Abstract
The spatial distribution of neutral genetic diversity is mainly influenced by barriers to dispersal. The nature of such barriers varies according to the dispersal means and capabilities of the organisms concerned. Although these barriers are often obvious on land, in the ocean they can be more difficult to identify. Determining the relative influence of physical and biotic factors on genetic connectivity remains a major challenge for marine ecologists. Here, we compare gene flow patterns of 7 littoral fish species from 6 families with a range of early-life-history traits sampled at the same geographic locations across common environmental discontinuities in the form of oceanic fronts in the Western Mediterranean. We show that these fronts represent major barriers to gene flow and have a strong influence on the population genetic structure of some fish species. We also found no significant relation between the early-life-history traits most commonly investigated (egg type, pelagic larval duration, and inshore-offshore spawning) and gene flow patterns, suggesting that other life-history factors should deserve attention. The fronts analyzed and the underlying physical mechanisms are not site-specific but common among the oceans, suggesting the generality of our findings.
Keywords: gene flow, microsatellite, ocean circulation, pelagic stages
The spatial distribution of neutral genetic diversity in aquatic environments, as in terrestrial ones, is mainly generated and maintained by barriers to dispersal. However, although these barriers are often obvious on land, habitat discontinuities in the ocean are much more difficult to distinguish (1). Identifying such barriers is essential in defining the scale of exchange among marine populations, and this information, in turn, is fundamental to our understanding of the dynamic and genetic structure of populations as well as in the management of marine species, including the implementation of marine reserves (2).
Most littoral marine species have restricted adult movement, so their pelagic juvenile stages represent their most important dispersal mechanism (2). In the sea, it is well recognized that movements of pelagic stages are influenced by oceanographic processes, such as upwelling systems, fronts, moving convergences, eddies, and counter currents that can lead to dispersal of hundreds of kilometres (3, 4). Thus, it has been assumed that many marine populations operate as genetically open systems (5–7). However, mounting genetic evidence shows that pelagic stages often fail to fully achieve their dispersal potential (8–11), suggesting that the relationship between dispersal potential and realized gene flow among marine populations is more complex than previously assumed. Assessing the influences that oceanographic factors and early-life-history traits have in determining gene flow remains a major challenge for marine ecologists. Oceanic fronts, sharp discontinuities of physical and biochemical variables, are generated by various physical processes, occur in all oceans, and are likely to represent barriers to faunal exchange (12).
Data on dispersal distances have been collected for relatively few species, representing a limited number of possible dispersal scenarios (13, 14). Thus, high resolution hydrodynamically-based dispersal models have been developed (2, 15). However, recent empirical studies show greater local retention of pelagic stages than predicted by advection models (16–18). Also, it has proved extremely difficult to measure the frequency with which long distance movements occur during such stages (19).
Significant progress has recently been made in our ability to accurately track pelagic stages (20–22), as well as in determining genetic connectivity patterns by combining oceanographic models and molecular genetic data (23–25). However, relatively few empirical studies have investigated how oceanographic conditions influence larval dispersal and ultimately gene flow among marine fishes with varying life-history strategies (7, 26, 27). Also, these few studies have not sampled the same species at the same locations nor have they tested the effects of life-history traits across a well-defined oceanographic barrier and compared the results against barrier-free locations. Thus, the general hypothesis that oceanic fronts may act as a barrier to dispersal for most littoral fish species irrespective of their early-life-history traits remains largely untested. To this end, we compare genetic connectivity patterns of 7 littoral fish species with varied dispersal potentials sampled at the same geographical locations in the Western Mediterranean. Also, we consider how such patterns can be explained by reference to preexisting knowledge of their early-life-history characteristics and of the fronts that may act as oceanographic barriers.
Specifically, we aim to test the following predictions: (i) populations separated by oceanic fronts belong to genetically differentiated subunits; (ii) populations not divided by fronts are less genetically differentiated; and (iii) the degree of connectivity between populations is related to early-life-history traits and dispersal capabilities.
The Species.
In this study, we consider 7 cooccurring species from 6 different families, representing a wide spectrum of the main early-life-history characteristics found in littoral fish (Table 1). Early-life-histories vary greatly among marine species. Although some species may spawn benthic eggs from which pelagic larvae hatch, other species may display life cycles where both the eggs and the larvae are pelagic, whereas other species may have evolved peculiar forms of parental care such as the use of brood pouches, mouthbrooding, or bearing live young (28, 29). Also, the pelagic larval duration (PLD) as well as the spatial distribution of pelagic stages (i.e., inshore-offshore) also shows great variability among marine species (30–34). Under this scenario, it is expected that species with extended pelagic phases and offshore distributed larvae would be more apt to disperse over greater distances that species with shorter (or no) pelagic phase and inshore distribution (35).
Table 1.
Life-history traits of the different species
| Species | Family | Egg type | Larvae distribution | PLD, days |
|---|---|---|---|---|
| D. vulgaris | Sparidae | P | O | 29–58 |
| M. surmuletus | Mullidae | P | O | 28–35 |
| S. cabrilla | Serranidae | P | O | 21–28 |
| O. melanura | Sparidae | P | O | 14–18 |
| T. delaisi | Tripterygiidae | B | I | 16–21 |
| A. imberbis | Apogonidae | MB | I | 18–24 |
| S. tinca | Labridae | B | I | 9–13 |
Three of the study species can be considered to have extended PLD with pelagic eggs and offshore larvae, suggesting a high dispersal potential: the Common 2-banded bream (Diplodus vulgaris), the Striped red mullet (Mullus surmuletus), and the Comber (Serranus cabrilla). An intermediate condition is shown by the Saddled bream (Oblada melanura), which spawns pelagic eggs and has offshore larvae but with a shorter pelagic duration. The Blackfaced blenny (Tripterygion delaisi) and the Cardinal fish (Apogon imberbis) are species with low potential for dispersal, producing benthic or mouth-brooded eggs, inshore larvae, and having shorter PLD. Last, the Peacock wrasse (Symphodus tinca) has the shortest PLD (also benthic eggs) and should have the least potential for dispersal.
The Fronts.
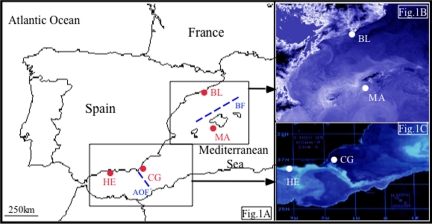
The Western Mediterranean is a well-studied geographical area considered as a “hot-spot” in marine biodiversity with a high level of endemism (36). Conservation strategies may benefit from the inclusion of ocean circulation and population genetic data. The Western Mediterranean is a small-scale ocean system, influenced by the inflow of Atlantic water through the strait of Gibraltar (12). The interaction between lighter Atlantic water and higher density Mediterranean water generates 2 oceanographic fronts, separated by ≈600 km, which may affect organismal dispersal (Fig. 1A). The Almeria-Oran front (AOF), situated ≈400 km East of the Strait of Gibraltar, is a sharp semi permanent boundary between surface water masses formed by the convergence of the Atlantic ocean and the Mediterranean sea. It is a thermohaline density front confined to the upper 300 m of the water column, and is characterized by strong density gradients (comparable with the Gulf Stream) and some upwelling areas (1.5 σT units, from 1,027.5 kg/m3 to 1,026 kg/m3 at the surface) (Fig. 1C) (37). The AOF has been reported to act as a barrier to gene flow in numerous species (38). East of the AOF, the Atlantic water flows north-eastward, reaching the Balearic Islands and forming a second density front in the northern part of the archipelago. This Balearic Front (BF) is a shelf/slope front, present in the upper 200 m, and is characterized by density differences of 0.5 σT units. This front is similar to shelf/slope fronts on many continental shelves around the world (12, 37, 39). Indeed, the Balearic Sea is a semienclosed basin with many similarities to other semienclosed seas, such as the South China or the Caribbean Seas (Fig. 1B) (39). The characteristics of each front provide a good opportunity to test their effects on gene flow among marine species with varying dispersal capabilities. Also, we compare the results with populations not separated by any front (NF, no front).
Fig. 1.
Sampling locations for the 7 species. (A) HE, Herradura; CG, Cabo de Gata; MA, Mallorca; BL, Blanes; and the locations of the fronts (dotted lines). (B) BF. (C) AOF.
Results
Estimates of genetic divergence between populations, measured as FST, showed significant differences across the AOF for all species except O. melanura (Table 2). Comparisons across the BF were also significant for 5 species, the exceptions being D. vulgaris and S. tinca. Genetic differentiation between populations from Cabo de Gata and Blanes (NF) was only significant for 4 species, and FST values were generally smaller than those obtained for the populations separated by the BF. By contrast, results from Cabo de Gata-Mallorca (NF) were quite similar to those of Mallorca-Blanes (separated by BF) (Table 2).
Table 2.
Pairwise FST values for the different species across AOF, BF, and NF
| Species | AOF | BF | NF | — | |
|---|---|---|---|---|---|
| HE-CG | MA-BL | CG-BL | CG-MA | Mean | |
| D. vul | 0.060 | 0.004 | 0.008 | 0.001 | 0.018 |
| M. sur | 0.021 | 0.134 | 0.001 | 0.127 | 0.071 |
| S. cab | 0.014 | 0.009 | 0.059 | 0.036 | 0.030 |
| O. mel | 0.001 | 0.17 | 0.052 | 0.155 | 0.095 |
| T. del | 0.018 | 0.027 | 0.038 | 0.034 | 0.029 |
| A. imb | 0.011 | 0.060 | 0.005 | 0.052 | 0.032 |
| S. tin | 0.033 | 0.002 | 0.001 | 0.001 | 0.009 |
| Mean | 0.023 | 0.058 | 0.023 | 0.058 | — |
| No. Sig | 6 | 5 | 4 | 5 | — |
Significant values (after Bonferroni correction) are shown in bold italics.
Among the study species, T. delaisi and S. cabrilla showed the most consistent structuring, with significant genetic differences between all populations compared (Table 2). In 2 species, A. imberbis and M. surmuletus, only Cabo de Gata-Blanes showed no significant difference; these populations were along continuous coastline and were not separated by either oceanic front. The only nonsignificant comparison for O. melanura was across the AOF, Herradura-Cabo de Gata. Significant differentiation was shown in 2 of the 4 comparisons of D. vulgaris: one pair separated by the AOF, and the other only by distance along continuous coastline (Cabo de Gata-Blanes). Surprisingly, the species expected to show the least gene flow between populations, having benthic eggs, inshore larvae, and the shortest pelagic larval period (Table 1); S. tinca, was found to be the species with the least evidence for population structure with only the populations separated by the AOF being significantly differentiated.
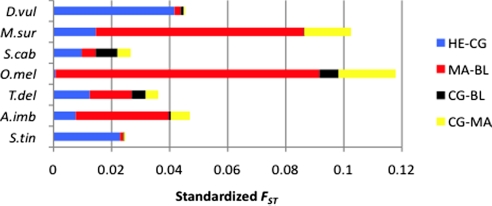
Regarding weighted genetic divergence by geographic distance (Fig. 2), for 3 species, the strongest genetic differentiation was found between those populations separated by the AOF, a pattern most clearly shown by D. vulgaris and S. tinca, and less so by S. cabrilla. In the other 4 species, A. imberbis, M. surmuletus, O. melanura, and T. delaisi, populations separated by the BF showed the greatest genetic differentiation. These species also showed the greatest overall levels of distance-adjusted differentiation (Fig. 2).
Fig. 2.
Standardized FST (genetic divergence per geographic unit) among pairwise adjacent sampling locations of D. vulgaris, M. surmuletus, S. cabrilla, T. delaisi, O. melanura, A. imberbis, and S. tinca species.
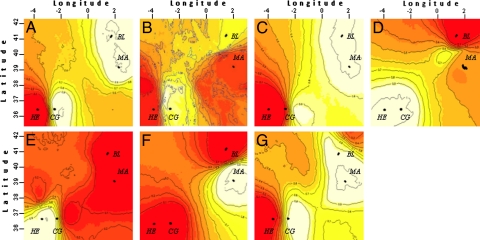
The geographically constrained Bayesian model suggested the existence of 2 genetically differentiated subunits within all species across the sampling area. The spatial location of this discontinuity differed among species. For D. vulgaris, M. surmuletus, and S. tinca, the genetic discontinuity was found between Herradura and Cabo de Gata, which is congruent with the location of the AOF (Fig. 3A, B, and G). For A. imberbis and O. melanura, a strong genetic discontinuity was observed between Mallorca and Blanes, locations separated by the BF (Fig. 3 F and D). For S. cabrilla and T. delaisi, in turn, the clearest genetic discontinuity was detected between Cabo de Gata and Mallorca, locations not separated by either of the fronts (Fig. 3 C and E).
Fig. 3.
Map of posterior probabilities of population membership and spatial location of genetic discontinuities for populations of D. vulgaris (A), M. surmuletus (B), S. cabrilla (C), O. melanura (D), T. delais (E), A. imberbis (F), and S. tinca (G) species. Contour lines indicate the spatial position of genetic discontinuities between populations. Lighter colours indicate higher probabilities of population membership.
Discussion
The results of this study allowed us to test 3 predictions, but with very different outcomes. First, we found that, as predicted, both the AOF and BF appeared to have a strong influence on population genetic structure of the study species. The second prediction, that populations not separated by fronts would be less genetically differentiated, was supported by the distance-adjusted FST figures and the Bayesian spatial plots, although a clear picture did not emerge from the raw FST data. Last, we found no clear relationship between the early-life-history traits investigated (egg type, PLD, and inshore-offshore spawning) and gene flow patterns. This finding suggests that genetic connectivity patterns cannot be readily predicted from the dispersal potential estimated from these early-life-history traits. Perhaps, other biophysical processes such as larval behavior, ontogeny, and mesoscale variability at short spatiotemporal scales may (also) influence genetic connectivity among marine fish populations.
The Effects of the AOF on the Genetic Structure of Littoral Fishes.
We estimated a significant effect of the AOF on genetic differentiation among 6 out of the 7 study species; although, the extent of the differentiation was different among species. Thus, it can be suggested that the AOF can restrict gene flow among populations of species with pelagic stages of various durations, whether they are largely spent inshore or offshore. In addition to the front itself, other environmental and biological factors may also contribute to the genetic structuring observed in this area. For example, flow patterns in the Alboran Sea (40, 41) coupled with frontal density formation may result in favourable conditions for egg and larval development, leading to behavioural and physiological responses to reduce transport and increase settlement, promoting retention.
D. vulgaris has pelagic eggs, offshore larvae, and a PLD of 29–58 days. By contrast, S. tinca has benthic eggs and inshore larvae with a PLD of a mere 9–13 days. However, these species showed similar levels of genetic differentiation across the AOF. Similarly, populations of T. delaisi and S. cabrilla on one hand, and those of A. imberbis and M. surmuletus on the other, showed very similar patterns of genetic differentiation despite the differences in their life-history traits (Table 1). The only species not showing significant genetic differentiation across the AOF was O. melanura, a species showing intermediate values for traits that might be expected to be associated with genetic structuring.
Our results agree with the majority of studies (38), which have shown evidence that significant population differences can be found across the AOF, even for species that have very long pelagic larval stages (≈4 months) (42). Some previous studies have also reported a lack of significant genetic differentiation for species with relatively weaker presumed dispersal capabilities, such as those with shorter PLDs (≈30 days) or benthic eggs (43). Similarly, in our study, O. melanura showed no significant genetic differentiation across the AOF, despite its presumably limited larval dispersal capabilities. However, in this species, it might be suggested that gene flow across the AOF could be facilitated by movements of the shoaling benthopelagic adults.
Our study indicated contrasting levels of divergence among the 2 sparid species, D. vulgaris and O. melanura. Different results have also been noted in previous studies of closely-related species within the area. Significant differences among populations of the anglerfish species Lophius budegassa have been found, but not among those of Lophius piscatorius, which has a qualitatively similar life history (42). Likewise, a gene flow break for 3 out of 5 sparid species investigated was found (43), and later it was shown that patterns were consistent within species irrespective of whether mitochondrial and nuclear (allozyme) markers were used (44). Thus, although the AOF can be an effective barrier to gene flow for most marine species, there are exceptions, which cannot be easily predicted from their early-life-history traits nor the genetic markers used. Explaining why closely-related species with similar life-history traits show strikingly different patterns of gene flow remains a challenge worthy of further research. The use of coupled biophysical models holds great promise in this regard (45, 46).
The Effects of BF on the Genetic Structure of Littoral Fishes.
Like the AOF, the BF also reduces gene flow in most species, except for D. vulgaris and S. tinca, species lying at opposite poles of the distribution of predicted dispersal capabilities. It was particularly surprising to find evidence for high levels of gene flow in the inshore-living S. tinca, which has benthic eggs and short larval duration. Not only are the Mallorca and Blanes populations separated by the front, but a littoral species might be expected to show little gene exchange between continental and insular populations. The mating and settlement behavior of the species might explain this apparently anomalous result. Like other species of the genus Symphodus, spawning takes place in nests built with branching algae that are often destroyed and transported by waves (47). Because such algal clumps are also suitable habitat for settlers (48), passive transportation through drifting algae could help homogenize gene pools across the BF. This hypothesis, which merits further research, highlights the possible importance that life-history traits other than PLD, may have on the connectivity of marine populations.
Likewise, environmental and biological factors in addition to the BF may have also contributed to the genetic structure observed in the other species. The effect of the BF may be intensified by the existence of habitat discontinuities between insular and continental populations. The oceanographic features of offshore habitats can differ substantially from those of coastal waters (36), and the survival of inshore-spawned larvae may be poor, compared with that of offshore-spawned larvae. Therefore, although it would appear that offshore larvae are also constrained by the front, the Balearic Islands display partial hydrodynamic interactions with the continent (49). This intermittent connection may allow for passive transport of early-life stages of some species with pelagic eggs and offshore larvae, such as D. vulgaris. Not only has this species the largest PLD in this study, but it reproduces in autumn, when the density gradients of the BF are reduced (39). This early-life-history trait suggests that the PLD and the seasonality of the fronts might be key elements allowing larvae, at least of this species, to reach the archipelago. Another habitat discontinuity that could pose a natural barrier is the deep-water trench of >1,000 m that separates the Balearic Islands from the continent (50). Restriction of gene flow by deep water has been shown for other marine species (51–53).
The Genetic Structure in the Absence of Fronts.
From inspection of the raw FST figures, it would appear that populations not separated by fronts are no less genetically differentiated (Table 2). However, the standardized FST values indicate that the fronts are associated with a substantial restriction in gene flow (Fig. 2), a pattern also evident from the maps of posterior probabilities (Fig. 3).
In the comparison between the continental populations of Cabo de Gata and Blanes, 4 species (D. vulgaris, S. cabrilla, O. melanura, and T. delaisi) were significantly different. This differentiation may have been a result of the considerable distance between the localities, suggesting isolation by distance. The species M. surmuletus, A. imberbis, and S. tinca showed no significant genetic differentiation. No common life-history characteristics readily explain the absence of genetic differentiation between these 2 localities. The adults of these species are relatively sedentary, suggesting that larval dispersal could be more relevant in connecting populations. We did not have information about all species differences in larval behavior or preferred position in the water column as such traits could influence the response of larvae to currents (54). For example, M. surmuletus larvae are neustonic (found in surface waters), whereas S. cabrilla larvae are typically found in deeper layers. This difference could affect their transport as much as their inshore-offshore gradient. Future studies may need to consider how these traits affect genetic connectivity.
The continental populations at Cabo de Gata and the insular populations at Mallorca were significantly differentiated in 5 out of the 7 species. In fact, the levels of differentiation for this comparison were more similar than those obtained between populations separated by the BF, Mallorca, and Blanes (mean FST = 0.058). Of course, Blanes is also much closer to Mallorca than Cabo de Gata is, so the standardised FST figures were much higher for the populations separated by the BF, for all species except S. cabrilla (Table 2, Fig. 2), consistent with the third prediction.
Genetic Differentiation and Early-Life-History Characteristics.
No evidence was found that a single life-history trait is strongly related to population genetic differentiation, whether in the presence or absence of oceanographic barriers. Some studies have found that early-life-history traits are poor predictors of population genetic structure (7, 55), whereas others have found a clear relationship between early-life-history and gene flow (29, 56). This discrepancy indicates that population structure cannot always be explained by simple predictors. Other biological characteristics may have additional and significant influence on the connectivity between populations, including larval and adult behavior, and reproductive and recruitment strategies (57). Also, small-scale hydrographic variability and local processes may have ecologically meaningful effects on population connectivity (45).
In summary, by using a multispecies approach, our results demonstrate that the spatial location of genetic discontinuities between populations differed among species. Biotic factors, such as egg type, PLD, and inshore-offshore spawning, proved to be poor predictors of population structure, showing not necessarily that they are unimportant, but rather that they may be insufficient in the absence of further knowledge. Also, our results highlight the relationship between gene flow patterns and barriers to dispersal such as ocean fronts. The fronts analyzed and the underlying physical mechanisms are not site-specific but common among the oceans, suggesting the generality of our findings. Oceanic fronts could isolate populations of many species, increasing their tendency to self-recruitment and increasing their vulnerability. This isolation could have important implications in conservation-management policies for marine resources (58), particularly in semienclosed areas, such as the Mediterranean, South China, or Caribbean Seas, considered as hot-spots for marine biodiversity (36). Therefore, the design of a network of marine reserves or any other conservation-management strategy should be influenced by the scale of genetic connectivity produced by the combination of oceanographic features and life histories of species.
Materials and Methods
Sample Collection and Genetic Analysis.
During 2004–2007, a total of 1,197 adults from 7 different marine fish species were sampled at the same 4 locations along the Spanish Mediterranean coast and the Balearic Islands (Fig. 1A). All samples were obtained by scuba diving. A mean of 47.5 individuals per site and species were analyzed [see supporting information (SI) Table S1]. A total of 63 species-specific microsatellite loci (mean 9 loci per species) were successfully amplified by PCR and analyzed to determine the genetic diversity and population structure of each species (see SI Text and Tables S2–S8). Population structure was investigated by using raw and distance-adjusted FST values, whereas Bayesian methods were used to seek spatial genetic discontinuities.
Spatial Genetic Structure.
The effect of the oceanographic fronts on gene flow levels within and among species was assessed through 2 different approaches, at both the population and individual levels. First, at the population level, pairwise values of FST and their estimated probabilities were calculated within species by 10,000 random permutations by using GENETIX v. 4.02 (59). Permuted FST values were compared with observed values to estimate the probability of randomly achieving a value greater than or equal to the observed. The alternative hypothesis of significant genetic differentiation was accepted if this probability was equal to or <0.05. Also, we standardized population FST values by dividing each pairwise value by its corresponding pairwise geographic distance. In this way, a genetic distance per geographic distance unit was obtained and used to evaluate the relative effect of each front on each species.
At the species level, spatial locations of genetic discontinuities (i.e., genetic boundaries) along the sampling area were determined by using the R package GENELAND (60). The program makes use of a geographically constrained Bayesian model that explicitly takes into account the spatial position of sampled multilocus genotypes without any prior information on the number of populations and degree of differentiation between them. Geographical coordinates were assigned to each individual by randomly choosing n spatial locations encompassed within a 10-km radius from each of the 4 sampling locations, by using the Mapping toolbox in MATLAB v.7. (Mathworks), where n equals the number of individuals per species sampled at a given location. Thus, the same geographic coordinates were used for individuals of all species sampled at the same site. The inference algorithm was launched by using the Dirichlet distribution as prior for allele frequencies with 20,000 MCMC iterations using spatial information. Then, the algorithm was rerun with an additional 20,000 MCMC iterations, fixing the value of K (i.e., number of populations) to that determined by the mode of the posterior distribution of the MCMC chain, and setting the Poisson processes equal to the number of sampled individuals per species.
Supplementary Material
Acknowledgments.
We thank J. Tintore and J. Salat for helpful discussions on the oceanography of the Western Mediterranean; Gary Carvalho and Bill Hutchinson for valuable comments and suggestions; and the contribution of the Department of Environment of the Balear Region and colleagues from the Instituto Español de Oceanografía for help in sampling some localities and species. This work was supported by Junta de Andalucia Grant 2003X880_1 and Consejo Superior de Investigaciones Científicas of the Ministerio de Ciencia e Innovación Grant 2007301022. The PhD studentship of J.A.G. was supported by Consejo Nacional de Ciencia y Tecnología. This work was also in part supported by Ministerio de Educación y Ciencia Grant CTM2007-66635 and Ministerio de Medio Ambiente Grant 119/2003. The authors are part of the Generalitat de Catalunya Research Groups 2005SGR-00277 (E.M. and J.C.-C.) and 2005SGR-00995 (M.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806804106/DCSupplemental.
References
- 1.Thorrold SR. Ocean Ecology: Don't Fence Me in. Curr Biol. 2006;16:638–640. doi: 10.1016/j.cub.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 2.Cowen RK, Paris CB, Srinivasan A. Scaling of connectivity in marine populations. Science. 2006;311:522–527. doi: 10.1126/science.1122039. [DOI] [PubMed] [Google Scholar]
- 3.Palumbi SR. Marine reserves and ocean neighborhoods: The spatial scale of marine populations and their management. Annu Rev Env Resour. 2004;29:31–68. [Google Scholar]
- 4.Sponaugle S, et al. Predicting self-recruitment in marine populations: Biophysical correlates and mechanisms. Bull Mar Sci. 2002;70:341–375. [Google Scholar]
- 5.Roberts CM. Connectivity and Management of Caribbean Coral Reefs. Science. 1997;278:1454–1457. doi: 10.1126/science.278.5342.1454. [DOI] [PubMed] [Google Scholar]
- 6.Scheltema RS. Evidence for trans - Atlantic transport of gastropod larvae belonging to the genus Cymatium. Deep-Sea Res Part I Oceanogr Res Pap. 1965;13:83–86. [Google Scholar]
- 7.Shulman MJ, Bermingham E. Early life histories, ocean currents and the population genetics of caribbean reef fishes. Evolution. 1995;49:897–910. doi: 10.1111/j.1558-5646.1995.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 8.Marko PB, Rogers-Bennett L, Dennis AB. MtDNA population structure and gene flow in lingcod (Ophiodon elongatus): Limited connectivity despite long-lived pelagic larvae. Mar Biol. 2007;150:1301–1311. [Google Scholar]
- 9.Froukh T, Kochzius M. Genetic population structure of the endemic fourline wrasse (Larabicus quadrilineatus) suggests limited larval dispersal distances in the Red Sea. Mol Ecol. 2007;16:1359–1367. doi: 10.1111/j.1365-294X.2007.03236.x. [DOI] [PubMed] [Google Scholar]
- 10.Barber PH, Palumbi SR, Erdman MV, Moosa MK. Sharp genetic brakes among populations of Haptosquilla pulchella (Stomatopoda) indicate limits of larval transport: Patterns, causes and consequences. Mol Ecol. 2002;11:659–674. doi: 10.1046/j.1365-294x.2002.01468.x. [DOI] [PubMed] [Google Scholar]
- 11.Jin-Xian L, Tian-Xiang G, Shi-Fang W, Ya-Ping Z. Pleistocene isolation in the Northwestern Pacific marginal seas and limited dispersal in a marine fish, Chelon haematocheilus (Temminck and Schlegel, 1845) Mol Ecol. 2007;16:275–288. doi: 10.1111/j.1365-294X.2006.03140.x. [DOI] [PubMed] [Google Scholar]
- 12.Millot C. Circulation in the Mediterranean Sea: Evidences, debates and unanswered questions. Sci Mar. 2005;69:5–21. [Google Scholar]
- 13.Sammarco PW, Andrews JC. The Helix Experiment: Differential Localized Dispersal and Recruitment Patterns in Great Barrier Reef Corals. Limnol Oceanogr. 1989;34:896–912. [Google Scholar]
- 14.Shanks AL, Grantham B, Carr MH. Propagule dispersal distance and the size and spacing of marine reserves. Ecol Appl. 2003;13:159–169. [Google Scholar]
- 15.Siegel DA, et al. The stochastic nature of larval connectivity among nearshore marine populations. Proc Natl Acad Sci USA. 2008;105:8974–8979. doi: 10.1073/pnas.0802544105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Almany GR, Berumen ML, Thorrold SR, Planes S, Jones GP. Local Replenishment of Coral Reef Fish Populations in a Marine Reserve. Science. 2007;316:742–744. doi: 10.1126/science.1140597. [DOI] [PubMed] [Google Scholar]
- 17.Carreras-Carbonell J, Macpherson E, Pascual M. High self-recruitment levels in a Mediterranean littoral fish population revealed by microsatellite markers. Mar Biol. 2007;151:719–727. [Google Scholar]
- 18.Gerlach G, Atema J, Kingsford MJ, Black KP, Miller-Sims V. Smelling home can prevent dispersal of reef fish larvae. Proc Natl Acad Sci USA. 2007;104:858–863. doi: 10.1073/pnas.0606777104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel DA, Kinlan BP, Gaylord B, Gaines SD. Lagrangian descriptors of marine larval dispersal. Mar Ecol Prog Ser. 2003;260:83–96. [Google Scholar]
- 20.Becker BJ, Levin LA, Fodrie FJ, McMillan PA. Complex larval connectivity patterns among marine invertebrate populations. Proc Natl Acad Sci USA. 2007;104:3267–3272. doi: 10.1073/pnas.0611651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patterson HM, Swearer SE. Long-distance dispersal and local retention of larvae as mechanisms of recruitment in an island population of a coral reef fish. Austral Ecol. 2007;32:122–130. [Google Scholar]
- 22.Paris CB, Cowen R. Direct evidence of a biophysical retention mechanism for coral reef fish larvae. Limnol Oceanogr. 2004;49:1964–1979. [Google Scholar]
- 23.Galindo HM, Olson DB, Palumbi SR. Seascape Genetics: A Coupled Oceanographic-Genetic Model Predicts Population Structure of Caribbean Corals. Curr Biol. 2006;16:1622–1626. doi: 10.1016/j.cub.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 24.Hohenlohe PA. Limits to gene flow in marine animals with planktonic larvae: Models of Littorina species around Point Conception, California. Biol J Linn Soc. 2004;82:169–187. [Google Scholar]
- 25.Baums IB, Paris CB, Cherubin LM. A bio-oceanographic filter to larval dispersal in a reef-building coral. Limnol Oceanogr. 2006;51:1969–1981. [Google Scholar]
- 26.Shulman MJ. What can population genetics tell us about dispersal and biogeographic history of coral-reef fishes? Austral Ecol. 1998;23:216–225. [Google Scholar]
- 27.Waples RS, Rosenblatt RH. Patterns of larval drift in southern California marine shore fishes inferred from allozyme data. Fish Bull. 1987;85:1–11. [Google Scholar]
- 28.Bonhomme F, Planes S. Some evolutionary arguments about what maintains the pelagic interval in reef fishes. Environ Biol Fishes. 2000;59:365–383. [Google Scholar]
- 29.Riginos C, Victor BC. Larval spatial distributions and other early life-history characteristics predict genetic differentiation in eastern Pacific blennioid fishes. Proc R Soc B. 2001;268:1931–1936. doi: 10.1098/rspb.2001.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johannes RE. Reproductive strategies of coastal marine fishes in the tropics. Environ Biol Fishes. 1978;3:65–84. [Google Scholar]
- 31.Kingsford MJ, Choat JH. Horizontal distribution patterns of presettlement reef fish: Are they influenced by the proximity of reefs? Mar Biol. 1989;10:285–297. [Google Scholar]
- 32.Knutsen H, et al. Transport of North Sea cod larvae into the Skagerrak coastal populations. Proc Biol Sci. 2004;271:1337–1344. doi: 10.1098/rspb.2004.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabatés A. Distribution pattern of larval fish populations in the Northwestern Mediterranean. Mar Ecol Prog Ser. 1990;59:75–82. [Google Scholar]
- 34.Macpherson E, Raventos N. Relationship between pelagic larval duration and geographic distribution in Mediterranean littoral fishes. Marine Ecology Progress Series. 2006;327:257–265. [Google Scholar]
- 35.Mora C, Chittaro PM, Sale PF, Kritzer JP, Ludsin SA. Patterns and processes in reef fish diversity. Nature. 2003;421:933–936. doi: 10.1038/nature01393. [DOI] [PubMed] [Google Scholar]
- 36.Macpherson E. Large-Scale Species-Richness Gradients in the Atlantic Ocean. Proc Biol Sci. 2002;269:1715–1720. doi: 10.1098/rspb.2002.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tintoré J, La Violette PE, Blade I, Cruzado A. A study of an intense density front in the eastern Alboran Sea: The Almeria-Oran front. J Phys Oceanogr. 1988;18:1384–1397. [Google Scholar]
- 38.Patarnello T, Volckaert FAMJ, Castilho R. Pillars of Hercules: Is the Atlantic-Mediterranean transition a phylogeographical break? Mol Ecol. 2007;16:4426–4444. doi: 10.1111/j.1365-294X.2007.03477.x. [DOI] [PubMed] [Google Scholar]
- 39.Pinot J-M, Tintore J, Gomis D. Quasi-synoptic mesoscale variability in the Balearic Sea. Deep-Sea Res Part I Oceanogr Res Pap. 1994;41:897–914. [Google Scholar]
- 40.Béranger K, Mortier L, Crépon M. Seasonal variability of water transport through the Straits of Gibraltar, Sicily and Corsica, derived from a high-resolution model of the Mediterranean circulation. Prog Oceanogr. 2005;66:341–364. [Google Scholar]
- 41.Viúdez A, Pinot JM, Haney RL. On the upper layer circulation in the Alborán Sea. J Geophys Res. 1998;103:21653–21666. [Google Scholar]
- 42.Charrier G, et al. Discrepancies in phylogeographical patterns of two European anglerfishes (Lophius budegassa and Lophius piscatorius) Mol Phylogenet Evol. 2006;38:742–754. doi: 10.1016/j.ympev.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Bargelloni L, et al. Discord in the family Sparidae (Teleostei): Divergent phylogeographical patterns across the Atlantic–Mediterranean divide. J Evol Biol. 2003;16:1149–1158. doi: 10.1046/j.1420-9101.2003.00620.x. [DOI] [PubMed] [Google Scholar]
- 44.Bargelloni L, et al. The Atlantic-Mediterranean transition: Discordant genetic patterns in two seabream species, Diplodus puntazzo (Cetti) and Diplodus sargus (L) Mol Phylogenet Evol. 2005;36:523–535. doi: 10.1016/j.ympev.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 45.Werner FE, Cowen RK, Paris CB. Coupled Biological and physics models. Present capabilities and necessary developments for future studies of population connectivity. Oceanography. 2007;20:54–69. [Google Scholar]
- 46.Cowen R, Sponaugle S. Larval Dispersal and Marine Population Connectivity. Annu Rev Mar Sci. 2009;1:443–466. doi: 10.1146/annurev.marine.010908.163757. [DOI] [PubMed] [Google Scholar]
- 47.Raventos N. Effects of wave action on nesting activity in the littoral five-spotted wrasse, Symphodus roissali (Labridae), in the northwestern Mediterranean Sea. Sci Mar. 2004;68:257–264. [Google Scholar]
- 48.Raventos N, Macpherson E. Environmental influences on temporal patterns of settlement in two littoral labrid fishes in the Mediterranean Sea. Estuarine, Coastal and Shelf. Science. 2005;63:479–487. [Google Scholar]
- 49.Jordi A, Orfila A, Basterretxea G, Tintoré J. Shelf-slope exchanges by frontal variability in a steep submarine canyon. Prog Oceanogr. 2005;66:120–141. [Google Scholar]
- 50.Palanques A, et al. General patterns of circulation, sediment fluxes and ecology of the Palamós (La Fonera) submarine canyon, northwestern Mediterranean. Prog Oceanogr. 2005;66:89–119. [Google Scholar]
- 51.Doherty P, Planes S, Mather P. Gene flow and larval duration in seven species of fish from the Great Barrier Reef. Ecology. 1995;76:2373–2391. [Google Scholar]
- 52.Stepien CA, Rosenblatt RH. Patterns of gene flow and genetic divergence in the northeastern Pacific Clinidae (Teleostei: Blennioidei), based on allozyme and morphological data. Copeia. 1991;4:873–896. [Google Scholar]
- 53.Shaw PW, Arkhipkin AI, Al-khairulla H. Genetic structuring of Patagonian toothfish populations in the Southwest Atlantic Ocean: The effect of the Antarctic Polar Front and deep-water troughs as barriers to genetic exchange. Mol Ecol. 2004;13:3293–3303. doi: 10.1111/j.1365-294X.2004.02327.x. [DOI] [PubMed] [Google Scholar]
- 54.Leis JM. Behaviour as input for modelling dispersal of fish larvae: Behaviour, biogeography, hydrodynamics, ontogeny, physiology and phylogeny meet hydrography. Mar Ecol Prog Ser. 2007;347:185–193. [Google Scholar]
- 55.Bernardi G, Findley L, Rocha-Olivares A. Vicariance and dispersal across Baja California in disjunct marine fish populations. Evolution. 2003;57:1599–1609. doi: 10.1111/j.0014-3820.2003.tb00367.x. [DOI] [PubMed] [Google Scholar]
- 56.Purcell J, Cowen R, Hughes C, Williams D. Weak genetic structure indicates strong dispersal limits: A tale of two coral reef fish. Proc R Soc B. 2006;273:1483–1490. doi: 10.1098/rspb.2006.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramon ML, Nelson PA, De Martini E, Walsh WJ, Bernardi G. Phylogeography, historical demography, and the role of post-settlement ecology in two Hawaiian damselfish species. Mar Biol. 2008;153:1207–1217. [Google Scholar]
- 58.Palumbi SR. Population genetics, demographic connectivity and the design of marine protected areas. Ecol Appl. 2003;13:146–158. [Google Scholar]
- 59.Belkhir K, Borsa P, Goudet J, Chikhi L, Bonhomme F. Genetix v. 4.01 Software under Windows for Population Genetics. Montpellier, France: Centre National de la Recherche Scientifique Unité Propre de Recherche 9060; 1997. (Translated from French). Laboratoire Génome, Populations, Interactions. [Google Scholar]
- 60.Guillot G, Mortier F, Estoup A. Geneland: A computer package for landscape genetics. Mol Ecol Notes. 2005;5:712–715. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.