Abstract
Understanding the principles of information processing in neural circuits requires systematic characterization of the participating cell types and their connections, and the ability to measure and perturb their activity. Genetic approaches promise to bring experimental access to complex neural systems, including genetic stalwarts such as the fly and mouse, but also to nongenetic systems such as primates. Together with anatomical and physiological methods, cell-type-specific expression of protein markers and sensors and transducers will be critical to construct circuit diagrams and to measure the activity of genetically defined neurons. Inactivation and activation of genetically defined cell types will establish causal relationships between activity in specific groups of neurons, circuit function, and animal behavior. Genetic analysis thus promises to reveal the logic of the neural circuits in complex brains that guide behaviors. Here we review progress in the genetic analysis of neural circuits and discuss directions for future research and development.
1. Introduction
The realization that individual neurons are the building blocks of the nervous system was a key conceptual leap in neuroscience (Cajal, 1911). This advance is analogous to the insight that the gene is the unit of operation in genetics and molecular biology (Morgan, 1911; Beadle and Tatum, 1941; Benzer, 1955; Jacob and Monod, 1961). However, studying individual genes is insufficient to understand cells. Similarly, studying single neurons is insufficient to comprehend how the brain works.
The mammalian brain consists of billions of neurons, including thousands of cell types, connected into circuits by trillions of synapses. The ultimate goal of neuroscience is to understand the principles organizing these complex circuits and thereby decipher how they process information and guide behavior. Recent developments suggest that genetic analysis will play a prominent role in dissecting neural circuits.
Informative analogies can be made between gene interaction networks that regulate complex biological processes and neural circuits (Figure 1). Remarkably, formal analysis has suggested that gene networks and neural circuits share basic organizational principles (Milo et al., 2002). In gene networks, the interactions of different proteins implement information processing, such as transducing cell surface signals to transcriptional response in the nucleus or orchestrating cell division. The networks can be adjusted by regulating the concentrations of individual components through transcription, translation, and degradation, or by regulating protein-protein interactions through posttranslational modifications. In the brain, individual neurons (in simple organisms) or groups of neurons of the same type (in vertebrates) act as the basic functional units. Their connection patterns and the strengths and properties of their functional interactions determine how neural circuits process information.
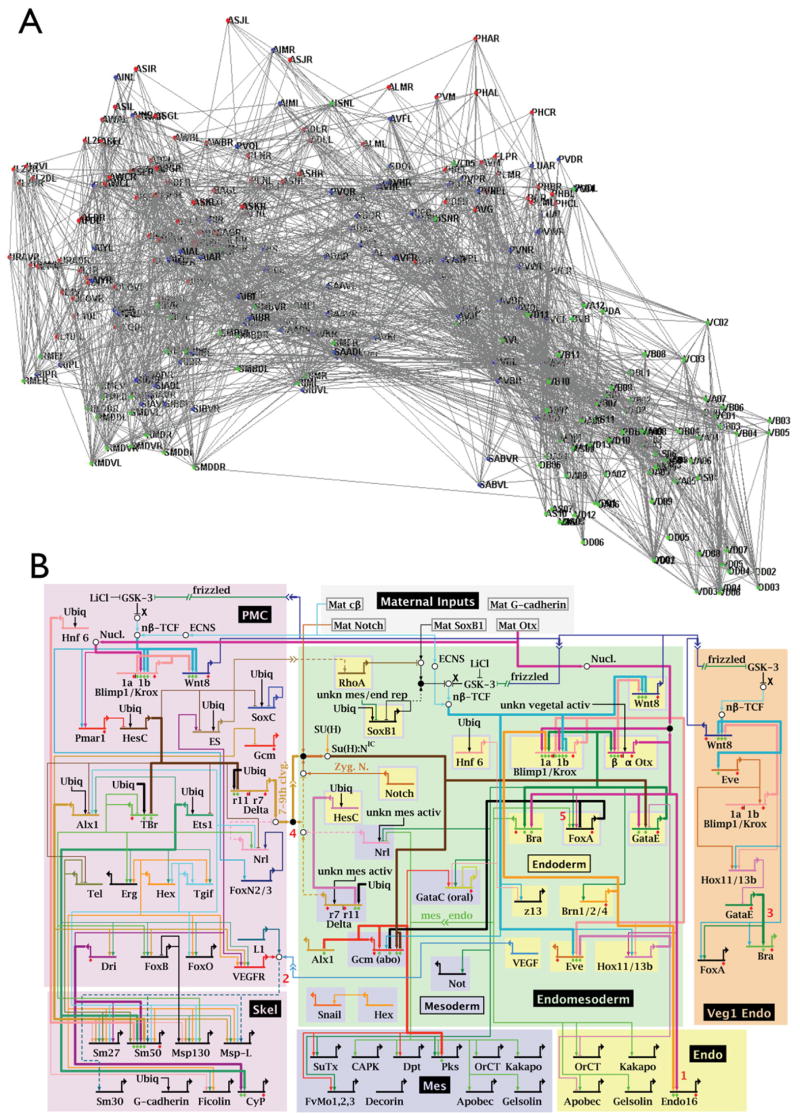
Figure 1. Neural and Gene Networks.
(A) Complete wiring diagram of connections among 302 neurons in C. elegans, reconstructed from serial-section EM. Depicted are individual neurons and their connections. For more details see http://www.wormatlas.org/handbook/nshandbook.htm/nswiring.htm. Courtesy of D. Chklovskii.
(B) Diagram of gene interaction network that orchestrates early endomesoderm development of sea urchin embryos. Depicted are individual genes and their regulatory relationships. For more details see http://sugp.caltech.edu/endomes/. Courtesy of E. Davidson.
Genetic analysis can decipher the logic of gene networks that underlie biological processes, including such complex phenomena as the embryonic patterning of multicellular organisms (Nüsslein-Volhard and Wieschaus, 1980). Systematic protein-protein and transcription factor-DNA interactions contribute to deciphering the gene networks. Similarly, systematic discovery of neuronal cell types and analysis of the connectivity between these cell types is necessary to establish the wiring diagram of neural circuits (Sections 2 and 3). Measurements of gene expression and posttranslational modifications of proteins are readouts of the state of the gene network. Similarly, the measurement of activity in defined neuronal cell types is critical to track the dynamic properties of neural circuits (Section 4). Finally, loss-of-function (LOF) and gain-of-function (GOF) experiments identify essential components of gene interaction networks, and establish causal relationships (necessity, sufficiency) between a gene and its contribution to the network’s function. Similarly, precise LOF and GOF experiments can reveal the contributions of individual neuronal cell types to the functional output of the circuits (Section 5).
Genetic analysis is promising to facilitate breakthroughs in our understanding of how neural circuits process information, and to establish causality between the activity in specific groups of neurons, the function of neural circuits, and animal behavior. In this primer we review recent progress in the development of tools that allow genetic dissection of neural circuits, and discuss their strengths and limitations in comparison to traditional methods. Examples are drawn largely from our areas of expertise, mainly the olfactory system in fruit flies and the cerebral cortex of mice and primates, but the concepts and techniques we discuss are applicable to other genetic or nongenetic model organisms.
2. Genetic Targeting of Cell Types
2a. What Is a Cell Type?
Although this important question is central to neural circuit analysis, the definition of cell type is complex and contentious, requiring in-depth review by itself. Here we discuss definitions of cell type with an emphasis on the practical aspects relevant to circuit analysis.
A cell type is usually the unit to be monitored and manipulated for circuit analysis. Historically, several overlapping parameters have been used to define cell types, including cell body location, developmental history, dendritic morphology, axonal projection, electrophysiological characteristics, gene expression pattern, and function. It is widely believed that a unique combination of these parameters defines each cell type. A logical definition of cell type is functional: neurons that perform the same function within the circuit belong to the same cell type. The limitation of this definition is that only in a few cases do we know the precise functions of neurons in brain circuits; indeed, discovering these functions is a major goal of neural circuit analysis.
In well-studied cell types, these different definitions converge. For example, olfactory receptor neurons that express a common odorant receptor, and therefore detect and transmit information about the same odorants, functionally belong to the same cell type. Their axons also project to a common glomerular target, connecting with the same sets of postsynaptic neurons. The correspondence among gene expression, axonal projection, connectivity, and function is excellent. The promoters corresponding to specific odorant receptors also provide a technically useful way to genetically access cell types (Buck and Axel, 1991; Mombaerts et al., 1996; Gao et al., 2000; Vosshall et al., 2000).
In general, defining cell types in invertebrate organisms with identified neurons is less ambiguous. Each of the 302 C. elegans neurons has a stereotyped lineage (Sulston et al., 1983), largely stereotyped connectivity (White et al., 1986; Chen et al., 2006) (Figure 1A), and probably function. Even individual neurons belonging to bilateral pairs can exhibit different gene expression patterns and functions (Troemel et al., 1999; Wes and Bargmann, 2001; Hobert et al., 2002).
Defining cell types becomes increasingly challenging as the nervous system’s complexity increases. Certain highly organized nervous tissues such as the vertebrate retina and cerebellum are viewed as having well-defined, discrete cell types. However, even in these “crystalline” structures, additional cell types are being defined based on more detailed studies of gene expression patterns, connectivity, and function (reviewed in Masland, 2001; Sillitoe and Joyner, 2007).
Nowhere is it more challenging to define cell types than in the mammalian cerebral cortex. Starting from classifications of spiny pyramidal and aspiny stellate cells based on Golgi staining, later studies revealed that these correspond largely (but not always) to glutamatergic excitatory neurons and GABAergic inhibitory neurons, respectively. While this basic dichotomy endures, we now know that there are dozens of subtypes of both excitatory and inhibitory cortical neurons. They differ in the locations of their cell bodies within distinct cortical layers, dendritic morphology, axonal projection, and spiking patterns. Even in this complex situation, gene expression profiles distinguish cell types with distinct morphologies and firing patterns (Sugino et al., 2006; N. Heintz, personal communications).
In summary, many parameters are currently used to define cell types. We suggest that as our understanding deepens, definitions based on distinct parameters will be refined and likely converge. For the purpose of dissecting neural circuits at present, useful operational definitions correspond to our abilities to use genetic tools to study neurons. These include, foremost, gene expression patterns, which yield enhancer/promoter elements to access specific cell types. Other useful definitions include axon projection patterns or cell-surface receptor expression, which allow targeting with viruses using specific injection sites or engineered tropism. In the rest of this section, we review methods that allow us to genetically access cell types as a prerequisite to dissecting their functions in neural circuits.
2b. Targeting Cell Types by Mimicking Endogenous Gene Expression
A common way to genetically target specific cell types is by mimicking endogenous gene expression. The simplest and most widely used method is to isolate cis-regulatory elements (enhancers and promoters) that specify such expression, and use these elements to drive the cDNA that encodes the desired protein as a transgene (Figure 2A). Often the appropriate cis-regulatory elements are 5′ to the endogenous promoter, although they can also be located in introns or 3′ to the transcription unit. These transgenesis methods can be extended to most organisms using electroporation of DNA (Fukuchi-Shimogori and Grove, 2001; Haas et al., 2001; Saito and Nakatsuji, 2001; Kitamura et al., 2008) or virus-mediated gene transduction (see Section 2h below).
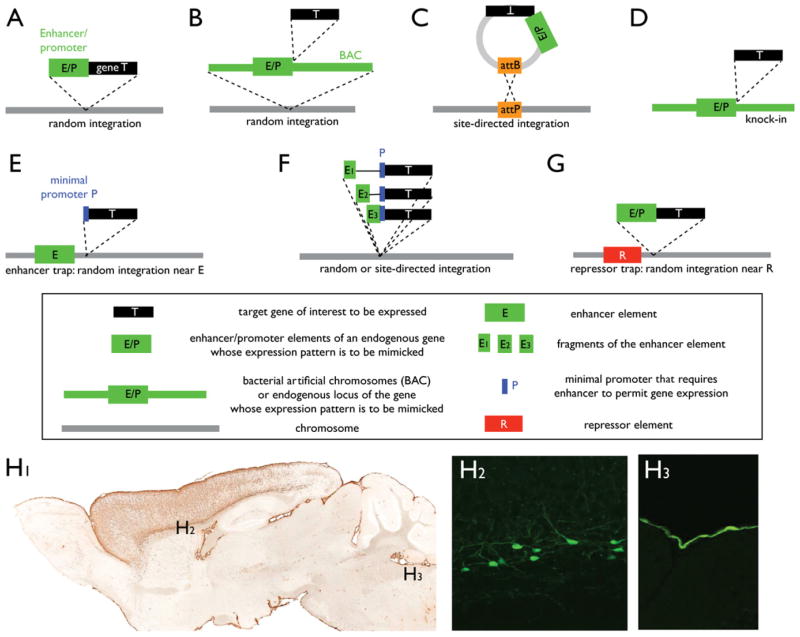
Figure 2. Methods for Targeting Gene Expression.
Box provides a glossary for the symbols in (A)–(G). See text for more details.
(A) Simple transgenic method to express the coding sequence of target gene of interest under the control of the enhancer/promoter of a gene whose expression is to be mimicked.
(B) Bacterial artificial chromosome (BAC)-mediated transgenic expression.
(C) Integrase-mediated, site-directed integration of a transgene at a defined chromosomal locus.
(D) Knockin of target gene of interest at the endogenous locus of a gene whose expression is to be mimicked.
(E) Enhancer trap method, which allows target gene of interest to be under the control of enhancer elements near its chromosomal integration site.
(F) Enhancer bashing to create subset expression patterns of an endogenous gene.
(G) Restriction of transgene expression is likely due to trapping of repressor elements and chromatin structures local to integration sites.
(H) Transgenic mouse expressing GFP under the control of the BAC for the connective tissue growth factor (ctgf). In the cerebral cortex a subpopulation of layer 6b neurons are labeled. The axons of these neurons span all cortical layers and their function is unknown. H2, cell bodies; H3, axonal projections. For more details see http://www.gensat.org/. Courtesy N. Heintz.
Cis-regulatory elements are often located tens or even hundreds of kilobases away from the gene they regulate, making it difficult to generate conventional transgenics that include all relevant elements. In addition, because the transgene integrates randomly in the genome, its expression will be influenced by local regulatory elements. Although expression patterns altered by integration effects can be very useful (see Section 2c below), for most applications it is desirable to minimize such effects.
Bacterial artificial chromosome (BAC)-mediated transgenics mimic the expression patterns of endogenous genes more faithfully (Figure 2B). Because BACs can span more than 100 kilo-bases of genomic sequence, they contain a relatively complete set of cis-regulatory elements (reviewed in Giraldo and Montoliu, 2001; Heintz, 2001). In addition, the expression of the transgene is buffered from the influence of enhancers and repressors surrounding the integration site. Recent development of recombineering technology (e.g., Warming et al., 2005) has made the construction of BACs nearly as convenient as conventional plasmids. However, perhaps because of random integration, coupled with its inability to guarantee that all cis-regulatory elements are included, BAC-mediated transgenesis may still not reliably recapitulate endogenous gene expression.
Site-specific integration of a transgene to a predetermined locus can also combat the effects of random transgene integration (Figure 2C). For instance, the phage ΦC31 integrase allows a foreign piece of DNA containing an attP site to integrate at the attB site previously inserted at a specific chromosomal location (Groth and Calos, 2004; Groth et al., 2004; Bischof et al., 2007).
The most faithful mimicry of endogenous gene expression is achieved using gene targeting (“knockin”). Here the target gene is inserted, via homologous recombination in embryonic stem cells, at the endogenous locus of the gene whose expression pattern is to be mimicked (Figure 2D). This can be achieved in mice and flies (Capecchi, 1989; Rong and Golic, 2000). Knockins typically disrupt expression of the endogenous gene. Although losing one copy of most genes usually does not result in detectable phenotypes, this is not always the case. A potential remedy is to use the internal ribosomal entry site (IRES) so that the endogenous and target gene can be expressed bicistronically from the same mRNA (e.g., Mombaerts et al., 1996). However, the expression levels often differ significantly for the open reading frames before and after the IRES. Another promising strategy is to link the open reading frames of the endogenous and target genes with the self-cleaving 2A peptide; the self-cleavage of the peptide results in equal expression of two proteins (e.g., Szymczak et al., 2004).
Targeting transgenes to specific neuronal populations is facilitated by comprehensive data on gene expression patterns. To address this need, large-scale in situ hybridization studies in the mouse have mapped the expression of transcription factors during critical stages of development (Gray et al., 2004) and the entire transcriptome in the adult brain (Lein et al., 2007). A large-scale BAC transgenic project (GENSAT) is providing complementary data on regulatory elements that may restrict gene expression to specific cell types (Gong et al., 2003) (Figure 2H).
2c. Targeting Cell Types by Enhancer Trap, Enhancer Bashing, and ‘Repressor Trap’
The systematic characterization of cis-regulatory elements of many genes will require tremendous effort. Alternative strategies are based on random insertion in the genome of target genes under the control of a minimal promoter. The transgene will then be expressed according to the specific pattern conferred by enhancers close to the integration site (Figure 2E). These enhancer trap methods have been spectacularly successful in flies (Bellen et al., 1989; Bier et al., 1989; Brand and Perrimon, 1993; Hayashi et al., 2002). They have also been applied to the mouse (Allen et al., 1988; Gossler et al., 1989; R. Davis [pronuclear injection], C. Lois [lentiviral transgenesis], personal communications) and zebrafish (Davison et al., 2007; Scott et al., 2007).
Often, expression of endogenous genes or enhancer traps is still too widespread to be useful. The expression of a gene is typically controlled by separate activators and repressors that bind at different sites of the cis-regulatory element. One strategy to target a subset of cells is to generate a series of DNA fragments corresponding to different parts of an endogenous enhancer/promoter element, and use these DNA fragments to drive target gene expression (e.g., Small et al., 1992) (Figure 2F). This “enhancer bashing” strategy is currently used to subdivide patterns of neural gene expression in flies (G. Rubin, personal communication).
Another useful way to restrict gene expression harnesses random integration effects. One starts with an enhancer/promoter that drives the expression of a target gene (Figure 2A). In particular lines of transgenic animals, the expression of the target gene is often limited to a subset of cells in which the enhancer/ promoter is normally active. For example, transgenes driven by the promoter of CAMKIIα, which is normally expressed in most excitatory forebrain neurons, can be restricted to specific cell types of the hippocampus and striatum (Tsien et al., 1996; Nakazawa et al., 2002; Kellendonk et al., 2006). A similar effect has also been observed using glutamic acid decarboxylase (GAD) promoters to drive GFP expression in several different transgenic mouse lines. Rather than expressing GFP in all GAD-positive inhibitory neurons, expression is restricted to diverse subsets of inhibitory neurons that are reproducible across animals within a single transgenic line (Oliva et al., 2000; Chattopadhyaya et al., 2004; Lopez-Bendito et al., 2004). Perhaps the most remarkable examples are thy-1-promoter-driven transgenes in mice. Endogenous thy-1 is expressed in many projection neurons (PNs), but thy-1-promoter-driven transgenes are often expressed in a subset of these neurons, ranging from nearly all to 0.1%, depending on the integration sites (Caroni, 1997; Feng et al., 2000; De Paola et al., 2003). These expression patterns are genetically heritable and thus very useful for experiments requiring sparse labeling of neurons with high concentrations of fluorescent protein (see Section 3a) (Trachtenberg et al., 2002; Grutzendler et al., 2002). Although the mechanisms for such mosaicism are unclear (see Discussion in Feng et al., 2000), the influence of local repressor elements, including chromatin structures at integration sites (which we term “repressor trap,” in analogy with enhancer trap; Figures 2G and 2E) likely plays a role.
2d. Binary Expression Strategies
In the methods described above, cis-regulatory elements directly drive the target gene expression (Figure 2). An alternative is to use binary expression strategies, which can have many advantages. For example, the Gal4/UAS system (Fischer et al., 1988; Brand and Perrimon, 1993) has changed the world for Drosophila biologists. In this strategy, a cis-regulatory element “A” is used to drive the yeast transcription factor Gal4 as a transgene. In a separate transgene, target gene “T” is under the control of Gal4-UAS (upstream activation sequence). When A-Gal4 and UAS-T transgenes are introduced into the same fly, T will be under the control of A (Figure 3A). Transcriptional amplification through the binary strategy can increase transgene expression level (at least in the case of the Gal4/UAS system in Drosophila). This is highly significant because the level of transgene expression often limits the usefulness of various effectors for circuit analysis (Sections 3–5). Another important advantage of this strategy is that one can create a library of Gal4 lines, each of which can be used to drive the expression of a battery of UAS-transgenes that encode proteins to label, measure activity, and inactivate or activate specific populations of neurons (see Sections 3–5 below). This combinatorial power is critical for neural circuit analysis.
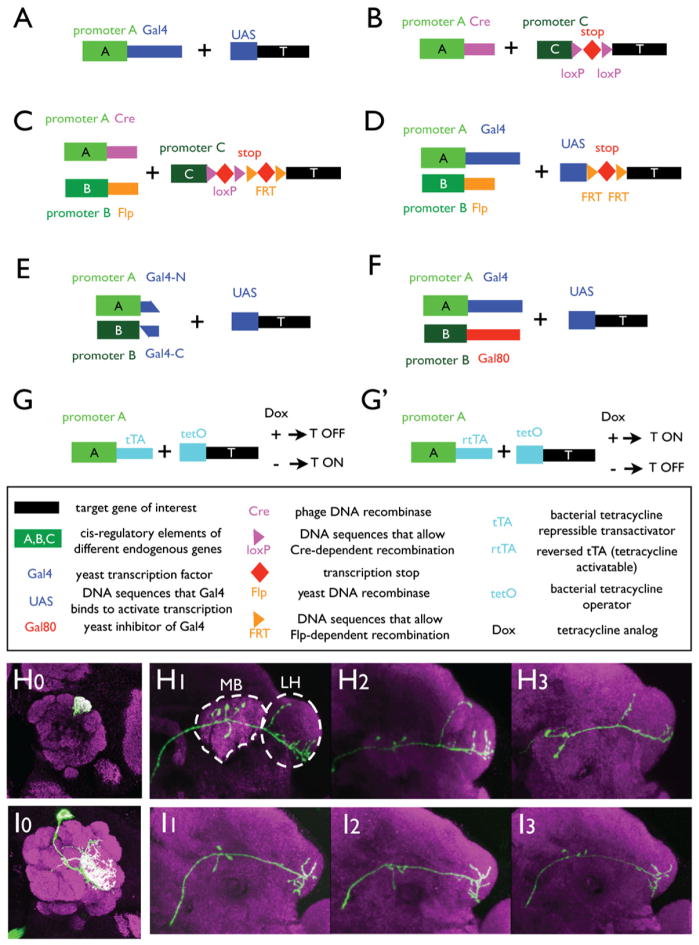
Figure 3. Binary and Intersectional Methods of Gene Expression.
Box below provides a glossary for the symbols in (A)–(G′).
(A) Yeast transcription factor Gal4 binds to UAS and activates target gene T expression in cells where promoter A is active. The same scheme applies to other transcription factor/binding site-based binary expression systems.
(B) Cre/loxP-mediated recombination removes the transcription stop, allowing target gene T to be expressed in cells that are active for both promoters A and C. Promoter C is often constitutive for general application; if promoter C is also specific, it can provide intersectional restrictions with promoter A. Cre can be replaced with a taxoxifen-inducible CreER to allow control of timing and amount of recombination. The same scheme also applies to other site-directed recombination systems, such as Flp/FRT.
(C) Combination of Cre/loxP and Flp/FRT recombination systems allow target gene of interest to be expressed in cells that are active for both promoters A and B (and C).
(D) The combination of Gal4/UAS and Flp/FRT allows the target gene of interest to be expressed in cells that are active for both promoters A and B. Gal4/UAS can be replaced with other binary expression systems; Flp/FRT can be replaced by other recombination systems.
(E) Intersectional method that utilizes the reconstitution of N- and C-terminal parts of Gal4.
(F) Target gene is expressed in cells that are active for promoter A but not promoter B, as Gal80 inhibits Gal4 activity.
(G and G′) Tetracycline-inducible transcription of target gene T. Dox, doxycycline, a tetracycline analog.
(H–I) Examples of restricting gene expression in genetically identified single cells using the MARCM method (see text) in Drosophila. Three olfactory projection neurons (PNs) from three individual flies that send dendrites to the DL1 glomerulus (H0) exhibit stereotyped axon termination patterns in higher olfactory centers, the mushroom body (MB), and particularly, the lateral horn (LH) (H1–H3). Likewise, three PNs that send dendrites to the VA1lm glomerulus (I0) exhibit stereotyped axon terminations (I1–I3) distinct from those of DL1 PNs. Green: mCD8-GFP that labels dendritic and axonal projections of single PNs; magenta: mAB nc82 staining that stains the neuropil structure. Modified from Marin et al., 2002.
The Gal4/UAS system is so effective that most of the enhancer trap screens in flies have been performed based on this strategy, and thousands of Gal4 lines have been characterized. Gal4/UAS has also been used in zebrafish (Davison et al., 2007; Sato et al., 2007a; Scott et al., 2007) and mice (Ornitz et al., 1991; Rowitch et al., 1999). Another binary expression system is based on lex-Aop (operator)-driven transgene expression by bacterial DNA-binding protein lexA fused with various eukaryotic transcription activation domains (Lai and Lee, 2006). Tetracycline-inducible transgene expression, a popular binary system in mice, additionally offers temporal regulation (see Section 2f below).
A distinct class of binary expression strategies is based on site-specific DNA recombination (Figure 3B). A cis-element A is used to drive the expression of a DNA recombinase. The target gene of interest T is under the control of a ubiquitous promoter “C,” but interrupted by a transcription stop flanked by two recombinase target sites. When these two transgenes are introduced into the same animal, the transcription stop is deleted in cells expressing the recombinase, triggering the expression of T. The bacteriophage recombinase Cre, which induces recombination between two loxP sites, has been widely applied in the mouse. Because the same strategy has been used for Cre/loxP-mediated conditional knockouts, many transgenic mice expressing the Cre re-combinase with different spatial and temporal patterns have been generated (reviewed in Nagy, 2000; Garcia-Otin and Guillou, 2006). Indeed, Cre drivers are being created as NIH-sponsored projects (e.g., http://www.mmrrc.org; http://www.gensat.org) (Gong et al., 2007). As with the fly Gal4/UAS system, a growing collection of transgenic Cre mouse lines and “floxed stop” alleles (Figure 3B) provides combinatorial power for experimental design.
A similar recombination strategy is based on the yeast Flippase/FLP recognition target (Flp/FRT). Flp/FRT was originally introduced for mosaic analysis in Drosophila (Golic and Lindquist, 1989) but has also been used for targeted expression of transgenes in flies (Struhl and Basler, 1993) and mice (Dymecki, 1996). Other recombination systems (Thomson and Ow, 2006) could also be exploited, particularly for intersectional gene expression.
2e. Intersectional Methods of Gene Expression
A cell type is usually not defined by expression of a single gene but rather by the combination of several genes. Intersectional methods become necessary to access these cell types for manipulating neural circuits with higher precision. For example, to manipulate a specific type of neuron at a specific location, it is possible to express a transgene at the intersection of two different sets of cis-regulatory elements (equivalent to the logic “and” gate), one specifying the location and the other cell type (reviewed in Dymecki and Kim, 2007). The Cre/loxP method discussed in the previous section is an example of an intersectional expression strategy (Figure 3B). One can replace the ubiquitous promoter C with a second tissue-specific promoter, such that gene T can only be expressed in cells in which both promoters are active.
A second intersectional method depends on the combination of both Cre/loxP and Flp/FRT recombination systems (Figure 3C). The target gene is turned on only in cells that express both Cre and Flp recombinases, which remove the double transcription stops before its cDNA. This method has been used in mice (Awatramani et al., 2003). Variations on this theme employ Gal4 and Flp transgenes to create intersections (Figure 3D) (e.g., Stockinger et al., 2005).
Another intersectional strategy is to split a transcription factor, such as Gal4, into N- and C-terminal half proteins; each half is not able to activate transcription, but together they reconstitute the function of Gal4. Thus, one can drive Gal4-N with promoter A, and Gal4-C with promoter B. Only in cells in which both A and B are active would UAS-T be expressed (Figure 3E) (Luan et al., 2006).
The intersectional methods discussed so far implement the logic gate “A and B.” Other logic can also be implemented. For example, Gal4 driven by promoter A is expressed in regions 1 and 2. To restrict expression to region 1, a second promoter B, whose expression covers region 2 but not 1, can be used to drive Gal80, a yeast inhibitor for Gal4 that also works in multicellular organisms (Lee and Luo, 1999). Thus, combining A-Gal4 and B-Gal80 can refine UAS-T expression by implementing the logic gate “A not B” (Figure 3F) (Suster et al., 2004; C. Potter and L.L., unpublished data).
2f. Temporal Control of Transgene Expression
It is often useful to control the timing of transgene expression in a cell type. A widely used tool in the mouse is the tetracycline-dependent promoter (Gossen and Bujard, 1992). The bacterial tetracycline-regulated transactivator (tTA) is driven by promoter A, which activates the expression of target gene T under the control of the tetO (operator) only in the absence of tetracycline (Figure 3G). A modification to the tTA has been made to reverse the direction of tetracycline control, such that the modified product, rtTA, is only active in the presence of tetracycline (Figure 3G′). Thus, one can use tetracycline to control the timing and to some extent the amount of transgene expression (reviewed in Berens and Hillen, 2004).
Another popular method for temporal regulation in mice uses CreER, a fusion between Cre and a modified estrogen-binding domain of the estrogen receptor, to control site-directed recombination (Figure 3B). This fusion protein is normally retained in the cytoplasm, but translocates into the nucleus to activate recombination upon the administration of tamoxifen, an estrogen analog (Feil et al., 1996). Conceptually similar modifications have been made to the transcription factor Gal4 in both mice and flies, rendering its activity controllable by drugs (Wang et al., 1994; Osterwalder et al., 2001; Roman and Davis, 2001).
A temporal regulation method used in flies involves expressing a temperature-sensitive mutation of Gal80, Gal80ts, to inhibit Gal4 expression by growing flies at permissive temperature. At the desired time flies are shifted to a restrictive temperature to inactivate Gal80ts, allowing Gal4-induced transgene expression (McGuire et al., 2003). Likewise, a heat-shock-promoter-driven Flp recombinase (hsFlp) can be used to induce gene expression after site-directed recombination (Figure 3B). In general, systems based on transcription factors (Gal4, tTA) are reversible, whereas systems based on recombination (CreER, hsFlp) are not.
2g. Refining Transgene Expression by Lineage and Birth Timing
In the absence of an absolute definition of cell types (Section 2a), and considering the paucity of specific promoters that target each defined cell type in most multicellular organisms, it is often useful to divide up existing broad expression patterns into smaller components. Cell lineage and birth timing have been used for this purpose.
For example, in the Drosophila olfactory system, each 2nd order olfactory PN projects dendrites to one of 50 glomeruli and relays a specific set of odorant information to higher brain centers. Thus, one can operationally define PNs that project dendrites to a specific glomerulus as a specific cell type. Endogenous genes or enhancer/promoter elements that are specific to individual PN types are yet to be discovered. However, the Mosaic Analysis with Repressible Cell Marker (MARCM) method, which combines Flp/FRT and Gal4/Gal80 (discussed above) to couple transgene expression with mitotic recombination (Lee and Luo, 1999), allows the separation of a PN-enhancer trap line into three lineages such that one can independently control gene expression in these three subsets. Further, by inducing mitotic recombination at defined times during development, it is possible to access single PNs reproducibly because of a close relationship between birth order and the dendrite target (Jefferis et al., 2001) (Figures 3H and 3I).
This approach requires an understanding of the biology of the neurons being investigated; in the case of Drosophila PNs, the cell types are specified by lineage and birth order. Similar useful relationships are being discovered in the mammalian CNS. For example, recent studies have revealed unexpected relationships between lineage and axonal projection patterns of cerebellar granule cells in the mouse (Zong et al., 2005), and birth order and physiological subtypes of cortical interneurons (Miyoshi et al., 2007).
2h. Targeting Transgene Expression with Viral Vectors
Viral vectors deliver genetic material and can thus employ many of the transgenic strategies described above. They can also be combined with transgenic animals by delivering transgenes such as Cre or tTA. Importantly, viral vectors allow genetic methods to be applied to species such as primates, where the production of transgenic lines is not practical. Finally, viral vectors can be used to target specific cell types based on injection sites and natural or engineered viral tropism.
Several recombinant vectors allow long-term gene expression without significant toxicity, including HIV-derived lentivirus, adeno-associated virus (AAV), and HSV amplicon vectors, among others (Table 1) (reviewed in Kootstra and Verma, 2003; Verma and Weitzman, 2005). These vectors have been refined over several years in order to overcome limitations of the parent viruses from which they were derived. They each naturally possess the ability to transduce nondividing cells—a crucial property for use in the nervous system. All of these vectors can be produced using helper virus-free systems, which insure that they act as delivery vehicles for genetic material without the potential to replicate or express genes that induce cytotoxic effects (Figure 4A).
Table 1.
Properties of Recombinant Viral Vectors Useful for Gene Delivery in the Adult Nervous System
| Adeno-Associated Virus (AAV) | Lentivirus | Herpes Simplex Virus (HSV) Amplicon | |
|---|---|---|---|
| Genetic material | single-stranded DNA | RNA | double-stranded DNA |
| Capacity for genetic material | ~5 kilobases | ~8 kilobases | ~150 kilobases |
| Speed of expression | weeks | weeks | days |
| Duration of expression | years | years | weeks to months, but elements can be added for persistent expression |
| Enveloped? | no | yes | yes |
| Natural tropism | many different serotypes are available, some with broad tropism, some very specific | usually pseudotyped with VSVg for broad tropism (natural tropism of HIV is for immune cells) | broad tropism for neurons |
| Engineered tropism | can alter capsid protein or pseudotype with existing capsid serotypes | can pseudotype with envelope protein from other naturally occuring viruses | can alter or delete existing envelope proteins or add envelope proteins from other viruses |
| Retrograde infection | yes, but variable | yes, but variable | yes |
The above table compares properties of three of the most commonly used viral vectors for studies of the adult nervous system. All of these vectors are able to transduce nondividing neurons and are generated using helper virus-free systems. For further details see text and reviews by Kootstra and Verma (2003) and Verma and Weitzman (2005).
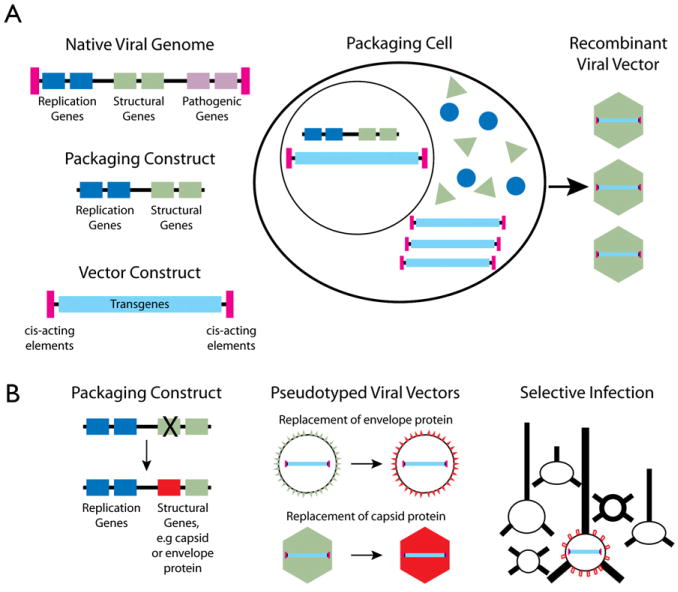
Figure 4. Virus-Mediated Gene Expression.
(A) Generation of helper virus-free viral vectors. The native viral genome includes coding sequences for genes that are pathogenic (pink) as well as genes required to produce viral proteins, including those which contribute to replication of genetic material (dark blue) and to the structure of the viral particle (green). These genes are flanked by cis-acting elements (magenta) that provide the origin of replication and signal for encapsidation. The packaging construct includes only genes required for replication and structural genes. The vector includes only the cis elements (magenta), which are required to incorporate it into the vector particles, plus the transgene cassette (light blue) that contains transcriptional regulatory elements (e.g., promoters) and coding sequences for transgenes. To produce recombinant vectors, packaging cells are transfected with the packaging construct and the vector construct. Replicated vector genomes are incorporated into virus particles, resulting in the generation of recombinant viral vector. After Verma and Weitzman (2005).
(B) Pseudotyping of viral vectors allows modification of viral tropism. Viral vectors are pseduotyped by modifying the packaging construct and replacing structural genes from the native virus (green) with structural genes from some other virus (red). For pseudotyping nonenveloped viruses, such as AAV, the relevant structural protein is capsid. For enveloped viruses such as lentivirus and HSV, the relevant structural proteins are envelope proteins. Selective infection can be achieved by pseudotyping with an envelope or capsid that interacts with cell surface receptors that are present only on a specific subset of cells.
Different vectors can preferentially transduce particular neuron types or glia in a species-specific manner. Although a growing number of publications report the ability of particular vectors to transduce cells in particular brain regions and species, few studies have investigated the cell types involved (but see Wu et al., 2006). Cell-type-specific differences in infectivity have the potential for targeting gene expression. These differences can be advantageous if they happen to limit expression to a particular cell type of interest, but are a limitation if the vector fails to infect a desired population.
Similar to the case of transgenesis, gene expression can be restricted to a subset of cells within the transduced population by incorporating cis-regulatory elements. For AAV and lentivirus, the small capacity of the vectors (~5 kilobases and ~8 kilobases, respectively) presents a major challenge, and there has been little success in generating such vectors which restrict expression to specific cell types by virtue of cis-regulation. HSV amplicon vectors can potentially incorporate cis-regulation more effectively because they have a capacity of more than 150 kilobases, sufficiently large to incorporate BACs (Wade-Martins et al., 2001).
The natural variation in viral tropism for particular cell types can be harnessed by designing vectors with particular tropism by pseudotyping (Figure 4B). Lentiviruses can take advantage of the tropism of virtually any enveloped virus, while AAV can incorporate capsid protein from other AAV serotypes. Beyond naturally occurring tropism, vectors can be engineered for uptake by cells expressing particular cell surface receptors. For example, an integrin binding site was inserted into the AAV2 capsid to allow uptake by cell types not normally transduced by AAV2 (Shi et al., 2006). Another strategy could potentially have even more far-reaching utility: AAV capsids can incorporate a sequence coding for the immunoglobulin-binding Z34C fragment of protein A, which mediates binding to antibodies. Mixing these vectors with antibodies against particular cell surface receptors allows the antibody to act as a bridge to facilitate transduction of cell types expressing those receptors (Gigout et al., 2005). This strategy could be used to target cell types which express a receptor for any genetically encodable ligand, such as neurotrophin or neuropeptide receptors. A conceptually similar strategy can be used in enveloped viruses, such as lentivirus (Snitkovsky and Young, 1998; Snitkovsky et al., 2001). Although these bridging strategies have worked in vitro, their utility in the brain is unknown.
Another useful strategy uses viral tropism to target cell types based on their axonal projections. For example, different types of cortical pyramidal neurons project axons to distinct distant targets. Viruses that can efficiently infect neurons through their axon terminals can therefore be injected into a particular target structure, resulting in the selective infection of neurons that have axons in that structure. This method has been successfully employed using HSV amplicon vectors, recombinant rabies virus, and adenovirus, as well as lentivirus pseudotyped with the rabies virus envelope protein (Mazarakis et al., 2001; Sandler et al., 2002; Tomioka and Rockland, 2006; Wickersham et al., 2007a).
2i. Concluding Remarks
The transgenesis and viral transduction methods described above allow genetic targeting of cell types in most organisms. These methods are critical to dissect the roles of specific cell types in neural circuits, including mapping connectivity (Section 3), measuring activity (Section 4), and inactivating and activating specific neurons (Section 5). In choosing methods for targeted gene expression, or initiating genetic approaches in a new organism, one must consider the complex trade-offs between genetic precision, technical ease, and the availability of existing resources. As these gene targeting methods are further refined, instructed by feedback from applications, they will yield increasing precision and ease for targeting gene expression.
3. Genetic Neuroanatomy
One of the great challenges in deciphering the brain’s wiring diagram is bridging the gap from the anatomy of single cells to their synaptic connections. The patterns of axonal and dendritic arborizations of single cells reveal their potential to be connected to one another: the axonal and dendritic arbors of two different types of cells must overlap for synapses to occur. However, additional experiments using transsynaptic markers, ultrastructural microscopy, or electrophysiology are required to determine whether the potential for synapses is in fact realized and what the strength and properties of the synapses are. Ultimately, all these approaches are likely to be complementary and each will benefit from genetic targeting.
In the four subsections below, we discuss (1) methods for observing the anatomy of single cells or populations of a single type; (2) methods for EM reconstruction of neural circuits; (3) trans-neuronal tracers; and (4) physiological assays to reveal functional connectivity.
3a. Early Neuroanatomy and Today’s ‘Second’ Renaissance
The first studies linking neural circuits to cell types originated more than one hundred years ago with the Golgi method (Golgi, 1873; Cajal, 1911). Until the early 1970s, Golgi staining and tract tracing with degeneration methods were essentially the only tools available. Cowan (1998) wrote, “Indeed, virtually all we knew of the organization of dendritic arbors and of ‘local circuit neurons’ until the late 1960s and 1970s had come from the analysis of Golgi-impregnated material.” This classical era was followed by the first “renaissance in morphological studies of the nervous system, due in large part to the introduction of a variety of new neuroanatomical methods” (reviewed in Cowan, 1998). These new methods included the development of anterograde and retrograde tracers, and intracellular labeling. During the last several decades it has been the creative exploitation of various combinations of these tools, often in concert with EM, that has provided the clearest links among cell types, connectivity, and function.
Although powerful, these methods require painstaking effort and are often limited by the need to perform several independent experiments to link different types of information. For example, particularly influential studies used intracellular recordings and dye filling to directly correlate cell function with morphology (Gilbert and Wiesel, 1979; Martin and Whitteridge, 1984; Anderson et al., 1993). But tying the measured functional properties to circuitry requires accumulation of data across separate animals. The link from function to circuitry rests on the reliability with which cell types can be identified, based solely on their morphological features. Bridging different levels of analysis can be improved by methods that allow reproducible access to the same cell type across experimental paradigms.
Genetic methods have already started what is likely to be a second renaissance for neuroanatomy. Relative to more traditional methods, they provide ease of reproducibility: morphological and functional analyses are all based on relatively uniform, genetically identified cell populations. Genetic methods also allow access to rare cell types. Perhaps as important, they can provide simpler assays that can be performed quickly and systematically, without specialized skills and equipment.
The first step in “genetic neuroanatomy” is to take advantage of the methods described in Section 2 to express markers in genetically defined neuronal types. Usually small cytosolic fluorescence proteins, such as GFP, serve as good markers to label the entire dendritic trees and axonal projections of neurons (e.g., Feng et al., 2000; Zong et al., 2005). In some cases GFP fused with membrane tags (Lee and Luo, 1999; De Paola et al., 2003), or with microtubule binding proteins (Giniger et al., 1993; Callahan and Thomas, 1994; Mombaerts et al., 1996), can help to label thin processes and long-distance axons. By using promoter elements or intersectional methods that allow transgene expression in desired cell types (Figure 2 and Figure 3), one can systematically trace bulk projections of genetically defined neurons (e.g., Gong et al., 2003) as an initial step in genetic neuroanatomy. Expression of tagged synaptic proteins can additionally mark synapses on these neurons (Jorgensen et al., 1995; De Paola et al., 2003; Jefferis et al., 2007). This is especially useful for invertebrate CNS neurons because it is often difficult to determine dendritic or axonal compartments based purely on morphology.
Most often, promoter elements or even intersectional methods label too many neurons at the same time to allow visualization of individual neurons. The repressor trap method (Figure 2F), best exemplified by the thy-1-XFP mice (Feng et al., 2000), provides a striking example of “genetic Golgi” in live cells (see Section 2c). The repressor trap method is random in that it relies on chance integration of transgenes into chromosomal loci that happen to repress the expression of markers in all but a small fraction of neurons. It is therefore difficult to predict the probability of targeting cell types at a desirable frequency. Similar limitations apply to viral methods (Dittgen et al., 2004).
A more systematic strategy of labeling a small fraction of neurons of a defined cell type relies on cell-type-specific expression of a transgene that is further activatable by recombinase-based excision. For instance, one can use either or both promoters to define what cell type expresses a cell marker (T) (Figure 3B). An inducible recombination system (for instance CreER in mice or heat-shock-inducible Flp in flies) allows control of the frequency of Cre/loxP or Flp/FRT induced recombination. With the limit of low recombination rates, only a small fraction of neurons will express the cell marker. This strategy has been successfully used both in flies (termed Flp-out) (Struhl and Basler, 1993; Ito et al., 1997; Wong et al., 2002) and in mice (Badea et al., 2003; Buffelli et al., 2003). Local injection of Cre-expressing virus into transgenic reporter mice, or reporter virus into Cre-expressing mice, could also be used to refine transgene expression temporally and spatially.
Another approach to sparsely labeling genetically defined population of neurons is based on site-specific interchromosomal recombination, as exemplified by the MARCM method in flies (Lee and Luo, 1999) and Mosaic Analysis with Double Markers (MADM) method in mice (Zong et al., 2005). Here again the sparseness can be controlled by the amount and duration of Flp or Cre expression. The cell type labeled is typically controlled by the promoters that drive the recombinase (both methods) or Gal4 (MARCM).
These methods of genetic neuroanatomy have been extensively used to study the Drosophila olfactory circuit. For example, using MARCM (Marin et al., 2002) or Flp-out (Wong et al., 2002), a large collection of individually labeled 2nd order olfactory PNs were sorted into classes based on their dendritic innervation of 1 of ~50 glomeruli in the fly antennal lobe. Systematic analysis of PN classes revealed striking stereotypy of axonal terminal arborization patterns (Figures 3H and 3I), implying the existence of a hard-wired spatial map in high olfactory centers (Marin et al., 2002; Wong et al., 2002). MARCM has been combined with high-resolution image registration methods to warp individually reconstructed neurons onto a common reference brain, allowing quantitative estimates of synaptic density maps (Jefferis et al., 2007) and potential connectivity between 2nd and 3rd order olfactory neurons (Jefferis et al., 2007; Lin et al., 2007).
Reconstructions of single neurons can typically only be achieved in sparsely labeled specimens. This limitation has recently been overcome in the Brainbow mice by expression of distinct mixtures of XFPs in individual neurons (Livet et al., 2007). Brainbow mice contain transgenes in which Cre/loxP recombination creates stochastic expression of multiple XFPs with varying concentrations. Neighboring neurons can be distinguished based on their color. For example, in these mice it is possible to reconstruct hundreds of nearby axons in the cerebellum.
3b. Electron Microscopy and Other Super-Resolution Methods
The structures of dendritic and axonal arbors, derived from optical microscopy, have provided the data for coarse estimates of wiring diagrams (Braitenberg and Schutz, 1991; Binzegger et al., 2004; Shepherd et al., 2005; Stepanyants and Chklovskii, 2005; Jefferis et al., 2007; Lin et al., 2007). These wiring diagrams are more or less explicitly based on Peter’s rule: where dendrites and axons overlap they will form synapses, roughly in proportion to the extent of the overlap. However, it is well established that neural circuits display exquisite specificity in their synaptic connections, beyond the shapes of dendrites and axons. Axons often target a particular cell type in a target region, while excluding other cell types (reviewed in Callaway, 2002). Even the connectivity between particular cell types is highly nonrandom (Shepherd and Svoboda, 2005; Song et al., 2005; Yoshimura et al., 2005). Making the leap from circuits based on overlaps of dendritic and axonal arbors to circuits based on synaptic connectivity is a major challenge.
The ultimate wiring diagram would consist of a connection matrix describing the synapses made between each neuron in individual animals. Currently only serial-section electron microscopy (EM) has sufficient contrast and resolution to trace the thinnest neuronal structures, including axons and spine necks (both can have diameters as small as 50 nm), and detect synapses. This technique relies on imaging thin (~50 nm) tissue sections, tracing membranes in single images, and reconstructing neuronal structures across multiple sections. Serial-section EM has been used to reconstruct the entire wiring diagram of C. elegans (White et al., 1986; Chen et al., 2006) (Figure 1A). The huge impact of this tour de force underscores the importance of complete wiring diagrams.
However, EM techniques do not scale easily to larger nervous systems. Applying current serial-section EM techniques to reconstructing one cubic millimeter of nervous tissue would require more than 10,000 person-years of effort. Automation of data acquisition and analysis will thus be required to reconstruct large tissue volumes. Although the data acquisition and data storage bottlenecks are being overcome (Denk and Horstman, 2004; Briggman and Denk, 2006) (http://www.mcb.harvard.edu/lichtman/ATLUM/ATLUM_web.htm), automated reconstructions of neuronal structure from EM data on the scale relevant to neural circuits (>106 μm3) remain to be demonstrated.
Serial-section EM poses especially difficult challenges for tracing and reconstructing neural circuits. Since tortuous and thin axons are densely packed in tissue, the slightest tissue imperfections in the sample and misalignment of successive sections could lead to errors in linking axons across successive sections. Because axons have to be traced across many thousands of sections, even small error rates in tracing axons across sections would severely degrade EM-based wiring diagrams.
Labeling individual axons with distinguishable markers could provide the necessary information for error correction. Such multiplexing is difficult to implement for EM, but could be achieved using fluorescence techniques. For example, in Brainbow mice up to 100 distinct axons can be distinguished by their color, based on differential expression of fluorescent proteins (Livet et al., 2007). Axonal profiles could thus be joined across sections based on their color. Furthermore, recently several fluorescence-based microscopy techniques have been described with resolution in the ~10 nm range (Betzig et al., 2006; Rust et al., 2006; Hell, 2007), which is sufficient for neural circuit reconstruction. A clever combination of EM and optical microscopy techniques (e.g., Micheva and Smith, 2007) may ultimately be required to reconstruct wiring diagrams at the synaptic level.
In the absence of large-volume reconstructions, EM could still have a large impact on wiring diagrams. For example, EM has been used to count synapses connecting pairs of recorded neurons. Similarly, EM could be used to verify synapses in putative connections between genetically defined neuronal populations. Synapses made specifically by these neuronal populations could be detected based on GFP (Trachtenberg et al., 2002) or HRP (Watts et al., 2004) expression.
3c. Genetic Methods for Transneuronal Labeling from Specific Cell Types
Transneuronal tracers, including chemicals, proteins, and neurotropic viruses, have been used extensively for studies of neural circuits (Schwab et al., 1979; Fujisawa and Jacobson, 1980; Itaya and van Hoesen, 1982; Kelly and Strick, 2000; Enquist, 2002). Typically they are injected into a particular brain region and then allowed to spread either anterogradely or retrogradely through the circuit. This approach can reveal multisynaptic links across what might otherwise appear to be relatively independent or distant structures (e.g., Hoshi et al., 2005). The timing of the spread can provide information about the numbers of synaptic steps between an injection site and a labeled structure. Here we focus our review on genetically encoded transneuronal methods, including neurotropic viruses, because they can reveal the connectivity between specific cell types.
Before describing these methods in detail it is important to comment on the concept of “synaptic specificity.” We reserve the term “transsynaptic” for methods in which it has been demonstrated that spread occurs only between neurons that have actual synaptic connections. By this definition the only current transsynaptic tracer is rabies virus (Ugolini, 1995a). We use “transneuronal” to describe methods for which synaptic specificity of spread has not been demonstrated. In most cases this simply reflects the fact that definitive experiments have not been conducted. In other cases it has been demonstrated that nonsynaptic spread can occur.
Genetically expressed transneuronal tracers include wheat germ agglutinin (WGA, a mostly anterograde tracer) and tetanus toxin C fragment (TTC, a retrograde tracer). These can be used to trace either the inputs to (TTC) or outputs from (WGA) populations of the neuron type that expresses the transgene. Targeting to a cell type can be achieved using any of the genetic methods described in Section 2 (Braz et al., 2002; Kinoshita et al., 2002; Sano et al., 2007). Even with temporal and cell-type-specific control of expression, these transneuronal tracers have important limitations.
First, low sensitivity tends to prevent detection when tracer is spreading from a small population of neurons or through small numbers of synaptic contacts. For example, it is not possible to detect WGA or TTC in connected cells when it is expressed in only a single neuron (E.M.C., unpublished data). Successful applications of WGA appear to be limited to cases where large populations of neurons express the tracer and make convergent inputs onto postsynaptic cells (Yoshihara et al., 1999). Similarly, TTC seems to label presynaptic neurons in cases where these neurons make divergent inputs onto a large number of TTC-expressing neurons (Maskos et al., 2002). A lack of labeling therefore cannot be interpreted as a lack of connectivity. Since only a small fraction of tracer is transferred across the synapse, the morphology and projection pattern of transneuronally labeled cells cannot be discerned. Efforts to amplify the signal by fusing tracers with transcription factors or recombinases need to solve the problem of allowing the transferred proteins to escape the endocytic compartment and have nuclear access.
Second, variability in the rate of spread complicates the interpretation of transneuronal labeling. Since both the extent and rate of spread appear to be dependent on the strength or number of synaptic contacts (or both), these tracers are likely to spread more quickly through strongly connected pathways. Thus, as additional circuit elements appear over time, it is not possible to distinguish weak direct connections from strong indirect connections.
Neurotropic viruses have the potential to overcome the limitations of transneuronal tracers. Because these viruses spread through the nervous system as part of their natural life cycle, they have evolved useful traits that can be further improved or selected by genetic engineering. Commonly used tracer viruses include alpha-herpes viruses and rabies virus. The alpha-herpes viruses include several different strains of herpes simplex virus (HSV) and pseudorabies virus (PRV, no relation to rabies virus). Most naturally occurring strains of PRV and HSV spread in both the anterograde and retrograde directions. But the most useful strains for tracing are variants that spread exclusively in the anterograde (Garner and LaVail, 1999) or retrograde (Ugolini, 1995a; Enquist, 2002) direction. The utility of a viral tracer is influenced by its cytotoxicity. Strains with reduced toxicity allow detection of spread to presynaptic neurons before the parent cells or the entire animal is killed (Enquist, 2002). Cytotoxicity may also be related to the degree to which transneuronal spread is synapse specific (see below).
Transneuronal spread can be controlled genetically to allow tracing of connections to a specific cell type. An important early example involved an attenuated strain of PRV in which the coding sequence for thymidine kinase (TK), which is necessary for replication, was replaced with a floxed stop followed by a GFP-IRES-TK cassette (DeFalco et al., 2001). This virus is unable to replicate and spread from infected neurons unless they express Cre. In contrast, Cre-expressing cells turn green and the recombined virus spreads to neurons presynaptic to the parent cells, which also turn green. The recombined virus continues to spread retrogradely across multiple synapses. Thus, as detailed above, it can be difficult to distinguish between weak direct connections and strong indirect connections.
Recent methods based on genetically modified rabies virus have demonstrated monosynaptic transsynaptic labeling (Wickersham et al., 2007b). This rabies variant had the coding sequence for the envelope protein (rabies glycoprotein [RG]) replaced with EGFP (Etessami et al., 2000). RG is required for rabies virus assembly and spread to presynaptic neurons. Thus, this virus allows in situ trans-complementation. Following infection of specific cells that also expresses RG in trans, RG is incorporated into nascent rabies particles, allowing them to spread to presynaptic cells where they both express EGFP and replicate to allow amplification. But because the presynaptic neurons do not express RG, the virus is unable to spread beyond this single synaptic step. By pseudotyping recombinant rabies with EnvA from an avian virus that cannot normally infect mammalian cells, it was possible to engineer specific cells to be susceptible to infection, by expressing the EnvA receptor, TVA, in the target cells. Using this method it is possible to label neurons that are presynaptic to a single parent cell (Wickersham et al., 2007b). It should be possible to use this approach in combination with the genetic targeting strategies described in Section 2. All that is necessary is to drive expression of RG and TVA in a specific cell type or in a single neuron and then infect those cells with the EnvA pseudotyped virus.
As noted above, a crucial consideration in designing and interpreting any experiment using transneuronal tracers is whether the spread of virus (or tracer) is limited to neurons that are synaptically coupled. For alpha-herpes viruses, including PRV, there is clear evidence that this is not always the case (Ugolini, 1995b). Spread of rabies virus may be restricted to synaptically connected neurons (Ugolini, 1995a), perhaps because of specific interactions between rabies viral components and synaptic specializations (Lafon, 2005). Another possible reason for these differences is that HSV (and PRV) can cause infected cells to lyse, potentially distributing viral particles indiscriminantly to both connected and unconnected neurons with nearby processes. In contrast, even wild-type rabies infection does not result in lysis of infected neurons. At any rate, future studies using viral vectors to label neurons from a single starting cell should allow quantitative assessment of the rate of false positives by using paired intracellular recordings to test for synaptic connections from labeled cells (e.g., Wickersham et al., 2007a).
A recently developed, light-level anatomical method for identifying synaptic connectivity does not fall neatly into any of the categories above. Feinberg et al. (2007) devised a system in which GFP is split into two parts, neither of which can fluoresce independently from the other. But when the two parts are fused to synaptic transmembrane proteins and then expressed in connected neurons, they can come into close apposition at the sites of synaptic contact. The combined proteins are then fluorescent, indicating not only that the neurons are connected, but also the location of the synaptic contacts. Like EM, this method can uniquely identify the locations of synaptic contacts, but it has the additional advantage of potentially identifying the neurons involved in the formation of those connections. This method is likely to prove very powerful on its own and in combination with other genetic and viral methods.
3d. Physiological Methods for Mapping Circuits
The perfect wiring diagram consists of a connection matrix describing the number of synapses made between any pair of neurons. However, in addition to its wiring, a circuit diagram will have to incorporate information about the integrative properties of particular cell types and the signs and strengths of the synapses that connect them. For example, consider the excitatory synapses impinging onto layer 4 spiny stellate neurons in the visual cortex. Although thalamocortical synapses make up only ~10% of the total, these inputs play a disproportionate role in driving their targets (Douglas and Martin, 2004). One of the mechanisms underlying this apparent discrepancy is that thalamocortical synapses are stronger and more reliable than intra-cortical synapses (Stratford et al., 1996).
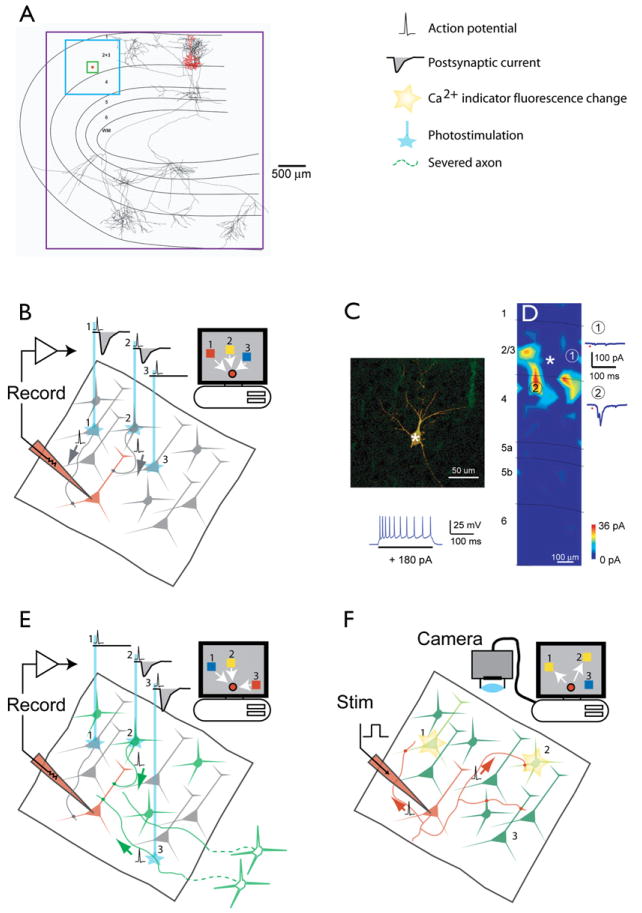
The most direct way to measure the strengths and properties of synapses is to stimulate one neuron while recording intracellularly from another neuron that potentially receives input from the stimulated cells. Neurons are typically then filled with dye and their anatomy studied to identify the type of cell that was stimulated and recorded. Multiple intracellular recordings have been used to estimate the connection probability and the synaptic strength between connected neurons in brain slices, with the goal of constructing local circuit diagrams (Gupta et al., 2000; Thomson and Bannister, 2003). These approaches have also been used to probe the dynamics of local circuit motifs in the neocortex (Galarreta and Hestrin, 1999; Gibson et al., 1999) and the hippocampus (Pouille and Scanziani, 2004). The main drawbacks with multiple dual intracellular recordings are that they are slow and inefficient, and they must be conducted in brain slices. Thus, they are limited to highly local circuits made by neurons with high connection probabilities (Holmgren et al., 2003) (Figure 5A).
Figure 5. Methods for Functional Circuit Mapping in Brain Slices.
(A) The spatial ranges of circuit mapping techniques. The boxes indicate the lengthscales accessible to different methods. Red box, 20 μm, 3D electron microscopy reconstructions; green box, 200 μm, paired recordings; blue box, 1000 μm, laser scanning photostimulation with glutamate uncaging and optical probing; purple box, potentially the entire brain, axon tracing and ChR2-assisted circuit mapping. The reconstruction is a layer 2/3 pyramidal neuron superposed on a schematic of the cat visual cortex (J. Hirsh, USC). Dendrites are in red; axons, in black.
(B) Glutamate uncaging mapping. The schematic shows a brain slice in which synaptic responses are recorded in a single neuron (red). Neurons are excited by photolysis of caged glutamate, typically by using a UV laser that is scanned over the brain slice (blue line). If glutamate is photoreleased near the soma (but not on distal dendrites or axons), it evokes action potentials. Postsynaptic whole-cell currents (or potentials) recorded in the recorded neuron are used to generate a map in a computer. This so-called “synaptic input map” is a quantitative representation of the spatial distribution of synaptic input to the recorded neuron.
(C and D) The use of glutamate uncaging mapping to measure the spatial distribution of excitatory inputs impinging onto genetically defined GABAergic interneurons (X. Xu and E.C., unpublished data). (C) Morphology of the recorded neuron in neocortical layer 2/3. GFP fluorescence is overlaid with Cy3 streptavidin labeling intracellularly injected biocytin (top). (Bottom) Firing pattern of the recorded neuron. (D) Synaptic input map showing hotspots of input from layer 4 and layer 2/3. Traces to the right are examples of excitatory postsynaptic currents evoked following stimulation at sites 1 and 2.
(E) ChR2-assisted circuit mapping. A specific subpopulation of neurons is targeted for expression of ChR2 (green). ChR2-positive neurons (2) and axons (3) are excited by a blue laser that is scanned over the brain slice (blue lines), whereas ChR2-negative neurons are not perturbed (1). Postsynaptic whole-cell currents (or potentials) are used to generate a map in a computer. ChR2-assisted circuit mapping has genetic specificity because ChR2 expression is necessary for exciting action potentials. Furthermore, since severed axons can be excited (3), connectivity between distal brain regions can be studied even in a brain slice.
(F) Optical probing. All neurons are bulk-loaded with Ca2+ indicator. One neuron is stimulated with brief bursts of action potentials. Postsynaptic neurons that fire action potentials can be detected using [Ca2+] imaging.
Another method for dissecting circuits in brain slices combines intracellular recordings from one postsynaptic neuron and photoactivation of groups of presynaptic neurons by glutamate uncaging (Callaway and Katz, 1993; Katz and Dalva, 1994) (Figure 5B). At each stimulation site in the slice, somata close to the laser spot are excited to fire action potentials. Importantly, axons of passage are not excited. The spatial resolution of stimulation is better than 100 μm, providing sublaminar and subcolumnar resolution (Dantzker and Callaway, 2000; Shepherd et al., 2003; Shepherd and Svoboda, 2005). The synaptic responses in the postsynaptic neuron are used as a measure of the strength of input arising from a particular location, corresponding to the position of the uncaging laser. Such glutamate uncaging mapping thus provides quantitative images of the spatial distribution of excitatory and inhibitory input impinging onto single recorded neurons (Dantzker and Callaway, 2000; Shepherd et al., 2003; Shepherd and Svoboda, 2005; Yoshimura et al., 2005). Glutamate uncaging mapping has been used to map intracortical (Dalva and Katz, 1994; Dantzker and Callaway, 2000; Shepherd et al., 2003; Shepherd and Svoboda, 2005; Yoshimura et al., 2005), intrathalamic (Deleuze and Huguenard, 2006), and thalamocortical (Bureau et al., 2006) circuits. Glutamate uncaging mapping can be combined with recording from genetically defined neurons in animals expressing XFPs in specific neuronal populations (Figures 5C–5F). Glutamate uncaging mapping has been applied to challenging preparations, such as monkey brain slices (Sawatari and Callaway, 2000), reflecting the efficiency of the technique.
Glutamate uncaging mapping is quantitative and efficient, but it suffers from two major drawbacks. First, only connections that are preserved in brain slices can be probed. Second, most cell types in the mammalian brain express glutamate receptors and are therefore excited by glutamate uncaging, making it possible to identify the locations, but not necessarily the types, of presynaptic neurons.
Both drawbacks can be overcome by replacing uncaging of glutamate with photoactivation of genetically encoded photosensitivity. In particular, expression of a 300 amino acid fragment of channelrhodopsin-2 (ChR2) is sufficient to produce rapid light-activated cationic photocurrents in heterologous cells (Nagel et al., 2003). The kinetics of the currents is similar to the fastest excitatory postsynaptic currents. Furthermore, neurons expressing ChR2 can be entrained to fire complex action potential trains (Boyden et al., 2005; Li et al., 2005) (see Section 5). In ChR2-assisted circuit mapping, photostimulation is combined with whole-cell recording. Circuits are mapped between presynaptic neurons, defined by ChR2 expression, and postsynaptic neurons, defined by targeted patching (Figure 5G). Remarkably, even ChR2-positive axons that are severed from their parent somata can be photostimulated to fire action potentials (Petreanu et al., 2007). This means that ChR2 can be used to map long-range projections. For example, ChR2 has been used to map callosal projections linking left and right somatosensory cortex (Petreanu et al., 2007). Similar approaches have been used to map circuits from the olfactory bulb to the olfactory cortex in vivo (Arenkiel et al., 2007). ChR2 thus allows the mapping of synaptic connectivity over all spatial scales in the brain.
Axonal photoexcitability degrades the spatial resolution of ChR2 mapping: when illuminating a particular spot in the brain slice, it may be difficult to distinguish excitation of a nearby soma and an axon of passage originating from a distant cell, perhaps from a different brain region. The excitability of ChR2-positive axons can therefore be a drawback for experiments that rely on estimating the location of stimulated neurons. Targeting of ChR2 to the soma and dendrite of genetically targeted cells will likely overcome this problem (Arnold, 2007).
Glutamate uncaging mapping and ChR2 mapping measure the inputs impinging onto a recorded neuron. It is also of interest to identify the postsynaptic targets of a given neuron. A promising approach involves stimulating one neuron with a recording electrode while measuring responses in multiple postsynaptic neurons using Ca2+ imaging (Kozloski et al., 2001) (optical probing, Figure 5H). However, since Ca2+ imaging reports spikes in postsynaptic neurons, this approach is biased to detect only the strongest connections in a circuit.
4. Genetic Neurophysiology
A central problem in neuroscience is deciphering how individual neurons encode information. This is traditionally addressed by electrophysiology. Extracellular recordings of single units have helped to reveal the basic principles of brain organization (Kuffler, 1953; Mountcastle, 1957; Hubel and Wiesel, 1959) and information processing (Adrian, 1932; Bialek et al., 1991; Meister et al.,1995). Single-unit techniques have also been used to analyze neuronal signals that couple sensory perception and action in awake, behaving animals (O’Keefe and Dostrovsky, 1971; Britten et al., 1992; Platt and Glimcher, 1999). Despite their great importance, extracellular recording methods have important drawbacks. Typically only one neuron is probed in a volume of neural tissue that contains thousands of other neurons. The cell type and location, and hence its relationship to the underlying neural circuit, are poorly defined. Since neurons are detected based on their responsiveness to particular stimuli, strongly responding neurons are usually selected for recording, causing strong biases in the sample under study.
In vivo intracellular recordings can avoid some of the shortcomings of single-unit methods (Gilbert and Wiesel, 1979; Somogyi et al., 1983; Wilson et al., 1983; Svoboda et al., 1997; Kamondi et al., 1998; Margrie et al., 2003; Brecht et al., 2004). Recordings in awake animals (Wilson and Groves, 1981; Brecht et al., 2004) and freely moving animals (Lee et al., 2006) are possible. It is even possible to introduce expression plasmids into individual electrophysiologically characterized neurons by electroporation (Kitamura et al., 2008). Since neurons are selected for intracellular recording based on their stable membrane potential, rather than strong spiking responses, the sample is less biased than single-unit methods. In addition, histological stains can be introduced through the recording pipette, allowing post hoc analysis of the structure and histochemistry of the recorded neuron. However, intracellular recordings are technically demanding, have relatively short durations, and typically only probe one neuron at a time. Optical and electrophysiological methods, in combination with genetic targeting, are beginning to overcome the drawbacks of classical electrophysiology.
4a. Electrophysiological Recordings from Genetically Defined Cell Types
Transgenic animals expressing XFPs in genetically defined neurons are becoming widely available. If the expression level is sufficiently high, XFP expression can be used for visually guided recordings. This method has already been used to record intracellularly from specific neuronal types in the fly antennal lobe (Schlief and Wilson, 2007) and the mouse neocortex in vitro (Oliva et al., 2000) and in vivo (Margrie et al., 2003). Recordings from genetically targeted cell types can be used to address questions that would be difficult for blind recordings. For example, one can record from genetically labeled 2nd order olfactory PNs in the fly antennal lobe that are postsynaptic to a specific class of olfactory receptor neurons with defective odorant receptors (Olsen et al., 2007; Root et al., 2007; Schlief and Wilson, 2007). These experiments are beginning to clarify how olfactory information is propagated from sensory to 2nd order neurons.
Another approach for recordings from genetically defined neuronal populations relies on expression of a protein sensitizer. For example, neurons expressing the light-activated channel ChR2 (Nagel et al., 2003) can fire action potentials with short latencies and small response jitter in response to illumination with blue light (Boyden et al., 2005; Li et al., 2005; Zhang and Oertner, 2006). The ChR2-positive neurons can thus be identified by virtue of their short-latency spikes in response to brief flashes of blue light (Arenkiel et al., 2007) or their light-evoked firing pattern (S. Lima and A. Zador, personal communication), even when using blind recording techniques. This approach should allow recording from specific neuronal populations using chronically implanted electrodes.
4b. Imaging Neuronal Activity
A major challenge of systems neurophysiology is recording the activity of multiple, perhaps all, neurons over time. Optical microscopy, together with fluorescent indicators of neuronal activity, is poised to have a major impact in this area. Direct imaging of membrane depolarization has been achieved with synthetic voltage-sensitive dyes (reviewed in Grinvald and Hildesheim, 2004). However, because of limited signal levels and nonspecific labeling of most membranes in the tissue, imaging with single-cell resolution has remained beyond reach except in thin preparations with large neurons (Baker et al., 2005; Briggman et al., 2005).
[Ca2+] imaging can be used as an alternative to voltage imaging to detect spiking activity. Action potentials open voltage-gated calcium channels. The resulting saw tooth-shaped [Ca2+] transients can be readily detected using [Ca2+] imaging methods (Helmchen et al., 1996; Svoboda et al., 1997). Under favorable conditions, using intracellular loading with indicator in brain slices, it is straightforward to detect individual spikes with firing rates at least up to 20 Hz (Helmchen et al., 1996).
To image populations of neurons, neural tissue is typically bulk-loaded with membrane permeable [Ca2+] indicators (Yuste et al., 1992; Stosiek et al., 2003) or dextran-conjugated indicators (O’Donovan et al., 1993; O’Malley et al., 1996; Nagayama et al., 2007). [Ca2+] imaging can track the dynamics of populations of individual neurons in vivo, in some instances with single-action-potential sensitivity (Kerr et al., 2005; Sato et al., 2007b). For example, [Ca2+] imaging has revealed the fine-scale organization of the developing zebrafish tectum (Niell and Smith, 2005), maps in the visual cortex (Ohki et al., 2005; Mrsic-Flogel et al., 2007) and somatosensory cortex (Sato et al., 2007b), and olfactory responses in the zebrafish (Yaksi and Friedrich, 2006) and rat olfactory bulb (Verhagen et al., 2007). However, measurements with bulk-loaded [Ca2+] indicators have inferior signal-to-noise ratios to those with pipette-loaded indicators; it is therefore challenging to relate fluorescence changes of [Ca2+] indicators to the number of spikes or spike timing (Svoboda and Yasuda, 2006; Yaksi and Friedrich, 2006; Sato et al., 2007b).
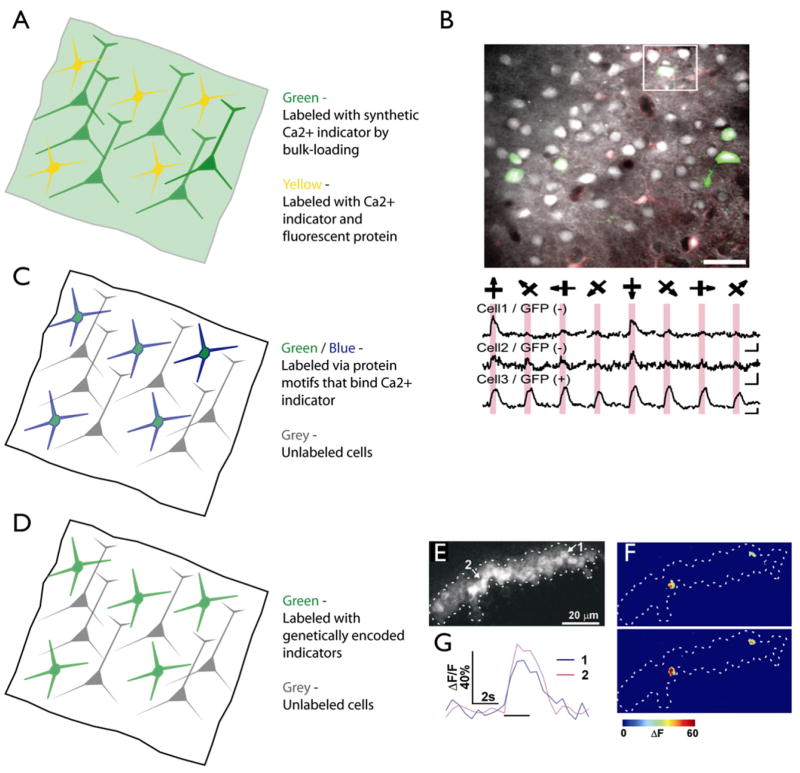
One major drawback of synthetic [Ca2+] indicators is that they do not distinguish between cell types in the labeled region. This problem is beginning to be overcome with three independent approaches, all making use of genetically identified cell types (Figure 6). First, similar to electrophysiological recordings, [Ca2+] imaging can be performed in animals expressing XFPs in a defined subset of neurons. XFP fluorescence can be separated from [Ca2+] indicator fluorescence using excitation or emission filters. One can then image the activity of XFP-positive and XFP-negative neurons in the same experiment (Figure 6A). This approach has been used to measure orientation selectivity of interneurons in the mouse visual cortex (Sohya et al., 2007) (Figure 6B), the activity of mouse spinal cord interneurons (Wilson et al., 2007), and odor responses in zebrafish mitral cells (Yaksi and Friedrich, 2006).
Figure 6. Strategies for Imaging Genetically Specified Neuronal Populations with [Ca2+] Indicators.
(A) All neurons are labeled nondiscriminately by bulk-loading with a [Ca2+] indicator (diffuse green). A genetically specified set of neurons express a fluorescent protein (yellow).
(B) [Ca2+] imaging in mice expressing GFP in GABAergic interneurons. (Top) Image showing neurons bulk-loaded with [Ca2+] indicator. GFP fluorescence is overlaid in green. (Bottom) Responses of GFP-negative and GFP-positive (GABAergic) neurons to oriented bars. Modifed from Sohya et al., 2007.
(C) A genetically specified subpopulation of neurons express a protein (such as tetracysteine motifs; blue) that makes them susceptible to labeling by modified versions of [Ca2+] indicators (such as biarsenicals; green).
(D) A genetically specified subpopulation of neurons express a genetically encoded [Ca2+] indicator (green).
(E–G) Imaging odor-evoked activity in Kenyon cells of the Drosophila mushroom body using genetically encoded [Ca2+] indicators (G-CaMP1.3) in vivo. (E) G-CaMP fluorescence showing the mushroom body. (F) Two responses to the same odor (difference image; 2 s after odor onset minus baseline). Two Kenyon cells show strong activity. (G) Time course of G-CaMP responses. Modified from Wang et al., 2004.
Second, an emerging class of methods relies on chemically modified synthetic [Ca2+] indicators that accumulate only in cells expressing certain structural motifs or enzymes. For example, FlAsH is a membrane-permeant biarsenical Ca2+ indicator (based on Calcium Green) (Tour et al., 2007) that binds to tetra-cysteine protein motifs (Griffin et al., 1998). Bulk-loaded FlAsH accumulates preferentially in neurons expressing tetracysteine moieties (Figure 6C). Moreover, FlAsH can be targeted to Ca2+ nanodomains close to the mouths of Ca2+ channels, where it presumably selectively detects Ca2+ entering the cell through these channels, rather than via other Ca2+ influx pathways (Tour et al., 2007). Applications to in vivo imaging will have to overcome toxicity and fluorescence background from unbound FlAsH.
A conceptually similar method relies on expression of beta-galactosidase, a nontoxic bacterial protein, and calcium indicators linked to a sugar moiety (S. Nirenberg, personal communication). The indicator is taken up by all cells nondiscriminately, but cleavage to produce the functional indicator occurs only in cells expressing beta-galactosidase.
Third, genetically encoded indicators of neuronal activity provide the most elegant solution to imaging genetically defined neuronal populations (Figure 6D). A variety of such protein-based probes have recently emerged as alternatives to synthetic indicators (reviewed in Miyawaki, 2005). Genetically encoded voltage indicators consist of fusions of fluorescent proteins and components of voltage-gated ion channels (Siegel and Isacoff, 1997; Sakai et al., 2001; Ataka and Pieribone, 2002) or voltage-dependent phosphatases (Dimitrov et al., 2007). Voltage-dependent conformational changes in the transmembrane voltage sensors are coupled either to changes in the brightness of individual XFPs (Siegel and Isacoff, 1997; Sakai et al., 2001; Ataka and Pieribone, 2002) or to changes in FRET between pairs or XFPs (Ataka and Pieribone, 2002; Dimitrov et al., 2007). Voltage indicators can be delivered to the plasma membrane of specific cell populations, potentially reducing the background due to labeling of other membranes. Although voltage indicator responses are currently much too small to be useful for routine measurements at the level of individual neurons, recent developments give reasons for optimism. For example, a hybrid sensor that combines a genetically encoded fluorescent probe (membrane-anchored GFP) with dipicrylamine, a synthetic voltage-sensing molecule that partitions into the plasma membrane, provides an unusually large voltage-dependent fluorescence signal and fast response times (Chanda et al., 2005).
Genetically encoded [Ca2+] indicators (GECIs) are based on fusions of fluorescent proteins and protein Ca2+ buffers, such as calmodulin and troponin, that undergo large conformational changes in response to Ca2+ binding. These Ca2+-dependent conformational changes are coupled either to changes in the brightness of individual XFPs (Baird et al., 1999; Nagai et al., 2001) or to changes in FRET between pairs of XFPs (Miyawaki et al., 1997; Heim and Griesbeck, 2004). GECIs have been used in combination with two-photon microscopy in Drosophila (Wang et al., 2003, 2004) (Figures 6E–6G) and mice (Hasan et al., 2004; Garaschuk et al., 2007). Current families of GECIs have relatively small dynamic ranges and slow response kinetics in vivo, likely limiting their applicability. For example, GECIs cannot be used to reliably count action potentials in cortical neurons (Mao et al., 2008). In addition, the nonlinear relationships between activity and Ca2+ concentration, and Ca2+ concentration and GECI fluorescence, can make the interpretation of GECI fluorescence signals challenging (Pologruto et al., 2004). However, the development of GECIs (Nagai et al., 2004; Palmer et al., 2006; Tallini et al., 2006; Garaschuk et al., 2007) and data analysis algorithms (Ramdya et al., 2006; Yaksi and Friedrich, 2006) are progressing rapidly, promising substantial advances over the next few years.
Other genetically encoded indicators couple primarily to synaptic activity (Takao et al., 2005; Yasuda et al., 2006). In particular, synapto-pHluorin, a pH-sensitive protein that reports synaptic vesicle fusion (Miesenbock et al., 1998), can be used to report the release of synaptic vesicles (Sankaranarayanan and Ryan, 2000). Synapto-pHluorin has been used to map the activity of olfactory neurons in the fly antennal lobe (Ng et al., 2002) and the mouse olfactory bulb (Bozza et al., 2004).
The confluence of advances in genetically encoded indicators, deep-tissue microscopy (reviewed in Flusberg et al., 2005; Helmchen and Denk, 2005; Svoboda and Yasuda, 2006), and genetic targeting techniques (Section 2) will allow the imaging of activity in genetically defined neuronal ensembles in behaving animals.
5. Genetic Manipulation
A major goal of neuroscience is to relate spike trains in specific neuronal populations to brain function and behavior. Recording neuronal activity (Section 4) is an important step, mainly to generate hypotheses about the meaning of particular patterns of activity. These hypotheses can only be tested by manipulating activity in defined neuronal populations. Classical LOF techniques, mainly surgical lesions and pharmacological manipulations, are invasive and lack specificity for particular cell types (but see Gray et al., 2001). It is also difficult to inactivate spatially distributed cell types. In the case of lesions, adaptive rewiring after the surgery complicates the interpretation of the functional effects. GOF experiments have mostly employed electrical microstimulation, leading to major discoveries about the neural basis of perception (Salzman et al., 1990; Romo et al., 1998). However, microstimulation excites excitatory and inhibitory neurons, as well as axons of passage. Therefore, the cell type and location responsible for the observed effects are not well known. Estimates of the number of stimulated cells are notoriously imprecise (Tehovnik et al., 2006).
The need to interfere with defined neuronal populations in intact neural circuits has long been recognized (Crick, 1988). This makes it necessary that such manipulation is genetically targeted and as precise as possible in space and time. Ideal systems for activation or inactivation share some common properties. They should be genetically encoded. They should be innocuous in the absence of inducer. Inactivation or activation should be rapidly inducible with small molecules, light, or other minimally invasive triggers. Induction should be rapidly reversible. Recently, a number of methods have been demonstrated that meet these requirements (summarized in Table 2 and Table 3). Below we highlight a subset of these methods, focusing on systems that are primed for LOF and GOF experiments with genetically targeted neurons.
Table 2.
Genetically Targeted Methods for Inducible Inactivation of Neuronal Function
| Method | Model Systems Tested | Inducible (Timescale) | Reversible (Timescale) | Target of Action | Inducer | Caveats | References |
|---|---|---|---|---|---|---|---|
| Shibirets | Dm | minutes | minutes | ST | temperature shift | not applicable to mammals and birds | Kitamoto, 2001 |
| MIST | Mm, MC, MBS | tens of minutes | hours | ST | small- molecule dimerizers | some dimerizer drugs may have poor BBB penetration; expression levels need to be high | Karpova et al., 2005 |
| Allatostatin receptor/ Allatostatin | R, F, M, Mm | minutes | minutes | Vm, Rm | peptide | Allatostatin does not penetrate the BBB and needs to be applied locally | Lechner et al., 2002; Gosgnach et al., 2006; Tan et al., 2006 |
| Halorhodopsin | MC, MBS, Ce | milliseconds | milliseconds | Vm | yellow light | High expression levels and light intensities required | Han and Boyden, 2007; Zhang et al., 2007 |
| Rhodopsin-4 | MC, Ch | milliseconds | tens of milliseconds | ST, Vm, Rm | blue light | multiple nonspecific effects | Li et al., 2005 |
| GABAA-R/ zolpidem | Mm, MBS | minutes | hours | Vm, Rm | small- molecule | requires gamma-2 knockin mice and receptors containing α1–α3 subunits of the GABAA receptor | Wulff et al., 2007 |
| 5-HT Receptor | Mm | hours | hours | Vm, Rm | small- molecule | limited to Htr1a knockout mice | Tsetsenis et al., 2007 |
| Inducible Expression of TeTxLc | Mm | days | weeks | ST | small- molecule | slow induction and reversal | Sweeney et al., 1995; Yamamoto et al., 2003 |
| Expression of K+ Channels | MC | days | weeks | Vm, Rm | small- molecule | slow induction and reversal; expression of K+ channels in vivo can have variable and paradoxical effects in the mammalian CNS | Johns et al., 1999 |
| GluCl/IVM | Mm, MC | tens of minutes | hours to days | Vm, Rm | small- molecule | the inducer drug is potentially toxic; slow reversibility | Slimko et al., 2002; Lerchner et al., 2007 |
| Expression of Tethered Toxins | Zf | NA | NA | Vm | NA | inducible expression has not been demonstrated | Ibanez-Tallon et al., 2004 |
Abbreviations: Ce, C. elegans; Dm, Drosophila melanogaster; Zf, zebrafish; Ch, chick; M, monkey; Mm, mouse; R, rat; F, ferret; MC, mammalian cultured neurons; MBS, mammalian brain slices; BBB, blood-brain barrier; Vm, membrane potential; Rm, membrane resistance; ST, synaptic transmission.
Table 3.
Genetically Targeted Methods for Inducible Neuronal Activation
| Method | Model Systems Tested | Inducible (Timescale) | Reversible (Timescale) | Inducer | Comments | References |
|---|---|---|---|---|---|---|
| ChR2 | Mm, MC, MBS, Dm, Ce, Ch | millseconds | milliseconds | blue light | high expression levels and light intensities required | Nagel et al., 2003; Boyden et al., 2005; Li et al., 2005 |
| chARGe | MC | seconds | seconds | light | multiple transgenes required | Zemelman et al., 2002 |
| ATP/Capsaicin Receptor | MC, Dm | milliseconds | seconds | UV light/caged ligand | activation by photorelease of caged ligands | Zemelman et al., 2003; Lima and Miesenbock, 2005 |
| RASSL | Mm | seconds | seconds | small-molecule | limited to situations where G-protein activation depolarizes neurons | Redfern et al., 1999; Zhao et al., 2003 |
| LiGluR | MC, Zf | milliseconds | milliseconds | Light/tethered small-molecule agonist | activation by cystein- conjugated tethered agonists | Szobota et al., 2007 |
| SPARK | MC | milliseconds | milliseconds | Light/tethered small-molecule agonist | inactivation by cystein- conjugated tethered agonists | Banghart et al., 2004 |
Abbreviations: Ce, C. elegans; Dm, Drosophila melanogaster; Zf, zebrafish; Ch, chick; Mm, mouse; MC, mammalian cultured neurons; MBS, mammalian brain slices.
5a. Methods for Inactivation
Reversible inactivation of genetically targeted neuronal populations has been achieved either by blocking synaptic transmission or by abolishing action potential generation. shibirets, a dominant temperature-sensitive mutation of Drosophila dynamin, is the prototypical inducible method for inactivation. In neurons expressing shibirets, inactivation is triggered by raising the temperature from room temperature to ~30°C. At elevated temperatures endocytosis of synaptic vesicles grinds to a halt, leading to the depletion of the synaptic vesicle pool and rundown of synaptic transmission. Induction and reversal occurs within a few minutes after the temperature shift (Koenig et al., 1983; Kitamoto, 2001). In Drosophila, shibirets has been used to dissect the circuits underlying memory formation, courtship behavior, and olfactory processing (Waddell et al., 2000; Dubnau et al., 2001; McGuire et al., 2001; Manoli et al., 2005; Stockinger et al., 2005). One concern with shibirets is that dynamin has many roles in membrane trafficking that extend beyond synaptic transmission. In addition, temperature shift as an inducer is not generalizable to higher vertebrates.
Many proteins in synaptic terminals are specific and essential for synaptic transmission (reviewed in Fernandez-Chacon and Sudhof, 1999). The intricately choreographed sequence of protein-protein interactions leading to vesicle fusion and vesicle recycling provides numerous potential protein targets for inducible inactivation. Inducible expression of tetanus toxin light chain (TeTxLc) has been used to inactivate synaptic transmission in flies (Sweeney et al., 1995) and mice (Yamamoto et al., 2003). But the time course of induction and reversal are slow (Table 2).
More recently, protein crosslinking induced by small-molecule dimerizers (Spencer et al., 1993) was used to develop Molecular Systems for Inactivation of Synaptic Transmission (MISTs) (Karpova et al., 2005). MISTs consist of modified synaptic proteins that can be crosslinked by the addition of small molecule dimerizers to block aspects of the synaptic vesicle cycle. In excitatory (Karpova et al., 2005) and inhibitory (D. Tervo, T. Sudhof, K.S., A. Karpova, unpublished data) MIST-positive neurons in vitro, application of dimerizer induces inactivation of synaptic transmission within tens of minutes. Reversal occurs over 1 hr. When targeted to Purkinje cells in transgenic mice, MISTs interfere inducibly and reversibly with performance in a cerebellum-dependent behavioral assay (Karpova et al., 2005).
MISTs have three drawbacks. First, it is not clear if dimerizers cross the blood-brain barrier (BBB), and in vivo applications so far have relied on intracerebroventricular (ICV) delivery. Second, in the wild-type background, MISTs compete with endogenous synaptic proteins; as a result, efficient inactivation of synaptic transmission requires high-level expression of the transgenes. Third, assessing the efficacy of MIST-dependent silencing in vivo can be challenging since the targets of the MIST-positive cells need to be known.
Other inducible systems silence neurons by manipulating the membrane potential or membrane conductance. The Drosophila allatostatin (AL) receptor (AlstR) has been expressed in mammalian neurons in vitro and in vivo. Application of the peptide AL opens G protein-coupled inward rectifier K+ channels, counteracting action potential generation. Inducible and rapidly reversible AL-dependent silencing has been demonstrated in vitro (Lechner et al., 2002) and in a variety of anesthetized animals, including the monkey (Tan et al., 2006). Induction and reversal is rapid, at ~10 min. The main drawback of the AlstR/AL system is that AL does not cross the BBB. As a consequence, its use in freely moving animals in vivo requires insertion of a catheter for ICV administration (Tan et al., 2008). Other methods relying on G protein-coupled receptors activated by small molecules could overcome this limitation (Armbruster et al., 2007).
Another strategy relying on changing the membrane conductance is based on expression of the ivermectin (IVM)-gated Cl− channel (GluCl). This system has the advantage of being induced by a compound that can cross the BBB. GluCl consists of α and β subunits which must be coexpressed to form a functional channel. Cultured neurons expressing GluCl can be silenced in an IVM-dependent manner (Slimko et al., 2002). IVM increases the Cl− conductance of the membrane, shunting action potential generation. GluCl has been engineered for reduced sensitivity to glutamate (Li et al., 2002). The GluCl/IVM system has been tested in amphetamine-dependent rotational behavior in mice. In mice expressing GluCl unilaterally in the striatum, systemic administration of IVM caused unidirectional rotation of the animal, indicating that striatal neurons were silenced (Lerchner et al., 2007).
The GluCl/IVM system has several potential problems as a silencing strategy. First, because IVM is a glutamate receptor agonist, the effective IVM concentrations that need to be administered are potentially toxic. Second, IVM-dependent silencing is only slowly reversible (over a period that takes approximately days), opening up the possibility of compensatory circuit plasticity. Third, two transgenes need to be expressed in the same neuronal population. The requirement for two transgenes can be advantageous if intersectional methods for targeting cell types are desirable (see Section 2).
Engineering of GABAA receptors (GABAA-Rs) has recently been used for cell-type-specific silencing (Wulff et al., 2007). The GABAA agonist zolpidem binds to the ubiquitous GABAA γ2 subunit to prolong the duration of inhibitory currents by allo-steric action. A single amino acid substitution, γ2 I77F, abolishes zolpidem binding while leaving GABA binding unperturbed. As a consequence, γ2 I77F knockin mice are insensitive to zolpidem. Zolpidem sensitivity can be reconstituted in genetically defined subsets of neurons by expression of wild-type γ2 protein. In animals expressing zolpidem-sensitivite GABAA-Rs in Purkinje cells, performance in the rotarod test is reduced within minutes after systemic administration of zolpidem. The virtues of this technique include the excellent pharmacokinetic properties of zolpidem. In addition, zolpidem is an FDA-approved sleeping aid (Ambien), alleviating concerns with toxicity.
The GABAA-R/zolpidem system has some limitations as a silencing strategy. First, targeting zolpidem-sensitive channels relies on replacement of the endogenous gene, which is practical only in mice. Second, zolpidem likely enhances inhibition without full silencing of the target neuron; this could result in ambiguous results. Third, zolpidem binds the interface of the γ2 and α1 subunits, with reduced affinity for α2 and α3 subunits, and no affinity for α4–α6 subunits. Silencing neurons in regions without α1 expression, such as the amygdala, could thus prove challenging.
So far we have discussed strategies that rely on genetically targeted proteinaceous receptors and small molecule or peptide ligands. These pharmacogenetic strategies are relatively slow: infusion of ligands to spatially extended networks of neurons requires seconds to hours, depending on the route of administration and the properties of the ligand. These times are slower than the dynamics of spike trains underlying many behaviors (which are on the scale of milliseconds). In contrast, light has proven to be an excellent trigger for rapid silencing methods.
Expression of vertebrate rhodopsin 4 in CNS neurons can couple light stimuli to opening of potassium channels, reducing action potential frequency (Li et al., 2005). Rhodopsin 4 also modulates other G protein-coupled channels, for example voltage-gated calcium channels involved in neurotransmitter release, opening up the possibility of light-gated modulation of short-term synaptic plasticity. However, without additional development the lack of functional specificity of rhodopsin 4 is a major drawback of this approach.
Another exciting recent method relies on expression of Natronomonas pharaonis halorhodospin (NpHR), a light-gated outward chloride pump. In neurons expressing NpHR, pulses of bright yellow light induce rapid hyperpolarization that can abolish action potential generation (Han and Boyden, 2007; Zhang et al., 2007). Individual action potentials in complex spike trains could potentially be eliminated using this method. NpHR expression in muscles and motoneurons in C. elegans can produce light-gated control of C. elegans locomotor behavior (Zhang et al., 2007). Since halorhodopsin is a pump, rather than a channel, high expression levels and light intensities are required for inducible silencing. Furthermore, light delivery to activate deep brain structures could be a limiting factor.
In summary, a diverse arsenal of inducible and reversible LOF systems is available (Table 2). Interestingly, each method has distinct properties that may make it especially suitable for particular types of experiments. Two-component systems (GluCl, Sph-StxTm-MIST) are ideal if intersectional methods are desirable to restrict expression to specific cell types. Systems induced by light (rhodopsin 4, halorhodopsin) or locally applied ligands (AlstR) may be preferable if spatially restricted activation is desirable. In contrast, if spatially extended neuronal populations are to be silenced, then rapidly diffusible agonists that can be applied systemically are preferred (GluCl, zolpidem-sensitive GABAA-Rs). Local administration of dimerizer to a particular axonal projection originating in MIST-positive neurons could be used to silence selected synaptic pathways.
5b. Methods for Activation
Ideal methods for activation require excellent temporal control. Expression of ligand-gated channels by themselves is not sufficient because long-lasting activation would ultimately inactivate membrane currents and cause neuronal silencing. In the context of an intact circuit, long-lasting activation could cause runaway excitation or strong feedback inhibition, complicating the interpretation of the responses to activation. For these reasons it is necessary to develop systems with temporal control on the order of milliseconds. Light is the only activation trigger for existing techniques that is sufficiently quick for this purpose.
Similar to LOF methods, the development of GOF methods has relied almost exclusively on bioprospecting. One early attempt relies on reconstituting in mammalian cells the signaling pathway coupling light and depolarization used in Drosophila photoreceptors (Zemelman et al., 2002). But this method requires expression of multiple gene products, and the temporal precision of activation is poor.
A related method relies on expression of ligand-gated channels that are mostly expressed in the PNS and whose native ligands are not found at high levels in the brain (Tobin et al., 2002). Uncaging of the caged ligand then allows activation of genetically targeted cells (Zemelman et al., 2003). Expression of the ATP-gated P2X2 channel, in the presence of caged ATP, has been used to activate flight control circuits in Drosophila (Lima and Miesenbock, 2005). An obstacle to using these methods is that the ligand needs to be injected into the brain. In addition, the channels that have been tested in this context have slow deactivation kinetics (on the scale of approximately seconds).
The development of ChR2 has overcome these problems (Nagel et al., 2003; Boyden et al., 2005; Li et al., 2005; Bi et al., 2006; Zhang and Oertner, 2006). The potential impact of ChR2 can hardly be overstated. ChR2 has already allowed fast optical control of genetically targeted neuronal populations in vivo (Nagel et al., 2005; Schroll et al., 2006; Arenkiel et al., 2007; Suh et al., 2007). When combined with behavioral studies, ChR2 photostimulation allows precise tests of hypotheses about how patterns of action potentials in genetically targeted neurons contribute to behavior. For example, photostimulation of ChR2-positive hypocretin neurons accelerated the waking of sleeping mice (Adamantidis et al., 2007). Calibrated photostimulation allowed a precise estimate of the number of action potentials in neocortical layer 2/3 neurons required to drive a decision task (Huber et al., 2008). In addition, ChR2 is already beginning to have an impact on neuroanatomy (Section 3d) and the identification of recorded neurons in vivo (Section 4a).
ChR2 may still have some drawbacks. For example, in some systems, including the CNS of adult flies, retinal may not be present at sufficient concentrations for ChR2 function. In addition, the spatiotemporal currents produced by ChR2 differ from the currents produced by synaptic input. This could be a problem for experiments probing dendritic integration.
These limitations have been addressed using engineered channels in combination with tethered, light-switchable agonists and antagonists (Banghart et al., 2004; Szobota et al., 2007). For the purposes of circuit analysis, the most powerful system is the light-based ionotropic glutamate receptor (LiGluR) (Szobota et al., 2007). A light-switchable agonist (MAG) is covalently tethered to a cysteine that is engineered into the ligand binding domain of a glutamate receptor. Near-UV light switches the MAG isomerization from trans to cis and leads to agonist binding and channel opening. Green light reverses the isomerization and closes the channel. In neurons expressing LiGluR, light pulses can trigger short, postsynaptic currents that resemble normal synaptic transmission as well as prolonged depolarizations. LiGluR requires that MAG is introduced into the tissue of interest. Since LiGluRs are also activated by endogenous glutamate release, their overexpression may alter normal neuronal and circuit properties. Thus, in an ideal experiment, LiGluR might be used in knockin experiments where the endogenous GluR is replaced by LiGluR.
In summary, a diverse arsenal of rapid GOF systems is available (Table 3). Because of its simplicity, for most applications ChR2 is the method of choice. However, other systems, such as P2X2/caged-ATP and LiGluR, may fill important niches, for example by providing access to systems that do not produce retinal or by mimicking synaptic currents.
5c. Forward Genetic Screens
Genetically encoded tools for LOF and GOF manipulations can be used to perform forward genetic screens to identify new circuit elements necessary and sufficient for eliciting particular behaviors. For example, in Drosophila one can use thousands of enhancer trap or enhancer dissection lines driving Gal4 (Section 2) to express shibirets (Section 5a) in subsets of neurons. By using behavioral assays, it is then possible to screen for cells which, when reversibly taken out of the circuit, lead to behavioral defects. Such screens could provide a list of essential neurons that constitute functional circuits.
6. Conclusions and Outlook
When can we say that we have understood a neural circuit? Our understanding of the circuit mechanisms underlying behavior is relatively advanced in select simple systems with identified neurons, such as the stomatogastric nervous system of crustaceans (STG) (Marder and Bucher, 2007). The STG generates rhythmic motor behaviors. The accessibility of all cell types for electrophysiological recordings is the cardinal feature that has made the STG circuit tractable. First, all neurons can be unambiguously identified using positional, morphological, and electrophysiological parameters. Second, the neurons are amenable to routine extracellular and intracellular recordings. Multiple intracellular recordings can be used to construct a circuit diagram. Third, intracellular methods have been critical to correlate firing patterns with motor output and to probe the effects of activating or silencing neurons on the network. Individual neurons can also be selectively removed from the circuit using photoablation (Miller and Selverston, 1979). These experimental approaches, combined with quantitative analysis and modeling, have allowed researchers to delineate the logic of the central pattern generators that cause rhythmic motor behavior.
Genetic analysis is promising comparable levels of access in systems that are orders of magnitude more complex than the STG. This includes systems in genetic model organisms such as fly and mouse, but novel gene transfer methods make these tools also applicable to other systems, including monkeys. By combining neuroanatomical, physiological, and functional manipulations, genetic analysis will facilitate systematic reverse engineering of neural circuits in classical experimental paradigms and open up powerful new paradigms. It will establish causality between patterns of activity in specific groups of neurons, the function of neural circuits, and animal behavior. Just as genetic analyses of individual genes and their interactions in the past few decades have been enormously fruitful in dissecting complex biological processes, genetic approaches we outline here, together with theoretical modeling, may reveal the logic of the neural circuits in complex brains that guide behaviors.
Acknowledgments
We thank members of our laboratories and our colleagues for useful discussions, and NIH (L.L., E.M.C., and K.S.) and HHMI (L.L. and K.S.) for research support.
References
- Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian E. The Mechanisms of Nervous Action. Philadelphia: University of Pennsylvania Press; 1932. [Google Scholar]
- Allen ND, Cran DG, Barton SC, Hettle S, Reik W, Surani MA. Transgenes as probes for active chromosomal domains in mouse development. Nature. 1988;333:852–855. doi: 10.1038/333852a0. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Martin KA, Whitteridge D. Form, function, and intracortical projections of neurons in the striate cortex of the monkey Macacus nemestrinus. Cereb Cortex. 1993;3:412–420. doi: 10.1093/cercor/3.5.412. [DOI] [PubMed] [Google Scholar]
- Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, Augustine GJ, Ehlers MD, Feng G. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold DB. Polarized targeting of ion channels in neurons. Pflugers Arch. 2007;453:763–769. doi: 10.1007/s00424-006-0155-5. [DOI] [PubMed] [Google Scholar]
- Ataka K, Pieribone VA. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys J. 2002;82:509–516. doi: 10.1016/S0006-3495(02)75415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat Genet. 2003;35:70–75. doi: 10.1038/ng1228. [DOI] [PubMed] [Google Scholar]
- Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23:2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Kosmidis EK, Vucinic D, Falk CX, Cohen LB, Djurisic M, Zecevic D. Imaging brain activity with voltage- and calcium-sensitive dyes. Cell Mol Neurobiol. 2005;25:245–282. doi: 10.1007/s10571-005-3059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat Neuro-sci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle GW, Tatum EL. Genetic control of biochemical reactions in Neurospora. Proc Natl Acad Sci USA. 1941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, O’Kane CJ, Wilson C, Grossniklaus U, Pearson RK, Gehring WJ. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- Benzer S. Fine structure of a genetic region in bacteriophage. Proc Natl Acad Sci USA. 1955;41:344–354. doi: 10.1073/pnas.41.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens C, Hillen W. Gene regulation by tetracyclines. Genet Eng (N Y) 2004;26:255–277. doi: 10.1007/978-0-306-48573-2_13. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bi A, Cui J, Ma YP, Olshevskaya E, Pu M, Dizhoor AM, Pan ZH. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron. 2006;50:23–33. doi: 10.1016/j.neuron.2006.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek W, Rieke F, de Ruyter van Steveninck RR, Warland D. Reading a neural code. Science. 1991;252:1854–1857. doi: 10.1126/science.2063199. [DOI] [PubMed] [Google Scholar]
- Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, et al. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Binzegger T, Douglas RJ, Martin KA. A quantitative map of the circuit of cat primary visual cortex. J Neurosci. 2004;24:8441–8453. doi: 10.1523/JNEUROSCI.1400-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Bozza T, McGann JP, Mombaerts P, Wachowiak M. In vivo imaging of neuronal activity by targeted expression of a genetically encoded probe in the mouse. Neuron. 2004;42:9–21. doi: 10.1016/s0896-6273(04)00144-8. [DOI] [PubMed] [Google Scholar]
- Braitenberg V, Schutz A. Anatomy of the Cortex. Vol. 18. Berlin: Springer Verlag; 1991. [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Braz JM, Rico B, Basbaum AI. Transneuronal tracing of diverse CNS circuits by Cre-mediated induction of wheat germ agglutinin in transgenic mice. Proc Natl Acad Sci USA. 2002;99:15148–15153. doi: 10.1073/pnas.222546999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht M, Schneider M, Sakmann B, Margrie TW. Whisker movements evoked by stimulation of single pyramidal cells in rat motor cortex. Nature. 2004;427:704–710. doi: 10.1038/nature02266. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Denk W. Towards neural circuit reconstruction with volume electron microscopy techniques. Curr Opin Neurobiol. 2006;16:562–570. doi: 10.1016/j.conb.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Abarbanel HD, Kristan WB., Jr Optical imaging of neuronal populations during decision-making. Science. 2005;307:896–901. doi: 10.1126/science.1103736. [DOI] [PubMed] [Google Scholar]
- Britten KH, Shadlen MN, Newsome WT, Movshon JA. The analysis of visual motion: a comparison of neuronal and psychophysical performance. J Neurosci. 1992;12:4745–4765. doi: 10.1523/JNEUROSCI.12-12-04745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–181. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Buffelli M, Burgess RW, Feng G, Lobe CG, Lichtman JW, Sanes JR. Genetic evidence that relative synaptic efficacy biases the outcome of synaptic competition. Nature. 2003;424:430–434. doi: 10.1038/nature01844. [DOI] [PubMed] [Google Scholar]
- Bureau I, von Saint Paul F, Svoboda K. Interdigitated paralemniscal and lemniscal pathways in the mouse barrel cortex. PLoS Biol. 2006;4:e382. doi: 10.1371/journal.pbio.0040382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal SR. Histology of the nervous system of man and vertebrates. Oxford: Oxford University Press, Inc; 1911. 1995 translation. [Google Scholar]
- Callahan CA, Thomas JB. Tau-beta-galactosidase, an axon-targeted fusion protein. Proc Natl Acad Sci USA. 1994;91:5972–5976. doi: 10.1073/pnas.91.13.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway EM. Cell type specificity of local cortical connections. J Neurocytol. 2002;31:231–237. doi: 10.1023/a:1024165824469. [DOI] [PubMed] [Google Scholar]
- Callaway EM, Katz LC. Photostimulation using caged glutamate reveals functional circuitry in living brain slices. Proc Natl Acad Sci USA. 1993;90:7661–7665. doi: 10.1073/pnas.90.16.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- Caroni P. Overexpression of growth-associated proteins in the neurons of adult transgenic mice. J Neurosci Methods. 1997;71:3–9. doi: 10.1016/s0165-0270(96)00121-5. [DOI] [PubMed] [Google Scholar]
- Chanda B, Blunck R, Faria LC, Schweizer FE, Mody I, Bezanilla F. A hybrid approach to measuring electrical activity in genetically specified neurons. Nat Neurosci. 2005;8:1619–1626. doi: 10.1038/nn1558. [DOI] [PubMed] [Google Scholar]
- Chattopadhyaya B, Di Cristo G, Higashiyama H, Knott GW, Kuhlman SJ, Welker E, Huang ZJ. Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J Neurosci. 2004;24:9598–9611. doi: 10.1523/JNEUROSCI.1851-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BL, Hall DH, Chklovskii DB. Wiring optimization can relate neuronal structure and function. Proc Natl Acad Sci USA. 2006;103:4723–4728. doi: 10.1073/pnas.0506806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan WM. The emergence of modern neuroanatomy and developmental neurobiology. Neuron. 1998;20:413–426. doi: 10.1016/s0896-6273(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Crick F. What Mad Pursuit: A Personal View of Scientific Discovery. New York: Basic Books; 1988. [Google Scholar]
- Dalva MB, Katz LC. Rearrangements of synaptic connections in visual cortex revealed by laser photostimulation. Science. 1994;265:255–258. doi: 10.1126/science.7912852. [DOI] [PubMed] [Google Scholar]
- Dantzker JL, Callaway EM. Laminar sources of synaptic input to cortical inhibitory interneurons and pyramidal neurons. Nat Neurosci. 2000;3:701–707. doi: 10.1038/76656. [DOI] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4–VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paola V, Arber S, Caroni P. AMPA receptors regulate dynamic equilibrium of presynaptic terminals in mature hippocampal networks. Nat Neurosci. 2003;6:491–500. doi: 10.1038/nn1046. [DOI] [PubMed] [Google Scholar]
- DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, Enquist L, Friedman JM. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- Deleuze C, Huguenard JR. Distinct electrical and chemical connectivity maps in the thalamic reticular nucleus: potential roles in synchronization and sensation. J Neurosci. 2006;26:8633–8645. doi: 10.1523/JNEUROSCI.2333-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Horstman H. Serial block-face scanning electron microscopy (SBFSEM) to reconstruct 3D tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D, He Y, Mutoh H, Baker BJ, Cohen L, Akemann W, Knopfel T. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS ONE. 2007;2:e440. doi: 10.1371/journal.pone.0000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittgen T, Nimmerjahn A, Komai S, Licznerski P, Waters J, Margrie TW, Helmchen F, Denk W, Brecht M, Osten P. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc Natl Acad Sci USA. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RJ, Martin KA. Neuronal circuits of the neocortex. Annu Rev Neurosci. 2004;27:419–451. doi: 10.1146/annurev.neuro.27.070203.144152. [DOI] [PubMed] [Google Scholar]
- Dubnau J, Grady L, Kitamoto T, Tully T. Disruption of neuro-transmission in Drosophila mushroom body blocks retrieval but not acquisition of memory. Nature. 2001;411:476–480. doi: 10.1038/35078077. [DOI] [PubMed] [Google Scholar]
- Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki SM, Kim JC. Molecular neuroanatomy’s “Three Gs”: a primer. Neuron. 2007;54:17–34. doi: 10.1016/j.neuron.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist LW. Exploiting circuit-specific spread of pseudorabies virus in the central nervous system: insights to pathogenesis and circuit tracers. J Infect Dis. 2002;186(Suppl 2):S209–S214. doi: 10.1086/344278. [DOI] [PubMed] [Google Scholar]
- Etessami R, Conzelmann KK, Fadai-Ghotbi B, Natelson B, Tsiang H, Ceccaldi PE. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J Gen Virol. 2000;81:2147–2153. doi: 10.1099/0022-1317-81-9-2147. [DOI] [PubMed] [Google Scholar]
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon PF. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci USA. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, VanHoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living animals. Neuron. 2007;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fernandez-Chacon R, Sudhof TC. Genetics of synaptic vesicle function: toward the complete functional anatomy of an organelle. Annu Rev Physiol. 1999;61:753–776. doi: 10.1146/annurev.physiol.61.1.753. [DOI] [PubMed] [Google Scholar]
- Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Cocker ED, Piyawattanametha W, Jung JC, Cheung EL, Schnitzer MJ. Fiber-optic fluorescence imaging. Nat Methods. 2005;2:941–950. doi: 10.1038/nmeth820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa H, Jacobson M. Transsynaptic labeling of neurons in the optic tectum of xenopus after intraocular [3H]proline injection. Brain Res. 1980;194:431–441. doi: 10.1016/0006-8993(80)91223-8. [DOI] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- Galarreta M, Hestrin S. A network of fast-spiking cells in the neocortex connected by electrical synapses. Nature. 1999;402:72–75. doi: 10.1038/47029. [DOI] [PubMed] [Google Scholar]
- Gao Q, Yuan B, Chess A. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat Neurosci. 2000;3:780–785. doi: 10.1038/77680. [DOI] [PubMed] [Google Scholar]
- Garaschuk O, Griesbeck O, Konnerth A. Troponin C-based biosensors: A new family of genetically encoded indicators for in vivo calcium imaging in the nervous system. Cell Calcium. 2007;42:351–361. doi: 10.1016/j.ceca.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Garcia-Otin AL, Guillou F. Mamm. Genome targeting using site-specific recombinases. Front Biosci. 2006;11:1108–1136. doi: 10.2741/1867. [DOI] [PubMed] [Google Scholar]
- Garner JA, LaVail JH. Differential anterograde transport of HSV type 1 viral strains in the murine optic pathway. J Neurovirol. 1999;5:140–150. doi: 10.3109/13550289909021996. [DOI] [PubMed] [Google Scholar]
- Gibson JR, Beierlein M, Connors BW. Two networks of electrically coupled inhibitory neurons in neocortex. Nature. 1999;402:75–79. doi: 10.1038/47035. [DOI] [PubMed] [Google Scholar]
- Gigout L, Rebollo P, Clement N, Warrington KH, Jr, Muzyczka N, Linden RM, Weber T. Altering AAV tropism with mosaic viral capsids. Mol Ther. 2005;11:856–865. doi: 10.1016/j.ymthe.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Gilbert CD, Wiesel TN. Morphology and intracortical projections of functionally characterised neurones in the cat visual cortex. Nature. 1979;280:120–125. doi: 10.1038/280120a0. [DOI] [PubMed] [Google Scholar]
- Giniger E, Wells W, Jan LY, Jan YN. Tracing neurons with a kinesin-β-galactosidase fusion protein. Rouxs Arch Dev Biol. 1993;202:112–122. doi: 10.1007/BF00636536. [DOI] [PubMed] [Google Scholar]
- Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- Golgi C. Sulla struttura della sostanza grigia del cervello. Gazetta medica lombarda. 1873;IV [Google Scholar]
- Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosgnach S, Lanuza GM, Butt SJ, Saueressig H, Zhang Y, Velasquez T, Riethmacher D, Callaway EM, Kiehn O, Goulding M. VI spinal neurons regulate the speed of vertebrate locomotor outputs. Nature. 2006;440:215–219. doi: 10.1038/nature04545. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossler A, Joyner AL, Rossant J, Skarnes WC. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989;244:463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- Gray PA, Janczewski WA, Mellen N, McCrimmon DR, Feldman JL. Normal breathing requires preBotzinger complex neurokinin-1 receptor-expressing neurons. Nat Neurosci. 2001;4:927–930. doi: 10.1038/nn0901-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- Grinvald A, Hildesheim R. VSDI: a new era in functional imaging of cortical dynamics. Nat Rev Neurosci. 2004;5:874–885. doi: 10.1038/nrn1536. [DOI] [PubMed] [Google Scholar]
- Groth AC, Calos MP. Phage integrases: biology and applications. J Mol Biol. 2004;335:667–678. doi: 10.1016/j.jmb.2003.09.082. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutzendler J, Kasthuri N, Gan WB. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- Gupta A, Wang Y, Markram H. Organizing principles for a diversity of GABAergic interneurons and synapses in the neocortex. Science. 2000;287:273–278. doi: 10.1126/science.287.5451.273. [DOI] [PubMed] [Google Scholar]
- Haas K, Sin WC, Javaherian A, Li Z, Cline HT. Single-cell electroporation for gene transfer in vivo. Neuron. 2001;29:583–591. doi: 10.1016/s0896-6273(01)00235-5. [DOI] [PubMed] [Google Scholar]
- Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS ONE. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan MT, Friedrich RW, Euler T, Larkum ME, Giese GG, Both M, Duebel J, Waters J, Bujard H, Griesbeck O, et al. Functional Fluorescent Ca(2+) Indicator Proteins in Transgenic Mice under TET Control. PLoS Biol. 2004;2:e163. doi: 10.1371/journal.pbio.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, Aigaki T, Matsuzaki F, Nakagoshi H, Tanimura T, et al. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 2002;34:58–61. doi: 10.1002/gene.10137. [DOI] [PubMed] [Google Scholar]
- Heim N, Griesbeck O. Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J Biol Chem. 2004;279:14280–14286. doi: 10.1074/jbc.M312751200. [DOI] [PubMed] [Google Scholar]
- Heintz N. BAC to the future: the use of bac transgenic mice for neuro-science research. Nat Rev Neurosci. 2001;2:861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Imoto K, Sakmann B. Ca2+ buffering and action potential-evoked Ca2+ signaling in dendrites of pyramidal neurons. Biophys J. 1996;70:1069–1081. doi: 10.1016/S0006-3495(96)79653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Johnston RJ, Jr, Chang S. Left-right asymmetry in the nervous system: the Caenorhabditis elegans model. Nat Rev Neurosci. 2002;3:629–640. doi: 10.1038/nrn897. [DOI] [PubMed] [Google Scholar]
- Holmgren C, Harkany T, Svennenfors B, Zilberter Y. Pyramidal cell communication within local networks in layer 2/3 of rat neocortex. J Physiol. 2003;551:139–153. doi: 10.1113/jphysiol.2003.044784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi E, Tremblay L, Feger J, Carras PL, Strick PL. The cerebellum communicates with the basal ganglia. Nat Neurosci. 2005;8:1491–1493. doi: 10.1038/nn1544. [DOI] [PubMed] [Google Scholar]
- Hubel DH, Wiesel TN. Receptive fields of single neurones in the cat’s striate cortex. J Physiol. 1959;148:574–591. doi: 10.1113/jphysiol.1959.sp006308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D, Petreanu L, Ghitani N, Ranade S, Hromadka T, Mainen ZF, Svoboda K. Optical microstimulation of a sparse subset of layer 2/3 barrel cortex neurons drives learning and behavior in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Tallon I, Wen H, Miwa JM, Xing J, Tekinay AB, Ono F, Brehm P, Heintz N. Tethering naturally occurring peptide toxins for cell-autonomous modulation of ion channels and receptors in vivo. Neuron. 2004;43:305–311. doi: 10.1016/j.neuron.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Itaya SK, van Hoesen GW. WGA-HRP as a transneuronal marker in the visual pathways of monkey and rat. Brain Res. 1982;236:199–204. doi: 10.1016/0006-8993(82)90046-4. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurons and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR, Jr, Luo L. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128:1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- Johns DC, Marx R, Mains RE, O’Rourke B, Marban E. Inducible genetic suppression of neuronal excitability. J Neurosci. 1999;19:1691–1697. doi: 10.1523/JNEUROSCI.19-05-01691.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen EM, Hartwieg E, Schuske K, Nonet ML, Jin Y, Horvitz HR. Defective recycling of synaptic vesicles in synaptotagmin mutants of Caenorhabditis elegans. Nature. 1995;378:196–199. doi: 10.1038/378196a0. [DOI] [PubMed] [Google Scholar]
- Kamondi A, Acsady L, Buzsaki G. Dendritic spikes are enhanced by cooperative network activity in the intact hioppocampus. J Neurosci. 1998;18:3919–3928. doi: 10.1523/JNEUROSCI.18-10-03919.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova AY, Tervo DG, Gray NW, Svoboda K. Rapid and reversible chemical inactivation of synaptic transmission in genetically targeted neurons. Neuron. 2005;48:727–735. doi: 10.1016/j.neuron.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Katz LC, Dalva MB. Scanning laser photostimulation: a new approach for analyzing brain circuits. J Neurosci Methods. 1994;54:205–218. doi: 10.1016/0165-0270(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Kellendonk C, Simpson EH, Polan HJ, Malleret G, Vronskaya S, Winiger V, Moore H, Kandel ER. Transient and selective overexpression of dopamine D2 receptors in the striatum causes persistent abnormalities in prefrontal cortex functioning. Neuron. 2006;49:603–615. doi: 10.1016/j.neuron.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Kelly RM, Strick PL. Rabies as a transneuronal tracer of circuits in the central nervous system. J Neurosci Methods. 2000;103:63–71. doi: 10.1016/s0165-0270(00)00296-x. [DOI] [PubMed] [Google Scholar]
- Kerr JN, Greenberg D, Helmchen F. Imaging input and output of neocortical networks in vivo. Proc Natl Acad Sci USA. 2005;102:14063–14068. doi: 10.1073/pnas.0506029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N, Mizuno T, Yoshihara Y. Adenovirus-mediated WGA gene delivery for transsynaptic labeling of mouse olfactory pathways. Chem Senses. 2002;27:215–223. doi: 10.1093/chemse/27.3.215. [DOI] [PubMed] [Google Scholar]
- Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Judkewitz B, Kano M, Denk W, Hausser M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nat Methods. 2008;5:61–67. doi: 10.1038/nmeth1150. [DOI] [PubMed] [Google Scholar]
- Koenig JH, Saito K, Ikeda K. Reversible control of synaptic transmission in a single gene mutant of Drosophila melanogaster. J Cell Biol. 1983;96:1517–1522. doi: 10.1083/jcb.96.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kootstra NA, Verma IM. Gene therapy with viral vectors. Annu Rev Pharmacol Toxicol. 2003;43:413–439. doi: 10.1146/annurev.pharmtox.43.100901.140257. [DOI] [PubMed] [Google Scholar]
- Kozloski J, Hamzei-Sichani F, Yuste R. Stereotyped position of local synaptic targets in neocortex. Science. 2001;293:868–872. doi: 10.1126/science.293.5531.868. [DOI] [PubMed] [Google Scholar]
- Kuffler SW. Discharge patterns and functional organization of mammalian retina. J Neurophysiol. 1953;16:37–68. doi: 10.1152/jn.1953.16.1.37. [DOI] [PubMed] [Google Scholar]
- Lafon M. Rabies virus receptors. J Neurovirol. 2005;11:82–87. doi: 10.1080/13550280590900427. [DOI] [PubMed] [Google Scholar]
- Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- Lechner HA, Lein ES, Callaway EM. A genetic method for selective and quickly reversible silencing of Mammalian neurons. J Neurosci. 2002;22:5287–5290. doi: 10.1523/JNEUROSCI.22-13-05287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AK, Manns ID, Sakmann B, Brecht M. Whole-cell recordings in freely moving rats. Neuron. 2006;51:399–407. doi: 10.1016/j.neuron.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lerchner W, Xiao C, Nashmi R, Slimko EM, van Trigt L, Lester HA, Anderson DJ. Reversible silencing of neuronal excitability in behaving mice by a genetically targeted, ivermectin-gated Cl- channel. Neuron. 2007;54:35–49. doi: 10.1016/j.neuron.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Li P, Slimko EM, Lester HA. Selective elimination of glutamate activation and introduction of fluorescent proteins into a Caenorhabditis elegans chloride channel. FEBS Lett. 2002;528:77–82. doi: 10.1016/s0014-5793(02)03245-3. [DOI] [PubMed] [Google Scholar]
- Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, Chiel H, Hegemann P, Landmesser LT, Herlitze S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Lin HH, Lai JS, Chin AL, Chen YC, Chiang AS. A map of olfactory representation in the Drosophila mushroom body. Cell. 2007;128:1205–1217. doi: 10.1016/j.cell.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Lopez-Bendito G, Sturgess K, Erdelyi F, Szabo G, Molnar Z, Paulsen O. Preferential origin and layer destination of GAD65-GFP cortical interneurons. Cereb Cortex. 2004;14:1122–1133. doi: 10.1093/cercor/bhh072. [DOI] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli DS, Foss M, Villella A, Taylor BJ, Hall JC, Baker BS. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436:395–400. doi: 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- Mao T, O’Connor D, Nakai J, Svoboda K. Characterization and subcellular targeting of GCaMP-type genetically-encoded calcium indicators. PloS ONE. 2008 doi: 10.1371/journal.pone.0001796. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Bucher D. Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol. 2007;69:291–316. doi: 10.1146/annurev.physiol.69.031905.161516. [DOI] [PubMed] [Google Scholar]
- Margrie TW, Meyer AH, Caputi A, Monyer H, Hasan MT, Schaefer AT, Denk W, Brecht M. Targeted whole-cell recordings in the mammalian brain in vivo. Neuron. 2003;39:911–918. doi: 10.1016/j.neuron.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Marin EC, Jefferis GSXE, Komiyama T, Zhu H, Luo L. Representation of the glomerular olfactory map in the Drosophila brain. Cell. 2002;109:243–255. doi: 10.1016/s0092-8674(02)00700-6. [DOI] [PubMed] [Google Scholar]
- Martin KA, Whitteridge D. Form, function and intracortical projections of spiny neurones in the striate visual cortex of the cat. J Physiol. 1984;353:463–504. doi: 10.1113/jphysiol.1984.sp015347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskos U, Kissa K, St Cloment C, Brulet P. Retrograde trans-synaptic transfer of green fluorescent protein allows the genetic mapping of neuronal circuits in transgenic mice. Proc Natl Acad Sci USA. 2002;99:10120–10125. doi: 10.1073/pnas.152266799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. Neuronal diversity in the retina. Curr Opin Neurobiol. 2001;11:431–436. doi: 10.1016/s0959-4388(00)00230-0. [DOI] [PubMed] [Google Scholar]
- Mazarakis ND, Azzouz M, Rohll JB, Ellard FM, Wilkes FJ, Olsen AL, Carter EE, Barber RD, Baban DF, Kingsman SM, et al. Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum Mol Genet. 2001;10:2109–2121. doi: 10.1093/hmg/10.19.2109. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Davis RL. The role of Drosophila mushroom body signaling in olfactory memory. Science. 2001;293:1330–1333. doi: 10.1126/science.1062622. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Meister M, Lagnado L, Baylor DA. Concerted signaling by retinal ganglion cells. Science. 1995;270:1207–1210. doi: 10.1126/science.270.5239.1207. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenbock G, Angelis DAD, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- Miller JP, Selverston A. Rapid killing of single neurons by irradiation of intracellularly injected dye. Science. 1979;206:702–704. doi: 10.1126/science.386514. [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Miyawaki A. Innovations in the Imaging of Brain Functions using Fluorescent Proteins. Neuron. 2005;48:189–199. doi: 10.1016/j.neuron.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescence indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Morgan TH. The origin of five mutations in the eye color in Drosophila and their modes of inheritance. Science. 1911;33:534–537. doi: 10.1126/science.33.849.534-a. [DOI] [PubMed] [Google Scholar]
- Mountcastle VB. Modality and topographic properties of single neurons of cat’s somatic sensory cortex. J Neurophysiol. 1957;20:408–434. doi: 10.1152/jn.1957.20.4.408. [DOI] [PubMed] [Google Scholar]
- Mrsic-Flogel TD, Hofer SB, Ohki K, Reid RC, Bonhoeffer T, Hubener M. Homeostatic regulation of eye-specific responses in visual cortex during ocular dominance plasticity. Neuron. 2007;54:961–972. doi: 10.1016/j.neuron.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci USA. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama S, Zeng S, Xiong W, Fletcher ML, Masurkar AV, Davis DJ, Pieribone VA, Chen WR. In vivo simultaneous tracing and Ca(2+) imaging of local neuronal circuits. Neuron. 2007;53:789–803. doi: 10.1016/j.neuron.2007.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- Nakazawa K, Quirk MC, Chitwood RA, Watanabe M, Yeckel MF, Sun LD, Kato A, Carr CA, Johnston D, Wilson MA, Tonegawa S. Requirement for hippocampal CA3 NMDA receptors in associative memory recall. Science. 2002;297:211–218. doi: 10.1126/science.1071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Roorda RD, Lima SQ, Zemelman BV, Morcillo P, Miesenbock G. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 2002;36:463–474. doi: 10.1016/s0896-6273(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Niell CM, Smith SJ. Functional imaging reveals rapid development of visual response properties in the zebrafish tectum. Neuron. 2005;45:941–951. doi: 10.1016/j.neuron.2005.01.047. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- O’Donovan MJ, Ho S, Sholomenko G, Yee W. Real-time imaging of neurons retrogradely and anterogradely labelled with calcium-sensitive dyes. J Neurosci Methods. 1993;46:91–106. doi: 10.1016/0165-0270(93)90145-h. [DOI] [PubMed] [Google Scholar]
- O’Keefe J, Dostrovsky J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 1971;34:171–175. doi: 10.1016/0006-8993(71)90358-1. [DOI] [PubMed] [Google Scholar]
- O’Malley DM, Kao YH, Fetcho JR. Imaging the functional organization of zebrafish hindbrain segments during escape behaviors. Neuron. 1996;17:1145–1155. doi: 10.1016/s0896-6273(00)80246-9. [DOI] [PubMed] [Google Scholar]
- Ohki K, Chung S, Ch’ng YH, Kara P, Reid RC. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature. 2005;433:597–603. doi: 10.1038/nature03274. [DOI] [PubMed] [Google Scholar]
- Oliva AA, Jr, Jiang M, Lam T, Smith KL, Swann JW. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci. 2000;20:3354–3368. doi: 10.1523/JNEUROSCI.20-09-03354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SR, Bhandawat V, Wilson RI. Excitatory interactions between olfactory processing channels in the Drosophila antennal lobe. Neuron. 2007;54:89–103. doi: 10.1016/j.neuron.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Moreadith RW, Leder P. Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc Natl Acad Sci USA. 1991;88:698–702. doi: 10.1073/pnas.88.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AE, Giacomello M, Kortemme T, Hires SA, Lev-Ram V, Baker D, Tsien RY. Ca2+ Indicators Based on Computationally Redesigned Calmodulin-Peptide Pairs. Chem Biol. 2006 doi: 10.1016/j.chembiol.2006.03.007. in press. [DOI] [PubMed] [Google Scholar]
- Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- Platt ML, Glimcher PW. Neural correlates of decision variables in parietal cortex. Nature. 1999;400:233–238. doi: 10.1038/22268. [DOI] [PubMed] [Google Scholar]
- Pologruto TA, Yasuda R, Svoboda K. Monitoring neural activity and [Ca2+] with genetically encoded Ca2+ indicators. J Neurosci. 2004;24:9572–9579. doi: 10.1523/JNEUROSCI.2854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouille F, Scanziani M. Routing of spike series by dynamic circuits in the hippocampus. Nature. 2004;429:717–723. doi: 10.1038/nature02615. [DOI] [PubMed] [Google Scholar]
- Ramdya P, Reiter B, Engert F. Reverse correlation of rapid calcium signals in the zebrafish optic tectum in vivo. J Neurosci Methods. 2006;157:230–237. doi: 10.1016/j.jneumeth.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Redfern CH, Coward P, Degtyarev MY, Lee EK, Kwa AT, Hennighausen L, Bujard H, Fishman GI, Conklin BR. Conditional expression and signaling of a specifically designed Gi-coupled receptor in transgenic mice. Nat Biotechnol. 1999;17:165–169. doi: 10.1038/6165. [DOI] [PubMed] [Google Scholar]
- Roman G, Davis RL. Molecular biology and anatomy of Drosophila olfactory associative learning. Bioessays. 2001;23:571–581. doi: 10.1002/bies.1083. [DOI] [PubMed] [Google Scholar]
- Romo R, Hernandez A, Zainos A, Salinas E. Somatosensory discrimination based on cortical microstimulation. Nature. 1998;392:387–390. doi: 10.1038/32891. [DOI] [PubMed] [Google Scholar]
- Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- Root CM, Semmelhack JL, Wong AM, Flores J, Wang JW. Propagation of olfactory information in Drosophila. Proc Natl Acad Sci USA. 2007;104:11826–11831. doi: 10.1073/pnas.0704523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH, S-Jacques B, Lee SM, Flax JD, Snyder EY, McMahon AP. Sonic hedgehog regulates proliferation and inhibits differentiation of CNS precursor cells. J Neurosci. 1999;19:8954–8965. doi: 10.1523/JNEUROSCI.19-20-08954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM. Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- Sakai R, Repunte-Canonigo V, Raj CD, Knopfel T. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur J Neurosci. 2001;13:2314–2318. doi: 10.1046/j.0953-816x.2001.01617.x. [DOI] [PubMed] [Google Scholar]
- Salzman CD, Britten KH, Newsome WT. Cortical microstimulation influences perceptual judgements of motion direction. Nature. 1990;346:174–177. doi: 10.1038/346174a0. [DOI] [PubMed] [Google Scholar]
- Sandler VM, Wang S, Angelo K, Lo HG, Breakefield XO, Clapham DE. Modified herpes simplex virus delivery of enhanced GFP into the central nervous system. J Neurosci Methods. 2002;121:211–219. doi: 10.1016/s0165-0270(02)00262-5. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan S, Ryan TA. Real-time measurements of vesicle-SNARE recycling in synapses of the central nervous system. Nat Cell Biol. 2000;2:197–204. doi: 10.1038/35008615. [DOI] [PubMed] [Google Scholar]
- Sano H, Nagai Y, Yokoi M. Inducible expression of retrograde transynaptic genetic tracer in mice. Genesis. 2007;45:123–128. doi: 10.1002/dvg.20273. [DOI] [PubMed] [Google Scholar]
- Sato T, Hamaoka T, Aizawa H, Hosoya T, Okamoto H. Genetic single-cell mosaic analysis implicates ephrinB2 reverse signaling in projections from the posterior tectum to the hindbrain in zebrafish. J Neurosci. 2007a;27:5271–5279. doi: 10.1523/JNEUROSCI.0883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TR, Gray NW, Mainen ZF, Svoboda K. The functional microarchitecture of the mouse barrel cortex. PLoS Biol. 2007b;5:e189. doi: 10.1371/journal.pbio.0050189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawatari A, Callaway EM. Diversity and cell type specificity of local excitatory connections to neurons in layer 3B of monkey primary visual cortex. Neuron. 2000;25:459–471. doi: 10.1016/s0896-6273(00)80908-3. [DOI] [PubMed] [Google Scholar]
- Schlief ML, Wilson RI. Olfactory processing and behavior downstream from highly selective receptor neurons. Nat Neurosci. 2007;10:623–630. doi: 10.1038/nn1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroll C, Riemensperger T, Bucher D, Ehmer J, Voller T, Erbguth K, Gerber B, Hendel T, Nagel G, Buchner E, Fiala A. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16:1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Schwab ME, Suda K, Thoenen H. Selective retrograde transsynaptic transfer of a protein, tetanus toxin, subsequent to its retrograde axonal transport. J Cell Biol. 1979;82:798–810. doi: 10.1083/jcb.82.3.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Shepherd GM, Pologruto TA, Svoboda K. Circuit analysis of experience-dependent plasticity in the developing rat barrel cortex. Neuron. 2003;38:277–289. doi: 10.1016/s0896-6273(03)00152-1. [DOI] [PubMed] [Google Scholar]
- Shepherd GMG, Stepanyants A, Bureau I, Chklovskii DB, Svoboda K. Geometric and functional organization of cortical circuits. Nat Neurosci. 2005;8:782–790. doi: 10.1038/nn1447. [DOI] [PubMed] [Google Scholar]
- Shepherd GMG, Svoboda K. Laminar and columnar organization of ascending excitatory projections to layer 2/3 pyramidal neurons in rat barrel cortex. J Neurosci. 2005;25:5670–5679. doi: 10.1523/JNEUROSCI.1173-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Fang G, Shi W, Bartlett JS. Insertional mutagenesis at positions 520 and 584 of adeno-associated virus type 2 (AAV2) capsid gene and generation of AAV2 vectors with eliminated heparin- binding ability and introduced novel tropism. Hum Gene Ther. 2006;17:353–361. doi: 10.1089/hum.2006.17.353. [DOI] [PubMed] [Google Scholar]
- Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–741. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- Sillitoe RV, Joyner AL. Morphology, molecular codes, and circuitry produce the three-dimensional complexity of the cerebellum. Annu Rev Cell Dev Biol. 2007;23:549–577. doi: 10.1146/annurev.cellbio.23.090506.123237. [DOI] [PubMed] [Google Scholar]
- Slimko EM, McKinney S, Anderson DJ, Davidson N, Lester HA. Selective electrical silencing of mammalian neurons in vitro by the use of invertebrate ligand-gated chloride channels. J Neurosci. 2002;22:7373–7379. doi: 10.1523/JNEUROSCI.22-17-07373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snitkovsky S, Young JA. Cell-specific viral targeting mediated by a soluble retroviral receptor-ligand fusion protein. Proc Natl Acad Sci USA. 1998;95:7063–7068. doi: 10.1073/pnas.95.12.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snitkovsky S, Niederman TM, Mulligan RC, Young JA. Targeting avian leukosis virus subgroup A vectors by using a TVA-VEGF bridge protein. J Virol. 2001;75:1571–1575. doi: 10.1128/JVI.75.3.1571-1575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohya K, Kameyama K, Yanagawa Y, Obata K, Tsumoto T. GABAergic neurons are less selective to stimulus orientation than excitatory neurons in layer II/III of visual cortex, as revealed by in vivo functional Ca2+ imaging in transgenic mice. J Neurosci. 2007;27:2145–2149. doi: 10.1523/JNEUROSCI.4641-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Kisvarday ZF, Martin KA, Whitteridge D. Synaptic connections of morphologically identified and physiologically characterized large basket cells in the striate cortex of cat. Neuroscience. 1983;10:261–294. doi: 10.1016/0306-4522(83)90133-1. [DOI] [PubMed] [Google Scholar]
- Song S, Sjostrom PJ, Reigl M, Nelson S, Chklovskii DB. Highly nonrandom features of synaptic connectivity in local cortical circuits. PLoS Biol. 2005;3:e68. doi: 10.1371/journal.pbio.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- Stepanyants A, Chklovskii DB. Neurogeometry and potential synaptic connectivity. Trends Neurosci. 2005;28:387–394. doi: 10.1016/j.tins.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Stockinger P, Kvitsiani D, Rotkopf S, Tirian L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci USA. 2003;100:7319–7324. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratford KJ, Tarczy-Hornoch K, Martin KA, Bannister NJ, Jack JJ. Excitatory synaptic inputs to spiny stellate cells in cat visual cortex. Nature. 1996;382:258–261. doi: 10.1038/382258a0. [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Suh GS, Ben-Tabou de Leon S, Tanimoto H, Fiala A, Benzer S, Anderson DJ. Light activation of an innate olfactory avoidance response in Drosophila. Curr Biol. 2007;17:905–908. doi: 10.1016/j.cub.2007.04.046. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Suster ML, Seugnet L, Bate M, Sokolowski MB. Refining GAL4-driven transgene expression in Drosophila with a GAL80 enhancer-trap. Genesis. 2004;39:240–245. doi: 10.1002/gene.20051. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Denk W, Kleinfeld D, Tank DW. In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature. 1997;385:161–165. doi: 10.1038/385161a0. [DOI] [PubMed] [Google Scholar]
- Sweeney ST, Broadie K, Keane J, Niemann H, O’Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- Szobota S, Gorostiza P, Del Bene F, Wyart C, Fortin DL, Kolstad KD, Tulyathan O, Volgraf M, Numano R, Aaron HL, et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–545. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single ’self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Takao K, Okamoto K, Nakagawa T, Neve RL, Nagai T, Miyawaki A, Hashikawa T, Kobayashi S, Hayashi Y. Visualization of synaptic Ca2+ /calmodulin-dependent protein kinase II activity in living neurons. J Neurosci. 2005;25:3107–3112. doi: 10.1523/JNEUROSCI.0085-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallini YN, Ohkura M, Choi BR, Ji G, Imoto K, Doran R, Lee J, Plan P, Wilson J, Xin HB, et al. Imaging cellular signals in the heart in vivo: Cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc Natl Acad Sci USA. 2006;103:4753–4758. doi: 10.1073/pnas.0509378103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EM, Yamaguchi Y, Horwitz GD, Gosgnach S, Lein ES, Goulding M, Albright TD, Callaway EM. Selective and quickly reversible inactivation of mammalian neurons in vivo using the Drosophila allatostatin receptor. Neuron. 2006;51:157–170. doi: 10.1016/j.neuron.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Tan W, Janczewski WA, Yang P, Shao XM, Callaway EM, Feldman JL. Rapid silencing of preBötzinger complex somatostatin-expressing neurons induces persistent apnea in adult awake rats. Nat Neurosci. 2008 doi: 10.1038/nn.2104. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehovnik EJ, Tolias AS, Sultan F, Slocum WM, Logothetis NK. Direct and indirect activation of cortical neurons by electrical microstimulation. J Neurophysiol. 2006;96:512–521. doi: 10.1152/jn.00126.2006. [DOI] [PubMed] [Google Scholar]
- Thomson AM, Bannister AP. Interlaminar connections in the neocortex. Cereb Cortex. 2003;13:5–14. doi: 10.1093/cercor/13.1.5. [DOI] [PubMed] [Google Scholar]
- Thomson JG, Ow DW. Site-specific recombination systems for the genetic manipulation of eukaryotic genomes. Genesis. 2006;44:465–476. doi: 10.1002/dvg.20237. [DOI] [PubMed] [Google Scholar]
- Tobin D, Madsen D, Kahn-Kirby A, Peckol E, Moulder G, Barstead R, Maricq A, Bargmann C. Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron. 2002;35:307–318. doi: 10.1016/s0896-6273(02)00757-2. [DOI] [PubMed] [Google Scholar]
- Tomioka R, Rockland KS. Improved Golgi-like visualization in retrogradely projecting neurons after EGFP-adenovirus infection in adult rat and monkey. J Histochem Cytochem. 2006;54:539–548. doi: 10.1369/jhc.5A6838.2005. [DOI] [PubMed] [Google Scholar]
- Tour O, Adams SR, Kerr RA, Meijer RM, Sejnowski TJ, Tsien RW, Tsien RY. Calcium Green FlAsH as a genetically targeted small-molecule calcium indicator. Nat Chem Biol. 2007;3:423–431. doi: 10.1038/nchembio.2007.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, Bargmann CI. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell. 1999;99:387–398. doi: 10.1016/s0092-8674(00)81525-1. [DOI] [PubMed] [Google Scholar]
- Tsetsenis T, Ma XH, Lo lacono L, Beck SG, Gross C. Suppression of conditioning to ambiguous cues by pharmacogenetic inhibition of the dentate gyrus. Nat Neurosci. 2007;10:896–902. doi: 10.1038/nn1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Ugolini G. Specificity of rabies virus as a transneuronal tracer of motor networks: transfer from hypoglossal motoneurons to connected second-order and higher order central nervous system cell groups. J Comp Neurol. 1995a;356:457–480. doi: 10.1002/cne.903560312. [DOI] [PubMed] [Google Scholar]
- Ugolini G. Transneuronal tracing with alpha-herpesviruses: A review of the methodology. In: Kaplitt MG, Loewy AD, editors. Viral Vectors. San Diego, CA: Academic Press; 1995b. pp. 293–317. [Google Scholar]
- Verhagen JV, Wesson DW, Netoff TI, White JA, Wachowiak M. Sniffing controls an adaptive filter of sensory input to the olfactory bulb. Nat Neurosci. 2007;10:631–639. doi: 10.1038/nn1892. [DOI] [PubMed] [Google Scholar]
- Verma IM, Weitzman MD. Gene therapy: twenty-first century medicine. Annu Rev Biochem. 2005;74:711–738. doi: 10.1146/annurev.biochem.74.050304.091637. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- Wade-Martins R, Smith ER, Tyminski E, Chiocca EA, Saeki Y. An infectious transfer and expression system for genomic DNA loci in human and mouse cells. Nat Biotechnol. 2001;19:1067–1070. doi: 10.1038/nbt1101-1067. [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Wang Y, O’Malley BW, Jr, Tsai SY, O’Malley BW. A regulatory system for use in gene transfer. Proc Natl Acad Sci USA. 1994;91:8180–8184. doi: 10.1073/pnas.91.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Guo H, Pologruto TA, Hannan F, Hakker I, Svoboda K, Zhong Y. Stereotyped odor-evoked activity in the mushroom body of drosophila revealed by green fluorescent protein-based ca2+ imaging. J Neurosci. 2004;24:6507–6514. doi: 10.1523/JNEUROSCI.3727-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol. 2004 doi: 10.1016/j.cub.2004.03.035. in press. [DOI] [PubMed] [Google Scholar]
- Wes PD, Bargmann CI. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature. 2001;410:698–701. doi: 10.1038/35070581. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007a;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007b;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CJ, Groves PM. Spontaneous firing patterns of identified spiny neurons in the rat neostriatum. Brain Res. 1981;220:67–80. doi: 10.1016/0006-8993(81)90211-0. [DOI] [PubMed] [Google Scholar]
- Wilson CJ, Chang HT, Kitai ST. Origins of post synaptic potentials evoked in spiny neostriatal projection neurons by thalamic stimulation in the rat. Exp Brain Res. 1983;51:217–226. doi: 10.1007/BF00237197. [DOI] [PubMed] [Google Scholar]
- Wilson JM, Dombeck DA, Diaz-Rios M, Harris-Warrick RM, Brownstone RM. Two-photon calcium imaging of network activity in XFP-expressing neurons in the mouse. J Neurophysiol. 2007;97:3118–3125. doi: 10.1152/jn.01207.2006. [DOI] [PubMed] [Google Scholar]
- Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Wulff P, Goetz T, Leppa E, Linden AM, Renzi M, Swinny JD, Vekovischeva OY, Sieghart W, Somogyi P, Korpi ER, et al. From synapse to behavior: rapid modulation of defined neuronal types with engineered GABA(A) receptors. Nat Neurosci. 2007;10:923–929. doi: 10.1038/nn1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaksi E, Friedrich RW. Reconstruction of firing rate changes across neuronal populations by temporally deconvolved Ca(2+) imaging. Nat Methods. 2006;3:377–383. doi: 10.1038/nmeth874. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Wada N, Kitabatake Y, Watanabe D, Anzai M, Yokoyama M, Teranishi Y, Nakanishi S. Reversible suppression of glutamatergic neurotransmission of cerebellar granule cells in vivo by genetically manipulated expression of tetanus neurotoxin light chain. J Neurosci. 2003;23:6759–6767. doi: 10.1523/JNEUROSCI.23-17-06759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda R, Harvey CD, Zhong H, Sobczyk A, van Aelst L, Svoboda K. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat Neurosci. 2006;9:283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, Mizuno T, Nakahira M, Kawasaki M, Watanabe Y, Kagamiyama H, Jishage K, Ueda O, Suzuki H, Tabuchi K, et al. A genetic approach to visualization of multisynaptic neural pathways using plant lectin transgene. Neuron. 1999;22:33–41. doi: 10.1016/s0896-6273(00)80676-5. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Dantzker JL, Callaway EM. Excitatory cortical neurons form fine-scale functional networks. Nature. 2005;433:868–873. doi: 10.1038/nature03252. [DOI] [PubMed] [Google Scholar]
- Yuste R, Peinado A, Katz LC. Neuronal domains in developing neocortex. Science. 1992;257:665–669. doi: 10.1126/science.1496379. [DOI] [PubMed] [Google Scholar]
- Zemelman BV, Lee GA, Ng M, Miesenbock G. Selective photostimulation of genetically chARGed neurons. Neuron. 2002;33:15–22. doi: 10.1016/s0896-6273(01)00574-8. [DOI] [PubMed] [Google Scholar]
- Zemelman BV, Nesnas N, Lee GA, Miesenbock G. Photochemical gating of heterologous ion channels: remote control over genetically designated populations of neurons. Proc Natl Acad Sci USA. 2003;100:1352–1357. doi: 10.1073/pnas.242738899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zhang YP, Oertner TG. Optical induction of synaptic plasticity using a light-sensitive channel. Nat Methods. 2006;4:139–141. doi: 10.1038/nmeth988. [DOI] [PubMed] [Google Scholar]
- Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]