Abstract
β-Defensins are small secreted antimicrobial and signaling peptides involved in the innate immune response of vertebrates. In humans, a cluster of at least 7 of these genes shows extensive copy number variation, with a diploid copy number commonly ranging between 2 and 7. Using a genetic mapping approach, we show that this cluster is at not 1 but 2 distinct genomic loci ≈5 Mb apart on chromosome band 8p23.1, contradicting the most recent genome assembly. We also demonstrate that the predominant mechanism of change in β-defensin copy number is simple allelic recombination occurring in the interval between the 2 distinct genomic loci for these genes. In 416 meiotic transmissions, we observe 3 events creating a haplotype copy number not found in the parent, equivalent to a germ-line rate of copy number change of ≈0.7% per gamete. This places it among the fastest-changing copy number variants currently known.
Humans differ in diploid DNA dosage of certain regions. The introduction of new technologies has allowed a more accurate appreciation of the true scale and frequency of this dosage variation, or copy number variation (CNV), in the human genome (1–4). It is clear that it contributes to variation in gene expression (5) and that it is associated with variable phenotypes (6), including infectious and autoimmune disease (7–9). Although there have been interesting recent studies on the mechanisms by which copy number may be altered in the germ line (10–13), there are relatively few loci that have been studied in detail, and it is likely that mutation rates and mechanisms will differ between individual loci. The CNV in human β-defensin genes involves a cluster of at least 7 β-defensins in 8p23.1, including genes encoding proteins known to have antimicrobial properties (14) but which also have other functions and effects (15, 16). For most of these genes there is little detailed information about function and expression (14), and the β-defensin CNV forms part of a complex spectrum of population and evolutionary dynamics in this region of the genome (17). Nearly all individuals have between 2 and 7 copies of this cluster per diploid genome (18–21), but some individuals can have up to 12 copies, with the highest-copy-number haplotypes forming the basis of the cytogenetic 8p23.1 euchromatic variant (18, 22). In the more frequent copy number range, low β-defensin copy number has been reported to be associated with Crohn's disease of the colon (23) and high copy number with predisposition to psoriasis (20). This common variation may be involved in predisposition to other inflammatory disorders, but progress in these association studies has been hampered by the difficulty of measuring copy number accurately for large numbers of samples (24, 25).
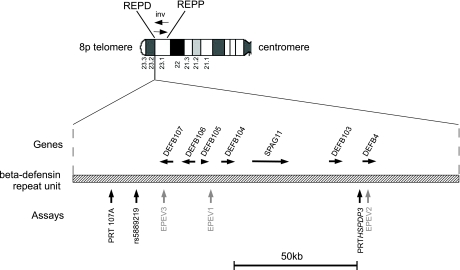
This region contains another large-scale structural polymorphism: a common inversion that involves nearly the whole chromosome band (26, 27), which is apparently mediated by recombination between clusters of repeats at the flanking proximal (REPP) and distal (REPD) regions (Fig. 1). REPP and REPD contain not only olfactory receptor gene repeats but also complex and polymorphic clusters of FAM90A genes (28). Approximately one-quarter of Europeans and one-third of Japanese are heterozygous for this inversion; in these heterozygotes unequal recombination can lead to the formation of the pathological rearrangement inv dup(8p) or its reciprocal product, +der (8) (26, 29).
Fig. 1.
8p23.1 genomic region showing an example of a β-defensin repeat. The 2 olfactory receptor/FAM90A repeat regions REPD and REPP are shown at either end of an inversion (inv) on the chromosome ideogram. An example of a partial β-defensin repeat, based on 1 of the 2 assembled versions in human genome assembly hg18, is shown below the ideogram. RefSeq genes, together with the copy number assays used in this article, are shown. The exact length of the copy number variable β-defensin unit is not known, and the region highlighted corresponds to the class V repeat identified previously (30).
The combination of frequent structural polymorphism involving extensive repeat-rich regions of unknown variable size makes the sequence of the REPD and REPP regions extremely difficult to assemble (30). Because of the large size of the repeats and the multiple individuals used to generate BAC libraries used in genome assemblies, even long Sanger sequencing reads and deep clone coverage have not, as yet, resolved a gap at this region in the human genome assembly. The REPD and REPP regions are very similar to other olfactory repeat regions, which are themselves copy number variable (31). This makes selecting clones for cytogenetic probes and accurately interpreting results difficult because of variable hybridization intensities against different olfactory repeat regions and extensive cross-hybridization. An accurate representation of the sequence of this region may be resolved only by constructing several alternative allelic assemblies and determining the frequency of those allelic assemblies in the population. It is likely that the uncertainty surrounding this region is responsible for misinterpretation of cytogenetic signals in some studies (32, 33).
We decided to use a genetic approach to investigate the structure and mutation processes at the β-defensin CNV, which does not a priori assume correct physical mapping of the region. Little is known about the processes generating and maintaining CNV; most new mutation is assumed, and in some cases has been demonstrated, to occur by nonallelic homologous recombination between flanking repeats generating reciprocal products with a loss and a gain of a repeat, respectively. Given the functional consequences of this region and its relevance to disease, it is clearly important that these questions are addressed.
Results
To examine mutation processes at the β-defensin CNV, we analyzed inheritance of the region in pedigrees for de novo mutation or recombination events. We determined copy numbers of the variable β-defensin CNV in Centre d'Etude du Polymorphisme Humain (CEPH) family members using the paralogue ratio test (PRT), combined with analysis of variant ratios at microsatellites EPEV-1 (18) and EPEV-3, and the multiallelic length polymorphism including the indel rs5889219. PRT is a development of the multiplex comparative PCR approach that uses a single primer pair to amplify both test and reference loci, leading to more accurate and robust copy number determination (24). Ratios of products from multiallelic length polymorphisms could be used to confirm copy number measurements; for example, amplification of 3 variants with yields in the ratio 2:2:1 strongly suggests a copy number of 5 (see, for example, the rs5889219 profile of 133306 in Fig. 2). Although not fully informative (for example, a 6-copy sample might have variants in the ratio 2:1, compatible with any multiple of 3), these ratio-based measurement methods provided valuable additional information by distinguishing different repeat identities in the analysis of segregation. By contrast, simple “copy-counting” methods like PRT give unambiguous information on copy number but provide no information on the individual identities of the repeat units.
Fig. 2.
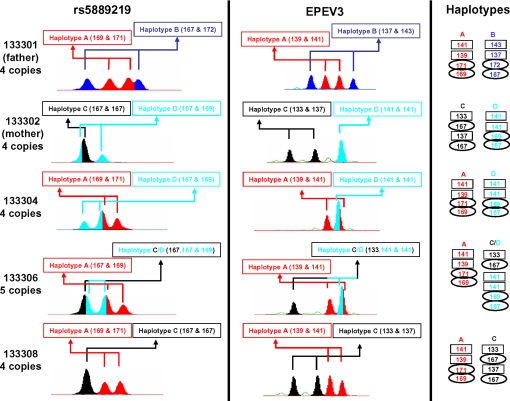
CEPH offspring 133306 inherits an altered copy number after recombination between maternal haplotypes. Paternal (red/blue) and maternal (black/cyan) haplotypes in the parents (133301 and 133302) were defined by analysis of grandparental genotypes and of segregation patterns into nonrecombinant offspring (e.g., 133304 and 133308) at the multiallelic indel rs5889219 (Left) and the microsatellite EPEV-3 (Center). Right shows an interpretation of the segregation pattern in which EPEV-3 alleles are shown in rectangles and rs5889219 alleles are shown within ellipses. The appearance of a recombinant C/D haplotype in offspring 133306 coincides with a maternal crossover in the interval, as deduced from CEPH segregation data (see Fig. 4 and Table S1).
We combined these approaches to establish the pattern of β-defensin transmission in 208 offspring from 26 CEPH families. Consistent with previous data (18–21), copy number among the parents of these 26 families varied between 2 and 7, with a modal copy number of 4 and a mean of 4.58. We observed that at least 24 offspring inherited recombinant haplotypes in which parental copies of the β-defensin repeat had been reassorted. In many instances the haplotype copy number was unchanged, but in some cases a new copy number was found in the recombinant chromosome; in Fig. 2, for example, child 133306 receives a recombinant maternal chromosome carrying 3 repeat units, whereas children inheriting nonrecombinant haplotypes show that the mother (133302) has 2 2-copy haplotypes. The data in Fig. 3 show that children 134106 and 134109 have each received different (reciprocal) recombinant maternal haplotypes, but with no change in the haplotype copy number. Most unexpectedly, mapping the positions of the crossover breakpoints using CEPH segregation data showed that these recombinations cannot all simply be occurring within a coherent block of β-defensin tandem repeats; the breakpoint of the maternal crossover in 133306, for example, is located proximal to rs2001329 at position chr8:11,024,269 on the March 2006 Genome Assembly, ≈3 Mb proximal to the assembly position of the β-defensin repeats (Fig. 4). In some transmissions, for example, the paternal chromosome of 1329208 [see supporting information (SI) Fig. S1], where parental repeats share many variants, the multiallelic markers typed were not sufficiently informative to specify that crossovers deduced from flanking markers necessarily involved reassortment of β-defensin repeats. For this reason, the rates we deduce for these reassortment processes are underestimates. In principle, somatic deletion, duplication, or rearrangement of β-defensin repeats in CEPH lymphoblastoid cell lines could lead to anomalous genotypes that mimic these recombinations. However, all 3 events that we document here that alter haplotype copy number not only accompany an unambiguous germ-line recombination event in 8p23.1, but also involve clearly identifiable repeat units from the other parental haplotype and so cannot be explained by simple cell line rearrangements deleting or duplicating existing copies.
Fig. 3.
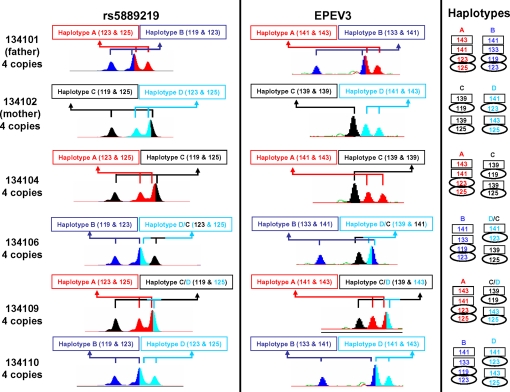
CEPH offspring 134106 and 134109 inherit different reciprocal maternal crossovers that do not alter copy number. Paternal (red/blue) and maternal (black/cyan) haplotypes in the parents (134101 and 134102) were defined by segregation patterns into nonrecombinant offspring, including 134104 and 134110 shown here, at the multiallelic indel rs5889219 (Left) and the microsatellite EPEV-3 (Center). The interpretation in Right shows that the crossovers are clearly demonstrated by EPEV-3 but that rs5889219 is not fully informative; reciprocal crossovers are inherited by 1341-06 (D/C) and 1341-09 (C/D), and corresponding crossovers can be observed on linkage analysis of markers in this region (see Fig. 4 and Table S1).
Fig. 4.
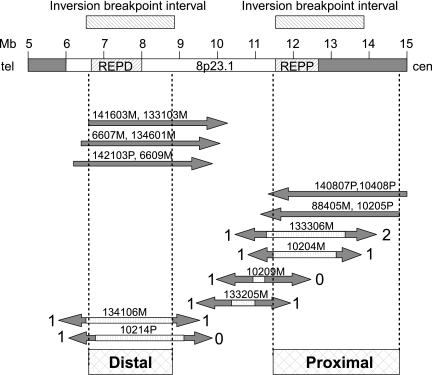
Genetic mapping of β-defensin repeats relative to crossover breakpoints in CEPH pedigrees, shown against genome assembly coordinates. The approximate locations of REPP and REPD are shown, as are the intervals containing the inversion endpoints. The crossovers indicate, at the top, selected unidirectional recombinants mapping the location of all repeat units on a haplotype, identified by family and individual name and parent of origin (e.g., “6607M” is the maternal haplotype of child 07 in family 66). Below are shown selected crossovers that map repeats to the proximal and/or distal sites; the numbers of repeats mapping in each direction are indicated, and the hashed segments indicate uncertainty in the placement of breakpoints because of the lack of informative markers. Based on these data, β-defensin repeats can be mapped to the distal and proximal intervals shown at the bottom.
These and other genetic mapping data, summarized in Fig. 4 and Dataset S1, and shown in full in Fig. S1 and Table S1, forced us to conclude that some of the β-defensin repeats segregating in these CEPH pedigrees could not be located at the established interval in the genome assembly (within REPD at approximately chr8:6,900,000–8,150,000) but instead mapped to another site more proximal on chromosome 8p. Analysis of crossover breakpoints in CEPH segregation data (Table S1) showed that this second site must be bounded by AFMb294yg5 (chr8:11,515,050) distally and AFM205tb10 (chr8:14,724,000) proximally, shown as the hashed region labeled “Proximal” in Fig. 4. We saw no examples of segregation inconsistent with location of β-defensin repeat units at one or both of these sites on chromosome 8p. One crossover in child 135009 involved a breakpoint between β-defensin repeats already mapped to the proximal site by crossovers in other children (135005, 135006, and 135008); the placement of the breakpoint in 135009 is consistent with the results of other mapping data and allows further refinement of the proximal site to a smaller interval by placing a proximal bound at chr8:12,880,230. We therefore conclude that, in addition to the known location in distal 8p23.1, variable numbers of β-defensin repeats can also be found at a location in proximal 8p23.1 (Fig. 4), probably within the REPP repeat region, which shares considerable sequence identity with REPD. Furthermore, these same data also demonstrate that reassortment of repeat units between haplotypes by germ-line crossover is the primary mechanism for the generation of new CNV in β-defensin genes, with ≈6% of transmitted haplotypes (24 of 416) bearing new combinations of repeat units and ≈0.7% undergoing a change in the total haplotype copy number (3 of 416: 95% confidence interval 0.2–1.85%).
The 2 β-defensin CNV locations correspond approximately to the end points of a large inversion polymorphism flanked by olfactory receptor repeats (26). Published genetic mapping data (27), confirmed by our own observations in this work, suggest that the distal breakpoint of this inversion maps to the assembly interval chr8:6,741,958–8,805,729 and the proximal boundary to the interval chr8:11,515,050–12,786,453. This same analysis has shown that the inverted state of this polymorphism is not the result of a “once-only” event, such that all inverted haplotypes are descended from a common ancestor, but rather appears to have been independently generated and/or to have reverted several times in human populations. Recent fosmid sequencing data also suggest that different inversion alleles exist with different breakpoints (3). We also find little clear stratification in the multiallelic marker genotypes of β-defensin repeat units between proximal and distal loci. Indeed, repeats bearing identical combinations of alleles (e.g., EPEV-1 169 bp/EPEV-3 139 bp/rs5889219 125 bp) can be found at both loci, suggesting that they are related by recent descent from a common ancestral repeat and have thus been recently exchanged between the sites.
This polymorphic inversion spanning the region will cause apparent levels of recombination between the loci to be dependent on inversion genotype; pairing may be suppressed in inversion heterozygotes, and any recombination produces chromosome structures that will impair the viability of offspring. This will have 2 effects: first, the contribution of allelic recombination across 8p23.1 to de novo generation of copy number haplotypes will vary from family to family based on the parental inversion genotypes. Similarly, the copy number haplotype diversity between populations will be influenced by the frequency of the 8p23.1 inversion in the populations. This emphasizes the close relationship between the 2 forms of structural variation, and we predict that a higher frequency of the inversion allele will decrease the rate of generation of copy number haplotype diversity in a population, although other factors such as selection and drift will influence the level of diversity observed.
If the β-defensin CNV and the inversion polymorphism are coupled in this way, selection for β-defensin copy number may be one of the factors influencing the dynamics of the inversion in human populations. However, although the details are likely to be complicated, it is expected that a mechanism of selection that depends on aggregate diploid copy number summed over 2 linked but distinct loci, between which there is a high but individually variable frequency of recombination, will be weaker and less deterministic than at a single, stable locus. Nevertheless, a stable distribution can emerge if there is selection against extreme copy numbers (34). Although case-control association studies (20, 23) at present constitute the only current evidence that selection may be operating on human β-defensin copy number, we note that recombination between distal and proximal sites is expected in at least 4% of transmitted chromosomes (sex-average). Furthermore, among parental chromosomes shown by genetic mapping to lack β-defensin region repeats at 1 of the 2 locations, null sites are found at both the proximal and distal locations; for example, 10202 haplotype D is null at the proximal location, and 133302 haplotype D is null at the distal location (see Fig. S1), such that completely null (zero-copy) haplotypes could be created by simple allelic recombination between such chromosomes. By contrast, in typing >1,500 unrelated individuals for β-defensin copy number (18–20, 24), we have observed only a single example of an individual with 1 copy, suggesting that zero-copy haplotypes are infrequent and may be very rare. However, a zero-copy haplotype is most likely to be found in combination with a 2-copy haplotype and thus will escape unambiguous detection by diploid copy number measurement alone; we do not yet have sufficiently precise data on frequencies of haplotypes with different copy numbers at each site to establish whether the observed frequency of 1-copy individuals represents a significant departure from the predictions of a neutral model. In the particular case of the known 1-copy individual, ethical conditions agreed before collection of the DNA sample prevented further investigation of any associated phenotype.
Discussion
We have used a genetic approach to map and characterize the β-defensin CNV region by identifying recombinant chromosomes in large pedigrees and subsequent linkage analysis. This shows unequivocally 2 separate loci for the β-defensin CNV region separated by several megabases of single-copy sequence, mapping to locations consistent with REPP and REPD repeat regions. Physical mapping data from radiation hybrid panel Genebridge 4, using polymorphic short tandem repeats within the β-defensin repeats as novel sequence-tagged sites, confirms 2 separate β-defensin CNV loci (ref. 35 and unpublished data).
At both proximal and distal β-defensin CNV loci, we have examples of 0, 1, or 2 copies. Simple meiotic crossing over between the loci at a frequency equivalent to the genetic distance between them can generate new copy number haplotypes very rapidly, as observed in this study. Given the evidence for recurrent inversion between the proximal and distal β-defensin loci discussed above, 1 simple mechanism for the creation of diversity in the placement of β-defensin repeats would be for frequent flipping of this sequence between different inversion states to sometimes include, and sometimes exclude, β-defensin repeats at each location. Although the copy-variable β-defensins have generally been treated as outside the inverted segment (28, 29), it is evident that at least at the distal locus they are located between complex nested series of repeat sequences (REPD) (17, 26, 29, 30), of which different elements may sponsor a variety of exchanges, including recombination between inverted repeats at REPP to mediate a change in inversion status. Despite the dynamic nature of the defensin repeats, it is clear that there are few if any repeats of variant gene composition; measurements of copy number within the defensin CNV have shown that the copy numbers of all genes in the repeat vary coordinately (18, 21).
Initial DNA sequence analysis of DEFB4 and DEFB103 in these CEPH pedigrees, using segregation in recombinant offspring to map sequence variants to the proximal or distal sites (our unpublished work), has not yet revealed any variants strongly associated with location at either site or any variants predicted to have an effect on gene function. Variation at more than one site may provide at least a partial explanation for the failure of attempts to find SNP variation in strong linkage disequilibrium with β-defensin copy number (1). Analyses based on integrated information from SNPs at both sites may be more successful, but it remains likely that frequent germ-line recombination of repeat units into new haplotypes will mean that even combined 2-locus prediction of β-defensin copy number from flanking SNP genotypes will be too inaccurate to be of practical use. This unusual mechanism of frequent diversification by crossing over between distinct sites is unlikely to be responsible for diversification at many other CNV loci and may even be unique to the β-defensin CNV. The mechanism does imply, however, that if an association is seen between β-defensin copy number and a clinical phenotype, such as psoriasis (20), it is very unlikely that copy number is indirectly associated (as a proxy for a directly associated sequence variant) and more probable that in such an association the effect is attributable to defensin gene copy number itself.
Materials and Methods
Pedigrees and Linkage Analysis.
We analyzed β-defensin copy number in DNA samples from 26 of the 2- and 3-generation CEPH reference pedigrees, including a total of 208 offspring. CEPH segregation data for loci on chromosome 8 were obtained from the CEPH genotype database v.10 (www.cephb.fr/cephdb/). The genotypes for rs2203837 in family 104 created an anomalous crossover breakpoint, and, after individual retyping, the genotypes for individuals 10407 and 10408 were corrected; otherwise CEPH database segregation data were used without further analysis or correction. Linkage analysis of markers on chromosome 8 was performed by using the CHROMPIC option of CRIMAP (36) and corresponding physical locations inferred against the noninverted orientation of the region as represented in the UCSC March 2006 Genome Assembly (hg18, NCBI Build 36).
Copy Number and Multiallelic Marker Analysis.
Copy number of the β-defensin repeat unit in CEPH family members was determined by combining different and complementary methods of analysis. A PRT used comparative PCR between the heat-shock protein pseudogene HSPDP3 and an unlinked reference locus on chromosome 5 as described (24) to produce an estimate of copy number per diploid genome. We used analysis of 3 multiallelic loci within each repeat, both to confirm the copy numbers determined by PRT and to distinguish repeat units in segregation analysis. These multiallelic markers were 2 microsatellites, EPEV-1 and EPEV-3, and a multiallelic indel at chr8:7,363,807–7,363,976. The (AG)n microsatellite EPEV-1 was typed as described (18); typing the (AC)n microsatellite EPEV-3 used primers HEX-EPEV3F (Hex)-GATACTGTGAACTACAGATCAC and EPEV3R CTGCCCTGATTCAGTATTGAAC.
The multiallelic indel at chr8:7,363,807–7,363,976 includes the dinucleotide indel rs5889219 and for clarity will be here referred to simply as “rs5889219.” However, PCR using the primers FAM-5DELF (Fam)-AAACCAATACCCTTTCCAAG and 5DELR TCTTTTGTTTCAGATTCAGATG produces a product of variable length including not only the 2-bp deletion at rs5889219 but also a 5-bp deletion, thus yielding 3 variant products of lengths 126, 129, and 131 bp. These products have a biased strand composition and on capillary electrophoresis with fluorescent detection migrate with mobilities corresponding to 119, 123, and 125 nt, respectively, which are the allele designations used in our genotype data. By using the alternative reverse primer 5DELR2 CCCCAATTCATTAGGGTTTTT, we also included length variation from an (A)7–9 mononucleotide array, so that 4 distinct products can be detected with apparent lengths on capillary electrophoresis of 167, 169, 171, and 172 nt. Additional information from this alternative assay at rs5889219 was useful in clarifying the segregation pattern in some families.
The microsatellites EPEV-1 and EPEV-3 underwent polymerase slippage during PCR amplification. To minimize the effect on variant dosage ratios, the yield of each product had to be corrected for the effects of this slippage. Where different length products were well-separated by electrophoresis, the slippage products from distinct variants could be simply added to the main peaks. If the main peaks were adjacent to one another, it was assumed that the slippage process affected the different variants equally in calculations reconstructing the true yield of each product from electrophoretic profiles.
In some families where additional information was required we also typed the microsatellite EPEV-2 as described (19) and measured copy number using the alternative PRT system PRT107A using primers FAM-PRT107AF (5′ FAM-AGCCTCATTTAACTTTGGTGC) and PRT107AR (GGCTATGAAGCAATGGCCTA).
Integration and Analysis of Data.
The pattern of segregation for HSPDP3 PRT and the multiallelic markers was generally consistent within a pedigree. In some families or individuals, the apparent copy number deduced from PRT was different from the very clear copy numbers indicated by multiallelic markers. In nearly all of these cases, which we attribute to the poor condition of some DNA samples (on the basis that alternative DNA stocks when available resolved many of the anomalies), we were able to verify the correct copy number by typing PRT107A in parental DNA samples. In no case did we observe any consistent evidence for variant repeat units containing only some of the regions assayed. The dataset collated for all families is summarized as Fig. S1 and tabulated in Dataset S1. Our genetic mapping data allow us to define the orientation of the inversion polymorphism only in parents of offspring who show a crossover in the interval. For 1 parent (136202) this allowed us to demonstrate that the parent was homozygous for the inverted orientation; in all other parents from whom crossovers were defined, segregation data indicated homozygosity for the noninverted orientation. For simplicity, data on crossovers from 136202 have been shown using coordinates based on the noninverted orientation, but with designation of proximal and distal markers in the orientation appropriate for inverted chromosomes (Table S1).
Supplementary Material
Acknowledgments.
We are grateful to Raquel Palla for her work in developing the PRT107A system, to Alessandro Doria for helpful discussions during the early stages of this work, to Liz Bailes for help with linkage software, to Ketty Kessler and Celia May for radiation hybrid panel mapping, and to Jess Tyson and Tamsin Majerus for comments on the manuscript. This work was supported by a Wellcome Trust Bioarchaeology Fellowship (Grant 071024 to E.J.H.). S.A.B. is supported by a scholarship from the Ministry of Higher Education, Malaysia.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809073106/DCSupplemental.
References
- 1.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khaja R, et al. Genome assembly comparison identifies structural variants in the human genome. Nat Genet. 2006;38:1413–1418. doi: 10.1038/ng1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 5.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry GH, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): Low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanciulli M, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 10.Holloway K, Lawson VE, Jeffreys AJ. Allelic recombination and de novo deletions in sperm in the human beta-globin gene region. Hum Mol Genet. 2006;15:1099–1111. doi: 10.1093/hmg/ddl025. [DOI] [PubMed] [Google Scholar]
- 11.Lam KWG, Jeffreys AJ. Processes of de novo duplication of human alpha-globin genes. Proc Natl Acad Sci USA. 2007;104:10950–10955. doi: 10.1073/pnas.0703856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JA, Carvalho CMB, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 13.Turner DJ, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganz T. Defensins: Antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 15.Niyonsaba F, Ogawa H, Nagaoka I. Human beta-defensin-2 functions as a chemotactic agent for tumour necrosis factor-alpha-treated human neutrophils. Immunology. 2004;111:273–281. doi: 10.1111/j.0019-2805.2004.01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Candille SI, et al. A beta-defensin mutation causes black coat color in domestic dogs. Science. 2007;318:1418–1423. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollox EJ, Barber JCK, Brookes AJ, Armour JAL. Defensins and the dynamic genome: What we can learn from structural variation at human chromosome band 8p23.1. Genome Res. 2008;18:1686–1697. doi: 10.1101/gr.080945.108. [DOI] [PubMed] [Google Scholar]
- 18.Hollox EJ, Armour JAL, Barber JCK. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am J Hum Genet. 2003;73:591–600. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollox EJ, et al. Beta-defensin genomic copy number is not a modifier locus for cystic fibrosis. J Neg Res BioMed. 2005;4:9. doi: 10.1186/1477-5751-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollox EJ, et al. Psoriasis is associated with increased β-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groth M, et al. High-resolution mapping of the 8p23.1 beta-defensin cluster reveals strictly concordant copy number variation of all genes. Hum Mutat. 2008;29:1247–1254. doi: 10.1002/humu.20751. [DOI] [PubMed] [Google Scholar]
- 22.Barber JCK, et al. Duplications and copy number variants of 8p23.1 are cytogenetically indistinguishable but distinct at the molecular level. Eur J Hum Genet. 2005;13:1131–1136. doi: 10.1038/sj.ejhg.5201475. [DOI] [PubMed] [Google Scholar]
- 23.Fellermann K, et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armour JAL, et al. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007;35:e19. doi: 10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarroll SA. Copy-number analysis goes more than skin deep. Nat Genet. 2008;40:5–6. doi: 10.1038/ng0108-5. [DOI] [PubMed] [Google Scholar]
- 26.Giglio S, et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broman KW, et al. In: Science and Statistics: A Festschrift for Terry Speed. Goldstein DR, editor. Beachwood, OH: Institute of Mathematical Statistics; 2003. pp. 237–245. [Google Scholar]
- 28.Bosch N, et al. Characterization and evolution of the novel gene family FAM90A in primates originated by multiple duplication and rearrangement events. Hum Mol Genet. 2007;16:2572–2582. doi: 10.1093/hmg/ddm209. [DOI] [PubMed] [Google Scholar]
- 29.Sugawara H, et al. Complex low-copy repeats associated with a common polymorphic inversion at human chromosome 8p23. Genomics. 2003;82:238–244. doi: 10.1016/s0888-7543(03)00108-3. [DOI] [PubMed] [Google Scholar]
- 30.Taudien S, et al. Polymorphic segmental duplications at 8p23.1 challenge the determination of individual defensin gene repertoires and the assembly of a contiguous human reference sequence. BMC Genomics. 2004;5:92. doi: 10.1186/1471-2164-5-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young JM, et al. Extensive copy-number variation of the human Olfactory receptor gene family. Am J Hum Genet. 2008;83:228–242. doi: 10.1016/j.ajhg.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milunsky JM, Huang XL. Unmasking Kabuki syndrome: Chromosome 8p22–8p23.1 duplication revealed by comparative genomic hybridization and BAC-FISH. Clin Genet. 2003;64:509–516. doi: 10.1046/j.1399-0004.2003.00189.x. [DOI] [PubMed] [Google Scholar]
- 33.Milunsky JM, et al. A re-examination of the chromosome 8p22–8p23.1 region in Kabuki syndrome. Clin Genet. 2008;73:502–503. doi: 10.1111/j.1399-0004.2008.00983.x. [DOI] [PubMed] [Google Scholar]
- 34.Crow JF, Kimura M. An Introduction to Population Genetics Theory. New York: Harper and Row; 1970. [Google Scholar]
- 35.Gyapay G, et al. A radiation hybrid map of the human genome. Hum Mol Genet. 1996;5:339–346. doi: 10.1093/hmg/5.3.339. [DOI] [PubMed] [Google Scholar]
- 36.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.