Abstract
The tumor suppressor p53 regulates downstream genes in response to many cellular stresses and is frequently mutated in human cancers. Here, we report the use of a crosslinking strategy to trap a tetrameric p53 DNA binding domain (p53DBD) bound to DNA and the X-ray crystal structure of the protein/DNA complex. The structure reveals that two p53DBD dimers bind to B form DNA with no relative twist and that a p53 tetramer can bind to DNA without introducing significant DNA bending. The numerous dimer-dimer interactions involve several strictly conserved residues thus suggesting a molecular basis for p53DBD-DNA binding cooperativity. Surface residue conservation of the p53DBD tetramer bound to DNA highlights possible regions of other p53 domain or p53 cofactor interactions.
The gene encoding the p53 tumor suppressor is commonly mutated in human cancer and its protein product has been extensively studied at both the cellular and molecular level. In response to DNA damage, cell stress and some oncogenic proteins, p53 induces cell cycle arrest or apoptosis(Horn and Vousden, 2007). p53 must retain its ability to oligomerize and bind specific DNA sequences to fulfill its function(Pietenpol et al., 1994). Through many structural studies, residues within the p53 DNA binding domain (p53DBD) that are crucial for domain stability, DNA binding and dimerization have been elucidated and correlated with known cancerous mutations(Cho et al., 1994; Ho et al., 2006; Kitayner et al., 2006). The in vivo form of p53 is a tetramer and while several models of four p53 core domains bound to a consensus sequence have been proposed (Ho et al., 2006; Kitayner et al., 2006; Nagaich et al., 1999; Pan and Nussinov, 2007) and one p53 tetramer bound to a discontinuous DNA duplex has been recently described (Kitayner et al., 2006), a p53 tetramer bound to a continuous cognate DNA site has not been previously reported.
The polypeptide chain of p53 has four distinct domains: a loosely folded N terminal transactivation domain (residues 1 – 44), a DNA binding core domain (residues 99 – 289), a tetramerization domain (residues 317 – 353), and a C terminal regulatory domain (residues 353 – 390)(Jeffrey et al., 1995). The DNA binding domain binds p53 consensus sequences that contains two half sites of the following motif: 5′ – Pu Pu Pu C (A/T) | (T/A) G Py Py Py – 3′, where Pu and Py stand for purine and pyrimidine respectively(El-Diery et al., 1992). The half sites may be separated by 0 to 20 basepairs, with a zero base-pair separation being the most common(El-Diery et al., 1992). The symmetry of the consensus sequence and the structure of the tetramerization domain determined in 1995 (Jeffrey et al., 1995) lead to the hypothesis that p53 is a dimer of dimers.
Recently, the structure of a mouse p53DBD dimer bound to DNA was determined through the use of structure based cross-linking (Ho et al., 2006). Covalent crosslinking has emerged as a useful tool to crystallize otherwise difficult protein-DNA complexes (He and Verdine, 2002; Verdine and Norman, 2003). Structure based crosslinking uses site-directed mutagenesis to introduce a crosslinkable cysteine residue and the convertible nucleoside approach to create a DNA oligomer bearing a disulfide linker for crosslinking with the engineered cysteine residue of the protein (He and Verdine, 2002; MacMillan and Verdine, 1991; Verdine and Norman, 2003). When the protein binds the DNA, the cysteine and linker come close enough to form a disulfide bond, thus trapping the protein/DNA complex in the form of a crosslinked product. This particular strategy, which requires some knowledge of the protein/DNA interface, has previously lead to high-resolution structures and a deeper understanding of HIV-1 reverse transcriptase(Huang et al., 1998) and MutM DNA glycosylase(Banerjee et al., 2006; Banerjee et al., 2005).
The mouse p53DBD dimer work relied on results from Cho et al.’s landmark X-ray crystal structure study of a p53DBD monomer bound to DNA in order to design an appropriate protein-DNA crosslink. Cys277 (human numbering) was reported to specifically contact Cyt18, the first pyrimidine within the provided DNA half site (Cho et al., 1994). Ho et al. modified the cytosine at this position by replacing the N4 amine with a cystamine and crosslinked wild type p53DBD to the new half site. The result of this work was an X-ray structure determination of a dimeric mouse p53DBD dimer bound to DNA and characterization of the core domain dimer interface (Ho et al., 2006). Further support for the ability of p53-DNA crosslinking to provide biologically meaningful results was provided by Shakked and coworkers with their structure of a human p53DBD dimer of dimers bound to a discontinuous DNA duplex, which showed analogous p53DBD dimer interactions on DNA (Kitayner et al., 2006).
Previous attempts to crystallize four p53DBDs bound to a continuous DNA consensus site have met with little success. This is underscored by the structure determined by Cho et al., whose asymmetric unit contained only one of three p53DBDs bound sequence specifically to a DNA half site (Cho et al., 1994). More recently, the dimer of p53DBD dimers bound to a discontinuous DNA duplex where the break in DNA is between two p53DBD dimer/DNA complexes was reported, but the DNA packed in the crystal lattice such that the DNA is out of register (by about two base-pairs) relative to a continuous DNA duplex (Kitayner et al., 2006). In order to visual a p53 core domain tetramer bound to a continuous cognate DNA duplex, we extended the previously utilized crosslinking strategy to trap a p53DBD tetramer bound to a full p53 consensus site containing the most common zero base-pair separation between the two DNA half sites and now report on its structure.
RESULTS AND DISCUSSION
Crystal structure of the p53DBD tetramer bound to DNA
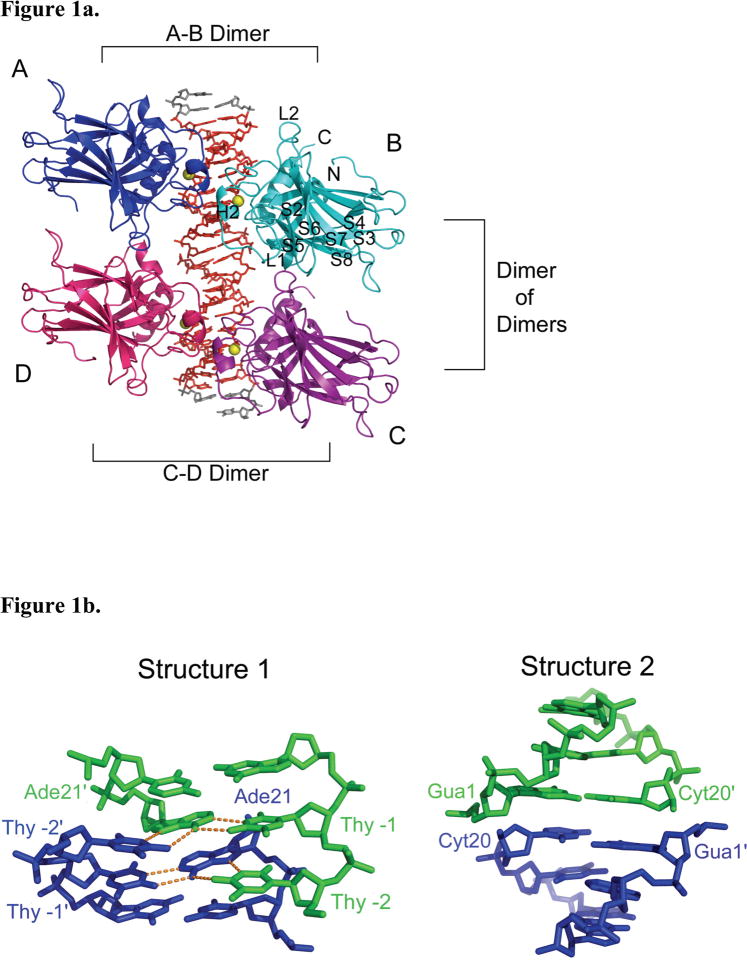
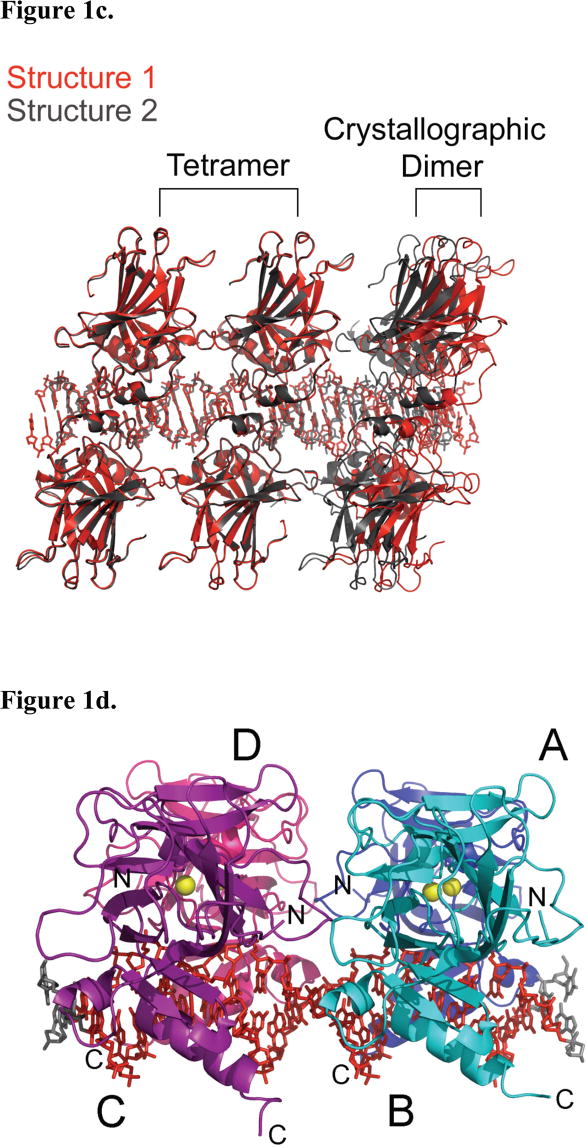
Two tetrameric p53DBD/DNA complex structures, referred to as structure 1 and structure 2, were determined to resolutions of 2.00 and 2.20 Å respectively (Table 1). While both protein/DNA complexes crystallized in spacegroup C2, the cell parameters and asymmetric units of these complexes differ, with structure 1 containing a p53DBD dimer bound to a DNA half site and structure 2 containing a p53DBD subunit bound to a DNA quarter site. As a result of these differences, the crystal packing contacts show significant differences between the two structures. The most significant difference lies along the direction of the DNA helical axis. The DNA in structure 1 forms end-end Hoogsteen base-pairs involving the overhanging 5′ thymine base of one DNA duplex with the Watson-Crick base-paired adenine of another duplex (Figure 1b). In contrast, the DNA duplex in structure 2 does not show electron density for the overhanging 5′ thymine base or the adjacent adenine-thymine base pair, which are presumably looped out of the DNA helix. As a result of this, two adjacent DNA duplexes of structure 2 stack end-to-end forming a pseudo continuous helix with the terminal DNA base-pairs from adjacent DNA duplexes about 3.6 Å apart (Figure 1b). The shorter ordered DNA duplex within structure 2 relative to structure 1 results in more extensive protein contacts between adjacent p53DBD tetramer/DNA complexes within the crystal lattice of structure 2 relative to structure 1 (Figure 1c). Despite these crystal packing differences between structure 1 and 2, the overall structures of the p53DBD tetramer/DNA complexes are essentially superimposable in the two crystal lattices (Figure 1c), arguing for the biological relevance of the crystallographic tetramer. Unless otherwise stated, discussion will focus on the higher resolution structure 1.
Table 1. Structure A.
Crystallographic statistics.
| Structure | 1 | 2 |
|---|---|---|
|
| ||
| Space Group | C2 | C2 |
|
| ||
| Cell Dimensions | ||
| a, b, c (Å) | 114.75, 68.016, 75.162 | 109.41, 68.10, 34.42 |
| A, β, γ (°) | 90.00, 111.12, 90.00 | 90.00, 104.17, 90.00 |
| Resolution (Å) | 2.00 | 2.20 |
| Rsyma | 0.083 [0.465] | 0.100 [0.277] |
| <I> / σ | 16.5 [3.6] | 14.7 [5.6] |
| Completeness | 99.1% [99.4%] | 99.6% [97.3%] |
| Redundancy | 4.9 [4.9] | 4.9 [4.4] |
|
| ||
| Refinement | ||
| Resolution Range (Å) | 70.186 – 2.00 | 57.354 – 2.20 |
| Reflections | 177,707 | 60,952 |
| Rworkb / Rfreec | 0.225 / 0.260 | 0.205 / 0.250 |
| Asymmetric Unit | Dimer + half site | Monomer + quarter site |
|
| ||
| Number of Atoms | ||
| Protein | 3034 | 1510 |
| DNA | 464 | 204 |
| Zinc | 2 | 1 |
| Water | 577 | 161 |
|
| ||
| Average B Factor (Å2) | 37.67 | 33.91 |
|
| ||
| RMSDs | ||
| Bond lengths (Å) | 0.00547 | 0.0108 |
| Bond angles (°) | 1.37 | 1.54 |
: Rsym = ΣhΣl |Ihj − Ih| / ΣhΣh,i, where Ih is the mean intensity of symmetry-related reflections.
: Rwork = Σ ||Fo| − |Fc|| / Σ |Fo|, Fo is observed structure factor amplitudes; Fc is calculated structure factor amplitudes.
: Rfree is calculated from the withheld 5% of data.
Figure 1.
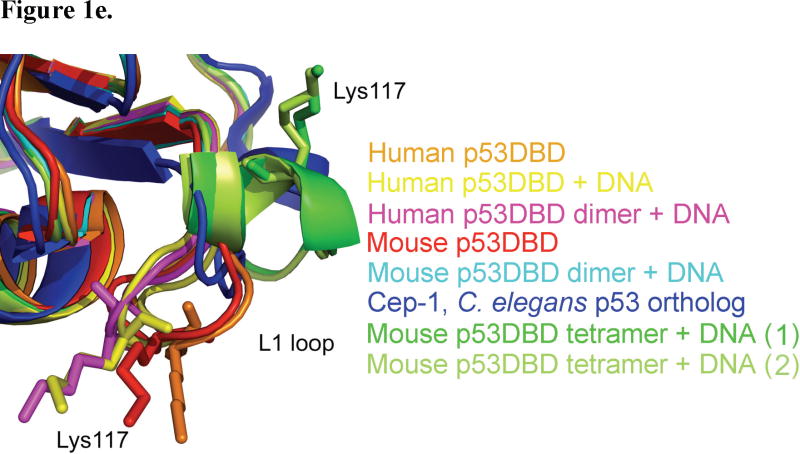
Structure 1 of the tetrameric p53DBD/DNA-complex and crystal packing. (a) Overall view of the tetramer in cartoon representation (DeLano, 2002). Subunits A (blue) and B (cyan) represent one dimer and subunits C (purple) and D (pink) represent the other. Zn2+ ions are shown as yellow spheres. The DNA consensus sequence is rendered in red and other bases are rendered in gray. (b) View of DNA base pairing between crystallographic DNA duplexes in structures 1 and 2 with involved bases numbered as in Figure 2a. One duplex is colored in green and the crystallographic duplex is colored in blue. Hydrogen bonds are colored in orange. (c) View of tetramer crystal packing in both structures 1 and 2. Structure 1 is colored red and structure 2 is colored gray. A tetramer is shown for both structures with a dimer from the adjacent crystallographic tetramer shown to the right. A clear gap is seen between structure 1 and its crystallographic dimer neighbor while no such gap is seen for structure 2. (d) View of the tetramer perpendicular to the DNA helical axis. (e) Close up view of the L1 loops from human (2OCJ, orange) and mouse (1HU8, red) p53DBD; human p53DBD (1TSR, yellow), human p53DBD dimer (2AC0, magenta), mouse p53DBD dimer (2GEQ, cyan) bound to DNA; Cep-1 p53 ortholog (1T4W, blue), mouse p53DBD tetramer bound to DNA structure 1 (green) and mouse p53DBD tetramer bound to DNA structure 2 (light green).
Structure of each p53DBD within the tetramer and DNA contacts
Within the tetramer, the structure of each of the p53DBD subunits is nearly identical to each other (Cα rmsd ≤ 0.105 Å) and to other p53DBD domain structures that have been determined (Cα rmsd ≤ 0.559 Å) (Cho et al., 1994; Wang et al., 2007; Zhao et al., 2001). Briefly, each domain is comprised of two anti-parallel βsheets that form an immunoglobulin-like β sandwich. Each strand is twisted resulting in two very different ends of the sandwich; one end is compact with short loops while the other end splays to the loop-sheet-helix motif, which contains a tetrahedrally bound Zn2+ ion, the L2 loop, and the H2 α helix (Figures 1a).
The major difference between each tetramer subunit and the previous structures lies within the L1 loop, residues 109 – 119 (Figure 1e). Both Cho et al. (human p53DBD monomer bound to DNA) and Kitayner et al. (human p53DBD dimer of dimers bound to a discontinuous DNA duplex) stress the importance of the L1 loop for DNA recognition since Lys120 (Lys117 in mouse) at the loop’s tip points into the major groove to make a base-specific DNA contact (Cho et al., 1994; Kitayner et al., 2006). In contrast, here Lys117 and the L1 loop in each subunit has moved nearly 15 Å away from the DNA and adopts some α helical structure.
The DNA contacts made by each subunit to the DNA are essentially as reported by Cho et al.(Cho et al., 1994). Three minor variations, which are also observed in the other p53DBD/DNA structures, are seen in each subunit. First, Lys117 (Lys120 in human p53) of the L1 loop has moved away from the DNA and towards the tetramerization interface as previously described in subunits B and D of structure 1 and all subunits of structure 2 (Figures 1e, 3b, and 3c). In subunits A and C of structure 1, this residue is disordered. Second, Arg280 (Arg283 in human p53), part of the H2 helix, is too far to contact the phosphate backbone. Third, Arg245 (Arg248 in human p53), the most frequently mutated residue in human cancers, adopts two different conformations in structure 1 and its DNA contact changes accordingly. In structure 2 and subunits A and C of structure 1, Arg245 sits in the minor groove and contacts the DNA as reported in Cho et al. However, in subunits B and D of structure 1, the same residue adopts a 90° bend and only contacts the backbone of one DNA strand. Taken together, analysis of the protein/DNA contacts within the tetrameric p53DBD bound to DNA reveals that the basic residues 117, 245 and 280 have variable roles in DNA recognition.
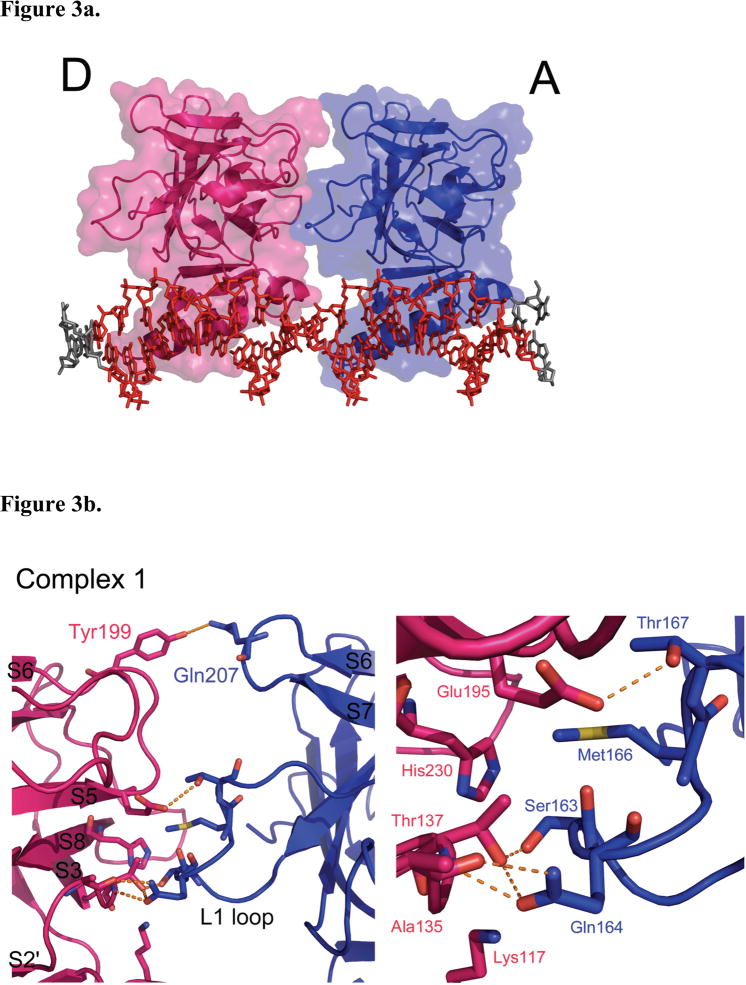
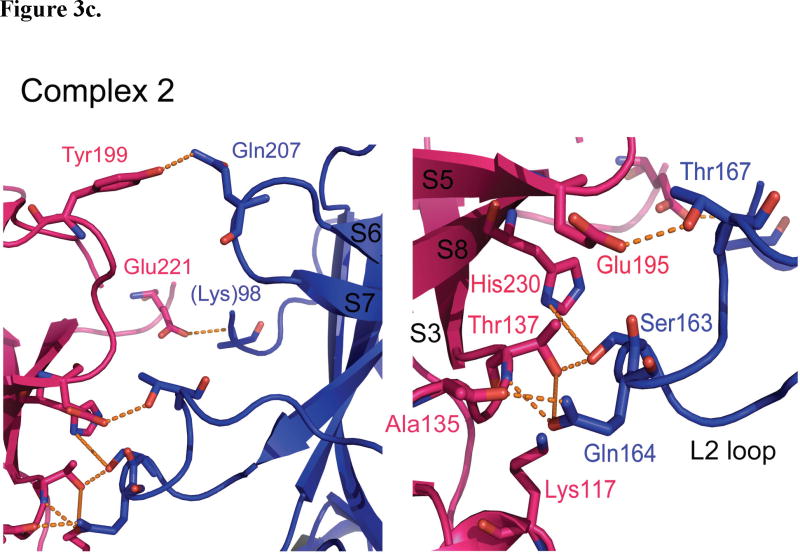
Figure 3.
p53DBD dimer-dimer formation. (a) View of subunits A (blue) and D (magenta) in surface representation for structure 1 (DeLano, 2002). Subunits B and C have been removed for clarity. (b) Overall view (left) and close up view (right) of the dimer-dimer interaction between subunits A and D for Structure 1. (c) Overall view (left) and close up view (right) of the dimer-dimer interaction between subunits A and D for structure 2. Hydrogen bonds are depicted as orange dashed lines. The same interactions are seen between subunits B and C for both structures.
Structure of each p53DBD dimer and tethers
Each dimer (A–B and C–D, Figures 1a and 1d) within the tetramer comes together as previously described by Shakked and coworkers. for 2AC0 (Kitayner et al., 2006). Key residues involved are Arg178, Pro174, His175, Gly241 and Met243. Also, the dimers show little structural variation from the previously reported mouse p53DBD dimer (Cα rmsd 0.776 Å(Ho et al., 2006)).
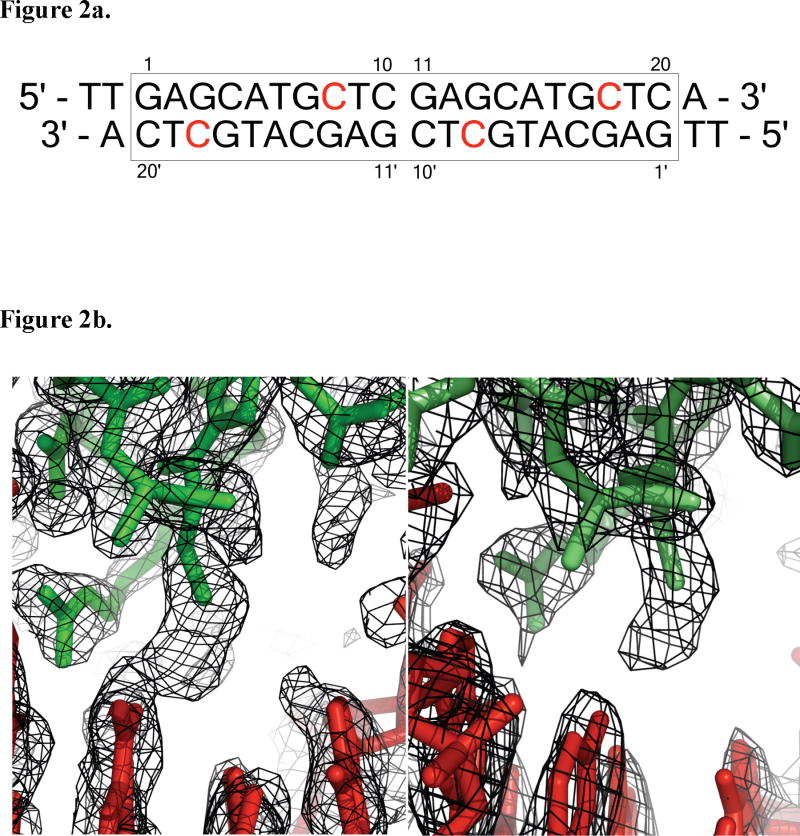
Each p53DBD subunit is tethered to its quarter site via the side chain sulfur of Cys274 to the modified sulfur-bearing linker attached to the N4 atom of the cytosine 3′ to the invariant guanine (Figure 2a). The extended tether has a length of 7.5 Å(Huang et al., 1998). Figure 2b shows simulated annealing composite omit maps for each p53DBD/DNA complex structure around residue Cys274. Structure 2 clearly shows a break in density, strongly indicating that the linking atoms are not rigid. Structure 1 shows connected density between the atoms but it lacks defined structure also supporting the tether’s flexibility. The distance between the crosslinked atoms is less than 4.1 Å and nearly all the DNA contacts previously reported are intact suggesting that the tether is flexible and does not perturb the p53/DNA interface.
Figure 2.
DNA used for crystallization and view of crosslink. (a) DNA sequence used for crystallization. The consensus sequence is boxed and crosslinked bases are in red. (b) Simulated annealing composite omit 2Fo−Fc map contoured at 1.0 σ (left) 1.3 σ (right) at the area of crosslinking. DNA consensus sequence atoms are colored red. Protein atoms are colored in green (structure 1, left) or light green (structure 2, right) (DeLano, 2002).
Taken together, the similarity of p53DBD-DNA interactions reported here to the DNA interactions with the monomeric p53DBD(Cho et al., 1994; Wang et al., 2007; Zhao et al., 2001) and dimeric p53DBD (Ho et al., 2006; Kitayner et al., 2006), and the lack of rigid tether density in the structures reported here, these data support the conclusion that the tether does not significantly disrupt the natural binding properties of the p53DBD to its consensus site.
Structure of the p53DBD tetramer and dimer-dimer contacts
The p53DBD tetramer binds to its DNA consensus sequence as a dimer-of-dimers as previously proposed (Figure 1a). Each dimer (A–B and C–D) binds to its respective half site with no relative twist and without significant DNA bending (Figures 1a and 1d). Each dimer-dimer interface buries an average of 665 Å2 of surface area and shows nearly the same interactions in each structure. The major interface involves the L2 loop from subunits A and C tucking against the S2′/S3, S5/S6 and S7/S8 loops from subunits B and D (Figure 3a) and is stabilized by both van der Waals forces and a network of hydrogen bonds (Figures 3b and 3c). The backbone nitrogen of D-Ala135 (S2′/S3) donates a hydrogen bond to A-Gln164 OE1 (L2), which in turn accepts a hydrogen bond from the side chain hydroxyl of D-Thr137 (S3). This hydroxyl also accepts a hydrogen bond from the side chain of A-Ser163 (L2). D-Gln164 NE2 also donates a hydrogen bond to the backbone oxygen of D-Ala135, while the side chain of A-Thr167 (L2) donates a hydrogen bond to OE1 of D-Glu195 (S5/S6). Completing this major contact region are two van der Waals interactions: D-His230 CE1 (S8) with A-Ser163 CB and D-His230 CD2 with A-Met166 CE (L2). In structure 2 (Figure 3c), the van der Waals contact involving A-Met166 CE is lost due to a different side chain conformation, but an additional hydrogen bond is present between D-His230 ND1 and the side chain of A-Ser163.
Both structures 1 and 2 show that D-Tyr199 (S5/S6) donates a hydrogen bond to AGln207 (S6/S7) near the top of each interface. Structure 2 has one additional hydrogen bond between D-Glu221 (S7/S8) and the backbone nitrogen of A-Lys98 (modeled as alanine in the structure). Overall, both structures have very similar dimer-dimer interfaces and involve nearly the same residues.
A sequence alignment of all p53DBD sequences available(Gasteiger et al., 2003) underscores the importance of these dimer-dimer interacting residues. Glu195 and Ala135 are strictly conserved, while Thr137 is strictly conserved in all species except chicken. Also, Gln207, Gln164 and Glu221 only show conservative mutations. This described interface is also consistent with previous studies implicating specific residues for dimer-dimer interactions (Cho et al., 1994; Ho et al., 2006; McClure and Lee, 1998; Pan and Nussinov, 2007; Pan and Nussinov, 2008) The most often sited tetramerization residues implicated for such interactions include Ser93, Ser96, Thr167, Ser163, Gln164 from one domain and Glu195, Thr137, and Glu221 from the other. All except for residues Ser93, Ser96 (disordered in these structures) and Gly196, each of the implicated dimer-dimer contact residues are observed to make contacts at the dimer-dimer interface of the structures reported here.
A curious area of the tetramerization interface involves the enigmatic Lys117 of the L1 loop. As previously stated, the L1 loop of each subunit has moved far from where other p53DBD and p53DBD/DNA structures have it placed (Figures 1e, 3b, and 3c). Due to this movement, Lys117 points up towards residues D-Thr137 and A-Ser163 but does not make any specific contacts with the opposite dimer. Several possibilities exist to explain this conformational change. Careful analysis around Cys274 shows that the tether can only loop in one direction, which is towards the L1 loop. In fact, the tether is likely blocking the DNA bases that Lys117 has previously been shown to contact. However, the tether is only 7.5 Å in length and the L1 loop has moved approximately 15 Å away, which is much too far to attribute only to steric hindrance by the tether. Alternatively, the current position of the L1 loop be stabilized by crystal contacts from a symmetry-related subunit or the presence of a citrate molecule, which sits close to the new conformation. Perhaps the most intriguing possibility is that the shift in position is induced by tetramerization itself. Lys117, which is strictly conserved across all species but shows low cancerous sensitivity(Ho et al., 2006), and although this residue is shown to make base specific contacts in several reported experimental p53/DNA complexes (Cho et al., 1994; Kitayner et al., 2006; Pan and Nussinov, 2008), but it also disordered in others (Ho et al., 2006; Kitayner et al., 2006; Pan and Nussinov, 2008). Within structure 1, Lys117 is only ordered in subunits B and D, the two p53DBD subunits in the crystal lattice that allow Lys117 to come close to the tetramerization interface. Lys117 is ordered in structure 2. Zupnick and Prives studied a K120A (Lys117 in mouse p53DBD) mutation for its effects on p53 function. They found that the mutant had strong DNA binding in vitro, but a weaker affinity in vivo. Overexpression in cells resulted in normal p53 function, however upon expression at normal cellular levels, the mutant showed defects in transactivation and apoptosis (Zupnick and Prives, 2006). It is interesting to speculate that Lys117 might alternate between DNA base contact and tetramerization contact, thus explaining its ambiguity. Clearly, additional studies will have to be carried out to resolve the role of Lys117 in the DNA binding properties of p53.
Comparison with other p53DBD tetramers
Two p53DBD tetramer complexes with DNA have been made available recently and vary in their structures. Ho et al. proposed a model p53DBD tetramer bound to DNA, which was generated by translating their crystallographic p53DBD dimer down the DNA to the next half site(Ho et al., 2006). This model predicted that the dimers come close at the L2 loop and S7/S8 regions, which is consistent with what we see in our current structures. A 40° DNA bend was also proposed in this model as well as models from Nagaich et al.(Nagaich et al., 1999) and Pan et al. (Pan and Nussinov, 2007). Interestingly, the present structures reveal that the DNA is relatively straight. Previous studies suggested that a straight p53DBD tetramer/DNA complex would not be possible due to steric clashes. To alleviate these clashes, it was proposed that the DNA would have to bend by as much as 50°(Balagurumoorthy et al., 1995; Nagaich et al., 1997a; Nagaich et al., 1999; Nagaich et al., 1997b; Pan and Nussinov, 2007). In contrast to these proposals, the two p53DBD tetramer/DNA structures reported here clearly show that a stable tetrameric complex can bind to straight, B-form DNA and maintain well-ordered dimer-dimer interfaces.
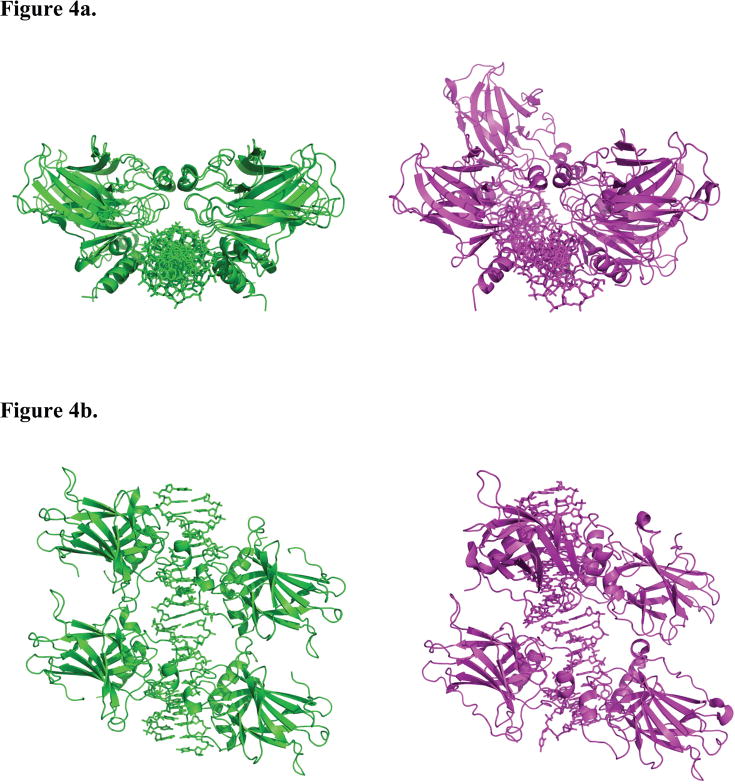
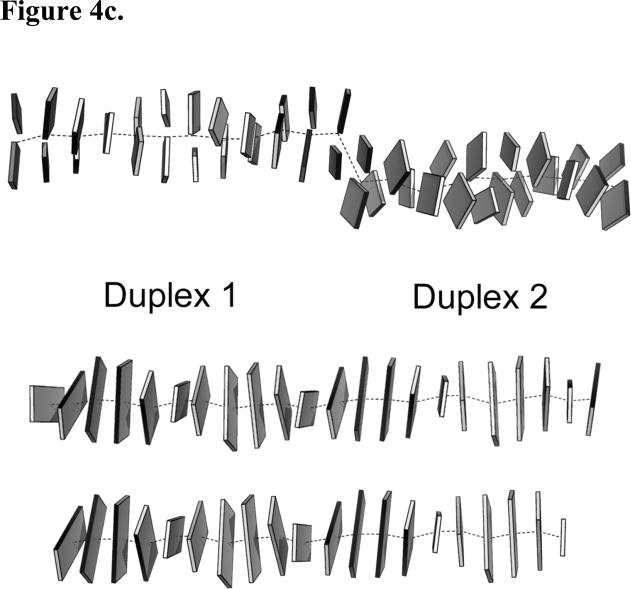
Shakked and coworkers reported on the structure of a p53DBD tetramer bound to discontinuous DNA where two p53DBD dimers bound to DNA came together end-to-end within their crystal lattices(Kitayner et al., 2006). These dimers are twisted 33° relative to each other in each structure (Figures 4a and 4b). While, the two DNA half sites appear to be out of register for each (Figure 4c), they can be considered approximate p53DBD tetramers and provide insight into alternative dimer-dimer contacts. Each tetramer shows its own set of limited dimer-dimer contacts. 2AC0, 2AHI, and 2ATA are quite similar in that one dimer-dimer interface is devoid of interaction and the other has limited hydrogen bonds and water-mediated interactions. 2ADY is unique in that both interfaces have the same five hydrogen bond interactions. Interestingly, as in the p53DBD tetramer/DNA complexes reported here, Ser166, Gln167, and Thr170 (Ser163, Gln164 and Thr167 in mouse p53DBD) are involved in hydrogen bonds in the p53DBD tetramer/DNA complexes reported by Shakked and coworkers but these residues mediate different hydrogen bonds within the two complexes.
Figure 4.
Superposition of the p53DBD tetramer/DNA complex reported here with the p53DBD tetramer bound to a discontinuous DNA duplex. (a) View down the helical axis of 2AC0 after superposition on the p53DBD tetramer/DNA complex (DeLano, 2002). 2AC0 is rendered in magenta and structure 1 is rendered in green. Each p53DBD tetramer/DNA complex is shown separately for clarity. The 33° twist of the back dimer relative to the front is easily seen for 2AC0. (b) View of the superposition oriented 90° from the view in A. (c), DNA from 2AC0 (upper), structure A (middle) and structure B (lower) crystal structures as Calladine-Drew schematics. Duplex 1 and 2 of 2AC0 are labeled and the central helical axes are the dotted lines. Duplex 2 is lower and rotated toward the viewer relative to duplex 1. All schematics were made with 3DNA and rendered in PyMOL (DeLano, 2002; Lu and Olson, 2003).
Multiple Tetramerization Modes for p53 on DNA
After comparison of the p53DBD tetramer/DNA models with the two complexes reported here, we hypothesize that dimer-dimer contacts within the p53DBD tetramer on DNA can vary in vivo. This hypothesis is supported by several lines of evidence. First, p53 response elements are varied across the genome due to different base-pair spacing between half sites and response elements which fulfill the consensus sequence requirement (El-Diery et al., 1992). Second, the current structure shows that the DNA need not be bent for four p53DBD domains to stably bind to it. Given that solution studies and molecular dynamics suggest that bent DNA complexes are possible (Nagaich et al., 1999) and the inherent flexibility of the DNA sequences has been studied (McNamara et al., 1990; Nagaich et al., 1994; Zhurkin et al., 1991), it is likely that other energy minima exist besides the one captured crystallographically here. Also, while several of the residues at the interface involved are conserved, cancerous mutations of the residues are low compared with DNA contacts or monomer-dimer contact residues within the p53DBD(Ho et al., 2006). Taken together, these observations suggest that perhaps the tetramerization interface is not fixed, but able to accommodate different contacts depending on different cellular conditions.
p53DBD DNA binding cooperativity
It has been shown that the predominantly monomeric p53DBD will bind to its consensus sequence DNA in a highly cooperative manner (Balagurumoorthy et al., 1995) and the basis for this cooperativity has been the subject of much speculation. McClure et al. studied how p53DBD dimers and tetramers could bind DNA oligomers by varying the placement of the consensus quarter sites. Results from this work showed that two p53DBD dimers will bind a full consensus site for a far longer period of time (t1/2 ~ 15 minutes) than one dimer alone (t1/2 ~ 30 seconds) (McClure and Lee, 1998). A similar experiment was performed with wild-type p53 (residues 94 – 360, tetrameric) and an L344A mutant construct that could only form dimers. The results showed that tetramers had a six-fold greater affinity for DNA over dimers (Weinberg et al., 2004). Correlating this data with the two structures reported here suggests that two dimers are more stably bound to DNA due to the numerous dimer-dimer contacts that are observed in the structures reported here. One dimer bound to DNA benefits from both stabilizing DNA and dimerization contacts (Klein et al., 2001), but according to McClure et al and supported by Weinberg et al, will still diffuse away from the DNA in a short period of time. With the added dimerization contacts, the complex is more stable, as evidenced by the increase in t1/2 and affinity.
Other suggestions have been reported for p53DBD cooperativity but are not supported by crystal structures (McClure and Lee, 1998). No conformational change is seen within the p53DBD upon tetramerization or DNA binding. While it is possible that other areas of the protein may undergo structural rearrangement during DNA binding, cooperativity is seen at the level of p53DBD binding to DNA alone, thus implicating determinants for DNA-binding cooperativity within the DNA-binding domain of p53. Also, cooperative DNA binding by p53 is largely influenced by its tetramerization domain in vivo. Taking these observations together with the p53DBD tetramer/DNA complexes reported here suggests the DNA-induced interactions that are observed between the two p53DBD dimers plays a major role in cooperative p53DBD binding to DNA.
p53DBD conservation and implication for function
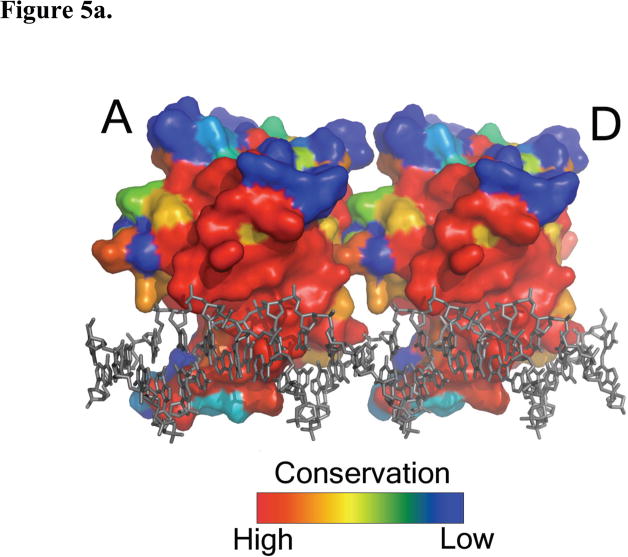
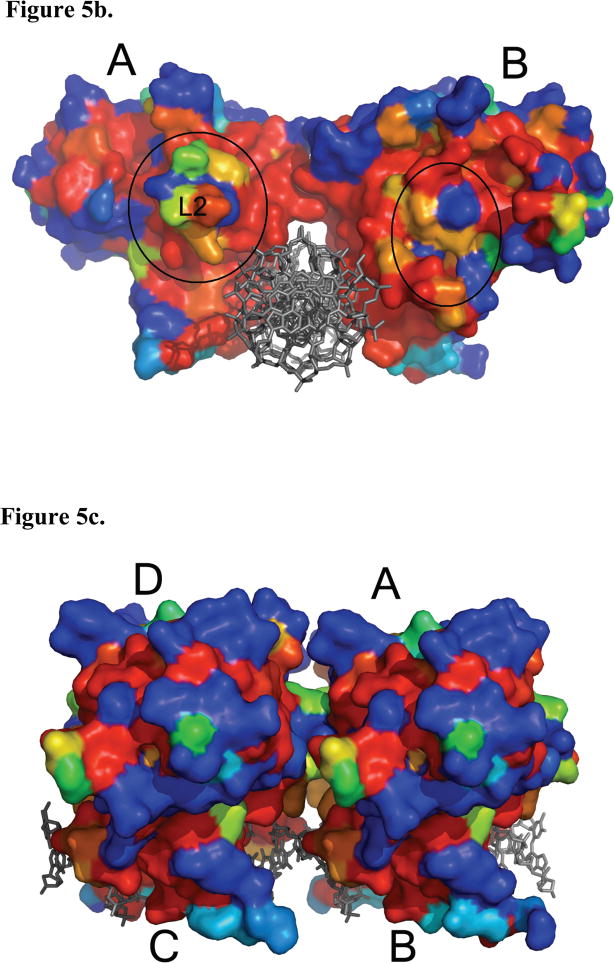
A surface conservation map of the p53DBD in the context of the p53DBD tetramer bound to DNA reveals several areas of high conservation and by inference functional importance. The highest degree of conservation maps to the DNA binding region of the monomer and the monomer-dimer interface (Figure 5a) highlighting the relative importance of these regions. In contrast, the dimer-dimer interface shows far less conservation (Figure 5b) arguing for a less significant role of p53DBD dimer-dimer contacts for DNA recognition. Subunit A shows a long groove of conservation that is set back from the L2 loop. A mirrored pattern on subunit B would suggest a strict dimer-dimer interaction, but this is not seen and further supports the model that multiple tetramerization modes can accommodate p53 binding to DNA.
Figure 5.
Surface map of amino acid conservation in p53DBD. (a) A sequence alignment was performed with all known sequences of p53 using ClustalW (Gasteiger et al., 2003) and mapped with ESPript (Gouet et al., 1999). View of subunits A and D with strictly conserved residues colored in red to nonconserved residues in blue (DeLano, 2002). The DNA is rendered in gray. Subunits B and C have been removed for clarity. Monomer-dimer interface is circled with a black, dashed line. (b) View along the helical axis of subunits A and B with subunits C and D removed for clarity. Dimer-dimer interface is circled with a black, dashed line. (c) Back view of subunits B and C showing the band of conserved residues that wrap around each subunit.
Intriguingly, a comparison of subunit A and B shows that one side of each subunit is more conserved than the other (Figure 5b). For subunits B and D, this highly conserved surface would face the solvent rather than the opposite dimer. It is possible that these residues participate in binding other domains of p53 or partner proteins of p53. This could also be the case for the ring of conserved amino acids that map around the back surface of each subunit (Figure 5c). Taken together, the sequence conservation on the surface to the p53DBD tetramer bound to DNA suggests other protein binding and perhaps regulatory surfaces.
Considering the proposed variability of the dimer-dimer contacts within the tetrameric p53DBD bound to DNA, one might ask why p53 binds DNA as a tetramer. This could simply be to increase the affinity of p53 for DNA. It is also possible that other regions of p53 interact with the DNA binding domain to support tetrameric binding to DNA. Correlating with this possibility, the N- and C- terminal ends of the p53DBD are located at the dimer-dimer interface (Figure 1d). Given that p53 has been reported to interact with many proteins, it is also possible that the p53DBD tetramer bound to DNA provides a unique protein contact surface. Taken together, these correlations suggest that the p53DBD domain may play a more active role in p53 function than simply providing a scaffold for DNA binding.
METHODS
Growth and purification of mouse p53DBD and purification of DNA containing unnatural nucleotides
This was carried out essentially as described in Ho et al.(Ho et al., 2006)
Purification of Crosslinked Tetramer
Crosslinking was performed at room temperature in 20 mM citrate, pH 6.1, 100 mM NaCl, and 0.5 mM TCEP. A 4:1 molar ratio of p53DBD to DNA was added to excess buffer and allowed to sit for three hours. The crosslinking reaction was loaded onto Q resin and higher order oligomers were eluted with a gradient of 100 mM to 500 mM NaCl. Tetrameric complexes eluted first since its negative charges were the least available for binding with the resin. Denaturing but non-reducing gels were run to assay purity. Tetramer purity was determined by the presence of only one band corresponding to one strand of DNA with two bound p53DBDs. These fractions were pooled and concentrated to 5 μM as determined by standard curve of known protein-DNA concentrations and frozen at −80°C until use.
Data Collection and Structure Refinement
Crystals were grown in 200 mM lithium citrate, 20% PEG 3350 by hanging drop vapor diffusion. Small, rectangular crystals appeared in two days and reached a maximum size of 160 × 30 × 30 uM after one week. Crystals were frozen in the well condition with 15 – 20% glycerol. Data collection was carried out on the 23-IDB beamline at the Advanced Photon Source synchrotron at Argonne National Laboratories. Data was indexed and scaled using HKL2000 (Otwinowski and Minor, 1997). The structures were solved by molecular replacement using PHASER with 2GEQ as a search model. For structure 1, the resulting solution was the p53DBD dimer bound to its half site in an asymmetric unit cell with a full tetramer containing a crystallographic two-fold axis. For structure 2, the resulting solution was the p53DBD monomer bound to its quarter site in an asymmetric unit. These models were refined in CNS using simulated annealing, minimization and individual B-factor refinement. For structure 1, medium NCS restraints were initially used for the two p53DBD domains in the asymmetric unit. The NCS restraints were gradually lowered and were eventually dropped completely. Between refinement cycles, the model was manually rebuilt using the program Coot(Emsley and Cowtan, 2004). Data collection and refinement statistics for structures 1 and 2 are summarized in Table 1.
References
- Balagurumoorthy P, Sakamoto H, Lewis MS, Zambrano N, Clore GM, Gronenborn AM, et al. Four p53 DNA-binding domain peptides bind natural p53-response elements and bend the DNA. PNAS USA. 1995;92:8591–8595. doi: 10.1073/pnas.92.19.8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Santos WL, Verdine GL. Structure of a DNA Glycosylase Searching for Lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal Structure of a p53 Tumor Suppressor-DNA Complex: Understanding Tumorigenic Mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002. [Google Scholar]
- El-Diery WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nature Genetics. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. ACta Crystallography Section D - Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Research. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- He C, Verdine GL. Trapping Distinct Structural States of a Protein/DNA Interaction through Disulfide Crosslinking. Chemistry and Biology. 2002;9:1297–1303. doi: 10.1016/s1074-5521(02)00283-1. [DOI] [PubMed] [Google Scholar]
- Ho WC, Fitzgerald MX, Marmorstein R. Structure of the mouse p53 core domain dimer bound to DNA. Journal of Biological Chemistry. 2006;281:20494–20502. doi: 10.1074/jbc.M603634200. [DOI] [PubMed] [Google Scholar]
- Horn H, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- Huang H, Chopra R, Verdine GL, Harrison SC. Structure of Covalently Trapped Catalytic Complex of HIV-1 Reverse Transcriptase: Implications for Drug Resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- Jeffrey PD, Gorina S, Pavletich NP. Crystal Structure of the Tetramerization Domain of the p53 Tumor Suppressor at 1.7 Angstroms. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- Kitayner M, Rosenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, et al. Structural Basis of DNA Recognition by p53 Tetramers. Molecular Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Klein C, Georges G, Kunkele KP, Huber R, Engh RA, Hansen S. High thermostability and lack of cooperative DNA binding distinguish the p63 core domain from the homologous tumor suppressor p53. Journal of Biological Chemistry. 2001;276:37390–37401. doi: 10.1074/jbc.M103801200. [DOI] [PubMed] [Google Scholar]
- Lu X-L, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Research. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan AM, Verdine GL. Engineering Tethered DNA Molecules by the Convertible Nucleoside Approach. Tetrahedron. 1991;47:2603–2616. [Google Scholar]
- McClure KG, Lee PWK. How p53 binds DNA as a tetramer. The EMBO Journal. 1998;17:3342–3350. doi: 10.1093/emboj/17.12.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara PT, Bolshoy A, Trifonov EN, Harrington RE. Sequence-dependent kinks induced in curved DNA. Journal of Biomolecular Structural Dynamics. 1990;8:529–538. doi: 10.1080/07391102.1990.10507827. [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Apella E, Harrington RE. DNA Bending is Essential for the Site-specific Recognition of DNA Reponse Elements by the DNA Binding Domain of the Tumor Suppressor Protein p53. The Journal of Biological Chemistry. 1997a;272:14842–14849. doi: 10.1074/jbc.272.23.14842. [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Bhattacharyya D, Brahmachari SK, Bansal M. CA/TG sequence at the 5′ end of oligo(A)-tracts strongly modulates DNA curvature. Journal of Biological Chemistry. 1994;269:7824–7833. [PubMed] [Google Scholar]
- Nagaich AK, Zhurkin VB, Durell SR, Jernigan RL, Apella E, Harrington RE. p53-induced DNA bending and twisting: p53 tetramer binds on the outer side of a DNA loop and increases DNA twisting. PNAS USA. 1999;96:1875–1880. doi: 10.1073/pnas.96.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich AK, Zhurkin VB, Sakamoto H, Gorin AA, Clore GM, Gronenborn AM, et al. Architectural Accomodation in the Complex of Four p53 DNA Binding Domain Peptides with the p21/waf1/cip1 DNA Response Element. The Journal of Biological Chemistry. 1997b;272:14830–14841. doi: 10.1074/jbc.272.23.14830. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997:276. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pan Y, Nussinov R. Structural Basis for p53 Binding-induced DNA Bending. The Journal of Biological Chemistry. 2007;282:691–699. doi: 10.1074/jbc.M605908200. [DOI] [PubMed] [Google Scholar]
- Pan Y, Nussinov R. p53-Induced DNA Bending: The Interplay between p53-DNA and p53-p53 Interactions. The Journal of Physical Chemistry B. 2008;112:6716–6724. doi: 10.1021/jp800680w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietenpol JA, Tokino T, Thiagaligam S, El-Diery WS, Kinzler KW, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. PNAS USA. 1994;91:1998–2000. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdine GL, Norman DPG. Covalent Trapping of Protein-DNA Complexes. Annual Reviews in Biochemistry. 2003;72:337–366. doi: 10.1146/annurev.biochem.72.121801.161447. [DOI] [PubMed] [Google Scholar]
- Wang Y, Rosengarth A, Luecke H. Structure of the human p53 core domain in the absence of DNA. Acta Crystallography. 2007;D63:276–281. doi: 10.1107/S0907444906048499. [DOI] [PubMed] [Google Scholar]
- Weinberg RL, Veprintsev DB, Fersht AR. Cooperative Binding of Tetrameric p53 to DNA. Journal of Molecular Biology. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- Zhao K, Chai X, Johnston K, Clements A, Marmorstein R. Crystal Structure of the Mouse p53 Core DNA-binding Domain at 2.7 A Resolution. Journal of Biological Chemistry. 2001;276:12120–12127. doi: 10.1074/jbc.M011644200. [DOI] [PubMed] [Google Scholar]
- Zhurkin VBBUN, Gorin AA, Jernigan RL. Static and statistical bending of DNA evaluated by Monte Carlo simulations. Proceedings of the National Academy of Sciences, USA. 1991;88:7046–7050. doi: 10.1073/pnas.88.16.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupnick A, Prives C. Mutational Analysis of the p53 Core Domain L1 Loop. The Journal of Biological Chemistry. 2006;281:20464–20473. doi: 10.1074/jbc.M603387200. [DOI] [PubMed] [Google Scholar]