Abstract
Principal components analysis and, more generally, the Singular Value Decomposition are fundamental data analysis tools that express a data matrix in terms of a sequence of orthogonal or uncorrelated vectors of decreasing importance. Unfortunately, being linear combinations of up to all the data points, these vectors are notoriously difficult to interpret in terms of the data and processes generating the data. In this article, we develop CUR matrix decompositions for improved data analysis. CUR decompositions are low-rank matrix decompositions that are explicitly expressed in terms of a small number of actual columns and/or actual rows of the data matrix. Because they are constructed from actual data elements, CUR decompositions are interpretable by practitioners of the field from which the data are drawn (to the extent that the original data are). We present an algorithm that preferentially chooses columns and rows that exhibit high “statistical leverage” and, thus, in a very precise statistical sense, exert a disproportionately large “influence” on the best low-rank fit of the data matrix. By selecting columns and rows in this manner, we obtain improved relative-error and constant-factor approximation guarantees in worst-case analysis, as opposed to the much coarser additive-error guarantees of prior work. In addition, since the construction involves computing quantities with a natural and widely studied statistical interpretation, we can leverage ideas from diagnostic regression analysis to employ these matrix decompositions for exploratory data analysis.
Keywords: randomized algorithms, singular value decomposition, principal components analysis, interpretation, statistical leverage
Modern datasets are often represented by large matrices since an m × n real-valued matrix A provides a natural structure for encoding information about m objects, each of which is described by n features. Examples of such objects include documents, genomes, stocks, hyperspectral images, and web groups. Examples of the corresponding features are terms, environmental conditions, temporal resolution, frequency resolution, and individual web users. In many cases, an important step in the analysis of such data is to construct a compressed representation of A that may be easier to analyze and interpret in light of a corpus of field-specific knowledge. The most common such representation is obtained by truncating the Singular Value Decomposition (SVD) at some number k ≪ min{m,n} terms. For example, Principal Components Analysis (PCA) is just this procedure applied to a suitably normalized data correlation matrix.
Recall the SVD of a general matrix A ∈ ℝm×n. Given A, there exist orthogonal matrices U = [u1u2…um] ∈ ℝm×m and V = [v1v2…vm] ∈ ℝn×n, where {ut}t=1m ∈ ℝm and {vt}t=1n ∈ ℝn are such that
where Σ ∈ ℝm×n, ρ = min{m,n}, σ1 ≥ σ2 ≥ … ≥ σρ ≥ 0, and diag(·) represents a diagonal matrix with the specified elements on the diagonal. Equivalently, A = UΣVT. The 3 matrices U, V, and Σ constitute the SVD of A (1)—the σi are the singular values of A and the vectors ui and vi are the i th left and the i th right singular vectors, respectively—and O(min{mn2,m2n}) time suffices to compute them.
The SVD is widely used in data analysis, often via methods such as PCA, in large part because the subspaces spanned by the vectors (typically obtained after truncating the SVD to some small number k of terms) provide the best rank-k approximation to the data matrix A. If k ≤ r = rank(A) and we define Ak = ∑t=1kσtutvtT, then
i.e., the distance, as measured by the Frobenius norm ∥·∥F, where ∥A∥F2 = ∑ijAij2, between A and any rank k approximation to A, is minimized by Ak (1).
Although the truncated SVD is widely used, the vectors ui and vi themselves may lack any meaning in terms of the field from which the data are drawn. For example, the eigenvector
being one of the significant uncorrelated “factors” or “features” from a dataset of people's features, is not particularly informative or meaningful. This fact should not be surprising. After all, the singular vectors are mathematical abstractions that can be calculated for any data matrix. They are not “things” with a “physical” reality.
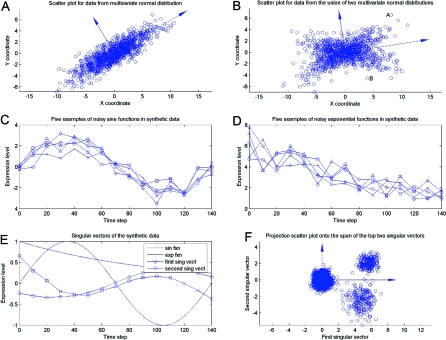
Nevertheless, data analysts often fall prey to a temptation for reification, i.e., for assigning a physical meaning or interpretation to all large singular components. In certain special cases, e.g., a dataset consisting of points drawn from a multivariate normal distribution on the plane, as in Fig. 1A, the principal components may be interpreted in terms of, e.g., the directions of the axes of the ellipsoid from which the data are drawn. In most cases, however, e.g., when the data are drawn from the union of 2 normals as in Fig. 1B, such reification is not valid. In this and other examples it would be difficult to interpret these directions meaningfully in terms of processes generating the data. Although reification is certainly justified in some cases, such an interpretative claim cannot arise from the mathematics alone, but instead requires an intimate knowledge of the field from which the data are drawn (2).
Fig. 1.
Applying the SVD to data matrices A. (A) 1,000 points on the plane, corresponding to a 1,000 × 2 matrix A (and the 2 principal components) drawn from a multivariate normal distribution. (B) 1,000 points on the plane (and the 2 principal components) drawn from a more complex distribution, in this case the union of 2 multivariate normal distributions. (C – F) A synthetic dataset considered by Wall et al. (3) to model oscillatory and exponentially decaying patterns of gene expression from Cho et al. (4), as described in the text. (C) Overlays of 5 noisy sine wave genes. (D) Overlays of 5 noisy exponential genes. (E) The first and second singular vectors of the data matrix (which account for 64% of the variance in the data), along with the original sine pattern and exponential pattern that generated the data. (F) Projection of the synthetic data on its top 2 singular vectors. Although the data cluster well in the low-dimensional space, the top 2 singular vectors are completely artificial and do not offer insight into the oscillatory and exponentially decaying patterns that generated the data.
To understand better the reification issues in modern biological applications, consider a synthetic dataset introduced by Wall et al. (3) to model oscillatory and exponentially decaying patterns of gene expression from Cho et al. (4). The data matrix consists of 14 expression level assays (columns of A) and 2,000 genes (rows of A), corresponding to a 2,000 × 14 matrix A. Genes have 1 of 3 types of transcriptional response: noise (1,600 genes); noisy sine pattern (200 genes); and noisy exponential pattern (200 genes). Fig. 1 C and D present the “biological” data, i.e., overlays of 5 noisy sine wave genes and five noisy exponential genes, respectively; Fig. 1E presents first and second singular vectors of the data matrix, along with the original sine pattern and exponential pattern that generated the data; and Fig. 1F shows that the data cluster well in the space spanned by the top 2 singular vectors, which in this case account for 64% of the variance in the data. Note, though, that the top 2 singular vectors display both oscillatory and decaying properties, and thus they are not easily interpretable as “latent factors” or “fundamental modes” of the original “biological” processes generating the data.
This is problematic when one is interested in extracting insight from the output of data analysis algorithms. For example, biologists are typically more concerned with actual patients than eigenpatients, and researchers can more easily assay actual genes than eigengenes. Indeed, after describing the many uses of the vectors provided by the SVD and PCA in DNA microarray analysis, Kuruvilla et al. (5) bluntly conclude that “While very efficient basis vectors, the (singular) vectors themselves are completely artificial and do not correspond to actual (DNA expression) profiles. … Thus, it would be interesting to try to find basis vectors for all experiment vectors, using actual experiment vectors and not artificial bases that offer little insight.”
These concerns about reification and interpreting linear combinations of data elements lie at the heart, conceptually and historically, of SVD-based data analysis methods. Recall that Spearman—a social scientist interested in models of human intelligence—invented factor analysis (2, 6). He computed the first principal component of a battery of mental tests, much as depicted in the first singular vector of Fig. 1B, and he invalidly reified it as an entity, calling it “g” or “general intelligence factor.” Subsequent principal components, such as the second singular vector of Fig. 1B, were reified as so-called “group factors.” This provided the basis for ranking of individuals on a single intelligence scale, as well as such dubious social applications of data analysis as the 11+ examinations in Britain and the involuntary sterilization of imbeciles in Virginia (2, 6). See Gould (2) for an enlightening and sobering discussion of invalid reification of singular vectors in a social scientific application of data analysis.
Main Contribution
We formulate and address this problem of constructing low-rank matrix approximations that depend on actual data elements. As with the SVD, the decomposition we desire (i) should have provable worst-case optimality and algorithmic properties; (ii) should have a natural statistical interpretation associated with its construction; and (iii) should perform well in practice. To this end, we develop and apply CUR matrix decompositions, i.e., low-rank matrix decompositions that are explicitly expressed in terms of a small number of actual columns and/or actual rows of the original data matrix. Given an m × n matrix A, we decompose it as a product of 3 matrices, C, U, and R, where C consists of a small number of actual columns of A, R consists of a small number of actual rows of A, and U is a small carefully constructed matrix that guarantees that the product CUR is “close” to A. Of course, the extent to which A ≈ CUR, and relatedly the extent to which CUR can be used in place of A or Ak in data analysis tasks, will depend sensitively on the choice of C and R, as well as on the construction of U.
To develop intuition about how such decompositions might behave, consider the previous pedagogical examples. In the dataset consisting of the union of 2 normal distributions, one data point from each normal distribution (as opposed to a vector sitting between the axes of the 2 normals) could be chosen. Similarly, in the synthetic dataset of Wall et al. (3), one could choose one sinusoid and one exponential function, as opposed to a linear combination of both. Finally, in the applications of Kuruvilla et al. (5), actual experimental DNA expression profiles, rather than artificial eigenprofiles, could be chosen. Thus, C and/or R can be used in place of the eigencolumns and eigenrows, but since they consist of actual data elements they will be interpretable in terms of the field from which the data are drawn (to the extent that the original data points and/or features are interpretable).
Prior CUR Matrix Decompositions
Within the numerical linear algebra community, Stewart developed the quasi-Gram–Schmidt method and applied it to a matrix and its transpose to obtain a CUR matrix decomposition (7, 8). Similarly, Goreinov, Tyrtyshnikov, and Zamarashkin developed a CUR matrix decomposition (a pseudoskeleton approximation) and related the choice of columns and rows to a “maximum uncorrelatedness” concept (9, 10).
Within the theoretical computer science community, much work has followed that of Frieze, Kannan, and Vempala (11), who randomly sample columns of A according to a probability distribution that depends on the Euclidean norms of those columns. If the number of chosen columns is polynomial in k and 1/ɛ (for some error parameter ɛ), then worst-case additive-error guarantees of the form
can be obtained, with high probability. Here PCA denotes the projection of A on the subspace spanned by the columns of C. Subsequently, Drineas, Kannan, and Mahoney (12) constructed an additive-error CUR matrix decomposition by choosing columns and rows simultaneously.* In 2 passes over the matrix A, they randomly construct a matrix C of columns, a matrix R of rows, and a matrix U such that
with high probability.
The additive-error algorithms of refs. 11 and 12 were motivated by resource-constrained computational environments, e.g., where the input matrices are extremely large or where only a very small fraction of the data is actually available, and in those applications they are appropriate. For example, this additive-error CUR matrix decomposition has been successfully applied to applications such as hyperspectral image analysis, recommendation system analysis, and DNA SNP analysis (13, 14). These additive-error matrix decompositions are, however, quite coarse in the worse case. Moreover, the insights provided by their sampling probabilities into the data are limited—the probabilities are often uniform due to data preprocessing, or they may correspond, e.g., simply to the degree of a node if the data matrix is derived from a graph.
Statistical Leverage and Improved Matrix Decompositions
To construct C (similarly R), we will compute an “importance score” for each column of A, and we will randomly sample a small number of columns from A by using that score as an importance sampling probability distribution. This importance score (see Eq. 3 below) depends on the matrix A, and it has a natural interpretation as capturing the “statistical leverage” or “influence” of a given column on the best low-rank fit of the data matrix. By preferentially choosing columns that exert a disproportionately large influence on the best low-rank fit (as opposed to procedures that sample columns that have larger Euclidean norm, or empirical variance, as in prior work), we will ensure that CUR is nearly as good as Ak at capturing the dominant part of the spectrum of A. In addition, by choosing “high statistical-leverage” or “highly influential” columns, we can leverage ideas from diagnostic regression analysis to apply CUR matrix decompositions as a tool for exploratory data analysis.
To motivate our choice of importance sampling scores, recall that we can express the j th column of A (denoted by Aj) exactly as
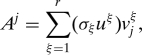 |
where r = rank(A) and where vjξ is the j th coordinate of the ξ th right singular vector. That is, the j th column of A is a linear combination of all the left singular vectors and singular values, and the elements of the j th row of V are the coefficients. Thus, we can approximate Aj as a linear combination of the top k left singular vectors and corresponding singular values as
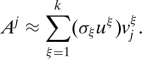 |
Since we seek columns of A that are simultaneously correlated with the span of all top k right singular vectors, we then compute the normalized statistical leverage scores:
 |
for all j = 1,…,n. With this normalization, it is straightforward to show that πj ≥ 0 and that ∑nj = 1 πj = 1, and thus that these scores form a probability distribution over the n columns.
Our main algorithm for choosing columns from a matrix—we will call it ColumnSelect—takes as input any m × n matrix A, a rank parameter k, and an error parameter ɛ, and then performs the following steps:
1. Compute v1,…,vk (the top k right singular vectors of A) and the normalized statistical leverage scores of Eq. 3.
2. Keep the jth column of A with probability pj = min{1,cπj}, for all j ∈{1,…,n}, where c = O(klogk/ɛ2).
3. Return the matrix C consisting of the selected columns of A.
With this procedure, the matrix C contains c′ columns, where c′ ≤ c in expectation and where c′ is tightly concentrated around its expectation. The computation of the column leverage scores uses the top k right singular vectors of A, and this computation is the bottleneck in the running time of ColumnSelect. It can be performed in time linear in the number of nonzero elements of the matrix A times a low-degree polynomial in the rank parameter k (1). We have proven that, with probability at least 99%, this choice of columns satisfies
where PC denotes a projection matrix onto the column space of C.† See ref. 15 for the proof of Eq. 4, which depends crucially on the use of Eq. 3. In some applications, this restricted CUR decomposition, A ≈ PCA = CX, where X = C+A, is of interest.
In other applications, one wants such a CUR matrix decomposition in terms of both columns and rows simultaneously. Our main algorithm computing a CUR matrix decomposition—we will call it AlgorithmCUR—is illustrated in supporting information (SI) Appendix, Fig. S0. This algorithm takes as input any m × n matrix A, a rank parameter k, and an error parameter ɛ, and then it performs the following steps:
1. Run ColumnSelect on A with c = O(k logk /ɛ2) to choose columns of A and construct the matrix C.
2. Run ColumnSelect on AT with r = O(k logk /ɛ2) to choose rows of A (columns of AT) and construct the matrix R.
3. Define the matrix U as U = C+AR+, where X+ denotes a Moore–Penrose generalized inverse of the matrix X (17).
As with our algorithm for selecting columns, the running time of AlgorithmCUR is dominated by computation of the column and row leverage scores. For this choice of C, U, and R, we will prove that
with probability at least 98%.‡. First, since U = C+AR+, it immediately follows that
Adding and subtracting CC+A and applying the triangle inequality for the Frobenius norm, we get
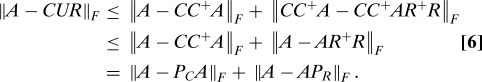 |
Inequality (6) follows since CC+ is a projection matrix and thus does not increase the Frobenius norm, and the last equality follows since PC = CC+ and similarly PR = R+R. Since AlgorithmCUR chooses columns and rows by calling ColumnSelect on A and AT, respectively, Eq. 5 follows by 2 applications of Eq. 4. Note that r = c for AlgorithmCUR, and also that the failure probability for AlgorithmCUR is at most twice the failure probability of ColumnSelect, since the latter algorithm may fail when applied to columns or when applied to rows, independently.
Although one might like to fix a rank parameter k and choose k columns and/or rows deterministically according to some criterion—e.g., such as to define a parallelepiped of maximal volume over all such parallelepipeds, or to span a subspace that “captures” a maximal amount of variance from A over all such subspaces—most such criteria would lead to intractable combinatorial optimization problems (9, 10, 18). Thus, AlgorithmCUR takes advantage of oversampling (choosing slightly more than k columns) and randomness as computational resources to obtain its strong provable approximation guarantees.
Note that the quantities in Eq. 3 are, up to scaling, equal to the diagonal elements of the so-called “hat matrix,” i.e., the projection matrix onto the span of the top k right singular vectors of A (19, 20). As such, they have a natural statistical interpretation as a “leverage score” or “influence score” associated with each of the data points (19–21). In particular, πj quantifies the amount of leverage or influence exerted by the j th column of A on its optimal low-rank approximation. Furthermore, these quantities have been widely used for outlier identification in diagnostic regression analysis (22, 23). Thus, using these scores to select columns not only is crucial for our improved worst-case bounds but also aids in exploratory data analysis.
Diagnostic Data Analysis Applications
Implementation of AlgorithmCUR is straightforward. We have applied this algorithm to data analysis problems in several application domains: Internet term-document data analysis (see Fig. 2 and SI Appendix, Figs. S2–S5); genetics (see Fig. 3); and social science (see SI Appendix, Fig. S1). In practice, we typically only need to sample a number of columns and/or rows that is a small constant, e.g., between 2 and 4, times the input rank parameter k. In addition, not only can we perform common data analysis tasks (such as clustering and classification) for which the basis provided by truncating the SVD is often used, but we can also use the normalized leverage scores to explore the data and identify whether there are any disproportionately “important” data elements. (Note that since CUR decompositions are low-rank approximations that use information in the top k singular subspaces, their domain of applicability is not expected to be broader than that of the SVD.)
Fig. 2.
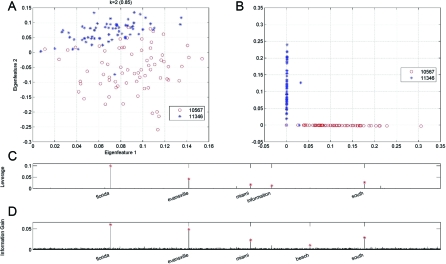
Application of ALGORITHMCUR to Internet term-document data. The matrix consists of 139 documents from TechTC on 2 topics: US:Indiana:Evansville (id:10567) and US:Florida (id:11346). (A) A projection of the documents on the 2D space spanned by the top 2 eigenterms. (B) A projection of the documents on the best 2D approximation to the space spanned by the 5 actual terms with the highest statistical leverage scores of Eq. 3. (C) The statistical leverage scores of the 15,170 terms, with the top 5 terms highlighted. (D) The information gain statistic of the 15,170 terms, with the top 5 terms highlighted.
Fig. 3.
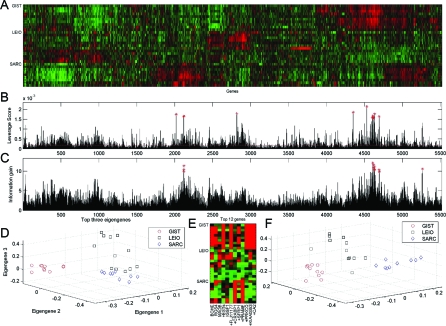
Application of ALGORITHMCUR to cancer microarray data: gene expression patterns of patients with soft tissue tumors analyzed with cDNA microarrays. (A) Raster plot of the data. (B) Statistical leverage scores for each gene. Red stars indicate the 12 genes with the highest leverage scores, and the red dashed line indicates the uniform leverage scores. (C) Information gain for each gene, with the highest scoring genes marked. (D) A projection of the 31 patients in the subspace spanned by the top 3 eigengenes of the full data matrix. (E) Raster plot of the 31 patients with respect to the top 12 genes from B. A (+) sign in front of the name of a gene indicates that this gene was also selected by the (supervised) information gain metric. (F) A 3D plot of the 31 patients in the span of the genes of E.
Internet term-document data are a common application of SVD-based techniques, often via latent semantic analysis (24, 25). The Open Directory Project (ODP) (26) is a multilingual open content directory of World Wide Web links. TechTC (Technion Repository of Text Categorization Datasets) is a publicly available benchmark set of term-document matrices from ODP with varying categorization difficulty (27) (Table 1). Each matrix of the TechTC dataset encodes information from ≈150 documents from 2 different ODP categories. To illustrate our method, we focused on 4 datasets such that the documents clustered well into 2 classes when projected in a low-dimensional space spanned by the top few left singular vectors.§ (See Table 1 for a description of the data and also Fig. 2 and SI Appendix, Figs. S3–S5.)
Table 1.
Four TechTC matrices that cluster well into the correct 2 topics by using k-means in the best low-dimensional space
| Categories (numeric ID, description) | no. docs × no. terms | k | PCC | 1/(no. terms) | High-leverage terms |
|---|---|---|---|---|---|
| 10567, US: Indiana: Evansville | 139 × 15170 | 2 | .85 | .000066 | florida (.099482), evansville (.042291), |
| 11346, US: Florida | south (.026892), miami (.016890), | ||||
| information (.011792) | |||||
| 11346, US: Florida | 125 × 14392 | 2 | .97 | .000069 | florida (.097158), nanaimo (.085653), |
| 22294, Canada: British Columbia: Nanaimo | south (.026414), miami (.014415), | ||||
| contact (.007828) | |||||
| 20186, US: Texas: Dallas | 130 × 12708 | 2 | .90 | .000079 | dallas (.079332), nanaimo (.071752), |
| 22294, Canada: British Columbia: Nanaimo | information (.007878), texas (.007788), | ||||
| contact (.007616) | |||||
| 22294, Canada: British Columbia: Nanaimo | 127 × 10012 | 2 | .88 | .000100 | nanaimo (.055424), taiwan (.019860), |
| 25575, Asia: Taiwan: Business and Economy | megahome (.004304), contact (.004113), | ||||
| distiller (.003906) |
The first 3 columns indicate the 2 topics of the documents in each matrix and its dimensions (each topic has a numeric ID, as well as a short description). The fourth column indicates our choice for the number k of significant principal components in the data. The fifth column indicates the Pearson correlation coefficient (PCC) between the ground truth and the 2 clusters returned by applying k-means clustering on the projected data. The sixth column indicates the uniform leverage scores for each term, i.e., each term is assigned the same score. The last column indicates the 5 terms with the highest leverage scores for each matrix, as well as their corresponding scores in parentheses. (Notice that the scores are many orders of magnitude larger than the uniform score.)
For example, consider the collection of 139 documents (each described with respect to 15,170 terms) on 2 topics: US:Florida (id:11346) and US:Indiana:Evansville (id:10567). The first topic has 71 documents, and the second has 68; the topics names are descriptive; and the sparsity of the associated document-term matrix is 2.1%. Projecting the documents on the top 2 eigenterms and then running k-means clustering on the projected data leads to a clustering which has a Pearson correlation coefficient of 0.85 with the (provided) ground truth, is illustrated in Fig. 2A. This class separation implies that the documents are semantically well-represented by low-rank approximation via the SVD, even though they are not numerically low rank. The singular vectors, however, are dense; they contain negative entries; and they are not easily interpretable in terms of natural languages or the ODP hierarchy.
Not only does a CUR matrix decomposition capture the Frobenius norm of the matrix (data not shown), but it can be used to cluster the documents. Fig. 2B shows the 139 documents projected on the best rank 2 approximation to the subspace spanned by the top five “highest-leverage” terms. (Two was chosen as the dimensionality of the low-dimensional space since the documents belong to one of 2 categories—the slowly decaying spectrum provides no guidance in this case. Five was chosen as the number of selected columns based on an analysis of the leverage scores, as described below, and other choices yielded worse results.) In this case, the class separation is quite pronounced—the Pearson correlation coefficient is 0.94, which is improved since CUR provides a low-dimensional space that respects the sparsity in the data. Of course, the data are much more axis-aligned in this low-dimensional space, largely because this space is the span of a small number of actual/interpretable terms, each of which is extremely sparse.
As an example of how we can leverage ideas from diagnostic regression analysis to explore the data, consider Fig. 2C, which shows the statistical leverage scores of all 15,170 terms. The leverage scores of the top 5 terms—florida (.099482), evansville (.042291), south (.026892), miami (.016890), and information (.011792), as seen in Table 1, are orders of magnitude larger than the uniform leverage scores, equal to 1/n, where n = 15,170 here. This, coupled with the obvious relevance of these terms to the task at hand, suggests that these 5 terms are particularly important or influential in this low-dimensional clustering/classification problem. Further evidence supporting this intuition follows from Fig. 2D, which shows the information gain (IG) statistic for each of the 15,170 terms, again with the top 5 terms highlighted. (Recall that the IG for the i th term is defined as IGi = |fi,1 − fi,2|, where fi,1 is the frequency of the i th term within the documents of the first category, and fi,2 is the frequency of the i th term within the documents of the second category. In particular, note that it is a supervised metric, i.e., the topic of each document is known prior to computing it.)
The truncated SVD has also been applied to gene expression data (3, 5, 28, 29) in systems biology applications (where one wants to understand the relationship between actual genes) and in clinical applications (where one wants to classify tissue samples from actual patients with and without a disease). See Fig. 3A for a raster plot of a typical such dataset (30), consisting of m = 31 patients with 3 different cancer types [gastrointestinal stromal tumor (GIST), leiomyosarcoma (LEIO), and synovial sarcoma (SARC)] with respect to n = 5520 genes.¶ For this data set, the top 2 eigengenes/eigenpatients capture 37% of the variance of the matrix, and reliable clustering can be performed in the low-dimensional space provided by the truncated SVD, as illustrated in Fig. 3D. That is, there is clear separation between the 3 different cancer types, and running k-means on the projected data perfectly separates the 3 different classes of patients.
Fig. 3B shows the statistical leverage scores of the 5,520 genes, with red stars indicating the top 12 genes whose leverage score is well above the average uniform leverage score of approximately 0.18 · 10−3. (Here, 12 was chosen by trial-and-error, and applying the IG metric to select the 12 genes that are most differentiating between the 3 cancer types leads to selecting 8 of these 12 high-leverage genes.) Again, the leverage scores are quite nonuniform; random sampling according to the probabilities of Eq. 3 easily recovers the highest scoring genes; and these genes are the most interesting and useful data points for the application of interest. Fig. 3E illustrates a raster plot of those 12 genes—notice that we select genes that are overexpressed for the gastrointestinal stromal tumor and the synovial sarcoma. The fact that our leverage scores pick these highly influential genes explains why the 3 different cancer types are reliably separated in the space spanned by these 12 genes, as demonstrated by Fig. 3F. That is, running k-means by using only these 12 genes leads to a perfect clustering of the 31 patients to their respective cancer types since these actual genes exert a disproportionately large influence on the best low-rank fit of the data matrix (presumably in part for reasons implicit in the data preparation).
Finally, and crucially for medical interpretation of this analysis in clinical settings, some of the selected genes are well-known to have expression patterns that correlate with various cancers, and thus they can be further studied in the lab. Most notably, PRKCQ has been associated with gastrointestinal stromal tumors; PRAME has been known to confer growth or survival advantages in human cancers; BCHE has been associated with colorectal carcinomas and colonic adenocarcinomas; and SFRP1 and CRABP1 have been associated with a wide range of cancers. There is no need for the reification of the artificial singular directions that offer little insight into the underlying biology (5).
From the perspective of diagnostic regression analysis, the extreme nonuniformity in “importance” of individual data elements is quite surprising. In those applications, one is taught to be wary of data points with leverage greater than 2 or 3 times the average value (20), investigating them to see whether they are errors or outliers. Moreover, there is no a priori reason that the nonuniformity in this statistic should correlate with nonuniformity in a supervised metric like information gain. In our experience, however, this nonuniformity is not uncommon. (It is, of course, far from ubiquitous. For example, Congressional roll call data (31, 32) are much more homogeneous; if k = 2 and n is the number of representatives, the highest leverage score for any representative is only 1.38k/n.) Most often, this phenomenon arises in cases where SVD-based methods are used for computational convenience, rather than because the statistical assumptions underlying its use are satisfied by the data. This suggests the use of these “leverage scores” more generally in modern massive dataset analysis. Intuitively, conditioned on being reliable, more “outlier-like” data points may be the most important and informative.
Conclusion
Although the SVD per se cannot be blamed for its misapplication, the desire for interpretability in data analysis is sufficiently strong so as to argue for interpretable low-rank matrix decompositions. Even when an immediate application of the truncated SVD is not appropriate, the low-rank matrix approximation thereby obtained is a fundamental building block of some of the most widely used data analysis methods, including PCA, multidimensional scaling, factor analysis, and many of the recently developed techniques to extract nonlinear structure from data. Thus, although here we have focused on the use of CUR matrix decompositions for the improved interpretability of SVD-based data analysis methods, we expect that their promise is much more general.
Supplementary Material
Acknowledgments.
We thank Deepak Agarwal, Christos Boutsidis, Gene Golub, Ravi Kannan, Kenneth Kidd, Jon Kleinberg, and Michael Jordan for useful discussions and constructive comments. This work was supported by National Science Foundation CAREER Award and a NeTS-NBD award (to P.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
J.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803205106/DCSupplemental.
The algorithms of refs. 11 and 12 provide worst-case additive-error guarantees since the “additional error” in Eqs. 1 and 2 is ɛ ∥A∥F, which is independent of the “base error” of ∥A − Ak∥F. This should be contrasted with a relative-error guarantee of the form Eq. 4, in which the “additional error” is ɛ ∥A − Ak∥F, for ɛ > 0 arbitrarily small, and thus the total error is bounded by (1 + ɛ) ∥A − Ak∥F. It should also be contrasted with a constant-factor guarantee of the form Eq. 5, in which the additional error is γ ∥A − Ak∥F, for some constant γ.
The randomness and failure probability in COLUMNSELECT and ALGORITHMCUR are over the choices made by the algorithm and not over the input data matrix. The quality of approximation bound Eq. 4 holds for any input matrix A, regardless of how A is constructed; its proof relies on matrix perturbation theory (15, 16). The arbitrarily chosen failure probability can be set to any δ > 0 by repeating the algorithm O(log(1/δ)) times and taking the best of the results.
This can be improved to 1 + ɛ by using a somewhat more complicated algorithm (15)
As is common with term-document data, the TechTC matrices are very sparse, and they are not numerically low-rank. For example, the top 2.5% and 5% of the nonzero singular values of these matrices capture (on average) 5.5% and 12.5% of the Frobenius norm, respectively. For data sets in which a low-dimensional space provided by the SVD failed to capture the category separation, CUR matrix decompositions performed correspondingly poorly.
The original data contained 46 patients from 6 different cancer types; here, we consider a subset of 31 patients with 1 of 3 cancer types. Linear dimensionality reduction techniques such as SVD could accurately separate patients suffering from gastrointestinal stromal tumor, leiomyosarcoma, and synovial sarcoma. Including patients suffering from the remaining 3 cancer types (liposarcoma, malignant fibrous histiocytoma, and schwannoma) led to several inaccurate classifications, even using all 5,520 genes. This is consistent with the finding that hierarchical clustering in all 5,520 genes resulted in 5 (instead of 6) clusters, with several incorrect assignments (30). To illustrate the use of low-rank CUR matrix decompositions, we focused on the subset of the data that were amenable to analysis with the truncated SVD. Similar, but noisier, results are obtained on the full dataset.
References
- 1.Golub GH, Van Loan CF. Matrix Computations. Baltimore: Johns Hopkins Univ Press; 1996. [Google Scholar]
- 2.Gould SJ. The Mismeasure of Man. New York: W. W. Norton; 1996. [Google Scholar]
- 3.Wall ME, Rechtsteiner A, Rocha LM. Singular value decomposition and principal component analysis. In: Berrar DP, Dubitzky W, Granzow M, editors. In A Practical Approach to Microarray Data Analysis. Boston: Kluwer; 2003. pp. 91–109. [Google Scholar]
- 4.Cho RJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 5.Kuruvilla FG, Park PJ, Schreiber SL. Vector algebra in the analysis of genomewide expression data. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-research0011. research0011.1-0011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blinkhorn SF. Burt and the early history of factor analysis. In: Mackintosh NJ, editor. In Cyril Burt: Fraud or Framed? New York: Oxford Univ Press; 1995. [Google Scholar]
- 7.Stewart GW. Four algorithms for the efficient computation of truncated QR approximations to a sparse matrix. Numer Math. 1999;83:313–323. [Google Scholar]
- 8.Berry MW, Pulatova SA, Stewart GW. College Park, MD: University of Maryland; 2004. Computing sparse reduced-rank approximations to sparse matrices. Technical Report UMIACS TR-2004-32 CMSC TR-4589. [Google Scholar]
- 9.Goreinov SA, Tyrtyshnikov EE, Zamarashkin NL. A theory of pseudoskeleton approximations. Linear Algebra and Its Applications. 1997;261:1–21. [Google Scholar]
- 10.Goreinov SA, Tyrtyshnikov EE. The maximum-volume concept in approximation by low-rank matrices. Contemp Math. 2001;280:47–51. [Google Scholar]
- 11.Frieze A, Kannan R, Vempala S. Fast Monte-Carlo algorithms for finding low-rank approximations. J ACM. 2004;51(6):1025–1041. [Google Scholar]
- 12.Drineas P, Kannan R, Mahoney MW. Fast Monte Carlo algorithms for matrices III: Computing a compressed approximate matrix decomposition. SIAM J Comput. 2006;36:184–206. [Google Scholar]
- 13.Mahoney MW, Maggioni M, Drineas P. Proceedings of the 12th Annual ACM SIGKDD Conference. New York: Association for Computing Machinery; 2006. Tensor-CUR decompositions for tensorbased data; pp. 327–336. [Google Scholar]
- 14.Paschou P, et al. Intra- and interpopulation genotype reconstruction from tagging SNPs. Genome Res. 2007;17:96–107. doi: 10.1101/gr.5741407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drineas P, Mahoney MW, Muthukrishnan S. Relative-error CUR matrix decompositions. SIAM J Matrix Anal Appl. 2008;30:844–881. [Google Scholar]
- 16.Stewart GW, Sun JG. Matrix Perturbation Theory. New York: Academic; 1990. [Google Scholar]
- 17.Nashed MZ, editor. Generalized Inverses and Applications. New York: Academic; 1976. [Google Scholar]
- 18.Garey MR, Johnson DS. Computers and Intractability: A Guide to the Theory of NP-Completeness. New York: Freeman; 1979. [Google Scholar]
- 19.Hoaglin DC, Welsch RE. The hat matrix in regression and ANOVA. Am Stat. 1978;32:17–22. [Google Scholar]
- 20.Chatterjee S, Hadi AS. Sensitivity Analysis in Linear Regression. Wiley: NewYork; 1988. [Google Scholar]
- 21.Chatterjee S, Hadi AS. Influential observations, high leverage points, and outliers in linear regression. Stat Sci. 1986;1:379–393. [Google Scholar]
- 22.Velleman PF, Welsch RE. Efficient computing of regression diagnostics. Am Stat. 1981;35:234–242. [Google Scholar]
- 23.Chatterjee S, Hadi AS, Price B. Regression Analysis by Example. New York: Wiley; 2000. [Google Scholar]
- 24.Deerwester ST, Dumais ST, Furnas GW, Landauer TK, Harshman R. Indexing by latent semantic analysis. J Am Soc Inf Sci. 1990;41:391–407. [Google Scholar]
- 25.Landauer TK, Laham D, Derr M. From paragraph to graph: Latent semantic analysis for information visualization. Proc Natl Acad Sci USA. 2004;101:5214–5219. doi: 10.1073/pnas.0400341101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Open Directory Project. Available at http://www.dmoz.org/. Accessed October 8, 2007.
- 27.Gabrilovich E, Markovitch S. Proceedings of the 21th International Conference on Machine Learning. New York: Association for Computing Machinery; 2004. Text categorization with many redundant features: using aggressive feature selection to make SVMs competitive with C4.5; pp. 41–48. [Google Scholar]
- 28.Alter O, Brown PO, Botstein D. Singular value decomposition for genomewide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holter NS, et al. Fundamental patterns underlying gene expression profiles: Simplicity from complexity. Proc Natl Acad Sci USA. 2000;97:8409–8414. doi: 10.1073/pnas.150242097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen T, et al. Molecular characterisation of soft tissue tumours: A gene expression study. Lancet. 2002;359:1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- 31.Poole KT, Rosenthal H. Patterns of congressional voting. Am J Political Sci. 1991;35:228–278. [Google Scholar]
- 32.Porter MA, Mucha PJ, Newman MEJ, Warmbrand CM. A network analysis of committees in the U.S. House of Representatives. Proc Natl Acad Sci USA. 2005;102:7057–7062. doi: 10.1073/pnas.0500191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.