Abstract
Dengue virus (DENV) and West Nile virus (WNV) are members of the Flavivirus genus of positive-strand RNA viruses. RNA sequences and structures, primarily in the untranslated regions, have been shown to modulate flaviviral gene expression and genome replication. Previously, we demonstrated that a structure in the DENV coding region (cHP) enhances translation start codon selection and is required for viral replication. Here we further characterize the role of the cHP in the DENV life cycle. We demonstrate that the cHP is required for efficient viral RNA synthesis in a sequence-independent manner. Viruses with a disrupted cHP are rescued by a spontaneous compensatory mutation that restabilizes the structure. Furthermore, the cHP, which is predicted to be conserved among arthropod-borne flaviviruses, is required for WNV replication. We propose that the cHP is a multifunctional determinant of flavivirus replication, functioning in both translation and RNA synthesis.
INTRODUCTION
Dengue virus (DENV) is the causative agent of dengue fever (DF) and its more severe and potentially lethal manifestation, dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS), which are prevalent in tropical and subtropical regions of the world. The four serotypes of DENV (DENV1-4) are transmitted to humans via two mosquito species: Aedes aegypti and Ae. albopictus. DENV is one of the leading causes of arboviral disease worldwide, with tens of millions of DF cases and 250,000–500,000 hospitalizations for DHF/DSS occurring annually, and over 3 billion people at risk for infection (Gubler, 1998). West Nile virus (WNV), which historically has been associated with rare and mild infections of humans, has recently emerged as a pathogen of significant concern to both public health and wildlife ecology, and has disseminated widely throughout North America in a short period of time. Infection with WNV can result in neuroinvasive disease and death in humans (Gubler et al., 2006). DENV and WNV are members of the Flaviviridae family of viruses, which includes hepatitis C virus (HCV), pestiviruses, and the flaviviruses Japanese encephalitis virus, yellow fever virus (YFV) and tick-borne encephalitis virus (Lindenbach and Rice, 2006).
Like all flaviviruses, DENV is an enveloped, non-polyadenylated positive-strand RNA virus with a type 1 cap structure located at the 5' end of the genome and 5' and 3' untranslated regions (UTRs) flanking a single open reading frame. The flavivirus replication cycle begins when the virus binds and enters a susceptible host cell by receptor-mediated endocytosis, leading to the release of the viral genome into the cytoplasm. The virus’ single open reading frame (ORF) is subsequently translated as a polyprotein from a start codon located at the 5' end of the region encoding the viral capsid protein (C). The polyprotein is co- and post-translationally cleaved into the three structural proteins (C; pre-membrane, prM; and envelope, E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5). Genomic RNAs (vRNAs) are synthesized by the viral replicase complex via a negative-sense intermediate, and newly transcribed vRNAs undergo late rounds of translation that are dependent upon RNA synthesis. vRNAs are then packaged into virions and released through the host secretory pathway (Clyde et al., 2006; Lindenbach and Rice, 2003; Lindenbach and Rice, 2006). The processes of input-strand translation, RNA synthesis, late translation and packaging are inferred to be coupled, the previous step being required for progression to the next, although the exact mechanisms are unknown. A number of cis elements, primarily in the untranslated regions of the genome, have been shown to be required for flavivirus replication, though the precise roles of many RNA structures and sequences that function in the life cycle of flaviviruses have yet to be characterized.
Translation of the DENV ORF is believed to occur under standard conditions by traditional cap-dependent initiation and scanning through the 5'UTR (Lindenbach and Rice, 2003), although a non-canonical end-dependent mechanism has also been described (Edgil et al., 2006). The start codon of many of the mosquito-borne flaviviruses is in a poor Kozak initiation context, which predicts that the 5' C start codon would be utilized inefficiently for initiation of translation. We have previously shown that an element of secondary structure in the C coding region, cHP, can enhance recognition of the 5' C start codon (Clyde and Harris, 2006). The cHP structure, which has been verified by solution-structure probing (C. Polacek, J. Foley and E. Harris, unpublished data), is predicted to be maintained among mosquito- and tick-borne flaviviruses regardless of start codon context and its ability to modulate start site selection. This observation, together with the finding that disruption of the DENV2 cHP element resulted in a viral replication defect far greater than could be explained by the decreased production of the full-length C protein, led us to hypothesize that the cHP element may serve an additional purpose, namely, to function as a cis element at another stage of the flaviviral life cycle in addition to enhancing start codon selection in mosquito-borne flaviviruses. Here, we have extended our analysis of the role of the cHP element in the DENV2 life cycle. Using a combination of reporter replicon and infectious clone variants, we show that an intact cHP structure is required for efficient RNA synthesis and functions independently of its sequence. In addition, we show that the cHP is required for replication of the related flavivirus, WNV.
RESULTS
The cHP element is required for efficient viral replication
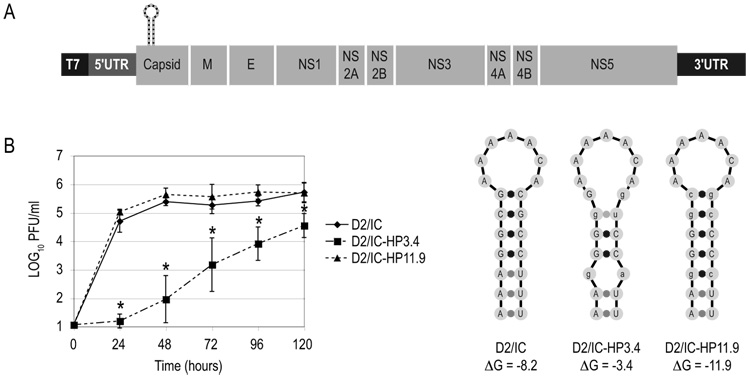
Previously, we had shown that destabilization of the cHP element (pD2/IC-HP3.4) resulted in a decrease in the production of infectious virus to below the limit of detection by plaque assay in human (Hep3B) and Ae. albopictus(C6/36) cells, and that compensatory mutations to restore the hairpin structure (pD2/IC-HP11.9) rescued viral output to WT levels (Clyde and Harris, 2006). Schematic diagrams of pD2/IC (WT), pD2/IC-HP3.4 and pD2/IC-HP11.9 are shown in Figure 1A. To determine whether there were any differences in the growth kinetics between the HP11.9 and WT viruses, BHK cell monolayers were transfected with infectious clone (D2/IC) RNAs, and virus titration was conducted at various timepoints post-transfection. As shown in Figure 1B, there is no significant difference in the growth kinetics of the HP11.9 virus versus the infectious clone-derived WT virus; therefore, a more stable hairpin does not impact viral output or alter the kinetics of infection. Whereas no virus had been previously detected from D2/IC-HP3.4 -transfected Hep3B and C6/36 cells (Clyde and Harris, 2006), in BHK cells, a low level of virus was produced as early as 24 hours post-transfection. However, throughout all timepoints, the level of HP3.4 virus that accumulated was significantly reduced compared to WT and HP11.9 and exhibited greater variability between experiments (Fig. 1B).
Figure 1. Disruption of the cHP element results in reduced viral titer.
BHK cell monolayers were transfected with infectious clone (D2/IC) RNAs containing a WT cHP sequence and mutations predicted to disrupt (HP3.4) and restore (HP11.9) cHP basepairing. Viral titer was assessed at 24, 48, 72, 96 and 120 hours post-transfection and normalized for transfection efficiency as determined by qRT-PCR at 2 hours post-transfection. (A) Schematic diagram of pD2/IC and pD2/IC-HP3.4 and pD2/IC-HP11.9 variants. (B) Kinetics of viral replication in BHK cells. In experiments where HP3.4 virus was below the limit of detection, titer was recorded as the limit of detection, 1.06 LOG10 PFU/ml. Data represent an average of at least 3 experiments, and error bars indicate SD. *, p < 0.002 relative to WT by Wilcoxon rank sum test.
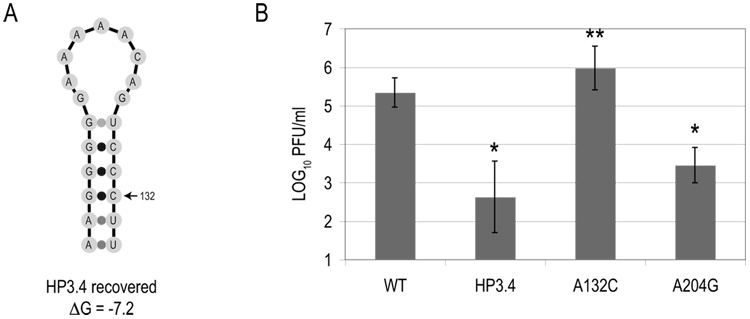
The production of virus from D2/IC-HP3.4-transfected BHK cells allowed us to determine whether selection for reversions or compensatory mutations to reform the cHP was occurring. Virus produced from D2/IC-HP3.4-transfected BHK cells from two separate experiments was passaged through C6/36 cells, and the resulting vRNA was sequenced. The C6/36-passaged virus harbored a compensatory mutation in the cHP that partially restored its structure (A132C; Fig. 2A) without reverting to the WT sequence. No other mutations were detected in the range of nt 92 – 918 of the vRNA, and there were no chromatographic peaks overlapping the C132 peak in the sequencing chromatogram (data not shown). The A132C substitution is predicted to increase the stability of the HP from −3.4 kcal/mol to −7.2 kcal/mol (Fig. 2A) and represents the most stable hairpin structure that could be formed by a single nucleotide substitution as computed by mfold. The substitution did not result in a change in amino acid sequence of the C protein from the input HP3.4 vRNA nor a substantial change in codon usage frequency in either Mesocricetus auratus(BHK) or Ae. albopictus (C6/36) (Nakamura et al., 2000).
Figure 2. A spontaneous compensatory mutation to restabilize the cHP arises in passaged virus.
BHK cell monolayers were transfected with in vitro-transcribed D2/IC RNA, and viral titers were assessed at 72 hours post-infection by plaque assay. (A) Sequence and structure of cHP in virus supernatant obtained from D2/IC-HP3.4-transfected BHK cells. A compensatory mutation at position 132 (A132C) was detected in four separate virus batches at 96 and 120 hours post-transfection and in virus passaged through C6/36 cells. ΔG values in kcal/mol. (B) Viral titers from infectious clone-transfected BHK cells. In experiments where variants were below the limit of detection, 1.06 LOG10 PFU/ml, titer was recorded as the limit of detection. Data represent an average of 3 experiments and error bars indicate SD. *, p < 0.003 relative to WT by Wilcoxon signed rank test. **, p < 0.003 relative to HP3.4 by Wilcoxon signed rank test.
In order to determine whether the A132C mutation was generated in BHK cells prior to passage in C6/36 cells, vRNA was extracted from supernatants from the 96h and 120h timepoints of D2/IC-HP3.4-transfected BHK cells. Sequencing chromatograms revealed a mixed population of A and C at position 132 (data not shown). Cloning and sequencing of RT products revealed that while the parental sequence was also recovered, the frequency of the A132C compensatory mutation increased with time from 18% (11 of 60 clones) to 39% (26 of 66 clones) at 96h and 120h, respectively, and correlated with higher viral titer (p < 0.02 between the two timepoints by Fisher’s exact test). A number of other substitutions were observed in one (0.8%) or in a single case, two (1.6%) of the cloned RT-PCR products. Their lack of accumulation indicates that these mutations did not undergo positive selection (p > 0.48). To determine whether the A132C mutation was unique in rescuing viral replication to WT levels, this single substitution was made in the HP3.4 infectious clone (pD2/IC-HP3.4-A132C). Additionally, another mutation that spontaneously arose was chosen that did not alter the coding sequence: A204G, which is located well downstream of the cHP. As shown in Figure 2B, the A204G infectious clone retains a defect in viral replication, while the A132C is the only mutation that restores viral replication to WT levels. Therefore, the impact of the A132C compensatory mutation on the stability of the cHP structure accounts for the rescue of viral replication and highlights the importance of a stable cHP structure in the viral life cycle.
The cHP element does not significantly affect the stability of DENV RNA
The steps in the viral life cycle most likely impacted by a change in RNA sequence or structure are the efficiency of translation, RNA synthesis, and genomic RNA packaging, as well as the overall stability of the RNA. To ascertain whether a reduction in the stability of the DENV2 RNA could be responsible for the observed decreased viral titer, the half-life (t1/2) of the D2/IC, D2/IC-HP3.4 and D2/IC-HP11.9 RNAs in BHK cells was determined. Non-replicating infectious clone variants of D2/IC, D2/IC-HP3.4 and D2/IC-HP11.9 were generated in which the NS5 RNA-dependent RNA polymerase (RdRP) active site was mutated from a conserved GDD motif to GVD (pD2/IC-RdRPmut). This mutation has been demonstrated to abolish flavivirus RdRP activity (Guyatt et al., 2001). BHK cell monolayers were transfected with the RdRPmut variants, and intracellular D2/IC RNA was measured over time by quantitative real-time RT-PCR (qRT-PCR). Half-life values, the R2 values of the exponential curves from which the t1/2 values were derived, and the p-values relative to WT are shown in Table 1. Although the D2/IC-HP11.9 RNA is somewhat, though not significantly, more stable (average t1/2 of 17.7 h) than both the WT D2/IC and D2/IC-HP3.4 RNAs, the t1/2 of the nonreplicating D2/IC-HP3.4 RNA (average t1/2 of 12.7 h) is similar to that of the WT D2/IC (average t1/2 of 12.6 h); thus, a reduction in vRNA stability cannot account for the defect in infectious virus production.
Table 1.
Calculation of RNA half-life of variants of D2/IC-RdRPmut.
| Expt. 1 | Expt. 2 | Expt. 3 | ||||||
|---|---|---|---|---|---|---|---|---|
| t1/2 (h) | R2 | t1/2 (h) | R2 | t1/2 (h) | R2 | Average t1/2 (hours) ± SD | Pa | |
| WT | 10.3 | 0.92 | 13.3 | 0.93 | 14.3 | 0.78 | 12.6 ± 2.1 | |
| HP3.4 | 14.5 | 0.97 | 12.5 | 0.98 | 11.0 | 0.87 | 12.7 ± 1.7 | 1 |
| HP11.9 | 21.0 | 0.58 | 12.9 | 0.95 | 19.1 | 0.82 | 17.7 ± 4.2 | 0.29 |
P-value relative to WT by Wilcoxon signed rank test
The cHP is required for late, but not early, translation of viral proteins
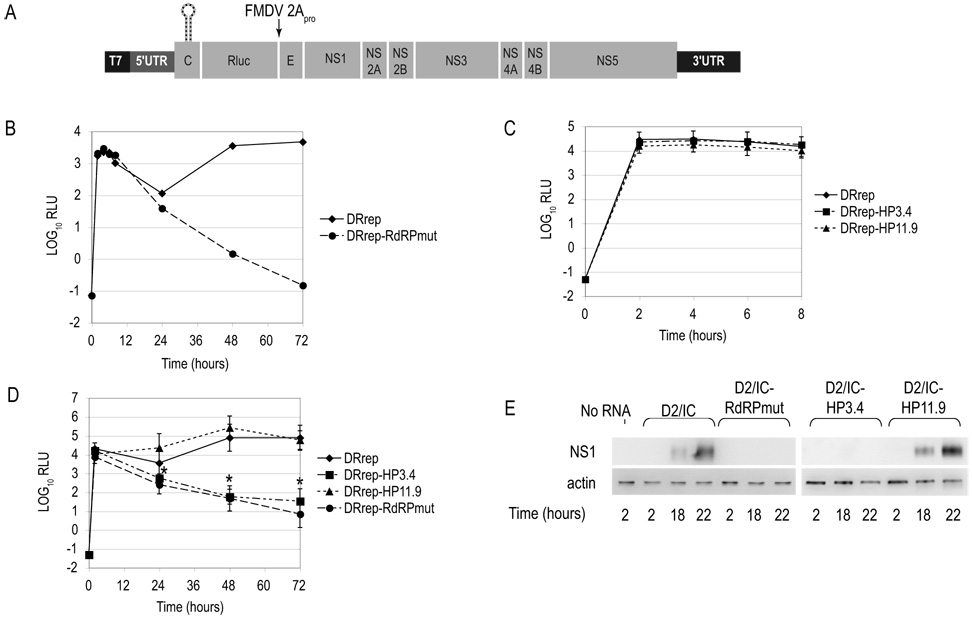
No differences were previously observed in the overall level of translation of reporter RNAs containing an intact or disrupted cHP, though usage of the first or second of multiple in-frame AUGs was affected (Clyde and Harris, 2006). However, in the context of a larger genome, the cHP element could conceivably function to enhance viral gene expression by recruiting or stabilizing initiation factors. Analysis of the role of the cHP in input-strand translation was accomplished with a newly constructed DENV2 reporter replicon, pDRrep (Fig. 3A). pDRrep is based on pD2/IC, with the majority of the structural gene coding region having been replaced by a Renilla luciferase (Rluc) gene. The Rluc coding sequence follows the first 72 nucleotides of the C coding region, which includes the cHP and cis elements required for RNA synthesis, and concludes with a FMDV 2Apro autoproteolytic cleavage site. Also included in the C region are the first two native, in-frame AUG codons of capsid, which ensures that any alteration of first start codon usage brought about by destabilizing or stabilizing the cHP does not impact the overall efficiency of translation; if the scanning initiation complex bypasses the first start codon, it will initiate at the next codon it locates with a similar degree of efficiency, provided that the two AUGs are sufficiently closely spaced. Following Rluc is the C-terminus of the E protein, included for proper targeting and processing of the nonstructural genes. The kinetics of DRrep relative to an RdRPmut version (DRrep-RdRPmut) are shown in Figure 3B. Translation of the input strand takes place during the first several hours, after which viral translation is reduced. By 48 hours, a second peak of translation occurs, which is dependent upon RNA synthesis. DRrep-RdRPmut, which cannot synthesize new RNA, undergoes efficient input strand translation at levels similar to the WT replicon, but lacks the second peak of translation (Fig. 3B). Note that the kinetics of the DENV replicon are slower than that of the virus, which reaches peak egress at 24 hours post-infection. This is similar to what has been reported with other replicons of both DENV and WNV (Alvarez et al., 2005; Holden et al., 2006; Lo et al., 2003; Tilgner and Shi, 2004).
Figure 3. cHP mutants do not progress to late rounds of translation.
BHK cells were transfected with a DENV2 reporter replicon and translation was assessed at the given timepoints by luciferase assay. (A) Schematic diagram of pDRrep, a reporter replicon of DENV2 strain 16681, in which most of the structural protein-coding region has been replaced by Renilla luciferase (Rluc). The N-terminus of C and C-terminus of E flank the Rluc gene. ΔG values are in kcal/mol. (B) Kinetics of translation of WT DRrep and DRrep containing an inactive RdRP (RdRPmut). BHK cell monolayers were transfected with replicon RNAs and assayed for Rluc activity at 2, 4, 6, 8, 24, 48 and 72 hours post-transfection. Data shown are representative of 5 experiments. (C) Early translation events in DRrep, DRrep-HP3.4 and DRrep-HP11.9 replicon-transfected cells. Transfected cells were assayed for Rluc activity at 2, 4, 6 and 8 hours post-transfection. RLU are normalized for transfection efficiency. Data are averages of 5 experiments and error bars indicate SD. (D) Late translation events in DRrep-transfected cells. Transfected cells were assayed for Rluc activity at 2, 24, 48 and 72 hours post-transfection. RLU are normalized for transfection efficiency. Data are averages of 6 experiments, and error bars indicate SD. (E) Early and late translation events in infectious clone-transfected cells. Transfected cells were lysed at 2, 18 and 22 hours post-transfection and the cytoplasmic fraction was separated by SDS-PAGE. Immunoblots directed against the DENV NS1 protein and actin are representative of experiments. RLU, relative luciferase units. *, p < 0.003 relative to WT by Wilcoxon signed rank test.
To establish whether input-strand translation was impacted by the HP3.4 mutations, BHK cells were transfected with DRrep, DRrep-HP3.4 and DRrep-HP11.9 and assayed for Rluc activity at 2, 4, 6 and 8 hours post-transfection. Compared to WT and HP11.9, no significant difference was seen with the HP3.4 variant at the step of input-strand translation (Fig. 3C); therefore, the defect in the production of infectious virus occurs after the initial expression of viral genes. Moreover, the lack of a significant difference in input-strand translation between WT DRrep and DRrep-HP11.9 demonstrates that first AUG selection per se does not influence the overall efficiency of translation, as the HP11.9 was shown to direct first AUG selection nearly 3-fold more efficiently than the WT cHP in mammalian cells (Clyde and Harris, 2006).
We next sought to determine whether the block in viral replication exhibited by D2/IC-HP3.4 occurs prior to virion assembly. To establish whether an intact cHP is required for progression to the second peak of translation that is dependent upon RNA synthesis, BHK cells were transfected with DRrep, DRrep-HP3.4 and DRrep-HP11.9 RNAs, and Rluc activity was assessed at 2, 24, 48 and 72 hours post-transfection. In contrast to its ability to efficiently direct input strand translation, the pDRrep-HP3.4 variant lacks the second peak of translation that is dependent upon RNA synthesis (Fig. 3D).
To determine whether the replicon behaves similarly to the full-length virus in this regard, BHK cells were transfected with infectious clone RNAs, and the levels of the viral nonstructural protein NS1 were assessed by immunoblot at 2, 18 and 22 hours post-transfection. Input-strand translation is below the limit of detection in this assay; therefore, visualization of viral proteins requires translation subsequent to RNA synthesis. Like DRrep-HP3.4, D2/IC-HP3.4-transfected BHK cells do not accumulate detectable levels of viral proteins as late as 22 hours post-transfection, similar to D2/IC-RdRPmut (Fig. 3E). In contrast, WT D2/IC and D2/IC-HP11.9 accumulate viral proteins at 18 and 22 hours post-transfection. Therefore, the cHP element is required prior to the step of assembly in both the replicon and full-length virus.
The cHP element is required for RNA synthesis
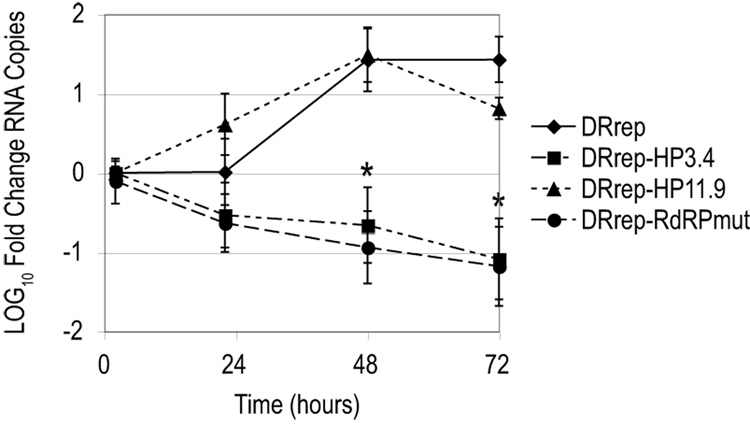
The lack of a second peak of translation by flavivirus reporter replicons is typically interpreted as a defect in vRNA synthesis (Alvarez et al., 2005; Holden et al., 2006; Lo et al., 2003). Accordingly, the DRrep-HP3.4 expression profile is similar to that of DRrep-RdRPmut, suggesting the cHP element may be required for RNA synthesis. To determine whether the lack of a second peak of translation in DRrep-HP3.4 parallels a defect in RNA accumulation, the level of replicon RNA was assessed by qRT-PCR at 2, 22, 48 and 72 hours post-transfection. As shown in Figure 4, levels of replicon RNA increase over time in cells transfected with WT DRrep and DRrep-HP11.9 RNAs. The lack of a significant increase in RNA at 24 hours in the WT replicon mirrors what was observed in D2/IC-transfected cells prior to virion release. This is likely due to the abundance of input RNA masking the de novo synthesis of vRNA, similar to what is seen in other DENV replicon systems (Alvarez et al., 2005). However, by 48 hours post-transfection, there is a clear increase in WT replicon RNA over input RNA. In contrast, replicon RNA does not accumulate in cells transfected with DRrep-HP3.4 or DRrep-RdRPmut (Fig. 4), thus confirming that an intact cHP element is required for efficient RNA synthesis.
Figure 4. The cHP element is required for efficient RNA synthesis.
BHK cell monolayers were transfected with DRrep RNA, and total replicon RNA was assayed over time by qRT-PCR. Values at 22, 48 and 72 hours post-transfection are expressed as increases or decreases relative to the level of input RNA at 2 hours post-transfection. Data are averages of 3 experiments. Error bars indicate SD. *, p < 0.03 relative to WT by Wilcoxon signed rank test.
The cHP sequence does not determine its function in RNA synthesis
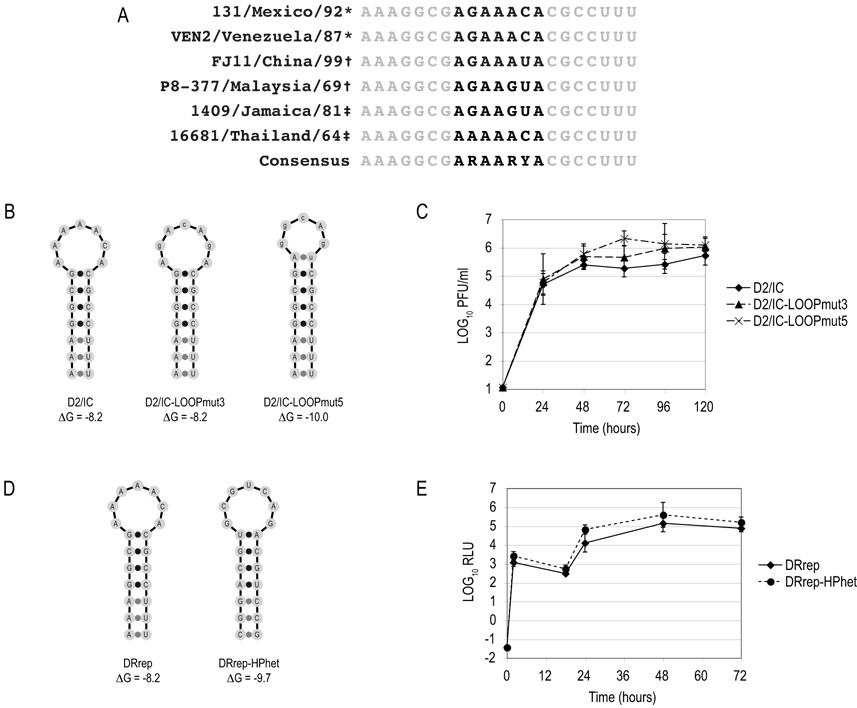
We had previously demonstrated that the cHP functions independently of sequence in translation start codon selection using reporter constructs, but had not investigated the dependence on sequence for replication of the full-length virus. Compensatory mutations made to restore basepairing in the cHP stem region in pD2/IC-HP11.9 did not result in a defect in viral replication, implicating the integrity of the stem region and not the sequence per se in the function of the cHP. However, alignment of different strains representing different genotypes of DENV2 show conservation of the stem sequence and partial conservation of the loop sequence (Fig. 5A). To determine whether the sequence of the loop is a determinant of cHP function in the DENV2 life cycle, mutations were made to 3 or 5 of the 7 loop nucleotides (pD2/IC-LOOPmut3 and pD2/IC-LOOPmut5, respectively; Fig. 5B). Loop mutations were designed to avoid major changes to the amino acid sequence of C (R9K/Q10N in LOOPmut3 and R9K/Q10N/S11T in LOOPmut5). As shown in Figure 5C, there is no significant difference in the growth kinetics of the LOOPmut3 and LOOPmut5 viruses versus the WT.
Figure 5. Hairpin sequence is not a determinant of cHP function in the DENV2 life cycle.
(A) Sequence of cHP from different strains of DENV2. *, American genotype; †, Malaysia/Indian Subcontinent genotype; ‡, Southeast Asian genotype. R, A or G; Y, C or U. (B) Schematic diagram of pD2/IC variants containing mutations to 3 (LOOPmut3) or 5 (LOOPmut5) of the 7 cHP loop nucleotides. (C) Kinetic analysis of viral replication in D2/IC-, D2/IC-LOOPmut3- and D2/IC-LOOPmut5-transfected BHK cell monolayers. Supernatants were harvested at 24, 48, 72, 96 and 120 hours post-transfection, and viral replication was assessed by plaque assay. PFU are normalized for transfection efficiency. Data are averages of at least 3 experiments. Error bars indicate SD. (D) Schematic of pDRrep-HPhet. (E) Early and late translation events in DRrep-HPhet-transfected cells. Transfected cells were assayed for Rluc activity at 2, 18, 24, 48 and 72 hours post-transfection. RLU are normalized for transfection efficiency as in 5C. Data are averages of 3 experiments and error bars indicate SD. RLU, relative luciferase units. ΔG values are in kcal/mol.
While neither stem nor loop mutations that maintained the C protein sequence had a deleterious effect on viral replication in the context of a stable cHP, it is possible that the remaining, unchanged nucleotides confer some sequence-specificity. To avoid a possible phenotype resulting from a drastic change to the C protein sequence in the virus, the sequence-dependence of the cHP was explored further using the replicon system, in which the region of C that is included functions only as a cis element. The cHP element was replaced with a hairpin of heterologous sequence (HPhet) that is predicted to adopt a structure of similar ΔG (−9.7 kcal/mol; Fig. 5D). As shown in Figure 5E, DRrep-HPhet was capable of efficient RNA synthesis, similar to WT. Thus, the cHP element functions independently of sequence in RNA synthesis.
The cHP performs the same function in WNV
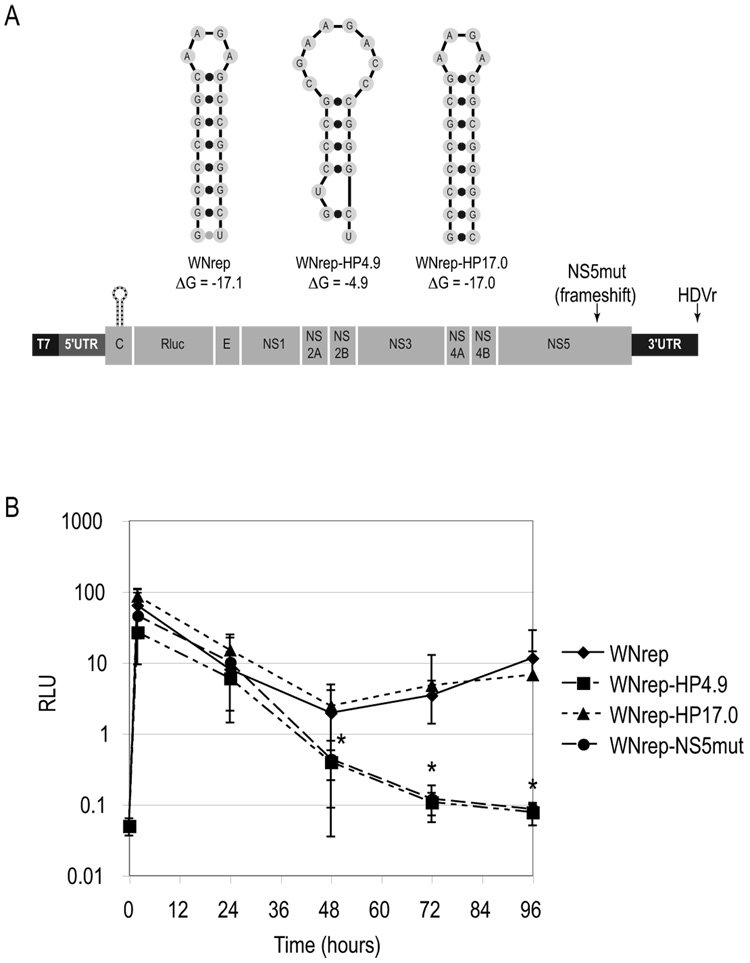
The cHP element was initially identified by computed secondary structure predictions of the 5' ends of numerous flaviviruses (Clyde and Harris, 2006). We reasoned that a structure maintained among diverse members of the Flavivirus genus would likely function in the same capacity among those viruses. In order to determine whether the cHP element functions at the step of RNA synthesis of other flaviviruses, we utilized a reporter replicon of WNV lineage I, a member of the Japanese encephalitis serogroup. This reporter replicon, RlucRep-HDVr (hereafter WNrep), which has been described previously (Tilgner and Shi, 2004), exhibits the same distinct peaks of translation corresponding to input-strand translation and RNA synthesis (Fig. 6B and Tilgner and Shi, 2004) as DRrep. A variant containing a single nucleotide frameshift in the RdRP gene (WNrep-NS5mut) lacks a second peak of translation, similar to DRrep-RdRPmut. The structures of WNrep variants are outlined in Figure 6A. Mutations were made to the WT WNV cHP (ΔG = −17.1 kcal/mol) to disrupt the secondary structure, creating a less stable hairpin (WNrep-HP4.9; ΔG = −4.9 kcal/mol), and compensatory mutations to restore basepairing generated a hairpin of similar stability to the WT (WNrep-HP17.0; ΔG = −17.0 kcal/mol). As shown in Figure 6B, all WNrep RNAs translate to similar efficiency upon entry into the cell. However, only the WNrep RNAs harboring an intact cHP (WT and HP17.0) progress to a second round of translation. WNrep-HP4.9 behaves similarly to WNrep-NS5mut (Fig. 6B). Therefore, we deduce that the cHP element functions in RNA synthesis in the life cycles of both West Nile virus and dengue virus.
Figure 6. The cHP is required for West Nile virus replication.
(A) Schematic diagram of WNrep and HP4.9, HP17.0 and NS5mut variants. ΔG values are in kcal/mol. (B) Early and late translation events in WNrep-transfected cells. Transfected cells were assayed for Rluc activity at 2, 24, 48, 72 and 96 hours post-transfection. Data are averages of at least 4 experiments and error bars indicate SD. RLU, relative luciferase units. *, p < 0.002 relative to WT by Wilcoxon rank sum test.
DISCUSSION
We had previously demonstrated that a coding-region hairpin element, cHP, modulates DENV start codon selection and is required for efficient production of infectious virus in human (Hep3B) and mosquito (C6/36) cells (Clyde and Harris, 2006). Here, we have extended our investigation of the role of the cHP in the DENV life cycle. Utilizing a reporter replicon system, we have demonstrated that the cHP mutant initially translates to WT levels in transfected cells, but does not progress to late, RNA synthesis-dependent rounds of translation or accumulate replicon RNA, pinpointing the defect to the step of RNA synthesis. The cHP was shown to operate independently of its sequence, which indicates that it likely functions in RNA synthesis as part of the overall topology of the 5' end, with sequence-specific elements present elsewhere. Finally, the cHP was demonstrated to be required for replication of the related flavivirus, West Nile virus.
Coding-region elements that are required for RNA synthesis have been characterized in other positive strand viruses. Among the best studied are the cis-acting replication elements (CRE) among Picornaviridae, originally identified in human rhinovirus-14 (HRV-14) (McKnight and Lemon, 1996; McKnight and Lemon, 1998). Since then, similar elements have been identified in HRV-2 (Gerber et al., 2001), poliovirus (Goodfellow et al., 2000), coxsackievirus B3 (van Ooij et al., 2006) and encephalomyocarditis viruses (Lobert et al., 1999) and have been shown to be required for viral RNA synthesis (Lobert et al., 1999; McKnight and Lemon, 1998; Murray and Barton, 2003; van Ooij et al., 2006; Yang et al., 2004). Within the Flaviviridae family, it has been demonstrated that HCV features an internal structure required for RNA synthesis, located near the 3' end of the genome in the NS5B-coding region (You et al., 2004). This element, 5BSL3.2, forms a pseudoknot with another stem-loop structure in the 3'UTR, SL2 (Friebe et al., 2005). The long-range interaction, independent of the location of 5BSL3.2 and of its sequence, is required for efficient HCV RNA synthesis.
A number of cis elements in the untranslated regions of flavivirus genomes have been shown to perform multiple roles in the viral life cycle. Structures in the 3'UTR, including the variable region (Alvarez et al., 2005; Chiu et al., 2005), dumbbell-shaped structures (DB1 and DB2) (Alvarez et al., 2005; Chiu et al., 2005) and 3' stem-loop (3'SL) (Holden and Harris, 2004; Holden et al., 2006; Tilgner et al., 2005; You et al., 2001; Yu and Markoff, 2005; Zeng et al., 1998) have been demonstrated to enhance both vRNA synthesis and translation. Both the 5'UTR and 3'UTR together have been shown to be required for translation when host cell cap-dependent translation has been shut off (Edgil and Harris, 2006; Edgil et al., 2006), in addition to playing a role in RNA synthesis (Filomatori et al., 2006b; Nomaguchi et al., 2003; Nomaguchi et al., 2004; You et al., 2001; You and Padmanabhan, 1999). The cHP, however, is the first element in the flavivirus coding region for which two distinct functions have been characterized: a role in translation start codon selection (Clyde and Harris, 2006) and, as shown here, a role in RNA synthesis. In the tick-borne flaviviruses, which have strong start codon contexts, a predicted cHP is located at the junction between the 5'UTR and the coding region, where it may function in RNA synthesis but not likely in translation start site selection. It is tempting to speculate that the cHP originally arose as a replication element that was later recruited for translation initiation site selection among the mosquito-borne viruses, which primarily harbor weak start codons.
Previously, we had shown that the cHP element enhanced recognition of the first AUG in translation of the viral polyprotein and that disruption of the structure resulted in a severe (at least 500 – 10,000-fold) defect in viral replication in Hep3B and C6/36 cells, below the limit of detection (Clyde and Harris, 2006). Here, the production of HP3.4 virus from BHK cells allowed the sequencing of the resulting viral genomes, revealing selection of the single most stabilizing substitution that was also a diversion from the WT sequence (Fig. 2). The recovery of virus containing the parental HP3.4 sequence in addition to the compensatory A132C mutation implies that a low level of RNA synthesis does occur from these templates, below the limit of detection by qRT-PCR (Fig. 4). The recovery of only A132C virus from C6/36 after infection with a mixed population of HP3.4 and HP3.4-A132C viruses supports our earlier finding that the parental HP3.4 virus does not replicate in this cell type. Detectable replication in BHK but not in Hep3B or C6/36 may reflect a greater permissiveness to RNA synthesis in BHK than in the other cell types and/or differences in host cell factors that participate in this process.
Surprisingly, it was not possible to define a sequence-specific element in the cHP, despite the conservation of the stem sequence and certain positions of the loop among diverse DENV2 isolates (Fig. 5). However, among different flaviviruses, the cHP sequence is poorly conserved (Clyde and Harris, 2006). The conservation within the serotype may simply reflect maintenance of the structure, where a substitution in the stem region would need to be compensated elsewhere in order to counteract a loss of viral fitness. Given the location of the structure in the polyprotein-coding region, multiple mutations would impact the protein sequence and are thus less likely to be phenotypically neutral.
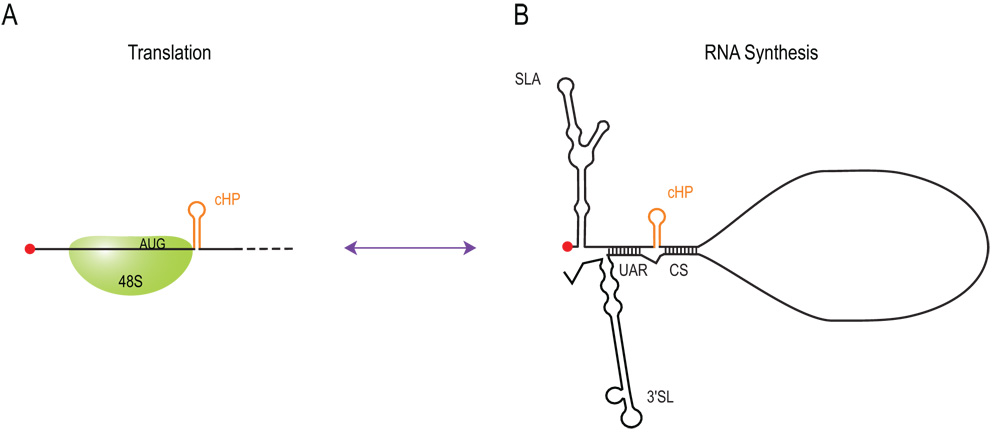
Our results here, combined with the results of others, suggest the following model for the role of the cHP in the viral life cycle, which is illustrated in Figure 7. During translation, the flavivirus vRNA is most likely not in a panhandle formation, given that basepairing between the cyclization sequences (CS) does not appreciably impact translation efficiency (Chiu et al., 2005; Edgil and Harris, 2006) or start codon selection (Clyde and Harris, 2006), and basepairing via the upstream of AUG region (UAR) would be unwound by the scanning initiation complex. The cHP element briefly stalls the scanning initiation complex over the AUG, allowing its recognition in the absence of a strong initiation context. When the 5' and 3' CS are unpaired, the cHP has an elongated stem structure that includes the first nt of the CS, as determined by solution structure probing of the 5' end (C. Polacek, J. Foley and E. Harris, unpublished data). Once the vRNA has switched from its role as an mRNA to a template for vRNA synthesis, cyclization decreases the length of the cHP stem slightly, according to the solution structure of the 5' end in the presence of the 3'UTR (C. Polacek, J. Foley and E. Harris, unpublished data). As a replication element, the cHP performs in a sequence-independent manner, which would imply that the overall topology of the 5' end is important for promoting RNA synthesis. It is also possible that selection of the first start codon per se by the scanning ribosome is important for RNA synthesis, perhaps by allowing rearrangement of RNP complexes around the start codon or by recruiting other factors to allow a shift in conformation necessary for RNA synthesis. Alternatively, if rounds of RNA synthesis are tightly coupled with rounds of translation late in the life cycle, the cHP could enhance RNA synthesis by enhancing translation late in viral infection in a manner analogous to the DLP of the alphaviruses Sindbis virus and Semliki Forest virus (Frolov and Schlesinger, 1994; Frolov and Schlesinger, 1996; Gorchakov et al., 2004; McInerney et al., 2005; Ventoso et al., 2006). However, results from our previous work indicate that first start codon selection does occur, albeit to a lesser extent, in the absence of an intact cHP (39% of WT for HP3.4), and thus we would expect a less severe defect in RNA synthesis from either of these scenarios than what we report here. Additionally, an infectious clone with a mutated first AUG produced higher viral titers than did the HP3.4 variant; therefore, start codon selection alone is unlikely to account wholly for the HP3.4 phenotype.
Figure 7. The cHP functions in translation and RNA synthesis of dengue virus.
(A) The cHP element functions in translation start site selection, probably by stalling the scanning 48S complex over the first AUG, enhancing its recognition as a start codon in the absence of a strong initiation context. The efficiency of first AUG selection is independent of the sequence of the cHP but is dependent on its position relative to the AUG and on its stability (Clyde and Harris, 2006). (B) Multiple structures at the 5' and 3' ends of flaviviruses have been shown to play an essential role in RNA synthesis (Alvarez et al., 2005; Filomatori et al., 2006a; Holden et al., 2006; Nomaguchi et al., 2003; Nomaguchi et al., 2004; Tilgner et al., 2005; You et al., 2001; You and Padmanabhan, 1999; Yu and Markoff, 2005; Zeng et al., 1998). The cHP element is also required for efficient RNA synthesis in a sequence-independent manner, possibly by forming part of the overall topology of the 5' end that initially binds the replicase complex or by stabilizing the cyclization interaction between the CS and UAR domains. Abbreviations: SLA, stem-loop A; UAR, upstream of AUG region; CS, cyclization sequence; 3'SL, 3' stem-loop.
The cis elements that participate in flaviviral RNA synthesis do so in a number of ways. The stem-loop structure of the 5'UTR (SLA) has been shown to directly bind the DENV NS5 RNA-dependent RNA polymerase (Filomatori et al., 2006b). The long-range basepairing interactions between the 5' and 3' CS domains and the 5' and 3' UAR domains contribute to RNA synthesis likely by bringing the 3'UTR into close proximity of the 5'UTR so that the replicase complex can be delivered to the 3' end for negative-strand synthesis. The exact function of the cHP in vRNA synthesis remains to be determined. Although disruption of stem-loop A (SLA) abrogated binding of NS5, the cHP element, which is located immediately upstream of the 5'CS, was included in the RNAs used in the binding assays (Filomatori et al., 2006b). Therefore, it is possible that the cHP participates in recruitment of the replicase complex to the 5' end of the positive strand in a SLA-dependent manner. Another possibility is that the cHP element somehow stabilizes or favors the panhandle structure that is formed via basepairing between the 5' and 3' ends of the genome.
RNA sequences and structures constitute important signaling elements in RNA viruses, assisting in the processes of translation, RNA synthesis and encapsidation. Here we have shown that a structure in the coding region of dengue virus type 2 serves a dual function in the viral life cycle: to enhance recognition of the first start codon of the viral polyprotein and to promote vRNA synthesis. The identification of two coding region cis elements, the 5'CS and cHP, in DENV and WNV raises the question of whether additional internal regulatory elements are present in flavivirus genomes. Experiments to define other coding region RNA elements involved in viral replication are currently underway. Delineation of RNA structures and their roles in the life cycles of RNA viruses can serve to broaden our understanding of the mechanisms viruses use to reproduce and to subvert or evade the host antiviral response and allow for the focused development of vaccines and antiviral therapies.
MATERIALS AND METHODS
DNA constructs
Variants HP3.4 and HP11.9 have been described previously (Clyde and Harris, 2006). All infectious clones are modifications of the WT pD2/IC-30P-A (pD2/IC) infectious cDNA of DENV2 Thai strain 16681 (Kinney et al., 1997). The pDRrep replicon is based on pD2/IC and was constructed similarly to a firefly luciferase (Fluc) reporter replicon described previously (Holden et al., 2006) except that the Fluc gene was replaced with a Renilla luciferase (Rluc; Promega) coding sequence and the 3' terminus contains an XbaI site that is identical to that of pD2/IC. The WNV replicons RlucRep-HDVr and RlucRep-NS5mut-HDVr (referred to here as WNrep and WNrep-NS5mut; gift of P.-Y. Shi, New York State Department of Health, Albany, NY) have been described previously (Tilgner and Shi, 2004). Mutations in the C or NS5 coding region of pD2/IC, pDRrep or WNrep were introduced by overlap-extension PCR or QuikChange (Stratagene, La Jolla, CA). Primer sequences are available upon request.
RNA templates
All pD2/IC, pDRrep and pWNrep DNA templates were generated by digestion with XbaI and were gel purified using the QIAquick Gel Extraction kit (Qiagen, Valencia, CA). DENV replicon (DRrep) and infectious clone (D2/IC) RNAs were generated by in vitro transcription with the RiboMax Large Scale RNA Production System (T7; Promega) with the following modifications to the manufacturer’s protocol: 5 mM each GTP, CTP and UTP, 1 mM ATP, and 5 mM m7G(5')ppp(5')A cap analog (New England Biolabs, Beverly, MA) were incubated for 4 hours at 30°C with the addition of 2 mM ATP after 30 min. WNrep RNAs were generated by in vitro transcription as above except that m7G(5')ppp(5')G cap analog (New England Biolabs), 5 mM ATP and 1mM GTP were utilized, with the addition of GTP to the reaction after 30 min. RNAs were subsequently treated with 80 U/ml TURBO DNase (Ambion, Austin, TX) for 15 min at 37°C, and unincorporated nucleotides were removed from DENV and WNV RNAs by size exclusion chromatography on Micro Bio-Spin P-30 Tris columns (Bio-Rad Laboratories, Hercules, CA).
Cell culture and transfection
BHK-21 (BHK) cells were grown in Minimal Essential Medium-Alpha (MEM-α; Gibco, Carlsbad, CA) containing 10 mM HEPES (Gibco) and 5% fetal bovine serum (FBS; HyClone, Logan, UT) at 37°C and 5% CO2. C6/36 cells were grown in Leibovitz’s L-15 medium (Gibco) containing 10 mM HEPES and 5% FBS at 28°C without CO2. Prior to transfection, cells were washed with Opti-MEM (Gibco), then were transfected with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. At 2 hours post-transfection, transfection medium was replaced with growth medium following extensive washing to remove non-internalized RNA. For DENV replicon and virus titration experiments, a duplicate well of transfected cells was harvested at 2 hours post-transfection, and total cellular RNA was extracted and subjected to qRT-PCR to control for transfection efficiency. In all transfection experiments, cells were grown to 50% confluence, and transfections were conducted in duplicate. For virus growth curve experiments, cells in 12-well plates were transfected with 1 µg D2/IC RNA/well. At each timepoint, 50% of the cell supernatant was harvested for plaque assay and replenished with fresh growth medium. For DRrep experiments, BHK cells in 48-well plates were transfected with 125 ng replicon RNA/well. For WNrep experiment, BHK cells in 48-well plates were transfected with 500 ng replicon RNA/well. For RNA stability experiments, cells in 48-well plates were transfected with 250 ng/well D2/IC-RdRPmut.
Virus titration by plaque assay
Infectious virus produced from D2/IC-transfected cells was quantified by standard plaque assay on BHK cells as described previously (Clyde and Harris, 2006; Diamond et al., 2000). Briefly, BHK cell monolayers were incubated for 2 hours with dilutions of D2/IC-transfected cell supernatant and overlaid with MEM-α (Sigma), 1% SeaPlaque low melting point agarose (Cambrex, Rockland, ME) and 5% FBS. After 6 days, wells were fixed with formaldehyde and stained with crystal violet, and plaque-forming units (PFU) per ml were calculated. Virus titers were normalized to transfection efficiency as determined by qRT-PCR at 2 hours post-transfection.
RT-PCR and quantitative RT-PCR
RNA was extracted from cells using the Mini RNA Isolation II Kit (Zymo Research, Orange, CA). Viral RNA was extracted from the supernatant of infected cells using the QIAamp Viral Mini RNA kit (Qiagen). Quantitative RT-PCR (qRT-PCR) was conducted using a modification to the method of Houng et al (2000) with a FAM-TAMRA-labeled probe using the TaqMan One-Step Master Mix (Applied Biosystems, Foster City, CA). Primer and probe sequences are as follows: DV10621F, 5'-CATATTGACGCTGGGAAAGA-3'; DV10723R, 5'- AGAACCTGTTGATTCAACAGCACC-3'; DV.P1-FAM-TAMRA, 5'-[6FAM]-CTGTCTCCTCAGCATCATTCCAGGCA-[TAMRA]-3'. For the purpose of determining transfection efficiency, target RNA was first normalized to cellular 18S RNA using the TaqMan VIC-MGB Primer Limited Eukaryotic 18S rRNA Endogenous Control (Applied Biosystems) in a parallel reaction. qRT-PCR was conducted on a Sequence Detection System 7300 (Applied Biosystems). For purposes of sequencing, viral cDNA was produced using the Qiagen One-Step RT-PCR kit. For sequencing of supernatants from BHK cells (Table 1), RT-PCR products were subcloned into pCR2.1-TOPO (Invitrogen), and plasmid DNA was isolated by alkaline lysis. Sequencing was conducted at the University of California, Berkeley DNA Sequencing Facility.
Calculation of RNA half-life
Total RNA from D2/IC-RdRPmut-transfected cells was extracted at 2, 4, 8, 24, 48 and 72 hours post-transfection, treated with DNase, and quantitated by qRT-PCR using the primer and probe set complementary to the 3' end described above. Values were graphed, and an exponential trendline was fitted to the data in Microsoft Excel 2004 for Macintosh. Half-life of each variant for each experiment was determined from the exponential trendline equation. R2 values for the line equations and t1/2 values from each experiment are shown in Table 1. P-values relative to WT were derived by Wilcoxon signed rank test in Mstat for Macintosh.
Luciferase assay
Cells were harvested at the indicated timepoints, and samples were analyzed with the Renilla Luciferase Assay System (Promega) according to the manufacturer’s instructions on a TD-20/20 luminometer (Turner BioSystems, Sunnyvale, CA). DRrep translation levels were normalized to relative transfection efficiency at 2 hours post-transfection as determined by qRT-PCR.
Immunoblot analysis
D2/IC-transfected BHK cells were lysed at the indicated timepoint in 125 mM NaCl, 50 mM Tris-Cl, pH 8.0, 10% glycerol and 1% NP-40, and the lysate was clarified by centrifugation. Samples were separated by SDS-PAGE on a 10% polyacrylamide gel and transferred to a nitrocellulose membrane. Immunoblots were incubated with anti-NS1 7E11 monoclonal antibody (gift of Dr. Robert Putnak, Walter Reed Army Institute of Research, Silver Spring, MD) followed by incubation with horseradish peroxidase-conjugated goat anti-mouse Ig (Jackson Immunoresearch, West Grove, PA). Blots were washed and reprobed with HRP-conjugated anti-actin monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Immunoblots were imaged on a ChemiDoc EQ system (Bio-Rad).
RNA sequences and structures
cHP sequences from diverse strains of DENV2 were chosen based on sequence availability and their classification into American, Malaysia/Indian Subcontinent and Southeast Asian genotypes (Rico-Hesse, 2003). GenBank accession numbers for the strains selected are as follows: 131/Mexico/92 (AF100469), P8-377/Malaysia/69 (U88237), 1409/Jamaica/81 (M20558), FJ11/China/99 (AF359579), VEN2/Venezuela/87 (AF100465), and 16681/Thailand/64 (U87411). Secondary structures were predicted using mfold (Mathews et al., 1999; Zuker, 2003). Free energy values were derived from independent folding of the relevant hairpin sequence and reflect efn2 refinement as described previously (Clyde and Harris, 2006). Codon usage was assessed for Mesocricetus auratus (BHK) and Aedes albopictus (C6/36) using the Codon Usage Database implementation of Codon Usage Tabulated from GenBank (Nakamura et al., 2000).
Statistical analysis
Wilcoxon signed rank, Wilcoxon rank sum and Fisher’s exact tests were performed in Mstat for Macintosh. Statistical significance was defined as p < 0.05.
ACKNOWLEDGMENTS
We thank Richard Kinney for providing pD2/IC, Katherine L. Holden and Theodore C. Pierson for providing pDRrep, and Pei-Yong Shi for providing RlucRep-HDVr and RlucRep-HDVr-NS5mut (WN). We are grateful to Melinda Lee for assistance with cloning, to Diana J. Flores for assistance with plaque assays, and to Jennifer L. Kyle and Luhua Zhang for assistance with qRT-PCR. We thank Scott Balsitis, Suman M. Paranjape, José Peña, Charlotta Polacek and Sondra and Milton J. Schlesinger for careful reading of this manuscript and/or helpful discussions.
Funding for this research was provided by NIH grant AI052324 (E.H.), as well as by the Russell R. Grossman Endowment (K.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Karen Clyde, Email: kclyde@berkeley.edu.
Julio Barrera, Email: Julio.Barrera@ucsf.edu.
REFERENCES
- Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3'UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339(2):200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Chiu WW, Kinney RM, Dreher TW. Control of translation by the 5'- and 3'-terminal regions of the dengue virus genome. J Virol. 2005;79(13):8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K, Harris E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J Virol. 2006;80(5):2170–2182. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K, Kyle JL, Harris E. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis. J Virol. 2006;80(23):11418–11431. doi: 10.1128/JVI.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol. 2000;74(11):4957–4966. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgil D, Harris E. End-to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res. 2006;119(1):43–51. doi: 10.1016/j.virusres.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgil D, Polacek C, Harris E. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J Virol. 2006;80(6):2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5' RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006a doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5' RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006b;20(16):2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Boudet J, Simorre JP, Bartenschlager R. Kissing-loop interaction in the 3' end of the hepatitis C virus genome essential for RNA replication. J Virol. 2005;79(1):380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Schlesinger S. Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J Virol. 1994;68(12):8111–8117. doi: 10.1128/jvi.68.12.8111-8117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Schlesinger S. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J Virol. 1996;70(2):1182–1190. doi: 10.1128/jvi.70.2.1182-1190.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber K, Wimmer E, Paul AV. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2A(pro) J Virol. 2001;75(22):10979–10990. doi: 10.1128/JVI.75.22.10979-10990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I, Chaudhry Y, Richardson A, Meredith J, Almond JW, Barclay W, Evans DJ. Identification of a cis-acting replication element within the poliovirus coding region. J Virol. 2000;74(10):4590–4600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Frolova E, Williams BR, Rice CM, Frolov I. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J Virol. 2004;78(16):8455–8467. doi: 10.1128/JVI.78.16.8455-8467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11(3):480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ, Kuno G, Markoff L. Flaviviruses. In: Fields BN, Knipe DM, Howley PM, Griffin DE, editors. Fields Virology. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2006. pp. 1153–1252. [Google Scholar]
- Guyatt KJ, Westaway EG, Khromykh AA. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J Virol Methods. 2001;92(1):37–44. doi: 10.1016/s0166-0934(00)00270-6. [DOI] [PubMed] [Google Scholar]
- Holden KL, Harris E. Enhancement of dengue virus translation: role of the 3' untranslated region and the terminal 3' stem-loop domain. Virology. 2004;329(1):119–133. doi: 10.1016/j.virol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Holden KL, Stein DA, Pierson TC, Ahmed AA, Clyde K, Iversen PL, Harris E. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3' stem-loop structure. Virology. 2006;344(2):439–452. doi: 10.1016/j.virol.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Houng HH, Hritz D, Kanesa-Thasan N. Quantitative detection of dengue 2 virus using fluorogenic RT-PCR based on 3'-noncoding sequence. J Virol Methods. 2000;86(1):1–11. doi: 10.1016/s0166-0934(99)00166-4. [DOI] [PubMed] [Google Scholar]
- Kinney RM, Butrapet S, Chang GJ, Tsuchiya KR, Roehrig JT, Bhamarapravati N, Gubler DJ. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology. 1997;230(2):300–308. doi: 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Flaviviridae: The Viruses and Their Replication. In: Fields BN, Knipe DM, Howley PM, Griffin DE, editors. Fields Virology. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2006. pp. 1101–1152. [Google Scholar]
- Lo MK, Tilgner M, Bernard KA, Shi PY. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3' untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J Virol. 2003;77(18):10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobert PE, Escriou N, Ruelle J, Michiels T. A coding RNA sequence acts as a replication signal in cardioviruses. Proc Natl Acad Sci U S A. 1999;96(20):11560–11565. doi: 10.1073/pnas.96.20.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288(5):911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol Biol Cell. 2005;16(8):3753–3763. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight KL, Lemon SM. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J Virol. 1996;70(3):1941–1952. doi: 10.1128/jvi.70.3.1941-1952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight KL, Lemon SM. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4(12):1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray KE, Barton DJ. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J Virol. 2003;77(8):4739–4750. doi: 10.1128/JVI.77.8.4739-4750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28(1):292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomaguchi M, Ackermann M, Yon C, You S, Padmanabhan R. De novo synthesis of negative-strand RNA by dengue virus RNA-dependent RNA polymerase in vitro: Nucleotide, primer, and template parameters. J Virol. 2003;77(16):8831–8842. doi: 10.1128/JVI.77.16.8831-8842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomaguchi M, Teramoto T, Yu L, Markoff L, Padmanabhan R. Requirements for West Nile virus (−)- and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J Biol Chem. 2004;279(13):12141–12151. doi: 10.1074/jbc.M310839200. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315–341. doi: 10.1016/s0065-3527(03)59009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner M, Deas TS, Shi PY. The flavivirus-conserved penta-nucleotide in the 3' stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology. 2005;331(2):375–386. doi: 10.1016/j.virol.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Tilgner M, Shi PY. Structure and function of the 3' terminal six nucleotides of the west nile virus genome in viral replication. J Virol. 2004;78(15):8159–8171. doi: 10.1128/JVI.78.15.8159-8171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij MJ, Vogt DA, Paul A, Castro C, Kuijpers J, van Kuppeveld FJ, Cameron CE, Wimmer E, Andino R, Melchers WJ. Structural and functional characterization of the coxsackievirus B3 CRE(2C): role of CRE(2C) in negative- and positive-strand RNA synthesis. J Gen Virol. 2006;87(Pt 1):103–113. doi: 10.1099/vir.0.81297-0. [DOI] [PubMed] [Google Scholar]
- Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20(1):87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Rijnbrand R, Watowich S, Lemon SM. Genetic evidence for an interaction between a picornaviral cis-acting RNA replication element and 3CD protein. J Biol Chem. 2004;279(13):12659–12667. doi: 10.1074/jbc.M312992200. [DOI] [PubMed] [Google Scholar]
- You S, Falgout B, Markoff L, Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5'- and 3'-terminal regions that influence RNA structure. J Biol Chem. 2001;276(19):15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- You S, Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3'-end of exogenous viral RNA templates requires 5'- and 3'-terminal complementary sequence motifs of the viral RNA. J Biol Chem. 1999;274(47):33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- You S, Stump DD, Branch AD, Rice CM. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol. 2004;78(3):1352–1366. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Markoff L. The topology of bulges in the long stem of the flavivirus 3' stemloop is a major determinant of RNA replication competence. J Virol. 2005;79(4):2309–2324. doi: 10.1128/JVI.79.4.2309-2324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Falgout B, Markoff L. Identification of specific nucleotide sequences within the conserved 3'-SL in the dengue type 2 virus genome required for replication. J Virol. 1998;72(9):7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]