Abstract
We report a previously undescribed quorum-sensing mechanism for triggering multicellularity in Bacillus subtilis. B. subtilis forms communities of cells known as biofilms in response to an unknown signal. We discovered that biofilm formation is stimulated by a variety of small molecules produced by bacteria—including the B. subtilis nonribosomal peptide surfactin—that share the ability to induce potassium leakage. Natural products that do not cause potassium leakage failed to induce multicellularity. Small-molecule-induced multicellularity was prevented by the addition of potassium, but not sodium or lithium. Evidence is presented that potassium leakage stimulates the activity of a membrane protein kinase, KinC, which governs the expression of genes involved in biofilm formation. We propose that KinC responds to lowered intracellular potassium concentration and that this is a quorum-sensing mechanism that enables B. subtilis to respond to related and unrelated bacteria.
Keywords: biofilm, quorum sensing
Traditionally viewed as solitary organisms, bacteria of many kinds are now known to form complex multicellular communities that are composed of specialized cell types (1). Examples of bacteria that exhibit multicellularity are the myxobacteria, which form elaborate fruiting bodies, and streptomycetes, which form an aerial mycelium similar in appearance to that of certain fungi (2, 3). It is known that the formation of these multicellular communities involves extensive intercellular communication during the course of development, but relatively little is known about the signals that trigger multicellular behaviors.
Another example of bacterial multicellularity is the formation of surface-associated communities known as biofilms (4), whose constituent cells are held together by an extracellular matrix often composed of exopolysaccharide and protein (5). The process of biofilm formation in diverse bacteria has been shown to specifically involve the recognition of and response to self-generated secreted small molecules, i.e., “quorum sensing” (4, 6). Here we report a new twist on quorum sensing signaling during biofilm formation. We have discovered that biofilm formation by the spore-forming bacterium Bacillus subtilis can be triggered in response to a variety of structurally unrelated natural products produced by diverse microorganisms, including a molecule produced by B. subtilis itself. We present evidence suggesting that these agents act by causing potassium leakage across the cytoplasmic membrane of B. subtilis. This leads to the activation of a protein kinase that sets in motion a chain of regulatory events that induced the expression of genes involved in the synthesis of the extracellular matrix.
Results and Discussion
Small-Molecule Natural Products That Cause Potassium Leakage Induce Biofilm Formation.
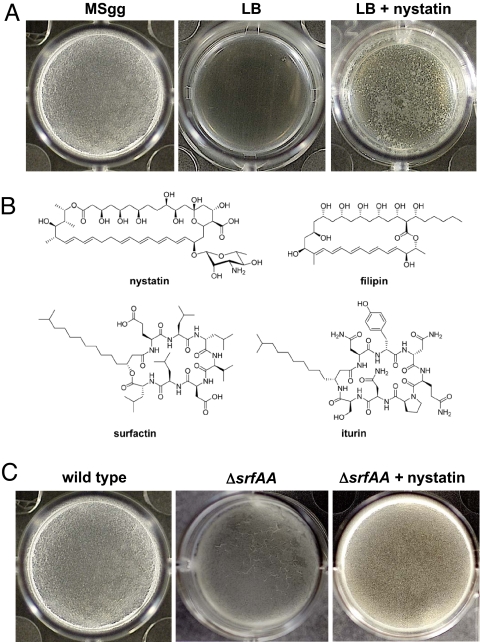
This discovery stemmed from our finding that robust floating biofilms (pellicles) formed under some conditions but not others. Incubation in the minimal defined medium MSgg induced biofilm formation whereas no biofilms were detected when cells were grown in the complex medium LB (Fig. 1A). This allowed us to screen for agents that induced biofilm formation in LB. After testing a relatively small panel of compounds, we found that the polyene polyketide nystatin, produced by the soil bacterium Streptomyces noursei, caused B. subtilis to form biofilms in LB medium (Fig. 1A). This result was surprising because nystatin is well-known to have antifungal activity but, to our knowledge, was not known to have effects on bacterial growth or physiology (7).
Fig. 1.
Nystatin and surfactin induce biofilm formation. (A) Nystatin induces B. subtilis biofilm formation. Images of culture wells (1.8-cm diameter) are shown. B. subtilis forms a pellicle when cultured in MSgg but not in LB. Supplementing LB medium with 60 μM nystatin leads to pellicle formation. (B) Chemical structures of the polyene polyketides nystatin and filipin and the N-acylated nonribosomal peptides surfactin and iturin. Nystatin and surfactin induce B. subtilis biofilm formation whereas filipin and iturin do not. (C) Surfactin does not induce biofilm formation as a surfactant but rather as a signaling molecule. A mutant unable to make surfactin displays defective pellicle morphology in MSgg medium (compare Left and Center). This phenotype can be reversed by the addition of nystatin, which is not a surfactant (see Right).
To decipher how B. subtilis senses nystatin, we attempted to define the properties of this polyene polyketide that might be important for its detection by B. subtilis by testing other small-molecule natural products for their ability to induce biofilm formation. Nystatin is known to insert into cellular membranes and forms pores that allow cation efflux (7). We first tested filipin (Fig. 1B). Structurally related to nystatin, filipin is also a polyene polyketide, but it differs from nystatin in its functionality. Filipin does not form pores in membranes; instead, it disrupts membrane structure (7). Filipin did not induce biofilm formation by B. subtilis when grown in LB medium, suggesting that the sensing mechanism does not detect all polyene polyketides and is not a general response to membrane disruption. We also tested a battery of nonspecific membrane disrupting detergents, and none of them triggered biofilm formation, adding further support to the notion that membrane disruption per se was not the biofilm-inducing signal [see supporting information (SI) Fig. S1]. We therefore hypothesized that the observed effects were due to nystatin's ability to induce cation leakage from the cytoplasm to the extracellular space.
To test this hypothesis we assayed several small-molecule natural products differing in both structure and functionality for the ability to induce biofilm formation. The natural products tested are listed in Table S1. None of the compounds test that induced biofilm formation had any effect on growth rate at the concentrations that were used. Amphotericin is structurally similar to nystatin and is also known to cause cation leakage; not surprisingly it also induced biofilm formation. In addition, the nonribosomally synthesized peptide gramicidin, which is structurally unrelated to nystatin but whose functionality is also to cause cation leakage, also induced biofilm formation (7–9). We tested 2 cyclic lipopeptides known to be produced by different strains of B. subtilis: surfactin and iturin (Fig. 1B). Although both molecules insert in membranes and form ion-conducting pores, they have different mechanisms. Surfactin-induced pores show some selectivity for potassium over other cations whereas iturin-induced pores show a slight selectivity for anions over cations (10, 11). In our assay, surfactin stimulated biofilm formation whereas iturin did not (Table S1). These results, coupled with the fact that potassium is the most abundant cytoplasmic cation, suggested that potassium leakage was the common effect of all of the small molecules tested that induced biofilm formation. This possibility was further supported by the finding that the potassium-selective ionophore valinomycin also induced biofilm formation (Table S1). Valinomycin shows high potassium selectivity but does not form membrane pores. Rather, it binds potassium ions directly, thus facilitating their diffusion through the membrane (12, 13). We also tested several other small-molecule natural products whose known physiological effects do not involve cation leakage, and none of them induced biofilm formation (see Table S1 for the complete list). In addition, the synthetic proton-selective ionophore CCCP, an uncoupler of the membrane potential (14), did not induce biofilm formation at sublethal concentrations. From these results we concluded that small-molecule natural products capable of causing potassium leakage are able to induce biofilm formation by B. subtilis in LB.
Surfactin's ability to stimulate biofilm formation is particularly intriguing, because this natural product is produced by the B. subtilis strain used in our studies, NCIB3610.* Importantly, surfactin is produced at very low levels in LB, perhaps explaining why biofilm formation is not stimulated under normal growth conditions in this medium. In contrast, surfactin is produced in large amounts in the minimal medium MSgg. The reasons behind this difference remain unknown. However, these observations raised the possibility that surfactin might act as an “autoinducer” or “quorum-sensing” signal made by B. subtilis under certain conditions that might regulate the expression of genes involved in biofilm formation.
As a first step in gaining a greater understanding of surfactin's role in biofilm formation, we wanted to determine whether the surfactant properties of surfactin were absolutely required in the process of biofilm formation by B. subtilis. In prior work we had shown that surfactin-defective mutants, grown in the minimal medium MSgg, yielded extremely thin and fragile biofilms (15). Discovering that both surfactin and nystatin could induce biofilm formation when B. subtilis was grown in LB led us to ask whether addition of nystatin to a mutant unable to make surfactin might reverse its defects in biofilm formation. This was an interesting test to perform because nystatin is not a surfactant. Fig. 1A already shows that in LB nystatin can induce biofilm formation using the WT strain, which still has the capacity to produce surfactin. By testing a surfactin defective mutant in MSgg we could address the requirement for surfactin's surfactant activity directly. As shown in Fig. 1C, when the mutant unable to make surfactin was grown in MSgg in the absence of surfactin it produces an extremely thin and very fragile biofilm - probably due to residual expression of extracellular matrix genes (see below). However, addition of nystatin reverses this phenotype, resulting in a biofilm identical to that produced by the WT. This result suggests that the surfactant properties of surfactin are not absolutely required for biofilm formation at least as far as the pellicle formation assay can detect. The result was also consistent with the hypothesis that surfactin might act as an autoinducer or quorum-sensing signaling molecule affecting the expression of genes involved in biofilm formation. To test this hypothesis we analyzed the effects of surfactin and nystatin on the expression of genes known to play key roles in biofilm formation.
Surfactin and Nystatin Activate the Regulatory Circuitry That Controls Matrix Production.
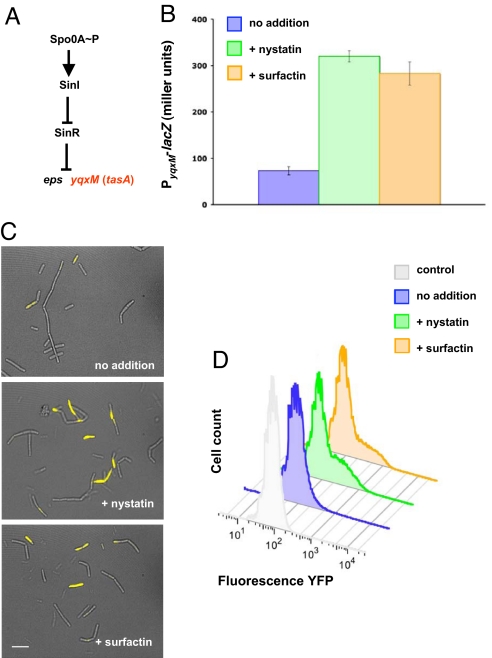
A defining step in the formation of B. subtilis biofilms is the synthesis of an extracellular matrix that holds the constituent cells together (16–19). The regulatory circuit controlling the synthesis of the extracellular matrix in this bacterium has been well-characterized (Fig. 2A). Two multigene operons, epsA-O and yqxM-sipW-tasA, direct the synthesis and export of an exopolysaccharide (EPS) and a protein component of the matrix (TasA), respectively (20). Both of these operons are repressed by the master regulator SinR, which is itself antagonized by SinI (19). In turn, the sinI gene is transcribed only when the level of the phosphorylated transcription factor Spo0A∼P rises above a threshold level (21). We hypothesized that the ability of surfactin and nystatin to induce biofilm formation was due to increased expression of the genes involved in matrix synthesis.
Fig. 2.
Nystatin and surfactin stimulate transcription of yqxM operon in a subpopulation of cells by inducing potassium leakage. (A) Schematic representation of the signaling pathway leading to matrix production. Spo0A is activated by phosphorylation and induces the expression of SinI, which antagonizes the repressor SinR. Once SinR is antagonized, the eps and yqxM operons are derepressed. (B) β-Galactosidase assay monitoring the expression of the yqxM operon using the transcriptional fusion PyqxM-lacZ. (C) Overlay of fluorescence (yellow) and transmitted light (gray) micrographs of WT B. subtilis harboring PyqxM-yfp. (Scale bar: 3 μm.) (D) Flow cytometry analysis of WT cells harboring PyqxM-yfp, untreated and in the presence of nystatin or surfactin. Untreated control cells that do not harbor any yfp gene show a single population of cells with very low relative fluorescence due to background. Cells harboring PyqxM-yfp fusion also show a single population but with slightly higher fluorescence (blue peak). In contrast, treatment with nystatin or surfactin (green and orange graphs, respectively) results in the appearance of a subpopulation of cells with high relative fluorescence, seen as the shoulder to the right of the main peaks.
To test this hypothesis, we analyzed the effects of surfactin and nystatin on matrix gene expression. Using a lacZ reporter gene, we observed that transcription from the promoter of the yqxM-sipW-tasA operon PyqxM increased upon treatment with either surfactin or nystatin (Fig. 2B). Even though the addition of these compounds led to a dramatic phenotypic effect vis-à-vis biofilm formation, the effect on gene transcription was modest; we observed only a 3- to 4-fold increase. However, this was because of the fact that matrix synthesis is induced in only a fraction of the cells, caused by a bistable switch that controls Spo0A∼P levels (22). This bistability of matrix gene expression was apparent upon microscopic observation of individual cells harboring the gene for the yellow fluorescent protein, yfp, under the control of PyqxM (Fig. 2C). Accordingly, flow cytometric quantification of the effects of surfactin and nystatin on matrix gene expression demonstrated that only a subpopulation of the cells is highly induced (Fig. 2D). These results demonstrate that, when cells are grown in LB, treatment with surfactin or nystatin induces the transcription of genes involved in biofilm formation. That surfactin can do this indicates that it is indeed an autoinducer.
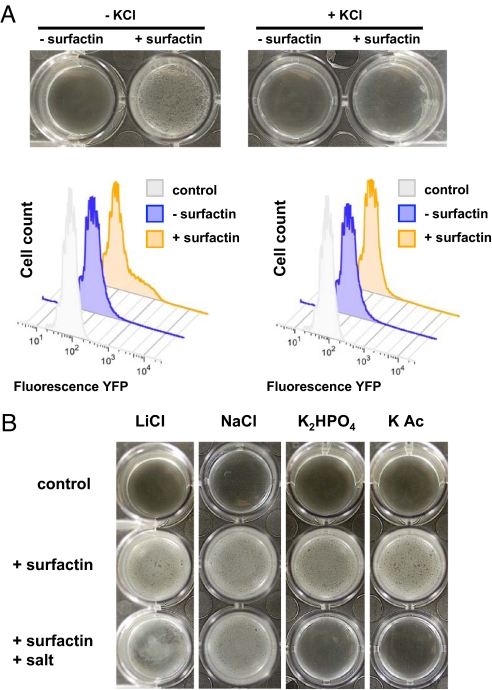
We next asked whether the induction of matrix gene expression by surfactin could be abrogated by increasing the extracellular potassium concentration. We reasoned that surfactin-mediated potassium leakage would be favored only when then extracellular concentration of potassium was significantly lower than the intracellular concentration. To address this question, we modulated the level of potassium that surfactin-treated B. subtilis encountered in LB medium. By increasing the external potassium concentration to 150 mM, a level that is close to the intracellular concentration, the surfactin-mediated efflux of potassium from the cytoplasm should be diminished insofar as the difference in potassium concentration between the inside and the outside of the cell has been reduced and thus the electrochemical gradient should no longer favor potassium release. We observed that addition of 150 mM KCl diminished the ability of surfactin to induce biofilm formation, suggesting that the condition being sensed was either the intracellular potassium concentration or its leakage from the cell, rather than the presence of surfactin per se. In addition, using flow cytometry we found that no subpopulation of cells expressing matrix genes could be observed if surfactin was added in the presence of 150 mM KCl (Fig. 3A). Importantly, this effect was specific for potassium because addition of 150 mM LiCl or 150 mM NaCl to LB had no effect on surfactin's ability to induce biofilm formation. In contrast, 2 other potassium salts (K2HPO4 and KAcetate) added at 150 mM to LB still reversed the effect of surfactin (Fig. 3B).
Fig. 3.
Increasing the extracellular potassium concentration abrogates the effects of surfactin. (A) Pellicle formation assay for surfactin-treated cells grown in LB in the absence or presence of 150 mM KCl. Pellicles and flow cytometry results are shown. (B) Specificity for extracellular potassium addition to counteract the effect of surfactin. To test the ion specificity of increasing the extracellular salt concentration supplemented to the LB growth medium, several other salts were tested in addition to KCl. The addition of NaCl or LiCl did not result in diminished biofilm formation. In contrast, other potassium salts (potassium phosphate and potassium acetate) had the same effect as KCl.
The Effect of Surfactin Is Sensed by the Membrane Histidine Kinase KinC.
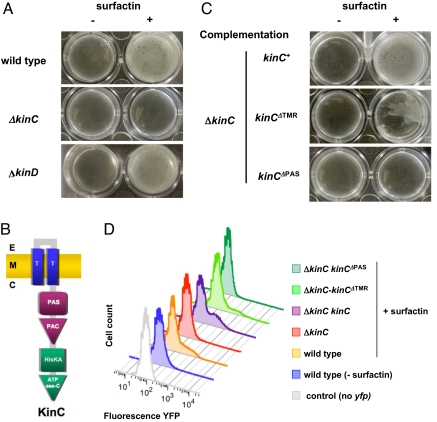
The activation of matrix gene expression by surfactin suggested that levels of Spo0A∼P are elevated through the action of a kinase able to sense the effects of potassium leakage. By screening kinase mutants for defects in biofilm formation we identified 2 kinases potentially involved in sensing the surfactin effects, KinC and KinD. This screen depended on the fact that matrix production results in a wrinkled colony phenotype when B. subtilis is grown on MSgg agar (19). Individual mutants lacking either KinC or KinD show subtle phenotypic differences, whereas the double mutant shows a more dramatic change in colony morphology. Independently, Kobayashi et al. (23) also reported that mutations in kinC and kinD resulted in defective pellicle formation in their assays. KinC and KinD both belong to the large family of bacterial sensor histidine kinases and are predicted to contain 2 membrane-spanning domains (24). We tested whether mutations in the genes encoding these kinases affected the ability of B. subtilis to respond to surfactin (Fig. 4A). Only the mutant lacking KinC failed to respond to surfactin; the mutant lacking KinD responded like the WT.
Fig. 4.
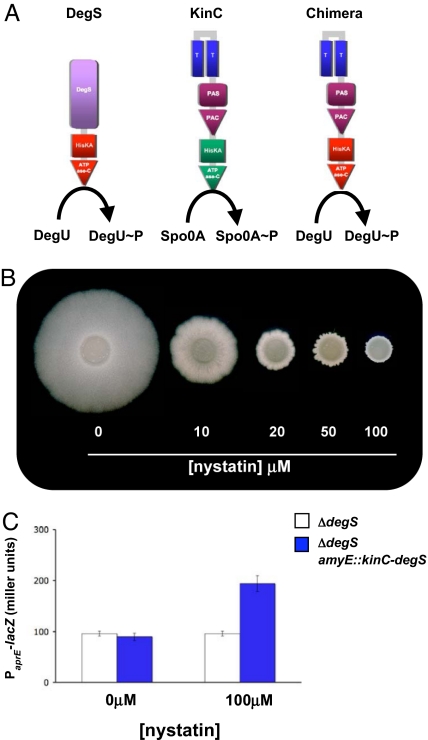
KinC is the histidine kinase involved in the response to surfactin. (A) Pellicle assays of the response to surfactin of the WT and mutants defective in KinC or KinD. (B) Schematic of the domain structure of KinC. E, extracellular milieu; M, cytoplasmic membrane; C, cytoplasm; T, transmembrane domain; PAS/PAC, sensor domains; HisKA, histidine kinase domain; ATPase C, ATPase domain. (C) Pellicle assays of the response to surfactin of the mutant defective in KinC harboring 3 different kinC alleles: WT kinC+, kinCΔTMR, and kinCΔPAS. (D) Flow cytometry of cells harboring the reporter construct PyqxM-yfp and the KinC alleles described in C.
KinC is predicted to have 2 membrane-spanning segments joined by only 7 extracellular residues and a PAS-PAC sensor domain (Fig. 4B) (25). To determine which of these domains played a role in sensing the effects of potassium leakage, we constructed three kinC alleles and tested them for their ability to complement the effects of the kinC mutation. One allele was the WT (kinC+), the second contained a deletion of the membrane-spanning region (kinCDTMR), and the third had the PAS-PAC domain deleted (kinCDPAS); all three mutant genes yielded detectable amounts of protein in a Western blot assay (Fig. S2). The results of these complementation experiments are shown in Fig. 4C. The WT kinC+ allele fully complemented the phenotype of kinC deficiency because the kinC+-complemented strain was once again able to respond to surfactin by making a biofilm in LB. Interestingly, deleting the membrane-spanning regions yielded a kinase still able to sense the action of surfactin, albeit poorly. The poor response may be due to the fact that this allele yielded the least amount of protein (see Fig. S2). In contrast, deleting the PAS-PAC domain abolished the complementation. The effects of the various kinC alleles on the expression of matrix genes was quantitated in flow cytometry, and the results are shown in Fig. 4D. The flow cytometry results were in complete agreement with the biofilm formation assay. All of these results indicate that KinC, in some way involving its PAS-PAC domain, is the critical kinase that senses the effect of surfactin and other compounds that result in potassium leakage.
To further define the regions of KinC involved in sensing the effects of potassium leakage, we constructed a hybrid histidine sensor kinase. By phosphorylating the transcription factor DegU, the cytoplasmic histidine sensor kinase DegS controls the swarming behavior or B. subtilis as well as its ability to produce several extracellular proteases (26, 27). We replaced the histidine kinase and ATPase domains of KinC with those of DegS (Fig. 5A). Production of this chimeric kinase within B. subtilis led to a strain in which both swarming and protease production were controlled by nystatin (Fig. 5 B and C). As controls, neither the WT strain nor the mutant lacking DegS altered their swarming or protease production in response to nystatin when they did not harbor the chimeric kinase (data not shown). Thus, the critical domain of KinC for sensing the effects of potassium leakage is its PAS-PAC domain.
Fig. 5.
The N-terminal portion of KinC is a modular element that responds to nystatin. (A) Schematic representation of DegS, KinC, and the KinC-DegS chimera. Domain designations are as in Fig. 4B. (B) Swarming assay of a DegS-deficient strain complemented by KinC-DegS. (C) Expression of aprE monitored by using the PaprE-lacZ reporter construct.
Heterologous Expression and Activation of the Signal Transduction Pathway in Listeria monocytogenes.
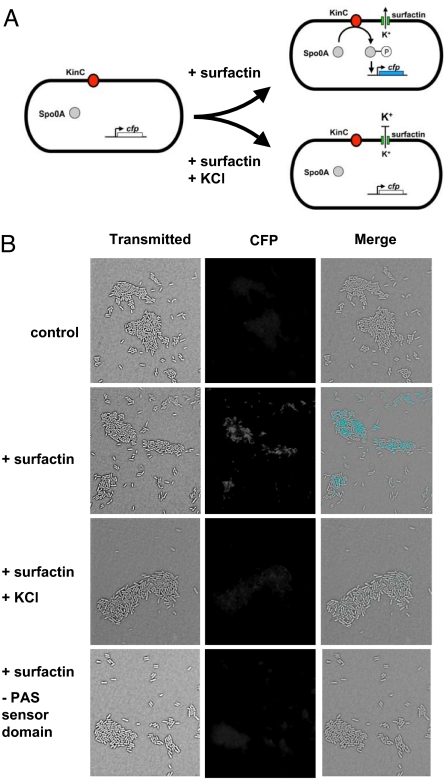
The simplest interpretation of our results so far is that KinC is a signal transduction protein that senses and responds to potassium leakage by phosphorylating Spo0A. As a further test of this idea, we introduced the B. subtilis genes for KinC and Spo0A together with a transcriptional reporter for Spo0A∼P-directed gene transcription, Pskf-cfp (21), into the Gram-positive bacterium L. monocytogenes. L. monocytogenes is related to B. subtilis but lacks orthologs of KinC, Spo0A, and the other components of the regulatory circuit for extracellular matrix production (Fig. 6A). The addition of surfactin to L. monocytogenes producing KinC and Spo0A resulted in activation of the Pskf-cfp reporter (Fig. 6B). Furthermore, surfactin-mediated Spo0A activation was inhibited by 100 mM KCl and depended on the PAS-PAC sensor domain of KinC (Fig. 6B). We conclude that KinC and Spo0A are the only B. subtilis-specific proteins needed to confer surfactin responsiveness in a heterologous host bacterium.
Fig. 6.
Reconstitution of the signal transduction pathway in the heterologous host L. monocytogenes. (A) Activation of the signaling system is monitored by expression of the reporter gene Pskf-cfp. (B) Surfactin induces expression of Pskf-cfp, and this induction is diminished by adding potassium in the medium. Signaling pathway activation requires the KinC PAS domain. Images are transmitted light (gray), fluorescence (white), and overlaid pictures, with fluorescence colored blue. (Scale bar: 3 μm.)
We have characterized a previously undescribed signal transduction mechanism in B. subtilis that is activated when the cells are treated with surfactin and other compounds, apparently because they cause intracellular potassium to leak out from the cell. We identified the kinase that, directly or indirectly, senses the effect of the compounds. We present evidence suggesting that KinC activation is not triggered by uncoupling of the membrane potential or cell envelope stress and does not result from compounds binding as ligands to the kinase, as might have been expected. Rather, the signal that activates the kinase is apparently related to the loss of potassium ions. Is the kinase sensing a decrease in the cytoplasmic concentration of potassium, or is the signal the actual flux of the ions? The fact that pore-forming molecules, such as surfactin, as well as the non-pore-former valinomycin induced biofilm formation suggests that the flux of potassium per se is not the signal. Rather, it is likely that the lowered intracellular concentration is somehow being sensed. Exactly how this might be occurring remains unknown. One possibility is that the intracellular concentration of potassium decreases, at least temporarily, enough to be sensed. But of course the cell has multiple potassium-uptake mechanisms that should quickly restore intracellular potassium (28–30). It should be noted that treatment with neither surfactin nor nystatin at the concentration we used (20 and 60 μM, respectively) leads to any noticeable decrease in growth rate—an indication that intracellular potassium homeostasis is not severely compromised. The transient effects of potassium leakage might be sensed indirectly, for example through a resulting decrease in intracellular pH as protons move into the cell or through the uptake of another cation to compensate for the transient leakage of potassium.
The results presented here argue that surfactin can act as an autoinducer or a quorum-sensing signal that indirectly activates KinC because of its ability to cause potassium leakage. Surfactin production is restricted to a subset of free-living Bacillus species (B. subtilis, Bacillus amyloliquefaciens, Bacillus licheniformis, and Bacillus pumilus). Notably, 3 other genetic elements are found exclusively in this Bacillus subgroup: (i) comQXP, the quorum-sensing system that regulates surfactin production; (ii) kinC; and (iii) the closely linked gene ktrC (see Fig. S3A) (31, 32). The phylogenetic conservation of surfactin production and KinC suggests an evolutionary connection between the two. Interestingly, ktrC encodes a putative potassium channel regulator (30). When we mutated ktrC we observed an increase in biofilm formation in MSgg that could still be suppressed by increasing concentrations of potassium (see Fig. S3B). Taken together, these results suggest a functional linkage among potassium, surfactin, and KinC.
All of the small molecules that induced biofilm formation are natural products from soil-dwelling bacteria. B. subtilis may thus have evolved a mechanism that allows it to sense the presence of many of its potential neighbors, not by producing a receptor that binds directly to each small molecule, but by sensing a common change in cell state that results from the action of structurally diverse compounds. As such, the sensing mechanism represents a previously undescribed form of sensing in which the organism, by ultimately sensing chemical function rather than a specific structure, can respond to its own signal as well as diverse compounds from other soil-dwelling organisms. Exactly what benefit B. subtilis gains by making an extracellular matrix and building a biofilm in response to other species of bacteria is not known, but perhaps biofilm formation represents a defensive strategy against antibiotics produced by competing bacterial species. The fact that 1 of the molecules identified as inducing biofilm formation (nystatin) is widely used in the treatment of fungal infections indicates that the specific therapeutic use of natural products does not necessarily reflect their native function.
An important challenge for the future will be to determine exactly how KinC is activated. It will also be of interest to determine whether other kinases somehow use ion leakage as their activating signals. A candidate for an organism that employs a similar sensing mechanism is the fungal plant pathogen Phytophthora palmivora, whose zoospores alter their behavior dramatically when treated with valinomycin without losing viability (33). It should now be possible to search among many kinases whose activating signals remain unknown to determine whether ion concentration as an activating signal is a widespread phenomenon.
Experimental Procedures
Strains, Media, and Culture Conditions.
B. subtilis strain NCIB3610 was incubated at 37 °C in LB medium. For pellicle formation experiments B. subtilis strain NCIB3610 was cultured in LB medium in Falcon Multiwell (24-well) plates at 30 °C for 8 h (19). All small molecules tested were added directly to each well. Pellicle formation was observed maximally with the addition of 60 μM nystatin or 20 μM surfactin. “Swarm agar” plate assays were performed as described (34).
Strain Construction, Reporters, and Protein Expression Constructs.
Strains used and generated in this work are listed in Table S2. Deletion mutants were generated by using long flanking homology PCR (35). Constructs generated were inserted by double recombination into neutral integration sites (amyE and lacA) in the genome of B. subtilis by inducing natural competence (36). Primers used are listed in Table S3. The different alleles of KinC and a chimera were created with long flanking homology PCR (using primers listed in Table S3). They were cloned in pKM008, a vector used for integration in amyE as well as the translational fusions to yfp used to determine protein expression by immunoblotting. Transcriptional fusions were inserted into the amyE locus by using pKM008 and pDG1663, respectively (37), or lacA locus using pDR183 vector (38). Constructions were transferred to NCIB3610 by SPP1 phage transduction as described previously (39). All β-galactosidase assays were performed as described previously (19).
The signaling pathway was reconstituted in L. monocytogenes M35303A by cloning the genes kinC and spo0A, each with its own promoter, into the plasmid pPL2 (40). Additionally, the transcriptional fusion Pskf-cfp was cloned divergently to kinC and spo0A in pPL2 to monitor levels of activation of Spo0A by phosphorylation (21). Additionally, a control plasmid containing only the reporter Pskf-cfp was constructed by using the same restriction sites. The integration of the plasmid into the genome of L. monocytogenes was performed according to the literature (40).
Microscopy and flow cytometry profiles were obtained as in Vlamakis et al. (41).
Supplementary Material
Acknowledgments.
We thank Y. Chai, A. McLoon, A. Earl, C. Aguilar, H. Vlamakis, and V. Murthy for discussions. This work was supported by National Institutes of Health Grants GM58213 (to R.K.), GM82137 (to R.K.), and GM18568 (to R.L.). D.L. was the recipient of a postdoctoral fellowship from the Fundación Séneca, Comunidad Autónoma de la Región de Murcia (Spain).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810940106/DCSupplemental.
It is important to note that NCIB3610 is a wild strain that produces surfactin. The commonly used laboratory strain 168 does not produce surfactin because of a mutation in the sfp gene.
References
- 1.Aguilar C, Vlamakis H, Losick R, Kolter R. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol. 2007;10(6):638–643. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claessen D, de Jong W, Dijkhuizen L, Wosten HA. Regulation of Streptomyces development: Reach for the sky! Trends Microbiol. 2006;14(7):313–319. doi: 10.1016/j.tim.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Kaiser D. Signaling in myxobacteria. Annu Rev Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- 4.Kolter R, Greenberg EP. Microbial sciences: The superficial life of microbes. Nature. 2006;441(7091):300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 5.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: The matrix revisited. Trends Microbiol. 2005;13(1):20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Davies DG, et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 7.Bolard J. How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim Biophys Acta. 1986;864(3–4):257–304. doi: 10.1016/0304-4157(86)90002-x. [DOI] [PubMed] [Google Scholar]
- 8.Daniele RP, Holian SK. A potassium ionophore (valinomycin) inhibits lymphocyte proliferation by its effects on the cell membrane. Proc Natl Acad Sci USA. 1976;73(10):3599–3602. doi: 10.1073/pnas.73.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katsu T, Kobayashi H, Fujita Y. Mode of action of gramicidin S on Escherichia coli membrane. Biochim Biophys Acta. 1986;860(3):608–619. doi: 10.1016/0005-2736(86)90560-2. [DOI] [PubMed] [Google Scholar]
- 10.Maget-Dana R, Ptak M, Peypoux F, Michel G. Pore-forming properties of iturin A, a lipopeptide antibiotic. Biochim Biophys Acta. 1985;815(3):405–409. doi: 10.1016/0005-2736(85)90367-0. [DOI] [PubMed] [Google Scholar]
- 11.Sheppard JD, Jumarie C, Cooper DG, Laprade R. Ionic channels induced by surfactin in planar lipid bilayer membranes. Biochim Biophys Acta. 1991;1064(1):13–23. doi: 10.1016/0005-2736(91)90406-x. [DOI] [PubMed] [Google Scholar]
- 12.Aqvist J, Alvarez O, Eisenman G. Ion-selective properties of a small ionophore in methanol studied by free energy perturbation simulations. J Phys Chem. 1992;96:10019–10025. [Google Scholar]
- 13.Marrone TJ, Merz KM. Molecular recognition of K+ and Na+ by valinomycin in methanol. J Am Chem Soc. 1995;117:779–791. [Google Scholar]
- 14.Heytler PG, Prichard WW. A new class of uncoupling agents—carbonyl cyanide phenylhydrazones. Biochem Biophys Res Commun. 1962;7:272–275. doi: 10.1016/0006-291x(62)90189-4. [DOI] [PubMed] [Google Scholar]
- 15.Straight PD, Willey JM, Kolter R. Interactions between Streptomyces coelicolor and Bacillus subtilis: Role of surfactants in raising aerial structures. J Bacteriol. 2006;188(13):4918–4925. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59(4):1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 17.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98(20):11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59(4):1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- 19.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55(3):739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 20.Branda SS, et al. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186(12):3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujita M, Gonzalez-Pastor JE, Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187(4):1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;67(2):254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi K, Kuwana R, Takamatsu H. kinA mRNA is missing a stop codon in the undomesticated Bacillus subtilis strain ATCC 6051. Microbiology. 2008;154(Pt 1):54–63. doi: 10.1099/mic.0.2007/011783-0. [DOI] [PubMed] [Google Scholar]
- 24.Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70(4):910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor BL, Zhulin IB. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63(2):479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amati G, Bisicchia P, Galizzi A. DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J Bacteriol. 2004;186(18):6003–6014. doi: 10.1128/JB.186.18.6003-6014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhamme DT, Kiley TB, Stanley-Wall NR. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol Microbiol. 2007;65(2):554–568. doi: 10.1111/j.1365-2958.2007.05810.x. [DOI] [PubMed] [Google Scholar]
- 28.Albright RA, Ibar JL, Kim CU, Gruner SM, Morais-Cabral JH. The RCK domain of the KtrAB K+ transporter: Multiple conformations of an octameric ring. Cell. 2006;126(6):1147–1159. doi: 10.1016/j.cell.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 29.Albright RA, Joh K, Morais-Cabral JH. Probing the structure of the dimeric KtrB membrane protein. J Biol Chem. 2007;282(48):35046–35055. doi: 10.1074/jbc.M704260200. [DOI] [PubMed] [Google Scholar]
- 30.Holtmann G, Bakker EP, Uozumi N, Bremer E. KtrAB and KtrCD: Two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J Bacteriol. 2003;185(4):1289–1298. doi: 10.1128/JB.185.4.1289-1298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeDeaux JR, Grossman AD. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177(1):166–175. doi: 10.1128/jb.177.1.166-175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77(2):207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 33.Appiah AA, van West P, Osborne MC, Gow NA. Potassium homeostasis influences the locomotion and encystment of zoospores of plant pathogenic oomycetes. Fungal Genet Biol. 2005;42(3):213–223. doi: 10.1016/j.fgb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49(3):581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 35.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12(3):259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 36.Hardwood CR, Cutting SM. Molecular Biological Methods for Bacillus. New York: Wiley; 1990. [Google Scholar]
- 37.Guerout-Fleury AM, Frandsen N, Stragier NP. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 38.Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55(6):1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- 39.Yasbin RE, Young FE. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14(6):1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol. 2002;184(15):4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22(7):945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.