Abstract
Myotubularin-related protein 6 (MTMR6) is a catalytically active member of the myotubularin (MTM) family, which is composed of 14 proteins. Catalytically active myotubularins possess 3-phosphatase activity dephosphorylating phosphatidylinositol-3-phoshate and phosphatidylinositol-3,5-bisphosphate, and some members have been shown to form homomers or heteromeric complexes with catalytically inactive myotubularins. We demonstrate that human MTMR6 forms a heteromer with an enzymatically inactive member myotubularin-related protein 9 (MTMR9), both in vitro and in cells. MTMR9 increased the binding of MTMR6 to phospholipids without changing the lipid binding profile. MTMR9 increased the 3-phosphatase activity of MTMR6 up to 6-fold. We determined that MTMR6 is activated up to 28-fold in the presence of phosphatidylserine liposomes. Together, MTMR6 activity in the presence of MTMR9 and assayed in phosphatidylserine liposomes increased 84-fold. Moreover, the formation of this heteromer in cells resulted in increased protein levels of both MTMR6 and MTMR9, probably due to the inhibition of degradation of both proteins. Furthermore, co-expression of MTMR6 and MTMR9 decreased etoposide-induced apoptosis, whereas decreasing both MTMR6 and MTMR9 by RNA interference led to increased cell death in response to etoposide treatment when compared with that seen with RNA interference of MTMR6 alone. Thus, MTMR9 greatly enhances the functions of MTMR6.
Myotubularin proteins are a family of 14 proteins with the canonical dual specificity protein tyrosine phosphatase active site CX5R motif (1–3). Eight members of the myotubularin family possess catalytic activity, dephosphorylating phosphatidylinositol 3-phosphate (PtdIns-3-P)4 and phosphatidylinositol 3,5-bisphosphate (PtdIns-3,5-P2) at the D-3 position, and six members are not catalytically active because they lack the conserved cysteine residue in the protein tyrosine phosphatase motif that is required for activity. Interest in this group of proteins originated from the genetic evidence linking myotubularin, the founding member of this family, to myotubular myopathy, an X-linked disorder characterized by severe hypotonia and generalized muscle weakness (4). Subsequently, mutations in MTMR2 and in its inactive binding partner MTMR13 were linked to a subset of Charcot-Marie-Tooth disease type 4B, a demyelinating neurodegenerative disorder (5, 6).
Despite near identical substrate specificity, biochemical and genetic evidence supports the hypothesis that myotubularin proteins are not redundant and have unique functions within cells (2, 7–9). The mechanisms by which loss of function of myotubularin proteins produce diseases are not known. Current evidence supports the hypothesis that each myotubularin protein regulates a specific pool of PtdIns-3-P and/or PtdIns-3,5-P2, which in turn regulates a variety of cellular functions. Differences in tissue expression and subcellular localization play a role in the specificity of different myotubularins (10–15).
The functions of myotubularin proteins are altered by the formation of heteromers between catalytically active and inactive members of the family. The initial biochemical purification of MTM1 demonstrated the presence of MTM1 homodimers and MTM1-3-phosphatase adapter protein (3PAP) heteromers (16), which was later described as MTMR12 (15, 17). MTMR2 was found to form heteromers with MTMR5 (13) and MTMR13 (18), and MTMR7 formed heteromers with MTMR9 (19). In each case, a catalytically active myotubularin protein interacted with an inactive protein. Heteromerization generated two important effects: increased catalytic activity of the active component (13, 15, 19, 20) and targeting of the heteromer to specific subcellular locations (15). Mutations in the inactive member MTMR13 result in a similar phenotype in patients as the mutations in its catalytically active binding partner MTMR2, indicating an indispensable role for the catalytically inactive subunit (21).
Myotubularin proteins can be grouped into subfamilies based on homology. Closely related MTMR6, MTMR7, and MTMR8 comprise such a subfamily. We have previously characterized the interaction between mouse MTMR7 and MTMR9 proteins (19). In this report, we characterize the interaction between human MTMR6 and MTMR9. MTMR6 and MTMR9 have been shown to form a heteromeric complex in mouse and Caenorhabditis elegans (19, 22). MTMR6 has been shown to inhibit the activity of a calcium-activated potassium channel (type KCa3.1) (23, 24). Two screening experiments implicate MTMR6 as a regulator of apoptosis. By RNA microarray analysis, increased MTMR6 expression was observed in B cell chronic lymphoid leukemia cells with increased resistance to irradiation-induced apoptosis (25), whereas in an RNA interference screen in HeLa cells, decreased MTMR6 expression promoted apoptosis (26).
Here we show that MTMR6 interacts with MTMR9 in vitro and in human cells. This interaction increases the phospholipid binding and enzymatic activity of MTMR6 in vitro. Co-expression of either subunit in cells dramatically increased the protein levels of the individual binding partners, suggesting that heteromer formation increases the stability of the proteins. Finally, MTMR9 was found to potentiate the effects of MTMR6 on apoptosis.
EXPERIMENTAL PROCEDURES
Reagents and Chemicals—All chemicals and reagents, unless specifically noted, were purchased from Sigma-Aldrich.
Cell Culture, Transfection, and Treatment—HeLa cells were maintained in culture using 10% fetal bovine serum in Dulbecco's modified Eagle's medium. Unless noted, transfection was conducted by using Lipofectamine 2000 (Invitrogen). RNAi transfections were done using a Nucleofector kit (Amaxa, Gaithersburg, MD). The RNAi duplexes were obtained from Ambion (Austin, TX), and sequences are: control RNAi (luciferase) duplex, sense 5′-CUUACGCUGAGUACUUCGAdTdT-3′; antisense, 5′-UCGAAGUACUCAGCGUAAGDTdT-3′; MTMR6 RNAi (R6-1), sense, 5′-GGAAGTCAATGGCACTAATgg-3′; antisense, 5′-TTTAGTGCCATTGACTTCCaa-3′; MTMR9 RNAi (R9-1), 5′-CAAAGGAGGTGGCTTTGA Tca-3′ and 5′-TCAAAGCCACCTCCTTTGgc-3′. The specificity and efficacy were determined using quantitative reverse transcription-PCR (data not shown). There was about 50% reduction upon the RNAi treatment in both MTMR6 and MTMR9, and no cross-reaction was detected.
Apoptosis was induced by treatment with 100 μm etoposide for the indicated times. Stably transfected cell lines overexpressing MTMR6 or MTMR9 were generated by transfecting MTMR6 or MTMR9 constructs, as described previously (27), into HEK-293 TRex cells (Invitrogen).
Cloning, Expression, and Purification of Human MTMR6 and MTMR9—Full-length human MTMR6 and MTMR9 were cloned by PCR using the Marathon-Ready human brain cDNA library (Clontech) as template. Primers were designed to clone the entire open reading frame according to the published DNA sequence for MTMR6 (NM_004685, GenBank™) and MTMR9 (NM_015458, GenBank). FLAG peptide fusion constructs were made by subcloning PCR products into pBak-Pak9-HF plasmid (BD Biosciences) for baculovirus expression and protein purification. HA peptide and FLAG peptide fusion constructs were generated by adding the HA peptide and FLAG peptide DNA sequences to the C terminus of both MTMR6 and MTMR9 followed by a stop codon in pcDNA4/TO plasmid. MTMR6 and MTMR9 were also subcloned into a pGEX-6p-1 vector (Amersham Biosciences) with a GST tag on the N terminus. All constructs were verified by DNA sequencing. Expression and purification of human MTMR6 and MTMR9 were conducted in Sf9 cells as described previously (19).
GST Binding Assays, Co-immunoprecipitation Assays, and Western Blot Analysis—For GST binding assays, GST, or a GST fusion protein (100 μg) was bound to glutathione beads (Amersham Biosciences) and incubated with 500 ng of MTMR9-FLAG or MTMR6-FLAG in assay buffer (25 mm Hepes, pH 7.2, 125 mm potassium acetate, 2.5 mm magnesium acetate, 1 mm dithiothreitol, 0.4% Triton X-100, and Complete Protease Inhibitor Mix (Roche Applied Science)) on a rotator at 4 °C overnight. For in vitro co-immunoprecipitation assays, MTMR9-FLAG (100 μg) was bound to anti-FLAG M2 affinity gel (Sigma) and incubated with GST-MTMR6 in assay buffer on a rotator at 4 °C overnight. The beads were then washed four times with assay buffer before 2× SDS-PAGE sample buffer was added. Co-immunoprecipitation assays using HeLa cells, transfected with both MTMR6 (HA-tagged) and MTMR9 (FLAG-tagged) for 24 h, were conducted as described (28). Rabbit polyclonal antibodies against MTMR6 and MTMR9 were raised against a peptide spanning amino acid residues 519–546 in MTMR6 and 211–231 in MTMR9.
Immunofluorescence Microscopy—Twenty-four hours after transfection, HeLa cells grown on coverslips were fixed as described previously (29) and then washed with Tris-buffered saline and solubilized with 0.5% Triton X-100 in phosphate-buffered saline for 10 min. Primary antibody and secondary antibody were diluted in phosphate-buffered saline, 0.1% Triton X-100, 5% BSA. Cells were incubated for 1 h with primary and for 30 min with secondary antibodies at 37 °C. Following antibody incubations, washes were performed by dipping coverslips into phosphate-buffered saline 30 times. Cells were mounted in Prolong mounting medium (Molecular Probes). Images were taken by using an Olympus IX70 inverted microscope and processed with the Metamorph software (Molecular Devices, Sunnyvale, CA).
Phospholipid Binding Using PIP Strip and PIP Array—PIP strip and PIP array membranes (Echelon Biosciences) were blocked in Tris-buffered saline-T (0.05% Tween 20 in Tris-buffered saline) + 3% fatty acid-free BSA (Sigma) for 1 h and then incubated with 0.5 μg/ml protein for 2 h at room temperature. In assays containing both MTMR6-FLAG and MTMR9-FLAG proteins, the two proteins were incubated together for 30 min at 4 °C before being applied to the membrane. The membrane was incubated with anti-FLAG antibody and then with horseradish peroxidase-conjugated anti-IgG mouse antibody (Pierce). Both primary and secondary antibodies were diluted in Tris-buffered saline-T + 3% fatty acid-free BSA, and the membranes were washed three times with Tris-buffered saline-T + 3% fatty acid-free BSA after each step. Bands were visualized using SuperSignal West Pico chemiluminescent substrate.
3-Phosphatase Activity Assay—For determination of pH optima, [32P]PtdIns-3-P (∼20,000 cpm/assay), unlabeled PtdIns-3-P, and PS were dried under nitrogen in a siliconized microcentrifuge tube and resuspended in 25 mm buffer (MES for pH 5.5, 6.0, and 6.5, MOPS for pH 7.0, 7.5, and 8.0, or CHES for pH 8.5 and 9.0), containing 125 mm sodium chloride, 1 mm dithiothreitol, and 50 μg/ml BSA followed by brief sonication. Enzyme assays were performed in a reaction volume of 50 μl containing 1 μm PtdIns-3-P and 100 μm PS. The activity assay was performed as described previously (30).
For kinetic studies, enzyme assays were performed in a reaction volume of 50 μl containing 0.1 μm PtdIns-3-P and 100 μm carrier lipids as described. Reactions were started by the addition of 100 ng of enzyme and carried out for 0, 10, 20, 30, 40, 50, and 60 s at 37 °C and were terminated by the addition of 500 μl of 10% trichloroacetic acid.
Protein Stability Assay—HeLa cells were transfected with either MTMR6 alone or MTMR6 + MTMR9 using Lipofectamine 2000 (Invitrogen). For stably transfected HEK-293 TRex cells overexpressing MTMR6 or MTMR9, 0.5 μg/ml tetracycline was added into the medium 6 h after the transient transfection was conducted with vector alone or MTMR6 or MTMR9. Twenty-four hours after transfection, cycloheximide (150 μg/ml) was added to halt protein synthesis. Cells were harvested at the indicated time points. Total protein concentrations of cell lysates were measured, and immunoblotting was conducted. Quantification was performed by using the ImageJ software.
Apoptotic Cell Staining—HeLa cells treated with RNAi were grown on 6-well plates. Thirty-six hours later, cells at 90–95% confluence were treated with 100 μm etoposide for 8 h. Apoptotic cells were detected using the APOPercentage apoptotic kit (Accurate Chemical, Westbury, NY) according to the manufacturer's instruction. The cells that are undergoing apoptosis selectively import the APOPercentage dye that accumulates on the cell.
FACS—Four million HeLa cells were transfected with a total of 4 μl of RNAi oligonucleotides (100 μm): control (luciferase) RNAi (4 μl), MTMR6 RNAi (2 μl of R6 RNAi and 2 μl of control RNAi), MTMR9 RNAi (2 μl of R9 RNAi and 2 μl of control RNAi), and MTMR6 +R9 RNAi (2 μl of R6 RNAi and 2 μl of MTMR9 RNAi). Apoptotic cells were detected with an annexin V-FITC apoptosis detection kit (Calbiochem) and analyzed on a BD FACScan.
RESULTS
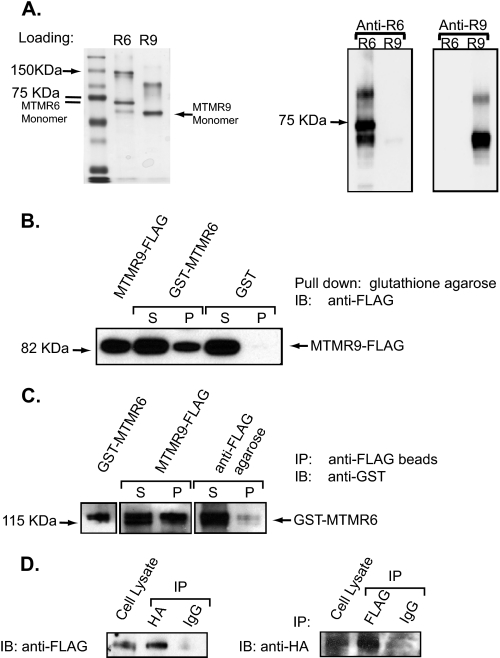
Human MTMR6 and MTMR9 Proteins Form a Heteromeric Complex—Previously, it was demonstrated that mouse MTMR7 interacts with MTMR9 resulting in enhanced 3-phosphatase activity of MTMR7, and MTMR6 interacts with MTMR9 in mouse and C. elegans (19, 22). In addition, MTM1 and MTMR2 have been shown to form homodimers as well as heteromers with catalytically inactive myotubularin proteins (12, 16). To investigate whether the human orthologs of MTMR6 and MTMR9 interact, we cloned by PCR the human MTMR6 (NM_004685) and MTMR9 (NM_015458) cDNAs based on sequences available from the GenBank data base, from a human whole brain cDNA library. Human MTMR6 and MTMR9 proteins were expressed in Sf-9 insect cells, and the respective proteins were purified using an anti-FLAG affinity column (Fig. 1A, left panel). Both MTMR6 and MTMR9 appear to form homodimers as the band corresponding to the molecular weight of a dimer is recognized by the MTMR6- and MTMR9-specific antibodies in a Western blot (Fig. 1A, right panel).
FIGURE 1.
Interaction between MTMR6 and MTMR9. A, purified FLAG-MTMR6 (R6) and FLAG-MTMR9 (R9) were analyzed by SDS-PAGE. The gels were either stained with Coomassie Blue (left panel) or transferred to nitrocellulose membrane and blotted with anti-MTMR6 or anti-MTMR9 antibody. B, purified GST-MTMR6 and MTMR9-FLAG were incubated with glutathione-conjugated beads. MTMR9-FLAG input was loaded in the left lane, and the unbound (S) and bound (P) fractions of MTMR9-FLAG to GST-MTMR6 and to GST were loaded as marked. IB, immunoblotting. C, purified MTMR9-FLAG and GST-MTMR6 were incubated with anti-FLAG beads. GST-MTMR6 (input), unbound (S), and bound (P) fractions of GST-MTMR6 to MTMR9-FLAG were visualized by anti-GST Western blot. Anti-FLAG beads incubated with GST-MTMR6 was used as a negative control. IP, immunoprecipitation. D, HeLa cells were co-transfected with MTMR6-HA and MTMR9-FLAG. Twenty-four hours after transfection, HA-tagged or FLAG-tagged proteins were immunoprecipitated with anti-HA and anti-FLAG polyclonal antibodies, respectively. Western blotting was done to analyze the immunoprecipitates using antibodies against FLAG or HA. Rabbit IgG was used as a negative control. Note: Samples in panels B and C were electrophoresed using the NuPAGE gel system (Invitrogen) in which the proteins migrate slightly differently than those on a standard Tris-glycine gel.
We verify that human MTMR6 directly interacts with MTMR9 in vitro by GST binding assay and co-immunoprecipitation experiments. GST-MTMR6 pulled down FLAG-MTMR9, suggesting that the heteromer forms between MTMR6 and MTMR9 (Fig. 1B). The reciprocal experiment using FLAG antibody demonstrated that GST-MTMR6 co-immunoprecipitated with FLAG-MTMR9 (Fig. 1C). To further confirm this interaction in the cell, we co-expressed HA-MTMR6 and FLAG-MTMR9 in HeLa cells and immunoprecipitated using anti-HA or anti-FLAG antibodies. Both MTMR6 and MTMR9 were detected in the immunoprecipitate (Fig. 1D). Thus, we conclude that MTMR6 directly binds to MTMR9 both in vitro and in cells.
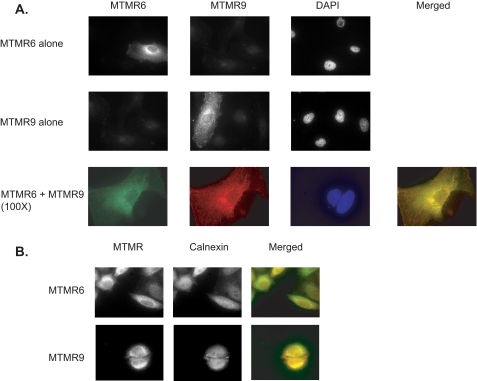
Cellular Co-localization of MTMR6 and MTMR9—Several lines of evidence suggest that the association between catalytically active and inactive members of the myotubularin family regulates the enzymatic profile by either altering the enzyme activity and/or subcellular localization (13, 15). To test whether MTMR6 could co-localize with MTMR9 or whether the subcellular distribution pattern of MTMR6 could be altered upon overexpression of MTMR9, we expressed MTMR6, MTMR9, or both in HeLa cells and performed immunofluorescence. As shown in Fig. 2A, MTMR6 has a very similar subcellular localization pattern to MTMR9 as both accumulate in the perinuclear region, although MTMR6 shows less punctate distribution in the cytosolic compartment when compared with MTMR9. When MTMR6 and MTMR9 were co-expressed, MTMR6 staining clearly overlapped with the MTMR9 staining. We conclude that MTMR6 co-localizes with MTMR9, and we did not observe any alteration of MTMR6 distribution upon the overexpression of MTMR9 or vice versa. We also investigated localization of both MTMR6 and MTMR9 with a panel of cytosolic organelle markers, including Lamp-1, γ-adaptin, early endosome antigen 1(EEA1) and calnexin. None of the tested markers overlapped perfectly with MTMR6 or MTMR9. As shown in Fig. 2B, both MTMR6 and MTMR9 staining partially co-localized with calnexin, suggesting that they may be enriched in the endoplasmic reticulum.
FIGURE 2.
MTMR6 co-localizes with MTMR9. A, HeLa cells were transfected with MTMR6-HA, MTMR9-FLAG, or both. After fixation, the cells were co-stained with anti-HA (rabbit) antibody and anti-FLAG monoclonal antibody conjugated with cy3 followed by Alexa Fluor 488-anti-rabbit antibody. The co-localization images are shown at high magnification (×100). 4′,6-Diamidino-2-phenylindole (DAPI) staining shows nuclei. B, HeLa cells were transfected with MTMR6-HA or MTMR9-FLAG. After fixation, the cells were co-stained with anti-HA (rabbit) polyclonal antibody (MTMR6 panel) or anti-FLAG (rabbit) polyclonal antibody (MTMR9 panel) and anti-calnexin antibody (mouse) monoclonal antibody followed by Alexa Fluor 488-anti-rabbit antibody and Alexa Fluor 555-anti-mouse antibody.
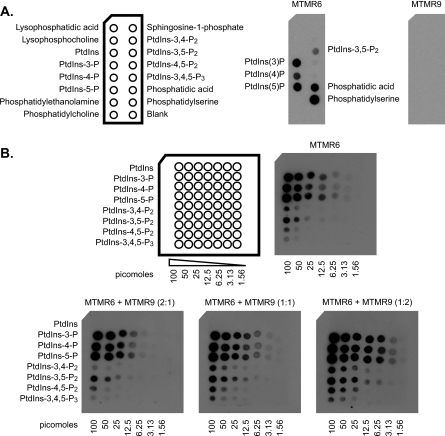
Phospholipid Binding of MTMR6 Is Increased by MTMR9—Given the interaction of MTMR6 with MTMR9, we determined whether the complex formation had an effect on lipid binding of MTMR6 and MTMR9. Lipid overlay assays were performed with purified recombinant MTMR6 and MTMR9 using PIP strips. MTMR6 bound all monophosphorylated phosphatidylinositols (PtdIns-3-P, PtdIns-4-P, and PtdIns-5-P), PtdIns-3,5-P2, phosphatidic acid, and PS; however, MTMR9 did not demonstrate any significant binding to phospholipids (Fig. 3). Similar results were observed using a PIP array. The affinity of MTMR6 binding to individual monophosphorylated phosphatidylinositols was similar to but significantly greater than that of polyphosphorylated phosphatidylinositols (Fig. 3A); of the polyphosphorylated phosphatidylinositols, MTMR6 bound to its substrate Ptd-3,5-P2 with the greatest affinity. When MTMR6 was preincubated with MTMR9 for 30 min, the lipid binding of MTMR6 was significantly increased in an MTMR9 concentration-dependent manner (Fig. 3B). Thus, the interaction of MTMR9 with MTMR6 significantly enhances the binding properties of MTMR6 toward negatively charged phospholipids.
FIGURE 3.
MTMR6 phospholipid binding is increased by MTMR9. A, recombinant MTMR6-FLAG (0.5 μg/ml) or MTMR9-FLAG proteins (0.5 μg/ml) were used to probe the PIP strips. Bound protein is indicated by Western blot with the appropriate antibody. Phospholipids bound by MTMR6 are indicated in the middle panel. MTMR9 did not bind to any lipid on the membrane (far right panel). A schematic diagram of the PIP Strip membrane with the phospholipids is indicated on the left. PtdIns-3,4-P2, phosphatidylinositol 3,4-bisphosphate; PtdIns-4,5-P2, phosphatidylinositol 4,5-bisphosphate; PtdIns-3,4,5-P2, phosphatidylinositol 3,4,5-trisphosphate. B, a schematic diagram of the PIP array membrane containing serial dilutions of the indicated phosphatidylinositol phosphates (in picomoles) is shown on the left. MTMR6-FLAG (0.5 μg/ml) and MTMR6-FLAG + MTMR9-FLAG were used to probe the PIP arrays. The ratio of MTMR6 to MTMR9 is as noted. Bound proteins were visualized by anti-FLAG antibody.
Catalytic Activity of MTMR6 Is Increased by MTMR9—We determined the pH optimum for MTMR6 catalytic activity to be 7.0, differing from the previously published pH optimum of 6.5 (31). The pH optimum of MTMR6 was not changed by the addition of MTMR9 (data not shown). To investigate the effect of the MTMR6 and MTMR9 interaction on the 3-phosphatase activity of MTMR6, we determined the first-order rate constant, as measured by the release of [32P]PO4 from PtdIns-3-P (Table 1). Given the strong lipid binding of MTMR6 to PS (Fig. 3), we initially examined the 3-phosphatase activity in the presence of increasing amounts of PS in liposomes. By increasing PS from 0 to 50% and then to 100%, the first-order rate constant of MTMR6 increased to 1.8- and 28.1-fold of the level without PS, respectively. In the presence of MTMR9, the catalytic activity of MTMR6 increased under both PS concentrations tested; the activity increased by 1.7-fold for 0% PS, 6.4-fold for 50% PS, and 3-fold for 100% PS. In the presence of MTMR9 and 100% PS, MTMR6 activity is increased by 84-fold when compared with that seen in 50% phosphatidylcholine, 50% phosphatidylethanolamine in the absence of MTMR9. These results show the requirement for PS and MTMR9 to achieve maximal catalytic activity of MTMR6.
TABLE 1.
First-order rate constant of MTMR6
PC, phosphatidylcholine; PE, phosphatidylethanolamine; ND, not determined.
|
First-order rate constant (×10-5 s-1)
|
|||
|---|---|---|---|
| 1% PtdIns-4-P | 1% PtdIns-5-P | ||
| 50% PC/50% PE | |||
| MTMR6 | 4.1 | ND | ND |
| MTMR6 + MTMR9 | 7.1 | ND | ND |
| 50% PC/50% PS | |||
| MTMR6 | 7.5 | 13.8 | 11 |
| MTMR6 + MTMR9 | 48.3 | 344 | 30 |
| 100% PS | |||
| MTMR6 | 115 | 212 | 288 |
| MTMR6 + MTMR9 | 344 | 481 | 565 |
Previously, Schaletzky et al. (31) demonstrated that PtdIns-5-P was an allosteric activator of MTM1, MTMR3, and MTMR6. Given that MTMR6 binds to both PtdIns-4-P and PtdIns-5-P (Fig. 3), we examined MTMR6 activity with and without MTMR9 in the presence of PtdIns-4-P or PtdIns-5-P (Table 1). PtdIns-4-P increased MTMR6 activity 1.8-fold in both 50% PS and 100% PS, and PtdIns-5-P increased MTMR6 activity 1.5- and 2.5-fold in 50 and 100% PS, respectively. In the presence of MTMR9, both PtdIns-4-P and PtdIns-5-P had an inhibitory effect on MTMR6 activity in 50% PS; the 3-phosphatase activity decreased 30 and 40% for PtdIns-4-P and PtdIns-5-P, respectively. However, both PtdIns-4-P and PtdIns-5-P increased MTMR6 activity in the presence of MTMR9 in 100% PS. These results underscore the importance of PS, allosteric modifying phospholipids, and MTMR9 in modulating MTMR6 activity.
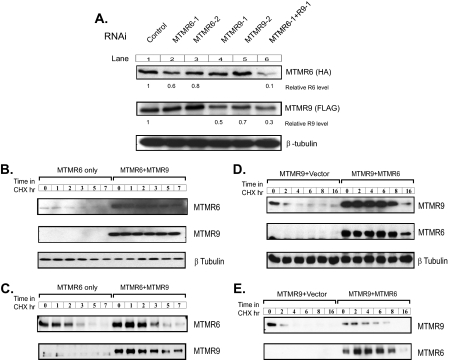
MTMR6 and MTMR9 Protein Stability Is Increased by the Formation of the Complex—In transfected cells, significantly higher levels of either MTMR6 or MTMR9 protein were always observed in the presence of its binding partner (Fig. 4, B and D, 0 time points). In other words, co-expression of both MTMR6 and MTMR9 resulted in much higher protein levels for both proteins than overexpression of MTMR6 or MTMR9 alone when the same amount of plasmid DNA was used for transfection and equal amounts of total protein were loaded. RNAi of MTMR6 reduced protein expression 20–40%, but a much greater reduction in the level of MTMR6 (up to 90%) was seen when RNAi of both proteins was carried out as seen in Fig. 4A (lanes 2 and 6). The levels of MTMR9 protein were also further reduced by a combination of RNAi oligonucleotides targeting both binding partners (Fig. 4A, compare lanes 4, 5, and 6). This also suggests that the formation of the MTMR6 and MTMR9 complex stabilizes the proteins, possibly by decreasing the degradation rates. To test this, we chased MTMR6 in cycloheximide-treated HeLa cells in the presence or absence of MTMR9 and observed a higher protein level and slower turnover rate of MTMR6 in cells co-expressing both proteins. Based on the intensity of Western blots, the half-life of MTMR6 was increased from ∼40 min to 4 h when MTMR9 was co-expressed in HeLa cells (Fig. 4, B and C). To rule out the possibility of signal over saturation on the Western blot, we artificially loaded 3-fold more total protein of the MTMR6 only samples and analyzed it after a very light exposure. As shown in Fig. 4C, in the absence of MTMR9, the level of MTMR6 decreased dramatically faster despite an almost identical MTMR6 protein level at the zero time point. We next asked whether MTMR9 stability was also enhanced when MTMR6 was present using a stably transfected TRex-293 cell line expressing recombinant MTMR9 under regulation of tetracycline. Six hours after the transient transfection with MTMR6, MTMR9 expression was turned on by the addition of tetracycline. Consistently, we observed more stable MTMR9 in the presence of MTMR6 (Fig. 4, D and E). Again, we artificially loaded 3-fold more total protein of the MTMR9 only samples to rule out the possibility of signal over saturation on the Western blot (Fig. 4E).
FIGURE 4.
Stability of MTMR6 and MTMR9 is increased by the formation of the complex. A, HeLa cells transfected with MTMR6-HA and MTMR9-FLAG for 24 h were treated with control or RNAi against MTMR6 (R6-1, R6-2), MTMR9 (R9-1, R9-2), or both (R6-1 + R9-1) for an additional 24 h. Cell lysates were analyzed by Western blotting for MTMR6 (HA) or MTMR9 (FLAG). β-tubulin levels are shown as a loading control. Relative R6 level, relative MTMR6 level; Relative R9 level, relative MTMR9 level. B, HeLa cells transfected with MTMR6-HA, or MTMR6-HA plus MTMR9-FLAG for 24 h. One hundred and fifty μg/ml cycloheximide (CHX) was added into the medium for the times indicated. The levels of MTMR6 and MTMR9 were detected by Western blotting with an anti-HA or anti-FLAG antibody, respectively. C, the same as B except with MTMR6 alone; the left six lanes contain 3-fold more total protein. D, HEK-293 cells stably transfected with MTMR9-FLAG were transiently transfected with MTMR6-HA. Six hours after the transfection, 0.5 μg/ml tetracycline was added into medium to induce the expression of MTMR9-FLAG. Eighteen hours later, cells were treated with cycloheximide for the indicated times. The levels of both MTMR9 and MTMR6 were detected by Western blotting with anti-FLAG or anti-HA antibody, respectively. Each sample contains equal amounts of total protein. E, the same as C except with MTMR9 alone; the left six lanes contain 3-fold more total protein.
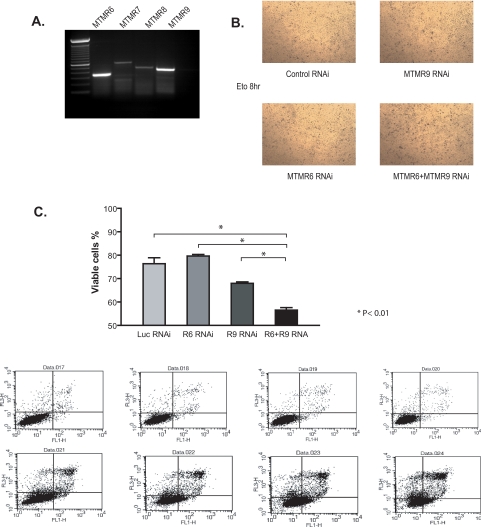
The Anti-apoptotic Function of MTMR6 Is Modulated by MTMR9—In B cell chronic lymphoid leukemia, increased resistance to irradiation-induced apoptosis was linked to increased expression of MTMR6 by RNA microarray analysis (25), and in HeLa cells, decreased expression of MTMR6 after RNA interference induced apoptosis (26). From these results, we postulated that MTMR6 regulates apoptosis, and we investigated whether MTMR9 could enhance this function of MTMR6. In HeLa cells in which both endogenous MTMR6 and MTMR9 are present (Fig. 5A), we selectively reduced the levels of MTMR6, MTMR9, and both by RNAi. Apoptotic cell death upon treatment with etoposide was measured by FACS analysis in cells depleted of MTMR6, MTMR9, or both MTMR6 and MTMR9 with RNAi. Down-regulation of MTMR6 alone did not have any significant effect on cell viability when compared with control RNAi as seen in Fig. 5C. However, when combined with MTMR9 RNAi, MTMR6 depletion evoked increased apoptosis as shown in Fig. 5, B and C. Interestingly, knocking down MTMR9 alone also led to a significant cell death when compared with control RNAi, implying that there might be other members of MTMR family involved in the cell death pathway that are also controlled by MTMR9. These data highlight the role of the catalytically inactive MTM members in regulating cellular functions through the interaction with active members.
FIGURE 5.
MTMR6 and MTMR9 regulate apoptosis. A, reverse transcription-PCR of MTMR subfamily members in HeLa cells (MTMR6, MTMR7, MTMR8, and MTMR9) analyzed by specific primers targeting each mRNA. B, APOPercentage staining of cells treated with equal amounts of oligonucleotides for: control RNAi, RNAi against MTMR6 + control RNAi, RNAi against MTMR9 + control RNAi, or RNAi against both MTMR6 plus MTMR9. HeLa cells were transfected with the indicated RNAi for 36 h and then treated with 100 μm etoposide (eto) to induce apoptosis for 8 h before labeling. Apoptotic cells are selectively labeled with APOPercentage dye showing as darker spots. C, HeLa cells treated with control RNAi, RNAi against MTMR6 (R6 RNAi) + control RNAi, RNAi against MTMR9 (R9 RNAi) + control RNAi, or RNAi against both MTMR6 plus MTMR9 (R6+R9 RNA). Twenty-four hours later, cells were treated with 100 μm etoposide to induce apoptosis for 16 h. Apoptotic cells were measured with annexin V-FITC, propidium iodide staining, and FACS analysis. The annexin V-FITC signal was detected by FL1 and propidium iodide fluorescence signal was detected by FL3 detector on a BD FACScan. The log of annexin V fluorescence is displayed on the x axis, and the log of propidium iodide fluorescence is on the y axis. Viable cells, defined as negative for both FITC and propidium iodide, are found in the lower left-hand quadrant of the dot plot. Quantification of FACS is shown as the bar graph, and results are expressed as the percentage of viable cells (76.4 ± 5.1 in control RNAi versus 56.5 ± 2.2 in MTMR6 + MTMR9 RNAi. n = 4, p < 0.01). Luc, luciferase.
DISCUSSION
In this report, we verify that human MTMR6 and MTMR9 form a functional heteromeric complex and characterize this interaction. Importantly, we found that MTMR9 influences MTMR6 function at multiple levels, including increasing its stability. We demonstrate that MTMR6 co-localizes with MTMR9 with a perinuclear cellular distribution. Complex formation also increased lipid binding and the catalytic activity of MTMR6. Finally, we show that heteromer formation played a role in regulating apoptosis.
MTMR6 was found to bind to all monophosphorylated phosphatidylinositols, with a weaker affinity for PtdIns and PtdIns-3,5-P2, a known myotubularin substrate. Interestingly, PS and phosphatidic acid also bind to MTMR6. MTMR9 did not bind lipids; however, complex formation between MTMR6 and MTMR9 enhanced the lipid binding of MTMR6 without changing the lipid binding profile. We obtained an MTMR6 binding profile different from that previously published possibly due the fact that Choudhury et al. (32) used only the pleckstrin homology/GRAM (glucosyltransferases, Rab-like GTPase activators, and myotubularins) domain, whereas in this study, the entire MTMR6 protein was used.
The catalytic activity of active myotubularin proteins increases when they are in a heteromeric complex with inactive binding partners. This increased activity was previously demonstrated for the MTMR7-MTMR9 complex, the MTM1-MTMR12 complex (15), the MTMR2-MTMR5 complex (13), and the MTMR2-MTMR13 complex (20). MTMR6 activity increased significantly with an increasing percentage of PS in substrate liposomes; elevating PS from 0 to 100% increased MTMR6 activity 28-fold. This is consistent with the strong PS binding observed in the lipid overlay assay, supporting the importance of PS in MTMR6 activity. The highest concentration of PS in the cell is at the inner leaflet of the plasma membrane with 15–30% PS (33–35); therefore, a site of increased MTMR6 activity may be the plasma membrane. This is consistent with the findings of Srivastava et al. (23) and Choudhury and co-workers (24, 32) that MTMR6 localized to the plasma membrane dephosphorylated PtdIns-3-P, thereby inhibiting a Ca2+-activated K+ channel, KCa3.1. Interestingly, from our high magnification immunofluorescence images, in addition to co-localizing with MTMR6, MTMR9 seems to distribute along the plasma membrane (Fig. 2). It would be of interest to characterize the effect of MTMR9 on the MTMR6-mediated regulation of the Ca2+-activated K+ channel.
MTMR9 increased MTMR6 activity under all conditions examined, resulting in 2–6-fold increases. The highest increase, 6.4-fold, was in 50% PS liposomes with the remaining conditions in the 2–4-fold range. Acting together, PS and MTMR9 can potentially increase MTMR6 activity 84-fold. Given these data, we postulate that MTMR9 functions to increase the lipid binding of MTMR6, recruiting MMTR6 to the membrane, and in the presence of both MTMR9 and local PS, significantly activating MTMR6, resulting in a decreased local pool of PtdIns-3-P.
In addition to increasing the enzymatic activity and lipid binding, we demonstrated that MTMR6 and MTMR9 are stabilized by complex formation with each other. The stability studies suggested that the turnover rate of both proteins was significantly decreased when the complex was formed. The half-life of MTMR6 is increased about 6-fold with concomitant overexpression of MTMR9. Reciprocally, the complex was also able to significantly protect MTMR9 from degradation. This phenomenon has not been previously described in the myotubularin family of proteins, and it would be interesting to investigate whether complex-forming subunits of other family members affect each other's turnover rate. Indeed, Robinson and Dixon (18) demonstrated a roughly 50% reduction in the level of MTMR2 protein in MTMR13–/– sciatic nerves, suggesting that the effect on stability could be a general phenomenon among myotubularin complex subunits. We have established that the MTMR6-MTMR9 interaction is critical for lipid binding, catalytic activity, and protein stability of MTMR6. We also found effects on apoptosis wherein depletion of both proteins promoted apoptosis. Under the conditions in this report, it seems that both need to be depleted to detect their role in the apoptotic pathway. Due to the effects on the stability of the protein complex subunits, it is possible that the efficiency of MTMR6 RNAi is higher when MTMR9 is also depleted, causing a further decrease in MTMR6 levels, lipid binding, and/or catalytic activity. Given that the MTMs have the same substrate specificity, it could also be possible that some other MTM member that binds to MTMR9 (such as MTMR7 or MTMR8) could compensate as well for the function of MTMR6 in the presence of MTMR9 and no longer keep the balance in the absence of MTMR9.
Acknowledgments
We thank Monita Wilson for assistance in baculovirus expression and protein purification, Marina Kisseleva for input on activity assays and microscopic technique, Alexander Ungewickell for critical analysis, and Peter Nicholas and Cecil Buchanan for general technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant [grant-num]HL 016634[/grant-num](to P. W. M.). This work was also supported by the American Heart Association Grant [grant-num]0475014N[/grant-num] (to S. C. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PtdIns, phosphatidylinositol; PtdIns-3-P, phosphatidylinositol 3-phosphate; PtdIns-4-P, phosphatidylinositol 4-phosphate; PtdIns-5-P, phosphatidylinositol 5-phosphate; PtdIns-3,5-P2, phosphatidylinositol 3,5-bisphosphate; MTM, myotubularin; MTMR, myotubularin-related protein; BSA, bovine serum albumin; GST, glutathione S-transferase; PS, phosphatidylserine; HA, hemagglutinin; FITC, fluorescein isothiocyanate; RNAi, RNA interference; FACS, fluorescence-activated cell sorter; MES, 4-morpholineethanesulfonic acid; MOPS, 4-morpholinepropanesulfonic acid; CHES, 2-(cyclohexylamino)-ethanesulfonic acid.
References
- 1.Begley, M. J., and Dixon, J. E. (2005) Curr. Opin. Struct. Biol. 15 614–620 [DOI] [PubMed] [Google Scholar]
- 2.Laporte, J., Bedez, F., Bolino, A., and Mandel, J. L. (2003) Hum. Mol. Genet. 12 Spec. No. 2, R285–R292 [DOI] [PubMed] [Google Scholar]
- 3.Taylor, G. S., Maehama, T., and Dixon, J. E. (2000) Proc. Natl. Acad. Sci. U. S. A 97 8910–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laporte, J., Hu, L. J., Kretz, C., Mandel, J. L., Kioschis, P., Coy, J. F., Klauck, S. M., Poustka, A., and Dahl, N. (1996) Nat. Genet. 13 175–182 [DOI] [PubMed] [Google Scholar]
- 5.Bolino, A., Muglia, M., Conforti, F. L., LeGuern, E., Salih, M. A., Georgiou, D. M., Christodoulou, K., Hausmanowa-Petrusewicz, I., Mandich, P., Schenone, A., Gambardella, A., Bono, F., Quattrone, A., Devoto, M., and Monaco, A. P. (2000) Nat. Genet. 25 17–19 [DOI] [PubMed] [Google Scholar]
- 6.Senderek, J., Bergmann, C., Weber, S., Ketelsen, U. P., Schorle, H., Rudnik-Schoneborn, S., Buttner, R., Buchheim, E., and Zerres, K. (2003) Hum. Mol. Genet. 12 349–356 [DOI] [PubMed] [Google Scholar]
- 7.Laporte, J., Blondeau, F., Buj-Bello, A., Tentler, D., Kretz, C., Dahl, N., and Mandel, J. L. (1998) Hum. Mol. Genet. 7 1703–1712 [DOI] [PubMed] [Google Scholar]
- 8.Clague, M. J., and Lorenzo, O. (2005) Traffic 6 1063–1069 [DOI] [PubMed] [Google Scholar]
- 9.Taylor, G. S., and Dixon, J. E. (2003) Methods. Enzymol. 366 43–56 [DOI] [PubMed] [Google Scholar]
- 10.Kim, S. A., Taylor, G. S., Torgersen, K. M., and Dixon, J. E. (2002) J. Biol. Chem. 277 4526–4531 [DOI] [PubMed] [Google Scholar]
- 11.Xue, Y., Fares, H., Grant, B., Li, Z., Rose, A. M., Clark, S. G., and Skolnik, E. Y. (2003) J. Biol. Chem. 278 34380–34386 [DOI] [PubMed] [Google Scholar]
- 12.Berger, P., Schaffitzel, C., Berger, I., Ban, N., and Suter, U. (2003) Proc. Natl. Acad. Sci. U. S. A 100 12177–12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, S. A., Vacratsis, P. O., Firestein, R., Cleary, M. L., and Dixon, J. E. (2003) Proc. Natl. Acad. Sci. U. S. A 100 4492–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenzo, O., Urbe, S., and Clague, M. J. (2005) J. Cell Sci. 118 2005–2012 [DOI] [PubMed] [Google Scholar]
- 15.Nandurkar, H. H., Layton, M., Laporte, J., Selan, C., Corcoran, L., Caldwell, K. K., Mochizuki, Y., Majerus, P. W., and Mitchell, C. A. (2003) Proc. Natl. Acad. Sci. U. S. A 100 8660–8665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caldwell, K. K., Lips, D. L., Bansal, V. S., and Majerus, P. W. (1991) J. Biol. Chem. 266 18378–18386 [PubMed] [Google Scholar]
- 17.Nandurkar, H. H., Caldwell, K. K., Whisstock, J. C., Layton, M. J., Gaudet, E. A., Norris, F. A., Majerus, P. W., and Mitchell, C. A. (2001) Proc. Natl. Acad. Sci. U. S. A 98 9499–9504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson, F. L., and Dixon, J. E. (2005) J. Biol. Chem. 280 31699–31707 [DOI] [PubMed] [Google Scholar]
- 19.Mochizuki, Y., and Majerus, P. W. (2003) Proc. Natl. Acad. Sci. U. S. A 100 9768–9773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger, P., Berger, I., Schaffitzel, C., Tersar, K., Volkmer, B., and Suter, U. (2006) Hum. Mol. Genet. 15 569–579 [DOI] [PubMed] [Google Scholar]
- 21.Azzedine, H., Bolino, A., Taieb, T., Birouk, N., Di Duca, M., Bouhouche, A., Benamou, S., Mrabet, A., Hammadouche, T., Chkili, T., Gouider, R., Ravazzolo, R., Brice, A., Laporte, J., and LeGuern, E. (2003) Am. J. Hum. Genet. 72 1141–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang, H., Li, Z., Skolnik, E. Y., and Fares, H. (2004) Mol. Biol. Cell 15 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivastava, S., Li, Z., Lin, L., Liu, G., Ko, K., Coetzee, W. A., and Skolnik, E. Y. (2005) Mol. Cell. Biol. 25 3630–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava, S., Ko, K., Choudhury, P., Li, Z., Johnson, A. K., Nadkarni, V., Unutmaz, D., Coetzee, W. A., and Skolnik, E. Y. (2006) Mol. Cell. Biol. 26 5595–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallat, L., Magdelenat, H., Merle-Beral, H., Masdehors, P., Potocki de Montalk, G., Davi, F., Kruhoffer, M., Sabatier, L., Orntoft, T. F., and Delic, J. (2003) Blood 101 4598–4606 [DOI] [PubMed] [Google Scholar]
- 26.MacKeigan, J. P., Murphy, L. O., and Blenis, J. (2005) Nat. Cell Biol. 7 591–600 [DOI] [PubMed] [Google Scholar]
- 27.Verbsky, J. W., Chang, S. C., Wilson, M. P., Mochizuki, Y., and Majerus, P. W. (2005) J. Biol. Chem. 280 1911–1920 [DOI] [PubMed] [Google Scholar]
- 28.Zou, J., Marjanovic, J., Kisseleva, M. V., Wilson, M., and Majerus, P. W. (2007) Proc. Natl. Acad. Sci. U. S. A 104 16834–16839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ungewickell, A., Hugge, C., Kisseleva, M., Chang, S. C., Zou, J., Feng, Y., Galyov, E. E., Wilson, M., and Majerus, P. W. (2005) Proc. Natl. Acad. Sci. U. S. A 102 18854–18859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kisseleva, M. V., Cao, L., and Majerus, P. W. (2002) J. Biol. Chem. 277 6266–6272 [DOI] [PubMed] [Google Scholar]
- 31.Schaletzky, J., Dove, S. K., Short, B., Lorenzo, O., Clague, M. J., and Barr, F. A. (2003) Curr. Biol. 13 504–509 [DOI] [PubMed] [Google Scholar]
- 32.Choudhury, P., Srivastava, S., Li, Z., Ko, K., Albaqumi, M., Narayan, K., Coetzee, W. A., Lemmon, M. A., and Skolnik, E. Y. (2006) J. Biol. Chem. 281 31762–31769 [DOI] [PubMed] [Google Scholar]
- 33.Biro, E., Akkerman, J. W., Hoek, F. J., Gorter, G., Pronk, L. M., Sturk, A., and Nieuwland, R. (2005) J. Thromb. Hemostasis 3 2754–2763 [DOI] [PubMed] [Google Scholar]
- 34.Zachowski, A. (1993) Biochem. J. 294 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vance, J. E., and Steenbergen, R. (2005) Prog. Lipid Res. 44 207–234 [DOI] [PubMed] [Google Scholar]