Abstract
SOX4 is a critical developmental transcription factor in vertebrates and is required for precise differentiation and proliferation in multiple tissues. In addition, SOX4 is overexpressed in many human malignancies, but the exact role of SOX4 in cancer progression is not well understood. Here we have identified the direct transcriptional targets of SOX4 using a combination of genome-wide localization ChIP-chip analysis and transient overexpression followed by expression profiling in a prostate cancer model cell line. We have also used protein-binding microarrays to derive a novel SOX4-specific position-weight matrix and determined that SOX4 binding sites are enriched in SOX4-bound promoter regions. Direct transcriptional targets of SOX4 include several key cellular regulators such as EGFR, HSP70, Tenascin C, Frizzled-5, Patched-1, and Delta-like 1 We also show that SOX4 targets 23 transcription factors such as MLL, FOXA1, ZNF281, and NKX3-1 In addition, SOX4 directly regulates expression of three components of the RNA-induced silencing complex (RISC), namely Dicer, Argonaute 1, and RNA Helicase A. These data provide new insights into how SOX4 impacts developmental signaling pathways and how these changes may influence cancer progression via regulation of gene networks involved in microRNA processing, transcriptional regulation, the TGFβ, Wnt, Hedgehog, and Notch pathways, growth factor signaling, and tumor metastasis.
Keywords: Prostate Cancer, SOX4, Transcription, Systems Biology, ChIP-chip
INTRODUCTION
The sex determining region Y-box 4 (SOX4) gene is a developmental transcription factor important for progenitor cell development and Wnt signaling (1, 2). SOX4 is a 47 kDa protein that is encoded by a single exon and contains a conserved high-mobility group (HMG) DNA binding domain (DBD) related to the TCF/LEF family of transcription factors that mediate transcriptional responses to Wnt signals. SOX4 directly interacts with β-catenin, but its precise role in the Wnt pathway is unknown (2). In adult mice, SOX4 is expressed in the gonads, thymus, T- and pro-B-lymphocyte lineages and to a lesser extent in the lungs, lymph nodes and heart (1). Embryonic knock-out of SOX4 is lethal around day E14 due to cardiac failure and these mice also showed impaired lymphocyte development (3). Tissue specific knock-out of SOX4 in the pancreas results in failure of normal development of pancreatic islets (4). SOX4 heterozygous mice have impaired bone development (5), whereas prolonged expression of SOX4 inhibits correct neuronal differentiation (6). These studies suggest a critical role for SOX4 in cell fate decisions and differentiation.
While SOX2 is known to be critical for maintenance of stem cells (7), SOX4 may specify transit-amplifying progenitor cells that are the immediate daughters of adult stem cells and have been proposed to be the population that gives rise to cancer stem cells. In humans, SOX4 is expressed in the developing breast and osteoblasts and is upregulated in response to progestins (8). SOX4 is upregulated at the mRNA and protein level in prostate cancer cell lines and patient samples and this upregulation is correlated with Gleason score or tumor grade (9). In addition, SOX4 is overexpressed in many other types of human cancers, including leukemias, melanomas, glioblastomas, medulloblastomas (10), and cancers of the bladder (11) and lung (12). A meta-analysis examining the transcriptional profiles of human cancers found SOX4 to be one of 64 genes upregulated as a general “Cancer Signature” (12), suggesting that SOX4 has a role in many malignancies. Furthermore, SOX4 cooperates with Evi1 in mouse models of myeloid leukemogenesis (13). Recently, we showed that SOX4 can induce anchorage-independent growth in prostate cancer cells (9). Consistent with the concept that SOX4 is an oncogene, three independent studies searching for oncogenes have found SOX4 to be one of the most common retroviral integration sites, resulting in increased mRNA (14-16).
Despite these findings, the role that SOX4 plays in carcinogenesis remains poorly defined. While the transactivational properties of SOX4 have been characterized (17), genuine transcriptional targets remain elusive. To date, three studies have used expression profiling of cells after either siRNA knockdown or overexpression of SOX4 to identify candidate downstream target genes (9, 11, 18). Very recently, 31 SOX4 target genes were confirmed by chromatin immunoprecipitation (ChIP) in a hepatocellular carcinoma cell line (19). While interesting, this study was limited by the fact that it focused on a specific tumor stage transition and did not use a genome-wide localization approach.
Here, we have performed a genome-wide localization analysis using a ChIP-chip approach to identify those genes that have SOX4 bound at their proximal promoters in human prostate cancer cells. We have identified 282 genes that are high-confidence direct SOX4 targets, including many genes involved in microRNA processing, transcriptional regulation, developmental pathways, growth factor signaling, and tumor metastasis. We have also utilized unique protein-binding DNA microarrays (PBMs) (20-22) to query the binding of recombinant SOX4 to every possible 8-mer. The PBM-derived SOX4 DNA binding data will further facilitate computational analyses of genomic SOX4 binding sites. These data provide new insights into how SOX4 impacts key growth factor and developmental pathways and how these changes may influence cancer progression.
MATERIALS AND METHODS
Cell Culture and Stable Cell Line Construction
All cell lines were cultured as described by ATCC except LNCaP cells which were cultured with T-Medium (Invitrogen, Carlsbad, CA). HA tagged SOX4 was cloned into the pHR-UBQ-IRES-eYFP-ΔU3 lentiviral vector (gift from Dr. Hihn Ly, Emory University) and stable cells isolated as previously described (23).
Chromatin Immunoprecipitation
Two 90% confluent P150s of both LNCaP-YFP and LNCaP-YFP/HA-SOX4 or RWPE-1-YFP and RWPE-1-YFP/HA-SOX4 cells were formaldehyde fixed, sonicated and ChIP assay performed as described previously (23). Anti-HA 12CA5 or mouse IgG was used to immunoprecipitate protein-DNA complexes overnight at 4°C and collected using Dynal M280 sheep anti-mouse IgG beads for 2 hours. Dynal beads were washed, protein-DNA complexes eluted and DNA purified as described previously (24). A detailed description of the ChIP-chip protocol can be found in the supplemental methods. Anti-HA 12CA5, Anti-Flag-M2 (Sigma-Aldrich, St. Louis, MI) or mouse IgG was used to immunoprecipitate protein-DNA complexes overnight at 4°C. All PCR primers used in ChIP-PCR can be found in Supplemental Table 7.
ChIP-chip Analysis
To determine the direct SOX4 target genes on a global scale we performed ChIP assays in triplicate from the LNCaP cell line stably expressing SOX4 and in duplicate from a control cell line that expressed YFP alone. Immunoprecipitated and input DNA were subjected to whole genome amplification, Cy3/Cy5 fluorescent labeling, and hybridization to the NimbleGen 25K human promoter array set. Input and immunoprecipitated DNA isolated from LNCaP-YFP and LNCaP-YFP/HA-SOX4 cells was amplified using linker-mediated PCR as described previously (25). Amplified DNA was labeled and hybridized in triplicate by NimbleGen Systems, Inc to their human 25K promoter array. This set consists of two microarrays that tile 4 kb of upstream promoter sequence and 750 bp of downstream intronic sequence on average, with a total genomic coverage of 110 Mb. Raw hybridization data was Z-score normalized and ratios of IP to Input DNA were determined for each sample. ChIPOTle software was used to determine enriched peaks using a 500 bp sliding window every 50 bp as previously described (23). NimbleGen microarray data are available from the GEO database, accession number GEO11915.
Luciferase Assays
PCR fragments representing the binding sites in the EGFR, ERBB2 and TLE1 genes were cloned in front of the pGL3-promoter luciferase construct (Promega, Fitchburg, WI). Primers sequences used can be found in Supplemental Table 7 LNCaP cells were transfected with 100 ng of a TK-Renilla construct, 500 ng of pGL3-promoter vector alone and with cloned inserts, as well as 500 ng of either a SOX4 or vector expression construct. Dual Luciferase assays were performed 48 hours post transfection according to the manufacturer’s guidelines (Promega, Fitchburg, WI). All assays were performed in triplicate on separate days.
qPCR
LNCaP cells were plated in 6-well culture dishes and grown to 90% confluency before transfection with 1 μg of SOX4 plasmid or vector control using Lipofectamine-2000 (Invitrogen, Carlsbad, CA). 24 hours post-transfection total RNA was harvested using the RNeasy kit (Qiagen, Valencia, CA) and reverse transcription performed using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA). qPCR was performed using SYBR Green I (Invitrogen, Carlsbad, CA) on a Biorad iCycler using 18s or β-actin as a control and data analyzed using the delta-Ct method (26). All primers used in this study are listed in Supplemental Table 7.
Microarray Analysis
Total RNA was isolated from three independent experiments of either vector control or SOX4 transfected LNCaP cells as described above. Each transfection was performed in triplicate and each sample was hybridized in duplicate creating six data points for each condition. Total RNA was submitted to the Winship Cancer Institute DNA Microarray Core facility (http://microarray.cancer.emory.edu/). All samples demonstrated RNA integrity (RIN) of 8.3 or greater using an Agilent 2100 Bioanalyzer. RNA was hybridized to the Illumina Human6 v2 Expression Beadchip that query roughly 47,000 transcripts with 48,701 probes, and after normalization significantly changed probes were calculated using Significance Analysis of Microarrays (SAM) software (27). Settings for SAM were: two-class unpaired (X4 vs Vector control), Imputation engine – 10 Nearest Neighbor, Permutations – 500, RNG seed – 1234567, Delta – 1.316, Fold Change – 1.5, False discovery rate – 0.749%. Microarray data are available in the GEO database, accession number GEO11915.
Immunoblotting
Cells were lysed in lysis buffer (0.137M NaCl, 0.02M TRIS pH 8.0, 10% Glycerol, and 1% NP-40), 50 μg total lysate separated by SDS-PAGE electrophoresis and transferred to nitrocellulose for immunoblotting. Immunoblots were probed with polyclonal rabbit SOX4 antisera described previously (9) and DICER (Santa Cruz, Santa Cruz, CA). To control for equal loading immunoblots were also probed with a mouse monoclonal antibody to protein phosphatase 2A (PP2A) catalytic subunit (BD Biosciences, San Jose, CA).
RESULTS
SOX4 Transcriptionally Activates EGFR
Using expression profiling to determine the genes whose mRNA levels change when SOX4 is either overexpressed, or eliminated using siRNA (9), we identified EGFR as a candidate SOX4 transcriptional target (Fig. 1A). Analysis of the promoter and first intron of EGFR and other family members with CONFAC software (28) revealed the presence of potential SOX4 binding sites within the first intron of EGFR and ERBB2 (Fig. 1B). CONFAC functions by identifying the conserved sequences in the 3 kb proximal promoter region and first intron of human-mouse ortholog gene pairs and then identifying transcription factor binding sites (TFBS), defined by position weight matrices from the MATCH software (29), that are conserved between the two species (28).
Figure 1.
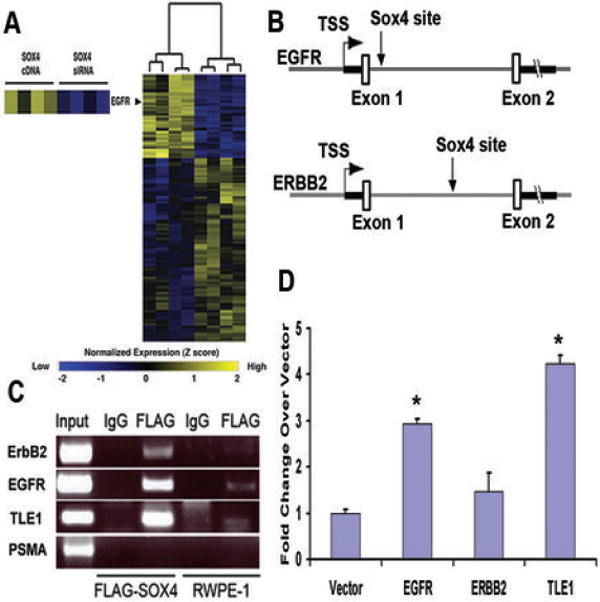
(A) Affymetrix U133A GeneChip microarray analysis of SOX4 overexpression and knockdown in LNCaP prostate cancer cells. Overexpression of SOX4 leads to increased EGFR expression while siRNA knockdown of SOX4 results in decreased EGFR expression. (B) Schematic showing the location of the SOX4 binding site in the first intron of the EGFR (Top) and ERBB2 (Bottom) genes. Arrows denote location of the SOX4 binding site. (C) ChIP assay of FLAG-SOX4 bound to the introns of EGFR, ERBB2 and TLE1 PSMA is shown as a negative control. SOX4 bound DNA is specifically amplified in the FLAG IP lane from FLAG-SOX4 expressing cells (lane 3) and not control cells (lane 5) or with a non-specific antibody (lanes 2 and 4). (D) Luciferase reporter assays with SOX4 binding sites showing activation in the presence of SOX4 compared to empty vector. * indicates p-value less than 0.01 by students T-test and error bars indicate 1 SD (n = 3 independent biological replicates performed on separate days).
While limited commercial antibodies exist for SOX4 and show activity in immunoblots, in our hands, none of them have been useful in a ChIP assay. Therefore, we employed epitope-tagged SOX4 as described in other SOX4 ChIP studies (9, 19). While the FLAG epitope tag was not tested directly for activity, a GST-SOX4 construct demonstrated binding to a known SOX4 motif and not a control motif (Supplemental Fig. 2B), validating that the epitope tag does not interfere with SOX4 binding. To determine if SOX4 directly bound the EGFR and ERBB2 enhancers, we performed ChIP analysis on RWPE-1 prostate cancer cells stably infected with FLAG-SOX4 or a control lentiviral vector. DNA representing the predicted SOX4 sites was specifically amplified from the FLAG-SOX4 cell line and not from the control cell line, indicating that SOX4 binds to intronic sequence of EGFR and ERBB2 (Fig. 1C). EGFR is expressed in RWPE-1 cells, but not in LNCaP cells, and SOX4 did not bind to these sequences in LNCaP cells (data not shown).
To characterize the transcriptional effect of SOX4 levels on the regions bound by SOX4 in ChIP assays, the amplified ChIP fragments were cloned in front of a minimal promoter luciferase reporter plasmid and tested in transient transfections in LNCaP cells. Compared to a vector control, SOX4 significantly increased transcription of the EGFR fragment 3-fold and the TLE1 positive control fragment roughly 4-fold. While not found significant, ERBB2 was activated 1.5-fold compared to the vector control (Fig. 1D). Consistent with microarray data, SOX4 transcriptionally activates the EGFR enhancer.
Genome-wide Localization Analysis
To determine the direct SOX4 target genes on a global scale we performed ChIP assays in triplicate from the LNCaP HA-SOX4 stable cell line and in duplicate from the control LNCaP-YFP cell line. Peaks (p < 0.001) that overlapped in at least two of the three data sets and were not present in the LNCaP-YFP cell line were called significant (Fig. 2A). Based on these parameters, we classified 3,600 significant, overlapping peaks as SOX4 target sequences. Since some transcription start sites (TSS) are quite close to each other (< 3 kb), it was not always possible to assign a unique gene to every peak. In addition, many genes had multiple peaks in their promoters, and thus we mapped the 3,600 peaks to 3,470 different genes (Supplemental Table 1).
Figure 2.
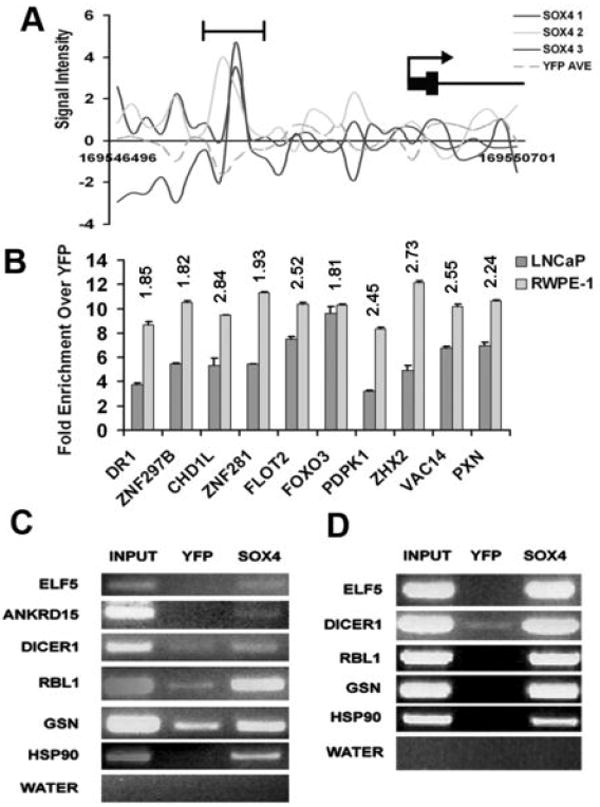
(A) Graph showing enrichment in the three HA-SOX4 lanes over the average of the two YFP replicates for the SOX4 target gene FMO4 Y-axis is the signal intensity across the genomic coordinates on the X-axis. (B) qPCR ChIP analysis of 10 randomly selected genes verified in both the RWPE-1 and LNCaP cell lines. Graph shows fold enrichment of the HA-SOX4 IP over the YFP negative control IP. Numbers above the bars represent the mean log2 of fold enrichment of ChIP-chip signal for the probes contained in the peak relative to YFP. Error bars indicate 1 SD (n = 3 technical replicates). (C) and (D) Genes that were verified by conventional ChIP assay. HA-SOX4 and YFP cells were subjected to conventional ChIP followed by PCR in both the LNCaP (C) and RWPE-1 (D) prostate cell lines. Six genes verified in the LNCaP cell lines and five in the RWPE-1 cell lines.
To verify the set of 3,600 SOX4 peaks, 28 candidate SOX4 target sites representing a range of p-values in promoters of genes of biological interest were chosen, primers were designed around the peaks, and enrichment was verified by conventional ChIP. Ten of these 28 candidates were analyzed by ChIP quantitative real-time PCR (qPCR) and 18 by ChIP-PCR. Overall, 24/28 (86%) of the candidate targets were confirmed, validating our dataset. All 10 of the peaks chosen to validate by qPCR were reproducibly enriched over the YFP control in both the LNCaP-HA-SOX4 cell line as well as the RWPE-1 cell line (Fig. 2B). Of the target sites validated by conventional PCR, 14 of 18 genes were confirmed in both the LNCaP and RWPE-1 cell lines while a mock, control PCR was negative (Fig. 2C and 2D and data not shown). The only exception was ANKRD15, which was enriched only in the LNCaP cell line and not in the RWPE-1 line.
Target Gene Expression Analysis
To determine whether SOX4 binding affects transcription of the 3,470 genes that have SOX4 bound at their promoters, we performed whole genome expression analysis on LNCaP cells after transfection with SOX4 or a control vector. To increase the likelihood of identifying direct SOX4 targets, total RNA was isolated at a relatively early timepoint (24 hours post-transfection) and hybridized to Illumina Human 6-v2 whole genome arrays. A total of 1,766 genes were changed at least 1.5-fold with a false discovery rate (FDR) of 0.749% (Fig. 3A, and Supplemental Table 2). Of those 1,766 genes, 244 were also direct SOX4 targets by ChIP-chip analysis (Fig. 3A, and Supplemental Table 3). Seven of these genes were confirmed by qPCR (Fig. 3B).
Figure 3.
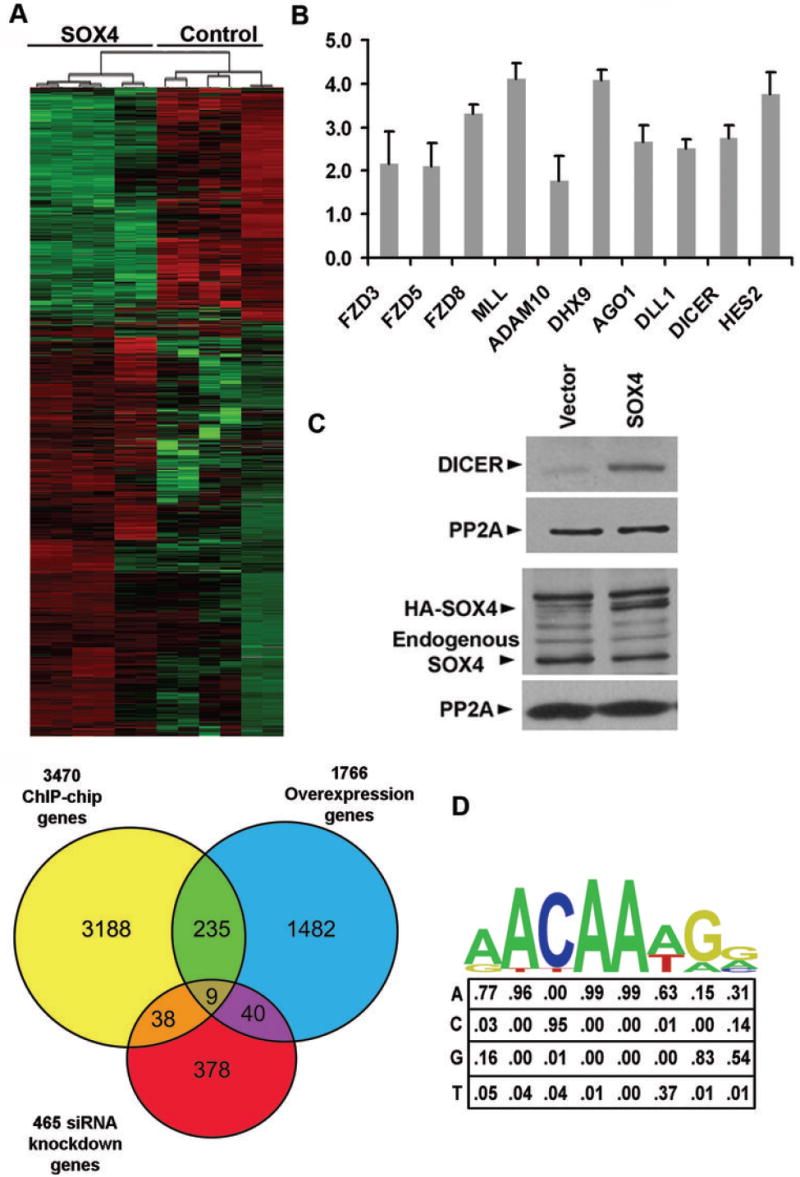
(A) Heat map (top) illustrating Illumina expression data of the 1,766 significant genes as determined by SAM analysis. Red indicates overexpressed and green denotes underexpressed genes. Venn diagram (bottom) depicts the overlap between 3,470 ChIP-chip SOX4 direct target genes, the Illumina expression data set of 1,766 genes, and the Affymetrix expression dataset of 465 genes. (B) qPCR expression analysis of SOX4 direct target genes after SOX4 overexpression in LNCaP cells. All ten genes were upregulated over a vector control transfection, similar to values determined by the Illumina array with a p-value less than 0.005 by students T-test. Error bars indicate 1 SD (n = 3 independent biological replicates performed on separate days). (C)DICER protein expression is upregulated by SOX4 HA-SOX4 or vector control were transfected into LNCaP cells and immunoblots were probed for DICER, SOX4, and PP2Ac as a loading control. (D) PBM-derived 8mer PWM for SOX4 displayed both graphically and numerically for each base position derived from incubation of recombinant GST-SOX4-DBD with a universal ‘all 8-mer’ double-stranded DNA protein-binding microarray. With stringent criteria (core similarity ≥ 0.85, matrix similarity ≥ 0.75) we find 60% of the peaks in the 282 high-confidence promoters contain SOX4 binding sites.
Our previous expression profiling of LNCaP cells after SOX4 siRNA knockdown (9) identified 465 downstream targets, and we confirmed that SOX4 regulates the expression of DICER, DLL1 and HES2 in LNCaP cells by qPCR (Fig. 3B). We further confirmed SOX4’s regulation of DICER at the protein level (Fig. 3C). Out of those 465 candidate targets, 47 genes overlapped with the 3,470 ChIP-chip targets, increasing the number of direct SOX4 targets to 282 genes (Fig. 3A and Supplemental Table 3). We classified these 282 genes bound by SOX4 in ChIP-chip and significantly changed by expression profiling as high confidence direct SOX4 target genes. Nine genes (PIK4CA, DHX9, BTN3A3, CDK2, MVK, ADAM10, RYK, ISG20, and DBI) overlapped in all three datasets. The transcription factor SON and purine biosynthetic enzyme GART, two genes on chromosome 21 that are transcribed in opposite directions and regulated by a bidirectional promoter, were affected in opposite ways. SON was activated by SOX4 1.8-fold as detected by SOX4 overexpression, while GART was increased almost 3-fold as determined by SOX4 siRNA knockdown, suggesting that SOX4 regulates the directionality of this promoter.
We next analyzed the p-values of the peaks in our ChIP-chip dataset, comparing the p-values of the genes that were altered by transient overexpression of SOX4 with those that were not (Supplemental Fig. 2). We found no difference in the distributions of the ChIP-chip p-values for those genes that were changed in expression profiling experiments and those that were not. Thus, based on our ChIP-chip validation experiments and the similar p-value distributions, we conclude that SOX4 is genuinely bound at the promoters of the 3,188 genes that did not change, but that SOX4 by itself is not limiting or sufficient to generate changes in transcription without corresponding changes in the cellular context, such as activation of co-factors or signaling pathways.
Novel SOX4 Position Weight Matrix (PWM)
To facilitate computational analyses of SOX4 DNA binding sites we sought to determine the DNA binding preferences of SOX4 using universal protein-binding microarrays (PBMs) (20). This universal PBM array allows recombinant SOX4 protein to interact with and bind every possible 8-mer, thus allowing in vitro binding site specificities to be calculated.
We generated an N-terminal, GST-SOX4-DBD fusion protein, expressed and purified it from E. coli, and tested for activity (Supplemental Fig. 3). The GST-SOX4-DBD was incubated with the protein binding microarray and a novel PWM (RWYAAWRV) was calculated from the PBM data (Supplemental Table 4) using the Seed-and-Wobble algorithm (Fig. 3D) (20). Three groups have previously reported similar binding site sequences for SOX4: AACAAAG (30), AACAAT (31) and WWCAAWG (19). Our PWM confirms the SOX4 core binding sequence of the previously known binding sites, but there are some differences in the specificity at the 1st and 7th positions and we find a bias towards A,C and G at the 8th position. These differences could be due to the fact that earlier reports used no more than 31 sequences to develop the binding motif while our study queried every possible 8-mer.
SOX4 Peaks Contain SOX4 Binding Sites
Using our newly derived PWM, we applied CONFAC software (28) to analyze the enriched sequences for the presence of SOX4 binding sites. We analyzed the sequences of the peaks in the promoters of our 282 high confidence genes against 10 sets of control promoter sequences to see if SOX4 sites were enriched in our target gene set. Control promoter peaks, of equal size to SOX4 peaks, were chosen randomly from sequences covered by the NimbleGen array and each control set contained equal total sequence coverage as our 282 high confidence peaks. With stringent criteria (core similarity ≥ 0.85, matrix similarity ≥ 0.75) we find 60% of the peaks contain SOX4 binding sites. SOX4 sites were significantly enriched relative to 10 sets of random promoter sequence, by Mann-Whitney U-test using Benjamini correction for multiple hypothesis testing (q < 0.0019).
To further characterize the SOX4 binding sites we searched the entire set of 3600 SOX4 peaks, and 10 equal sets of random promoter sequence for the presence of PBM-bound k-mers (here, ungapped 8-mers). The specificity of PBM k-mers can be quantified by the enrichment score (ES), which ranges from -0.5 to 0.5 (32). We analyzed the enrichment of PBM k-mers with 0.45 > ES > 0.40 (moderate) and ES > 0.45 (stringent). While both SOX4-bound peaks and random promoter sequence contained moderate and stringent k-mers, SOX4 peaks contained significantly more stringent (p = 0.0002) and moderate (p = 1.08 × 10-5) k-mers by two-tailed Mann-Whitney test (Supplemental Fig. 4).
To investigate interaction with protein partners that may increase SOX4’s affinity for ‘poor’ matching sites in vivo, we searched for enrichment of co-occuring TFBS in the SOX4 peaks. We applied CONFAC software to search the sequences for the presence of co-occurring transcription factor binding sites within the same peak (Table 1). Using the same criteria as above, we determined that the E2F family had the most frequently co-occurring motif (similar to TTTCGCGC, q-value =1.78 × 10-11). Interestingly, Ingenuity Pathway Analysis (IPA) identified Cell Cycle as a functionally enriched process in the 3,470 SOX4 target genes (p = 0.00916), suggesting that part of SOX4’s function is to control the expression of genes involved in cell-cycle progression.
Table 1.
Benjamini corrected q-values for co-occurring transcription factor binding sites.
| Transcription Factor | Family | Benjamini Corrected q-value |
|---|---|---|
| E2F4 | E2F | 1.78E-11 |
| E2F1 | E2F | 3.06E-11 |
| PAX5 | Paired Box | 2.07E-10 |
| WHN | Forkhead | 2.94E-10 |
| SMAD3 | SMAD | 1.82E-09 |
| SMAD4 | SMAD | 3.33E-09 |
| MYC | MYC | 6.25E-09 |
| NFKAPPAB | NF-κB | 2.95E-08 |
| LEF1/TCF1 | LEF | 1.12E-06 |
CONFAC analysis identified other significant TFBS motifs enriched in the SOX4 peaks (Table 1), including those for transcription factors in the TGFβ, Wnt, and NF-κB pathways. SOX4 modulates Wnt signaling via interaction with β-catenin and the TCF4 transcription factor (2), suggesting a possible role for SOX4 in transcriptionally modulating Wnt signals. We confirmed the recent report that SOX4 cooperates with constitutively active β-catenin to activate TOP-Flash luciferase reporters (2), and found that SOX4 synergistically induces activation of these constructs, further highlighting a role for SOX4 in the Wnt pathway (Supplemental Fig. 5).
SOX4 Target Genes
In order to determine the biological processes and functions of the SOX4 targets we performed a Gene Ontology analysis using DAVID software (33) on the 282 high confidence SOX4 targets. Among the SOX4 targets were 23 transcription factors (Table 2), and DAVID analysis determined that the top annotations were transcription (p = 3.7×10-18), transmembrane (p = 5.59×10-10) and protein phoshorylation/dephosphorylation (p = 3.5×10-18/6.6×10-7). These findings are paralleled by expression profiling of SOX4 overexpression in HU609 bladder carcinoma cells where top annotated functions were signal transduction and protein phosphorylation (11).
Table 2.
DAVID analysis identified 23 transcription factors present in our high confidence SOX4 target genes. GO Term: transcription, DNA dependent (p = 3.7×10-18).
| Entrez ID | Symbol | Microarray Fold Change |
|---|---|---|
| 196528 | ARID2 | 1.99 |
| 2001 | ELF5 | -2.65 |
| 3169 | FOXA1 | -2.47 |
| 2976 | GTF3C2 | -3.12 |
| 64412 | GZF1 | 2.42 |
| 84458 | LCOR | 2.41 |
| 4173 | MCM4 | 1.55 |
| 58508 | MLL3 | 2.06 |
| 10933 | MORF4L1 | 2.07 |
| 8031 | NCOA4 | 2.64 |
| 4784 | NFIX | -2.83 |
| 4824 | NKX3-1 | -4.53 |
| 7799 | PRDM2 | 2.48 |
| 5933 | RBL1 | 1.80 |
| 55509 | SNFT | -2.32 |
| 6722 | SRF | -2.03 |
| 54816 | SUHW4 | -1.93 |
| 9412 | SURB7 | -2.24 |
| 9338 | TCEAL1 | -1.57 |
| 7718 | ZNF165 | 1.53 |
| 7738 | ZNF184 | 1.66 |
| 23528 | ZNF281 | 1.71 |
| 30834 | ZNRD1 | -1.63 |
Commercial Ingenuity Pathway Analysis (IPA) software1 identified biological pathways and functions that are enriched in our 282 high confidence targets, as well as the 1,766 significant genes identified by SAM analysis, and the 3,470 unique genes that had SOX4 bound at their promoters in ChIP-chip. As anticipated, among the most significant annotations were cell cycle, cancer, and tissue development. In the significant expression data set of 1,766 genes we observed an upregulation of three Frizzled family receptors, FZD3, FZD5 and FZD8, as well as the downstream transcription factor TCF3 Overall, IPA analyses discovered key components of the EGFR, Notch, AKT-PI3K, microRNA, and Wnt-β-catenin pathways as SOX4 regulatory targets. Based on these findings, we built SOX4 regulatory networks found in prostate cancer cells (Fig. 4 and Supplemental Fig. 6). SOX4 target genes comprise key pathway components such as ligands (DLL1 and NGR1), receptors (FZD5 and PTCH1), an AKT regulatory kinase (PDPK1), and downstream transcription factors (FOXO3 and HES2). In addition, SOX4 activates expression of tenascin C (TNC), an extracellular matrix protein that is a target of TGFβ signaling (34) and β-catenin (35). In addition, SOX4 regulates three components of the RISC complex, DICER, AGO1, and RHA/DHX9 (Supplemental Table 3). We confirmed these data by qPCR (Fig. 3B) and by western blot for DICER (Fig. 3C).
Figure 4.
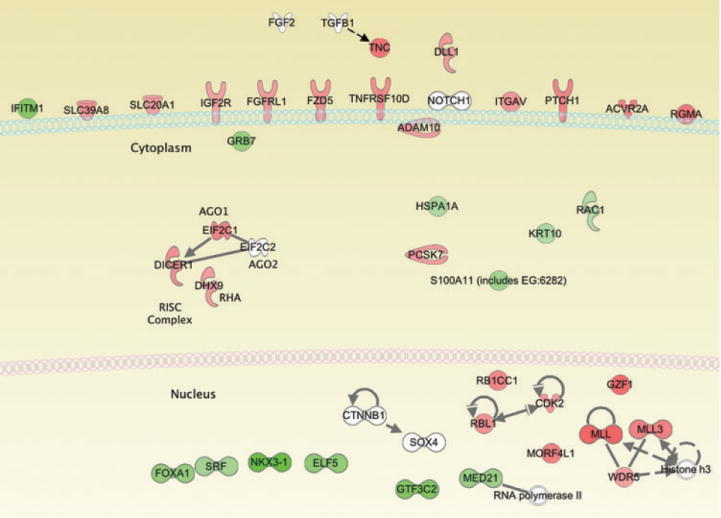
IPA analysis of direct target genes graphically illustrating the cellular location of the SOX4 transcriptional target genes. SOX4 regulates a host of nuclear and membrane localized proteins as well as multiple components of the RISC complex. Red indicates target genes upregulated by SOX4, green denotes downregulated genes and white represents genes for which no expression change was detected.
Gene Set Enrichment Analysis (GSEA) (36) and GSEA Leading Edge analysis (37) of these gene sets identified TGFβ-induced SMAD3 direct target genes (Supplemental Table 5) as enriched in SOX4 target genes. SOX4 is upregulated by TGFβ-1 treatment (4, 38) and we found SMAD4 sites are significantly enriched in the SOX4 ChIP-chip peaks (Table 1), suggesting that SOX4 impacts key developmental and growth factor signaling pathways in prostate cancer cells at both the transmembrane signaling and transcriptional levels.
DISCUSSION
While many studies have identified SOX4 as a crucial developmental transcription factor that is often overexpressed in many types of malignancies, little is known of what SOX4 regulates in cancer cells. We have utilized a ChIP-chip approach to report the first genome-wide localization analysis of SOX4 and mapped 3,600 binding peaks that represent 3,470 unique genes possibly under the transcriptional control of SOX4 We have also identified 1,766 genes that respond to increased SOX4 levels by whole genome expression profiling. Integration of these datasets mapped 282 high-confidence direct targets in the SOX4 transcriptional network. In addition, we have utilized protein-binding microarrays to determine a novel PWM specific for SOX4 and show that our ChIP-chip predicted peaks are significantly enriched for SOX4 binding sites. These data provide several new insights into the roles that SOX4 plays in the cell.
SOX4 Direct Target Genes
Although only 10% of the significant differentially expressed genes overlapped with the ChIP-chip data, this is likely a conservative estimate, because the NimbleGen 25K promoter array only queries proximal promoter sequences, and none more than 1 kb downstream of the TSS. We found that SOX4 binds EGFR and ERBB2 in the first intron over 20 kb downstream of the TSS (Fig. 1D), and unsurprisingly we did not detect EGFR or ERBB2 in our ChIP-chip experiment. Thus, more of the 1,900 genes that responded to changes in SOX4 mRNA levels (but were not detected by ChIP-chip) could still be direct targets. Excellent candidates would be the 40 genes that responded to SOX4 on both microarray platforms, such as the IL6 receptor, SOX12, and NME1 (Supplemental Table 6). While 3,600 is a fairly large number of SOX4 bound regions, some background can be expected. Nevertheless, we were able to validate 24 out of 28 (86%) candidate binding sites chosen adding confidence to our data set. In fact, an even higher number of over 4,200 genomic binding sites had been previously observed for c-Myc in ChIP-PET whole genome studies (39). Whole genome tiling arrays or ChIP-seq could provide additional binding sites that may show more overlap with the Illumina expression data set.
Conversely, many of the bound genes may not respond to changes in SOX4 mRNA levels alone, but to multiprotein activator complexes of which SOX4 is only one component. Furthermore, the stability of SOX4 bound to a promoter could be greater than unbound SOX4, limiting the effects observed by siRNA knockdown. In different cell types or cellular contexts, SOX4 may activate a different subset of these genes. Of the 31 SOX4 target genes reported by Liao et al (19) only six are represented in our NimbleGen data set and three found to be changed in our Illumina expression profiling data set. The small overlap could be due to the fact that those genes were identified in hepatocellular carcinomas, while we have examined prostate cancer cells. Interestingly, DKK was one of the six genes that overlapped in both data sets, further implicating SOX4 in the Wnt pathway. Since SOX4 is known to interact with β-catenin and other co-activators, it may be poised at many of these promoters to enable responses to developmental signals from the Wnt or TGFβ pathway.
Receptor and Signaling Regulation
Our data suggest that SOX4 regulates cellular differentiation through a variety of transcription factors and receptors. SOX4 is upregulated in response to numerous external ligands ranging from TGFβ (38) and BMP-6 (40) to parathyroid hormone and progesterone (8). Previous work has shown that SOX4 directly signals from IL-5Rα (41) and here we have shown that SOX4 directly regulates the EGFR receptor (Fig. 1). Membrane receptors in the SOX4 transcriptional network also include Frizzled family members FZD3, FZD5, FZD8; the Hedgehog receptor PTCH-1; the Notch ligand DLL1; TRAIL decoy receptor TNFRSF10D; and other growth factor receptors such as FGFRL1 and IGF2R. DAVID analysis also revealed protein phosphorylation/dephosphorylation (p = 3.5×10-18/6.6×10-7) and transcription (p = 3.7×10-18) are enriched annotations, identifying 23 transcription factors that are direct targets of SOX4 This evidence suggests that SOX4 regulates signaling events both at the external “input” level as well as the internal “output” or transcription level. This regulation could be direct, as with IL- 5Rα, or through the transcriptional targets SOX4 activates.
Transcription Factors and SOX4
Here we have reported DNA binding specificity data for SOX4, which will improve computational analyses for SOX4 specific binding sites. Our data confirm the known SOX family core-binding motif and adds new specificity at the 1st, 7th and 8th positions. While crystal structure evidence from SOX2 has shown the importance of the core-binding motif, it is possible that the specificity for SOX4 is enhanced outside of the core motif at the extra positions. A limitation of these data is that we did not assess how other DNA binding proteins influence the sequences to which SOX4 can bind. The enrichment of SMAD4 sites is particularly interesting in light of the GSEA results, which suggest that SOX4 regulates many TGFβ target genes, including Tenascin C. Thus, we hypothesize that SOX4 may physically interact with SMAD4 in response to TGFβ signals. Experiments to test this hypothesis are underway. Nevertheless, evidence points to a role for SOX4 in modulating other transcriptional programs via hierarchical regulation of 23 downstream transcription factors.
SOX4 and Cancer
Based on the target genes we identified, SOX4 appears to influence cancer progression in several ways. First, it plays a key role in activation of, and response to, developmental pathways such as Wnt, Notch, Hedgehog, and TGFβ. Second, SOX4 inhibits differentiation via repression of transcription factors such as NKX3.1, and activation of MLL and MLL3, two histone H3 K4 methyltransferases that induce activation of HOX gene expression (42). MLL methyltransferase complexes also facilitate E2F activation of S phase promoters, facilitating cell cycle progression. Activation of MLL also suggests a mechanism for SOX4’s role in myeloid leukemogenesis, since MLL is a critical oncogene that is often translocated or amplified in this disease (43). Thirdly, SOX4 targets growth factor receptors such as EGFR, FGFRL1, and IGF2R, enhancing proliferative signals in tumors and potentially activating the PI3K-AKT pathway. Mice heterozygous for NKX3.1 and PTEN in the prostate develop prostate adenocarcinomas and metastases to the lymph node (44). Thus, our data suggest that SOX4 may promote prostate cancer progression directly through NKX3.1 repression and indirectly through PI3K-AKT activation. Finally, SOX4 appears to promote metastasis via upregulation of tenascin C. Recently, both SOX4 and tenascin C were shown to enhance metastasis of breast cancer cells to the lung (45), as has the TGFβ pathway which activates their expression (46). Other metastasis-associated SOX4 target genes include Integrin αV and Rac1 Rac1 was recently shown to control nuclear localization of β-catenin in response to Wnt signals (47).
SOX4 regulates components of the RISC Complex and small RNA pathway
MicroRNAs (miRNAs) are small noncoding RNA species that regulate the translation and stability of mRNA messages for hundreds of downstream target genes via partial complementarity to short sequences in the 3’UTRs of messenger RNAs. The RNA-induced silencing complex (RISC), which is composed of Argonaute 1 (AGO1) or Argonaute 2 (AGO2), TRBP, and Dicer processes miRNAs from precursors (pre-miRNAs) to their mature form, cleaves target mRNAs, and participates in translational inhibition. RNA Helicase A (RHA/DHX9) interacts with the RISC complex and participates in loading of small RNAs into the RISC complex (48). We observed that three components of the RISC complex, DICER, AGO1, and RHA/DHX9, are high-confidence direct targets of SOX4 (Supplemental Table 3) and we confirmed these data by qPCR (Fig. 3B). Dicer has been independently observed to be overexpressed in prostate cancers (49).
In addition, we observed that Toll-like Receptor 3 (TLR3), which binds to double-stranded RNAs, induces gene silencing, and can induce apoptosis (50), was induced 2.8-fold upon overexpression of SOX4 This induction may be indirect, since TLR3 was not detected by ChIP-chip, but we cannot exclude the possibility that SOX4 may directly regulate TLR3 from a distal or intronic enhancer.
Our observation that SOX4 targets three genes important in small RNA processing is of particular interest in light of SOX4’s role in development and cancer progression. MicroRNAs have been implicated in numerous physiological processes from development to oncogenesis. MiRNAs can also act as suppressors of breast cancer metastasis via targeting of tenascin C and SOX4 (45), and as promoters of breast cancer metastasis (51). The finding that SOX4 can affect expression of multiple components of the RISC complex also provides insight into why long-term loss of SOX4 induces widespread apoptosis (9, 18). In summary, these data shed light on the mechanisms and pathways through which SOX4 may exert its effects during development and cancer progression. Further studies are necessary to elucidate the precise role of SOX4 in the functioning of these pathways.
Supplementary Material
Acknowledgments
The authors thank Dr. Maja Ordanic-Kodani at the Winship Cancer Institute Microarray Core Facility for performing the Illumina microarray labeling and hybridization, Robert Karaffa at the Emory University FlowCore for cell sorting, Dr. Hinh Ly for the IRES-eYFP lentiviral vector, and Dr. Anita Corbett for pGEX-4T-1 plasmid.
This research was supported by NCI R01 CA106826 to CSM. MFB and MLB were supported by grant RO1 HG003985 from NIH/NHGRI to MLB. CDS and CDM were supported by DOD CDMRP Prostate Cancer Predoctoral Training Fellowship PC060145 and Post-doctoral Training Fellowship PC060114, respectively.
Footnotes
References
- 1.Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 2.Sinner D, Kordich JJ, Spence JR, et al. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27(22):7802–15. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ya J, Schilham MW, de Boer PA, Moorman AF, Clevers H, Lamers WH. Sox4-deficiency syndrome in mice is an animal model for common trunk. Circ Res. 1998;83(10):986–94. doi: 10.1161/01.res.83.10.986. [DOI] [PubMed] [Google Scholar]
- 4.Wilson ME, Yang KY, Kalousova A, et al. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes. 2005;54(12):3402–9. doi: 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
- 5.Nissen-Meyer LS, Jemtland R, Gautvik VT, et al. Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. J Cell Sci. 2007;120(Pt 16):2785–95. doi: 10.1242/jcs.003855. [DOI] [PubMed] [Google Scholar]
- 6.Hoser M, Baader SL, Bosl MR, Ihmer A, Wegner M, Sock E. Prolonged glial expression of Sox4 in the CNS leads to architectural cerebellar defects and ataxia. J Neurosci. 2007;27(20):5495–505. doi: 10.1523/JNEUROSCI.1384-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Graham JD, Hunt SM, Tran N, Clarke CL. Regulation of the expression and activity by progestins of a member of the SOX gene family of transcriptional modulators. J Mol Endocrinol. 1999;22(3):295–304. doi: 10.1677/jme.0.0220295. [DOI] [PubMed] [Google Scholar]
- 9.Liu P, Ramachandran S, Ali-Seyed M, et al. Sex-Determining Region Y Box 4 is a Transforming Oncogene in Human Prostate Cancer Cells. Cancer Res. 2006;46(8):4011–9. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 10.Lee CJ, Appleby VJ, Orme AT, Chan WI, Scotting PJ. Differential expression of SOX4 and SOX11 in medulloblastoma. J Neurooncol. 2002;57(3):201–14. doi: 10.1023/a:1015773818302. [DOI] [PubMed] [Google Scholar]
- 11.Aaboe M, Birkenkamp-Demtroder K, Wiuf C, et al. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66(7):3434–42. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- 12.Rhodes DR, Yu J, Shanker K, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A. 2004;101(25):9309–14. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyd KE, Xiao YY, Fan K, et al. Sox4 cooperates with Evi1 in AKXD-23 myeloid tumors via transactivation of proviral LTR. Blood. 2006;107(2):733–41. doi: 10.1182/blood-2003-05-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T, Shen H, Akagi K, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32(1):166–74. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 15.Lund AH, Turner G, Trubetskoy A, et al. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat Genet. 2002;32(1):160–5. doi: 10.1038/ng956. [DOI] [PubMed] [Google Scholar]
- 16.Shin MS, Fredrickson TN, Hartley JW, Suzuki T, Agaki K, Morse HC., 3rd High-throughput retroviral tagging for identification of genes involved in initiation and progression of mouse splenic marginal zone lymphomas. Cancer Res. 2004;64(13):4419–27. doi: 10.1158/0008-5472.CAN-03-3885. [DOI] [PubMed] [Google Scholar]
- 17.Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36(9):3101–17. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25(41):5626–39. doi: 10.1038/sj.onc.1209566. [DOI] [PubMed] [Google Scholar]
- 19.Liao YL, Sun YM, Chau GY, et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008 doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 20.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24(11):1429–35. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc Natl Acad Sci U S A. 2001;98(13):7158–63. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee S, Berger MF, Jona G, et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36(12):1331–9. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCabe CD, Spyropoulos DD, Martin D, Moreno CS. Genome-wide analysis of the homeobox C6 transcriptional network in prostate cancer. Cancer Res. 2008;68(6):1988–96. doi: 10.1158/0008-5472.CAN-07-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odom DT, Zizlsperger N, Gordon DB, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303(5662):1378–81. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren B, Robert F, Wyrick JJ, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290(5500):2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karanam S, Moreno CS. CONFAC: Automated Application of Comparative Genomic Promoter Analysis to DNA Microarray Datasets. Nucleic Acids Res. 2004;32(Web Server issue):W475–84. doi: 10.1093/nar/gkh353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31(13):3576–9. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Wetering M, Oosterwegel M, van Norren K, Clevers H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. Embo J. 1993;12(10):3847–54. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wotton D, Lake RA, Farr CJ, Owen MJ. The high mobility group transcription factor, SOX4, transactivates the human CD2 enhancer. J Biol Chem. 1995;270(13):7515–22. doi: 10.1074/jbc.270.13.7515. [DOI] [PubMed] [Google Scholar]
- 32.Berger MF, Badis G, Gehrke AR, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133(7):1266–76. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 34.Pearson CA, Pearson D, Shibahara S, Hofsteenge J, Chiquet-Ehrismann R. Tenascin: cDNA cloning and induction by TGF-beta. Embo J. 1988;7(10):2977–82. doi: 10.1002/j.1460-2075.1988.tb03160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beiter K, Hiendlmeyer E, Brabletz T, et al. beta-Catenin regulates the expression of tenascin-C in human colorectal tumors. Oncogene. 2005;24(55):8200–4. doi: 10.1038/sj.onc.1208960. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23(23):3251–3. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 38.Ruebel KH, Leontovich AA, Tanizaki Y, et al. Effects of TGFbeta1 on gene expression in the HP75 human pituitary tumor cell line identified by gene expression profiling. Endocrine. 2008;33(1):62–76. doi: 10.1007/s12020-008-9060-3. [DOI] [PubMed] [Google Scholar]
- 39.Zeller KI, Zhao X, Lee CW, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103(47):17834–9. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ylostalo J, Smith JR, Pochampally RR, et al. Use of differentiating adult stem cells (marrow stromal cells) to identify new downstream target genes for transcription factors. Stem Cells. 2006;24(3):642–52. doi: 10.1634/stemcells.2005-0270. [DOI] [PubMed] [Google Scholar]
- 41.Geijsen N, Uings IJ, Pals C, et al. Cytokine-specific transcriptional regulation through an IL-5Ralpha interacting protein. Science. 2001;293(5532):1136–8. doi: 10.1126/science.1059157. [DOI] [PubMed] [Google Scholar]
- 42.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10(5):1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 43.Chowdhury T, Brady HJ. Insights from clinical studies into the role of the MLL gene in infant and childhood leukemia. Blood cells, molecules & diseases. 2008;40(2):192–9. doi: 10.1016/j.bcmd.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Abate-Shen C, Banach-Petrosky WA, Sun X, et al. Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 2003;63(14):3886–90. [PubMed] [Google Scholar]
- 45.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Padua D, Zhang XH, Wang Q, et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133(1):66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, Long F. Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell. 2008;133(2):340–53. doi: 10.1016/j.cell.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26(4):523–37. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 49.Ambs S, Prueitt RL, Yi M, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68(15):6162–70. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salaun B, Coste I, Rissoan MC, Lebecque SJ, Renno T. TLR3 can directly trigger apoptosis in human cancer cells. J Immunol. 2006;176(8):4894–901. doi: 10.4049/jimmunol.176.8.4894. [DOI] [PubMed] [Google Scholar]
- 51.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


