Abstract
The deduced protein product of open reading frame slr0946 from Synechocystis sp. strain PCC 6803, SynArsC, contains the conserved sequence features of the enzyme superfamily that includes the low-molecular-weight protein-tyrosine phosphatases and the Staphylococcus aureus pI258 ArsC arsenate reductase. The recombinant protein product of slr0946, rSynArsC, exhibited vigorous arsenate reductase activity (Vmax = 3.1 μmol/min · mg), as well as weak phosphatase activity toward p-nitrophenyl phosphate (Vmax = 0.08 μmol/min · mg) indicative of its phosphohydrolytic ancestry. pI258 ArsC from S. aureus is the prototype of one of three distinct families of detoxifying arsenate reductases. The prototypes of the others are Acr2p from Saccharomyces cerevisiae and R773 ArsC from Escherichia coli. All three have converged upon catalytic mechanisms involving an arsenocysteine intermediate. While SynArsC is homologous to pI258 ArsC, its catalytic mechanism exhibited a unique combination of features. rSynArsC employed glutathione and glutaredoxin as the source of reducing equivalents, like Acr2p and R773 ArsC, rather than thioredoxin, as does the S. aureus enzyme. As postulated for Acr2p and R773 ArsC, rSynArsC formed a covalent complex with glutathione in an arsenate-dependent manner. rSynArsC contains three essential cysteine residues like pI258 ArsC, whereas the yeast and E. coli enzymes require only one cysteine for catalysis. As in the S. aureus enzyme, these “extra” cysteines apparently shuttle a disulfide bond to the enzyme's surface to render it accessible for reduction. SynArsC and pI258 ArsC thus appear to represent alternative branches in the evolution of their shared phosphohydrolytic ancestor into an agent of arsenic detoxification.
Arsenate and related compounds (e.g., arsenite, antomite) are naturally occurring, broadly acting toxins frequently encountered at biologically deleterious concentrations in the environment (reviewed in reference 44). The strong chemical parallels between phosphorous and arsenic, which reside in the same column of the periodic table, limit the ability of phosphate transport systems to discriminate between this vital nutrient and arsenate (45), thus exacerbating the latter's toxic potential. Microorganisms combat the collateral importation of arsenate by a two-step mechanism in which this compound is first reduced to arsenite. Although arsenite is a more potent toxicant than arsenate, the former can be selectively banished from the cell's interior through the intervention of a dedicated, inducible transporter.
While the basic strategy for conferring arsenical resistance is broadly conserved among microorganisms, the arsenate reductases responsible for catalyzing the conversion of arsenate to arsenite are not. To date, three “detoxifying” arsenate reductases have been identified and characterized in molecular detail: Acr2p from the microbial eukaryote Saccharomyces cerevisiae (2), ArsC encoded by plasmid R773 from the gram-negative bacterium Escherichia coli (R773 ArsC) (7), and ArsC encoded by plasmid pI258 from the gram-positive bacterium Staphylococcus aureus (pI258 ArsC) (20). It should be noted that some microorganisms can utilize arsenate as a terminal electron acceptor for the generation of cellular energy (30). However, their “respiratory” arsenate reductases differ radically in both structure and function from the detoxifying arsenate reductases (24) and therefore will not be considered further herein.
Acr2p, R773 ArsC, and pI258 ArsC are the exemplars of three distinct, independent families of arsenate reductases that differ from one another in several of their physical and catalytic properties (reviewed in reference 37). Recombinant Acr2p forms a homodimer (38), while R773 ArsC (31) and pI258 ArsC (19) are monomeric. The E. coli (15) and yeast (38) enzymes obtain their reducing equivalents from glutathione and glutaredoxin, whereas pI258 ArsC requires thioredoxin (19, 21). While R773 ArsC displayed only a slight hint of sigmoidal kinetic behavior with respect to arsenate (28), Acr2p exhibited strong positive cooperativity (36), and the saturation curve for pI258 ArsC is strikingly biphasic (19, 33). The S. aureus enzyme contains three catalytically essential cysteine residues (32), while R773 ArsC (28) and Acr2p (36) each possess only one. While genes encoding homologs of both R773 ArsC (4, 11, 39, 47, 51) and pI258 ArsC (6, 46, 52) have been identified in a variety of bacterial organisms, in general, the functional characteristics of their protein products have yet to be examined in detail.
Perhaps not surprisingly given the chemical similarity of arsenate and phosphate, Acr2p from S. cerevisiae and pI258 ArsC from S. aureus both are descended from protein phosphatases (14, 48). Acr2p is a derivative of the Cdc25 family of dual-specific protein phosphatases that are responsible for the dephosphorylation of adjacent phosphotyrosine and phosphothreonine residues on cyclin-dependent protein kinases during the eukaryotic cell cycle (40). pI258 ArsC, on the other hand, is derived from a family of protein-tyrosine phosphatases sometimes referred to as the low-molecular-weight protein-tyrosine phosphatases (LMW PTPs) (42). Both pI258 ArsC (56) and a homologous protein from Bacillus subtilis (1) exhibit vestigial phosphohydrolase activity toward the small phosphomonoester p-nitrophenyl phosphate (pNPP).
Despite their discrete origins, Cdc25 and the LMW PTPs (along with a third protein phosphatase family, the conventional protein-tyrosine phosphatases) have converged upon a common catalytic mechanism and active-site motif, Cys-Xaa5-Arg, sometimes referred to as the P-loop (reviewed in references 18 and 57). The defining feature of this mechanism is the formation of a covalent phosphoenzyme intermediate with the conserved active-site cysteine. The P-loop arginine participates in the binding of phosphoester substrates and the stabilization of the phosphoenzyme intermediate. The Cys-Xaa5-Arg active-site signature sequence also is conserved among the respective arsenate-reducing derivatives of these protein phosphatases, where the conserved cysteine forms the analogous thioester with arsenate (reviewed in reference 37). Intriguingly, while R773 ArsC from E. coli exhibits no recognizable sequence similarity with either the yeast or S. aureus arsenate reductases, it too has converged upon a catalytic mechanism involving the formation of an arsenocysteine intermediate (31).
During the course of our continuing investigations into the protein-serine/threonine/tyrosine phosphorylation-dephosphorylation events that take place in Synechocystis sp. strain PCC 6803, we encountered three open reading frames [ORFs] in the cyanobacterium's genome whose deduced protein products contained the Cys-Xaa5-Arg-Ser/Thr-Xaa85-105-Asp-Pro signature sequence that is conserved among the LMW PTPs and their homologs (48). When analysis of the expressed protein product of one of these ORFs, slr0946, revealed it to be a relatively inefficient phosphohydrolase, we explored the possibility that it might be a pI258 ArsC-like arsenate reductase. While this did, indeed, prove to be the case, the catalytic mechanism of the arsenate reductase from Synechocystis sp. strain PCC 6803, SynArsC, was observed to deviate in many important respects from that of the S. aureus prototype.
MATERIALS AND METHODS
Materials.
Glutaredoxin 1, glutathione reductase, thioredoxin, and thioredoxin reductase were all from Sigma (St. Louis, Mo.). Chelating Sepharose Fast Flo was from Pharmacia (Uppsala, Sweden). The pET101/D-TOPO cloning kit was from Invitrogen (Carlsbad, Calif.). Oligonucleotides were from Life Technologies, Inc. (Frederick, Md.). [35S]glutathione was from Perkin-Elmer Life Sciences (Boston, Mass.). All general laboratory reagents and culture media were from Sigma or Fisher (Pittsburgh, Pa.), unless otherwise noted.
Standard procedures.
Protein concentrations were determined by the method of Bradford (3) using premixed reagent and a standardized solution of bovine serum albumin, both from Pierce (Rockford, Ill.). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (25). Gels were stained with Coomassie blue as described by Fairbanks et al. (13).
Growth of the organism and isolation of genomic DNA.
The cyanobacterium Synechocystis sp. strain PCC 6803 was cultivated in 200 ml of BG11 medium (43) at a temperature of 27°C with continuous shaking at 100 rpm and continuous lighting at 10 microeinsteins m2 s−1. Upon reaching the late exponential phase of growth, the cells were harvested by centrifugation, washed three times with type 1 reagent-grade water (resistivity ≥ 17.3 Mohm cm−1), and stored at −80°C until needed. Cells were lysed, and genomic DNA was isolated as described by Li et al. (26).
Cloning of ORF slr0946 and expression of its recombinant protein product.
ORF slr0946 was cloned using the materials provided in the pET101/D-TOPO cloning kit following the manufacturer's protocols. Briefly, slr0946 was amplified by PCR using genomic DNA (20 ng) as template and 10 pmol each of a forward and a reverse oligonucleotide primer. The sequences of the forward and reverse primers were, respectively, 5′-CACCGTGAAAAAGGTAATGTTCGTTTGC-3′ and 5′-GCTAATTTTGGCGATCAGGTTTTCCACC-3′. The resulting PCR product was then ligated into vector PET101/D-TOPO, which added oligonucleotides encoding a C-terminal extension to the full-length sequence of the wild-type protein that contained a hexahistidine sequence and a recognition epitope for the anti-Xpress antibody. Fusion to the C terminus was selected to avoid potential interference with the predicted active-site sequence of the protein, which was located near the extreme N terminus of the protein. The resulting plasmid was used to transform One Shot TOP 10 chemically competent E. coli cells. The orientation and sequence of the cloned gene were verified by DNA sequencing of the isolated plasmid. This plasmid was then used to transform E. coli BL21Star (DE3) One Shot cells. The transformed cells were cultured at 37°C in 200 ml of Luria-Bertani medium containing 0.1 mg of ampicillin/ml until the optical density at 600 nm reached 0.6 to 0.8. Isopropyl-β-d-thiogalactopyranoside was then added to a final concentration of 1 mM, and the cells were harvested 3 h later.
Site-directed mutagenesis of ORF slr0946 was performed using Promega's (Madison, Wis.) Gene Editor in vitro site-directed mutagenesis system according to the manufacturer's instructions. For the mutagenic alteration of Cys8, Cys13, Cys80, and Cys82 to Ser, the following primers were used: 5′-AATGTTCGTTAGCAAACGTAAT-3′, 5′-ACGTAATTCCAGTCGCTCCCA-3′, 5′-TAATTTCCCTTAAGTGGCTGTGG-3′, and 5′-CCTTTGTGGCAGTGGGGTTAA-3′, respectively.
Purification of rSynArsC.
The cell pellet harvested as described above was resuspended in 5 ml of 50 mM Tris-HCl, pH 7.5, containing 10 mM imidazole, 250 mM NaCl, and 5% (vol/vol) glycerol (lysis buffer). Lysozyme (10 mg) was added, and the suspension was placed on ice for 30 min. The cells then were lysed by sonic disruption, and the lysate was clarified by centrifugation at 10,000 × g for 20 min. The supernatant liquid was applied to a 0.5- by 5-cm column of chelating Sepharose Fast Flo that had been charged with NiSO4 and subsequently equilibrated with lysis buffer. The column was then washed in lysis buffer, and adherent proteins were eluted with lysis buffer containing 150 mM imidazole. Protein-containing fractions were then pooled, dialyzed versus 20 mM Tris-HCl (pH 7.5) containing 0.5 mM EDTA and 1 mM dithiothreitol (DTT), and stored at 4°C. Unless otherwise noted, recombinant SynArsC (rSynArsC) was dialyzed versus 20 mM Tris-HCl (pH 7.5) containing 0.5 mM EDTA to remove DTT prior to each experiment.
Assay of arsenate reductase activity.
Arsenate reductase activity was measured using a coupled assay system that measures the arsenate-dependent oxidation of NADPH (15). Briefly, the recombinant protein product of ORF slr0946 (rSynArsC; 25 μg unless otherwise indicated) was incubated at 37°C in 1.0 ml of 100 mM Tris-HCl (pH 7.5) containing, unless otherwise indicated, 0.1 mg of bovine serum albumin/ml, 0.25 mM NADPH, 0.2 μM yeast glutathione reductase, 8 mM reduced glutathione, 40 mM sodium arsenate, and 4 μM E. coli glutaredoxin 1. Oxidation of NADPH was monitored spectrophotometrically at a wavelength of 340 nM. The quantity of NADPH oxidized was calculated using an extinction coefficient of 6,200 M−1 cm−1. Kinetic constants were extrapolated using a Hanes-Woolf plot.
Assay of phosphatase activity.
The phosphatase activity of rSynArsC initially was assayed using pNPP as substrate. Briefly, 25 μg of rSynArsC was incubated for 30 min at 37°C in 200 μl of 100 mM Tris-HCl (pH 7.5) containing 5 mM DTT. The concentration of pNPP was varied over a range of 10 to 250 mM. The reaction was terminated by the addition of 400 μl of 0.5 M NaOH, and the absorbance of the resulting solution was determined at a wavelength of 410 nm. The quantity of p-nitrophenol produced was calculated using an extinction coefficient of 17,800 M−1 cm−1. Kinetic constants were extrapolated using a Hanes-Woolf plot. Inclusion of DTT in the assay buffer resulted in an approximately twofold increase in reaction rate with little effect on the apparent affinity for substrate.
The assay for protein-tyrosine phosphatase activity employed similar incubation conditions to those described above, with the exception that reduced carboxyamidomethylated and maleylated lysozyme that had been phosphorylated on tyrosine residues, at a final concentration of 3 μM protein-bound [32P]phosphate, was substituted for pNPP. The phosphorylation of reduced carboxyamidomethylated and maleylated lysozyme, using [γ-32P]ATP and the lyn protein-tyrosine kinase, and the determination of the quantity [32P]phosphate hydrolyzed following incubation with rSynArsC both were performed as described in reference 41. The estimated limit of detection of this assay was ≥0.55 pmol/min · mg.
Detection of protein-glutathione complexes.
The majority of the dissolved oxygen was purged from the solutions used for this experiment by the alternate application of vacuum and nitrogen gas. rSynArsC and mutationally altered versions thereof (24 to 30 μg) were incubated for 15 min at 37°C with 4 μCi of [35S]glutathione (4 mM final concentration) in a 100-μl volume of 50 mM Tris-HCl (pH 7.5) in the presence or absence of arsenate (20 mM). The incubation was terminated by the addition of 3 volumes of ice-cold acetone. The precipitated protein was collected by centrifugation at 12,000 × g for 3 min and washed three times with ice-cold acetone. The washed pellet was air dried and resuspended in 30 μl of water, and 10 μl of 4× SDS sample buffer was added. The protein was resolved from residual free glutathione by SDS-PAGE on a 15% (wt/vol) acrylamide gel. Radiolabeled species were visualized by electronic autoradiography. The sections of the gel containing rSynArsC were carefully excised, and the quantity of 35S radioactivity bound thereto was determined by liquid scintillation counting.
Detection of free sulfhydryl groups.
The number of free sulfhydryl groups on rSynArsC and mutagenically altered versions thereof was determined by the method of Ellman (12).
Phylogenetic analysis.
The DNA-derived amino acid sequence of SynArsC was aligned with those of other known or potential arsenate reductases and bacterial LMW PTPs. Next, this information was converted into a distance matrix from which a phylogenetic tree was constructed using the neighbor-joining method, all using Clustal W version 1.7 (53). The seed number for random number generation and the number of bootstrap trials were set to 111 and 1,000, respectively.
Cloning and expression of E. coli glutaredoxin 1.
The procedures used for cloning the grxA gene from E. coli K-12 (GenBank accession number M13449 [17]), altering its sequence via site-directed mutagenesis, and expressing and purifying the resulting recombinant protein products were identical to those used with ORF slr0946 with the following modifications. First, genomic DNA from E. coli K-12 was used as a template for the original PCR amplification step, and the sequences of the forward and reverse primers were 5′-CACCATGCAAACCGTTATTTTGGTCG-3′ and 5′-GGCGTCCAGATTTTCTTTCACCCATG-3′, respectively. For the mutagenic alteration of Cys11 to Ser, the following primer was used: 5′-CACAGTAAGGGGAACCCGAACGA-3′. For the mutagenic alteration of Cys14 to Ser, the following primer was used: 5′-TTGCACGCACAGAGTAAGGGCAAC-3′.
RESULTS
The protein product of ORF slr0946 hydrolyzes pNPP at a low but measurable rate.
The deduced protein product of ORF slr0946 contains the Cys-Xaa5-Arg-Ser/Thr-Xaa85-105-Asp-Pro signature sequence that is universally conserved among the LMW PTPs (Fig. 1). Although we recognized that these sequence features also were shared by pI258 ArsC from S. aureus (48), genome context appeared to argue against this possibility. The genes for the detoxifying arsenate reductases in other microorganisms were located in physically contiguous operons that also encoded the membrane pump responsible for extruding arsenite as well as the repressor that modulates their expression in response to arsenical compounds (2, 4, 6, 7, 11, 20, 46, 47, 51, 52). Since no obvious candidates for ORFs encoding either of these key partner proteins were evident in the vicinity of slr0946, we initially annotated its deduced protein product as an LMW PTP (48).
FIG. 1.
DNA-derived amino acid sequence of Slr0946 (22), the predicted product of ORF slr0946, aligned with that of ArsC from S. aureus plasmid pI258 (20) and the putative arsenate reductase from B. subtilis (52). Amino acid identities are indicated by colons, while amino acids with similar properties are indicated by dots. The P-loop conserved between S. aureus ArsC and LMW PTPs (48) is underlined. The catalytically essential cysteine residue within the P-loop of LMW PTPs is denoted by an asterisk, while the conserved, catalytically essential arginine is denoted by the @ symbol. The conserved Asp-Pro sequence, in which the Asp serves as a catalytically important acid-base in LMW PTPs, is denoted by a pair of arrowheads (∧∧). The two distal cysteines, Cys82 and Cys89, that participate in the catalytic disulfide bond cascade in S. aureus pI258 ArsC (32, 33) are denoted by # signs.
The full-length protein product of ORF slr0946 was expressed as a recombinant fusion protein in E. coli containing a C-terminal histidine tag and purified to apparent electrophoretic homogeneity by metal-chelate chromatography. The recombinant protein, which eluted as a homodimer when analyzed by gel filtration chromatography (data not shown), exhibited clearly detectable phosphohydrolase activity toward pNPP, a general substrate for many phosphohydrolases, including the LMW PTPs, in vitro. However, kinetic analyses indicated that its maximal catalytic efficiency for the hydrolysis of pNPP was 1 to 2 orders of magnitude lower and its Km was 1 to 2 orders of magnitude higher than those reported for LMW PTPs from other bacteria (Table 1). Moreover, when the protein was challenged with a commonly used phosphoprotein substrate for protein-tyrosine phosphatases, reduced carboxyamidomethylated and maleylated lysozyme that had been phosphorylated with the lyn protein-tyrosine kinase (41), no protein-tyrosine phosphatase activity was detected (data not shown).
TABLE 1.
Comparison of the phosphohydrolase activity of the recombinant protein product of ORF slr0946, rSlr0946, with other detoxifying arsenate reductases and several bacterial LMW PTPs
| Organism and enzyme | Vmax (μmol/ min/mg) | Km (mM) | Reference |
|---|---|---|---|
| Arsenate reductases | |||
| Synechocystis rSlr0946 (rSynArsC) | 0.08 | 77 | This paper |
| S. aureus ArsC | 0.04 | 146 | 56 |
| B. subtilis ArsC | 0.04a | NAb | 1 |
| LMW PTPs | |||
| S. coelicolor A3(2) PtpA | 4.85 | 0.75 | 27 |
| S. aureus PtpA | 33.6 | 1.2 | 50 |
| S. aureus PtpB | 1.2 | 1.5 | 50 |
| E. coli Wzb | 4.6 | 1 | 54 |
| A. johnsonii Ptp | 9.7 | 5 | 16 |
Rate of pNPP hydrolysis measured at a single substrate concentration of 70 mM.
NA = not applicable.
The protein product of ORF slr0946 possesses arsenate reductase activity.
Continued annotation of the genome of Synechocystis sp. strain PCC 6803 (22) revealed that an ORF immediately adjacent to slr0946, slr0945, resembled arsH. arsH is a gene of unknown function found in a few arsenate-resistant bacteria (39, 47). In pathogenic strains of Yersinia, however, its presence in either cis or trans was essential for arsenate resistance (39). The revelation that ORF slr0946 was juxtaposed with a potential arsenate resistance gene, coupled with the lackluster performance of its recombinant protein product as a phosphatase, stimulated us to directly assess whether it might be an arsenate reductase.
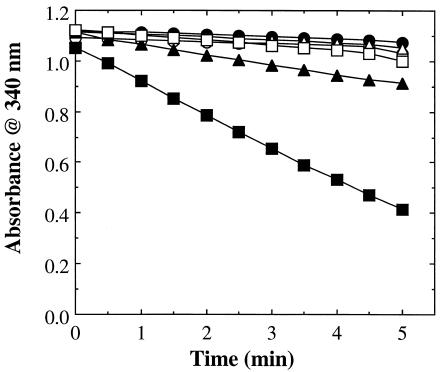
When the recombinant protein was assayed using thioredoxin—the source of reducing equivalents for the homologous arsenate reductase from S. aureus, pI258 ArsC (19, 21), thioredoxin reductase, and NADPH—no arsenate reductase activity could be detected. However, when glutathione and glutaredoxin—the source of reducing equivalents for the dissimilar arsenate reductases Acr2p (38) and R773 ArsC (15)—and glutathione reductase were substituted for thioredoxin and thioredoxin reductase, NADPH was oxidized at a relatively rapid rate (Fig. 2). Product formation in this coupled enzyme system was linearly dependent upon time and the quantity of protein present. Pretreatment of the protein with 1 mM DTT had no apparent effect on the observed reaction rate. In the absence of arsenate, little or no NADPH oxidation could be detected, even when an equivalent quantity of either phosphate or sulfate was included, indicating that the protein product of ORF slr0946 was acting as an arsenate reductase. We therefore designated the native protein SynArsC and the recombinantly produced fusion protein used for our experiments as rSynArsC. rSynArsC required glutaredoxin for activity (Fig. 2). Significant levels of arsenate-dependent NADPH oxidation, i.e., ≥25% of maximum, were detectable over a wide range of pH values, i.e., 6.0 to 9.5, with highest activity being achieved around pH 7.5 in this coupled enzyme system.
FIG. 2.
rSlr0946 possesses arsenate reductase activity. The arsenate reductase activity of the recombinant protein product of ORF slr0946, rSlr0946 (25 μg), was measured spectrophotometrically as described in Materials and Methods (open circles). Also shown are the changes in absorbance observed in control assays in which either the recombinant protein (closed circles), arsenate (open triangles), glutathione (closed triangles), glutaredoxin (open squares), or glutathione reductase (closed squares) was omitted.
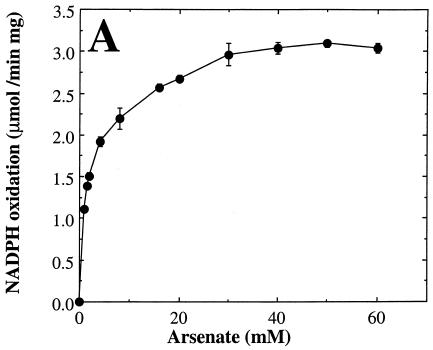
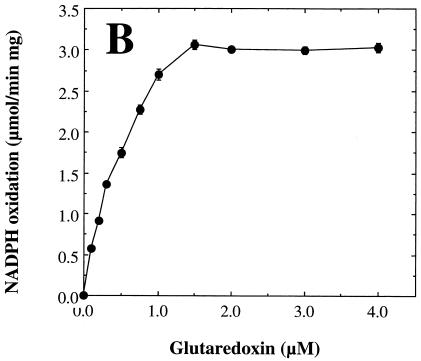
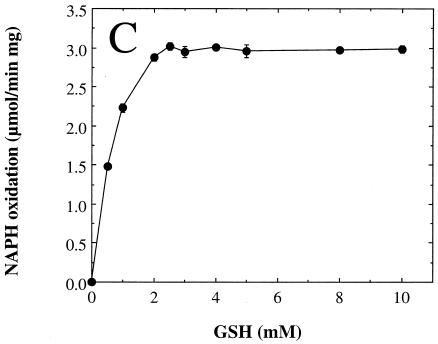
We next examined the dependence of the measured rate of arsenate reduction by rSynArsC upon the concentration of the individual components of the coupled assay system in order to determine optimal, saturating levels for each (Fig. 3). The arsenate reductase activity of rSynArsC exhibited simple saturation kinetics with respect to both arsenate (Fig. 3A) and glutaredoxin (Fig. 3B) over the entire concentration range examined. Kinetic analysis indicated that rSynArsC was a much better arsenate reductase than phosphohydrolase (Table 2). Its Vmax for the reduction of arsenate was roughly one-fifth that of its sequence homolog, pI258 ArsC, comparable to that of R773 ArsC, and an order of magnitude greater than that of Acr2p (Table 2). The Km of rSynArsC for arsenate, 1.25 mM, was 20-fold higher than that reported for pI258 ArsC, although it was significantly lower than that of either Acr2p or R773 ArsC (Table 2). The apparent Km of rSynArsC for glutaredoxin, 0.43 μM, was comparable to that of Acr2p, 0.6 μM (38).
FIG. 3.
The arsenate reductase activity of rSynArsC exhibits simple saturation kinetics. The arsenate reductase activity of rSynArsC (25 μg) was measured as described in Materials and Methods, with the exception that the levels of arsenate (A), glutaredoxin (Gx) (B), or reduced glutathione (GSH) (C) were varied as indicated. Shown is the average rate of arsenate-dependent NADPH reduction as a function of the concentration of the indicated assay component for duplicate assays, ± the average deviation from the mean.
TABLE 2.
Comparison of rSynArsC with detoxifying arsenate reductases from other microorganismsa
| Organism | Group | Protein | Class | Subunit structure | Reductant | HAsO42−Km (mM) | Gx Km (μM) | Vmax (μmol/min/mg) | Optimal pH | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Synechocystis sp. strain PCC 6803 | Cyanobacteria | rSynArsC | LMW PTP-like | Homodimer | GSH/Gx | 1.25 | 0.43 | 3.1 | 7.5 | This paper |
| E. coli plasmid R773 | Gram-negative bacteria | ArsC | Unique | Monomer | GSH/Gx | 8 | NR | 0.8-1.5 | 6.3-6.8 | 15 |
| S. aureus plasmid pI258 | Gram-positive bacteria | ArsC | LMW PTP-like | Monomer | Thioredoxin | 0.068 | NA | 14.5 | 7.5 | 33 |
| S. cerevisiae | Yeast | Acr2p | Cdc25-like | Homodimer | GSH/Gx | 35 | 0.6 | 0.3-0.4 | 6.5-8.5 | 38 |
Abbreviations: Gx, glutaredoxin; GSH, reduced glutathione; NA, not applicable; NR, not reported. The references listed were the sources for the reported Vmax, Km, and optimal pH values.
Mutagenic alteration of cysteine residues 8, 13, 80, or 82.
The roles of four of the five cysteine residues in rSynArsC, specifically those that appeared to be physically and/or functionally conserved between this cyanobacterial arsenate reductase and its homologs from S. aureus and B. subtilis, were examined using site-directed mutagenesis. As expected, mutagenic alteration of Cys8, which corresponds to the conserved active-site cysteine that forms an arsenocysteine intermediate in prototypical arsenate reductases (31) and the phosphocysteine intermediate in homologous LMW PTPs (9, 55), to serine resulted in the complete loss of both arsenate-dependent NADPH oxidation and pNPP hydrolysis (Table 3). Mutagenic alteration of Cys13, which is conserved in the pI258 ArsC from S. aureus and its homolog from B. subtilis (Fig. 1) as well as in many bacterial LMW PTPs (27), to serine had little or no effect on arsenate reductase activity (Table 3). Similar behavior was reported for pI258 ArsC (32). Substitution of Cys13 by serine did, however, abolish the ability of rSynArsC to hydrolyze pNPP. This finding was not wholly unexpected, as substitution of the equivalent cysteine residue in a bovine LMW PTP has been variously reported to either reduce (10) or eliminate (8, 9) its phosphohydrolase activity.
TABLE 3.
Effect of mutagenically produced alterations on the chemical and catalytic properties of rSynArsCa
| Mutagenic alteration | Arsenate reductase activity (μmol/min/mg) | Phosphatase activity
|
[35S]GSH bound (mol/mol) | Reactivity w/DTNB (A412) | |
|---|---|---|---|---|---|
| Vmax (μmol/min/mg) | Km (mM) | ||||
| None | 3.1 | 0.08 | 77 | 0.06 | 0.22 |
| Cys8 to Ser | NDb | ND | ND | ND | 0.15 |
| Cys13 to Ser | 1.8 | ND | ND | 0.04 | 0.15 |
| Cys80 to Ser | ND | 0.07 | 274 | 0.05 | 0.32 |
| Cys82 to Ser | ND | 0.08 | 98 | 0.05 | 0.31 |
The first column lists the various forms of rSynArsC, designated by the amino acid substitution, if any, that was produced by recombinant means. The right-hand columns give the arsenate reductase activity of each enzyme form, as measured under saturating conditions for all reactants with rSynArsC, the kinetic constants of the enzymes for hydrolysis of pNPP (phosphatase activity), the number of moles of [35S]glutathione bound per mole of protein, and the absorbance at 412 nm of the free thionitrobenzoate released when the enzyme was incubated with DTNB. Presented are the results of representative experiments. For further details, see Materials and Methods.
ND = not detectable, below the limits of detection for the relevant assay.
pI258 ArsC from S. aureus and its homolog from B. subtilis contain a pair of cysteine residues, Cys82 and Cys89, that are not conserved among their phosphohydrolytic counterparts, the LMW PTPs (Fig. 1). These cysteines are essential for the arsenate reductase activity of pI258 ArsC (32). In the enzyme from S. aureus, the catalytic arsenocysteine intermediate is reduced by electrons generated via the formation of a disulfide bond between the active-site cysteine residue, Cys10, and Cys82 (34). Cys10 is restored to its initial, reduced state via an internal disulfide transfer involving Cys89. The fully reduced, catalytically competent form of the enzyme is regenerated when thioredoxin reduces the resulting Cys82-Cys89 disulfide. The presumed function of the last step in this internal disulfide cascade is to render the S-S bond more accessible to and/or readily recognized by thioredoxin (56).
In contrast to pI258 ArsC, rSynArsC required glutathione and glutaredoxin for activity, like the structurally dissimilar arsenate reductases Acr2p and R773 ArsC. These latter enzymes require only a single, active-site cysteine for catalysis (28, 36). SynArsC, however, contains another cysteine residue, Cys80, whose position within the protein's amino acid sequence precisely corresponds to that of one of the conserved cysteines, Cys82, that participates in the internal disulfide cascade in pI258 ArsC and its homologs (Fig. 1). While no exact duplicate of the third essential cysteine from the S. aureus enzyme, Cys89, is present in SynArsC, the latter protein does contain another cysteine, Cys82, in close proximity to Cys80. Their close juxtaposition suggests that Cys80 and Cys82 could readily form an internal disulfide analogous to the Cys82-Cys89 disulfide of pI258 and B. subtilis ArsC. It was therefore asked whether either Cys80 or Cys82 in SynArsC was essential for catalysis. As can be seen in Table 3, mutagenic alteration of either cysteine to serine was sufficient to abolish the arsenate reductase activity of rSynArsC. The effect of these mutagenic alterations appears to have been discrete in nature, as they had little or no effect upon the ability of the enzyme to hydrolyze pNPP (Table 3).
Cys80 and Cys82 can form a disulfide bond in rSynArsC.
rSynArsC and mutagenically altered variants thereof were incubated with (5,5′-dithiobis) nitrobenzoic acid (DTNB, also known as Ellman's reagent), which reacts with accessible thiol groups to form a mixed disulfide with the concomitant release of the chromophore thionitrobenzoate (12). As can be seen from Table 3, substitution of either Cys8 or Cys13 with serine resulted in a net decrease in the quantity of free thionitrobenzoate produced, as expected. By contrast, replacement of either Cys80 or Cys82 with serine results in an increase of slightly more than one-third in the quantity of free thionitrobenzoate produced, despite the fact that the total number of cysteine residues in the protein has been reduced from five to four. Since mutagenic alteration of either partner in a disulfide bond would be expected to leave the second cysteine in a free, reduced state, these observations suggest that Cys80 and Cys82 are so linked in rSynArsC.
rSynArsC forms a stable complex with glutathione.
In the glutathione-glutaredoxin-dependent arsenate reductases Acr2pand R773 ArsC, catalysis is thought to proceed via a covalent cysteinyl-arseno-glutathione intermediate that subsequently breaks down to form free arsenite and a mixed disulfide between glutathione and the active-site cysteine (29, 31, 49). Glutaredoxin then reduces the mixed disulfide to regenerate the free, reduced form of the enzyme and a mixed disulfide between glutaredoxin and glutathione. In order to better understand the catalytic mechanism of rSynArsC, [35S]glutathione was used to determine whether this cyanobacterial arsenate reductase could form a stable complex with glutathione.
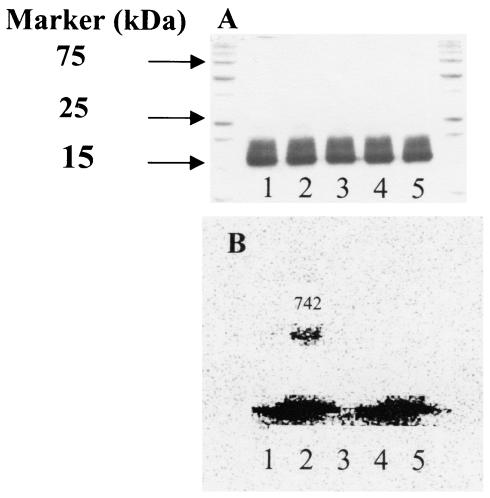
When rSynArsC was incubated with [35S]glutathione and sodium arsenate and then analyzed by SDS-PAGE in the absence of reducing agents, an SDS-stable complex with glutathione could be detected (Fig. 4). While the proportion of the enzyme molecules that bound the radiolabeled glutathione was relatively small, ranging from 4 to 11% over several experiments (Table 3), complex formation was dependent upon the presence of both arsenate and the active-site cysteine residue, Cys8 (Fig. 4). Mutagenic alteration of either of the other essential cysteines, Cys80 or Cys82, to serine had little discernible effect on complex formation, suggesting that—at least initially—the glutathione moiety binds to the enzyme via Cys8, as predicted (Table 3).
FIG. 4.
rSynArsC binds glutathione in an arsenate-dependent manner. rSynArsC or a form in which the predicted active-site cysteine, Cys8, was mutagenically altered to serine was incubated with [35S]glutathione (4 mM) in either the presence or absence of arsenate as described in Materials and Methods. (A) SDS-polyacrylamide gel stained with Coomassie blue. (B) Electronic autoradiogram of the same gel taken prior to staining. The outer lanes of the gel contain prestained standards, the molecular masses of certain of which are indicated at the left on panel A. Lane 1 contains rSynArsC that was incubated with [35S]glutathione (reduced) in the absence of arsenate, while lane 2 contains rSynArsC that was incubated with [35S]glutathione in the presence of arsenate. The number given above the upper band in lane 2 lists the number of counts per minute of radioactivity present in the band, as determined by electronic autoradiography (estimated efficiency, ≈2%), relative to that in the corresponding regions of the other lanes. Lane 3 contains rSynArsC, while lanes 4 and 5 contain the Cys8-to-Ser variant that was incubated with [35S]glutathione in the absence and presence of arsenate, respectively. The radioactive material at the bottom of the gel is free [35S]glutathione.
Glutaredoxin reduces a mixed disulfide between SynArsC and glutathione.
The immediate effect of reducing arsenate to arsenite in detoxifying arsenate reductases is to leave the enzyme in an oxidized state, which must subsequently be reduced to ready the enzyme for the next round of catalysis. In Acr2p and R773 ArsC, glutaredoxin reduces the mixed disulfide formed between the active-site cysteine residue and glutathione, while in pI258 ArsC thioredoxin reduces the internal disulfide formed between Cys82 and Cys89 via the enzyme's internal disulfide cascade (1, 56). SynArsC displays features consistent with both mechanisms, i.e., the participation of glutathione and glutaredoxin characteristic of Acr2p and R773 ArsC and the requirement for three, rather than one, cysteine residues characteristic of pI258 ArsC.
Glutaredoxin is capable of reducing both mixed protein-glutathione disulfides and internal protein disulfides. However, while the former requires the presence of only one of the enzyme's two active-site cysteine residues, Cys11 in the case of glutaredoxin 1 from E. coli, the commercially available form of which was used in the assays described above, the reduction of internal protein disulfides requires the participation of both Cys11 and Cys14 (5). We therefore cloned the grxA gene from E. coli K-12 and used it to produce recombinant forms, as histidine-tagged fusion proteins, of both glutaredoxin 1 and mutagenically altered forms in which either Cys11 and Cys14 were replaced with serine. While our recombinantly produced fusion protein was somewhat less efficient than the equivalent quantity of commercial glutaredoxin 1 in supporting catalysis, replacement of Cys14 by Ser had little effect on the rate of arsenate-dependent NADPH oxidation (1.10 ± 0.05 μmol/min · mg versus 1.23 ± 0.02 μmol/min · mg with the unaltered glutaredoxin 1), while mutagenic alteration of Cys11 abolished activity below the limits of detection. These results indicate that glutaredoxin reduces a mixed SynArsC-glutathione disulfide and not an internal cysteine-cysteine disulfide.
DISCUSSION
Analysis of the catalytic capabilities of the recombinant gene product of ORF slr0946 from Synechocystis sp. strain PCC 6803 indicated that it is an arsenate reductase. As has been shown to be the case for the homologous arsenate reductases from S. aureus (56) and B. subtilis (1), with which it shares 36 and 45% sequence identity, respectively (Fig. 1), rSynArsC also has retained weak, vestigial phosphohydrolase activity indicative of its descent from the LMW PTP family of protein phosphatases. Intriguingly, while SynArsC is clearly a sequence homolog of pI258 ArsC, its catalytic mechanism combines certain features of this S. aureus enzyme with others that mimic the structurally dissimilar arsenate reductases from yeast, Acr2p, and E. coli, R773 ArsC.
Like Acr2p (38) and R773 ArsC (15), rSynArsC utilizes glutathione and glutaredoxin as its immediate source of reducing equivalents, in direct contrast to pI258 ArsC, which uses thioredoxin (19, 21). As has been postulated for Acr2p and R773 ArsC (29, 31), rSynArsC forms a complex with glutathione. While only small quantities of the complex were detected, its formation was strictly dependent on the presence of both the active-site cysteine residue and arsenate, attributes consistent with catalytic relevance. As is also the case with Acr2p and R773 ArsC, glutaredoxin restores SynArsC to its reduced, catalytically ready state by reducing a mixed disulfide between the side chain of a cysteine residue and glutathione.
The behavior of rSynArsC deviates from that of the yeast and E. coli enzymes in at least one important respect. Whereas Acr2p (36) and R773 ArsC (28) require only a single active-site cysteine residue for catalysis, mutagenic alteration of any one of three cysteine residues in rSynArsC eliminated its ability to reduce arsenate at a measurable rate. The two “extra” cysteine residues, i.e., Cys8o and Cys82, appear to play specific, essential roles in the catalysis of arsenate reduction, since (i) their replacement with serine had little or no effect on the ability of the enzyme to hydrolyze pNPP and (ii) a similarly positioned pair of cysteine residues play key roles in the catalytic mechanism of pI258 ArsC (1, 32, 56).
In pI258 ArsC and its homolog from B. subtilis, the three essential cysteines participate in an internal disulfide cascade (56) that also has been referred to as a redox relay (1). The first step in this cascade is the attack of the sulfhydryl group of Cys82 in pI258 ArsC on the arsenocysteine intermediate formed between arsenate and Cys10, the active-site cysteine. The resulting disulfide constitutes the immediate source of the reducing equivalents necessary to convert arsenate to arsenite. An internal disulfide transfer then occurs that results in the formation of a disulfide bond between Cys82 and Cys89, regenerating the reduced active-site cysteine. The Cys82-Cys89 disulfide, in turn, is reduced by thioredoxin to complete the catalytic cycle. Presumably, the last step in this internal disulfide cascade renders the S-S bond accessible to and/or places it in an environmental context recognizable by thioredoxin. The essential nature of Cys8o and Cys82 in rSynArsC suggests that an analogous disulfide cascade takes place in this cyanobacterial arsenate reductase to translocate a mixed cysteine-glutathione disulfide to the enzyme's surface.
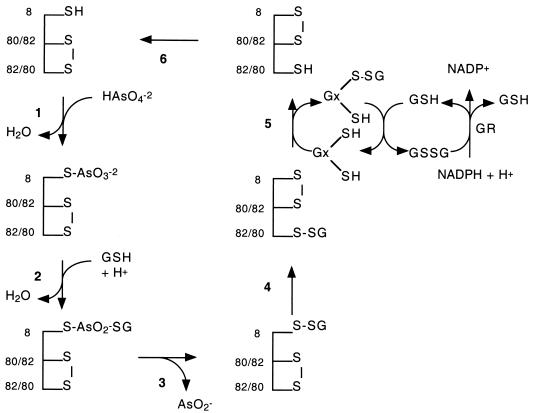
Figure 5 displays a possible mechanism for SynArsC that accommodates all of the experimental observations reported herein. Cys8o and Cys82 initially are in an oxidized, disulfide-bonded state, as was indicated by titrations with DTNB. Steps 1 to 3 are postulated to reprise the initial steps of the proposed reaction mechanisms of Acr2p and R773 ArsC. Specifically, an arsenoenzyme intermediate is formed in step 1 that then is attacked in step 2 by glutathione to form a cysteinyl-arseno-glutathione intermediate. This intermediate then breaks down, in step 3, to yield arsenite and a mixed disulfide between the active-site cysteine residue, Cys8, and glutathione. Steps 4 to 6 represent a modified version of the internal disulfide cascade that takes place in pI258 ArsC. In step 4, an internal disulfide exchange reaction takes place between the mixed disulfide between Cys8 and glutathione and the internal disulfide between Cys8o and Cys82 that shifts the mixed disulfide with glutathione from within the depths of the active-site pocket to a point nearer the enzyme's surface. The mixed disulfide then is reduced, in step 5, by glutaredoxin. In step 6, a second internal disulfide transfer event restores the active-site cysteine to its reduced, reactive state with the concomitant restoration of the original disulfide bond between Cys8o and Cys82.
FIG. 5.
Schematic description of a potential catalytic mechanism for SynArsC from Synechocystis sp. strain PCC 6803. Shown is a potential catalytic mechanism for the SynArsC arsenate reductase consistent with the experimental observations reported herein. Abbreviations: GR, glutathione reductase; GSH, reduced glutathione; GSSG, oxidized glutathione; Gx, glutaredoxin, Cys8, the active-site cysteine on rSynArsC; Cys80/82, either Cys80 or Cys82; Cys82/80, either Cys82 or Cys80.
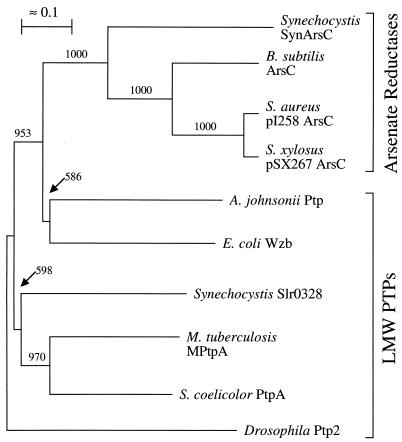
Regardless of whether the scheme presented in Fig. 5 proves correct in every detail, it is clear that SynArsC employs a unique combination of mechanistic steps to reduce arsenate to arsenite. How might this novel mechanism have arisen? One possible explanation is that SynArsC initially mimicked the catalytic mechanism of its sequence homolog from S. aureus. (Intriguingly, the source of reducing equivalents for the B. subtilis homolog of pI258 ArsC has yet to be determined.) However, SynArsC continued to refine its catalytic mechanism until it obtained its contemporary form. The reciprocal also may have occurred, in which case the cyanobacterial enzyme may be a molecular fossil reflecting an intermediate stage in the development of the catalytic mechanism now employed by pI258 ArsC. A third possibility is that cyanobacterial and S. aureus enzymes are the products of two independent transformations of LMW PTPs into arsenate reductases. This last possibility appears to be the least likely, as these arsenate reductases are more closely related to each other than they are to bacterial LMW PTPs (Fig. 6). Regardless, the addition of SynArsC to the present library of arsenate reductases and their ancestral LMW PTPs opens exciting new opportunities to explore the mechanisms by which enzymes evolve new catalytic capabilities.
FIG. 6.
Phylogenetic relationship between SynArsC and several known or likely arsenate reductases as well as known LMW PTPs from bacterial organisms. The phylogenetic relationships between known or likely pI258 ArsC-like arsenate reductases and known LMW PTPs from bacterial organisms were analyzed using CLUSTAL W version 1.7 (53) as described in Materials and Methods. Known or predicted arsenate reductases utilized for this analysis included the product of ORF slr0946 from Synechocystis sp. strain PC6803 (Synechocystis SynArsC [this manuscript]), ArsC from the skin element of B. subtilis (B. subtilis ArsC [52]), ArsC encoded by plasmid pI258 from S. aureus (20), ArsC encoded by plasmid pSX267 from Staphylococcus xylosus (46), and ArsC encoded by plasmid R773 from E. coli (E. coli R773 ArsC [7]). LMW PTPs utilized for comparison include Wzb from E. coli (54), Ptp from Acinetobacter johnsonii (16), the product of ORF slr0328 from Synechocystis sp. strain PC6803 (A. Mukhopadhyay and P. J. Kennelly, unpublished observations), MPtpA from M. tuberculosis (23), PtpA from Streptomyces coelicolor A3(2) (27), and Ptp2 from Drosophila melanogaster (35). The last named was used as the outgroup for these analyses. The scale bar indicates 1 base substitution per 10 amino acid residues. Bootstrap values above 500 (out of 1,000) are indicated at the corresponding nodes.
Acknowledgments
This work was supported by grant number GM55067 from the National Institutes of Health (P.J.K.) and an Honors Research Fellowship from Emory and Henry College Friends of the Sciences (J.D.H.).
REFERENCES
- 1.Bennett, M. S., Z. Guan, M. Laurberg, and X.-D. Su. 2001. Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA 98:13577-13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bobrowicz, P., R. Wysocki, G. Owsianik, A. Goffeau, and S. Ulaszewski. 1997. Isolation of three contiguous genes, ACR1, ACR2, and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae. Yeast 13:819-828. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and simple method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bruhn, D. F., J. Li, S. Silver, F. Roberto, and B. P. Rosen. 1996. The arsenical resistance operon of IncN plasmid R46. FEMS Microbiol. Lett. 139:149-153. [DOI] [PubMed] [Google Scholar]
- 5.Bushweller, J. H., F. Aslund, K. Wuthrich, and A. Holmgren. 1992. Structural and functional characterization of the mutant Escherichia coli glutaredoxin (C14→S) and its mixed disulfide with glutathione. Biochemistry 31:9288-9293. [DOI] [PubMed] [Google Scholar]
- 6.Butcher, B. G., S. M. Deane, and D. R. E. Rawlings. 2000. The chromosomal arsenical resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenical resistance to Escherichia coli. Appl. Environ. Microbiol. 66:1826-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C.-M., T. K. Misra, S. Silver, and B. P. Rosen. 1986. Nucleotide sequence of the structural gene of an anion pump. The plasmid-encoded arsenical resistance operon. J. Biol. Chem. 261:15030-15038. [PubMed] [Google Scholar]
- 8.Chiarugi, P., R Marzocchini, G. Raugei, C. Pazzagli, A. Berti, G. Camici, G. Manao, G. Cappugi, and G. Ramponi. 1992. Differential role of four cysteines on the activity of a low Mr phosphotyrosine protein phosphatase. FEBS Lett. 310:9-12. [DOI] [PubMed] [Google Scholar]
- 9.Cirri, P., P. Chiarugi, G. Camici, G. Manao, G. Raugei, G. Cappugi, and G. Ramponi. 1993. The role of Cys12, Cys17, and Arg18 in the catalytic mechanism of low-Mr cytosolic phosphotyrosine protein phosphatase. Eur. J. Biochem. 214:647-657. [DOI] [PubMed] [Google Scholar]
- 10.Davis, J. P., M.-M. Zhou, and R. L. Van Etten. 1994. Kinetic and site-directed mutagenesis studies of the cysteine residues of bovine low molecular weight phosphotyrosyl protein phosphatase. J. Biol. Chem. 269:8734-8740. [PubMed] [Google Scholar]
- 11.Diorio, C., J. Cai, J. Marmor, R. Shinder, and M. S. DuBow. 1995. An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J. Bacteriol. 177:2050-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellman, G. L. 1959. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82:70-77. [DOI] [PubMed] [Google Scholar]
- 13.Fairbanks, G., T. L. Steck, and D. F. H. Wallace. 1971. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry 10:2606-2617. [DOI] [PubMed] [Google Scholar]
- 14.Fauman, E. B., J. P. Cogswell, B. Lovejoy, W. J. Rocque, W. Holmes, V. G. Montana, H. Piwnica-Worms, M. J. Rink, and M. A. Saper. 1998. Crystal structure of the catalytic domain of human cell cycle control phosphatase, Cdc25A. Cell 93:617-625. [DOI] [PubMed] [Google Scholar]
- 15.Gladysheva, T. B., K. L. Oden, and B. P. Rosen. 1994. Properties of the arsenate reductase of plasmid R773. Biochemistry 33:7288-7293. [DOI] [PubMed] [Google Scholar]
- 16.Grangreasse, C., P. Doublet, C. Vincent, E. Vaganay, M. Riberty, B. Duclos, and A. J. Cozzone. 1998. Functional characterization of the low-molecular-mass phosphotyrosine-protein phosphatase of Acinetobacter johnsonii. J. Mol. Biol. 278:339-347. [DOI] [PubMed] [Google Scholar]
- 17.Hoog, J.-O., H. von Bahr-Lindstrom, H. Jornvll, and A. Holmgren. 1986. Cloning and expression of the glutaredoxin (grx) gene of Escherichia coli. Gene 43:13-21. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, M. D., and J. M. Denu. 2001. Molecular reactions of protein phosphatases—insights from structure and chemistry. Chem. Rev. 101:2313-2340. [DOI] [PubMed] [Google Scholar]
- 19.Ji, G., E. A. E. Garber, L. G. Armes, C.-M. Chen, J. A. Fuchs, and S. Silver. 1994. Arsenate reductase of Staphylococcus aureus plasmid pI258. Biochemistry 33:7294-7299. [DOI] [PubMed] [Google Scholar]
- 20.Ji, G., and S. Silver. 1992. Regulation and expression of the arsenic resistance operon from Staphylococcus aureus plasmid pI258. J. Bacteriol. 174:3684-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji, G., and S. Silver. 1992. Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc. Natl. Acad. Sci. USA 89:9474-9478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneto, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanaba, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 30:109-136. [DOI] [PubMed] [Google Scholar]
- 23.Koul, A., A. Choidas, M. Treder, A. K. Tyagi, K. Drlica, Y. Singh, and A. Ullrich. 2000. Cloning and characterization of secretory tyrosine phosphatases from Mycobacterium tuberculosis. J. Bacteriol. 182:5425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krafft, T., and J. M. Macy. 1998. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur. J. Biochem. 255:647-653. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Li, R., H. J. Debella, and W. W. Carmichael. 2001. Isolates identifiable as Athrospira maxima and Athrospira fusiformis (Oscillatoriales, Cyanobacteria) appear identical on the basis of a morphological study in culture and 16S rRNA sequences. Phycologia 40:367-371. [Google Scholar]
- 27.Li, Y., and W. R. Strohl. 1996. Cloning, purification, and properties of a phosphotyrosine protein phosphatase from Streptomyces coelicolor A3(2). J. Bacteriol. 178:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, J., T. B. Gladysheva, L. Lee, and B. P. Rosen. 1995. Identification of an essential cysteine residue in the ArsC arsenate reductase of plasmid R773. Biochemistry 34:13472-13476. [DOI] [PubMed] [Google Scholar]
- 29.Liu, J., and B. P. Rosen. 1997. Ligand interactions of the ArsC arsenate reductase. J. Biol. Chem. 272:21084-21089. [DOI] [PubMed] [Google Scholar]
- 30.Macy, J. M., K. Nunan, K. D. Hagen, D. R. Dixon, P. J. Harbour, M. Cahill, and L. I. Sly. 1996. Chrysiogenes arsenatis gen. nov., sp. nov., a new arsenate-respiring bacterium isolated from gold mine wastewater. Int. J. Syst. Bacteriol. 46:1153-1157. [DOI] [PubMed] [Google Scholar]
- 31.Martin, P., S. DeMel, J. Shi, T. Gladysheva, D. L. Gatti, B. P. Rosen, and B. F. P. Edwards. 2001. Insights into the structure, solvation, and mechanism of the ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure 9:1071-1081. [DOI] [PubMed] [Google Scholar]
- 32.Messens, J., G. Hayburn, A. Desmyter, G. Laus, and L. Wyns. 1999. The essential catalytic redox couple in arsenate reductase from Staphylococcus aureus. Biochemistry 38:16857-16865. [DOI] [PubMed] [Google Scholar]
- 33.Messens, J., J. C. Martins, E. Brosens, K. Van Belle, D. M. Jacobs, R. Willem, and L. Wyns. 2002. Kinetics and active site dynamics of Staphylococcus aureus arsenate reductase. J. Biol. Inorg. Chem. 7:146-156. [DOI] [PubMed] [Google Scholar]
- 34.Messens, J., J. C. Martins, K. Van Belle, E. Brosens, A. Desmyter, M. De Geiter, J.-M. Wieruszeski, R. Willem, L. Wyns, and I. Zegers. 2002. All intermediates of the arsenate reductase mechanism, including an intracellular dynamic disulfide cascade. Proc. Natl. Acad. Sci. USA 99:8506-8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, D. T., R. Read, J. Rusconi, and R. L. Cagan. 2000. The Drsophila primo locus encodes two low-molecular-weight tyrosine phosphatases. Gene 243:109. [DOI] [PubMed] [Google Scholar]
- 36.Mukhopadhyay, R., and B. P. Rosen. 2001. The phosphatase C(X)5R motif is required for catalytic activity of Saccharomyces cerevisiae Acr2p arsenate reductase. J. Biol. Chem. 276:34738-34742. [DOI] [PubMed] [Google Scholar]
- 37.Mukhopadhyay, R., B. P. Rosen, L. T. Phung, and S. Silver. 2002. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol. Rev. 26:311-325. [DOI] [PubMed] [Google Scholar]
- 38.Mukhopadhyay, R., J. Shi, and B. P. Rosen. 2000. Purification and characterization of Acr2p, the Saccharomyces cerevisiae arsenate reductase. J. Biol. Chem. 275:21149-21157. [DOI] [PubMed] [Google Scholar]
- 39.Neyt, C., M. Iriarte, V. H. Thi, and G. R. Cornelis. 1997. Virulence and arsenate resistance in yersiniae. J. Bacteriol. 179:612-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nilsson, I., and I. Hoffmann. 2000. Cell cycle regulation by the Cdc25 phosphatase family. Prog. Cell Cycle Res. 4:107-114. [DOI] [PubMed] [Google Scholar]
- 41.Potts, M., H. Sun, K. Mockaitis, P. J. Kennelly, D. Reed, and N. K. Tonks. 1993. A protein-tyrosine/serine phosphatase encoded by the genome of the cyanobacterium Nostoc commune UTEX 584. J. Biol. Chem. 268:7632-7635. [PubMed] [Google Scholar]
- 42.Ramponi, G., and M. Stefani. 1997. Structural, catalytic, and functional properties of low Mr phosphotyrosine protein phosphatases. Evidence of a long evolutionary history. Int. J. Biochem. Cell Biol. 29:279-292. [DOI] [PubMed] [Google Scholar]
- 43.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Genetic assignments, strain histories, and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 44.Rosen, B. P. 2002. Biochemistry of arsenic detoxification. FEBS Lett. 529:86-92. [DOI] [PubMed] [Google Scholar]
- 45.Rosenberg, H., R. G. Gerdes, and K. Chegwidden. 1977. Two systems for the uptake of phosphate in Escherichia coli. J. Bacteriol. 131:505-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenstein, R., A. Peschel, B. Wieland, and F. Gotz. 1992. Expression and regulation of the antomite, arsenite, and arsenate resistance operon of Staphylcoccus xylosus plasmid pSX267. J. Bacteriol. 174:3676-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryan, D., and E. Colleran. 2002. Arsenical resistance in the IncHI2 plasmids. Plasmid 47:234-240. [DOI] [PubMed] [Google Scholar]
- 48.Shi, L., M. Potts, and P. J. Kennelly. 1998. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 22:229-253. [DOI] [PubMed] [Google Scholar]
- 49.Shi, J., A. Vlamis-Gardikas, F. Aslund, A. Holmgren, and B. P. Rosen. 1999. Reactivity of glutaredoxins 1, 2, and 3 from Escherichia coli shows that glutaredoxin 2 is the primary hydrogen donor to ArsC-mediated arsenate reduction. J. Biol. Chem. 274:36039-36042. [DOI] [PubMed] [Google Scholar]
- 50.Soulat, D., E. Vaganay, B. Duclos, A.-L. Genestier, J. Etienne, and A. J. Cozzone. 2002. Staphylococcus aureus contains two low-molecular-mass phosphotyrosine phosphatases. J. Bacteriol. 184:5194-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki, K., N. Wakao, T. Kimura, K. Sakka, and K. Ohmiya. 1998. Expression and regulation of the arsenic resistance operon of Acidophilium multivorum AIU 301 plasmid pKW301 in Escherichia coli. Appl. Environ. Microbiol. 64:411-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takemaru, K.-I., M. Mizuno, T. Sato, M. Takeuchi, and Y. Kobayashi. 1995. Complete nucleotide sequence of a skin element excised by DNA rearrangement during sporulation in Bacillus subtilis. Microbiology 141:323-327. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity and progressive multiple sequence weighting, position specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vincent, C., P. Doublet, C. Gangreasse, E. Vaganay, A. J. Cozzone, and B. Duclos. 1999. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a protein-tyrosine phosphatase, Wzb. J. Bacteriol. 181:3472-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wo, Y.-Y. P., M.-M. Zhou, P. Stevis, J. P. Davis, Z.-Y. Zhang, and R. L. Van Etten. 1992. Cloning, expression, and catalytic mechanism of the low molecular weight phosphotyrosyl protein phosphatase from bovine heart. Biochemistry 31:1712-1721. [DOI] [PubMed] [Google Scholar]
- 56.Zegers, I., J. C. Martins, R. Willem, L. Wyns, and J. Messens. 2001. Arsenate reductase from S. aureus plasmid pI258 is a phosphatase drafted for redox duty. Nat. Struct. Biol. 8:843-847. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Z.-Y. 1998. Protein-tyrosine phosphatases: biological function, structural characteristics, and mechanisms of catalysis. Crit. Rev. Biochem. Mol. Biol. 33:1-52. [DOI] [PubMed] [Google Scholar]