Abstract
Paleogenomic research has shown that modern humans, Neanderthals, and their most recent common ancestor have displayed less genetic diversity than living great apes. The traditional interpretation that low levels of genetic diversity in modern humans resulted from a relatively recent demographic bottleneck cannot account for similarly low levels of genetic diversity in Middle Pleistocene hominins. A more parsimonious hypothesis proposes that the effective population size of the human lineage has been low for more than 500,000 years, but the mechanism responsible for suppressing genetic diversity in Pleistocene hominin populations without similarly affecting that of their hominoid contemporaries remains unknown. Here we use agent-based simulation to study the effect of culturally mediated migration on neutral genetic diversity in structured populations. We show that, in populations structured by culturally mediated migration, selection can suppress neutral genetic diversity over thousands of generations, even in the absence of bottlenecks or expansions in census population size. In other words, selection could have suppressed the effective population size of Pleistocene hominins for as long as the degree of cultural similarity between regionally differentiated groups played an important role in mediating intraspecific gene flow.
Keywords: culturally mediated migration, human evolution, Middle Pleistocene, structured populations, agent-based simulation
Modern humans display less genetic diversity than great apes, a puzzling finding given our much larger census population size (1, 2). Interestingly, recent studies have shown that modern humans are not the only hominins characterized by comparatively low levels of genetic diversity. The variability of Neanderthal mitochondrial DNA is on par with that found in modern humans (3–5). More importantly, the effective population size of the common ancestor of modern humans and Neanderthals was recently estimated at 9,999 (95% CI: 9,603–10,335)*, concurring with Noonan et al.'s (6) assumption that the effective population size of the common ancestor was similar to that of modern humans, ≈104. Why are all 3 of these Pleistocene hominin populations characterized by levels of genetic diversity that are lower than those found in extant great apes?
Modern human genetic diversity has previously been explained as resulting from a relatively recent demographic expansion from a small population (7, 8) that probably exhibited geographic structure (9, 10). It is worth noting that some genetic loci do not match the expectations of this bottleneck scenario (9, 11–15). At any rate, its timing—sometime between 30,000 and 130,000 years ago (9)—is too recent to account for the low level of genetic diversity in the modern human–Neanderthal common ancestor, unless the population bottleneck was very long (7). Others have interpreted characteristics of modern human genetic diversity as resulting from a geographic expansion that started from a single origin in subSaharan Africa and unfolded via successive colonization events by small groups of founders (16). Although the serial-founder effect provides a powerful explanation for some aspects of modern human genetic diversity, including the negative correlation between haplotype heterozygosity and distance from eastern Africa, it does not preclude the possibility that hominin genetic diversity was low before expansion out of Africa. Furthermore, to what extent the serial-founder effect applies to the population ancestral to modern humans and Neanderthals remains unclear. We investigate an alternative hypothesis, which proposes that the effective population size of the human lineage reached its current level more than 500,000 years ago, before the population ancestral to Neanderthals and modern humans split (7, 17, 18). Thinking about which factors could have suppressed Middle Pleistocene hominin genetic diversity without similarly affecting their hominoid contemporaries led us to investigate the effects of cultural differentiation in a metapopulation—a population structured by partially isolated subpopulations or groups (11, 19–21).
Culture refers to information acquired from conspecifics through learning or imitation that can lead to variation in behavior (22). Although culture is not unique to humans, ours is one of very few species for which cultural traits can play major roles as adaptations that directly affect fitness and as markers of social identity that do not directly affect fitness. The antiquity of the latter role in hominin societies is unknown, although pigments—possibly used as markers of social group identity—have been found in archaeological sites that date to ≈270,000 years ago in Africa (23) and slightly more recently in Europe (24). What is known is that gene flow between extant human groups is often mediated by cultural traits such as language, dress, diet code, caste, class, and religion. We use the term culturally mediated migration (CMM) to refer to the general mechanism whereby individuals can only migrate to groups that surpass a given level of cultural familiarity. Culturally transmitted variation is not thought to play a similar role in mediating gene flow between intraspecific groups in nonhuman primates. Could a primitive form of CMM explain the comparatively low genetic diversity estimates shared by modern humans, Neanderthals, and their most recent common ancestor?
To address this question without attributing modern human behaviors to Middle Pleistocene hominins a priori, we use an agent-based model that incorporates the dual inheritance of genetic and cultural variation to explore how a rudimentary version of CMM affects neutral gene diversity in spatially explicit populations (see Methods and SI Text). During the course of each simulation, selection and/or drift winnow the genetic and cultural variation created by mutation and innovation, respectively (see SI Text). Within this evolutionary framework, we test the hypothesis that increasing the severity of CMM—i.e., raising the cultural similarity threshold (CST) to migration—will significantly lower the average gene diversity of selectively neutral, partially linked loci in simulated populations. When CST = 0, simulated populations are structured by geographic distance only. With each increase in CST, each migrant requires prospective destination groups to surpass a higher threshold of cultural similarity. We also examine how variables associated with cultural evolution, such as innovation rate (ι) and proportion of neutral cultural traits, affect the relationship between CST and neutral gene diversity. An analysis of how total average gene diversity (H) is partitioned within subpopulations (HS) and between subpopulations (H–HS) allows us to explain our data in terms of selection among members of different culturally defined groups (see Methods).
Results
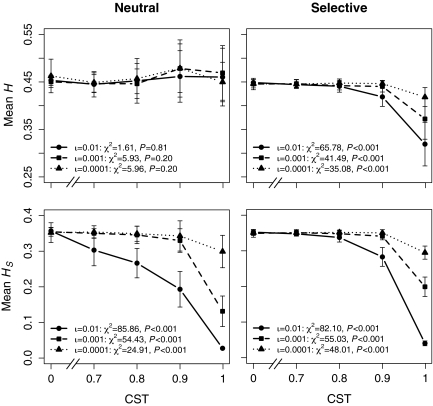
We collected data from 2 complementary sets of simulations, neutral and selective, to demonstrate that the effect of CMM on genetic diversity is driven by selection among members of different culturally defined groups. In neutral simulations, relative fitness does not vary among individuals, regardless of which cultural or genetic variants they possess. This uniformity in fitness results in neutral evolution. As expected under these conditions, increasing CST has no effect on total average gene diversity for all innovation rates tested, even though CST has a significant effect on within-group diversity (Fig. 1). That is, if no cultural or genetic variant is more advantageous than any other, total average gene diversity is unaffected by the cultural barriers to migration that significantly lower within-group gene diversity.
Fig. 1.
The effect of CST on average gene diversity in neutral (Left) and selective (Right) simulations. Each data point represents the mean (±1 SD) of 20 unique simulation runs per parameter setting. Text within the graphs provides the results of Kruskal–Wallis tests for the effect of CST on the average gene diversity of the entire metapopulation (Upper) and the effect of CST on the average gene diversity within subpopulations (Lower) under three different innovation rates (ι).
In selective simulations, relative individual fitness is calculated as a function of the variants each individual expresses at selective cultural and genetic loci. This function imbues heritable variation with differential fitness. As before, increasing CST has a significant effect on within-group diversity in selective simulations, but here it also affects total average gene diversity (Fig. 1). This important difference (with selection, increasing CST significantly decreases the average gene diversity of the metapopulation) holds true even after controlling for number of migrations. Furthermore, increasing the innovation rate or the proportion of neutral cultural traits allows for lower CST to suppress total average gene diversity (Table 1).
Table 1.
Innovation rate and proportion of neutral cultural traits affect onset CST, and increased CST suppresses gene diversity even under equivalent levels of gene flow
| Innovation rate | Proportion of neutral traits | Onset CST | Total migrations | Mean H |
|---|---|---|---|---|
| 0.01 | 0.7 | 0.8 | 0.58 | <0.001 |
| 0.001 | 0.7 | 0.99 | 0.12 | <0.001 |
| 0.0001 | 0.7 | 0.99 | 0.30 | <0.001 |
| 0.01 | 0.3 | 0.9 | 0.80 | <0.001 |
| 0.001 | 0.3 | 0.99 | 0.76 | <0.001 |
| 0.0001 | 0.3 | 0.99 | 0.08 | <0.001 |
Onset CST is the lowest CST value at which CMM significantly lowers mean H in comparison to that obtained in the absence of CMM (i.e., CST = 0). For simulations in which CST = 0, migration rate (ν) was adjusted such that the actual number of migrations per run is equivalent to the same value in simulations in which CST > 0. Total migrations and mean H are given as P values, which are provided by Wilcoxon rank sum tests that compare each set of 20 simulations in which CST = 0 to the corresponding set of 20 simulations in which CST = onset CST and ν = 0.05.
Because metapopulation size remains nearly constant (1,000 ± 20) throughout each simulation run, fluctuations in census size do not play a role in reducing average gene diversity. Our model assumptions are purposefully conservative. When they have an effect, it is of maintaining or increasing genetic variation within groups rather than between them (see SI Text). How does CST suppress neutral genetic diversity despite a nearly constant census population size and a conservative experimental design?
In a panmictic population, a selectively advantageous mutation evolves to fixation with a probability and at a rate that share a simple relationship to population size and the strength of selection. The manner in which a favorable mutation spreads through a structured population is not so simple (25). In a structured population, gene flow between subpopulations is required for an advantageous mutation to spread beyond the boundaries of the group in which it first appears. However, CMM can inhibit the spread of beneficial mutations by restricting gene flow to short cultural distances. One consequence of cultural isolation is that offspring inherit only those novel, beneficial mutations that spread to fixation within, but not beyond, the culturally defined boundaries of the group into which they are born. Another is that, when migration between groups is rare, the fate of each beneficial mutation—its frequency in the metapopulation—depends upon the rate at which its carrier's group fissions relative to other groups. Variance in groups' fission rates depends on how relative individual fitness is partitioned within and between groups. A group-level selective sweep, whereby 1 group (and its daughter and granddaughter groups) fissions more rapidly than other groups, requires low within-group variance and high between-group variance in relative individual fitness (26, 27). As long as these conditions persist, members of the group(s) that has accrued the most favorable mutations will contribute disproportionately more offspring to the metapopulation (28, 29). Although the selective sweep of a set of favorable mutations occurs at a higher level of the biological hierarchy in a structured population than the level at which a single favorable mutation sweeps through a panmictic population, the sweeps reduce average gene diversity in both cases (27).
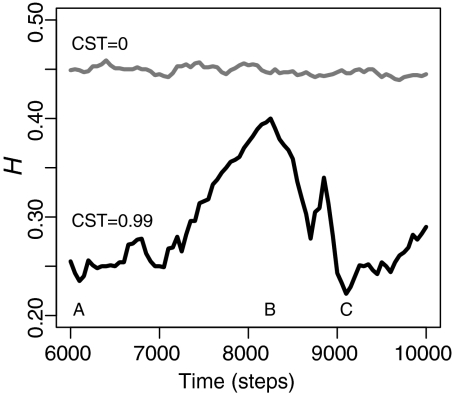
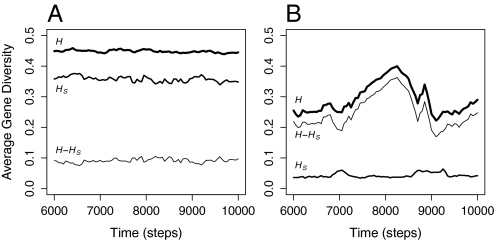
Our simulation results illustrate that, under sufficiently high CST, selection among members of different groups suppresses the average gene diversity of neutral loci over thousands of generations via a recursive cycle composed of 2 phases (Fig. 2). Genetic (and cultural) diversity increases during the collection phase, although almost entirely at the between-group level (Fig. 3). The resulting combination of low within-group variance and high between-group variance in relative individual fitness predictably leads to a reduction in total average gene diversity, as a relatively small proportion of the metapopulation produces disproportionately more offspring during the sweep phase. Between-group variance in relative individual fitness also decreases as the daughter (and granddaughter) groups of the initially successful group account for a greater proportion of the total number of groups. The average gene diversity of a metapopulation is low after a sweep because a large proportion of its individuals coalesce to a single individual who lived in the initially successful group. However, because such sweeps are rarely exhaustive, descendents of other groups often persist as viable members of the gene pool. As a result, the metapopulation can preserve older variation at some neutral genetic loci while reflecting relatively shallow coalescent events at others—2 seemingly contradictory signals found in modern human DNA that have proven difficult to reconcile without invoking hybridization or multiregional continuity (e.g., 14, 15; but see 30).
Fig. 2.
A recursive cycle suppresses genetic diversity in the presence of culturally mediated migration. Total average gene diversity (H) is plotted over the final 4,000 time steps of 2 selective simulations with identical parameter settings, except for CST. When CST = 0, H experiences relatively small perturbations but remains nearly constant through time. When CST = 0.99, we see lower H values and a recursive cycle that is generally characterized by a relatively long collection phase (time point A to time point B) followed by a much shorter sweep phase (time point B to time point C).
Fig. 3.
Total, within-group, and between-group gene diversity through time. (A) When CST = 0, the majority of the total average gene diversity is explained by diversity found within subpopulations. (B) When CST is much higher, between-group differences account for the majority of the total gene diversity through both the collection phase and the sweep phase. Each of the runs presented here, which are the same as those presented together in Fig. 2, is representative of a larger set of simulations.
Discussion
Sewall Wright's (28, 29) shifting balance theory describes how drift among geographically isolated groups in a metapopulation can yield novel, selectively advantageous gene combinations that would be difficult, if not impossible, for a panmictic population of similar size to discover given the same amount of time. Wright explained how the global frequency of favorable gene combinations will then increase via selective emigration, whereby carriers contribute more than their share of new migrants to adjacent groups (29). After this sweep, the process starts over.
Many aspects of our model are similar. During the collection phase, culturally structured populations accumulate a greater diversity of gene combinations more quickly than panmictic populations of similar size; selectively advantageous sets of mutations sweep through a culturally structured population via a form of selective emigration; and the end of each sweep phase marks the beginning of a new collection phase. However, the fact that in our model migration is mediated by cultural similarity in addition to geographic distance yields important differences. First, under relatively high CST, most migrations occur when daughter groups move to adjacent (possibly empty) cells following group fission events rather than when a solitary émigré moves from one previously established group to another. Second, genetic isolation can be maintained between culturally dissimilar groups regardless of their geographic proximity. Both allow CMM to support higher between-group and lower within-group variance in relative individual fitness than geographic distance alone. This support, in turn, facilitates longer, more comprehensive selective sweeps that yield even lower levels of average gene diversity. Another important difference is that cultural isolation can occur more quickly than geographic isolation, especially when innovation rates are high and/or a sizable proportion of cultural traits are neutral. In sum, culturally mediated population structure increases both the amplitude and the periodicity of what Wright first identified as a recursive process by which geographic population structure can facilitate relatively rapid and sustained evolution in natural populations.
Whitehead and colleagues (31) have shown that cultural hitchhiking suppresses genetic diversity in structured populations when genes and cultural traits are transmitted symmetrically, gene flow is low, intergroup cultural transmission is rare, and cultural evolution is rapid. However, their model of Late Pleistocene humans assumes that cultural traits—not genes—directly and substantially affect reproductive fitness, such that “culturally advanced” groups outcompete others. We relax this assumption on both counts by allowing for a proportion of cultural traits to be selectively neutral and for a proportion of genetic loci to directly affect fitness. Our more general model shows that cultural hitchhiking is not the only avenue through which cultural variation can influence genetic diversity. That is, when CST is sufficiently high, selection can suppress neutral genetic diversity even when cultural evolution is relatively slow, a large proportion of the cultural traits are neutral, and the direct fitness effects of nonneutral cultural traits are small.
Note that CMM can be invoked only for species in which cultural differentiation among regional subpopulations plays a role in mediating gene flow, such as matrilineal whales (32). Is there any empirical evidence for cultural differentiation among hominin groups as early as the Middle Pleistocene? In short, there is no unequivocal archaeological evidence for culturally mediated population structure this early in the Pleistocene record. However, archaeologists have begun to report levels of temporal and regional variability in Middle Pleistocene culture material that contradict the traditional notion of a staid and geographically homogenous technology (33–36). One plausible explanation is that this variability reflects cultural differentiation among regional subpopulations. Of course, it is possible that Middle Pleistocene hominins communicated social group identity by means other than nonperishable (or even perishable) culture material. Recent sequencing of the Neanderthal FOXP2 gene has found it to look modern human-like rather than chimp-like (37). This discovery may imply that the most recent common ancestor of modern humans and Neanderthals had already developed a vocal repertoire more complex than previously thought—a potentially salient cultural marker of community identity that is archaeologically invisible.
Other genetic data provide additional evidence for what could be signs of ancient culturally mediated population structure. Recent research on complete mitochondrial DNA sequences of modern humans has uncovered a 133,000- to 155,000-year-old population subdivision in subSaharan Africa, along with evidence that the resulting subpopulations remained maternally isolated during the following 50,000–100,000 years (38). Although it is currently unknown whether, or to what extent, this maternal population structure was culturally mediated, the finding supports the growing consensus that the Pleistocene hominin presence in Africa was a collection of partially isolated subpopulations rather than a single, geographically continuous population. Finally, the surprisingly high number of matrilineal lineages found for late Middle Pleistocene and Late Pleistocene human populations in subSaharan Africa, exemplified by the cooccurrence of at least 5 evolutionarily viable mitochondrial lineages by ≈150,000 years ago and >40 distinct mitochondrial lineages ≈60,000 years ago, may best illustrate the prominent role that population structure could have played since much earlier in the Pleistocene (38).
In sum, we are not implying that the census population size of hominins was constant during the Pleistocene or that the details of relatively recent demographic and geographic expansions within and then out of Africa did not affect some aspects of hominin genetic diversity. In fact, such effects may help explain the mismatch between 1 prediction of our model of Middle Pleistocene hominins—a high degree of between-group diversity—and the observation that a large majority (>90%) of the genetic diversity in living humans is now contained within continental groups (e.g., 16). Nonetheless, we have found that CMM could have suppressed effective population size for thousands of generations even in the absence of fluctuations in census population size and without the aid of a serial-founder effect. Given that living nonhuman primates do not engage in CMM but modern humans do, that among Pleistocene hominins comparatively low genetic diversity is not unique to modern humans, and that CMM can suppress average gene diversity to a greater extent than geographic distance alone, we propose that CMM might provide the mechanism that suppressed the neutral genetic diversity of (at least some) Middle Pleistocene hominin populations without similarly affecting their hominoid contemporaries. If this is true, then the comparatively low levels of genetic diversity displayed by modern humans, Neanderthals, and their common ancestor may have something to teach us about the influential role that culture played in structuring hominin populations during the Middle Pleistocene.
Methods
In our agent-based model, individuals live on a torus composed of regularly spaced square cells. Individuals that occupy the same cell are considered members of the same group. Each individual carries 100 cultural traits and 1,000 microsatellite, or short tandem repeat (STR), loci, proportions of each of which are selectively neutral (ε and η, respectively). Selectively neutral cultural variants are passed via unbiased cultural transmission as follows: For each neutral trait, an offspring simply adopts the variant displayed by 1 of its 2 parents. Selective cultural variants, however, are passed via biased cultural transmission as follows: For each selective trait, an offspring adopts the variant that has the highest fitness among those displayed by either of its parents and a “teacher” that is chosen randomly from among all members of groups within search radius τ of the offspring's group (including the offspring's group but excluding the offspring itself). Genetic variants are passed via sexual reproduction with recombination, as if contained on a single chromosome. At birth, each cultural trait undergoes single stepwise innovation with probability ι and each genetic locus undergoes single stepwise mutation with probability μ (see SI Text for details).
During each time step (see Fig. S1), each individual attempts to move out of its current group with probability ν. According to the spatially explicit, culturally mediated migration rule, a prospective emigrant can only move to a neighboring group (i.e., one located within the search radius) whose cultural similarity to its own group meets or exceeds the user-defined cultural similarity threshold (CST). Let i represent a prospective emigrant, and I the prospective emigrant's current group. One randomly chosen individual j, from each neighboring group, serves as a representative of the Jth's group's cultural identity. For each cultural trait l, the cultural variant expressed by j (jl) is compared to the set of cultural variants expressed by all members of I at l (Il). If the proportion of cultural traits in which jl is represented in Il exceeds CST, then i adds J to its list of possible destination groups. When multiple groups satisfy CST, i emigrates to one of them chosen at random. If none of the groups satisfies CST, i will not migrate during that time step.
Although migration between groups is nonrandom, mates are chosen randomly from within groups. Reproductive success is based on relative individual fitness (39). Selective cultural traits and selective genetic loci contribute additively and independently to one's relative fitness. There is no epistasis between cultural traits, between genetic loci, or between cultural traits and genetic loci. This is a major difference from models based on Wright's shifting balance theory. Death is stochastic and unrelated to one's relative fitness; that is, each individual experiences a constant probability of death per time step (δ).
To maintain a nearly constant population size, the relative fitness values of all individuals are weighted by a coefficient that is sensitive to the difference between the current population size and a target population size. As is the case in biological populations of modern humans, great apes, and (as presumably was the case in) now extinct Pleistocene hominins, simulated groups do not maintain a constant size or contribute equally to their metapopulation's gene pool. When the number of individuals in a group exceeds the maximum group size (ϕ), the group fissions and half of its members (chosen randomly) colonize 1 of the 8 surrounding cells. In the rare case wherein each of the surrounding cells is inhabited by a greater number of individuals, the group does not fission during that time step. See Table S1 for the values used for all of the parameters introduced above.
Each simulation is initiated with a metapopulation that is characterized by high levels of total and within-group genetic (and cultural) diversity. Each simulation is run for 10,000 time steps, which equate to ≈3,000 overlapping generations. The average gene diversity (H) of a population is defined as
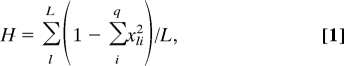 |
where xli is the frequency of the ith allele at the lth locus, q is the total number of alleles expressed at the lth locus, and L is the total number of loci examined; and average gene diversity found within subpopulations (HS) is defined as
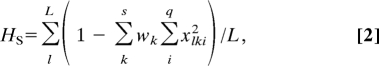 |
where xlki is the frequency of the ith allele at the lth locus in the kth subpopulation, wk is the size of the kth subpopulation in relation to the total population size, s is the number of subpopulations, q is the number of alleles expressed at the lth locus, and L is the total number of loci examined (40). We calculate H and HS for neutral genetic loci only, at an interval of 50 time steps. Only data collected during the final 1,000 time steps of each run are used to calculate mean H and mean HS to avoid the transient, nonequilibrium conditions of the simulation's initialization. See SI Text for a complete model description. The commented source code is freely available upon request.
Supplementary Material
Acknowledgments.
We thank Robert Boyd, Matina Donaldson, Anne Fischer, Ed Green, Steve Kuhn, Michael Lachmann, Sean Myles, Svante Pääbo, Susan Ptak, Amelie Schmolke, Mark Stoneking, Tim Weaver, and 2 anonymous reviewers for comments and contributions. This work was supported by the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This estimate includes the following assumptions: human–chimp coalescent time = 6.5 million years, modern human–Neanderthal divergence time (based on nuclear DNA) = 830,000 years, modern human–Neanderthal split time = 400,000 years, generation time = 20 years, modern human effective population size = 10,000, simple split model, standard neutral model, and mutations accumulate according to a Poisson distribution (to obtain 95% CI) (S. Ptak, R. E. Green, and S. Pääbo, personal communication).
This article contains supporting information online at www.pnas.org/cgi/content/full/0809194105/DCSupplemental.
References
- 1.Gagneux P, et al. Mitochondrial sequences show diverse evolutionary histories of African hominoids. Proc Natl Acad Sci USA. 1999;96:5077–5082. doi: 10.1073/pnas.96.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaessmann H, Wiebe V, Weiss G, Pääbo S. Great ape DNA sequences reveal a reduced diversity and expansion in humans. Nat Genet. 2001;27:155–156. doi: 10.1038/84773. [DOI] [PubMed] [Google Scholar]
- 3.Green RE, et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krings M, et al. A view of Neandertal genetic diversity. Nat Genet. 2000;26:144–146. doi: 10.1038/79855. [DOI] [PubMed] [Google Scholar]
- 5.Serre D, et al. No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol. 2004;2:313–317. doi: 10.1371/journal.pbio.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noonan JP, et al. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harpending HC, et al. Genetic traces of ancient demography. Proc Natl Acad Sci USA. 1998;95:1961–1967. doi: 10.1073/pnas.95.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers AR, Harpending HC. Population growth makes waves in distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 9.Harpending HC, Rogers AR. Genetic perspectives on human origins and differentiation. Annu Rev Genomics Hum Genet. 2000;1:361–385. doi: 10.1146/annurev.genom.1.1.361. [DOI] [PubMed] [Google Scholar]
- 10.Harpending HC, Sherry ST, Rogers AR, Stoneking M. The genetic structure of ancient human populations. Curr Anthropol. 1993;34:483–496. [Google Scholar]
- 11.Harding RM, McVean G. A structured ancestral population for the evolution of modern humans. Curr Opin Genet Dev. 2004;14:667–674. doi: 10.1016/j.gde.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Harris EE, Hey J. X chromosome evidence for ancient human histories. Proc Natl Acad Sci USA. 1999;96:3320–3324. doi: 10.1073/pnas.96.6.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris EE, Hey J. Human demography in the Pleistocene: Do mitochondrial and nuclear genes tell the same story? Evol Anthropol. 1999;8:81–86. [Google Scholar]
- 14.Hawks J, Hunley K, Lee S-H, Wolpoff M. Population bottlenecks and Pleistocene human evolution. Mol Biol Evol. 2000;17:2–22. doi: 10.1093/oxfordjournals.molbev.a026233. [DOI] [PubMed] [Google Scholar]
- 15.Hey J. Mitochondrial and nuclear genes present conflicting portraits of human origins. Mol Biol Evol. 1997;14:166–172. doi: 10.1093/oxfordjournals.molbev.a025749. [DOI] [PubMed] [Google Scholar]
- 16.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 17.Li W-H, Sadler LA. Low nucleotide diversity in man. Genetics. 1991;129:513–523. doi: 10.1093/genetics/129.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahata N, Satta Y. Footprints of intragenic recombination at HLA loci. Immunogenet. 1998;47:430–441. doi: 10.1007/s002510050380. [DOI] [PubMed] [Google Scholar]
- 19.Charlesworth B, Charlesworth D, Barton NH. The effects of genetic and geographic structure on neutral variation. Annu Rev Ecol Evol Syst. 2003;34:99–125. [Google Scholar]
- 20.Goldstein DB, Chikhi L. Human migrations and population structure: What we know and why it matters. Annu Rev Genomics Hum Genet. 2002;3:129–152. doi: 10.1146/annurev.genom.3.022502.103200. [DOI] [PubMed] [Google Scholar]
- 21.Wakeley J. Metapopulation models for historical inference. Mol Ecol. 2004;13:865–875. doi: 10.1111/j.1365-294x.2004.02086.x. [DOI] [PubMed] [Google Scholar]
- 22.Boyd R, Richerson PJ. Culture and the Evolutionary Process. Chicago: Univ of Chicago Press; 1985. [Google Scholar]
- 23.Barham LS. Systematic pigment use in the middle Pleistocene of south-central Africa. Curr Anthropol. 2002;43:181–190. [Google Scholar]
- 24.D'Errico F. The invisible frontier. A multiple species model for the origin of behavioral modernity. Evol Anthropol. 2003;12:188–202. [Google Scholar]
- 25.Barton NH. The probability of fixation of a favoured allele in a subdivided population. Genet Res. 1993;62:149–157. [Google Scholar]
- 26.Wang J, Caballero A. Developments in predicting the effective size of subdivided populations. Heredity. 1999;82:212–226. [Google Scholar]
- 27.Whitlock MC, Barton NH. The effective population size of a subdivided population. Genetics. 1997;146:427–441. doi: 10.1093/genetics/146.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. Proc Sixth Int Congr Genet. 1932;1:356–366. [Google Scholar]
- 29.Wright S. The genetical structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 30.Fagundes NJR, et al. Statistical evaluation of alternative models of human evolution. Proc Natl Acad Sci USA. 2007;104:17614–17619. doi: 10.1073/pnas.0708280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitehead H, Richerson P, Boyd R. Cultural selection and genetic diversity in humans. Selection. 2002;3:115–125. [Google Scholar]
- 32.Whitehead H. Cultural selection and genetic diversity in matrilineal whales. Science. 1998;282:1708–1711. doi: 10.1126/science.282.5394.1708. [DOI] [PubMed] [Google Scholar]
- 33.Clark JD. In: The Origin of Modern Humans and the Impact of Chronometric Data. Aitken MJ, Stringer CB, Mellars P, editors. Princeton: Princeton Univ Press; 1993. pp. 201–215. [Google Scholar]
- 34.Masao FT. In: Continuity or Replacement? Controversies in Homo sapiens Evolution. Brauer G, Smith FH, editors. Rotterdam: Balkema; 1992. pp. 99–109. [Google Scholar]
- 35.McBrearty S. In: Human Roots: Africa and Asia in the Middle Pleistocene. Barham LS, Robson-Brown K, editors. Bristol, United Kingdom: West Acad Specialist; 2001. pp. 81–97. [Google Scholar]
- 36.McBrearty S, Brooks A. The revolution that wasn't: A new interpretation of the origin of modern human behavior. J Hum Evol. 2000;39:453–563. doi: 10.1006/jhev.2000.0435. [DOI] [PubMed] [Google Scholar]
- 37.Krause J, et al. The derived FOXP2 variant of modern humans was shared with Neandertals. Curr Biol. 2007;17:1908–1912. doi: 10.1016/j.cub.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Behar DM, et al. The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson DS. What is wrong with absolute individual fitness? Trends Ecol Evol. 2004;19:245–248. doi: 10.1016/j.tree.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Univ Press; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.