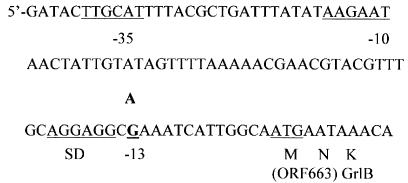Abstract
We report for the first time low-level quinolone resistance mediated by decreased expression of topoisomerase IV in Staphylococcus aureus. A single-step mutant of wild-type S. aureus strain ISP794, P18 selected by using twice the MIC of premafloxacin, had four- and four- to eightfold greater MICs of premafloxacin and ciprofloxacin, respectively, than the wild type. Sequencing of parEC and gyrBA with their promoter regions revealed a point mutation (G→A) 13 bp upstream of the start codon of parE. Genetic linkage studies showed that there was a high level of correlation between the mutation and the resistance phenotype, and allelic exchange confirmed the contribution of the mutation to resistance. Decreased expression of ParE and decreased steady-state levels of parEC transcripts in P18 and in resistant allelic exchange mutants were observed. The steady-state levels of gyrBA and topB transcripts were increased in P18 but not in two resistant allelic exchange mutants, and sequencing upstream of either gene did not reveal a difference between ISP794 and P18. The steady-state levels of topA transcripts were similar in the various strains. Growth competition experiments performed at 30, 37, and 41°C with a susceptible allelic exchange strain and a resistant allelic exchange strain suggested that loss of fitness was associated with reduced levels of ParE at 41°C. However, P18 had a growth advantage over ISP794 at all temperatures, suggesting that a compensatory mechanism was associated with the increased levels of gyrBA and topB transcripts. Thus, reduced levels of ParE appear to be compatible with cell survival, although there may be a fitness cost during rapid cell multiplication, which might be overcome by compensatory mechanisms without reversion of the resistance phenotype.
Quinolone antimicrobial agents interact with two type II topoisomerases, DNA gyrase (composed of two subunits, GyrA and GyrB) and topoisomerase IV (composed of two subunits, ParC and ParE), in Staphylococcus aureus and many other bacteria. Both of these enzymes are essential, but they have distinct and overlapping functions in the cell. DNA gyrase, the only enzyme that introduces negative supercoils into DNA, can also remove negative and positive supercoils, which accumulate ahead of the replication fork and in areas of gene transcription by RNA polymerase (48, 49). Although the gyrase can also resolve catenanes accumulating behind the replication fork, it does so substantially less efficiently than topoisomerase IV (55). Thus, the principal function of topoisomerase IV is the decatenation of interlinked replicated daughter chromosomes (1), an activity that it performs more efficiently than gyrase. Topoisomerase IV can also remove both negative and positive superhelical twists in vitro (28).
Type I topoisomerases, which are not targets of quinolones or other antimicrobial agents in clinical use, have additional roles in the cell that complement those of the quinolone target, type II enzymes. Topoisomerase I, encoded by the topA gene, opposes the negative supercoiling activity of DNA gyrase, removing negative supercoils from circular DNA and preventing excessive supercoiling. DNA supercoiling is under tight homeostatic control, involving reciprocal regulation of gene expression for DNA gyrase and topoisomerase I (13, 44). Topoisomerase III, the second type I topoisomerase, which is encoded by the topB gene, appears to play an important role in stabilization of the bacterial genome, and topB deletion mutants have increased rates of spontaneous deletions and RecA-independent recombination (43). Topoisomerase III, originally identified by its DNA-relaxing activity, is in fact inefficient in this function (41) and has been found to function as a decatenase in vitro (15) and, when overexpressed, in the removal of precatenanes in vivo (41), thus potentially complementing the function of topoisomerase IV.
The overlapping functions of different topoisomerases potentially allow compensation for a defect in one enzyme by changes in expression of another. For example, hyperexpression of gyrase may complement defects in topoisomerase IV (28), hyperexpression of topoisomerase IV may complement defects in topoisomerase I (27), and hyperexpression of topoisomerase III may complement defects in topoisomerase IV (41). Several organisms, including, Treponema pallidum, Mycobacterium tuberculosis, Mycobacterium smegmatis, Mycobacterium leprae, and Helicobacter pylori, do not contain genes encoding the subunits of topoisomerase IV (17, 42). Thus, in these species a gyrase or possibly another topoisomerase must perform the decatenation functions performed by topoisomerase IV in other species (17).
Quinolone antimicrobial agents act on type II topoisomerases by forming a ternary complex with the enzyme and DNA and by stabilizing a reversible enzyme reaction intermediate, the cleavable or cleavage complex, which can be converted to a form from which topoisomerase-mediated double-stranded DNA breaks are released. The lethal activity of quinolones is thought to result from the release of lethal, double-stranded breaks in DNA, an event that appears to require an additional poorly defined cellular function after formation of the ternary complex (9, 16, 34). Thus, quinolones convert type II topoisomerases to potent poisons which form DNA lesions that create double-stranded breaks in the genome (2, 29, 31).
Resistance to quinolones in bacteria is due primarily either to chromosomal mutations in the type II topoisomerases that make the enzymes less sensitive to inhibition by quinolones or to alterations that affect the access of the drug to the topoisomerase targets, such as increased expression of efflux pumps alone or (in gram-negative bacteria) in combination with reductions in the outer membrane porin diffusion channels (17, 18). Because for quinolones it is the DNA lesion formed by ternary complexes with the enzyme and drug that mediates quinolone activity, reductions in the levels of DNA lesions due to reductions in the amounts of a drug target enzyme could contribute to drug resistance. In this study, we observed for the first time low-level quinolone resistance mediated by decreased expression of topoisomerase IV in S. aureus. We also evaluated the consequences of reduced expression of topoisomerase IV for the bacterial cell.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and other materials.
The bacterial strains and plasmids used are shown in Table 1. Escherichia coli strains were grown in Luria-Bertani medium, and S. aureus strains were grown in brain heart infusion (BHI) medium, Trypticase soy medium (Becton Dickinson and Co., Sparks, Md.), or a minimal medium developed for S. aureus (51). Most strains were grown at 37°C; the only exceptions were S. aureus strains with the thermosensitive plasmids derived from pCL52.1, which were grown at 30°C. Lysostaphin was obtained from AMBI Products Inc., New York, N.Y., premafloxacin was obtained from Pharmacia & Upjohn Pharmaceuticals, Kalamazoo, Mich., and ciprofloxacin was obtained from Bayer Corp., Westhaven, Conn. Ampicillin, nalidixic acid, novobiocin, spectinomycin, tetracycline, and ethidium bromide were obtained from Sigma Chemical Co., St. Louis, Mo. Spectinomycin and ampicillin were used at a concentration of 50 μg/ml, and tetracycline was used at a concentration of 5 μg/ml, except during integration of plasmid pCL52.1 into the chromosome, when it was used at a concentration of 3 μg/ml. All primers used in this study were synthesized at the Tufts Core Facility, Boston, Mass., and are listed in Table 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or descriptiona | Source or reference |
|---|---|---|
| S. aureus strains | ||
| ISP794 | 8325 pig-131 | 45 |
| RN4220 | 8325-4 r− | 30 |
| MT5224c4 | 8325 nov (gyrB142) hisG15 pig 131 parC542 (Ser80Phe) | 46 |
| P18 | 8325 pig-131 flqA (parE), spontaneous premafloxacin resistance | This study |
| ISP2133 | 8325 pig-131 trp-489 Ω(chr::Tn917lac)2 | 46 |
| ISP2134 | 8325 pig-131 thrB484 Ω(chr::Tn917lac)1 | 46 |
| ISP225 | Ps55 | 46 |
| DIP18-55 | 8325 pig-131, susceptible allelic exchange transformant | This study |
| DIP18-6 | 8325 pig-131 flqA (parE), resistant allelic exchange transformant | This study |
| DIP18-35 | 8325 pig-131 flqA (parE), resistant allelic exchange transformant | This study |
| E. coli DH5α | F−φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 phoA hsdR17 (rK− mK−) supE44 λ−thi-1 gyrA96 relA1 | GIBCO-BRL |
| Plasmids | ||
| PGEM3-zf(+) | 3,199-bp cloning vector, Apr | Promega |
| pCL52.1 | 8,119-bp plasmid containing the replicon of pGB2, Spr (E. coli), and the temperature-sensitive replicon of pE194, Tcr | 35 |
| pTrcHisA | 4.4-kb expression vector, Apr | Invitrogen |
Apr, ampicillin resistance; Spr, spectinomycin resistance; Tcr, tetracyclineresistance.
TABLE 2.
Primers used in this study
| Primer for PCR and sequencing | Sequence (5′-3′) | Position of the 5′ nucleotide |
|---|---|---|
| parE primersa | ||
| GRLB1 | CGCCGATAAGATAACTTAGTACTA | 137 |
| GRLB1R | GCTCTTGACGCTCTTTACCAC | 1036 |
| GRLB2 | TAAGTGAACGACTACAAGAGTCTGC | 947 |
| GRLB2R | ACCTCCCGCAGAATCACCTTC | 1692 |
| GRLB3 | GCAAGGGAAGCTGCACGTAAAGC | 1537 |
| GRLB3R | GGCAATGCACGCTCTTGAATA | 2470 |
| parC primersa | ||
| GRLA1 | AGTTGAAGATGAAGTGCGTT | 2196 |
| GRLA1R | ACCTTGAATAATACCACCAG | 3041 |
| GRLA2 | GCTGAAGAGTTATTACGTGA | 2757 |
| GRLA2R | ACGCAAGATACCGTACTAGTA | 4028 |
| GRLA3 | GTTATGGTGCCTAGTGAAGAA | 3861 |
| GRLA3R | TAACGATAAGTACTTGGTCAGC | 4881 |
| Primers for upstream region of gyrBb | ||
| UPGB1 | AGGTGACGACTCGGTAACGC | 1,793 bp upstream of start site of gyrB |
| UPGB1R | AGGATATTCACGTCAGGATGACAT | 1,039 bp upstream of start site of gyrB |
| UPGB2 | TATCCCAGAATTACCTGAAGATG | 1,178 bp upstream of start site of gyrB |
| UPGB2R | ATAGGATCGACTTCAGAGAGAG | 127 bp downstream of start site of gyrB |
| gyrB primersc | ||
| GYRB1 | TAGACGATGTACTCAGTGAAT | 2 |
| GYRB1R | GTGGCATATCCTGAGTTATAT | 1004 |
| GYRB2 | CTATCACTATGAGGGCGGTA | 858 |
| GYRB2R | GCACGAGCAACGATAACACTC | 2252 |
| gyrA primers | ||
| GYRA1 | CCTCAATCAAGAATAAATGAACGA | 2164 |
| GYRA1R | TATCACGTAACAGTTCAAGTGT | 2566 |
| GYRA2 | GGTTCAATGGATGGAGATGG | 2482 |
| GYRA2R | CTGTATCTGACTCACGAATC | 3361 |
| GYRA3 | CGTACTGGTGTGCGTGTCGTT | 3046 |
| GYRA3R | CTGGCGTACGTTTACCATAAC | 4336 |
| GYRA4 | CAACGGGTGTGAAAGGTAT | 4211 |
| GYRA4R | GCCACCATCAAGACTTATC | 5027 (8) |
| norA promoter primersd | ||
| DMBL3 | CAATGCCGCTGACAGATGTA | 27 |
| DMBR3 | TTGTCAGCTAGCGTACCACCAA | 550 |
| Primers for upstream region of topBe | ||
| UPTB1 | GTTATGGCTGATATAGGTGGTG | 3,085 bp upstream of start site of topB |
| UPTB1R | CAAAACGCAATAACACATCATGAC | 2,259 bp upstream of start site of topB |
| UPTB2 | GGGCTTTTATTTATGACATAGTTG | 2,446 bp upstream of start site of topB |
| UPTB2R | ACTTGTTCATGTTCTATTCTCA | 1,922 bp upstream of start site of topB |
| UPTB3 | GACGACGATCTTTTCTTTCTCC | 2,047 bp upstream of start site of topB |
| UPTB3R | GGGGAGCACTTGCAAAAATA | 1,215 bp upstream of start site of topB |
| UPTB4 | CGTCGTACCACGAATT | 1,368 bp upstream of start site of topB |
| UPTB4R | AGGCATTCAACAATACAAACCA | 528 bp upstream of start site of topB |
| UPTB5 | TGTAATTTGCTGAAAGCACGA | 686 bp upstream of start site of topB |
| UPTB5R | CTAACGCCCACGTGACAATA | 122 bp downstream of start site of topB |
| parE probe primersa | ||
| GLBP | CAGGTTTTAGAGGGGTTAGAA | 421 |
| GLBPR | TGTCGGTTTACCTGATTTATG | 654 |
| parC probe primersa | ||
| GRLAP | GATACGACACTCGAACCAAT | 2817 |
| GRLA1R | ACCTTGAATAATACCACCAG | |
| gyrB probe primersc | ||
| GYBP | CTATCACTATGAGGGCGGTATTA | 858 |
| GYBPR | GTATCTTCACCAGAAAGTCTATC | 1124 |
| gyrA probe primers | ||
| GYRA3 | CGTACTGGTGTGCGTGTCGTT | |
| GYRA2R | CTGTATCTGACTCACGAATC | |
| topA probe primersb | ||
| TOPAP | GCGTAATCCAGCAAACCCATT | 747 bp from start site |
| TOPAPR | CACCTTGTTTCCCTGATGCTTT | 1018 bp from start site |
| topB probe primersb | ||
| TOPB3 | AAGCCAATCCGTCGATTATG | 370 bp from start site |
| TOPB3R | CGTTCCTGTCTGAACACGTC | 582 bp from start site |
Selection and characterization of mutant P18.
Mutant P18 was selected from wild-type S. aureus strain ISP794 on BHI agar containing twice the MIC of premafloxacin (0.008 μg/μl), as previously described (20). The mutant was purified once on BHI agar containing the same level of premafloxacin and then on BHI agar without premafloxacin to ensure a pure culture prior to storage at −70°C.
MICs of premafloxacin, ciprofloxacin, novobiocin, nalidixic acid, and ethidium bromide were determined by the agar dilution method. MICs of all antimicrobials and chemicals were determined in duplicate at least twice on Trypticase soy agar (TSA) containing serial twofold dilutions of the antibiotics; when the changes in MIC were only twofold, MICs were determined three times. Growth was scored after 24 and 48 h of incubation at 37°C.
PCR amplification for sequence analysis.
Chromosomal DNAs of S. aureus strains were extracted with an Easy-DNA kit (Invitrogen, Carlsbad, Calif.) following lysis with lysostaphin at a concentration of 0.1 mg/ml and were used as templates for PCRs. PCRs for parC, parE, gyrA, gyrB, the promoter region of parE, a 1,897-bp region upstream of gyrB, a 3,085-bp region upstream of topB, and the promoter region of norA were performed by using Vent DNA polymerase (New England Biolabs, Beverly, Mass.). An annealing temperature of 55°C and an extension time of 1 min for each 1,000 bp of PCR product were used for the reactions. PCR products were purified with a Compass kit (American Bioanalytical, Natick, Mass.). Automated DNA sequencing of the PCR products was performed by using an automated ABI 3100 DNA sequencer (Tufts Core Facility).
DNA transformation.
For transformation in S. aureus, high-molecular-weight (HMW) DNA was prepared by the method of Stahl and Pattee (45), and cells were made competent for transformation as described by Lindberg et al. (36), except that the original inoculum was grown in Trypticase soy broth (TSB) at 35°C overnight in a shaking water bath. Bacteriophage Φ55 was maintained on strain ISP225 and was used to render the S. aureus strains competent for transformation.
Correlation of resistance phenotype and genotype in genetic crosses.
Erythromycin-resistant transformants of ISP794, which had been transformed with a derivative of mutant P18 containing a Ω(chr::Tn917lac)2 insertion (encoding erythromycin resistance) linked to parEC, were screened for changes in the MIC of premafloxacin. Ten resistant transformants (MIC of premafloxacin, 0.032 μg/ml) and 10 susceptible transformants (MIC of premafloxacin, 0.008 μg/ml) were selected for PCR amplification and sequencing of the relevant promoter region and the structural gene of parE. The PCR primers used for amplification of the fragment encompassing the mutation in P18 were GRLB1 and GRLB1R (Table 2). An annealing temperature of 58°C and an extension time of 1 min were used for the reactions. DNA sequencing of the PCR products was performed to determine the presence of the mutation in resistant and susceptible transformants.
Cloning and allelic exchange.
The PCR primers used to amplify a 652-bp internal fragment of parE together with the 378-bp upstream region and the conditions used for PCRs have been described previously (21). Cloning into the thermosensitive shuttle vector pCL52.1 and the allelic exchange procedure were performed as previously described (21). The resultant colonies following the allelic exchange procedure were screened for susceptibility to tetracycline at a concentration of 3 μg/ml and for resistance to premafloxacin and ciprofloxacin at concentrations of 0.008 and 0.25 μg/ml, respectively.
Expression of ParE.
Cell lysates were prepared from 20-ml overnight cultures of ISP794, P18, a resistant allele exchange strain (DIP18-35), and a susceptible allelic exchange strain (DIP18-55) in TSB incubated at 37°C by pelleting the cells, resuspending them in 500 μl of phosphate-buffered saline (pH 7.4), and lysing them with lysostaphin at a concentration of 0.1 mg/ml in the presence of a complete protease inhibitor cocktail (Roche Molecular Biochemicals, Mannheim, Germany) for 1 min at 37°C, followed by 10 min on ice. Equal amounts of total protein were loaded in the wells of sodium dodecyl sulfate-12% polyacrylamide gels for electrophoresis, and the gels were blotted onto nitrocellulose membranes (NitroBind; 0.45-μm pore size; Osmonics Inc., Westborough, Mass.). Experiments were repeated several times and were performed with different cell lysates to ensure that the differences seen were not due to chance differences in loading. The methods used for overexpression and purification of histidine-tagged ParE and GyrB proteins have been described previously (23). Purified ParE protein was used as a positive control, and purified GyrB was used to show that there was no cross-reactivity between anti-ParE antibodies and GyrB. Polyclonal rabbit antisera against two epitopes of ParE (1:500; MNKQNNYSDDSIQC-amide and acetyl-CSEVQVLENDQFDEEFI-OH) prepared by QCB Division, Biosource International, Inc., Hopkinton, Mass., were used for Western blot analysis. Anti-rabbit immunoglobulin G peroxidase conjugate (1:5,000; Sigma-Aldrich, Inc., St. Louis, Mo.) was used as the secondary antibody.
To quantify changes in the levels of protein on Western blots, densitometry was performed by scanning the photographs and performing an analysis with a Power Macintosh G3 computer by using the public domain NIH Image program (version 1.61) (developed at the National Institutes of Health and available at http://rsb.info.nih.gov/nih-image/). The values were expressed in arbitrary units. The fold change in the signal intensity was determined after the background value was subtracted.
RNA analysis.
Following growth of strains to the mid-logarithmic phase (optical density at 600 nm [OD600], 0.6) in BHI medium and lysis with lysostaphin at a concentration of 0.1 μg/ml at 37°C for 20 min, total S. aureus RNA was extracted with an RNeasy mini kit (Qiagen, Valencia, Calif.). The concentration of total RNA was determined spectrophotometrically by measuring the absorbance at 260 nm. For Northern blot analysis, 8 μg of total RNA was electrophoresed in a 1% agarose-0.66 M formaldehyde gel in morpholinepropanesulfonic acid (MOPS) by using a NorthernMax kit (Ambion Inc., Austin, Tex.) and was blotted onto Hybond-N+ membranes (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom) with a vacuum blotter (Bio-Rad Laboratories, Hercules, Calif.). Visualization of the rRNA bands in ethidium bromide-stained agarose gels was used to validate the equivalence of total RNA added in the lanes prior to transfer to the blotting membranes. DNA probes for topoisomerase genes amplified from the ISP794 chromosome and labeled with psoralen-biotin were used for detection of specific RNA molecules (BrightStar psoralen-biotin nonisotopic labeling and detection kits; Ambion). The primers used for generation of probes are shown in Table 2. An annealing temperature of 55°C and an extension time of 1 min for each 1,000 bp were used for the PCRs. PCR products were extracted with a Compass kit before labeling. The probes for parE, parC, gyrB, gyrA, topA, and topB mRNA were 234, 225, 266, 316, 272, and 213 bp long, respectively. Northern blots were hybridized with probes overnight, washed, and autoradiographed with Hyperfilm ECL (Amersham Pharmacia Biotech). A Northern blot analysis for each gene was performed at least three times with at least two different RNA samples.
To quantify changes in levels of RNA on Northern blots, densitometry was performed by scanning the radiographs and performing an analysis with a Power Macintosh G3 computer by using the public domain NIH Image program as described above for protein quantification on Western blots. The values were expressed in arbitrary units, and the fold change in the signal intensity was determined after the background value was subtracted.
Fitness of the mutant.
S. aureus strains grown on TSA overnight were pelleted and resuspended in normal saline to an OD600 of 0.1; this was followed by 1:100 dilution into BHI broth, TSB, or supplemented defined medium (51). Separate cultures were grown at 30, 37, and 41°C with shaking, and the OD600 was measured hourly for up to 8 h. Colony counts were determined by plating appropriate dilutions of cultures on TSA at 0, 4, and 8 h. The biological cost of resistance was studied by performing competition experiments. Overnight cultures of a resistant allelic exchange transformant (DIP18-35) and a susceptible allelic exchange transformant (DIP18-55) on TSA were resuspended in TSB to an OD600 of 0.1, and 250 μl of each resuspension was used to inoculate 25 ml of TSB, resulting in a final concentration of ∼106 CFU/ml. Following growth for 4 h, the cultures were diluted 1:100 in fresh TSB and grown overnight. Sampling on TSA was performed at various times, and ≥100 colonies from each time were screened for resistance to premafloxacin. Competition experiments were performed at 30, 37, and 41°C in duplicate at least three times. Competition assays were also performed in duplicate twice for wild-type strain ISP794 and the original mutant P18 as described above. The competition results were expressed as the ratio of the number of sensitive colonies to the number of resistant colonies at each time examined (5).
RESULTS
Characterization of P18.
The MICs of premafloxacin and ciprofloxacin were fourfold greater for mutant P18 than for wild-type strain ISP794 (Table 3); this difference in MICs was similar to the differences observed for other first-step mutants with parEC mutations selected with premafloxacin (20). The MICs of novobiocin (which was used to screen for some parE mutations [11, 12, 20, 21, 23, 24]) and nalidixic acid (which was used to screen for some gyrA mutations [23]) were not different for the mutant and the wild type. Because other premafloxacin-selected, first-step mutants of S. aureus had had mutations in parE or parC (20), we first sequenced the quinolone resistance-determining regions of the parE and parC structural genes of mutant P18. After we found no mutations in these regions, we determined the complete sequences of the parE and parC genes and unexpectedly found a single point mutation (G→A) 13 bp upstream of the ATG start codon of parE, between the start codon and a putative ribosomal binding site (52) (Fig. 1). The complete sequences of the gyrA and gyrB genes, including the 179 bp upstream of gyrB, were also determined, and no additional mutation was found.
TABLE 3.
Characteristics of the single-step mutant P18 and wild-type strain ISP794
| Strain | MIC (μg/ml) of:
|
Mutations
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Premafloxacin | Ciprofloxacin | Novobiocin | Nalidixic acid | Ethidium bromide | grlA | parE | gyrA | gyrB | norA promoter | |
| ISP794 | 0.004-0.008 | 0.125-0.25 | 0.064-0.128 | 64-128 | 2.0-4.0 | |||||
| P18 | 0.016-0.032 | 1.0 | 0.064 | 128 | 4.0 | None | −13 (G→A) | None | None | None |
FIG. 1.
Nucleotide sequence upstream of ParE. The −35 and −10 regions of the putative promoter, the presumptive Shine-Dalgarno (SD) sequence, the start codon for ParE, and the G→A mutation at position −13 are underlined (52).
The MIC of ethidium bromide (which was used to screen for overexpression of the NorA efflux pump [26]) for mutant P18 was not different from the MIC for ISP794, suggesting that there is not overexpression of NorA or other as-yet-unidentified efflux pumps that contributes to the resistance phenotype. Sequencing of the promoter region of norA confirmed that there was no mutation.
Genetic linkage of quinolone resistance loci.
To define the relationship of the novel mutation to the quinolone resistance phenotype, genetic crosses in which HMW chromosomal DNA was used were performed as previously described (20). The Tn917 transposon insertions (chr::Tn917lac)2 and Ω(chr::Tn917lac)1, encoding erythromycin resistance (Ermr), have been shown previously to be linked to parC and parE, which are contiguous genes in S. aureus (12, 20, 21, 40, 46). Incrossing experiments were performed to assess whether the proportion of transformants that regained the susceptible phenotype was consistent with the known linkages of these Tn917 insertions and parE+ and to generate transformants in which Tn917 insertions were linked to the mutant parE allele; such transformants were then used to prepare HMW DNA to demonstrate by transformational outcrossing that the mutant phenotype and the mutant parE allele were linked to the Tn917 insertions and to each other. Thus, first HMW chromosomal DNA from strain ISP2133 Ω(chr::Tn917lac)2 parC+ parE+ or ISP2134 Ω(chr::Tn917lac)1 parC+ parE+ was used to transform mutant P18, with selection for Ermr. One Ermr transformant (P183315) that contained Ω(chr::Tn917lac)2 linked to parEC and that retained the premafloxacin resistance phenotype of P18 was then used to prepare HMW chromosomal DNA for a transformational outcrossing experiment with wild-type strain ISP794 (parC+ parE+).
In the incrossing experiments (Table 4), the linkages of the susceptible phenotype (associated with parE+) to Ω(chr::Tn917lac)2 and Ω(chr::Tn917lac)1 were similar to those previously identified for established resistance mutations in parC (20, 21, 46) and parE (12, 21) and were also shown in parallel experiments to be similar to the linkage to parC (Ser80Phe). In an outcrossing experiment (Table 4), as in the incrossing experiments, the linkage of the resistance phenotype to Ω(chr::Tn917lac)2 was again similar to the linkages reported for known parEC resistance mutations (12, 20, 21), establishing that the resistance phenotype was genetically linked to these Tn917 insertions in a manner consistent with the hypothesis that the phenotype was due to the mutant parE allele.
TABLE 4.
Genetic linkage of Tn917 insertions and parEC mutations and resistance by transformation: incrossing of parC+ and parE+ and outcrossing of parE
| Expt | Donor
|
Recipient
|
MICs (μg/ml) for recipient
|
MICs (μg/ml) for susceptible or resistant transformantsa
|
% Susceptible or resistant transformants (no. of transformants susceptible or resistant/total no. of transformants)a | ||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | Genotype | Strain | Genotype | Premafloxacin | Ciprofloxacin | Premafloxacin | Ciprofloxacin | ||
| Incrossing | ISP2133 | trp-489 Ω(chr::Tn917lac)2 | P18 | parE −13 (G→A) | 0.016-0.032 | 1.0 | 0.008 | 0.25 | 25 (28/112) |
| MT5224c4 | parC (Ser80Phe) | 0.032 | 1.0 | 0.008 | 0.25 | 21 (8/38) | |||
| ISP2134 | thrB494 Ω(chr::Tn917lac)1 | P18 | parE −13 (G→A) | 0.016-0.032 | 1.0 | 0.008 | 0.25 | 6 (6/104) | |
| MT5224c4 | parC (Ser80Phe) | 0.032 | 1.0 | 0.008 | 0.25 | 5 (8/171) | |||
| Outcrossing | P183315 | parE (G→A at nucleotide position −13) Ω(chr::Tn917lac)2 | ISP794 | Wild type | 0.004-0.008 | 0.25 | 0.016-0.032 | 1.0 | 14 (11/176) |
In the incrossing experiment the transformants examined were susceptible transformants, and in the outcrossing experiment the transformants examined were resistant transformants.
To establish the concordance of the resistance phenotype and the presence of the mutant parE allele, direct DNA sequencing of the amplicons encompassing parE and its upstream region was performed, and this revealed the presence of the mutant allele in 10 of 10 resistant outcrossed transformants and the presence of the wild-type allele in 10 of 10 susceptible outcrossed transformants. Thus, the presence of the novel mutation strongly correlated with resistance and appeared to be sufficient to account for the resistance phenotype.
Proof of the role of the novel mutation obtained by allelic exchange.
To obtain definitive proof of the role of the novel mutation in resistance in mutant P18, allelic exchange of the mutation in the parent strain was performed. Following introduction of the mutation into wild-type S. aureus strain ISP794, the MICs of ciprofloxacin (0.5 to 1.0 μg/ml) and premafloxacin (0.016 μg/ml) for both the P18 mutant and the genetically constructed resistant strains were found to be the same and were fourfold higher than the MICs for the wild-type strain. The presence of the desired mutation in two resistant allelic exchange transformants was also confirmed by direct DNA sequencing of PCR products performed with the primers used for correlation of the resistance phenotype and genotype in outcrossed transformants.
Levels of ParE in wild-type and resistant strains.
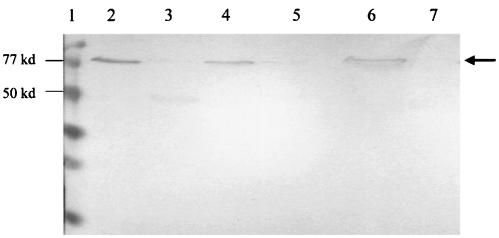
A novel resistance mutation near the ribosomal binding site of parE was predicted to decrease the expression of ParE. To test this hypothesis, Western blot analyses of cell extracts (Fig. 2) from wild-type strain ISP794 (lane 2) and mutant P18 (lane 3), as well as susceptible (DIP18-55) (lane 4) and resistant (DIP18-35) (lane 5) transformants generated during the allelic exchange procedure, were performed by using anti-ParE antisera. Reduced amounts of an immunoreactive, 79,000-kDa band were found in the resistant strain (P18 and DIP18-35) extracts compared to the amounts in the extracts from the wild-type parent strain and the susceptible transformant, which is consistent with the presence of reduced amounts of ParE in the mutant (Fig. 2). As determined by scanning densitometry, the reductions were 3.3-fold for P18 and 3.0-fold for DIP18-35 compared to the amounts in the wild-type parent strain. Similar reductions in the amounts of ParE were seen in logarithmically growing cells (data not shown) and in the stationary-phase cells used for the experiment whose results are shown in Fig. 2. Purified histidine-tagged ParE gave a positive signal, and purified histidine-tagged GyrB gave no signal, indicating the specificity of the antisera for ParE. Thus, the point mutation 13 bp upstream of the start codon of parE was associated with decreased expression of the ParE protein. An additional faint, 50-kDa, immunoreactive band was obtained with the P18 mutant. The nature of this band is uncertain, but we speculate that it could represent translation of an aborted parE transcript or a degradation product of ParE.
FIG. 2.
Western blot analysis of ParE in wild-type S. aureus ISP794, mutant P18, and susceptible and resistant allelic exchange transformants. Cell lysates of ISP794 (lane 2), P18 (lane 3), DIP18-55 (lane 4), and DIP18-35 (lane 5) were used. The control purified proteins used were ParE-His (lane 6) and GyrB-His (lane 7). The protein size markers in lane 1 were low-range sodium dodecyl sulfate-polyacrylamide gel electrophoresis standards (Bio-Rad). ParE expression (arrow) was decreased in both the original mutant and the allelic exchange clone containing the mutation at position −13. kd, kilodaltons.
Transcriptional levels of parE, parC, gyrB, gyrA, topA, and topB.
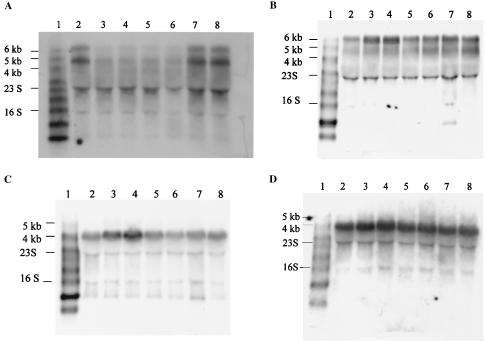
Northern blots of RNA from ISP794, P18, two resistant allelic exchange strains (DIP18-35 and DIP18-6), and a susceptible allelic exchange strain (DIP18-55), as well as a mutant with a common parC structural gene mutation known to cause quinolone resistance (MT522c4, Ser80Phe), were probed with psoralen-biotin-labeled DNA probes for parE, parC, gyrB, gyrA, topA, and topB. parE and parC are adjacent genes in S. aureus (total length, 4,393 bp), and the initiation codon of parC overlaps the stop codon of parE (10, 52). Although each gene has a putative promoter and a putative ribosomal binding site (52), parC expression appears to depend on the promoter of parE (11). Northern analysis for parE and parC revealed polycistronic expression; a ∼4.8-kb transcript corresponding to the transcription of parEC and a transcript slightly larger than 6 kb were found. The levels of the RNAs of both parE and parC were decreased in both the P18 mutant (3.3- and 2.9-fold, respectively) and the two allelic exchange transformants (DIP18-35 and DIP18-6) (3.3-fold) (Fig. 3A), suggesting that effects at the level of transcription or mRNA stability contributed to the decreased levels of ParE in the constructs. Since the 4.8- and 6-kb transcripts changed concordantly, we speculate that the 6-kb transcript could represent a transcriptional readthrough of the parC transcriptional stop sequence at the end of parC rather than the result of readthrough of additional putative promoters upstream of parE.
FIG. 3.
Northern blot analysis of topoisomerase gene expression. The following RNA samples were used: RNA marker (biotinylated RNA Millennium marker; Ambion) (lane 1), wild-type strain ISP794 (lane 2), original mutant P18 (lanes 3 and 4), resistant allelic exchange transformant DIP18-35 (lane 5), resistant allelic exchange transformant DIP18-6 (lane 6), susceptible allelic exchange transformant DIP18-55 (lane 7), and parC mutant MT522c4 (lane 8). (A) parE mRNA. Northern hybridization for parC mRNA produced identical results (data not shown). (B) gyrA mRNA. Northern hybridization for gyrB mRNA produced similar results (data not shown). (C) topB mRNA. (D) topA mRNA.
Since defects in topoisomerase IV may be compensated for by overexpression of gyrase, we determined the levels of gyrB and gyrA mRNAs (Fig. 3B). In S. aureus, gyrB and gyrA are adjacent genes, and there is a putative promoter upstream of gyrB (within the recF gene) and a putative transcription terminator just downstream of gyrA (8). gyrBA (from the start codon of gyrB to the stop codon of gyrA) is 4,635 bp long (25). Northern analysis of both gyrB and gyrA revealed two transcripts, a ∼4.8-kb transcript and a more predominant transcript that was slightly larger than 6 kb, indicating that polycistronic transcription occurred. The smaller transcript presumably corresponded to gyrBA, and the larger transcript corresponded to gyrBA together with the upstream gene recF, which was in the same open reading frame (as determined by using the open reading frame finder at the National Center for Biotechnology Information website). The steady-state levels of both gyrBA transcripts were increased in the P18 mutant (∼2-fold increase, as determined by densitometry) but, unexpectedly, were not increased in the resistant allelic exchange mutants, suggesting that there may have been an additional mutation either in the promoter region of gyrB or in a regulatory gene that caused increased transcription of gyrase in the P18 mutant. Since there is a putative transcription terminator just downstream of gyrA and since recF is in the same open reading frame as gyrBA, we sequenced a region upstream of gyrBA, including the promoter region of recF, to determine if any mutation was present. Sequencing of the 1,793 bp upstream of the start codon of gyrB (including the 1,113 bp of recF and 671 bp upstream of recF) revealed no differences between ISP794 and P18.
Levels of topB mRNA, encoding topoisomerase III, which, when overexpressed, has been shown to be able to act as the principal cellular decatenase in E. coli (41), were also determined. Similar to the results for gyrase, expression of topB was increased in the P18 mutant (approximately 1.7-fold increase, as determined by densitometry) but not in the allelic exchange mutants (Fig. 3C). The main transcript for topB was ∼4.1 kb long, and there was a weaker transcript at ∼5.1 kb, whereas the gene itself is 2,136 bp long in S. aureus (4, 32). Sequencing of the 3,085 bp upstream of the start codon of topB was performed for ISP794 and P18, and no mutation was found in P18.
Steady-state levels of topA mRNA, encoding topoisomerase I, were also determined by Northern analysis, and no differences were found among ISP794, P18, and the allelic exchange mutant (Fig. 3D).
In vitro fitness of resistant strains.
There was no detectable difference in the separately measured growth rates of ISP794, P18, resistant allelic exchange transformant DIP18-35, and susceptible allelic exchange transformant DIP18-55, as determined by both OD600 and viable cell counting during growth at 30, 37, or 41°C in rich media (BHI broth and TSB) or supplemented defined medium (data not shown).
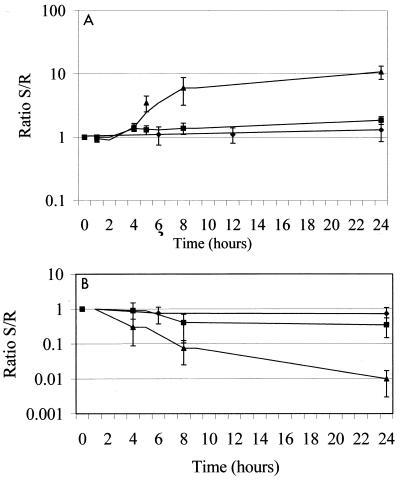
Since RNA analysis had indicated that there may have been compensatory increases in expression of DNA gyrase and topoisomerase III in the P18 mutant but not in the allelic exchange mutant, we postulated that a more subtle growth defect might result from the presence of reduced levels of ParE and that compensatory changes had occurred in the P18 mutant either upon selection or upon subsequent passage that were not present in the subsequently constructed allelic exchange mutant. Thus, we performed growth competition experiments with resistant (DIP18-35, mutation present) and susceptible (DIP18-55, no mutation present) transformants from the allele exchange experiment. No significant growth advantage for either strain (i.e., the ratio of the cell counts for the two strains remained stable and near 1 over time) was identified at 30 or 37°C. In contrast, for growth at 41°C at 24 h the competition ratio was approximately 10, with DIP18-55 outnumbering DIP18-35, suggesting that there was a loss of fitness at increased temperatures in the cells with reduced levels of ParE (Fig. 4A).
FIG. 4.
Growth competition experiments performed at 30°C (♦), 37°C (▪), and 41°C (▴). Ratio S/R, ratio of the susceptible strain to the resistant strain. (A) Competition between a resistant allelic exchange transformant (DIP18-35) and a susceptible allelic exchange transformant (DIP18-55). (B) Competition between wild-type strain ISP794 and resistant mutant P18. Similar patterns were seen when the data were expressed per generation of growth on the x axis.
Competition assays with the original mutant P18 and wild-type strain ISP794 were then performed to determine if the increased levels mRNAs of the genes encoding gyrase and topoisomerase III were associated with compensation for the growth defect seen in allelic exchange mutant DIP18-35. Interestingly, P18 outcompeted ISP794 at all temperatures, but the differences were most striking at 37 and 41°C, at which the competition ratios were 0.35 and 0.01, respectively (Fig. 4B). Thus, although both P18 and DIP18-35 had the quinolone resistance mutation upstream of parE and reduced levels of ParE, the growth defect found in DIP18-35 was not present in P18, which distinctively exhibited increased levels of gene transcripts coding for gyrase and topoisomerase III.
DISCUSSION
Quinolones are often referred to as topoisomerase poisons due to their ability to trap a covalent topoisomerase-DNA complex as a topoisomerase-quinolone-DNA ternary complex and subsequent generation of double-stranded breaks in DNA that ultimately lead to cell death (9, 14, 31). With agents that act in this manner, reductions in the amounts of a drug target can have the effect of reducing the number of poisonous complexes formed. Hence, a reduction in the amount of the topoisomerase IV in a cell could reduce drug action by reducing the number of ternary complexes, which are precursors of the lethal DNA lesions, thus leading to relative quinolone resistance. Low-level resistance to chemotherapeutic drugs that act as cellular poisons by targeting eukaryotic topoisomerase II has in fact been associated with decreased levels of this topoisomerase, in support of this principle (33).
Premafloxacin is an exceptionally potent quinolone which was initially evaluated for veterinary use (50, 56). The primary target of premafloxacin in S. aureus is topoisomerase IV, and selection of novel mutations outside the quinolone resistance-determining region suggested novel interactions of premafloxacin with topoisomerase IV (20). During selection of single-step mutants of S. aureus with premafloxacin to determine its target enzyme, we selected a mutant with a four- to eightfold increase in resistance to quinolones and no mutation in the structural genes of the two type II topoisomerases (DNA gyrase and topoisomerase IV). A point mutation (G→A) was found 13 bp upstream of the ATG start codon of parE, just downstream of the putative ribosomal binding site. Genetic linkage studies showed that this mutation was correlated with the resistance phenotype, and its contribution to the resistance phenotype was firmly established by allelic exchange.
This mutation, near the putative ribosomal binding site, was predicted to decrease expression of topoisomerase IV, primarily ParE. However, since ParC expression is also dependent on the promoter of ParE, decreased expression of both subunits of topoisomerase IV could occur. Western blot analysis of ParE revealed reduced amounts of ParE both in the original mutant and resulting from allelic exchange, thus providing the first example of low-level resistance to quinolones in bacteria due to reduced amounts of the target enzyme. We did not have access to specific antisera against S. aureus ParC and thus did not measure ParC levels directly. Reductions in the levels of parE and parC transcripts, however, suggest that the levels of the topoisomerase IV holoenzyme were similarly reduced. We do not yet know if the reductions in the levels of transcripts were due to decreased transcription (from changes in promoter strength or changes in the binding site for a regulatory factor) or decreased stability of the mutant mRNA.
Since topoisomerase IV is an essential enzyme and likely acts as the principal cellular decatenase under normal conditions, we evaluated the effects of reduced expression of this enzyme on cellular growth and sought evidence for compensatory mechanisms, which are often present when resistance mutations have negative effects on bacterial fitness (6, 7, 39). The most sensitive method for detecting differences in fitness between strains is pairwise competition experiments, which by repeated measurements allow detection of fitness differences of <1% (3). The reduced levels of topoisomerase IV found in the allelic exchange mutant were associated with reductions in competitive growth in laboratory media at 41°C but not at 37 or 30°C compared to the growth of strains generated in the same experiment that lacked the mutation. Thus, it appears that the level of reduction of topoisomerase IV in this mutant has a detectable fitness cost only at high temperatures in laboratory media, conditions under which more rapid cell multiplication and DNA replication may result in the most stringent need for decatenase function in the cell. Interestingly, no discernible differences in colony size were observed for the exchange mutant and the parent strain grown on agar at any of the three temperatures studied. It is also possible that additional fitness costs might be detected under different conditions of growth of this pair of strains.
Surprisingly, however, we found no growth defect in the original P18 mutant compared to its parent strain, suggesting that it had acquired compensating mechanisms. We thus evaluated the possibility that altered expression of the other cellular topoisomerases may be associated with the lack of a growth defect in P18. The levels of the gyrBA and topB transcripts were increased in the original mutant but not in the two resistant allelic exchange transformants. Since DNA gyrase and topoisomerase III have both been shown to have decatenase functions as purified enzymes and under certain conditions in the cell, overexpression of either enzyme might compensate at least in part for downregulation of topoisomerase IV. We thus speculate that the increased levels of transcripts of both DNA gyrase and topoisomerase III reflected a compensatory mechanism that accounted for the absence of a growth defect and even an apparent growth advantage in mutant P18. Furthermore, these data suggest that a physiological compensatory mechanism was not responsible for the apparent increased levels of expression of gyrase and topoisomerase III in P18, since the levels of reduction of ParE were similar in both P18 and the resistant allelic exchange mutant, the latter of which exhibited no increase in gyrase or topoisomerase III transcripts. Thus, the compensatory mechanism was likely genetic and extragenic to parCE. In addition, no mutations were found upstream of gyrBA or topB. Thus, the genetic basis of the apparent compensatory mechanisms remains to be defined, but increased expression from both gyrBA and topB suggests the possibility that there is a regulatory mechanism that has not been defined yet. It is possible that a mutation(s) in addition to the mutation in parE was selected in P18 upon exposure to premafloxacin, which, like other quinolones, is likely to induce error-prone SOS DNA repair (37, 54). We have described previously selection of a parC gyrA double mutant from a single plating on premafloxacin (20). Thus, either a premafloxacin-induced mutation or a spontaneous mutation overcame a more subtle growth defect during multiplication of P18 on laboratory media.
It is noteworthy that no change in the level of topA transcripts was seen in P18 or the allelic exchange mutants. Thus, the presumed compensatory effects in P18 do not affect all topoisomerases. Furthermore, although there is reciprocal regulation of DNA gyrase and topoisomerase I expression mediated by the DNA supercoiling dependence of the gyrase and topoisomerase I gene promoters (38, 44, 47), the level of the DNA gyrase gene transcripts in P18 was not associated with a reciprocal reduction in the level of the topoisomerase I gene transcripts. We speculate that the weak DNA relaxation activity of topoisomerase III when the enzyme is overexpressed may counterbalance the increase in DNA supercoiling expected to occur with increases in the level of gyrase. The absence of mutations upstream of the gyrase and topoisomerase III genes further suggests that an undefined trans-acting compensatory mutation(s) may affect the levels of expression of these enzymes.
Reduced levels of ParE in the allelic exchange mutants appear to be compatible with cell survival without increased expression of gyrase or topoisomerase III. Thus, we speculate that S. aureus tolerates reduced levels of topoisomerase IV and that this tolerance may account for the broader range of quinolone resistance mutations found for this drug target compared to the mutations found for DNA gyrase. In addition to the mutation in P18, we found a range of quinolone resistance mutations outside the classical quinolone resistance-determining regions of ParC and ParE (20-24), and purified topoisomerase IV reconstituted with some of the mutant subunits exhibited reduced catalytic function (X. Zhang and D. Hooper, unpublished observations). Although DNA gyrase and topoisomerase IV are both essential enzymes and there are lethal null mutations in the genes encoding subunits of these enzymes, the cell may tolerate incomplete reductions in the amounts of the two enzymes differently. Since DNA gyrase is the only cellular enzyme that can effect negative supercoiling of DNA, mutations that cause quinolone resistance and also produce incomplete reductions in the amount of active gyrase may be more poorly tolerated than mutations that reduce the functional levels of topoisomerase IV.
This difference between topoisomerase IV and gyrase further suggests that the range of first-step resistance mutations in the drug target may be greater for quinolones that target primarily topoisomerase IV than for quinolones that act on DNA gyrase as their primary target. In a recent survey, the reported alleles of gyrA associated with resistance in E. coli (gyrase is the primary target of most quinolones in E. coli) comprised 8 codons and resulted in 17 amino acid changes, and the reported alleles of parC associated with resistance in S. aureus (topoisomerase IV is the primary target of most quinolones in S. aureus) comprised 15 codons and resulted in 23 amino acid changes (19). Importantly, our data also provide another example of how bacteria are able to overcome reduced fitness associated with chromosomal antibiotic resistance mutations by acquisition of extragenic compensatory mechanisms without reversion of the resistance phenotype (6, 7, 39), a phenomenon that may be of increasing clinical importance as multidrug resistance increases. Also of concern is the observation that overcompensation appeared to occur such that the compensated P18 mutant grew more readily than the parent strain, a phenomenon that has also been reported for the virulence properties of antibiotic-resistant rpsL and gyrA mutants of Salmonella enterica serovar Typhimurium that developed extragenic mutations that overcompensated for the growth defects of the original mutants (6). Thus, like adaptations to the costs of resistance in E. coli, adaptations to the costs of resistance in S. aureus may further enhance fitness in unexpected ways.
Acknowledgments
We thank Xiamei Zhang for providing a parE clone for overexpression and purification of ParE and for providing purified GyrB, Chia Lee for providing plasmid pCL52.1, and Que-Chi Truong-Bolduc for her assistance with the RNA experiments and densitometry.
This work was supported by Public Health Service grant R01 AI23988 from the National Institutes of Health to D.C.H.
REFERENCES
- 1.Adams, D. E., E. M. Shekhtman, E. L. Zechiedrich, M. B. Schmid, and N. R. Cozzarelli. 1992. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell 71:277-288. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, V. E., T. D. Gootz, and N. Osheroff. 1998. Topoisomerase IV catalysis and the mechanism of quinolone action. J. Biol. Chem. 273:17879-17885. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, D. I., J. Bjorkman, and D. Hughes. 2001. Fitness and virulence of antibiotic resistant bacteria. In D. Hughes and D. I. Andersson (ed.), Antibiotic development and resistance. Taylor and Francis, New York, N.Y.
- 4.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 5.Björkholm, B., M. Sjölund, P. G. Falk, O. G. Berg, L. Engstrand, and D. I. Andersson. 2001. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. USA 98:14607-14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Björkman, J., D. Hughes, and D. I. Andersson. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorkman, J., I. Nagaeu, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479-1482. [DOI] [PubMed] [Google Scholar]
- 8.Brockbank, S. M., and P. T. Barth. 1993. Cloning, sequencing, and expression of the DNA gyrase genes from Staphylococcus aureus. J. Bacteriol. 175:3269-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drlica, K., and X. L. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 11.Fournier, B., and D. C. Hooper. 1998. Effects of mutations in GrlA of topoisomerase IV from Staphylococcus aureus on quinolone and coumarin activity. Antimicrob. Agents Chemother. 42:2109-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier, B., and D. C. Hooper. 1998. Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolone and coumarin activity. Antimicrob. Agents Chemother. 42:121-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heddle, J. G., T. Lu, X. L. Zhao, K. Drlica, and A. Maxwell. 2001. gyrB-225, a mutation of DNA gyrase that compensates for topoisomerase I deficiency: investigation of its low activity and quinolone hypersensitivity. J. Mol. Biol. 309:1219-1231. [DOI] [PubMed] [Google Scholar]
- 14.Hiasa, H. 2002. The Glu-84 of the ParC subunit plays critical roles in both topoisomerase IV-quinolone and topoisomerase IV-DNA interactions. Biochemistry 41:11779-11785. [DOI] [PubMed] [Google Scholar]
- 15.Hiasa, H., and K. J. Marians. 1994. Topoisomerase III, but not topoisomerase I, can support nascent chain elongation during theta-type DNA replication. J. Biol. Chem. 269:32655-32659. [PubMed] [Google Scholar]
- 16.Hooper, D. C. 1998. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin. Infect. Dis. 27:S54-S63. [DOI] [PubMed] [Google Scholar]
- 17.Hooper, D. C. 1999. Mechanisms of quinolone resistance. Drug Resist. Updates 2:38-55. [DOI] [PubMed] [Google Scholar]
- 18.Hooper, D. C. 2002. Fluoroquinolone resistance among Gram-positive cocci. Lancet Infect. Dis. 2:530-538. [DOI] [PubMed] [Google Scholar]
- 19.Hooper, D. C. 2003. Mechanisms of quinolone resistance, p. 41-67. In D. C. Hooper and E. Rubinstein (ed.), Quinolone antimicrobial agents. ASM Press, Washington, D.C.
- 20.Ince, D., and D. C. Hooper. 2000. Mechanisms and frequency of resistance to premafloxacin in Staphylococcus aureus: novel mutations suggest novel drug-target interactions. Antimicrob. Agents Chemother. 44:3344-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ince, D., and D. C. Hooper. 2001. Mechanisms and frequency of resistance to gatifloxacin in comparison to AM-1121 and ciprofloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ince, D., X. Zhang, and D. C. Hooper. 2003. Activity of and resistance to moxifloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:1410-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ince, D., X. Zhang, L. C. Silver, and D. C. Hooper. 2002. Dual targeting of DNA gyrase and topoisomerase IV: target interactions of garenoxacin (BMS-284756, T3811ME), a new desfluoroquinolone. Antimicrob. Agents Chemother. 46:3370-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ince, D., X. Zhang, L. C. Silver, and D. C. Hooper. 2003. Topoisomerase targeting with and resistance to gemifloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:274-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, H., H. Yoshida, M. Bogaki-Shonai, T. Niga, H. Hattori, and S. Nakamura. 1994. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2014-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaatz, G. W., and S. M. Seo. 1995. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 39:2650-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato, J., Y. Nishimura, R. Imamura, H. Niki, S. Hiraga, and H. Suzuki. 1990. New topoisomerase essential for chromosome segregation in E. coli. Cell 63:393-404. [DOI] [PubMed] [Google Scholar]
- 28.Kato, J., H. Suzuki, and H. Ikeda. 1992. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J. Biol. Chem. 267:25676-25684. [PubMed] [Google Scholar]
- 29.Khodursky, A. B., and N. R. Cozzarelli. 1998. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J. Biol. Chem. 273:27668-27677. [DOI] [PubMed] [Google Scholar]
- 30.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 31.Kreuzer, K. N., and N. R. Cozzarelli. 1979. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J. Bacteriol. 140:424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 33.Larsen, A. K., and A. Skladanowski. 1998. Cellular resistance to topoisomerase-targeted drugs: from drug uptake to cell death. Biochim. Biophys. Acta Gene Struct. Express. 1400:257-274. [DOI] [PubMed] [Google Scholar]
- 34.Levine, C., H. Hiasa, and K. J. Marians. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta Gene Struct. Express. 1400:29-43. [DOI] [PubMed] [Google Scholar]
- 35.Lin, W. S., T. Cunneen, and C. Y. Lee. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J. Bacteriol. 176:7005-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindberg, M., and R. P. Novick. 1973. Plasmid-specific transformation in Staphylococcus aureus. J. Bacteriol. 115:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mamber, S. W., B. Kolek, K. W. Brookshire, D. P. Bonner, and J. Fung-Tomc. 1993. Activity of quinolones in the Ames Salmonella TA102 mutagenicity test and other bacterial genotoxicity assays. Antimicrob. Agents Chemother. 37:213-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menzel, R., and M. Gellert. 1983. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell 34:105-113. [DOI] [PubMed] [Google Scholar]
- 39.Nagaev, I., J. Björkman, D. I. Andersson, and D. Hughes. 2001. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol. Microbiol. 40:433-439. [DOI] [PubMed] [Google Scholar]
- 40.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1996. Quinolone resistance mutations in topoisomerase IV: relationship of the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nurse, P., C. Levine, H. Hassing, and K. J. Marians. 2003. Topoisomerase III can serve as the cellular decatenase in Escherichia coli. J. Biol. Chem. 278:8653-8660. [DOI] [PubMed] [Google Scholar]
- 42.Schmutz, E., A. Muhlenweg, S. M. Li, and L. Heide. 2003. Resistance genes of aminocoumarin producers: two type II topoisomerase genes confer resistance against coumermycin A(1) and clorobiocin. Antimicrob. Agents Chemother. 47:869-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schofield, M. A., R. Agbunag, M. L. Michaels, and J. H. Miller. 1992. Cloning and sequencing of Escherichia coli mutR shows its identity to topB, encoding topoisomerase III. J. Bacteriol. 174:5168-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snoep, J. L., C. C. Van der Weijden, H. W. Andersen, H. V. Westerhoff, and P. R. Jensen. 2002. DNA supercoiling in Escherichia coli is under tight and subtle homeostatic control, involving gene-expression and metabolic regulation of both topoisomerase I and DNA gyrase. Eur. J. Biochem. 269:1662-1669. [DOI] [PubMed] [Google Scholar]
- 45.Stahl, M. L., and P. A. Pattee. 1983. Confirmation of protoplast fusion-derived linkages in Staphylococcus aureus by transformation with protoplast DNA. J. Bacteriol. 154:406-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trucksis, M., J. S. Wolfson, and D. C. Hooper. 1991. A novel locus conferring fluoroquinolone resistance in Staphylococcus aureus. J. Bacteriol. 173:5854-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tse-Dinh, Y. C. 1985. Regulation of the Escherichia coli DNA topoisomerase I gene by DNA supercoiling. Nucleic Acids Res. 13:4751-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, J. C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3:430-440. [DOI] [PubMed] [Google Scholar]
- 49.Wang, J. C., and A. S. Lynch. 1993. Transcription and DNA supercoiling. Curr. Opin. Genet. Dev. 3:764-768. [DOI] [PubMed] [Google Scholar]
- 50.Watts, J. L., S. A. Salmon, M. S. Sanchez, and R. J. Yancey. 1997. In vitro activity of premafloxacin, a new extended-spectrum fluoroquinolone, against pathogens of veterinary importance. Antimicrob. Agents Chemother. 41:1190-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkinson, B. J. 1997. Biology, p. 1-38. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 52.Yamagishi, J. I., T. Kojima, Y. Oyamada, K. Fujimoto, H. Hattori, S. Nakamura, and M. Inoue. 1996. Alterations in the DNA topoisomerase IV grlA gene responsible for quinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1157-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida, H., M. Bogaki, S. Nakamura, K. Ubukata, and M. Konno. 1990. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J. Bacteriol. 172:6942-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ysern, P., B. Clerch, M. Castano, I. Gibert, J. Barbe, and M. Llagostera. 1990. Induction of SOS genes in Escherichia coli and mutagenesis in Salmonella typhimurium by fluoroquinolones. Mutagenesis 5:63-66. [DOI] [PubMed] [Google Scholar]
- 55.Zechiedrich, E. L., and N. R. Cozzarelli. 1995. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 9:2859-2869. [DOI] [PubMed] [Google Scholar]
- 56.Zerva, L., S. A. Marshall, and R. N. Jones. 1996. Premafloxacin: a projected veterinary use fluoro-quinolone with significant activity against multi-resistant Gram-positive human pathogens. J. Antimicrob. Chemother. 38:742-744. [DOI] [PubMed] [Google Scholar]