Abstract
Skeletal muscle adapts to physiological demands by altering a number of programs of gene expression, including those driving mitochondrial biogenesis, angiogenesis, and fiber composition. Recently, the PGC-1 transcriptional coactivators have emerged as key players in the regulation of these adaptations. Many signaling cascades important in muscle physiology impinge directly on PGC-1 expression or activity. In turn, the PGC-1s powerfully activate many of the programs of muscle adaptation. These findings have implications for our understanding of muscle responses to physiological conditions like exercise, as well as in pathological conditions such as cachexia, dystrophy, and peripheral vascular disease.
Introduction
Muscle converts chemical energy into physical work. The tasks demanded of muscle vary widely, ranging from rapid lifting of heavy weights to pumping blood without fault for decades. Moreover, these tasks change over time. To accommodate these changing conditions, muscle is both diverse in composition and highly plastic: the makeup and function of muscle can adapt to external factors like activity or hormonal exposure. This is true in both health and disease. Endurance exercise, for example, renders muscle more oxidative and resistant to fatigue. Conversely, disuse and systemic catabolic diseases lead to atrophy, with profound loss of muscle function. Understanding the molecular pathways underlying this plasticity is therefore of great interest.
Changes in metabolic programs are at the core of muscle plasticity. Great inroads have been made over the past decade into understanding the molecular pathways that drive these changes. The PGC-1 transcriptional co-activators, in particular, have emerged as an area of great interest in muscle bioenergetics because these proteins are dominant regulators of oxidative metabolism in many tissues. PGC-1α and β powerfully regulate broad and comprehensive genetic programs, including the activation of fatty acid oxidation, oxidative phosphorylation, and numerous attendant activities needed to maintain functional mitochondria. This review will focus on the role of PGC-1 coactivators in muscle metabolism and plasticity.
Muscle Composition
Skeletal muscle gains much of its heterogeneity during development. Muscle is composed of thousands of fibers, each of which is a syncitium of hundreds of cells stretching from one tendon to another. The tasks faced by groups of fibers or muscles ranges from continuous, low-level activities such as gravitational tasks (maintenance of posture) to sudden bursts of intense activity. To achieve such varied functions, fibers with different bioenergetic and biophysical properties exist [1,2].
There are four predominant types of fibers in adult mouse skeletal muscle: I, IIA, IIX, and IIB, named after the myosin heavy chain (MHC) that is primarily expressed. Myosin ATPases drive the kinetics of the actin/myosin cycling machinery; along with ancillary sarcomeric proteins and sarcolemnal proteins that regulate Ca++ handling, MHCs thus dictate the biophysical properties of a fiber. MHC type IIB fibers, historically called “fast glycolytic” (FG) fibers, have a rapid and powerful stroke amenable to rapid bursts of activity. This comes at the expense of high ATP requirements, and large and fast fluxes of Ca++. MHC type I fibers, historically “slow oxidative” (SO) fibers, on the other hand, have a slower contraction/relaxation profile that is more energetically efficient. Type I fibers are also rich in mitochondrial networks, and favor fatty acid oxidation, a far more efficient source of ATP than anaerobic glycolysis. Type IIA and IIX fibers, historically “fast oxidative/glycolytic” (FOG) fibers, have intermediate biophysical properties but tend to also be oxidative and rich in mitochondria.
Muscles containing significant type I, IIA, and IIX fibers can thus maintain a steady supply of ATP, preserve glycogen stores, and resist fatigue. These muscles, such as the soleus and deep gastrocnemius, are useful for constant but low-power tasks such as locomotion. Conversely, type IIB-rich muscles like the superficial quadriceps tire more easily but provide fast and powerful force, useful for sporadic bursts of intense work. In certain organisms, fiber types segregate between muscles. In fowl, for example, oxidative fibers are found almost exclusively in the hindlimbs, where high concentrations of myoglobin and cytochromes render these fibers red (or brown when cooked, hence “dark meat”). In land-dwelling mammals, deeper portions of muscles tend to be richer in oxidative fibers, consistent with their role in ambulation.
Muscle Plasticity
Even though the baseline fiber type composition of muscle is largely determined during development, adult muscle retains the capacity for substantial plasticity [1,2]. Physical activity, or lack thereof, have particularly profound effects. Endurance training induces expansion of the mitochondrial compartment, significant angiogenesis, and a fast-to-slow fiber type switch. This leads to improved endurance performance and resistance to fatigue, to the benefit of, for example, marathon runners. Resistance training, on the other hand, induces significant muscle hypertrophy, a slow-to-fast fiber transformation, and a switch to the use of glycolysis as the favored source of energy. These effects reproduce across vast swaths of phylogeny, though the extent of plasticity varies significantly, dependent on species and exercise program.
A series of elegant, and now classic, experiments demonstrated that nerve input is the primary determinant of fiber-type conversions [3]. When the motor nerves innervating fast-twitch and slow-twitch fibers are crossed, the phenotypes of the fibers are reversed within a few weeks (including fiber composition and mitochondrial content). Similarly, tonic low-frequency electrical pacing of fast-twitch nerves triggers a fast-to-slow transition. Thus, the amplitude and frequency of electrical stimulation is a primary determinant of fiber identity. Various other signals are superimposed, most notably thyroid hormone activity.
Disease can also have profound effects on skeletal muscle. Pathological muscle atrophy, or cachexia, often accompanies chronic diseases, such renal failure and heart failure [4]. Disuse, as seen in prolonged bedrest, and age-related sarcopenia, reduce both muscle mass and function [5]. The loss of muscle innervation, such as occurs in ALS also induces serious atrophy. These atrophy programs do not simply reverse the hypertrophy program; specific proteases and enzymes that target proteins to proteosomal degradation are activated by atrophic signals[6]. Interestingly, different fiber types display varying sensitivities to atrophy; oxidative fibers, for example, resist atrophy in the face of denervation.
Alterations in skeletal muscle can, in turn, affect the development and progression of disease. The presence of cachexia in chronic diseases like heart failure is invariably an ominous prognostic factor [4]. Both the consequent frailty and changes in systemic metabolism likely contribute to poor outcomes. Muscle is also the primary depot for glucose clearance from the blood, and insulin resistance in skeletal muscle is a primary cause of Type II Diabetes Mellitus. Endurance training, and the associated changes in musculature, protects against cardiovascular disease, Type II Diabetes, and a number of other diseases. Indeed, physical activity is a major predictor of health[7]. The interaction between muscle plasticity and these diseases remains poorly understood. However, significant progress has been made over the past decade in uncovering the molecular mechanisms that regulate muscle plasticity. The PGC-1 coactivators appear to be key players in this process.
The PGC-1 coactivators
Coactivators are proteins that dock on transcription factors and alter chromatin structure and the transcription machinery to stimulate gene expression. Most transcription factors likely bind to one or more coactivators to initiate transcription. Recently, coactivators have emerged as potent regulatory targets of physiological stimuli and hormones [8]. PGC-1α is the best-studied example of such a regulated coactivators [9]. PGC-1α was first identified as a cold-inducible PPAR-γ binding protein in brown fat. Since then, it has become apparent that PGC-1α can bind to, and coactivate, most members of the nuclear receptor family, as well as many other transcription factors. It is likely that PGC-1α gains much of its specificity by coactivating different transcription factors in different contexts.
PGC-1α activates transcription by recruiting several proteins that have histone acetyltransferase activity, including CBP, p300, and SRC-1 [10], as well as the Mediator Protein Complex, which is thought to recruit RNA polymerase II [11]. In addition, PGC-1α contains an RNA-binding motif, RRM, and has been implicated in processing many mRNA's whose transcription it initiates [12]. The activity of PGC-1α can be modulated by numerous post-translational events, including phosphorylation by the p38 MAPK and AMPK [13-16], acetylation by the longevity gene SIRT1 [17,18], and methylation[19]. PGC-1α has two structural homologues, PGC-1β and the more distant PGC-1 Related Coactivator (PRC), which were both originally identified by sequence homology to PGC-1α, and which have been less extensively studied.
PGC-1s and oxidative metabolism
The PGC-1 coactivators have a variety of biological activities in different tissues, including muscle, and most of these activities are linked to oxidative metabolism. Both PGC-1α and β are expressed at high levels in oxidative tissues such as the heart, brain, kidney, and muscle [20,21]. When expressed ectopically, PGC-1α and β induce mitochondrial biogenesis and increase cellular respiration in cell culture. PGC-1α -/- and PGC-1β -/- animals have abnormal skeletal and cardiac energetics [22,23] [24-26] and PGC-1α -/- mice develop accelerated heart failure when stressed [27]. When transgenically expressed in skeletal muscle, both PGC-1α and PGC-1β induce marked mitochondrial biogenesis [28-30]. The intense induction of oxygen-containing cytochromes and myoglobin renders normally white muscle throughout the transgenic mice now red. The transgenic muscles have increased oxidative capacity[30] and resistance to fatigue[28], and the transgenic mice have improved aerobic work performance [29,31].
How the PGC-1s activate mitochondrial biogenesis has been studied in some detail (reviewed in [9]). The PGC-1s activate nuclear-encoded genes involved in mitochondrial biogenesis by coactivating at least three transcription factors, NRF-1 and 2 and ERRα. These factors interact with and activate regulatory regions in many mitochondrial genes encoded in the nucleus. Genes of fatty acid oxidation, a largely mitochondrial process, are regulated by coactivation of the nuclear receptor PPARα. PGC-1α also activates the expression of Tfam and factors B1 and B2 [9,32]; these factors are key regulators of proliferation and transcription of the mitochondrial genome. Thus the PGC-1 coactivators orchestrate the activation of hundreds of genes encoded by the nucleus, as well as the proliferation and transcription of the mitochondrial genome, all of which is critical for the generation of these complex organelles. Genetic studies in mice have shown that various tissues lacking either PGC-1α or β have significant defects in mitochondria, but the animals still have functional mitochondria [23-26,33]. Deletion of both PGC-1s, tested thus far only in the context of brown fat cells, leads to near complete loss of mitochondria [34].
PGC-1s and fiber types
In most of their tissue-specific roles, the PGC-1 coactivators increase a core program of mitochondrial biogenesis and respiration, as well as ancillary programs that go along with increased respiration in each tissue [9]. The expression of PGC-1α in white fat cells, for example, also gives them many of the properties of brown fat cells, including increased mitochondrial biogenesis and expression of UCP-1. In the liver, fasting induces PGC-1α, leading to gluconeogenesis and β-oxidation of fatty acids. Similarly, not only do the PGC-1s activate the mitochondrial program in skeletal muscle, but genes encoding myofibrillar proteins characteristic of oxidative fibers are also induced. Mice transgenically expressing PGC-1α increase the proportion of fibers expressing MHC I and IIA [28], while interestingly, mice expressing PGC-1β almost uniformly switch to oxidative IIX fibers [29]. The PGC-1 coactivators thus drive a complex phenotypic switch, including a core program of mitochondrial biogenesis and an ancillary program of myofibrillar content (and angiogenesis, see below).
Precisely which transcription factors mediate the activation of specific fiber-type genes by the PGC-1s remains incompletely understood. The MEF2 family of transcription factors has been implicated in fiber-type determination and is activated by exercise [35]. In cell culture experiments, PGC-1α coactivates MEF2's to induce type I fiber genes like troponin I slow and myoglobin [28]. However, PGC-1β is an equally robust inducer of MEF2 in these assays, yet PGC-1β has very different effects on fiber composition in vivo [29]. Clearly, other factors must exist to confer specificity on these programs. One possible candidate is PPARβ, a lipid-sensitive nuclear receptor. Muscle-specific ablation of PPARβ causes a slow-to-fast fiber switch, and muscle-specific overexpression of PPARβ has the converse effect [36-38]. The effect of PGC-1s and PPARβ together on fiber-specific promoters, however, has not been evaluated.
It is important to highlight that the PGC-1s are probably not the only determinants of fiber type identity. Animals lacking PGC-1α specifically in skeletal muscle have reductions in type I and IIa fibers, but they retain a fair number of these fibers [39]. Interestingly, animals lacking PGC-1α throughout the body have normal amounts of type I and IIA fibers [23], underscoring the existence of PGC-1α-independent compensatory mechansims. The fiber content of animals lacking PGC-1β (or lacking both α and β) has not been reported.
PGC-1α and exercise
Endurance training induces mitochondrial biogenesis and a fast-to-slow fiber type switch in skeletal muscle. Both PGC-1α and β are more highly expressed in oxidative fibers [28,29], and PGC-1α is preferentially expressed in type I and IIA-rich muscle beds like the soleus or deep gastrocnemius [28]. PGC-1α expression in human and rodent skeletal muscle is strongly induced by exercise (e.g., [40-42]), while PGC-1β expression appears unaffected. These observations, combined with the powerful ability of PGC-1α to induce mitochondrial biogenesis and fiber type switching, have led to the notion that PGC-1α mediates many of the genomic effects of exercise.
If so, what are the upstream signals? A number of signaling pathways activated during exercise can impinge on PGC-1α. Muscle contraction generates powerful Ca++ fluxes. The amplitude and frequency of these fluxes activates various signaling pathways, including those regulated by the calcineurin phosphatase and the calmodulin-modulated kinase, both of which can affect PGC-1α expression and activity [43]. Calcineurin, in particular, is activated by low-amplitude low-frequency Ca++ transients typical of locomotion or endurance training. Transgenic expression of calcineurin in skeletal muscle induces a fast-to-slow fiber switch [44,45]. Conversely, mice in which calcineurin isoforms have been deleted, as well as animals or isolated muscles treated with a calcineurin inhibitor, reveal either a loss of slow fibers or a block in activity-dependent fast-to-slow fiber switch [46-48]. It is therefore likely that calcineurin's effect on gene expression occurs in large part through PGC-1α. This may in part occur through activation of the MEF2 transcription factors, known targets of Ca++ signaling. Exercise induces MEF2 activity [35], and induction of the PGC-1α promoter by contraction requires intact MEF2 binding sites[49].
Exercise also affects levels of high-energy phosphates like ATP and ADP, leading to the activation of the AMP-sensitive AMP Kinase (AMPK) [50]. Recently, AMPK has been implicated in fiber transformations and mitochondrial biogenesis induced by exercise and other stimuli [51-55]. At least some of the genomic effects of AMPK in skeletal muscle occur through PGC-1α [14], suggesting that PGC-1α may transduce exercise-induced AMPK signals. The adrenergic system, via cyclic AMP and other second messengers, also probably contributes to exercise-mediated skeletal muscle plasticity. β-adrenergic stimulation potently induces PGC-1α expression in skeletal muscle, and β-adrenergic antagonists partially block the induction of PGC-1α by exercise [56].
The p38 MAPK has also been shown to be activated by exercise and likely contributes to the acute induction of PGC-1α expression [57]. In addition, p38 MAPK directly phosphorylates PGC-1α in cell culture, leading to both stabilization of PGC-1α protein and dissociation from the p160myb repressor [13,16], though whether this occurs in skeletal muscle in response to exercise remains to be shown. The production of reactive oxygen species (ROS) also increases during exercise, and ROS can induce PGC-1α [58], suggesting that ROS may also contribute to the induction of PGC-1α by exercise[59]. Finally, hypoxia has long been presumed to be a stimulus for genetic change during intermittent but sufficiently extreme exercise, and PGC-1α is inducible by hypoxia in skeletal muscle cells [60]. The effects of all these pathways on PGC-1β have not been studied extensively.
PGC-1α thus appears to represent an important nexus for translating extracellular information into changes in gene expression. It seems highly likely that at least some of these pathways impinge on the expression or function of PGC-1α (or PGC-1β) during exercise, contributing in turn to ensuing phenotypic changes, including mitochondrial biogenesis, fiber type switching, and angiogenesis (see below). Formally demonstrating this idea will require the use of mice lacking PGC-1α. PGC-1α -/- mice are hyperactive and lean, and thus difficult to study without numerous confounding issues [33]. Mice lacking PGC-1α specifically in skeletal muscle have moderately decreased performance capacity at baseline [39], but the effects of endurance training in these mice have not been reported. Mice lacking both PGC-1α and PGC-1β, if viable, will also be of great interest.
PGC-1α and angiogenesis
Peripheral vascular disease is a leading cause of morbidity and the most common cause of limb amputation in the industrialized world. Chronic ischemia can have profound effects on muscle makeup. Since oxidative phosphorylation is by far the most efficient way to produce ATP, the robust output of ATP in muscle vitally depends on the efficient delivery of both oxygen and nutrients. Angiogenesis is therefore a critical homeostatic response to chronic ischemia. Angiogenesis is a complex process whereby new blood vessels are formed from preexisting vessels [61,62]; it differs from vasculogenesis, the de novo formation of vessels during embryogenesis. The process is in large part coordinated by soluble factors emanating from tissues in need of neovascularization. VEGF, the best studied of these factors, is a powerful activator of endothelial proliferation, permeability, and migration, and is critical for almost all forms of neovascularization [63]. The angiogenic response to ischemia, however, is frequently insufficient, and great efforts are therefore underway to find ways to stimulate angiogenesis in ischemic tissue.
What gene regulatory events orchestrate angiogenesis has been the subject of intense study for decades. The best understood process is that involving the transcription factor Hypoxia Inducible Factor-1α (HIF-1α). In the presence of oxygen, HIF-α is inhibited by hydroxylation and targeted for degradation in the presence of oxygen [64,65]. Low oxygen halts these processes, and the stabilized HIF-1α heterodimerizes with the ubiquitous HIF-1β to activate a number of genes involved in the hypoxic response. Among these are VEGF and other soluble factors that coordinate a strong angiogenic response to hypoxia.
Recently, PGC-1α has emerged as an important and novel inducer of angiogenesis in skeletal muscle[60,66]. PGC-1α strongly induces expression of VEGF, and other angiogenic factors like PDGFB and angiopoietin 2, in cultured muscle cells and skeletal muscle in vivo. Transgenic expression of PGC-1α in skeletal muscle dramatically increases capillary density. Furthermore, PGC-1α is induced by lack of nutrients and oxygen, and the full induction of VEGF under these conditions requires PGC-1α. This suggested that PGC-1α may play a role in the angiogenic response to ischemia. Indeed, PGC-1α -/- mice show a striking failure to normally reconstitute blood flow to the limb after an ischemic insult. Conversely, PGC-1α transgenesis accelerates the recovery of blood flow during limb ischemia. These findings strongly implicate PGC-1α in the regulation of angiogenesis in muscle in response to ischemia. Surprisingly, the mechanism by which PGC-1α induces VEGF does not appear to involve the canonical HIF pathway. Instead, PGC-1α coactivates the orphan nuclear receptor ERRα on both the promoter and a newly-identified enhancer in the first intron of the VEGF gene[60]. Thus, PGC-1α and ERRα control a novel, HIF-independent angiogenic pathway that delivers needed oxygen and substrates in the face of ischemia.
It is likely that PGC-1α also plays a role in angiogenesis under physiologic conditions, in addition to pathologic ischemia. Microvascular density, maximum blood flow, and oxidative capacity are remarkably coupled in skeletal muscle under normal circumstances. Oxidative muscles contain rich capillary networks, and more capillaries abut oxidative than glycolytic fibers. In fact, the ratio of capillary surface area to that of the inner mitochondrial membrane in any given fiber appears to be tightly maintained at about 1:200 [67]. It is presumed that this tight coupling reflects the increased need for oxygen delivery to mitochondria, and the reduced effective diffusion distance created by the presence of metabolically active mitochondria. To maintain this tight relationship, endurance exercise triggers angiogenesis in skeletal muscle to match the mitochondrial biogenesis [67]. Similarly, chronic electrical stimulation of fast-twitch muscles at a frequency naturally occurring in slow-twitch muscles dramatically increases capillary density, along with increasing oxidative capacity. Few data exist to address the molecular mechanisms underlying this process. As already stated, PGC-1α expression is strongly induced by exercise in rodents and humans, and PGC-1α is a powerful inducer of both mitochondrial biogenesis and angiogenesis in skeletal muscle[30,60]. It seems likely, therefore, that PGC-1α plays a key role in both exercise-induced angiogenesis and mitochondrial proliferation, thereby tightly coordinating the delivery of oxygen and nutrients with their consumption by mitochondria. This remains to be proven.
PGC-1α and disease
The large mass of proteins that make up the myofibrillar apparatus of skeletal muscle is a critical reservoir for amino acids in the body. This reservoir can be tapped for energy needs in times of want, such as prolonged food deprivation, or in various pathologic conditions, such as cancer, sepsis, and renal and heart failure. Whereas protein breakdown is normal in response to prolonged fasting, wasting and cachexia associated with chronic disease can be debilitating. Indeed, skeletal muscle wasting is a poor prognostic factor in many chronic diseases like cancer and heart failure. Breakdown of the myofibrillar proteins is initiated, at least in part, by a set of E3-ubiquitin ligases, including Murf1 and Atrogin-1/MAFbx, which target proteins for degradation by the proteosome [6]. Expression of these ligases is strongly induced under various catabolic conditions, both physiologic and pathologic, by the translocation of the transcription factor FoxO3 to the nucleus, where it activates the Murf1 and Atrogin-1 genes. Anabolic signals, such as insulin or Insulin Growth Factor, antagonize this process.
It has long been known that different muscle fibers have different sensitivities to catabolic signals. Glycolytic fibers, for example, atrophy faster during fasting or sepsis than do oxidative fibers [68,69]. Conversely, physical activity is one of the most effective ways to prevent loss of muscle mass in pathologic catabolism. Catabolic states like cancer, sepsis, and renal failure, strongly suppress PGC-1α expression in skeletal muscle in vivo [70,71], while exercise increases PGC-1α levels. These observations suggested that PGC-1α may protect against atrophy. Denervation of the mouse hindlimb leads to marked atrophy, modeling the wasting seen in human motor neuron diseases like ALS. Transgenic expression of PGC-1α in skeletal muscle completely blocked this atrophic response [71]. Similarly, atrophy can be induced by direct gene delivery of constitutively active FoxO3 into skeletal muscle, thereby inducing Murf1 and Atrogin-1. Again, co-delivery of PGC-1α completely blocked atrophy in this setting [71].
PGC-1α may also protect against drug-induced myopathy. Statins are widely used for the treatment of elevated cholesterol, and muscle breakdown is their most significant, and sometimes deadly, side effect. Treating cells or animals with statins induces the atrophic program, including expression of Atrogin-1 [72]. Strikingly, increased expression of PGC-1α powerfully blocked the induction of Atrogin-1 and the ensuing muscle damage in this setting as well [72]. Thus, PGC-1α appears to have powerful anti-atrophic functions.
How PGC-1α expression is regulated in catabolic states, and how PGC-1α protects against atrophy, remain unclear. PGC-1α, normally a potent coactivator of transcription, is in this case efficiently suppressing the activity of FoxO3 on the Murf1 and Atrogin-1 genes. The presence of excess PGC-1α reduces the FoxO3 occupancy on the Atrogin-1 promoter[71], suggesting that PGC-1α is not acting as a direct inhibitor of transcription. Innervation is a powerful anabolic signal, and PGC-1α induces components of the neuromuscular junction (NMJ) [73]. PGC-1α may therefore in part block catabolic signals indirectly by increasing anabolic signaling from the NMJ. In addition, the mitochondrial biogenesis induced by PGC-1α may account for part of the protection by improving Ca++ handling and/or cellular energetics.
PGC-1α also confers protection against muscular dystrophy. Duchenne's muscular dystrophy (DMD) is a universally fatal disease caused by mutations in the gene dystrophin. The absence of dystrophin leads to skeletal and cardiac muscle degeneration, and invariably death by young adulthood. Mice deficient in the dystrophin gene (mdx mice) recapitulate many of the attributes of DMD. Strikingly, transgenic expression of PGC-1α in mdx mice markedly improves muscle degeneration and improves muscle function[73]. Again, how PGC-1α protects against dystrophy is not clear. Some mechanisms likely overlap with the anti-atrophic program. PGC-1α also induces utrophin, a dystrophin homolog that can compensate for the absence of dystrophin [73,74].
Skeletal muscle is the primary site of glucose uptake in response to insulin, and muscle resistance to the actions of insulin is a major causative factor in type II diabetes. The role of the PGC-1s in insulin resistance appears complex. PGC-1α induces GLUT4, the dominant glucose transporter in skeletal muscle [75]. Gene expression of both PGC-1s, as well as the entire program of oxidative phosphorylation, is coordinately repressed in skeletal muscle of patients with type II diabetes [76,77]. Strikingly, this is true even of unaffected family members, suggesting an early, possibly causal relationship. On the other hand, mice transgenically expressing PGC-1α in skeletal muscle are not sensitized to insulin signaling [31], and mice lacking PGC-1α in skeletal muscle are not resistant to insulin action [33,78]. The role of PGC-1α in muscle insulin resistance thus remains equivocal. Interestingly, the absence of PGC-1α in skeletal muscle does lead to pancreatic β-cell dysfunction and low insulin levels [78]. This implies the existence of important, PGC-1α-dependent, cross-talk between muscle and β-cells. How this occurs is not clear, though the presence of low-grade inflammation in mice lacking PGC-1α in skeletal muscle may account for some of the effect [78].
Future directions
Many unanswered questions clearly require attention. Though many pathways can impinge on PGC-1α expression or activity, which of these operate in skeletal muscle in response to external cues is still unclear. For example, how deprivation of oxygen and nutrients induces PGC-1α, and precisely what pathways induce PGC-1α during exercise, remains only partly understood. How the PGC-1s integrate these multiple incoming signals is also in need of further study. For example, lack of oxygen/nutrients somehow induces PGC-1α to activate its angiogenic program, but not that of mitochondrial biogenesis. Post-translational modifications of the PGC-1s are likely to play an important role. The presumed role of PGC-1α in endurance training-induced angiogenesis, mitochondrial biogenesis, and fiber type conversion, also awaits formal confirmation. Finally, the effects wrought by PGC-1α on muscle appear to have important effects on distant tissues; what these effects are, and how the organ cross-talk occurs, is only beginning to be examined. A possible anti-inflammatory role of the PGC-1s is particularly enticing [78].
Despite these many unanswered questions, what is clear is the powerful ability of PGC-1α and β to re-program numerous aspects of skeletal muscle. The improvements in exercise performance, induction of angiogenesis, and protection against atrophy and dystrophy are particularly noteworthy. The ability of the PGC-1s to orchestrate large programs of gene expression makes them appealing nodal points for pharmaceutical intervention. For example, human clinical trials that use VEGF for the treatment of chronic limb ischemia have yielded disappointing result [62,79,80], likely in part because VEGF alone cannot generate fully functional vessels. PGC-1α, on the other hand, appears to activate a complete and well-orchestrated angiogenic program, including the induction of VEGF as well as a number of other angiogenic factors like angiopoietin 2 and PDGFB[60]. Thus, independent of their precise role in physiological muscle plasticity, drugs that induce one or both PGC-1s could improve many muscular diseases, ranging from peripheral vascular disease to Duchenne's muscular dystrophy. Efforts to look for such drugs are underway [81].
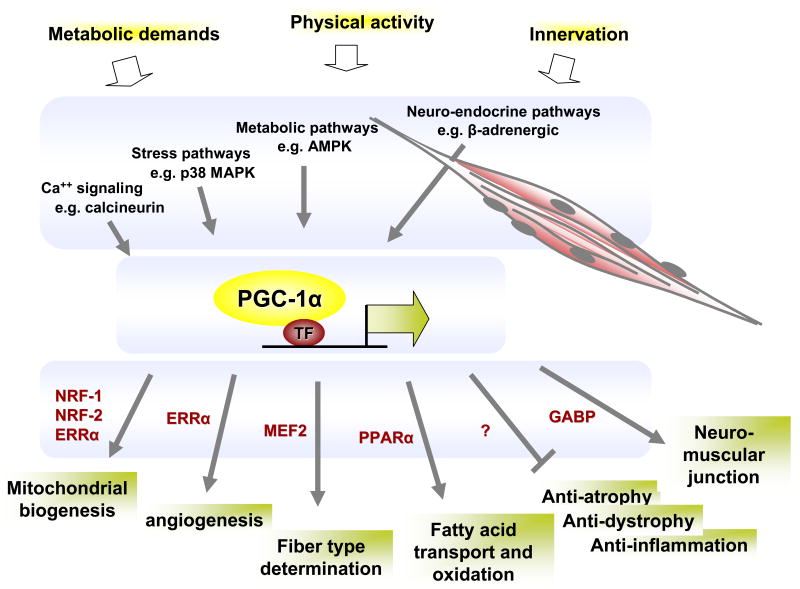
Figure 1.
Schematic of the role of PGC-1α in muscle plasticity. Physiological cues (top) are transduced via key signaling cascades and impinge on the expression of PGC-1α mRNA and the function of PGC-1α protein. In turn, PGC-1α interacts with, and coactivates, a number of transcription factors (examples of which are shown in red), leading to the activation of a number of broad genetic programs (bottom).
Acknowledgments
Many important contributions were unfortunately not covered due to space limitations. Dr. Stew Lecker provided critical reading of the manuscript. The author has no conflict of interests.
Highlighted publications
* Wende etal [30]: The authors generated a double transgenic mouse that induces PGC-1α expression in skeletal muscle in response to removal of doxycycline from the chow. They demonstrate that transient induction of PGC-1α in adult skeletal muscle induces mitochondrial biogenesis, respiration, fatty acid oxidation, and glycogen sparing.
** Hanai etal [72]: The authors show that statins, drugs widely used for the treatment of hypercholesterolemia, induce Atrogin-1 and atrophy in skeletal muscle cells in vitro and in zebrafish muscle in vivo. Ectopic expression of PGC-1α in cells or animals blocked both atrophy and the induction of Atrogin-1, demonstrating that PGC-1α can protect against statin-induced myopathy.
** Handschin etal [78]: Deletion of PGC-1α strictly in skeletal muscle led to hypophagia, hypermetabolism, resistance to weight gain induced by high fat diet, and impaired pancreatic β-cells. These observations demonstrate the existence of important PGC-1α-dependent cross-talk between skeletal muscle and other organs, most notably the endocrine pancreas.
** Arany etal [60]: This manuscript demonstrates that PGC-1α is a powerful inducer of angiogenesis in skeletal muscle, likely in large part by direct induction of VEGF and other angiogenic factors. Moreover, PGC-1α is induced by nutrient/oxygen deprivation, and PGC-1α -/- mice have a slowed recovery of blood flow after ischemic injury to the limb, indicating that PGC-1α plays an important role in angiogenesis in vivo.
** Sandri etal [71]: The authors show that transgenic expression of PGC-1α in skeletal muscle protects against atrophy caused by muscle denervation. Similarly, expression of PGC-1α by transient transfection of intact skeletal muscle protects against the induction of Atrogin-1 and the atrophy induced by ectopic expression of FOXO3. Thus, PGC-1α appears to have potent anti-atrophic activities.
** Handschin etal [73]: PGC-1α induces the expression of numerous genes encoding components of the neuromuscular junction, likely in large part by coactivating the transcription factor GA-Binding Protein. In addition, transgenic expression of PGC-1α in skeletal muscle ameliorates the muscle degeneration observed in mdx mice, a model of Duchenne's muscular dystrophy. These observations demonstrate that PGC-1α has anti-dystrophy properties.
* Gerhadt-Hines etal [17]: Continuing work previously done in hepatocytes, the authors demonstrate that PGC-1α in skeletal muscle is directly deacetylated, and activated, by the Sirt1 histone deacetylase. This may provide an important link between metabolic regulation in skeletal muscle and aging-related pathologies.
* Uldry etal [34]: The reduction of both PGC-1α and PGC-1β in brown fat cells in culture led to profoundly reduced mitochondrial content and activity. This provided the only evidence to date that the PGC-1s are not only powerful activators of mitochondrial biology, but in fact appear necessary for normal mitochondrial function.
* Akimoto etal [49]: Cell culture models of skeletal muscle are severely limited, lacking innervation, 3-dimensional organization, complete differentiation, and many other attributes of normal skeletal muscle. The authors develop imaging techniques for visualizing activity from the PGC-1α promoter in skeletal muscle in live mice and in real-time. This approach is likely to lead to interesting insights into the regulation of PGC-1α expression in skeletal muscle.
* Jager etal [14]: The AMP Kinase responds to increases in AMP by activating programs that favor the generation of ATP. The authors show AMPK directly phosphorylates PGC-1α, and that activation of a number of genes by AMPK requires PGC-1α. Hence AMPK, a central metabolic sensor, transduces some signaling to the genome via PGC-1α.
* Miura etal [56]: The authors show that β-adrenergic agonists are powerful inducers of PGC-1α expression in skeletal muscle. Moreover, the induction of PGC-1α by exercise is partially dependent of β-adrenergic signaling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pette D. The adaptive potential of skeletal muscle fibers. Can J Appl Physiol. 2002;27:423–448. doi: 10.1139/h02-023. [DOI] [PubMed] [Google Scholar]
- 2.Schiaffino S, Sandri M, Murgia M. Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology (Bethesda) 2007;22:269–278. doi: 10.1152/physiol.00009.2007. [DOI] [PubMed] [Google Scholar]
- 3.Pette D, Vrbova G. What does chronic electrical stimulation teach us about muscle plasticity? Muscle Nerve. 1999;22:666–677. doi: 10.1002/(sici)1097-4598(199906)22:6<666::aid-mus3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 4.Anker SD, Steinborn W, Strassburg S. Cardiac cachexia. Ann Med. 2004;36:518–529. doi: 10.1080/07853890410017467. [DOI] [PubMed] [Google Scholar]
- 5.Thomas DR. Loss of skeletal muscle mass in aging: examining the relationship of starvation, sarcopenia and cachexia. Clin Nutr. 2007;26:389–399. doi: 10.1016/j.clnu.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Cao PR, Kim HJ, Lecker SH. Ubiquitin-protein ligases in muscle wasting. Int J Biochem Cell Biol. 2005;37:2088–2097. doi: 10.1016/j.biocel.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Erikssen G, Liestol K, Bjornholt J, Thaulow E, Sandvik L, Erikssen J. Changes in physical fitness and changes in mortality. Lancet. 1998;352:759–762. doi: 10.1016/S0140-6736(98)02268-5. [DOI] [PubMed] [Google Scholar]
- 8.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 9.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman BM. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 11.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 12.Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 13.Fan M, Rhee J, St-Pierre J, Handschin C, Puigserver P, Lin J, Jaeger S, Erdjument-Bromage H, Tempst P, Spiegelman BM. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knutti D, Kressler D, Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc Natl Acad Sci U S A. 2001;98:9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, Spiegelman BM. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 17.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. Embo J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 19.Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 21.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 22.Leone T, Lehman J, Finck B, Schaeffer PJ, W A, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLOS. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, et al. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci U S A. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lelliott CJ, Medina-Gomez G, Petrovic N, Kis A, Feldmann HM, Bjursell M, Parker N, Curtis K, Campbell M, Hu P, et al. Ablation of PGC-1beta results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4:e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vianna CR, Huntgeburth M, Coppari R, Choi CS, Lin J, Krauss S, Barbatelli G, Tzameli I, Kim YB, Cinti S, et al. Hypomorphic mutation of PGC-1beta causes mitochondrial dysfunction and liver insulin resistance. Cell Metab. 2006;4:453–464. doi: 10.1016/j.cmet.2006.11.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-gamma coactivator 1alpha. Proc Natl Acad Sci U S A. 2006;103:10086–10091. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 29.Arany Z, Lebrasseur N, Morris C, Smith E, Yang W, Ma Y, Chin S, Spiegelman BM. The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Wende AR, Schaeffer PJ, Parker GJ, Zechner C, Han DH, Chen MM, Hancock CR, Lehman JJ, Huss JM, McClain DA, et al. A role for the transcriptional coactivator PGC-1alpha in muscle refueling. J Biol Chem. 2007;282:36642–36651. doi: 10.1074/jbc.M707006200. [DOI] [PubMed] [Google Scholar]
- 31.Calvo JA, Daniels TG, Wang X, Paul A, Lin J, Spiegelman BM, Stevenson SC, Rangwala SM. Muscle-specific expression of PPAR{gamma} coactivator-1{alpha} improves exercise performance and increases peak oxygen uptake. J Appl Physiol. 2008;104:1304–1312. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
- 32.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Wu H, Rothermel B, Kanatous S, Rosenberg P, Naya FJ, Shelton JM, Hutcheson KA, DiMaio JM, Olson EN, Bassel-Duby R, et al. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. Embo J. 2001;20:6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luquet S, Lopez-Soriano J, Holst D, Fredenrich A, Melki J, Rassoulzadegan M, Grimaldi PA. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. Faseb J. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- 37.Schuler M, Ali F, Chambon C, Duteil D, Bornert JM, Tardivel A, Desvergne B, Wahli W, Chambon P, Metzger D. PGC1alpha expression is controlled in skeletal muscles by PPARbeta, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metab. 2006;4:407–414. doi: 10.1016/j.cmet.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 40.Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. Faseb J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 41.Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, Dohm GL, Yan Z, Newgard CB, Muoio DM. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem. 2005;280:33588–33598. doi: 10.1074/jbc.M507621200. [DOI] [PubMed] [Google Scholar]
- 42.Norrbom J, Sundberg CJ, Ameln H, Kraus WE, Jansson E, Gustafsson T. PGC-1alpha mRNA expression is influenced by metabolic perturbation in exercising human skeletal muscle. J Appl Physiol. 2004;96:189–194. doi: 10.1152/japplphysiol.00765.2003. [DOI] [PubMed] [Google Scholar]
- 43.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J Biol Chem. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 45.Olson EN, Williams RS. Remodeling muscles with calcineurin. Bioessays. 2000;22:510–519. doi: 10.1002/1521-1878(200011)22:11<1049::AID-BIES14>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 46.Parsons SA, Millay DP, Wilkins BJ, Bueno OF, Tsika GL, Neilson JR, Liberatore CM, Yutzey KE, Crabtree GR, Tsika RW, et al. Genetic loss of calcineurin blocks mechanical overload-induced skeletal muscle fiber type switching but not hypertrophy. J Biol Chem. 2004;279:26192–26200. doi: 10.1074/jbc.M313800200. [DOI] [PubMed] [Google Scholar]
- 47.Parsons SA, Wilkins BJ, Bueno OF, Molkentin JD. Altered skeletal muscle phenotypes in calcineurin Aalpha and Abeta gene-targeted mice. Mol Cell Biol. 2003;23:4331–4343. doi: 10.1128/MCB.23.12.4331-4343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akimoto T, Li P, Yan Z. Functional interaction of regulatory factors with the Pgc-1{alpha} promoter in response to exercise by in vivo imaging. Am J Physiol Cell Physiol. 2008 doi: 10.1152/ajpcell.00104.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakamoto K, Goodyear LJ. Invited review: intracellular signaling in contracting skeletal muscle. J Appl Physiol. 2002;93:369–383. doi: 10.1152/japplphysiol.00167.2002. [DOI] [PubMed] [Google Scholar]
- 51.Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, Shulman GI. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci U S A. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rockl KS, Hirshman MF, Brandauer J, Fujii N, Witters LA, Goodyear LJ. Skeletal muscle adaptation to exercise training: AMP-activated protein kinase mediates muscle fiber type shift. Diabetes. 2007;56:2062–2069. doi: 10.2337/db07-0255. [DOI] [PubMed] [Google Scholar]
- 53.Bergeron R, Ren JM, Cadman KS, Moore IK, Perret P, Pypaert M, Young LH, Semenkovich CF, Shulman GI. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am J Physiol Endocrinol Metab. 2001;281:E1340–1346. doi: 10.1152/ajpendo.2001.281.6.E1340. [DOI] [PubMed] [Google Scholar]
- 54.Winder WW, Holmes BF, Rubink DS, Jensen EB, Chen M, Holloszy JO. Activation of AMP-activated protein kinase increases mitochondrial enzymes in skeletal muscle. J Appl Physiol. 2000;88:2219–2226. doi: 10.1152/jappl.2000.88.6.2219. [DOI] [PubMed] [Google Scholar]
- 55.Suwa M, Nakano H, Kumagai S. Effects of chronic AICAR treatment on fiber composition, enzyme activity, UCP3, and PGC-1 in rat muscles. J Appl Physiol. 2003;95:960–968. doi: 10.1152/japplphysiol.00349.2003. [DOI] [PubMed] [Google Scholar]
- 56.Miura S, Kawanaka K, Kai Y, Tamura M, Goto M, Shiuchi T, Minokoshi Y, Ezaki O. An increase in murine skeletal muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to exercise is mediated by beta-adrenergic receptor activation. Endocrinology. 2007;148:3441–3448. doi: 10.1210/en.2006-1646. [DOI] [PubMed] [Google Scholar]
- 57.Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280:19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 58.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 59.Kramer HF, Goodyear LJ. Exercise, MAPK, and NF-kappaB signaling in skeletal muscle. J Appl Physiol. 2007;103:388–395. doi: 10.1152/japplphysiol.00085.2007. [DOI] [PubMed] [Google Scholar]
- 60.Arany Z, Foo SY, Ma Y, Ruas J, Bommi-Reddy A, Girnun GD, Cooper M, Laznik D, Chinsomboon J, Rangwala S, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1α. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 61.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 62.Jain RK. Molecular regulation of vessel maturation. Nat Med. 2003;9:685–693. doi: 10.1038/nm0603-685. [DOI] [PubMed] [Google Scholar]
- 63.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 64.Semenza GL. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda) 2004;19:176–182. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- 65.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 66.Fraisl P, Baes M, Carmeliet P. Hungry for blood vessels: linking metabolism and angiogenesis. Dev Cell. 2008;14:313–314. doi: 10.1016/j.devcel.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 67.Hudlicka O, Brown M, Egginton S. Angiogenesis in skeletal and cardiac muscle. Physiol Rev. 1992;72:369–417. doi: 10.1152/physrev.1992.72.2.369. [DOI] [PubMed] [Google Scholar]
- 68.Li JB, Goldberg AL. Effects of food deprivation on protein synthesis and degradation in rat skeletal muscles. Am J Physiol. 1976;231:441–448. doi: 10.1152/ajplegacy.1976.231.2.441. [DOI] [PubMed] [Google Scholar]
- 69.Tiao G, Lieberman M, Fischer JE, Hasselgren PO. Intracellular regulation of protein degradation during sepsis is different in fast- and slow-twitch muscle. Am J Physiol. 1997;272:R849–856. doi: 10.1152/ajpregu.1997.272.3.R849. [DOI] [PubMed] [Google Scholar]
- 70.Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. Faseb J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 71.Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci U S A. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanai J, Cao P, Tanksale P, Imamura S, Koshimizu E, Zhao J, Kishi S, Yamashita M, Phillips PS, Sukhatme VP, et al. The muscle-specific ubiquitin ligase atrogin-1/MAFbx mediates statin-induced muscle toxicity. J Clin Invest. 2007;117:3940–3951. doi: 10.1172/JCI32741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Handschin C, Kobayashi YM, Chin S, Seale P, Campbell KP, Spiegelman BM. PGC-1alpha regulates the neuromuscular junction program and ameliorates Duchenne muscular dystrophy. Genes Dev. 2007;21:770–783. doi: 10.1101/gad.1525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bogdanovich S, Perkins KJ, Krag TO, Khurana TS. Therapeutics for Duchenne muscular dystrophy: current approaches and future directions. J Mol Med. 2004;82:102–115. doi: 10.1007/s00109-003-0484-1. [DOI] [PubMed] [Google Scholar]
- 75.Michael LF, Wu Z, Cheatham RB, Puigserver P, Adelmant G, Lehman JJ, Kelly DP, Spiegelman BM. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc Natl Acad Sci U S A. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 77.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Handschin C, Choi CS, Chin S, Kim S, Kawamori D, Kurpad AJ, Neubauer N, Hu J, Mootha VK, Kim YB, et al. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J Clin Invest. 2007;117:3463–3474. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carmeliet P. VEGF gene therapy: stimulating angiogenesis or angioma-genesis? Nat Med. 2000;6:1102–1103. doi: 10.1038/80430. [DOI] [PubMed] [Google Scholar]
- 80.Henry TD, Annex BH, McKendall GR, Azrin MA, Lopez JJ, Giordano FJ, Shah PK, Willerson JT, Benza RL, Berman DS, et al. The VIVA trial: Vascular endothelial growth factor in Ischemia for Vascular Angiogenesis. Circulation. 2003;107:1359–1365. doi: 10.1161/01.cir.0000061911.47710.8a. [DOI] [PubMed] [Google Scholar]
- 81.Arany Z, Wagner BK, Ma Y, Chinsomboon J, Laznik D, Spiegelman BM. Gene expression-based screening identifies microtubule inhibitors as inducers of PGC-1alpha and oxidative phosphorylation. Proc Natl Acad Sci U S A. 2008;105:4721–4726. doi: 10.1073/pnas.0800979105. [DOI] [PMC free article] [PubMed] [Google Scholar]