Abstract
Dendritic growth is pivotal in the neurogenesis of cortical neurons. The sodium pump, or Na,K-ATPase, is an evolutionarily conserved protein that, in addition to its central role in establishing the electrochemical gradient, has recently been reported to function as a receptor and signaling mediator. Although a large body of evidence points toward a dual function for the Na,K-ATPase, few biological implications of this signaling pathway have been described. Here we report that Na,K-ATPase signal transduction triggers dendritic growth as well as a transcriptional program dependent on cAMP response element binding protein (CREB) and cAMP response element (CRE)-mediated gene expression, primarily regulated via Ca2+/calmodulin-dependent protein (CaM) kinases. The signaling cascade mediating dendritic arbor growth also involves intracellular Ca2+ oscillations and sustained phosphorylation of mitogen-activated protein (MAP) kinases. Thus, our results suggest a novel role for the Na,K-ATPase as a modulator of dendritic growth in developing neurons.
Keywords: Calcium signaling; CREB; Dendritic growth; Na,K-ATPase signaling; Neuronal development
Dendrites characterize neuronal cells from other cell types and allow neurons to form networks that are responsible for different functions in the brain. The growth of dendrites is a highly dynamic process that involves a series of orchestrated signaling events that take place during neurogenesis (1–3). In the developing cortex, several phenomena and signaling mechanisms have been demonstrated, including layer-specific dendritic growth and branching activated by neurotrophins (4), restricted dendritic growth via Notch1 signaling (5), and dendritic development via intracellular calcium (Ca2+) signaling (6).
The sodium pump, or Na,K-ATPase, belongs to a family of evolutionarily ancient enzymes that catalyze active transport of cations through hydrolysis of adenosine triphosphate (ATP) across the cell membrane in all mammalian cells (7). In the central nervous system, mutations in Na,K-ATPase are known to cause neuronal dysfunction and neurodegeneration in Drosophila (8), familial hemiplegic migraine in humans (9), and rapid-onset dystonia-Parkinsonism (10). In addition to maintaining the electrochemical gradient, the Na,K-ATPase has been shown to act as a receptor and signal transducer. Previous reports indicate that Na,K-ATPase signal transduction stimulates proliferation in smooth muscle cells (11) and apoptosis in prostate cancer cells (12). Ouabain, an endogenously synthesized hormone (13, 14), is Na,K-ATPase's ligand and forms a hormone-receptor complex capable of inducing mitogen-activated protein (MAP) kinase phosphorylation (15, 16) and intracellular Ca2+ signaling (17–19).
In developing neurons, Ca2+ signaling has been shown to control proliferation, differentiation, and dendritic growth (6, 20–22) by acting on MAP kinases (23, 24) and Ca2+/calmodulin-dependent protein kinases (CaM kinases) (25, 26). Both of these kinases are known to control the transcription factor cAMP response element binding protein (CREB), which regulates cell proliferation and differentiation in a range of cell types in developing vertebrates (27–29). Because the CREB-activating pathways can also be triggered by Na,K-ATPase signaling, we investigated whether the Na,K-ATPase is involved in neuronal differentiation.
We report here that signal transduction activated by the Na,K-ATPase indeed modulates dendritogenesis in developing cortical neurons. This biological process involves a transcriptional program dependent on CREB-mediated and cAMP response element (CRE)-mediated gene expression via CaM kinases as well as MAP kinase phosphorylation and intracellular Ca2+ signaling. These data provide evidence for a novel signaling pathway by which the Na,K-ATPase stimulate dendritic growth in the developing brain and may thus play an important role in circuits wiring.
Results
Na,K-ATPase Receptor Activation Triggers Dendritic Growth in Cortical Neurons.
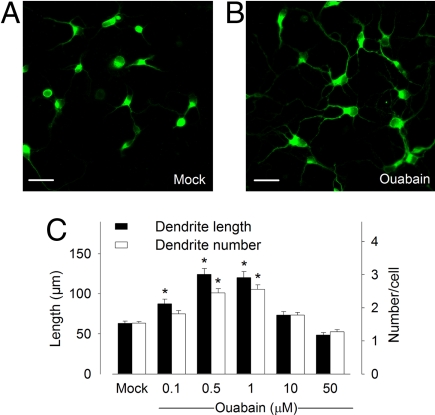
The effect of Na,K-ATPase signal transduction on dendritic growth was studied in cortical neurons prepared from embryonic day (E) 18 rats. To visualize dendritic morphology of individual cells we immunolabeled cortical neurons with a MAP2 antibody at E18 + 4 days in vitro (DIV). This approach reveals the detailed dendritic morphology of cortical neurons in primary culture and can be applied to study the role of diverse signaling pathways in regulating dendritic growth (30). Neurons exposed to the steroid hormone ouabain (1 μmol/l) for 48 hours showed distinctive morphological changes observed as significantly enhanced dendritic growth (Figs. 1A and 1B). The effect of ouabain on Na,K-ATPase varies in a dose-dependent manner (31, 32); therefore, cortical neurons were exposed to graded concentrations of ouabain, and dendritic arbor growth was examined. As shown in Fig. 1C, both the length and number of dendrites significantly increased when cells were treated with ouabain concentrations of 0.5 μmol/l and 1 μmol/l. The dendritic lengths were extended almost 2-fold and the number of dendrites increased by ≈100% after 1 μmol/l ouabain for 48 hours compared with mock treatment. In contrast, ouabain concentrations of 10 μmol/l or higher did not enhance dendritic growth in cortical neurons (Fig. 1C). Consequently, the optimal concentration for dendritogenesis, 1 μmol/l ouabain, was used for all subsequent experiments.
Fig. 1.
Na,K-ATPase signal transduction triggers dendritic arbor growth in cortical neurons. (A and B) Confocal images of E18 cortical neurons in culture at 4 DIV immunolabeled with anti-MAP2 antibody treated with mock (A) versus 1 μmol/l ouabain (B) for 48 hours. Scale bars, 40 μm. (C) Quantification of length and arborization of MAP2-positive dendrites treated with various concentrations of ouabain, as indicated in the figure. Results are pooled from 10 randomly selected fields of view times three cultures. * P < 0.05 versus mock.
Na,K-ATPase Signal Transduction Occurs Without Changing the Resting Membrane Potential.
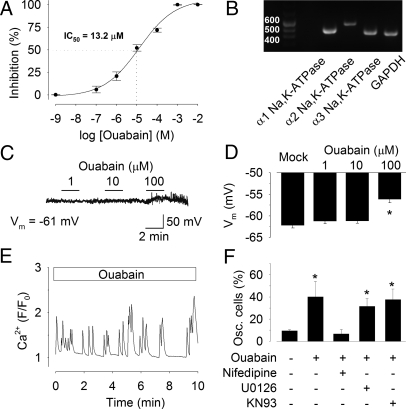
Ouabain is a well-established catalytic inhibitor of the Na,K-ATPase. To determine the extent to which ouabain inhibits Na,K-ATPase activity in cortical neurons, we carried out 86Rb+-flux experiments. Uptake of 86Rb+ is correlated with K+ uptake and thus reflects the turnover rate of the pump. The results show that cortical neuronal Na,K-ATPase had an IC50 of 13.2 μmol/l (95% confidence interval) for ouabain and that the optimal concentration for dendritic growth, 1 μmol/l ouabain, inhibited active 86Rb+ uptake only by 20.7 ± 4.8% (n = 7) (Fig. 2A). Previous studies have shown that Na,K-ATPase isoforms have different affinities for ouabain (31) as well as cell-type-specific and developmental-specific expression patterns (33). The α3- and α2-isoforms of Na,K-ATPase have a higher affinity to ouabain as compared with the rather insensitive α1-isoform (31, 32). Thus, we determined the identity of the α-subunit isoforms expressed in rat cortical neurons in primary culture by reverse transcription-polymerase chain reaction (RT-PCR) using sequence-specific primers. mRNA (cDNA) from three Na,K-ATPase α-subunit isoforms, including the neuron-specific α3-isoform, were detected in rat cortical neurons (Fig. 2B).
Fig. 2.
Na,K-ATPase signaling occurs without changing the resting membrane potential. (A) Effect of ouabain on active 86Rb+ transport by Na,K-ATPase. Cortical neurons at E18 + 4 DIV were incubated for 30 minutes with indicated concentrations of ouabain. The IC50 for the reduction in 86Rb+ transport was 13.2 μmol/l ouabain (n = 7). (B) Expression of α-subunit Na,K-ATPase isoform-specific mRNA (cDNA) in E18 cortical neurons at 4 DIV. Amplification of the GAPDH-specific signal was performed as control. (C) Whole-cell patch-clamp recording of a cortical neuron at E18 + 4 DIV stimulated with 1, 10, or 100 μmol/l ouabain (resting membrane potential, −61 mV). (D) Quantitative data of the membrane potential in cortical neurons at E18 + 4 DIV stimulated with 1, 10, or 100 μmol/l ouabain. * P < 0.05 versus mock. (E) Representative single-cell Ca2+ recording of a Fluo-3/AM-loaded E18 cortical neuron in culture at 4 DIV treated with ouabain 1 μmol/l, 6 hours before initiating measurements. Ratio F/F0 represents fluorescence intensity over baseline. (F) Quantification of the percentage of Na,K-ATPase signal-transduced Ca2+ oscillating cells after treatment with the inhibitors nifedipine, U0126, and KN93, respectively. *P < 0.05 versus mock.
To rule out an effect of ouabain on the resting membrane potential, whole-cell patch-clamp recordings were made from cortical neurons. Ouabain at a concentration of 1 μmol/l or 10 μmol/l had no significant effect on the resting membrane potential of cortical neurons (Figs. 2C and 2D). At a higher concentration of 100 μmol/l, at which enzyme activity is inhibited by 72.2 ± 2.2% (n = 7), ouabain depolarized the membrane potential from −62.2 ± 0.7 mV to −56.1 ± 0.8 mV (n = 12) (Figs. 2C and 2D). These results demonstrate that ouabain does not alter the resting membrane potential at concentrations that promote dendritogenesis.
Calcium signaling exerts a profound influence on dendritic growth during development (6). We therefore examined the effect of Na,K-ATPase signal transduction on Ca2+ signaling in cortical neurons. Intracellular Ca2+ dynamics were imaged in neuronal cells loaded with the Ca2+-sensitive fluorescent dye Fluo-3/AM. When cortical neurons were exposed to 1 μmol/l ouabain for 6 hours, the treatment that elicited a robust effect on dendritic growth, slow intracellular Ca2+ oscillations were induced (Fig. 2E). Intracellular Ca2+ signaling was also observed in untreated cells, 9.5 ± 0.9% (n = 147) but was significantly increased in neurons treated with ouabain, 40.1 ± 13.8% (n = 129) (Fig. 2F). These Ca2+ oscillations were strongly suppressed by the L-type Ca2+ channel blocker nifedipine, 6.7 ± 4.0% (n = 140) (Fig. 2F and [supporting information (SI) Fig. S1A]). Neither inhibition of MAP kinases by U0126 nor CaM kinases by KN93 abolished ouabain-induced Ca2+ oscillations in cortical neurons (below and Fig. 2F).
MAP Kinase Phosphorylation Is Stimulated by Na,K-ATPase Signaling.
Activation of MAP kinases plays an important role, largely through regulation of gene transcription, in a number of cellular processes including cell proliferation, DNA synthesis, differentiation, and survival (23, 34, 35). In neurons, MAP kinases have been shown to regulate dendritic growth and morphology (24, 36). To assess whether Na,K-ATPase signal transduction activates p42/44 MAP kinase (referred to as ERK1/2) in cortical neurons, we exposed neurons to ouabain and monitored ERK1/2 phosphorylation using Western blotting. When cells were examined after 10 minutes of ouabain treatment no ERK1/2 phosphorylation was observed (data not shown). Cortical neurons exposed to ouabain for 4 hours and 6 hours, however, showed a sustained and significant ERK1/2 phosphorylation, which was abolished using the selective MEK inhibitor U0126 (Fig. 3A). Sustained activation of MAP kinases has previously been associated with physiological phenomena such as gene transcription and dendritic growth (34, 37). We then tested whether MEK and MAP kinases had any effect on the Ca2+ oscillatory response evoked by treating cells with ouabain for 6 hours. U0126 did not block ouabain-induced Ca2+ oscillations (Fig. S1B) and had no significant effect on the number of responding cells, 31.6 ± 6.9% (n = 118), when compared with ouabain treatment alone (Fig. 2F). To determine the effect of Ca2+ signaling induced by Na,K-ATPase signal transduction on ERK1/2 activation, we next examined cells preincubated with the Ca2+ signaling inhibitor nifedipine. Cortical neurons exhibited a reduced level of Na,K-ATPase signal transduced ERK1/2 phosphorylation after nifedipine pretreatment (Fig. 3B).
Fig. 3.
MAP kinase activation is triggered by Na,K-ATPase signal transduction. (A-C) Western blot of Na,K-ATPase signal-transduced phosphorylated 42/44 MAP kinase (pERK1/2) in cortical neurons at E18 + 4 DIV treated with 1 μmol/l ouabain for 6 hours plus MAP kinase inhibitor U0126 (A), L-type Ca2+ channel blocker nifedipine (B), and CaM kinase inhibitor KN93 (C). Each bar represents the fold increase of phospho-ERK1/2 relative to mock after normalizing against total nonphosphorylated ERK1/2. *P < 0.05 versus mock; †P < 0.05 versus ouabain-treated cells.
The family of CaM kinases is a principal downstream target of Ca2+ signaling that is known to regulate neuronal differentiation and dendritic growth (25, 26, 29). To test whether these kinases were involved in mediating Na,K-ATPase signal transduction in cortical neurons, cells were stimulated with ouabain in the absence or presence of the general CaM kinase inhibitor KN93. After this treatment, phosphorylation of ERK1/2 was partially, yet not significantly (P = 0.3, one-way analysis of variance [ANOVA]), inhibited in cortical neurons (Fig. 3C). Next we investigated whether CaM kinases were involved in the ouabain-induced Ca2+ oscillations. Pretreatment with KN93 did not abolish Na,K-ATPase signal transduced Ca2+ oscillations, 37.7 ± 9.6% (n = 134) (Fig. 2F and Fig. S1C). These results indicate that activation of MAP kinases by Na,K-ATPase signal transduction involves the synergistic actions of MEK, Ca2+ oscillations, and CaM kinases.
Na,K-ATPase Signal Transduction Triggers CREB Transcription Through CaM Kinases.
The transcription factor CREB is implicated in neuronal differentiation (26, 27), where it is strongly regulated via CaM kinases (28, 38). To determine the effect of Na,K-ATPase signal transduction on CREB activation we first carried out immunocytochemistry experiments using an antibody specific for Ser-133-phosphorylated CREB. After ouabain treatment for 6 hours, immunostaining revealed a marked increase of phosphorylated CREB within the nucleus (Fig. 4A). The nuclear localization of CREB was confirmed by co-labeling cortical neurons with the nuclear stain TO-PRO3 (Fig. 4A). The role of Na,K-ATPase signal transduction on CREB activation was further investigated with Western blot experiments using the Ser-133 phospho-CREB antibody. These experiments showed that CREB was strongly Ser-133 phosphorylated by Na,K-ATPase signal transduction (Fig. 4B).
Fig. 4.
CREB activation induced by Na,K-ATPase signal transduction is mediated via CaM kinases. (A) Confocal images of phospho-Ser-133 CREB-immunostained (green) E18 cortical neurons in culture at 5 DIV treated with mock (left panel) and with ouabain 1 μmol/l (right panel) for 6 hours. Nuclear staining with TO-PRO3 (blue). (B) Levels of CREB phosphorylation as detected by phospho-Ser-133 CREB Western blot of cortical neurons at E18 + 5 DIV treated with 1 μmol/l ouabain for 6 hours, preincubated with MAP kinase inhibitor U0126, CaM kinase inhibitor KN93, and L-type Ca2+ channel blocker nifedipine (Nif), respectively. (C) CREB-mediated transcription, measured using a CRE-luciferase reporter gene transfected into E18 cortical neurons in culture at 3 DIV, of Na,K-ATPase signal transduction in cortical neurons at E18 + 5 DIV treated with mock versus 1 μmol/l ouabain for 6 hours, preincubated with MAP kinase inhibitor U0126, CaM kinase inhibitor KN93, and L-type Ca2+ channel blocker nifedipine (Nif), respectively. Each bar represents the fold increase relative to mock after normalizing against Renilla. *P < 0.05 versus mock.
Next we examined which signaling cascades were involved in the Na,K-ATPase signal-transduced CREB Ser-133 phosphorylation. Inhibiting MAP kinases using U0126 reduced the immuno signal detected by the Ser-133 phospho-CREB antibody (Fig. 4B). CREB Ser-133 phosphorylation was also downregulated by the CaM kinase blocker KN93 (Fig. 4B). CaM kinases are activated by Ca2+ signaling; consequently, when Ca2+ oscillations were blocked with nifedipine, a reduction in the CREB Ser-133 phosphorylation was observed (Fig. 4B). This set of experiments suggests that each of these pathways is partially involved in CREB Ser-133 phosphorylation via Na,K-ATPase signal transduction. Although phosphorylation of CREB at Ser-133 is required for inducing CREB-dependent transcription, there are instances in which Ser-133 phosphorylation is not sufficient for target gene activation (28). To further investigate CREB activation induced by Na,K-ATPase signal transduction, cortical neurons cells were transfected with a CRE-luciferase reporter gene that is activated by endogenous CREB. A significant 3-fold increase in CRE-luciferase activity was observed when cells were exposed to ouabain for 6 hours (Fig. 4C). Whereas U0126 and nifedipine resulted in only a marginal reduction in ouabain-induced CRE-luciferase activity, KN93 completely abolished the CREB activation triggered by Na,K-ATPase signal transduction (Fig. 4C). Taken together these results suggest that CREB activation in cortical neurons triggered by Na,K-ATPase signal transduction predominantly occurs through a CaM kinase-dependent mechanism.
Dendritic Growth Induced by Na,K-ATPase Signaling Is Orchestrated by MAP Kinases, CaM Kinases, and Extracellular Ca2+ Influx.
Finally, we examined the impact of Ca2+ signaling, MAP kinases, and CaM kinases on Na,K-ATPase signal-transduced dendritic growth in cortical neurons. A partial decrease in ouabain-triggered dendritic length and arborization was observed in response to nifedipine treatment, suggesting that L-type channel-mediated Ca2+ influx plays a role in dendritic growth (Fig. 5B). Inhibition of MAP kinase signaling with U0126 significantly attenuated dendritogenesis in ouabain-treated cells (Fig. 5C). Although nifedipine and U0126 both significantly decreased ouabain-induced neurite outgrowth, neither inhibitor was able to completely abrogate the Na,K-ATPase signal-transduced effects on dendritic growth and arborization in cortical neurons. Inhibition of CaM kinases with KN93, however, resulted in complete inhibition of ouabain-induced dendritic branching (Fig. 5D). These observations support a model in which Na,K-ATPase signal transduction triggers dendritic growth via a signaling pathway that involves extracellular Ca2+ influx as well as MAP kinase and CaM kinase-modulated CREB transcription.
Fig. 5.
Dendritic growth in cortical neurons induced by Na,K-ATPase signal transduction involves crosstalk between Ca2+ signaling, MAP kinases, and CaM kinases. (A-D) Confocal images of E18 cortical neurons at 4 DIV immunolabeled with anti-MAP2 antibody treated with 1 μmol/l ouabain (A) together with L-type Ca2+ channel inhibitor nifedipine (Nif) (B), MAP kinase inhibitor U0126 (C), and CaM kinase inhibitor KN93 (D) for 48 hours. Scale bars, 40 μm. (E) Quantification of length and arborization of MAP2-positive dendrites treated with 1 μmol/l ouabain together with nifedipine, U0126, and KN93, respectively. Results are pooled from 10 randomly selected fields of view times three cultures. *P < 0.05 versus mock; †P < 0.05 versus ouabain-treated cells.
Discussion
The findings presented in this report indicate that Na,K-ATPase signal transduction induces dendritogenesis in cortical neurons via activation of a transcriptional program that involves CREB- and CRE-mediated gene expression, primarily through a CaM kinase-dependent signaling pathway. This signaling event is induced by exposing cells to a concentration of ouabain that does not affect the resting membrane potential. Persistent Ca2+ oscillations and sustained MAP kinase phosphorylation also participate in this signaling cascade, as blocking L-type Ca2+ channels or MEK suppresses downstream effects.
Ouabain is an endogenous steroid hormone that is present in mammalian tissues; it has been isolated from hypothalamus (39), adrenals (40), and human plasma (41). Evidence indicates that circulating levels of ouabain and ouabain-like factors are elevated during pregnancy and in newborn infants (13, 32), thereby suggesting a developmental role for this signaling molecule. Our results, which show that Na,K-ATPase, the receptor of ouabain, stimulates dendritic growth in rat cortical neurons, support such a biological function for ouabain. Concentrations of endogenous ouabain in the embryonic human brain during the course of development are unknown but are predicted to lie in the subnanomolar-to-nanomolar range, as previously reported (13, 32). Indeed, nanomolar concentrations of ouabain stimulates neurite outgrowth in human neuroblastoma cells (Fig. S2). Furthermore, in the brain, there are multiple Na,K-ATPase isoforms that each have cell-type-specific and developmental-specific expression patterns (33, 42) as well as different ouabain affinities (31, 42). These spatial and temporal expression patterns of Na,K-ATPase isoform remain largely unknown but may be involved in the development and wiring of the brain. As such, the extent to which the different α-subunit isoforms contribute to the overall cellular effects of ouabain remain to be elucidated.
The mechanism by which Na,K-ATPase triggers signal transduction remains disputed, with two principal hypotheses being proposed. One mechanism suggests that partial inhibition of the Na,K-ATPase leads to an increased intracellular Na+ concentration that transactivates Na+/Ca2+-exchangers, which thereby produce cytosolic Ca2+ waves that are sufficient to elicit downstream signaling events (18, 43). The other mechanism suggests that inhibition of Na,K-ATPase pumping activity is not in itself required for a signaling event to occur; instead, Na,K-ATPase is proposed to function via a microdomain where ouabain-induced signaling occurs via physical association of Na,K-ATPase with a host of binding partners. Na,K-ATPase has been reported to associate with the inositol 1,4,5-trisphosphate receptor to generate intracellular Ca2+ oscillations (44) and to form a complex with the epidermal growth factor receptor, leading to MAP kinase activation (16). Dendritic growth via Na,K-ATPase signal transduction might be modulated by either or both of these signaling pathways.
Using electrophysiological recordings it was shown that 1 μmol/l ouabain, the optimal concentration for inducing dendritogenesis in cortical neurons, apparently does not change the resting membrane potential. Ouabain at 100 μmol/l, a concentration that did not induce dendritic growth, however, did depolarize the membrane potential. Thus, ouabain-induced depolarization alone does not appear to be sufficient to trigger dendritic growth in cortical neurons. These data, together with the 86Rb+-flux experiments, suggest that inhibition of Na,K-ATPase pumping activity per se is not the mechanism responsible for triggering this biological process. The observation that Na,K-ATPase signal transduction is not a fast signaling event is further strengthened by the experiments on Ca2+ signaling and MAP kinases, which both show delayed responses. Persistent Ca2+ oscillations and sustained ERK1/2 phosphorylation were detected after 6 hours of pretreatment with 1 μmol/l ouabain. Delayed and sustained phosphorylation of MAP kinases has previously been implicated in neuronal differentiation (45). These observations indicate that the signaling mechanism by which dendritic growth is induced in cortical neurons does not depend on changes in the resting membrane potential but, rather, involves a slow process that takes hours to activate.
Analysis of Ser-133 phosphorylation of CREB- and CRE-mediated gene expression in cortical neurons treated with ouabain gave rise to somewhat disparate results. This might be caused by the many phosphorylation sites on the CREB protein that differentially regulate total CREB activity (27, 28, 46). For example, phosphorylation at Ser-133 increases CREB activity, whereas Ser-142 phosphorylation inhibits CREB (38). Therefore we cannot directly correlate Ser-133 phosphorylation of CREB with CRE-dependent transcription. Phosphorylated CREB is also known to bind the Ca2+-dependent CREB binding protein (CBP) to form a molecular complex that determines CRE transcriptional activity (47). The differential ability of CaM kinases and MAP kinases to activate CBP (48) may be the molecular distinction that underlies the distinct biological effects of these kinases. This difference may explain why inhibiting MAP kinases decreased Ser-133 phosphorylation of CREB but failed to block CRE activation induced by Na,K-ATPase signaling. Furthermore, CREB activation was previously demonstrated to occur via an interplay between MAP kinases and CaM kinases, in which MAP kinases were involved in a more sustained phosphorylation of CREB whereas the fast onset of CREB phosphorylation was triggered by CaM kinases (34, 46, 49). Taken together, our results suggest that Na,K-ATPase receptor activation leads to CREB activation and dendritic growth via crosstalk between the MAP kinase and CaM kinase pathways.
Our findings suggest that Na,K-ATPase signal transduction stimulates dendritic growth in cortical neurons via activation of a transcriptional program that involves CREB- and CRE-mediated gene expression. The signaling pathway is primarily regulated via CaM kinases, but crosstalk with MAP kinase phosphorylation and Ca2+ oscillations also participates in this event. This type of mechanism may provide a novel basis for neuronal differentiation through Na,K-ATPase signal transduction and induction of a specific program of gene expression that is critical for the developing nervous system.
Materials and Methods
Cell Cultures.
Cerebral cortical neurons in primary culture were prepared from Sprague-Dawley rat fetuses at embryonic day (E)18. The cerebral cortices were dissected into Hank's balanced salt solution (HBSS). HBSS was removed and tissues were treated with 0.25% trypsin at 37 °C for 15 minutes. Tissues were then treated with 0.1% DNase I at 37 °C for 5 minutes. Cells were diluted to 106 cells/ml in Dulbecco's modified Eagle's medium (DMEM; Sigma) containing 10% fetal bovine serum (FBS), 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mmol/l l-glutamine, and were seeded on plates precoated with 40 μg/ml poly-l-lysine. After 3 hours, DMEM was replaced with neurobasal medium (GIBCO) containing B27 Supplement (GIBCO), 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mmol/l l-glutamine. Half of the cell culture medium was replaced after 3 DIV with neurobasal medium containing the supplements specified above and aracytine 10−7 mol/l.
Reagents.
Reagents and concentrations, unless otherwise stated, were as follows,: ouabain (1, 10, 100 μmol/l, Sigma), nifedipine (50 μmol/l, Sigma), KN93 (20 μmol/l, Tocris), and U0126 (20 μmol/l, Cell Signaling).
86Rb+ Uptake Assay.
Cortical neurons at E18 + 4 DIV were rinsed with phosphate buffered saline solution (PBS) and incubated with PBS containing indicated ouabain concentrations for 30 minutes at 37 °C. To each well 86Rb+ was added at ≈1.5 μCi/ml, and incubation was continued for another 10 minutes in 37 °C. Uptake was thereafter inhibited by 2 mmol/l ouabain, and this value was taken as the maximal rate of active uptake. The incubation was stopped by rinsing the plate four times with PBS plus BaCl2 (5 mmol/l). Cells were extracted with 0.3 ml of 1 mol/l NaOH for 10 minutes, and samples were counted in a scintillation counter. Each data point represents the average of the radioactivity present in four separate wells.
RT-PCR.
Total RNA was extracted from cultured rat embryonic cortical neurons at E18 + 4 DIV using TRIzol reagent (Invitrogen). Single-strand cDNA was synthesized from 1 μg total RNA by random primers and SSII-RT (Invitrogen) and the cDNA was then amplified by RT-PCR with TaqDNA polymerase (Invitrogen). The PCR reactions were performed as follows: 94 °C for 30 seconds, 60 °C for 30 seconds, and 72 °C for 60 seconds, repeated for 45 cycles. The PCR products were separated on a 1.5% agarose gel, and ethidium bromide was used for visualization.
Calcium Imaging.
Calcium experiments on cortical neurons at E18 + 4 DIV were carried out in artificial cerebrospinal fluid containing the following: 125 mmol/l NaCl, 25 mmol/l NaHCO3, 1.25 mmol/l NaH2PO4, 5 mmol/l KCl, 2 mmol/l CaCl2, 1 mmol/l MgSO4, and 20 mmol/l d-glucose bubbled with 95% O2 and 5% CO2 to reach a final pH of 7.3. Cells were incubated for 15–30 minutes at 37 °C with 5% CO2 in 5 μmol/l Fluo-3/AM (Molecular Probes, Invitrogen). Coverslips were mounted in a temperature controlled (37 °C) chamber (Warner Instruments) and clamped onto a Zeiss Axiovert 100M microscope, equipped with a C-Apochromat 40X/1.2NA water immersion objective (Zeiss), connected to a Lambda LS xenon-arc lamp (Sutter), Lambda 10–3 filter-wheel (Sutter), and a smartShutter (Sutter). Images were acquired at 0.2 Hz with an EMCCD camera Cascade II:512 (Photometrics) controlled by the acquisition software MetaFluor (Molecular Devices). Perfusion (1 ml/min) with 95% O2-5% CO2 bubbled artificial cerebrospinal fluid was used for Ca2+ recordings on cortical neurons.
Electrophysiology.
Cortical neurons in culture at E18 + 4 DIV were recorded in the whole-cell configuration using a patch-clamp amplifier (AxoPatch 200A, Axon Instruments). Neurons had an average resting membrane potential of −62.2 ± 0.7 mV and were continuously perfused with extracellular solution containing the following: 100 mmol/l NaCl, 2 mmol/l KCl, 1.2 mmol/l MgCl2, 2 mmol/l CaCl2, 10 mmol/l glucose, and 10 mmol/l HEPES, with pH adjusted to 7.6 using NaOH, and 223 mOsm. The pipettes were filled with a solution containing: 90 mmol/l K-gluconate, 5 mmol/l KCl, 0.1 μmol/l CaCl2, 10 mmol/l HEPES, 4 mmol/l Mg2ATP, 0.3 mmol/l Na4GTP, and 5 mmol/l phosphocreatine Na2 with pH adjusted to 7.4 using KOH, and 217 mOsm. Solutions and drugs were applied through a gravity-driven microperfusion system, with the tip placed close to the recorded cell.
Western Blot and Immunocytochemistry.
Cortical neurons at E18 + 4 DIV were exposed to various treatments. The cells were lysed using modified RIPA buffer for 20 min at 4 °C. Protein concentration was determined using a BCA protein assay (Pierce) and equal amounts of cellular protein (≈10–20 μg) were separated on a 10% sodium dodecyl sulfate gel electrophoresis, followed by a transfer to a nitrocellulose membrane. Membranes were blocked in 5% skim milk in Tris-buffered saline solution plus 0.5% Tween-20 for 1 hour before incubation with primary antibodies (Ab) (1:1000) (ERK1/2, Phospho-ERK1/2, Ser-133 CREB, and Phospho-Ser-133 CREB, all from Cell Signaling) overnight at 4 °C and further incubation with horseradish peroxidase-conjugated secondary Ab (1:5000) (Amersham) for 1 hour. Immunoreactive bands were visualized using an enhanced chemiluminescence kit (Amersham).
Immunocytochemical staining of phospho-Ser-133 CREB and MAP2 was performed according to standard protocol, using fixation by 4% paraformaldehyde in 1 hour. After blocking with 1% bovine serum albumen, cells were incubated with phospho-Ser-133 CREB Ab (1:100, Cell Signaling) or MAP2 Ab (1:400, Chemicon) overnight and then with Alexa 488 secondary Ab (1:2000, 1:200; Molecular Probes) for 1 hour, together with 0.3% Triton X-100. Nucleus was stained with TO-PRO-3 (1:200; Molecular Probes) for 10 minutes. Slides were mounted using the Prolong Antifade Kit (Molecular Probes) and scanned in a Zeiss LSM510 confocal microscope equipped with a C-Apochromat 40X/1.2 water immersion objective (Zeiss).
Luciferase Assay.
For CREB activity assay, cortical neurons were transfected at E18 + 3 DIV with pCRE-luciferase reporter (Stratagene) and pRL-TK Renilla reporter (Promega) using Lipofectamine 2000 (Invitrogen). After 48 hours, cells were treated with KN93, U0126, or nifedipine for 15 minutes followed by ouabain for 6 hours. Thereafter cells were harvested, and luciferase activity was measured using the dual-luciferase reporter assay system (Promega).
Data Analysis.
Dendritic growth was analyzed from 10 fields of view per cell culture using ImageJ (National Institutes of Health, Bethesda, MD) together with the plug-in NeuronJ. For each field, the average dendritic length and/or the number of dendrites per cell was calculated. When recording, intracellular Ca2+, cells were considered oscillating when at least three Ca2+ transients were observed exceeding 50% of the baseline. Data are presented as mean ± SE of a minimum of three experiments unless otherwise indicated. Statistical significance was accepted at P < 0.05 as determined by analysis of variance, followed by Bonferroni's post hoc test for multiple comparisons.
Supplementary Material
Acknowledgments.
This study was supported by the Swedish Research Council (Dnr 2005–6682 and DBRM), the Foundation for Strategic Research (CEDB), Knut och Alice Wallenbergs Stiftelse (CLICK), Åke Wibergs Stiftelse, Jeanssons Stiftelser, Magnus Bergvalls Stiftelse, Fredrik och Ingrid Thurings Stiftelse, and the Swedish Society for Medical Research. The authors thank Drs. Anita Aperia, Ruani Fernando, Ola Hermanson, Carlos Ibáñez, and the staff of the Molecular Neurobiology Unit, Department of Medical Biochemistry and Biophysics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809253106/DCSupplemental.
References
- 1.Cline HT. Dendritic arbor development and synaptogenesis. Curr Opin Neurobiol. 2001;11:118–126. doi: 10.1016/s0959-4388(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 2.Jan YN, Jan LY. The control of dendrite development. Neuron. 2003;40:229–242. doi: 10.1016/s0896-6273(03)00631-7. [DOI] [PubMed] [Google Scholar]
- 3.Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- 4.McAllister AK, Katz LC, Lo DC. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 5.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 6.Konur S, Ghosh A. Calcium signaling and the control of dendritic development. Neuron. 2005;46:401–405. doi: 10.1016/j.neuron.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan JH. Biochemistry of Na,K-ATPase. Annu Rev Biochem. 2002;71:511–535. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- 8.Palladino MJ, Bower JE, Kreber R, Ganetzky B. Neural dysfunction and neurodegeneration in Drosophila Na+/K+ ATPase alpha subunit mutants. J Neurosci. 2003;23:1276–1286. doi: 10.1523/JNEUROSCI.23-04-01276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Fusco M, et al. Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump alpha2 subunit associated with familial hemiplegic migraine type 2. Nat Genet. 2003;33:192–196. doi: 10.1038/ng1081. [DOI] [PubMed] [Google Scholar]
- 10.de Carvalho Aguiar P, et al. Mutations in the Na+/K+-ATPase alpha3 gene ATP1A3 are associated with rapid-onset dystonia Parkinsonism. Neuron. 2004;43:169–175. doi: 10.1016/j.neuron.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 11.Abramowitz J, et al. Ouabain- and marinobufagenin-induced proliferation of human umbilical vein smooth muscle cells and a rat vascular smooth muscle cell line, A7r5. Circulation. 2003;108:3048–3053. doi: 10.1161/01.CIR.0000101919.00548.86. [DOI] [PubMed] [Google Scholar]
- 12.McConkey DJ, Lin Y, Nutt LK, Ozel HZ, Newman RA. Cardiac glycosides stimulate Ca2+ increases and apoptosis in androgen-independent, metastatic human prostate adenocarcinoma cells. Cancer Res. 2000;60:3807–3812. [PubMed] [Google Scholar]
- 13.Bagrov AY, Shapiro JI. Endogenous digitalis: Pathophysiologic roles and therapeutic applications. Nat Clin Pract Nephrol. 2008;4:378–392. doi: 10.1038/ncpneph0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheiner-Bobis G, Schoner W. A fresh facet for ouabain action. Nat Med. 2001;7:1288–1289. doi: 10.1038/nm1201-1288. [DOI] [PubMed] [Google Scholar]
- 15.Dmitrieva RI, Doris PA. Ouabain is a potent promoter of growth and activator of ERK1/2 in ouabain-resistant rat renal epithelial cells. J Biol Chem. 2003;278:28160–28166. doi: 10.1074/jbc.M303768200. [DOI] [PubMed] [Google Scholar]
- 16.Haas M, Askari A, Xie Z. Involvement of Src and epidermal growth factor receptor in the signal-transducing function of Na+/K+-ATPase. J Biol Chem. 2000;275:27832–27837. doi: 10.1074/jbc.M002951200. [DOI] [PubMed] [Google Scholar]
- 17.Aizman O, Uhlen P, Lal M, Brismar H, Aperia A. Ouabain, a steroid hormone that signals with slow calcium oscillations. Proc Natl Acad Sci USA. 2001;98:13420–13424. doi: 10.1073/pnas.221315298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwamoto T, et al. Salt-sensitive hypertension is triggered by Ca2+ entry via Na+/Ca2+ exchanger type-1 in vascular smooth muscle. Nat Med. 2004;10:1193–1199. doi: 10.1038/nm1118. [DOI] [PubMed] [Google Scholar]
- 19.Yuan Z, et al. Na/K-ATPase tethers phospholipase C and IP3 receptor into a calcium-regulatory complex. Mol Biol Cell. 2005;16:4034–4045. doi: 10.1091/mbc.E05-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spitzer NC, Root CM, Borodinsky LN. Orchestrating neuronal differentiation: patterns of Ca2+ spikes specify transmitter choice. Trends Neurosci. 2004;27:415–421. doi: 10.1016/j.tins.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Weissman TA, Riquelme PA, Ivic L, Flint AC, Kriegstein AR. Calcium waves propagate through radial glial cells and modulate proliferation in the developing neocortex. Neuron. 2004;43:647–661. doi: 10.1016/j.neuron.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Zheng JQ, Poo MM. Calcium signaling in neuronal motility. Annu Rev Cell Dev Biol. 2007;23:375–404. doi: 10.1146/annurev.cellbio.23.090506.123221. [DOI] [PubMed] [Google Scholar]
- 23.Bonni A, et al. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 24.Wu GY, Deisseroth K, Tsien RW. Spaced stimuli stabilize MAPK pathway activation and its effects on dendritic morphology. Nat Neurosci. 2001;4:151–158. doi: 10.1038/83976. [DOI] [PubMed] [Google Scholar]
- 25.Fink CC, et al. Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron. 2003;39:283–297. doi: 10.1016/s0896-6273(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 26.Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 27.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 28.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 29.Wayman GA, et al. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. doi: 10.1016/j.neuron.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Vaillant AR, et al. Signaling mechanisms underlying reversible, activity-dependent dendrite formation. Neuron. 2002;34:985–998. doi: 10.1016/s0896-6273(02)00717-1. [DOI] [PubMed] [Google Scholar]
- 31.Kim JH, Sizov I, Dobretsov M, von Gersdorff H. Presynaptic Ca2+ buffers control the strength of a fast post-tetanic hyperpolarization mediated by the alpha3 Na(+)/K(+)-ATPase. Nat Neurosci. 2007;10:196–205. doi: 10.1038/nn1839. [DOI] [PubMed] [Google Scholar]
- 32.Schoner W, Scheiner-Bobis G. Endogenous and exogenous cardiac glycosides: Their roles in hypertension, salt metabolism, and cell growth. Am J Physiol Cell Physiol. 2007;293:509–536. doi: 10.1152/ajpcell.00098.2007. [DOI] [PubMed] [Google Scholar]
- 33.Wetzel RK, Arystarkhova E, Sweadner KJ. Cellular and subcellular specification of Na,K-ATPase alpha and beta isoforms in the postnatal development of mouse retina. J Neurosci. 1999;19:9878–9889. doi: 10.1523/JNEUROSCI.19-22-09878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dolmetsch RE, Pajvani U, Fife K, Spotts JM, Greenberg ME. Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science. 2001;294:333–339. doi: 10.1126/science.1063395. [DOI] [PubMed] [Google Scholar]
- 35.Whitmarsh AJ, Davis RJ. A central control for cell growth. Nature. 2000;403:255–256. doi: 10.1038/35002220. [DOI] [PubMed] [Google Scholar]
- 36.Markus A, Zhong J, Snider WD. Raf and akt mediate distinct aspects of sensory axon growth. Neuron. 2002;35:65–76. doi: 10.1016/s0896-6273(02)00752-3. [DOI] [PubMed] [Google Scholar]
- 37.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 38.Kornhauser JM, et al. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron. 2002;34:221–233. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 39.Kawamura A, et al. On the structure of endogenous ouabain. Proc Natl Acad Sci USA. 1999;96:6654–6659. doi: 10.1073/pnas.96.12.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider R, et al. Bovine adrenals contain, in addition to ouabain, a second inhibitor of the sodium pump. J Biol Chem. 1998;273:784–792. doi: 10.1074/jbc.273.2.784. [DOI] [PubMed] [Google Scholar]
- 41.Hamlyn JM, et al. A circulating inhibitor of (Na+ + K+)ATPase associated with essential hypertension. Nature. 1982;300:650–652. doi: 10.1038/300650a0. [DOI] [PubMed] [Google Scholar]
- 42.Richards KS, Bommert K, Szabo G, Miles R. Differential expression of Na+/K+-ATPase alpha-subunits in mouse hippocampal interneurones and pyramidal cells. J Physiol. 2007;585:491–505. doi: 10.1113/jphysiol.2007.144733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blaustein MP, Lederer WJ. Sodium/calcium exchange: Its physiological implications. Physiol Rev. 1999;79:763–854. doi: 10.1152/physrev.1999.79.3.763. [DOI] [PubMed] [Google Scholar]
- 44.Miyakawa-Naito A, et al. Cell signaling microdomain with Na,K-ATPase and inositol 1,4,5-trisphosphate receptor generates calcium oscillations. J Biol Chem. 2003;278:50355–50361. doi: 10.1074/jbc.M305378200. [DOI] [PubMed] [Google Scholar]
- 45.Kolkova K, Novitskaya V, Pedersen N, Berezin V, Bock E. Neural cell adhesion molecule-stimulated neurite outgrowth depends on activation of protein kinase C and the Ras-mitogen-activated protein kinase pathway. J Neurosci. 2000;20:2238–2246. doi: 10.1523/JNEUROSCI.20-06-02238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu GY, Deisseroth K, Tsien RW. Activity-dependent CREB phosphorylation: convergence of a fast, sensitive calmodulin kinase pathway and a slow, less sensitive mitogen-activated protein kinase pathway. Proc Natl Acad Sci USA. 2001;98:2808–2813. doi: 10.1073/pnas.051634198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu SC, Chrivia J, Ghosh A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- 48.Impey S, et al. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–244. doi: 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 49.Hardingham GE, Arnold FJ, Bading H. Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat Neurosci. 2001;4:261–267. doi: 10.1038/85109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.