Abstract
The mechanism underlying enhancer-blocking by insulators is unclear. We explored the activity of human β-globin HS5, the orthologue of the CTCF-dependent chicken HS4 insulator. An extra copy of HS5 placed between the β-globin locus control region (LCR) and downstream genes on a transgene fulfills the classic predictions for an enhancer-blocker. Ectopic HS5 does not perturb the LCR but blocks gene activation by interfering with RNA pol II, activator and coactivator recruitment, and epigenetic modification at the downstream β-globin gene. Underlying these effects, ectopic HS5 disrupts chromatin loop formation between β-globin and the LCR, and instead forms a new loop with endogenous HS5 that topologically isolates the LCR. Both enhancer-blocking and insulator-loop formation depend on an intact CTCF site in ectopic HS5 and are sensitive to knock-down of the CTCF protein by siRNA. Thus, intrinsic looping activity of CTCF sites can nullify LCR function.
Keywords: beta-globin genes, insulator, locus control region, epigenetics, transcription regulation
Chromatin insulators are thought to establish domains within which proper enhancer-gene interactions occur: these domains can be visualized in Drosophila cells where a protein complex including Su(Hw) forms loops that tether gypsy insulators to the nuclear lamina (1, 2). Insulators can also interfere with enhancer-gene interaction when placed between the two elements, but the molecular mechanisms underlying enhancer-blocking are not well understood.
In vertebrates, the protein CTCF is associated with enhancer-blocking (3). On maternal chromosomes, the CTCF-dependent imprinting control region (ICR) in the Igf2/H19 locus is thought to form a small loop with a second site, DMR1, upstream of Igf2 that includes Igf2 and restricts access to the gene by enhancers it shares with H19 (4, 5). Because detection of CTCF at DMR1 is dependent on its interaction at ICR, it is not clear whether CTCF or other proteins interact at DMR1 and participate in the long-range interactions. Other data indicate the ICR directly contacts Igf2 and the enhancers (6), suggesting that a complex series of chromosomal interactions may be involved in enhancer-blocking at this locus. CTCF also mediates the enhancer-blocking activity of the chicken β-globin insulator, 5′HS4, which forms the upstream border of the globin locus (7). Although a prediction (8), enhancer-blocking through loop formation by interacting CTCF insulator sites has not been demonstrated.
Locus control regions (LCRs) are complex enhancers that activate genes over long distances through their ability to establish close contacts with target promoters (9). For example, the β-globin LCR and active globin genes come into proximity to form a chromatin loop in erythroid cells (10, 11), an interaction that requires the erythroid factors GATA-1 and erythroid Kruppel-like factor (EKLF) (12, 13). The human β-globin LCR is composed of four DNase I hypersensitive sites, HS1 to HS4, far upstream of the structural genes. HS5, 3-Kb upstream of HS4, is more widely detected, has no enhancer activity, and is orthologous to the chicken HS4 insulator. HS5 and 3′HS1, downstream of the locus, are both sites of CTCF interaction and loop together in early erythroid cells before globin genes are expressed (14, 15). However, the function of HS5 in vivo is not clear. Chromosomal deletion of mouse HS5 had no significant effect on β-globin expression or on silent odorant receptor genes located downstream of HS5, indicating that HS5 is not required as an insulator at its endogenous location (16, 17). In contrast, an ectopic globin gene placed upstream in a human transgenic globin locus, and separated from the LCR by HS5, failed to be activated, although some evidence suggested the blocking varied in a developmental stage-specific fashion (18, 19).
Earlier studies suggested that human HS5 could function as a transcriptional enhancer-blocker when placed between the LCR and the globin genes on a transgene (20). We used this system to investigate the mechanism underlying enhancer-blocking. Ectopic human HS5 diminished recruitment of pol II, histone acetylation, and transcription activator NF-E2 to the target β-globin gene but not to the LCR. However, ectopic HS5 caused a redistribution of the erythroid factors GATA-1 and EKLF away from LCR HS2 to HS4 and interrupted looping between the LCR and β-globin gene. Loss of enhancer-looping coincided with the formation of a new loop between ectopic and endogenous HS5 that included the LCR. These results show that chromatin conformation can be reconfigured by an insulator to regulate LCR activity in chromosomes.
Results and Discussion
CTCF is Recruited to a Copy of Human HS5 Placed Between the β-globin LCR and Downstream Genes.
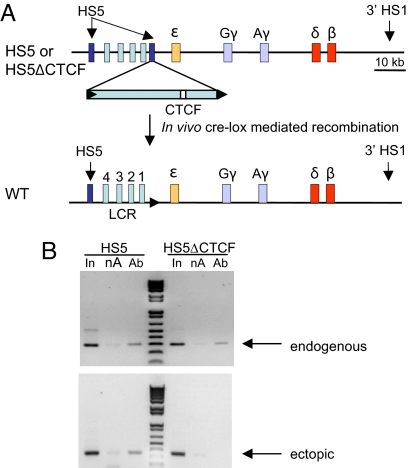
We investigated the insulator function of HS5 in mice carrying a single copy of a complete β-globin locus transgene with a copy of HS5 placed between the LCR and ε-globin, containing either an intact (HS5) or mutated CTCF site (ΔCTCF) or no HS5 (WT) (Fig. 1A) (20). CTCF occupies endogenous HS5 and 3′HS1 sites flanking the mouse and human β-globin loci in vivo (14, 21). ChIP and real-time quantitative PCR (qPCR) indicated that CTCF was recruited to these sites on the human transgene in anemic spleen erythroid cells of all three lines of mice [supporting information (SI) Fig. S1] but not in brain cells (data not shown). To distinguish CTCF recruitment specifically to endogenous or ectopic HS5, we used nested PCR to uniquely amplify a fragment from each HS5 copy and then amplify the CTCF site therein (Fig. 1B). CTCF interacted at ectopic HS5 but was absent when the CTCF motif was deleted, while binding to the endogenous HS5 site was retained in both mouse lines.
Fig. 1.
Ectopic HS5 recruits CTCF. (A) A copy of human HS5 (2.6 kbp) was floxed and introduced between HS1 and ε-globin on a human β-globin YAC (A201F4.3, inverted orientation) by homologous recombination in S. cerevisiae (20). The purified YAC DNA was used to generate lines of transgenic mice carrying either an intact transgene (HS5) or an HS5-containing transgene with a deleted CTCF site (HS5ΔCTCF). HS5 was then removed by expression of Cre recombinase, leaving a pseudo-WT control locus altered only by retention of the 34-bp Lox site. The CTCF motif within ectopic HS5 is represented by the open rectangle. (B) Adult anemic spleen chromatin was sonicated to 800- to 1000-bp fragments before performing ChIP. Conventional PCR was carried out with nested primers to amplify uniquely the endogenous and ectopic copies of HS5 (see Table S1 for primer sequences). PCR products were resolved on a 1.5% agarose gel and visualized with ethidium bromide. Ab, anti-CTCF; In, input; nA, no antibody.
On transgenes in mice, the human ε- and γ-globin genes are expressed predominantly in the blood islands of the murine yolk sac early in development. β-globin transcription becomes predominant during mouse fetal liver erythropoiesis and in adult life, along with a minor contribution from δ-globin. Ectopic HS5 reduced globin gene transcription at all stages of mouse development as determined by RT-qPCR (Fig. S2), substantially recapitulating earlier work using semiquantitative PCR (20). Deletion of the HS5 CTCF site restored γ- and β-globin expression, although it had little effect on ε-globin expression. We focused our attention on HS5 enhancer blocking of β-globin expression in adult spleen, which becomes more than 90% erythroid in animals made anemic by treatment with phenylhydrazine.
RNA Polymerase II Recruitment and Histone Acetylation at Active Globin Genes Are Reduced by Ectopic HS5 in a CTCF-Dependent Manner but Are Unchanged at the LCR.
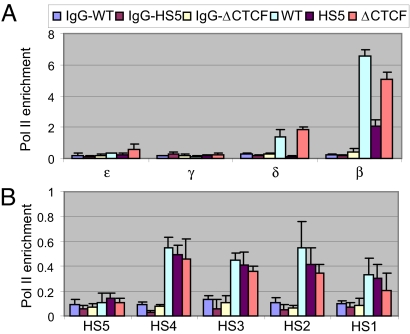
RNA polymerase II (pol II) is recruited to the β-globin LCR HSs early in erythroid differentiation and subsequently transferred to promoters of active genes (22), and this process might be blocked by an insulator. Alternatively, pol II might be recruited to the β-globin gene independent of the LCR (23) and enhancer-blocking might impede pol II release from the promoter. ChIP assays with an antibody against pol II were used to distinguish between these possibilities. Detection of pol II was reduced at the δ- and β-globin genes in HS5 mice but was restored in HS5ΔCTCF mice (Fig. 2A), in parallel with restoration of transcription. In contrast, pol II recruitment to the LCR was unaffected by HS5 insertion (Fig. 2B). Because ectopic HS5 impaired recruitment of pol II to a target globin gene but not to the LCR, we conclude that enhancer-blocking involves a step downstream of pol II recruitment to the LCR.
Fig. 2.
RNA polymerase II recruitment to transcribed globin genes is reduced by HS5 in a CTCF-dependent manner but recruitment to the LCR is unaffected. ChIP was carried out with antibodies to RNA pol II or control IgG and anemic spleen chromatin. Real-time qPCR was carried out with primers and TaqMan probes recognizing (A) individual globin genes or (B) LCR HS sites. For sequences of primers and TaqMan probes, see Tables S2 and S3. Note that the HS5 primers and probe recognize both the endogenous and ectopic copies of HS5. The mouse actin signal was used to normalize results between experiments. Error bars represent SEM and Student's t Test indicates P < 0.02, when comparing the adult globin promoters in WT versus HS5 mice.
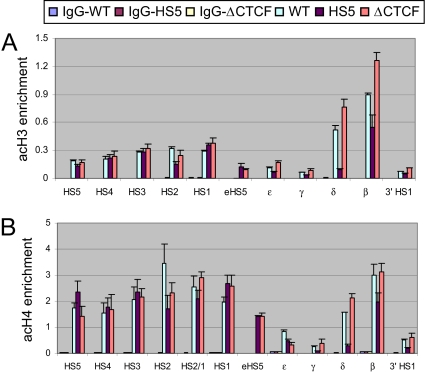
The LCR and the transcribed globin genes in mouse erythroid cells exist within extended but discontinuous regions of acetylated H3 and H4 and di-methylated H3 K4 marks of active chromatin (21). ChIP assays showed that domains characterized by these epigenetic marks form on a human transgene in erythroid cells in mice (Fig. 3). Ectopic HS5 diminished the levels of acH3 and acH4 at the δ- and β-globin genes in a CTCF-dependent fashion, but had no notable influence on histone acetylation in the LCR. Similar results were obtained with an antibody to di-methylated H3 K4 (Fig. S3). We conclude that formation of a domain of acetylated histones normally encompassing the transcribed globin genes is impeded by ectopic HS5 dependent on CTCF, but that the LCR is unaffected, similar to recruitment of pol II in HS5 mice.
Fig. 3.
Histone H3 and H4 acetylation at adult δ- and β-globin genes is reduced by ectopic HS5 in a CTCF-dependent manner, while acetylation across the LCR is not reduced. ChIP assays and real-time qPCR analysis was carried out as described in the legend to Fig. 2, using antibodies against (A) di-acetylated histone H3 and (B) tetra-acetylated H4. Note that the eHS5 primer recognizes the unique junction between ectopic HS5 and sequences upstream of ε-globin. The results were normalized to the mouse actin signal and are shown ± SEM. P < 0.04 when comparing the adult globin promoters in WT versus HS5 mice.
Activator and Coactivator Recruitment to Active Globin Promoters Is Reduced by Ectopic HS5 in a CTCF-Dependent Manner but the Effect on Recruitment to the LCR Is Variable.
The results thus far show that HS5 fulfills the classic prediction that an enhancer-blocker will not directly affect the enhancer but will only interfere with the signal it sends to a target gene. Because pol II and histone modifications were reduced at the β-globin gene by ectopic HS5, we asked whether the distribution of activators and coactivators involved in their recruitment to the locus was affected. ChIP was used to examine recruitment of CBP, a histone acetyltransferase important in erythroid development (24). CBP binding to the β-globin gene promoter was diminished in HS5 mice and restored after deletion of the CTCF site (Fig. 4A). However, CBP recruitment to the LCR was unaffected by HS5 enhancer-blocking, consistent with the histone acetylation patterns in HS5 mice.
Fig. 4.
Activator and coactivator recruitment to the β-globin gene promoter is reduced by HS5 in a CTCF-dependent manner, but HS5 affects LCR recruitment variably. ChIP assays and real-time qPCR analysis was carried out as described in the legend to Fig. 2 using antibodies against (A) CBP, (B) NF-E2, (C) EKLF, or (D) GATA-1. mNDN, mouse necdin control amplicon. The mouse β-globin promoter signal was used to normalize data between experiments and the results are shown ± SEM. Values between P < 0.001 and P < 0.05 were obtained when comparing the adult globin promoters in WT versus HS5 mice and P < 0.03 when comparing HS2 and HS3 (C) and HS2–4 (D).
NF-E2, GATA-1, and EKLF are erythroid transcription activators known to be recruited to the β-globin LCR and gene, and all three factors have been shown to interact with or be important for recruitment of CBP to the LCR (24–29). ChIP assays with antibodies to these proteins revealed that, in each case, activator recruitment to the β-globin promoter was compromised by HS5 and restored after deletion of the HS5 CTCF site (Fig. 4 B–D). NF-E2 interaction in the LCR was not affected by HS5. However, ectopic HS5 reduced recruitment of GATA-1 and EKLF to HS2 to HS4 with some augmentation of GATA-1 at HS1.
Thus, enhancer-blocking by HS5 affects recruitment of all of the activators investigated to the β-globin gene. In contrast, recruitment of pol II, active histone modifications, CBP, and NF-E2 to the LCR was unaffected by enhancer-blocking, while there was diminished detection of GATA-1 and EKLF at LCR HS2 to HS4.
HS5 Enhancer Blocking Involves Interference with the β-globin/LCR Enhancer Loop and Formation of an Insulator Loop.
GATA-1 and EKLF are required for looping between the LCR and β-globin gene, which occurs during gene activation in erythroid cells, but NF-E2 is not (12, 13, 30). LCR HS2 and HS3, where ectopic HS5 enhancer blocking diminished GATA-1 and EKLF recruitment, appear to form the closest interactions with the β-globin gene when enhancer loop formation occurs (10). Therefore, we next asked whether specific reduction of GATA-1 and EKLF at HS2 to HS4 within the LCR by ectopic HS5 corresponded to a reduction in LCR/β-globin looping. A chromosome conformation capture (3C) experiment was performed using a primer within an LCR HS2 to HS4 fragment as the anchor for PCR analysis of proximity to different regions of the locus in the three lines of transgenic mice (31).
The normal proximity in erythroid cells between the LCR and β-globin gene was reduced by half in HS5 mice (Fig. 5A). Deletion of the CTCF site in ectopic HS5 restored proximity, consistent with restoration of transcription. There was no alteration of LCR proximity with other regions across the locus in HS5 mice, nor was the expected proximity between HS5 and 3′HS1 changed (Fig. S4). We conclude that enhancer blocking by HS5 disrupts physical interaction between the LCR and β-globin gene in erythroid cells in a CTCF-dependent fashion. The HS5-dependent redistribution away from HS2 to HS4 of GATA-1 and EKLF, factors required to establish or maintain long-range contacts between the LCR and gene, is consistent with the idea that contact between LCRs and their target promoters augments local transcription-factor concentrations (32, 33).
Fig. 5.
Ectopic HS5 interferes with loop formation between the β-globin LCR and gene and establishes close proximity with endogenous HS5. (A) Chromatin from adult spleen was cross-linked with formaldehyde, digested with EcoRI, and re-ligated. After reversal of cross-links, ligation efficiencies between the anchor primer in the LCR HS2 to HS4 fragment and positions across the locus were determined. To compare results from different mouse lines, the data were normalized to an interaction in the mouse ERCC gene. The strongest cross-linking frequency was set to 1. (B) Chromatin from adult spleen was treated as above but was digested with Apo I and religated. The data were normalized to an interaction in the mouse CalR gene and the strongest cross-linking frequency was set to 1. See Table S4 for primer sequences. The results of multiple chromatin preparations with each enzyme are shown ± SEM. P < 0.01 when comparing proximity of the LCR to β-globin in WT versus HS5 mice.
Enhancer blocking by HS5 might involve a direct interaction with the LCR or with the β-globin gene as was reported for the CTCF-dependent Igf2/H19 ICR (6), or HS5 might regulate enhancer looping by forming a competing loop that topologically isolates the LCR away from its target genes. In vitro and on episomes, forced loop formation restricted an enhancer's activity (34, 35). To distinguish among these mechanisms, we increased the sensitivity of the 3C assay by cleavage of chromatin into smaller fragments by Apo I, which permitted use of an anchor primer downstream of HS1 that is shared among the three lines of mice and is adjacent to the 3′ end of the HS5 insert in HS5 and ΔCTCF mice, or to the residual loxP site in pseudo-WT mice.
We observed a striking interaction between ectopic HS5 and endogenous HS5 specifically in erythroid cells of HS5 mice (not in brain cells, data not shown), indicative of formation of a new loop (Fig. 5B). This interaction, which did not occur in the WT transgenic locus, was sensitive to removal of the CTCF site in ectopic HS5. We did not observe close contacts between ectopic HS5 and LCR HS2 to HS4 or with β-globin. These results indicate that an insulator loop around an LCR can circumscribe its activity and reveal an intrinsic interaction and looping capability of CTCF-insulator sites. We were unable to demonstrate an insulator loop around the globin genes in HS5 mice, likely for technical reasons. There was only a low signal for proximity between ectopic HS5 and 3′HS1 (see Fig. 5B), and examination of the corresponding ligation products by gel electrophoresis revealed a smear rather than a tight band. However, because endogenous HS5 maintains close proximity with both 3′HS1 and ectopic HS5 in HS5 mice, we expect a loop between ectopic HS5 and 3′HS1 exists.
Reduction of CTCF Protein by siRNA Reduces Insulator Loop Formation and Restores β-globin Transcription.
To further probe the CTCF dependence of the insulator loop, we reduced CTCF protein by siRNA. Spleen cells of phenylhydrazine-treated HS5 mice were transfected with a CTCF siRNA or a control siRNA and maintained in culture for 3 days. Western blot analysis demonstrated decreased CTCF protein in cells treated with the CTCF siRNA compared with control siRNA-treated cells (Fig. 6A). Correspondingly, 3C analysis showed that knock down of CTCF reduced proximity between ectopic and endogenous HS5 in HS5 mice, whereas a control siRNA did not, indicating that insulator loop formation requires CTCF (Fig. 6B). We next asked whether reducing insulator loop formation between endogenous and ectopic HS5 compromised enhancer blocking. Reduced insulator looping after 3 days of CTCF siRNA treatment corresponded with restoration of β-globin transcription to levels similar to those in WT and ΔCTCF mice (Fig. 6C and see Fig. S2 for comparison).
Fig. 6.
Reduction of CTCF by siRNA decreases loop formation between endogenous and ectopic HS5 and restores β-globin transcription. (A) Proteins were extracted from spleen cells of mice with an intact ectopic HS5, 3 days after transfection with control siRNA (siCTRL) or siRNA directed against CTCF (siCTCF). Western blot analysis was carried out and α-tubulin served as the loading control. (B) Spleen cells of HS5 mice were transfected with control siRNA or CTCF-directed siRNA and incubated for 3 days before isolation of chromatin for 3C analysis, to assess proximity between ectopic HS5 and other regions of the locus. Apo I cleavage was used and the data were normalized to an interaction in the mouse CalR gene, with the strongest cross-linking frequency set to 1. The results are shown ± SEM. P < 0.02 when comparing proximity between the two HS5 sites −/+ CTCF knock-down. (C) RNA was prepared from spleen cells of adult HS5 mice that were untransfected (CTRL) or at 2 or 3 days (d) after transfection with control siRNA (siCTRL) or siRNA directed against CTCF (siCTCF). RT-qPCR was carried out with a probe for the human β-globin gene and the data were normalized to mouse 18S RNA and are shown ± SEM. P < 0.03 when comparing β-globin transcripts before and 3 days after CTCF knock-down. (D) A model of ectopic HS5 loop formation with endogenous HS5 to block LCR activation of β-globin. In the active β-globin gene cluster, endogenous HS5 and 3′HS1 form a loop within which globin genes may interact with LCR HS4–1 for activation. The insertion of an ectopic copy of HS5 results in LCR/enhancer blocking, reducing LCR/gene interactions and resulting in formation of a new loop including the LCR and excluding the downstream genes. Although transcription factors can still be enriched at the LCR, without direct interaction between LCR and target genes, efficient communication of transcription “signals” are significantly compromised.
Together, these results show that β-globin HS5 displays intrinsic, portable enhancer-blocking activity manifest through formation of a new CTCF-dependent loop, apparently of greater stability than that formed between the LCR and β-globin gene. This loop includes the LCR and excludes the downstream genes, providing experimental evidence that such a loop would, in fact, restrict the activity of an LCR in vivo (Fig. 6D). The results demonstrate the feasibility of manipulating LCR function through reconfiguring chromatin loops at an endogenous locus.
Why might an insulator loop predominate in erythroid cells in which the protein factors known to participate in enhancer-looping are abundant? The formation of alternative loops with different regulatory outcomes at the Kit locus can be dynamically affected by the predominance of different GATA family members during hematopoietic differentiation (36). However, the alternative enhancer or insulator loops we observed formed within the same cellular milieu. Recent evidence shows that the cohesin complex, that mediates sister chromatid exchange by formation of a protein ring around the DNA strands, functionally associates with CTCF sites (37–39). ChIP experiments revealed that the cohesin complex is recruited to human HS5 and 3′HS1 in mouse erythroid cells (Fig. S5). We propose that cohesin may be one of the components contributing to the strength or stability of interaction between CTCF sites.
It remains unclear how topological isolation of the LCR and genes prevents their productive interaction. One possibility is that conformational changes within the insulator loop result in physical masking of the LCR. However, we do not favor this mechanism because pol II and at least some activators and coactivators can still access the LCR normally within the insulator loop, and the LCR HSs remain strongly sensitive to DNase I (data not shown). It will be important to test whether a gene isolated within the LCR loop can be activated by it, as would be predicted. Alternatively, as proposed for Igf2 in the H19 imprinted locus (5), it might be the structure of the loop sequestering the globin genes that restricts the access of the LCR. However, we prefer the interpretation that loss of enhancer-looping through stable formation of the insulator loop is the salient feature of enhancer-blocking. Because of the intrinsic propensity of CTCF sites to loop together, we cannot formally rule out the possibility that ectopic HS5, in the absence of participation in a loop, is sufficient for enhancer-blocking.
The function of the abundant CTCF sites genome-wide is intriguing and unknown (40, 41). Their widespread distribution suggests a general role in chromosome organization or tethering. The intrinsic enhancer-blocking potential of CTCF sites, such as HS5 may, perforce, result in their evolutionary exclusion from locations where they would compromise long-range regulatory interactions by competitive loop formation, which would explain their presence flanking coordinately regulated gene loci (42). It will be important to analyze the enhancer-blocking capability and interactions participated in by CTCF sites found throughout the genome to further understand their function.
Materials and Methods
Transgenic Mice.
Mice carrying a WT human β-globin gene locus transgene, or a transgene with an extra copy of HS5 with or without an intact CTCF site, have been described previously (20). The ectopic HS5 inserts were flanked by loxP sites. Single copy transgenic lines were selected because, technically, this configuration is ideal to achieve correct Cre-lox recombination and to avoid possible variation in gene-expression levels among multicopy transgenics. The sequence within HS5 that is italicized was deleted to produce the HS5ΔCTCF locus: TGCTGTTATGACCACTAGAGGGAAG AAGATACC. All experiments were performed under a protocol approved by the National Institutes of Health Animal Welfare Committee.
Preparation of Animal Cells.
Definitive erythroid cells were collected from E14.5 fetal liver or from adult anemic spleen. Brain cells were collected from E14.5 embryos as nonerythroid control cells. The tissues were first cut into small pieces before being passed through needles of different sizes (19GA, 23GA, and 25GA sequentially) to obtain single-cell suspensions.
Nested PCR.
Two forward primers were designed flanking the CTCF site in HS5 and were used in nested PCR with a reverse primer outside and downstream of ectopic HS5, to uniquely amplify this copy. Because the orientation of the two HS5 sites is inverted, the same two primers flanking the HS5 CTCF sites could be used as reverse primers, together with a forward primer in sequences outside and upstream to uniquely amplify endogenous HS5 (see Table S1 for primer sequences and illustration). PCR products were resolved on a 1.5% agarose gel and visualized with ethidium bromide.
RNA Preparation and Reverse Transcription.
RNA was purified from E9.5 yolk sacs, E14.5 fetal liver, and adult anemic spleen cells with Triazol reagent (Invitrogen). RNA (1 μg) was treated with DNaseI before reverse transcription with SuperScript First-Strand Kit (Invitrogen). cDNA was then diluted to 100 μl, and 5 μl was used as template for quantitative real-time PCR.
Chromatin Immunoprecipitation.
ChIP assays were carried out essentially as described (43), with some modifications. Briefly, 20 to ≈50 million cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Nuclei were prepared and digested with 200U of micrococcal nuclease at 37 °C for 5 min, followed by sonication to an average chromatin fragment size of 200 to 500 bp. Precleared chromatin was incubated with antibodies overnight at 4 °C and immunoprecipitated with protein A/G Plus agarose beads (Santa Cruz Biotechnology). Immunoprecipitated material was extensively purified after reversal of the cross-links. DNA was diluted in 200 μl of TE for real-time PCR.
Quantitative Real-Time PCR.
Real-Time PCR using TaqMan chemistry was performed to quantify enriched DNA from ChIP and cDNA from reverse transcription on an ABI Prism 7900HT (PE Applied Biosystems). The threshold was set to cross a point at which PCR amplification was linear, and the number of cycles (Ct) required to reach the threshold was collected and analyzed using Microsoft Excel. The analyses were performed in duplicate using multiple experimental samples. Primer and probe sequences appear in Tables S2 and S3.
Chromatin Conformation Capture Assay.
The 3C assay was performed as described (11, 12), with minor modification. Briefly, formaldehyde-fixed nuclei were digested with EcoRI overnight, followed by ligation with T4 DNA ligase at 16 °C for 4 h. Cross links were reversed and DNA was extensively purified. Specific ligation between two fragments was confirmed by sequencing the PCR products. Primer efficiency and ligation efficiency were determined as described (11). Quantification of ligated products was performed by real-time qPCR with published primers for EcoRI 3C (15) and new primers for ApoI 3C (Table S4). To compare results from different mouse lines, the results were normalized to an interaction in the mouse ERCC gene for EcoRI 3C and the mouse CalR for ApoI 3C samples. The use of single-copy transgenic lines avoids the possible complication of inter-copy interactions.
siRNA Knock-Down of CTCF.
Erythroid cells were isolated from HS5 mice and cultured in IMDM media with 20% FBS, insulin, erythropoietin, and transferrin. Short interfering RNA (siRNA) mediated knock-down of CTCF protein was carried out using mouse CTCF ON-TARGETplus SMART pool siRNA (Dharmacon). siCONTROL Non-Targeting siRNA (Dharmacon) was used as negative control. The siRNAs (5 μM) were delivered into erythroid cells using the Amaxa Biosystems Nucleofector/Cell Line Nucleofector Kit V. Cells were harvested after 48 or 72 h for Western blotting, RNA expression analysis, and Apo I 3C experiments.
Western Blotting.
Proteins were extracted from adult anemic spleen cells of HS5 mice that had been transfected with control siRNA or siRNA directed against CTCF. Antibodies used for Western blotting were rabbit anti-CTCF (Millipore), mouse anti α-tubulin (Sigma), and appropriate anti-rabbit or anti-mouse secondary antibodies conjugated to horseradish peroxidase for detection (Western Lightning Chemiluminescence Kit, Perkin-Elmer Life Sciences).
Antibodies.
Antibodies to CBP, GATA-1, pol II, and NF-E2 were obtained from Santa Cruz Biotechnology. Antibodies to CTCF, acH3, acH4 and H3 K4me2 were from Millipore. Rad21 and EKLF antibodies were from AbCam and SMC1 antibodies were from Bethyl Laboratories
Supplementary Material
Acknowledgments.
We thank Drs. Elissa Lei, Vasily Studitsky, and Rohinton Kamakaka for critical comments on the manuscript. This research was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808506106/DCSupplemental.
References
- 1.Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol. 2003;162:565–574. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capelson M, Corces VG. Boundary elements and nuclear organization. Biol Cell. 2004;96:617–629. doi: 10.1016/j.biolcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 5.Kurukuti S, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon YS, et al. Analysis of the H19ICR insulator. Mol Cell Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 8.Pant V, et al. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol Cell Biol. 2004;24:3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean A. On a chromosome far, far away: LCRs and gene regulation. Trends Genet. 2006;22(1):38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Carter D, Chakalova L, Osborne CS, Dai Y, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 11.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 12.Vakoc CR, et al. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 13.Drissen R, et al. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palstra RJ, et al. The β-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 16.Bender MA, et al. Description and targeted deletion of 5′ hypersensitive site 5 and 6 of the mouse β-globin locus control region. Blood. 1998;92:4394–4403. [PubMed] [Google Scholar]
- 17.Farrell CM, et al. A large upstream region is not necessary for gene expression or hypersensitive site formation at the mouse β-globin locus. Proc Natl Acad Sci USA. 2000;97:14554–14559. doi: 10.1073/pnas.97.26.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanimoto K, Liu Q, Bungert J, Engel JD. Effects of altered gene order or orientation of the locus control region on human beta-globin gene expression in mice. Nature. 1999;398:344–348. doi: 10.1038/18698. [DOI] [PubMed] [Google Scholar]
- 19.Wai AW, et al. HS5 of the human beta-globin locus control region: a developmental stage-specific border in erythroid cells. EMBO J. 2003;22:4489–4500. doi: 10.1093/emboj/cdg437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanimoto K, et al. Human beta-globin locus control region HS5 contains CTCF- and developmental stage-dependent enhancer-blocking activity in erythroid cells. Mol Cell Biol. 2003;23:8946–8952. doi: 10.1128/MCB.23.24.8946-8952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulger M, et al. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol Cell Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SI, Bultman SJ, Jing H, Blobel GA, Bresnick EH. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol Cell Biol. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawado T, Halow J, Bender MA, Groudine M. The β -globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 2003;17:1009–1018. doi: 10.1101/gad.1072303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Bieker JJ. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng X, Reginato MJ, Andrews NC, Lazar MA. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung HL, Kim AY, Hong W, Rakowski C, Blobel GA. Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J Biol Chem. 2001;276:10715–10721. doi: 10.1074/jbc.M007846200. [DOI] [PubMed] [Google Scholar]
- 28.Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol Cell Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Im H, et al. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci USA. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kooren J, et al. Beta-globin active chromatin hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J Biol Chem. 2007;282:16544–16552. doi: 10.1074/jbc.M701159200. [DOI] [PubMed] [Google Scholar]
- 31.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 32.Chambeyron S, Bickmore WA. Does looping and clustering in the nucleus regulate gene expression? Curr Opin Cell Biol. 2004;16:256–262. doi: 10.1016/j.ceb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Fraser P. Transcriptional control thrown for a loop. Curr Opin Genet Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Ameres SL, et al. Inducible DNA-loop formation blocks transcriptional activation by an SV40 enhancer. EMBO J. 2005;24:358–367. doi: 10.1038/sj.emboj.7600531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bondarenko VA, Jiang YI, Studitsky VM. Rationally designed insulator-like elements can block enhancer action in vitro. EMBO J. 2003;22:4728–4737. doi: 10.1093/emboj/cdg468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jing H, et al. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 39.Stedman W, et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Dillon N, Sabbattini P. Functional gene expression domains: defining the functional unit of eukaryotic gene regulation. Bioessays. 2000;22:657–665. doi: 10.1002/1521-1878(200007)22:7<657::AID-BIES8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 43.Song S-H, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range β-globin locus control region function. Mol Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.