Abstract
Understanding the diverse activities of the multisubunit core promoter recognition complex TFIID in vivo requires knowledge of how individual subunits contribute to overall functions of this TATA box-binding protein (TBP)/TBP-associated factor (TAF) complex. By generating altered holo-TFIID complexes in Drosophila we identify the ETO domain of TAF4 as a coactivator domain likely targeted by Pygopus, a protein that is required for Wingless-induced transcription of naked cuticle. These results establish a coactivator function of TAF4 and provide a strategy to dissect mechanisms of TFIID function in vivo.
Keywords: naked cuticle, Pygopus, TATA box-binding protein-associated factor, transcription, coactivator
The activator-mediated recruitment of TFIID to the core promoter has been proposed to be a critical and early step leading to transcription initiation by RNA polymerase II (RNAP II). The TFIID complex, comprising the TATA-binding protein (TBP) and TBP-associated factors (TAFs), contains both promoter recognition activity and coactivator functions that have been primarily characterized by in vitro experiments (1). Various sequence-specific DNA-binding transcription factors have been reported to target TFIID subunits, thereby leading to recruitment or stabilization of TFIID binding to the promoter. For example, the human transcription factor Sp1 targets the N-terminal glutamine-rich domains of TAF4 that results in enhanced transcription from synthetic GC-box-containing promoters in reconstituted in vitro reactions (2). Indeed, many instances of direct activator–TAF interactions have been observed in vitro although such coactivator mechanisms have remained poorly characterized in vivo (3–6). Thus, the specific role of putative TAF coactivator domains potentiating transcription at a native promoter in vivo has remained elusive.
Investigating the role of individual subunits within a multiprotein complex in cells and organisms presents a difficult challenge. Knockouts, temperature-sensitive alleles, and RNAi experiments targeting single subunits often lead to the destabilization of the entire complex or parts of the complex (see refs. 7–13). Although these approaches can be useful for assessing global contributions of the multisubunit complex in vivo, a detailed mechanistic analysis of individual subunits requires the ability to assay specific subunit function in the context of the holo-complex. This can be achieved by first identifying structural subunits or domains to segregate them from those with functional domains.
We previously reported the presence of a structural domain in the C-terminal region (CTR) of TAF4 that is necessary and sufficient to nucleate the assembly of the holo-TFIID complex in the absence of full-length, endogenous TAF4 (11). Here, we exploit this knowledge by using a combination of overexpression and RNAi-mediated depletion to generate altered TFIID complexes in Drosophila tissue culture cells and in flies to investigate the specific role of TAF4 in directly providing coactivator functions in the context of TFIID in vivo.
Results and Discussion
TAF4 Is Required for Wingless (Wg)-Regulated naked cuticle (nkd) Expression.
Having previously identified the CTR of TAF4 as being necessary and sufficient to nucleate the assembly of the TFIID complex (11), we postulated that the remaining N-terminal region of TAF4 would be free to perform other functions, possibly coactivation. To begin dissecting the putative coactivator properties of the TAF4 N-terminal region, we developed a cell-based assay in which the Wg signaling pathway is activated, either by treating the cells with a partially purified Wg preparation (see Materials and Methods) or transfecting the cells with a constitutively active Armadillo (Arm*) (14), which stimulates expression of the well-established downstream Wg target gene nkd (15) (for a review of Wg signaling see refs. 16 and 17). We found that for some experiments the Arm* transfection strategy yielded more consistent and reproducible results, so in certain experiments Arm* was used to activate the Wg pathway rather than Wg protein. Thus, activation of the Wg signaling pathway followed by analysis of nkd expression should allow us to assess TAF4 activity at an inducible endogenous promoter in its natural chromatin environment.
To first determine whether the Wg signaling pathway is intact in our Drosophila S2R+ tissue culture cells, as reported (18), we analyzed the Wg response by assaying Armadillo stabilization by immunoblotting (Fig. S1A) and up-regulation of nkd by quantitative reverse transcription–PCR (qRT-PCR). We also tested the dependence of nkd activation on many known components of the Wg pathway (Fig. S1B). We found that our assay system requires all tested components of the Wg pathway and suggest that S2R+ cells faithfully recapitulate the Wg signaling pathway observed in the organism.
Having established a highly-inducible, cell-based assay for activated transcription, we next asked whether this transcriptional activation depends on TFIID. RNAi-mediated depletion of individual subunits resulted in varying degrees of reduced activation (Fig. 1A). The most dramatic effects were seen when TAF1, TAF4, and TBP were depleted. These data indicate that, in stark contrast to the Drosophila Metallothionein A (MtnA) promoter (12), nkd highly depends on TFIID for full activation. These studies also identify TAF4, along with TAF1 and TBP, as potential target coactivators required for nkd transcriptional activation. For this study, we focused our attention on the TAF4 subunit of TFIID, which because of its role in nucleating complex assembly and stability (11), has been difficult to study mechanistically in vivo. To ensure that the dsRNA targeting TAF4 and the ensuing effects are specific, we made 2 additional dsRNAs targeting either the 5′ UTR or the 3′ UTR. As shown in Fig. 1B, all 3 dsRNAs targeting TAF4 reduced the ability of the cells to activate nkd in response to Wg signaling, suggesting the loss of activation is not likely caused by off-target effects. Importantly, Armadillo stabilization was not affected by TAF4 depletion (Fig. 1C). This result indicates that the defect caused by reduced TAF4 levels lies downstream of pathway activation, and likely at the level of transcription, thus suggesting that nkd is a bona fide TFIID-dependent gene.
Fig. 1.
nkd is a TFIID-dependent gene in S2R+ cells. (A) TFIID dependence of nkd was determined by depleting the indicated subunits by RNAi and analyzing nkd expression by qRT-PCR. (B) Three different dsRNAs targeting different regions of TAF4 reduced the level of nkd activation. (C) Armadillo stabilization was not affected by TAF4 depletion. Each dsRNA directed against TAF4 was able to efficiently deplete TAF4 levels as determined by α-TAF4 immunoblotting. β-tubulin served as a loading control.
We next sought to determine whether activation of nkd in the fly also requires TAF4. To do this, we expressed an inverted repeat that generates an siRNA targeting TAF4 in a stripe along the anterior–posterior boundary, perpendicular to the endogenous nkd expression along the dorsal–ventral boundary of the larval wing imaginal disk by using the dpp-Gal4 driver/UAS system. Endogenous nkd transcript was detected by in situ hybridization with an nkd antisense probe. As a negative control we expressed GFP in the same pattern. We also expressed an inverted repeat construct targeting Armadillo as a positive control. As shown in Fig. 2A, GFP had no effect on nkd expression, whereas depletion of TAF4 greatly diminished nkd levels at the intersection of the dpp-driven TAF4 RNAi and nkd expression pattern. As expected, depletion of Arm also severely down-regulated nkd levels (Fig. 2A) as did ectopic expression of Axin, a known negative regulator of Wg pathway (data not shown). Importantly, expression of Wg was unaffected in each of the experiments (Fig. 2A). These data suggest that in both our cell culture system and flies the TAF4 subunit of TFIID plays a role likely as a coactivator in the Wg-induced activation of nkd.
Fig. 2.
nkd is a TAF4-dependent gene in larval wing imaginal discs. (A) TAF4 RNAi in larval imaginal wing discs reduced expression of nkd. (Upper) Wg expression. (Lower) Endogenous nkd staining in discs expressing either GFP, an Arm-inverted repeat, or a TAF4-inverted repeat in a perpendicular pattern. Arrowhead indicates intersection between endogenous nkd expression and dpp-Gal4 driven expression of the indicated constructs. (Scale bars: 100 μm.) (B) TAF4 and RNAP II occupancy of the nkd promoter was analyzed by ChIP in cells treated with either buffer or Wg for 2 h. Immunoprecipitated chromatin was quantified by qRT-PCR relative to control serum.
TFIID and RNAP II Occupancy at the nkd Promoter.
To gain additional insight into the mechanism of activation of the nkd gene, we used ChIP to determine the occupancy of TFIID and RNAP II at the nkd promoter in the repressed, uninduced, and Wg-stimulated state. Interestingly, both TFIID and RNAP II were already easily detectable at the nkd promoter before Wg stimulation. Wg treatment resulted in a >50-fold activation of nkd expression but only a 2-fold increase in TAF4/TFIID and RNAP II occupancy at the promoter (Fig. 2B). This pattern of preinduction occupancy is reminiscent of the Wg activator complex component Pygopus, which is preloaded at the Wg-response elements (WREs) of nkd before Wg signaling (19). This induction method is in contrast to what we previously observed at the Drosophila MtnA promoter where both TFIID and RNAP II are recruited primarily to the promoter postinduction (12). Also, Pangolin, the Drosophila homolog of TCF/LEF, is assumed to preoccupy the nkd WREs given that dsRNA targeting Pangolin resulted in derepression of nkd (Fig. S1B). Consistent with our RNAP II ChIP data is an unbiased, genomewide ChIP-microarray analysis that also identified RNAP II at the nkd promoter in the absence of Wg stimulation (20). These findings suggest that TFIID and RNAP II may be recruited to the promoter before Wg stimulation and hence in the absence of Armadillo. Furthermore, it appears that the Groucho corepressor, which is recruited by Pangolin in the absence of signaling, likely functions at a step after preinitiation complex formation to repress nkd transcriptional activation effectively by allowing the formation of a “preloaded” preinitiation complex (PIC) that is inactive.
The N-Terminal Domain of TAF4 Is Required for nkd Activation.
RNAi-mediated depletion of TAF4 results in the destabilization and subsequent degradation of the majority of TFIID subunits (11). Consequently, it is impossible to conclude whether the effect on nkd activation seen in TAF4 RNAi cells is caused by a specific coactivator function of TAF4 or the wholesale destruction of the entire TFIID complex. Because we previously mapped a “structural” domain in the CTR of TAF4 that is sufficient to nucleate the TFIID complex (11), we developed a strategy combining TAF4 RNAi with overexpression of this structural domain to specifically probe the potential function of the TAF4 N-terminal domain in the context of a holo-TFIID complex. This approach allowed us to circumvent any confounding effects of TFIID disassembly and destabilization. It seemed likely that the TAF4 CTR is sufficient to nucleate the assembly of a functional, but altered, complex because we found that the resulting TAF4 CTR-containing TFIID is sufficient to rescue a G2/M cell cycle arrest seen after depletion of TAF4 (data not shown). To determine whether the N-terminal domain is required for nkd activation, we generated Flag-tagged stable cell lines constitutively expressing either full-length TAF4 or the TAF4 CTR. These cells were each treated with dsRNA targeting the 5′ and 3′ UTRs of TAF4 to deplete the endogenous TAF4. Under these conditions, we were able to assay holo-TFIID complexes containing a Flag-tagged TAF4 CTR in the absence of endogenous TAF4 protein. After 72 h, the cells were split; one half was transfected with a control empty vector, and the other half was transfected with Arm* to activate nkd. After 48 h, nkd levels were determined by qRT-PCR. As seen in Fig. 3A, the cell line expressing full-length TAF4 supported robust activation of nkd, whereas in contrast the TAF4 CTR failed to mediate nkd activation. Importantly, the expression level of transfected Arm* remained constant in each cell line, suggesting that the defect in nkd activation was not caused by loss of Arm* (Fig. 3B). These data identify a specific coactivator function found in the N-terminal domain of TAF4 that is required for activation of nkd. These results also imply that there are likely multiple roles for TFIID at the nkd promoter: one revealed when TAF1 and TBP are depleted, and another performed by the N-terminal domain of TAF4.
Fig. 3.
The TAF4 CTR does not rescue TAF4 RNAi for nkd activation. (A) Stable cell lines expressing either full-length TAF4 (TAF4 FL) or the TAF4 CTR were treated with dsRNA targeting the TAF4 UTRs. The cells were split and transfected with either empty vector or Arm* to activate nkd transcription. The levels of nkd were analyzed as before. (B) Transfected Arm* levels were equivalent in TAF4FL and TAF4 CTR cell lines as determined by α-V5 immunoblotting. Endogenous TAF4 levels were depleted efficiently in both cells. β-Tubulin was included as a loading control. (C) Overexpression of the TAF4 CTR, but not full-length TAF4, reduced nkd expression when coexpressed with Arm*. (D) Transfected Arm* levels were equivalent as determined by immunoblotting with the α-V5 antibody. β-Tubulin served as a loading control.
The TAF4 CTR Is a Dominant Negative for nkd Activation.
Because the TAF4 CTR is not sufficient to rescue nkd activation in a TAF4 RNAi background, and we previously observed that the TAF4 CTR is able to destabilize endogenous TAF4 when overexpressed (11), we postulated that it should act as a dominant negative when overexpressed. To test this hypothesis, we coexpressed either control empty actin expression vector, full-length TAF4, or the TAF4 CTR before and after induction with Arm*. Whereas full-length TAF4 had a modest stimulatory effect, the TAF4 CTR significantly reduced activation of nkd, suggesting that the TAF4 CTR is in fact able to act as a dominant negative (Fig. 3C). Expression levels of transfected Arm* were slightly elevated in the full-length TAF4 samples, whereas control and TAF4 CTR samples expressed similar levels, suggesting that the dominant negative effect is specific for nkd, and not due to a reduction in Arm* levels (Fig. 3D). This finding further supports the notion of a coactivator function in the N-terminal domain of TAF4, consistent with previous in vitro transcription experiments (2).
The ETO Domain of TAF4 Is Required for nkd Activation.
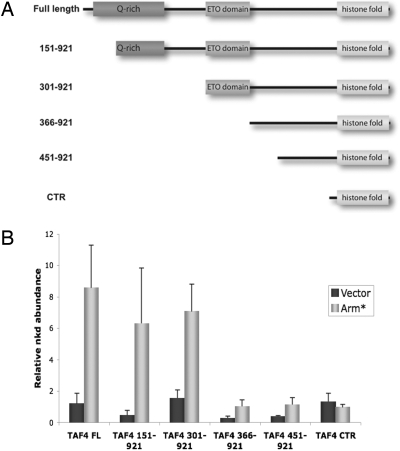
The N-terminal region of TAF4 consists of 2 loosely conserved, adjacent glutamine-rich domains that have been implicated in coactivation and a highly conserved, metazoan-specific ETO domain (also called conserved region I), recently shown to bind a hydrophobic peptide sequence found in some activators (21, 22). The intervening sequences are not conserved among metazoans. To determine which portion of the TAF4 N-terminal region mediates transcription activation of nkd we generated a series of stable cell lines constitutively expressing Flag-tagged N-terminal truncations (Fig. 4A). The expression levels of TAF4 derivatives in each cell line were confirmed by immunoblotting and determined to be roughly equivalent (data not shown). As reported above, full-length Flag-tagged TAF4 was able to mediate robust activation of nkd. Deletion of the first (amino acids 151–921) or both (amino acids 301–921) glutamine-rich domains resulted in a slight decrease in nkd activation (Fig. 4B). The next truncation, coding for residues 366–921, which removes the highly conserved ETO domain, dramatically reduced nkd activation. The largest truncation (amino acids 451–921), which removes the nonconserved sequence between the ETO domain and the histone fold, behaved similarly to the amino acids 366–921 construct. As seen before, the TAF4 CTR failed to support activation of nkd. These data suggest that sequences encompassing the ETO domain of TAF4 likely constitute a coactivator target required for nkd activation. Although it is possible that the functional coactivation domain also relies on sequences between the ETO domain and histone fold, we believe this is unlikely given the lack of sequence conservation. Thus, the ETO domain may represent a novel coactivation domain of TAF4 that is required for activation of the Wg target gene nkd. These studies also suggest that the N-terminal region of TAF4 likely contains multiple coactivator domains that include the glutamine-rich domains and the newly reported ETO.
Fig. 4.
The TAF4 ETO domain is required for nkd activation. (A) Stable cell lines were generated by expressing either full-length TAF4 or an N-terminal truncation depicted here. (B) The N-terminal coactivator activity mapped to the ETO domain. Experiments with each stable cell line were performed as in Fig. 3A.
The TAF4 ETO Domain Interacts with Pygopus.
Having mapped the nkd coactivator activity of TAF4 to the ETO domain, we next sought to investigate the mechanism by which it functions. Because we detected low levels of TAF4 at the nkd promoter before Wg stimulation by ChIP, we reasoned that the ETO domain might interact with a component of the Wg activator/coactivator machinery that is present at WREs in unstimulated cells. To test this idea, we generated a GST fusion of the ETO domain to use in pull-down assays (Fig. 5A). We transiently expressed V5-tagged Pangolin or Pygopus, two factors known to be present at the nkd WRE before Wg signaling, in S2R+ cells. Because it is possible that the ETO domain could make additional contacts after Wg stimulation, we also tested for an interaction with Armadillo. Lysates were made and incubated with either GST- or GST-ETO-bound glutathione Sepharose beads. After extensive washing, bound proteins were resolved by SDS/PAGE and analyzed by immunoblotting with the α-V5 antibody. Remarkably, whereas GST-ETO failed to interact efficiently with either Pangolin or Armadillo, the ETO domain specifically pulled Pygopus out of the whole-cell extracts, suggesting a strong and specific interaction between the two proteins (Fig. 5B). As a step toward further characterizing the interaction between the TAF4 ETO domain and Pygopus, we generated V5-tagged Pygo constructs encoding either the N-terminal 200 amino acids (Pygo 1-200) or the remaining C-terminal portion of the protein (Pygo 201-815) to use in GST pull-down experiments. As shown in Fig. 5C, the N-terminal Pygo 1-200 polypeptide interacted with the ETO domain of TAF4, whereas the Pygo 201-815 construct failed to interact. This N-terminal region of Pygopus contains a conserved NPFxD sequence that is required for full Pygopus function (23). We sought to determine whether this NPFxD motif is mediating the interaction with TAF4. We generated an AAAxA mutant Pygo 1-200 construct and tested its interaction with the TAF4 ETO domain in a GST pull-down experiment. As shown in Fig. 5D, mutation of the NPFxD motif had little effect on binding to the ETO domain of TAF4. This result suggests that a sequence different from the NPFxD motif within the N-terminal region of Pygopus is able to mediate its interaction with TAF4, and that a second coactivation domain likely resides within this region. Because the above experiments were performed in whole-cell lysates derived from Drosophila cells, it is possible that an unidentified protein present in these lysates mediates the interaction between Pygopus and TAF4. This becomes more important given the recent finding that Pygopus can be coimmunoprecipitated from Drosophila embryo extracts with an antibody against the MED13 subunit of the Mediator complex (24). To test whether the interaction between Pygopus and TAF4 is direct, we performed a GST pull-down experiment using a lysate prepared from bacteria expressing V5 tagged Pygo 1-200. As shown in Fig. 5E, bacterially-expressed Pygo 1-200 is also efficiently pulled down specifically by the ETO domain of TAF4. These results suggest that the interaction between TAF4 and Pygopus is direct and does not require a secondary factor, such as the Mediator complex. Taken together, these findings suggest that the ETO domain may function as a direct target coactivator of the Wg signaling pathway by providing an interaction interface for Pygopus. Future studies will be necessary to dissect the specific transcriptional activation mechanism by which the interaction between Pygopus and the TAF4 ETO domain leads to up-regulation of Wg target genes upon stimulation.
Fig. 5.
The ETO domain interacts with Pygopus in a GST pull-down experiment. (A) (Left) A schematic representation of the GST fusion protein used in the pull-down experiments. (Right) A Coomassie-stained gel loaded with protein molecular weight markers (M), GST, or GST-ETO beads. (B) GST pull-down experiment using either GST or GST-ETO beads incubated with lysates made from S2R+ cells transfected with V5 tagged Arm*, Pangolin, or Pygopus. Bound proteins were analyzed by α-V5 immunoblotting. (C) GST pull-down experiment performed as in B using lysates made from S2R+ cells transfected with V5 tagged Pygopus constructs encoding either residues 1–200 or 201–815. (D) GST pull-down experiment performed as in B using lysates made from S2R+ cells transfected with either wild-type Pygo 1–200 (wt) or a Pygo 1–200 construct with the NPFxD motif mutated to AAAxA (mut). (E) GST pull-down experiment performed as in B using lysates made from bacteria transformed with a plasmid encoding V5 tagged Pygo 1–200.
Concluding Remarks.
Here, we have identified nkd as a TFIID-dependent gene in both Drosophila tissue culture cells and larval imaginal discs and have mapped one component of this dependence to a small, conserved region of the TAF4 subunit. This finding differs from our previous work with the copper-inducible Mtn A gene, which did not strictly require TFIID for transcriptional activation (12). Induction of nkd also differs from MtnA in that both TFIID and RNAP II are preloaded at the promoter before stimulation, and occupancy of both at the promoter is only moderately increased upon activation even though transcription is up-regulated >50-fold after induction by Wg. By contrast, MtnA shows a dramatic recruitment of both TFIID and RNAP II in a copper-dependent manner, and the fold recruitment correlates closely with fold activation. These differences highlight 2 distinct mechanisms used by Drosophila cells to induce transcription upon a physiological stimulus.
Much of the work investigating coactivator activities associated with TAF4 had previously focused on the glutamine-rich domains (2, 25–27). Using an unbiased cell-based assay measuring activated transcription from an endogenous promoter we have identified a coactivator element in the TAF4 ETO domain that operates within the context of a holo-TFIID complex in vivo. We further show that the ETO domain binds directly to Pygopus, part of the dedicated Wg transcription activation machinery. Consistent with a conserved role for TAF4 in Wnt signaling, we found that depletion of TAF4 in SW480 human cells, which have constitutively active Wnt signaling (28), leads to a down-regulation of the Wnt target gene LEF-1 (29, 30) (Fig. S2A).
Interestingly, a recent report (24) suggests that an N-terminal region of Pygopus targets the MED12 and MED13 subunits of the Mediator coactivator complex. This result, taken together with the Pygopus–TAF4 interaction reported here, suggests there may be cross-talk between TFIID and Mediator at the nkd promoter, reminiscent of such a relationship reported for the MtnA promoter in Drosophila (12). It will be interesting to determine whether the Mediator complex is recruited before or after signaling. It is tempting to speculate that Mediator, recruited through a direct interaction with Armadillo via its MED12 subunit (31) and stabilized at WREs by the Pygopus N-terminal homology domain (NHD) (24), could serve as the “trigger” to activate the preloaded PIC reported here by a currently unknown mechanism. Such a mechanism would involve the recruitment of elongation factors, because Mediator has recently been found to bind the elongation factor pTEFb (32). It is not clear whether Mediator binding to Pygopus would interfere with its binding to TAF4, nor is it evident whether this dual binding would occur simultaneously or sequentially.
Deletion of the glutamine-rich domains of TAF4 resulted in only a marginal reduction in our nkd activation assay, suggesting that TAF4 contains at least 2 distinct coactivator domains that can be used in a gene- or activator-specific manner. It is likely that, similar to TAF4, many other TAF subunits contain multiple coactivation domains, thus greatly increasing the ability of TFIID to respond to the plethora of diverse activation signals metazoan organisms must accommodate. Our Wg signaling assay combined with overexpression and RNAi knock-down in cells and flies should prove valuable in studying other important coactivator functions in vivo.
Materials and Methods
Plasmids.
The TAF4 constructs used were generated by using PCR with appropriate oligos to generate fragments suitable for cloning into a vector containing the actin 5C promoter and the 3X Flag sequence. The GST-TAF4 ETO plasmid was generated by using PCR with appropriate oligos to generate a fragment suitable for cloning into the pGex2TK vector. Copper-inducible Pangolin and Pygopus constructs were generated by using PCR to amplify full-length Pangolin or Pygopus from S2R+ cDNA and cloned into the pMT-V5/His vector (Invitrogen). The Pygopus 1-200 and 201-815 constructs were generated by cloning a PCR fragment containing appropriate region into the pMT-V5/His vector. Quikchange mutagenesis was used to generate the NPFxD to AAAxD mutation in pMT-Pygo 1–200. Oligonucleotide sequences used to generate each construct are available on request.
Cell Culture, Transfections, and RNAi.
SW480 cells were obtained from ATCC and grown in Liebovitz's L-15 medium (ATCC) plus 10% FBS. Control, TAF4, and β-catenin siRNAs were obtained from Dharmacon and transfected by using Dharmafect 1 reagent (Dharmacon) according to the manufacturer's recommendations.
Wg Purification.
Purification of Wg was adapted from a previously described protocol (33). Briefly, Wg protein was partially purified from the culture medium of S2 cells expressing Wg under control of the tubulin promoter. Triton X-100 was added to the medium from ≈4 L of culture to a final concentration of 1%. This material was loaded onto a HiPrep 16/10 Heparin FF column (GE Health Sciences). The column was washed with 10 column volumes PBS plus 1% CHAPS. Wg protein was eluted with PBS plus 1% CHAPS and 1 M NaCl. Fractions containing Wg were identified by immunoblotting with 4D4 anti-Wg mAb (University of Iowa Developmental Studies Hybridoma Bank, Iowa City). These fractions were pooled, and glycerol was added to ≈15%. The partially purified Wg samples can be stored at −80 °C for >1 year without any detectable loss of activity.
qRT-PCR.
Total RNA was isolated by using TRI reagent, digested with DNase I, phenol/chloroform-extracted, and resuspended in H2O. cDNA was made by using SuperScript II (Invitrogen) following the manufacturer's recommendations from 2.5 μg of total RNA. qRT-PCR was performed by using 2 μL of cDNA in a 20-μL reaction with iQ SYBR Green qPCR mix (Bio-Rad) and a MJ Research Opticon 2 instrument. C(T) values were normalized to rpb1 and averaged, then graphed relative to the control in each experiment. Error bars indicate SD of 3 reactions. Oligonucleotide sequences are available on request.
Wg Signaling Assay.
Approximately 106 S2R+ cells were plated in 2 mL of culture medium in a 6-well plate and treated with 15 μL of either buffer (PBS plus 1% CHAPS and 1 M NaCl) or partially purified Wg. After 2 h, cells were harvested and analyzed by either immunoblotting for Armadillo stabilization or qRT-PCR.
Immunoblotting.
Immunoblots were performed by using standard procedures. The anti-Armadillo antibody N2 7A1 antibody was obtained from the University of Iowa Developmental Studies Hybridoma Bank. The human and fruit fly anti-TAF4 antibodies and anti-β-tubulin antibody have been described (11). The anti-β-catenin antibody was purchased from Santa Cruz Biotechnology.
Drosophila Larval Wing Disk in Situ Hybridization.
The nkd in situ hybridization was carried out by using standard methods with a hybridization temperature of 65 °C. The probe has been described (15). Oligonucleotide sequences used to generate the Wg probe are available on request. The sheep antidigoxigenin antibody was purchased from Roche. The donkey anti-sheep Alexa Fluor 488 antibody was purchased from Invitrogen. Discs were dissected from wandering third-instar lavae from crosses between male dpp-Gal4 driver flies and virgin female UAS-GFP, UAS-TAF4 RNAi, or UAS-Arm RNAi flies.
Fly Stocks.
The following stocks were obtained from the Bloomington Drosophila Stock Center at Indiana University: dpp-Gal4 (1553), UAS-GFP (1521), and UAS-axin-GFP (7224). The TAF4 (11764) and Arm (12540) UAS-inverted repeat flies were obtained from the Vienna Drosophila RNAi Center.
Cell Culture, Transfections, and RNAi.
S2R+ cells were grown at 25 °C in M3+BPYE medium (Drosophila Genomics Resource Center) plus 10% heat inactivated FBS. RNAi was performed by adding 15 μg of dsRNA/106 cells to the culture medium and incubating for 72 h. Oligonucleotide sequences used to generate each RNAi constructs are available on request. Stable cell lines were generated as described (11). Transfections were performed by using Effectene reagent (Qiagen) following the manufacturer's recommendations.
ChIP.
ChIPs were performed essentially as described (12). The antibodies used have been described (12).
GST Pull-Down.
GST or GST-TAF4 ETO was used to saturate Glutathione Sepharose beads (GE Health Sciences). For experiments using Pygopus fragments GST-ETO was cross-linked to the Glutathione Sepharose beads by treatment with 20 mM dimethylpimelimidate for 30 min. Twenty-five microliters of saturated beads was incubated with cell lysates made as described (11) from 3 × 106 S2R+ cells transfected with a pMT-V5 (Invitrogen) construct encoding either Pangolin, Pygopus, or Arm*. After 4 h at 4 °C the beads were washed 5 times with 20 mM Hepes (pH 7.6), 0.1 mM EDTA, 1 mM DTT, 250 mM NaCl, and 0.1% Triton X-100. Bound proteins were eluted with 50 μL of SDS sample buffer and analyzed by SDS/PAGE followed by immunoblotting.
Supplementary Material
Acknowledgments.
We thank M. Marr, Y. Fong, P. Hu, and M. Deato for comments on the manuscript; K. Wharton (University of Texas Southwestern, Dallas) for the plasmid used to generate the nkd probe; K. Cadigan (University of Michigan, Ann Arbor) for the Arm* plasmid; K. Wharton and K. Suyama for advice on wing disk in situ hybridizations, and M. Haggart for technical assistance.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811914106/DCSupplemental.
References
- 1.Albright SR, Tjian R. TAFs revisited: More data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 2.Hoey T, et al. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 3.Goodrich JA, et al. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 4.Thut CJ, Chen JL, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 5.Amrolia PJ, et al. The activation domain of the enhancer binding protein p45NF-E2 interacts with TAFII130 and mediates long-range activation of the α- and β-globin gene loci in an erythroid cell line. Proc Natl Acad Sci USA. 1997;94:10051–10056. doi: 10.1073/pnas.94.19.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang CM, Roeder RG. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 7.Reese JC, Zhang Z, Kurpad H. Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J Biol Chem. 2000;275:17391–17398. doi: 10.1074/jbc.M001635200. [DOI] [PubMed] [Google Scholar]
- 8.Yan Z, et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H, Kim S, Ishii S, Boyer TG. Mediator modulates Gli3-dependent Sonic hedgehog signaling. Mol Cell Biol. 2006;26:8667–8682. doi: 10.1128/MCB.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beall EL, Bell M, Georlette D, Botchan MR. Dm-myb mutant lethality in Drosophila is dependent upon mip130: Positive and negative regulation of DNA replication. Genes Dev. 2004;18:1667–1680. doi: 10.1101/gad.1206604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright KJ, Marr MT, 2nd, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci USA. 2006;103:12347–12352. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marr MT, 2nd, Isogai Y, Wright KJ, Tjian R. Coactivator cross-talk specifies transcriptional output. Genes Dev. 2006;20:1458–1469. doi: 10.1101/gad.1418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen WC, et al. Systematic analysis of essential yeast TAFs in genomewide transcription and preinitiation complex assembly. EMBO J. 2003;22:3395–3402. doi: 10.1093/emboj/cdg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang M, et al. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J. 2006;25:2735–2745. doi: 10.1038/sj.emboj.7601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng W, et al. naked cuticle encodes an inducible antagonist of Wnt signaling. Nature. 2000;403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
- 16.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Stadeli R, Hoffmans R, Basler K. Transcription under the control of nuclear Arm/β-catenin. Curr Biol. 2006;16:R378–R385. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 18.Yanagawa S, Lee JS, Ishimoto A. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J Biol Chem. 1998;273:32353–32359. doi: 10.1074/jbc.273.48.32353. [DOI] [PubMed] [Google Scholar]
- 19.de la Roche M, Bienz M. Wingless-independent association of Pygopus with dTCF target genes. Curr Biol. 2007;17:556–561. doi: 10.1016/j.cub.2007.01.063. [DOI] [PubMed] [Google Scholar]
- 20.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, et al. Conserved region I of human coactivator TAF4 binds to a short hydrophobic motif present in transcriptional regulators. Proc Natl Acad Sci USA. 2007;104:7839–7844. doi: 10.1073/pnas.0608570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei Y, et al. A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription. Nat Struct Mol Biol. 2007;14:653–661. doi: 10.1038/nsmb1258. [DOI] [PubMed] [Google Scholar]
- 23.Stadeli R, Basler K. Dissecting nuclear Wingless signaling: Recruitment of the transcriptional coactivator Pygopus by a chain of adaptor proteins. Mech Dev. 2005;122:1171–1182. doi: 10.1016/j.mod.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Carrera I, et al. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc Natl Acad Sci USA. 2008;105:6644–6649. doi: 10.1073/pnas.0709749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rojo-Niersbach E, Furukawa T, Tanese N. Genetic dissection of hTAF(II)130 defines a hydrophobic surface required for interaction with glutamine-rich activators. J Biol Chem. 1999;274:33778–33784. doi: 10.1074/jbc.274.47.33778. [DOI] [PubMed] [Google Scholar]
- 26.Saluja D, Vassallo MF, Tanese N. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/mcb.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanese N, et al. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc Natl Acad Sci USA. 1996;93:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goyette MC, et al. Progression of colorectal cancer is associated with multiple tumor suppressor gene defects but inhibition of tumorigenicity is accomplished by correction of any single defect via chromosome transfer. Mol Cell Biol. 1992;12:1387–1395. doi: 10.1128/mcb.12.3.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filali M, et al. Wnt-3A/β-catenin signaling induces transcription from the LEF-1 promoter. J Biol Chem. 2002;277:33398–33410. doi: 10.1074/jbc.M107977200. [DOI] [PubMed] [Google Scholar]
- 30.Hovanes K, et al. β-Catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 31.Kim S, Xu X, Hecht A, Boyer TG. Mediator is a transducer of Wnt/β-catenin signaling. J Biol Chem. 2006;281:14066–14075. doi: 10.1074/jbc.M602696200. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 33.Willert K, et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.