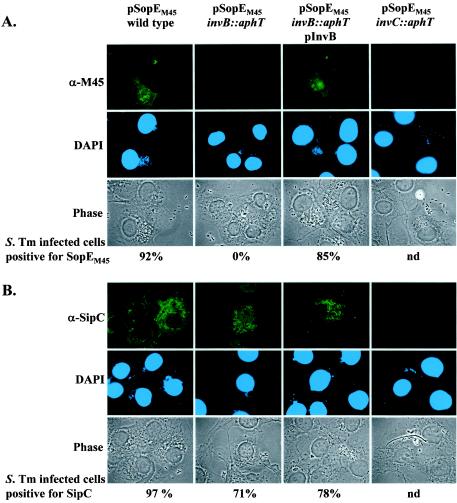
The role of invB in the SPI-1-dependent translocation of SopE into host cells was analyzed by immunofluorescence microscopy. To do this, COS-7 tissue culture cells were infected with wild-type Salmonella serovar Typhimurium SL1344, the secretion-deficient mutant SB161 (ΔinvG), and the invB mutant strain M568 harboring the sopEM45 expression plasmid pM438. The translocated effector proteins SopEM45 and SipC were detected by using the anti-M45 antibody (Fig. 6A) and an anti-SipC antiserum (Fig. 6B). SopEM45 could be detected in 92% of all cells infected with SL1344(pM438) but not in cells infected with M568(pM438) or SB161(pM438). The translocation defect of the invB mutant strain M568(pM438) could be restored (85% of all infected cells were SopEM45 positive) with an invB expression vector in M568 (pM438 and pM249). Together with the data shown in Fig. 4 and 5A, these results demonstrate that invB plays a specific role in SPI-1-dependent transport of SopE but not in transport of SipC.
FIG. 6.
Immunofluorescence analysis of invB-dependent translocation of SopEM45 into COS-7 tissue culture cells. (A) Salmonella serovar Typhimurium strains SL1344(pM438), M568(pM438), M568(pM438, pM249), and SB161(pM438) were used to infect COS-7 cells at an MOI of 60 bacteria/cell for 2 h (see Materials and Methods). The cells were fixed, permeabilized, and stained with a mouse anti-M45 (α-M45) antibody, with an anti-mouse FITC conjugate, and with DAPI. (B) As a control, COS-7 cells were infected with the strains described above but were stained to detect translocation of the effector protein SipC by using an anti-SipC (α-SipC) antiserum. For quantification of effector protein translocation at least 100 cells infected with more than five bacteria were evaluated (see Materials and Methods). S. Tm, Salmonella serovar Typhimurium; nd, not determined.
SopE2 secretion also depends on invB.
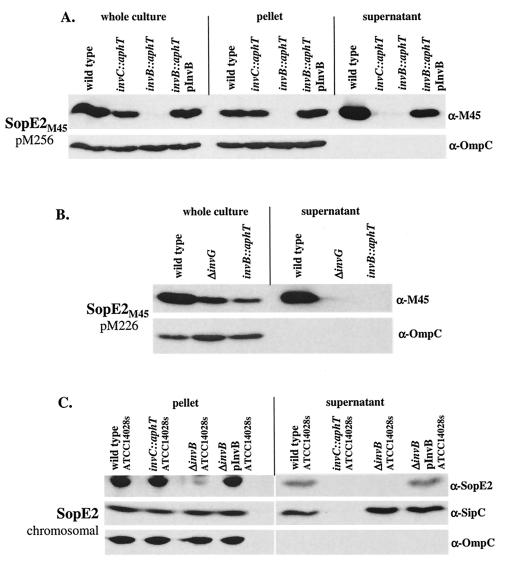
The Salmonella serovar Typhimurium effector protein SopE2 is 69% identical to SopE at the amino acid level (2, 66). SopE2 and SopE act as guanine nucleotide exchange factors for small Rho GTPases of the host cell and are transported via the SPI-1 TTSS (20, 30, 66). Based on these similarities, we speculated that SopE2 also requires invB for secretion. To investigate this, we transformed pM256 (sopE2 promoter; SopE2 tagged C terminally with the M45 epitope, Cmr) into wild-type Salmonella serovar Typhimurium SL1344, SB566 (invC::aphT), M568 (invB::aphT), and M568 carrying the invB expression vector pM249 (Ampr). Secretion of SopE2M45 from these strains was analyzed by Western blotting (Fig. 7A). SopE2M45 was detected in the culture supernatants of wild-type Salmonella serovar Typhimurium SL1344(pM256) and the invB-complemented strain M568(pM256, pM249) but not in supernatants from the secretion-deficient strain SB566(pM256) or the invB mutant M568(pM256) (Fig. 7A). Again, the complementation was incomplete, which might be attributable to a slight polar effect of the invB::aphT mutation. Nevertheless, it was striking that the intracellular SopE2M45 pools were much smaller in M568(pM256) than in SB566(pM256). This suggested that invB affects the cytosolic stability or expression of SopE2M45. However, the low cytoplasmic levels of SopE2M45 in M568(pM256) did not allow us to draw conclusions about a potential role of invB in the transport of this effector protein via the SPI-1 TTSS.
FIG. 7.
Western blot analysis of invB-dependent expression and secretion of SopE2M45 in SL1344 derivative strains and of SopE2 in ATCC 14028s derivatives. (A) SL1344(pM256), SB566(pM256), M568(pM256), and M568(pM256, pM249) were grown under SPI-1-inducing conditions (see Materials and Methods). One hundred microliters of a whole culture, pelleted bacteria from 100 μl of a culture, and proteins recovered from 1 ml of a culture supernatant were analyzed by Western blotting by using an anti-M45 (α-M45) antibody. α-OmpC, anti-OmpC antibody. (B) SL1344(pM226), SB161(pM226), and M568(pM226) were grown under SPI-1-inducing conditions (see Materials and Methods). One hundred microliters of a whole culture and proteins recovered from 500 μl of a culture supernatant were separated by SDS-PAGE and analyzed by Western blotting by using an anti-M45 (α-M45) antibody. α-OmpC, anti-OmpC antibody. (C) CS401, M622, PB502, and PB502(pM250) were grown under SPI-1-inducing conditions. Pelleted bacteria from 200 μl of a culture and proteins recovered from 1 ml of a culture supernatant were analyzed by Western blotting by using anti-SopE2 (α-SopE2) and anti-SipC (α-SipC) antisera. The blots were reprobed by using an anti-OmpC (α-OmpC) antibody to control the amount of protein loaded.
In order to analyze the role of invB in SopE2M45 secretion, the amount of cytosolic SopE2M45 was increased by using a pBAD derivative sopE2M45 expression plasmid (pM226), which has a higher copy number than the pACYC184 derivative pM256. pM226 was transformed into wild-type Salmonella serovar Typhimurium SL1344, SB161 (ΔinvG), and M568 (invB::aphT). Analysis of SopE2M45 secretion by Western blotting performed with the monoclonal anti-M45 antibody revealed that the cytoplasmic levels of SopE2M45 were higher and easily detectable even in M568 when we used pM226 (Fig. 7B) instead of pM256 (Fig. 7A). However, we still could not detect SopE2M45 in the culture supernatant of M568(pM226) (Fig. 7B). These results indicated that invB plays a role not only in the stabilization of intracellular SopE2M45 but also in transport of SopE2M45 via the SPI-1 TTSS.
Furthermore, we studied the InvB dependence of SopE2 secretion in Salmonella serovar Typhimurium ATCC 14028s. ATCC 14028s carries the sopE2 gene but lacks sopE. Using a polyclonal anti-SopE2 antiserum, we detected SopE2 in the culture supernatant of the wild-type ATCC 14028s strain but not in the supernatants of isogenic derivatives with a disrupted SPI-1 TTSS apparatus (invC::aphT) or a disrupted invB gene (ΔinvB) (Fig. 7C). SopE2 secretion by the latter strain could be complemented to wild-type levels by transformation with the invB expression vector pM250 (Fig. 7C and Table 1). The analysis of this strain verified that SipC secretion does not require invB (5) (see above) and that intracellular SopE2 levels are severely reduced in invB mutants (Fig. 7). Taken together, these results indicated that InvB affects intracellular SopE2 levels and transport of this effector protein via the SPI-1 TTSS.
Differential effect of invB on the cytosolic stability of SopE and SopE2.
The data presented above showed that the cytosolic pool of SopE2M45 was smaller in M568 (invB::aphT) than in Salmonella serovar Typhimurium mutants with a disrupted SPI-1 TTSS apparatus (SB161 and SB566) (Fig. 7A and B). This effect was not observed with the chromosomally encoded SopE protein or the plasmid-encoded SopEM45 protein (Fig. 4A and 5A). This indicated that the invB mutation might have a specific effect on the cytosolic SopE2M45 level.
Chaperones can control cytosolic effector protein levels by three different mechanisms: (i) facilitation of secretion or translocation into host cells via the TTSS (9); (ii) modulation of gene expression (11); and (iii) binding and protection of the effector protein from degradation and aggregation in the cytoplasm (47). To explain the reduced intracellular levels of SopE2M45 in M568 (invB::aphT) compared to the levels in SB161 and SB566, we explored the latter two possibilities.
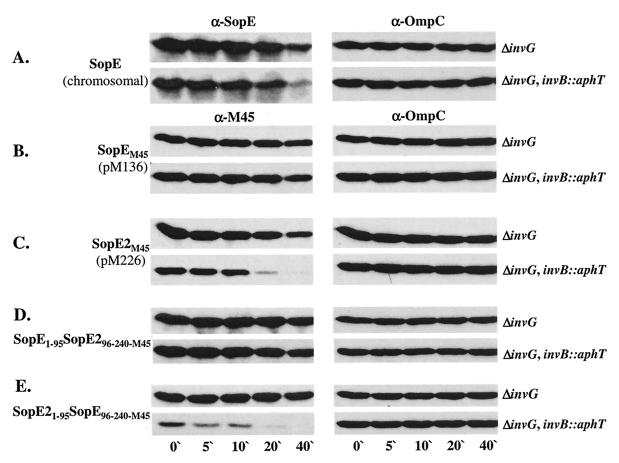
First, we analyzed the effect of invB on effector protein stability. In order to measure protein degradation of cytosolic effector proteins, it is important to examine protein stability in secretion-deficient strains and to compare the results. Therefore, we compared the stabilities of wild-type SopE, SopEM45 (pM136), and SopE2M45 (pM226) in SB161 (ΔinvG) and the isogenic invB mutant M590 (ΔinvG invB::aphT). The strains were grown under SPI-1-inducing conditions (see Materials and Methods), and de novo protein biosynthesis was stopped by addition of chloramphenicol. Aliquots were removed at different times, and degradation of cytosolic SopE, SopEM45, or SopE2M45 was analyzed by Western blotting (Fig. 8A to C). In the invB mutant M590 (ΔinvG invB::aphT) SopE2M45 was almost completely degraded after 20 min (Fig. 8C). However, in the presence of invB (SB161 [ΔinvG]), SopE2M45 was only partially degraded during the course of the assay (40 min). SopEM45 was much more stable than SopE2M45 in both Salmonella strains (Fig. 8B and C). It should be noted that the stability of the chromosomally encoded wild-type SopE was somewhat lower than that of the plasmid-encoded SopEM45 in both SB161 (ΔinvG) and M590 (ΔinvG invB::aphT) (Fig. 8A). However, even in this case the effect of invB on the stability of the cytoplasmic effector protein pool was much less pronounced than the effect observed with SopE2M45.
FIG. 8.
Effect of an invB mutation on the stability of SopE, SopEM45, and SopE2M45. SB161 (ΔinvG) and M590 (ΔinvG invB::aphT) or the same strains carrying pM136 (B), pM226 (C), pM416 (D), or pM417 (E) were expressed under secretion-inducing conditions (see Materials and Methods). After 4 h of subculturing 100-μl aliquots were removed (zero time), and chloramphenicol was added to the remaining cultures. One-hundred-microliter aliquots were removed after 5, 10, 20, and 40 min and separated by SDS-PAGE, and the protein levels were analyzed by Western blotting by using an anti-SopE (α-SopE) antiserum (A) and a monoclonal anti-M45 (α-M45) antibody (B to E). The blots were reprobed by using an anti-OmpC (α-OmpC) antibody to control the amount of protein loaded.
In the next analysis, we studied the role of invB in the transcription of sopEM45 and sopE2M45. To do this, we inserted a lacZ transcriptional reporter gene cassette downstream of the sopEM45 ORF in pM136 (pM185) and downstream of the sopE2M45 ORF in pM226 (pM687) (Table 1). These plasmids were transformed into wild-type Salmonella serovar Typhimurium SL1344, SB161, and M590. The bacteria were grown under identical conditions as described above. We found that both plasmids yielded identical levels of lacZ expression in all three Salmonella serovar Typhimurium strains (Fig. 9) (see Materials and Methods). Therefore, transcription of sopE2M45 and sopEM45 was not affected by disruption of invB or the SPI-1 type III secretion apparatus.
FIG. 9.
Effect of an invB deletion on transcription of sopEM45 and sopE2M45. Transcription of sopEM45 and sopE2M45 was measured by using transcriptional lacZ reporter constructs (Table 1). SL1344 (wild type) and the secretion-deficient strains SB161 (ΔinvG) and M590 (ΔinvG invB::aphT) harboring the sopEM45 (pM185) and sopE2M45 (pM687) expression vectors, respectively, were grown under SPI-1-inducing conditions. One-hundred-microliter samples were removed and assayed for β-galactosidase activity. Values are β-galactosidase units; standard deviations were determined from the results of three independent experiments.
In conclusion, the data show that invB does not affect transcription of sopEM45 and sopE2M45. Rather, invB seems to affect cytosolic effector protein pools by stabilizing cytosolic SopE2 (Fig. 8C) and by mediating secretion of SopE2 and SopE via the SPI-1 TTSS (Fig. 4A, 5A, and 7A and B).
N-terminal 95 aa determine different cytosolic stabilities of SopEM45 and SopE2M45.
Considering the high levels of functional and structural similarities, it was interesting to observe that SopE2M45 is less stable than SopEM45 in the cytosol of SB161 (ΔinvG) and M590 (ΔinvG invB::aphT). We wondered whether this difference is attributable to sequence variations in the N-terminal regions of the two proteins. The N-terminal region is sufficient to mediate secretion via the SPI-1 TTSS (Hardt and Galán, unpublished observations) and, based on data for the SptP-SicE complex from Salmonella serovar Typhimurium and for the Yersinia pYV-encoded TTSS, is expected to harbor the chaperone binding site (4, 58, 64).
We swapped the N-terminal 95 aa of SopEM45 and SopE2M45. The stabilities of the hybrid proteins in the cytosol of M590 (ΔinvG invB::aphT) and SB161 (ΔinvG) were compared to the stabilities of SopEM45 and SopE2M45 (Fig. 8B to E). We observed only minimal degradation of SopEM45 and SopE1-95SopE296-240M45 in SB161 (ΔinvG) and in the invB mutant strain M590 (ΔinvG invB::aphT) (Fig. 8B and D). SopEM45 and SopE1-95SopE296-240M45 were equally stable in the invB+ and invB strains over the entire course of the experiment.
In contrast, SopE2M45 and SopE21-95SopE96-240M45 were rapidly degraded in the absence of invB (Fig. 8C and E, lower panels). These data indicated that the N-terminal 95 aa of SopEM45 and SopE2M45 determine the different stabilities of the M45-tagged effector protein in the absence of invB.
Role of invB in SopE2 translocation.
We used immunofluorescence microscopy to analyze the invB dependence of SopE2 translocation into host cells. To infect COS-7 tissue culture cells, we used the strains that were used for the secretion assay whose results are shown in Fig. 7A (MOI, 60 bacteria/cell). Staining with a monoclonal anti-M45 antibody revealed that wild-type Salmonella serovar Typhimurium and the complemented invB strain [M568(pM249, pM256)] were able to translocate detectable amounts of SopE2M45 into host cells (Fig. 10A). In contrast, no SopE2M45 was detected inside the COS-7 cells infected with the invB mutant M568(pM256) or a mutant with a defective SPI-1 TTSS apparatus [SB566(pM256)] (Fig. 10). However, we cannot exclude the possibility that the failure to detect SopE2M45 translocation by M568(pM256) in this assay was due to the severely reduced intracellular SopE2M45 levels (Fig. 7A). In order to increase the amounts of SopE2M45, we repeated the translocation experiment using the medium-copy-number sopE2M45 expression vector pM226 (Fig. 7). The results confirmed that SopE2M45 is translocated by wild-type strain SL1344(pM226) but not by the invB mutant M568(pM226) or the secretion-deficient mutant SB161(pM226) (Fig. 10C). Overall, these data are in line with the notion that invB is required for translocation of SopE2M45 into host cells.
FIG.10.
Immunofluorescence microscopy analysis of invB-dependent translocation of SopE2M45 into COS-7 tissue culture cells. (A) SL1344(pM256), M568(pM256), M568(pM256, pM249), and SB566(pM256) were used to infect COS-7 cells for 2 h at an MOI of 60 bacteria/cell. Cells were fixed, permeabilized, and stained with an anti-M45 (α-M45) antibody, with an anti-mouse FITC conjugate, and with DAPI. (B) As a control, COS-7 cells were infected with the strains described above but were stained for translocation of the effector protein SipC by using an anti-SipC (α-SipC) antiserum. To quantify SopE2M45 and SipC translocation, at least 100 cells infected with more than five bacteria were scored as described in Materials and Methods. nd, not determined. (C) COS-7 cells were infected with SL1344(pM226), M568(pM226) and SB161(pM226) for 2 h at an MOI of 60 bacteria/cell. Fixed cells were stained with an anti-M45 antibody, an anti-mouse-FITC conjugate, and DAPI.
Role of invB in Salmonella serovar Typhimurium host cell invasion.
The translocation of effector proteins via the SPI-1 TTSS allows Salmonella serovar Typhimurium to invade host cells. We wanted to investigate the role of invB in this process.
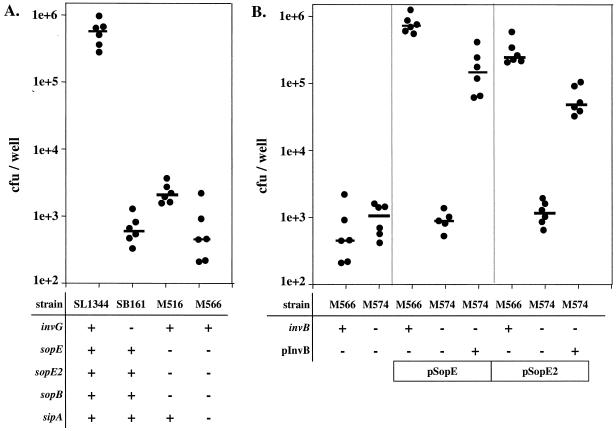
SopB/SigD, SopE, and SopE2 cooperate to mediate tissue culture cell invasion, and a sopB sopE sopE2 triple mutant (M516) was approximately 100-fold less invasive than the isogenic wild-type strain (Fig. 11A) (30, 49, 76). Recent reports have indicated that Sip/SspA might also play a role in host cell invasion (77, 78). Consistent with this, we found that a sopB sopE sopE2 sipA quadruple mutant (M566) was slightly less invasive than the triple mutant (M516) (Fig. 11A). The invasiveness of M566 did not differ significantly from that of a Salmonella serovar Typhimurium mutant with a defective SPI-1 TTSS apparatus (SB161 [ΔinvG]) (P > 0.05) (Fig. 11A). This indicated that invasion of COS-7 tissue culture cells is mediated principally by two different groups of SPI-1 effector proteins which depend on two different chaperones; translocation of SopE, SopE2 (this study), and Sip/SspA (5) is dependent on InvB, while translocation of SopB/SigD depends on the chaperone PipC/SigE (12).
FIG. 11.
Role of invB in SopE/SopE2-mediated host cell invasion. (A) SL1344 (wild type), SB161 (ΔinvG), M516 (ΔsopB sopE sopE2), and M566 (ΔsopB ΔsopE ΔsopE2 ΔsipA) were grown under SPI-1-inducing conditions and used to infect COS-7 tissue culture cells (MOI, 15 bacteria/cell) for 50 min, and the number of internalized bacteria was measured by the gentamicin protection assay (see Materials and Methods). The numbers of CFU per well from six independent experiments are shown. The bars indicate medians. (B) M566 (ΔsopB ΔsopE ΔsopE2 ΔsipA) and M574 (ΔsopB ΔsopE ΔsopE2 ΔsipA invB::aphT) carrying one or more of plasmids pSB1130, pM149, and pM249 were grown under SPI-1-inducing conditions (see Materials and Methods). COS-7 cells were infected as described above. The numbers of intracellular bacteria were determined in at least five independent experiments. The bars indicate medians.
Next, we analyzed the role of invB in SopE- and SopE2-mediated host cell invasion. To do this, we transformed sopE and sopE2 expression vectors (pSB1130 and pM149, respectively) into the quadruple mutant strain M566 (ΔsopB ΔsopE ΔsopE2 sipA) and the isogenic mutant M574 (ΔsopB ΔsopE ΔsopE2 ΔsipA invB::aphT). While sopE and sopE2 expression vectors were able to complement the host cell invasion defect of M566 (P < 0.05), no significant complementation was observed in invB strain M574 (Fig. 11B) (P > 0.05). Significant complementation was observed only when M574 carried an additional invB expression vector (pM249) (Fig. 11B) (P < 0.05). A slight polar effect of the invB::aphT mutation might be the reason for the incomplete complementation by pM249 (Fig. 11B). Nevertheless, these data further support the notion that invB is required for SopE- and SopE2-mediated host cell invasion.
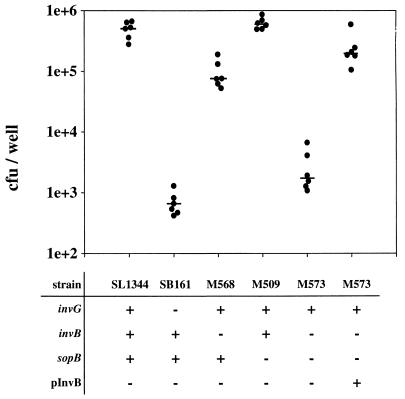
Interestingly, the invB mutant M568 is only slightly less invasive than the isogenic wild-type strain SL1344 (Fig. 12). To test whether the invasiveness of M568 is attributable to the translocation of SopB, we constructed an invB sopB double mutant (M573) (see Materials and Methods). Indeed, we found that the invasiveness of M573 is about as low as that of SB161, an isogenic strain with a disrupted SPI-1 TTSS apparatus (Fig. 12). The invasiveness of M573 could be restored with an invB expression vector (pM249). These data suggested that SopB/SigD translocation is not impaired in invB strains and provided further evidence that disruption of invB with the aphT cassette did not have a dramatic generalized effect on effector protein translocation. Rather, InvB seems to play the role of a chaperone for a specific subset of effector proteins, including Sip/SspA (5), SopE, and SopE2 (this study).
FIG. 12.
Role of invB in host cell invasion by a Salmonella serovar Typhimurium sopB mutant. SL1344, SB161, M568, M509, M573, and M573(pM249) were grown under SPI-1-inducing conditions (see Materials and Methods). COS-7 tissue culture cells were infected (MOI, 15 bacteria/cell) for 50 min. Extracellular bacteria were killed by addition of gentamicin. COS-7 cells were lysed, and the numbers of intracellular bacteria were determined. The data from six independent experiments are shown. The bars indicate medians.
DISCUSSION
The Salmonella effector proteins SopE and SopE2 are translocated into host cells via the SPI-1 TTSS (2, 31, 66, 75). Previously, it was not clear how these proteins are recognized by this TTSS and whether chaperones are involved. Here, we show that SopE can be copurified with the SPI-1-encoded chaperone InvB and that SopE and SopE2 require InvB for secretion and translocation via the SPI-1 TTSS, as speculated in a recent review (53). It has been reported previously that InvB is also required for efficient secretion and translocation of the SPI-1-encoded effector protein Sip/SspA (5). We confirmed this finding. Therefore, InvB seems to act as a chaperone for at least three different Salmonella serovar Typhimurium effector proteins, although Sip/SspA and SopE/SopE2 do not exhibit any significant sequence similarities.
Similarly, Spa15, the InvB homolog of Shigella flexneri, acts as a chaperone for at least three known or suspected effector proteins with unrelated sequences (54), IpgB1, OspC3, and IpaA. IpaA exhibits sequence similarity with Sip/SspA from Salmonella serovar Typhimurium (35, 40). In contrast to other type III chaperones which exhibit little sequence similarity, invB/spa15-like genes are present in a wide variety of bacteria, including Yersinia enterocolitica (ysaK [29]) and Sodalis glossinidius (invB [10]), and also on an Edwardsiella ictaluri plasmid (16). Based on this wide distribution, an unusual gene locus embedded between functional genes of the TTSS apparatus, and the ability to interact with multiple unrelated effector proteins, it has been proposed that InvB/Spa15 may represent a new family of chaperones (54, 55).
We found that disruption of invB has different consequences for the stability of the cognate effector proteins; SopEM45 was stable in the cytosol of an invB mutant, whereas SopE2M45 was not. Similarly, in S. flexneri Spa15 was required for the stability of IpgB1 but not for the stability of IpaA in the bacterial cytosol (54). The reason for the differential requirement of InvB/Spa15 for effector protein stabilization in the bacterial cytoplasm is still not clear.
In a domain swap experiment we mapped the regions of SopEM45 and SopE2M45 that determine the resistance to degradation in the absence of InvB to the N-terminal 95 aa. This region overlaps the proposed InvB/Spa15 binding regions of Sip/SspA (aa 1 to 158), OspC3 (aa 26 to 141), and IpgB (aa 23 to 190) (5, 54). Only the proposed Spa15 binding site of IpaA maps to a completely different region of the effector protein (aa 263 to 365) (54). This interaction domain has been mapped in yeast two-hybrid screening analyses. It has not been determined whether this interaction also occurs in the bacterial cytoplasm and whether IpaA might have another chaperone binding site closer to its N terminus. Overall, however, the data suggest that the binding site for the InvB/Spa15 family of chaperones covers aa 23 to 95 of the effector proteins. This region overlaps the known binding regions of other type III chaperones (19, 21, 36, 61, 65, 72).
Expression of the Salmonella serovar Typhimurium SPI-1 TTSS is restricted to specific locations in the host by an elaborate cascade of regulators, including PhoP/Q, SirA/BarA, InvF, HilA, HilD, SirC, and SprB (45). Several transcriptional regulators have been identified which ensure proper coexpression of effector proteins encoded within and outside of SPI-1 (11-13, 15, 17, 70). In addition, we show here that some effector proteins encoded within and outside of SPI-1 have the same chaperones for secretion and translocation into host cells. Therefore, Salmonella spp. possess at least two different mechanisms to coordinate effector protein expression and delivery: coregulation of transcription and sharing of secretion chaperones. The data show that effectors encoded outside of SPI-1 are directly controlled by SPI-1 itself.
The existence of tight regulatory networks which allow accurate functional integration of additional effector proteins is an important aspect of the horizontal gene transfer of type III effector proteins. Reassortment of effector protein repertoires has been discussed as a major driving force in the evolution of Salmonella spp., the adaptation to new hosts, and the emergence of new epidemic strains (17, 18, 49, 50, 67). For example, Salmonella serovar Typhimurium lysogens carrying the sopE-encoding phage SopEφ caused a major epidemic in England and the former East Germany in the 1970s and 1980s (50). It is thought that acquisition of the sopE gene by lysogenic conversion with SopEφ was an important step in the emergence of the epidemic strain. The data presented here explain why the phage-encoded effector protein SopE can actually function properly after lysogenic conversion of naïve Salmonella strains: the regulators and the chaperone InvB are already present (24, 56), no matter which Salmonella strain is infected by the phage (50). In conclusion, the functional control of effector proteins encoded in different genetic elements by SPI-1 regulators and chaperones might play an important role in the evolution of Salmonella host interactions because it ensures proper functional integration of newly acquired proteins into existing effector protein repertoires.
Acknowledgments
We thank Shiva P. Singh for providing the anti-OmpC antibody, Rene Brunisholz for performing the matrix-assisted laser desorption ionization mass spectrometry analysis, Cosima Pelludat for discussions and comments on the manuscript, and Jorge E. Galán for continued support.
This work was funded in part by a grant from the Deutsche Forschungsgemeinschaft (DFG) to W.-D.H.
REFERENCES
- 1.Anderson, D. M., and O. Schneewind. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140-1143. [DOI] [PubMed] [Google Scholar]
- 2.Bakshi, C. S., V. P. Singh, M. W. Wood, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2000. Identification of SopE2, a Salmonella secreted protein which is highly homologous to SopE and involved in bacterial invasion of epithelial cells. J. Bacteriol. 182:2341-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, J. C., and C. Hughes. 2000. From flagellum assembly to virulence: the extended family of type III export chaperones. Trends Microbiol. 8:202-204. [DOI] [PubMed] [Google Scholar]
- 4.Birtalan, S. C., R. M. Phillips, and P. Ghosh. 2002. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol. Cell 9:971-980. [DOI] [PubMed] [Google Scholar]
- 5.Bronstein, P. A., E. A. Miao, and S. I. Miller. 2000. InvB is a type III secretion chaperone specific for SspA. J. Bacteriol. 182:6638-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, L. W., D. M. Anderson, and O. Schneewind. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24:757-765. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, L. W., and O. Schneewind. 2000. Type III machines of Gram-negative bacteria: delivering the goods. Trends Microbiol. 8:214-220. [DOI] [PubMed] [Google Scholar]
- 8.Collazo, C. M., and J. E. Galan. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64:3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 10.Dale, C., S. A. Young, D. T. Haydon, and S. C. Welburn. 2001. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl. Acad. Sci. USA 98:1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darwin, K. H., L. S. Robinson, and V. L. Miller. 2001. SigE is a chaperone for the Salmonella enterica serovar Typhimurium invasion protein SigD. J. Bacteriol. 183:1452-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichelberg, K., C. C. Ginocchio, and J. E. Galan. 1994. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J. Bacteriol. 176:4501-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichelberg, K., W. D. Hardt, and J. E. Galan. 1999. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol. Microbiol. 33:139-152. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez, D. H., L. Pittman-Cooley, and R. L. Thune. 2001. Sequencing and analysis of the Edwardsiella ictaluri plasmids. Plasmid 45:52-56. [DOI] [PubMed] [Google Scholar]
- 17.Figueroa-Bossi, N., and L. Bossi. 1999. Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 33:167-176. [DOI] [PubMed] [Google Scholar]
- 18.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-272. [DOI] [PubMed] [Google Scholar]
- 19.Francis, M. S., M. Aili, M. L. Wiklund, and H. Wolf-Watz. 2000. A study of the YopD-LcrH interaction from Yersinia pseudotuberculosis reveals a role for hydrophobic residues within the amphipathic domain of YopD. Mol. Microbiol. 38:85-102. [DOI] [PubMed] [Google Scholar]
- 20.Friebel, A., H. Ilchmann, M. Aepfelbacher, K. Ehrbar, W. Machleidt, and W. D. Hardt. 2001. SopE and SopE2 from Salmonella typhimurium activate different sets of Rho GTPases of the host cell. J. Biol. Chem. 276:34035-34040. [DOI] [PubMed] [Google Scholar]
- 21.Fu, Y., and J. E. Galan. 1998. Identification of a specific chaperone for SptP, a substrate of the centisome 63 type III secretion system of Salmonella typhimurium. J. Bacteriol. 180:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 23.Galan, J. E., and R. Curtiss, III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galan, J. E., and R. Curtiss, III. 1991. Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect. Immun. 59:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galan, J. E., C. Ginocchio, and P. Costeas. 1992. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J. Bacteriol. 174:4338-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groisman, E. A., and H. Ochman. 1997. How Salmonella became a pathogen. Trends Microbiol. 5:343-349. [DOI] [PubMed] [Google Scholar]
- 27.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 28.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haller, J. C., S. Carlson, K. J. Pederson, and D. E. Pierson. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436-1446. [DOI] [PubMed] [Google Scholar]
- 30.Hardt, W. D., L. M. Chen, K. E. Schuebel, X. R. Bustelo, and J. E. Galan. 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93:815-826. [DOI] [PubMed] [Google Scholar]
- 31.Hardt, W. D., H. Urlaub, and J. E. Galan. 1998. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 33.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 34.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hueck, C. J., M. J. Hantman, V. Bajaj, C. Johnston, C. A. Lee, and S. I. Miller. 1995. Salmonella typhimurium secreted invasion determinants are homologous to Shigella Ipa proteins. Mol. Microbiol. 18:479-490. [DOI] [PubMed] [Google Scholar]
- 36.Jackson, M. W., J. B. Day, and G. V. Plano. 1998. YscB of Yersinia pestis functions as a specific chaperone for YopN. J. Bacteriol. 180:4912-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones, M. A., M. W. Wood, P. B. Mullan, P. R. Watson, T. S. Wallis, and E. E. Galyov. 1998. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect. Immun. 66:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaniga, K., J. C. Bossio, and J. E. Galan. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13:555-568. [DOI] [PubMed] [Google Scholar]
- 39.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 40.Kaniga, K., D. Trollinger, and J. E. Galan. 1995. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 177:7078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaniga, K., S. Tucker, D. Trollinger, and J. E. Galan. 1995. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 177:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lloyd, S. A., A. Forsberg, H. Wolf-Watz, and M. S. Francis. 2001. Targeting exported substrates to the Yersinia TTSS: different functions for different signals? Trends Microbiol. 9:367-371. [DOI] [PubMed] [Google Scholar]
- 43.Lloyd, S. A., M. Norman, R. Rosqvist, and H. Wolf-Watz. 2001. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol. Microbiol. 39:520-531. [DOI] [PubMed] [Google Scholar]
- 44.Lloyd, S. A., M. Sjostrom, S. Andersson, and H. Wolf-Watz. 2002. Molecular characterization of type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol. Microbiol. 43:51-59. [DOI] [PubMed] [Google Scholar]
- 45.Lucas, R. L., and C. A. Lee. 2000. Unravelling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol. Microbiol. 36:1024-1033. [DOI] [PubMed] [Google Scholar]
- 46.Luo, Y., M. G. Bertero, E. A. Frey, R. A. Pfuetzner, M. R. Wenk, L. Creagh, S. L. Marcus, D. Lim, F. Sicheri, C. Kay, C. Haynes, B. B. Finlay, and N. C. Strynadka. 2001. Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat. Struct. Biol. 8:1031-1036. [DOI] [PubMed] [Google Scholar]
- 47.Menard, R., P. Sansonetti, C. Parsot, and T. Vasselon. 1994. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell 79:515-525. [DOI] [PubMed] [Google Scholar]
- 48.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 49.Mirold, S., K. Ehrbar, A. Weissmüller, R. Prager, H. Tschäpe, H. Rüssmann, and W. D. Hardt. 2001. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5, and sopE2. J. Bacteriol. 183:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mirold, S., W. Rabsch, H. Tschape, and W. D. Hardt. 2001. Transfer of the Salmonella type III effector sopE between unrelated phage families. J. Mol. Biol. 312:7-16. [DOI] [PubMed] [Google Scholar]
- 52.Obert, S., R. J. O'Connor, S. Schmid, and P. Hearing. 1994. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol. Cell. Biol. 14:1333-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Page, A. L., and C. Parsot. 2002. Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 46:1-11. [DOI] [PubMed] [Google Scholar]
- 54.Page, A. L., P. Sansonetti, and C. Parsot. 2002. Spa15 of Shigella flexneri, a third type of chaperone in the type III secretion pathway. Mol. Microbiol. 43:1533-1542. [DOI] [PubMed] [Google Scholar]
- 55.Parsot, C., C. Hamiaux, and A. L. Page. 2003. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6:7-14. [DOI] [PubMed] [Google Scholar]
- 56.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prager, R., S. Mirold, E. Tietze, U. Strutz, B. Knüppel, W. Rabsch, W. D. Hardt, and H. Tschäpe. 2000. Prevalence and polymorphism of genes encoding translocated effector proteins among clinical isolates of Salmonella enterica. Int. J. Med. Microbiol. 290:605-617. [DOI] [PubMed] [Google Scholar]
- 58.Ramamurthi, K. S., and O. Schneewind. 2002. Type III protein secretion in Yersinia species. Annu. Rev. Cell Dev. Biol. 18:107-133. [DOI] [PubMed]
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Santos, R. L., S. Zhang, R. M. Tsolis, R. A. Kingsley, L. G. Adams, and A. J. Baumler. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 3:1335-1344. [DOI] [PubMed] [Google Scholar]
- 61.Schesser, K., E. Frithz-Lindsten, and H. Wolf-Watz. 1996. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178:7227-7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh, S. P., S. R. Singh, Y. U. Williams, L. Jones, and T. Abdullah. 1995. Antigenic determinants of the OmpC porin from Salmonella typhimurium. Infect. Immun. 63:4600-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sory, M. P., A. Boland, I. Lambermont, and G. R. Cornelis. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. USA 92:11998-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stebbins, C. E., and J. E. Galan. 2001. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414:77-81. [DOI] [PubMed] [Google Scholar]
- 65.Stebbins, C. E., and J. E. Galan. 2001. Structural mimicry in bacterial virulence. Nature 412:701-705. [DOI] [PubMed] [Google Scholar]
- 66.Stender, S., A. Friebel, S. Linder, M. Rohde, S. Mirold, and W. D. Hardt. 2000. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36:1206-1221. [DOI] [PubMed] [Google Scholar]
- 67.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tucker, S. C., and J. E. Galan. 2000. Complex function for SicA, a Salmonella enterica serovar Typhimurium type III secretion-associated chaperone. J. Bacteriol. 182:2262-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 70.Wattiau, P., and G. R. Cornelis. 1993. SycE, a chaperone-like protein of Yersinia enterocolitica involved in Ohe secretion of YopE. Mol. Microbiol. 8:123-131. [DOI] [PubMed] [Google Scholar]
- 71.Wattiau, P., S. Woestyn, and G. R. Cornelis. 1996. Customized secretion chaperones in pathogenic bacteria. Mol. Microbiol. 20:255-262. [DOI] [PubMed] [Google Scholar]
- 72.Woestyn, S., M. P. Sory, A. Boland, O. Lequenne, and G. R. Cornelis. 1996. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol. Microbiol. 20:1261-1271. [DOI] [PubMed] [Google Scholar]
- 73.Wood, M. W., M. A. Jones, P. R. Watson, S. Hedges, T. S. Wallis, and E. E. Galyov. 1998. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol. Microbiol. 29:883-891. [DOI] [PubMed] [Google Scholar]
- 74.Wood, M. W., M. A. Jones, P. R. Watson, A. M. Siber, B. A. McCormick, S. Hedges, R. Rosquist, T. S. Wallis, and E. E. Galyov. 2000. The secreted effector protein of Salmonella dublin, SopA, is translocated into eukaryotic cells and influences the induction of enteritidis. Cell. Microbiol. 2:293-303. [DOI] [PubMed] [Google Scholar]
- 75.Wood, M. W., R. Rosqvist, P. B. Mullan, M. H. Edwards, and E. E. Galyov. 1996. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol. Microbiol. 22:327-338. [DOI] [PubMed] [Google Scholar]
- 76.Zhou, D., L. M. Chen, L. Hernandez, S. B. Shears, and J. E. Galan. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248-260. [DOI] [PubMed] [Google Scholar]
- 77.Zhou, D., M. S. Mooseker, and J. E. Galan. 1999. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc. Natl. Acad. Sci. USA 96:10176-10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou, D., M. S. Mooseker, and J. E. Galan. 1999. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 283:2092-2095. [DOI] [PubMed] [Google Scholar]