Abstract
RNA viruses exhibit increased mutation frequencies relative to other organisms. Recent work has attempted to exploit this unique feature by increasing the viral mutation frequency beyond an extinction threshold, an antiviral strategy known as lethal mutagenesis. A number of novel nucleoside analogs have been designed around this premise. Herein, we review the quasispecies nature of RNA viruses and survey the antiviral, biological and biochemical characteristics of mutagenic nucleoside analogs, including clinically-used ribavirin. Biological implications of modulating viral replication fidelity are discussed in the context of translating lethal mutagenesis into a clinically-useful antiviral strategy.
Keywords: error catastrophe, error threshold, extinction catastrophe, lethal mutagenesis, mutational robustness, nucleoside analogs, quasispecies, ribavirin
Although significant effort has been expended in developing effective clinical therapies for viral infections, the number of approved antiviral drugs is quite limited. A comprehensive review in 2004 identified only 37 drugs licensed for the treatment of viral infections, 19 of which were for the treatment of HIV infection [1]. Recent approvals have expanded the list of antivirals targeting HIV to 25 distinct chemical entities [201]. Other targets were limited to HBV, HCV, herpesvirus and influenza virus [1]. Of the anti-viral compounds currently in clinical use, only one, the nucleoside analog ribavirin, exhibits broad-spectrum antiviral activity against both DNA- and RNA-based viruses [2]. The limited number of available treatments as well as the emergence of new viral diseases has led to a need for novel antiviral agents. Furthermore, RNA viruses have extraordinarily high mutation rates (discussed later) that can often lead to the rapid emergence of virus strains resistant to clinically employed antivirals.
Recently, a new antiviral strategy aimed at exploiting the elevated mutation rate of RNA viruses has begun to be explored. ‘Lethal mutagenesis’ aims to increase the viral mutation rate beyond a biologically-tolerable threshold, resulting in reduced viral fitness and, potentially, extinction. Importantly, resistance to lethal mutagenesis can be generated through an increase in viral replication fidelity, an adaptation that has been shown to affect viral spread and pathogenesis, and should result in virus populations that are less able to evolve resistance against classical antivirals targeting viral enzymes and structural proteins [3,4]. Thus, lethal mutagenesis may not only be effective in reducing viral load, but may also result in the selection of virus populations with reduced pathogenicity and capacity for resistance. Herein we review progress on development of lethal mutagenesis as a novel antiviral strategy, with an eye towards future milestones that may allow for effective translation of this strategy into a clinical setting.
Biological basis of lethal mutagenesis
RNA viruses replicate with a high mutation frequency
RNA viruses display extraordinarily high mutation frequencies compared with DNA-based organisms. The mutation rate for RNA viruses is generally accepted to be in the range of 10−5 mutations per incorporated nucleotide, compared with 10−8–10−11 per base pair for DNA-based organisms [5]. This results in approximately one mutation per genome per replication cycle for RNA viruses, and 0.1 mutations per genome per replication cycle for retroviruses. The equivalent value for DNA-based microbes is 0.003 per genome per replication cycle, even though the genome size is orders of magnitude larger [6]. Clearly, RNA viruses are subject to extensive mutation during replication. This phenomenon is likely due to the absence of a proofreading activity associated with most viral RNA-dependent RNA polymerases (RdRp), the enzymes responsible for replication of RNA genomes [7], although recent work suggests that coronaviruses may possess some proofreading activity [8] (which may be a requirement for RNA viruses to have a genome longer than 10,000 nucleotides).
The extremely high mutation rate of RNA viruses leads to the generation of a quasispecies, an extremely heterogeneous population that hovers around a most-fit ‘master sequence’ [5,9–11]. Most individual genomes in the population will have a sequence differing from the consensus sequence by one or more nucleotide changes. Studies on the chemical mutagenesis of poliovirus (PV) and vesicular stomatitis virus demonstrated that there was an upper limit to the increase in mutation frequency of RNA viruses [12]. Mutation frequencies at a single site could only be increased 1.1–2.8-fold, even in the presence of a mutagen that was sufficient for a 99% reduction in virus viability. Loeb and colleagues, utilizing the retrovirus HIV, also demonstrated significant decreases in virus viability when the mutation frequency was increased only modestly [13]. Serial passage of HIV in the presence of a mutagen that increased the mutation rate only threefold was sufficient for extinction of the virus population. Subsequently, Loeb and Mullins coined the term ‘lethal mutagenesis’ to describe an antiviral strategy based on increasing the viral mutation rate to a level beyond which the population is no longer genetically viable [14,15].
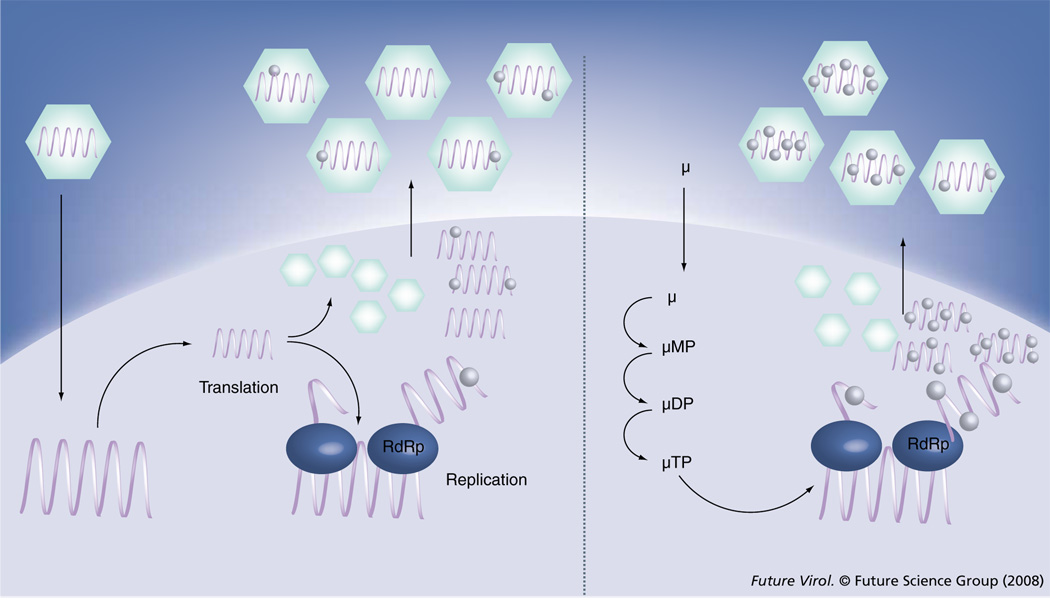
The mechanism of lethal mutagenesis is illustrated for a typical single-stranded, positive-sense RNA virus in Figure 1, but can similarly be applied to RNA viruses with other replication strategies, as well as retroviruses (although retroviral mutation must occur either through incorporation of a mutagen into DNA by reverse transcriptase, or through incorporation into viral RNA by cellular RNA polymerase II). Viral RNA is replicated following delivery of the genome to the cytoplasm and production of necessary viral proteins, including the RdRp. Replication is error-prone, resulting in the generation of approximately one mutation per progeny genome (left panel). Treatment with a nucleoside mutagen (µ) allows for intracellular accumulation of mono-, di- and triphosphorylated forms (µMP, µDP and µTP). The triphosphory lated analog µTP functions as an ambiguous substrate for the RdRp, inducing additional mutations into the progeny genomes. This results in more deleterious mutations per genome and therefore a lower average fitness of the viral population.
Figure 1. Lethal mutagenesis as an antiviral strategy against RNA viruses.
Treatment with a nucleoside mutagen (µ) increases the number of mutations per genome, decreasing the fitness of the progeny viral population. µMP, µDP and µTP represent mono-, di- and triphosphorylated forms of the nucleoside, respectively. RdRp: RNA-dependent RNA polymerase.
Theory of lethal mutagenesis
Whereas the quasispecies theory has been invoked to explain the antiviral activity of mutagens, the theoretical underpinnings of quasispecies theory, as it applies to the evolution of virus populations, are still quite thin and subject to heated debate [16–19]. Recent interest in lethal mutagenesis as an antiviral strategy has stimulated efforts to explain the scientific theory behind lethal mutagenesis to virologists.
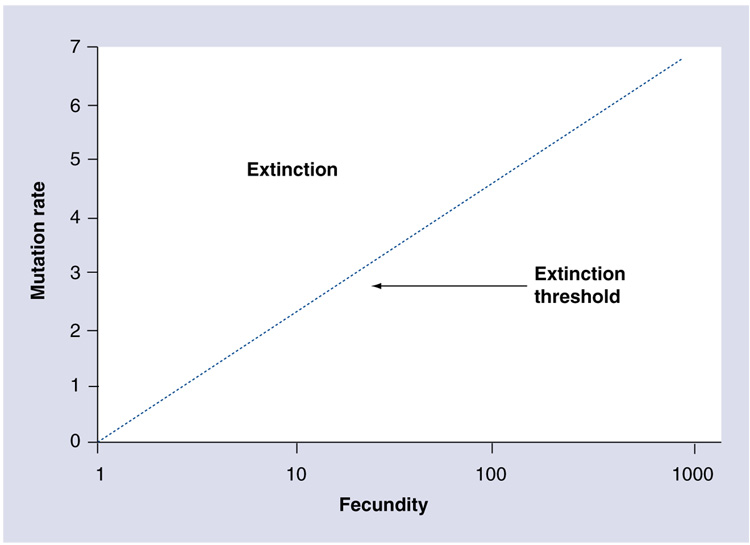
Bull, Sanjuan and Wilke have described the theory of lethal mutagenesis [20]. Population genetics theory shows that the extinction threshold is dependent not just upon mutation rate but also fecundity, the maximal number of progeny released and therefore a measure of viral fitness (Figure 2) [20]. Experimental support for this concept was demonstrated in foot-and-mouth disease virus, as low viral load or low fitness increased the frequency of viral extinction [21]. Thus, lethal mutagenesis has both genetic and environmental components. Extinction occurs when the mutation rate reduces average viral fitness below the point where the population can replenish itself.
Figure 2. The extinction threshold is dependent upon mutation rate and fecundity.
The dotted line represents the threshold between population extinction (above) and population survival (below). The log-linear relationship indicates that large increases in fecundity are required to overcome small changes in mutation rate.
Adapted from [20]
Importantly, Bull, Sanjuan and Wilke note that there is no distinct genetic signature of lethal mutagenesis to differentiate it from mutagenesis that does not lead to lethality [20]. This has led to difficulty in reconciling theoretical breakthroughs with empirical observations of virus populations subjected to mutagenic agents, particularly since mutation rate and fecundity are difficult parameters to accurately assess in vivo. Additionally, it has been proposed that defective genomes may function through ‘lethal defection’ to drive viral populations to extinction at lower mutation rates [22].
An important concept to arise from this look at RNA virus population genetics is the idea of ‘survival of the flattest’ [23–25]. Using ‘digital organisms’, Wilke et al. showed that high fitness populations can be outcompeted by lower fitness populations at high mutation rates, if the lower fitness population lies in a flatter region of the fitness landscape [24,26]. In other words, a high but narrow fitness peak can be inferior to a lower but broader fitness plateau, as mutations on the fitness plateau will only have a minimal effect on overall fitness of the population owing to support from sequences of similar fitness connected on the fitness landscape [23,24]. Survival of the flat-test should lead to the evolution of ‘mutational robustness’ in populations subjected to high error rates [27]. Mutational robustness refers to the ability of a population to minimize the effect of high mutation frequency by favoring sequences that are more resistant to mutation [23,27].
Lethal mutagenesis as an antiviral strategy
Given that linear increases in mutation rate result in exponential reduction in viral fecundity (Figure 2), even slight increases in mutagenesis may be sufficient to drive a viral population into extinction. RNA viruses should be particularly sensitive to such a ‘genetic meltdown’ since their genomes are extraordinarily dense with information. Viral genomes encode for proteins as well as RNA structural signals for translation and replication, and their small sizes have resulted in numerous strategies for maximizing genetic information, including transcriptional slippage [28] and programmed ribosomal frameshifting [29]. The number of allowed mutations per replication cycle should therefore be relatively small. Hence, RNA viruses likely exist at or near the extinction threshold and may be particularly susceptible to minor increases in mutation frequency.
Crotty et al., using PV as a model, demonstrated experimentally that small changes in mutation frequency can result in rapid reductions in viral fecundity [30]. Natural PV replication results in approximately 1.5 mutations per genome based on sequence ana lysis of capsidcoding regions. When PV was grown in the presence of a mutagen, a dramatic decline in specific infectivity of PV genomic RNA was seen when the mutation frequency exceeded two mutations per genome. Furthermore, an approximate fourfold increase in mutation rate was sufficient to cause a 20-fold reduction in specific infectivity. This observation provides additional support for the notion that only a small increase in mutagenesis is necessary to allow for a potent antiviral effect.
Subsequent work has demonstrated extinction of virus populations through the application of chemical mutagens. FMDV was driven into extinction by the combination of a mutagen and an antiviral inhibtor [31], and more recent work has shown that treatment with a mutagen alone is sufficient for extinction [32,33]. The mutagenic nucleobase 5-fluorouracil causes an increase in mutation frequency of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) and is able to drive the virus population into extinction [34].
The aforementioned experiments have suggested that the viral quasispecies might represent a novel antiviral target to be exploited. While most current antiviral agents target viral proteins, this approach instead targets the genetic information itself. While the viral polymerase is the ultimate mediator of antiviral activity, lethal mutagenic nucleosides would ideally not affect polymerase function as do traditional polymerase inhibitors or chain-terminating nucleosides. Instead, the eventual goal would be development of a ‘stealth nucleoside’ [35] that would not be discriminated against by the viral polymerase but would corrupt the viral genetic information when incorporated into the RNA genome.
Nucleoside analogs as antiviral lethal mutagens
The antiviral activity of ribavirin
Ribavirin (1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide) is a synthetic purine analog first reported in 1972 (Figure 3) [2]. The nucleoside has broad-spectrum antiviral activity and is effective in cultured cells against a wide range of both RNA and DNA viruses. Clinically, it is used in combination with IFN-α for treatment of HCV infection [36–38] and as a monotherapy for treatment of Lassa fever virus [39,40], respiratory syncitial virus [41] and some herpesvirus infec-tions [42]. It has also been used experimentally as a treatment for numerous other viruses, most recently for SARS-associated coronavirus [43].
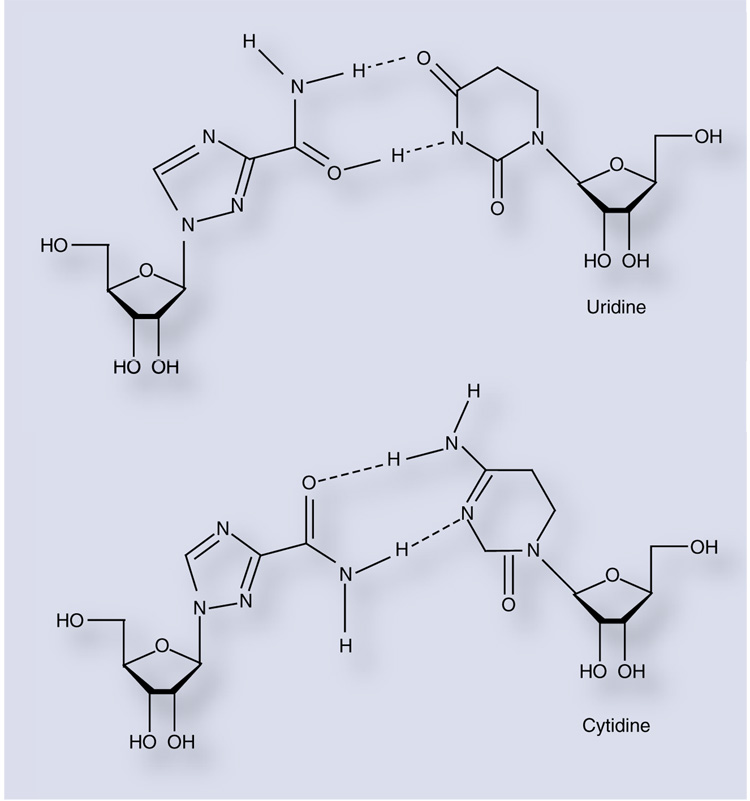
Figure 3. Ribavirin can act as an ambiguous purine analog.
Rotation of the exocyclic carboxamide allows for base pairing with either uridine (top) or cytidine (bottom).
Ribavirin is clinically administered as the nucleoside and, upon entering the cell, is rapidly converted to ribavirin monophosphate (RMP) by cellular adenosine kinase. Further phosphory lation by cellular kinases leads to the accumulation of ribavirin triphosphate (RTP). RTP is the primary form of ribavirin found in treated cells, although RMP is also present at levels 20–100-fold lower than the triphosphate [44,45].
While ribavirin has exhibited antiviral activity against a wide range of DNA and RNA viruses in the laboratory, its mechanism of action has remained elusive (reviewed in [46]). For 30 years, the antiviral activity of ribavirin was thought to be due to inhibition of cellular inosine 5′-monophosphate dehydrogenase (IMPDH) [47], although polymerase inhibition, immune modulation and inhibition of 5′ capping of viral RNA have also been implicated as potential mechanisms [46]. More recently, it has become apparent that ribavirin can induce transition mutations in viral genomes via ambiguous base-pairing during or subsequent to incorporation by the viral RdRp. Crotty and colleagues were able to demonstrate RTP incorporation by PV 3Dpol using an in vitro primer-extension assay [48]. RMP was incorporated at approximately the same rate as an incorrect nucleotide opposite either cytidine or uridine, and a template containing ribavirin was able to direct incorporation of both cytidine and uridine at approximately the same efficiency. This promiscuous base-pairing capacity is presumably due to rotation of the carboxamide moiety of the ribavirin pseudobase resulting in two distinct hydrogen-bonding con- figurations (Figure 3).
Evidence implicating lethal mutagenesis as the mechanism of action of ribavirin has subsequently been extended to other virus species, including HCV [49–51], GB virus B [52], FMDV [32], West Nile virus [53] and Hantaan virus [54,55]. However, the mechanism of action of ribavirin in vivo is still hotly debated [56–59]. Thus, despite extensive efforts into understanding the phenomenon of lethal mutagenesis in vitro, little is known of the potential of lethal mutagenesis as a bona fide clinical antiviral strategy.
Modifications to ribavirin
The broad-spectrum activity of ribavirin is unparalleled among antiviral drugs. This has led to considerable interest in the development of structural analogs which may exhibit increased activity or greater selectivity with regard to host cell functions and metabolism. Viramidine (Figure 4A) is an aminated prodrug that is rapidly converted to ribavirin upon absorption [60,61]. Levovirin, the l-analog of ribavirin, is not phosphorylated in vitro and lacks mutagenic and antiviral activity [59,62].
Figure 4. Analogs of ribavirin.
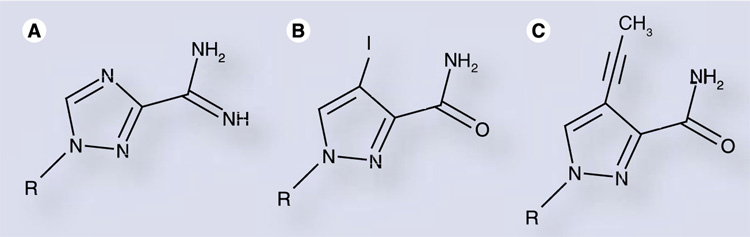
(A) Viramidine is an aminated prodrug that is rapidly converted to ribavirin intracellularly. (B) Iodo- and (C) propynyl-substituted ribavirin analogs demonstrate efficient incorporation by poliovirus 3Dpol compared with ribavirin [63].
More recently, Moriyama et al. synthesized a number of ribavirin analogs containing hydrophobic groups at the 4-position of a pyrazole ring of the nucleobase (modified from the 1,2,4-triazole of ribavirin) [63]. While two of these analogs (Figure 4B & C) demonstrated incorporation rates similar to ribavirin when PV 3Dpol was employed (within threefold, opposite either natural pyrimidine), antiviral activity was not observed. The basis for this result is unclear, but may be due to inefficient intracellular phosphorylation or reduced mutagenicity as a result of constrained rotation of the carboxamide moiety owing to steric hindrance from the substitution at the adjacent 4-position.
Novel nucleoside analogs as lethal mutagens
A novel nucleoside analog could improve upon the antiviral activity of ribavirin in a number of ways [64]. As ribavirin is incorporated very slowly by viral polymerases (at about the rate of an incorrect nucleotide) [48], a nucleoside which has a more efficient rate of incorporation may have greater antiviral effects. Additionally, a nucleoside with different templating properties, for instance a nucleoside that can mimic either cytidine or uridine, could exploit the lower cellular concentration of competing pyrimidines [65]. A pyrimidine analog might be most effective if administered in combination with a purine analog such as ribavirin, allowing for all possible genomic sites to be targeted. Novel nucleoside analogs may also possess other desirable properties such as increased cellular phosphorylation, leading to a more rapid accumulation in the target cell, or improved discrimination by cellular enzymes, providing greater specificity for the antiviral response over off-target cellular effects. Finally, ribavirin treatment causes significant clinical side effects owing to its pleiotropic biological properties [66]. Development of a nucleoside analog that does not have such secondary activities might eliminate the undesirable effects of clinical treatment.
Nucleosides that can induce mutagenesis through direct incorporation into nucleic acid can be divided into two general categories depending on the properties of the attached nucleobase. The first are those containing nucleobases with minimal hydrogen-bonding capacity that can be incorporated opposite any of the naturally occurring nucleosides, a class of molecules known as universal bases [67]. The second category includes nucleosides containing bases with ambiguous hydrogen-bonding properties that allow favorable hydrogen-bonding interactions with two or more of the natural nucleobases, such as ribavirin.
Universal nucleosides
A number of nonhydrogen-bonding nucleoside analogs have been developed, mostly as deoxyribonucleosides [67]. In general, these have hydrophobic, aromatic bases that are stable in nucleic acid duplexes due primarily to base-stacking interactions. Because this interaction is not dependent upon hydrogen bonding, these bases show little discrimination in ‘pairing’ opposite any of the naturally occurring nucleobases. This type of nucleoside analog has shown utility for use in degenerate primers and probes [67,68]. Unfortunately, these analogs have generally been poor substrates for enzymes.
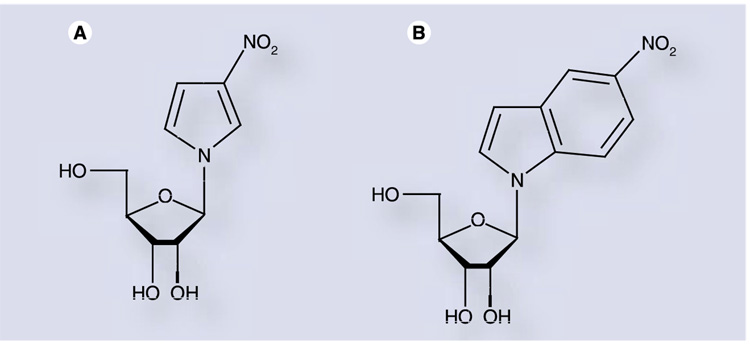
One such universal ribonucleoside, 1-ribofuranosyl- 3-nitropyrrole nucleoside (3-NPN; Figure 5A), was synthesized and investigated as a lethal mutagen against PV [69]. The deoxyribo-nucleoside containing this base is known to hybridize to all four bases when in a DNA duplex [70]. However, 3-NPN did not exhibit any significant antiviral effect against PV [69]. Primer-extension assays using PV 3Dpol and the triphosphorylated form of 3-NPN demonstrated that this nucleoside did in fact possess mutagenic properties, being incorporated as an analog of either adenosine or uridine. However, it did not exhibit true ‘universal’ characteristics, as it was not incorporated opposite all four natural bases. More significantly, 3-NPN was incorporated approximately 100-fold slower than ribavirin in vitro. Therefore, 3-NPN will be incorporated much less than once per genome during replication, making this compound an ineffective antiviral mutagen.
Figure 5. Ribonucleoside analogs with ‘universal’ nucleobases.
3-nitropyrrole nucleoside (A: 3-NPN) and 5-nitroindole nucleoside (B: 5-NINDN) have promiscuous templating properties, although low efficiency of polymerase incorporation prevents their use as lethal mutagens [69,71].
To overcome the shortcomings of 3-NPN, a second-generation analog with an increased aromatic system and enhanced π-stacking interactions was designed, 5-nitroindole nucleoside (5-NINDN; Figure 5) [71]. A deoxyribonucleoside containing the 5-nitroindole base was shown to have increased duplex stability relative to 3-nitropyrrole and was an efficient universal substrate for DNA polymerases, although it demonstrated chain-terminating properties [72,73]. The triphosphate of 5-NINDN was indeed found to be templated approximately equally well by all four natural bases when acting as a substrate for the PV RdRp [71]. While incorporated ten-fold more rapidly than the 3-NPN triphosphate, 5-NINDN triphosphate was still tenfold slower than ribavirin for PV RdRp incorporation [70]. However, chain termination was not observed with the ribonucleoside (as opposed to the results observed with the deoxyribonucleoside), and the nucleotide, while incorporated too slowly to be an effective mutagen, proved to be an inhibitor of the PV polymerase, with a Ki on the order of 30 µM [71,74].
The results obtained with 3-NPN and 5-NINDN suggest that nonhydrogen-bonding analogs may not be effective as lethal mutagens. Hydrogen-bonding interactions may be essential in stabilizing the nucleoside triphosphate substrate in the active site of the polymerase in order to allow incorporation into nucleic acid on a bio-logically relevant timescale. However, the particular structures of the 3-NPN and 5-NINDN nucleobases may have unique properties that prevent effective incorporation relative to other potential universal bases. Furthermore, the particular polymerase tested (PV 3Dpol) may be less efficient in utilizing nonhydrogen-bonding bases than polymerases from other viruses. Therefore, while the potential of nonhydrogen-bonding universal bases to function as lethal mutagens cannot be ruled out, the currently available data suggest that ‘universal’ nucleosides may be poor choices as mediators of viral mutagenesis.
Ambiguously hydrogen-bonding nucleosides
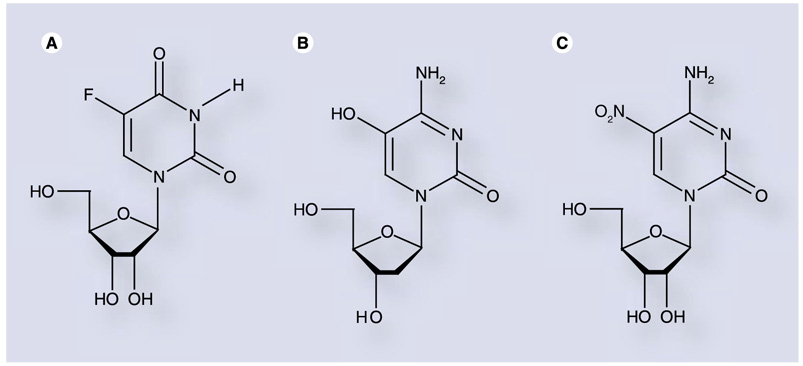
The second major class of mutagenic bases includes those that exhibit ambiguous hydrogen-bonding properties. In this case, hydrogen-bonding interactions are present but unique patterns of hydrogen bond donors and acceptors are displayed depending on the configuration of the molecule. Hydrogen-bonding patterns can vary based on factors such as rotation, ionization or tautomerization of the base moiety. Ribavirin is an example of this class of molecule, as rotation of the exocyclic carboxamide allows the nucleoside to mimic either of the naturally occuring purines (Figure 3) [48]. Similarly, 5-fluorouridine (Figure 6A) can substitute for either of the naturally occur-ring pyrimidines [75]. Furthermore, the mutagenic activity of 5-hydroxy-2′-deoxycytidine (Figure 6B) against HIV is likely due to increased prevalence of the imino tautomer, which allows effective base pairing with adenosine as well as guanosine [76].
Figure 6. Pyrimidine analogs with mutagenic nucleobases.
(A) 5-fluorouridine and (B) 5-hydroxy-2′-deoxycytidine have been shown to act as mutagenic nucleoside analogs. Substitution of a nitro moiety at the 5-position of cytidine (C) results in a nucleotide polymerase inhibitor [77].
To further investigate the antiviral properties exhibited by 5-hydroxy-2′-deoxycytidine, Harki et al. synthesized a series of 5-substituted cytidine ribonucleoside analogs and evaluated them for antiviral activity against the RNA viruses PV and coxsackievirus B3(CVB3) [77]. One analog in particular, 5-nitrocytidine (Figure 6C), exhibited potent antiviral activity, but through a polymerase inhibition mechanism rather than mutagenesis [77], as incorporation of this nucleo-tide was nearly 2000-fold slower than cytidine 5′-triphosphate (CTP).
Researchers at Koronis Pharmaceuticals (Redmond, WA, USA) described a novel mutagenic deoxynucleoside analog, 5-aza-5,6- dihydro-2′-deoxycytidine (KP-1212; Figure 7), which demonstrates potent antiviral activity against HIV in vitro [78,79]. The related ribonucleoside, 5-aza-5,6-dihydrocytidine, has been clinically investigated in oncology [80], although no evaluation of its antiviral activity has been published. KP-1212 was shown to inhibit HIV with an EC50 of 10 nM and therapeutic index of 100,000 [78]. Tautomerization of the nucleobase between the imino and amino form presumably allows base pairing with either of the natural purines (Figure 7), and this compound increased the mutation frequency of proviral HIV-1 DNA 50–100% with no evidence of evolution of resistance or genotoxicity to the host. KP-1461, a prodrug of KP-1212, is currently in Phase IIa clinical trials as a monotherapy for the treatment of HIV-1 infection in treatment-experienced patients harboring virus with significant resistance to standard antiretrovirals [81]. This trial has been suspended by Koronis to investigate a discrepancy in preclinical in vitro data and to examine Phase IIa clinical results [202]. Suspension was not requested by the US FDA and was not due to safety concerns or observation of drug-related adverse events [202].
Figure 7. KP-1212 is a mutagenic deoxyribonucleoside analog with antiretroviral activity.
Tautomerization of the nucleobase allows for alternative base pairing with either guanosine (top) or adenosine (bottom).
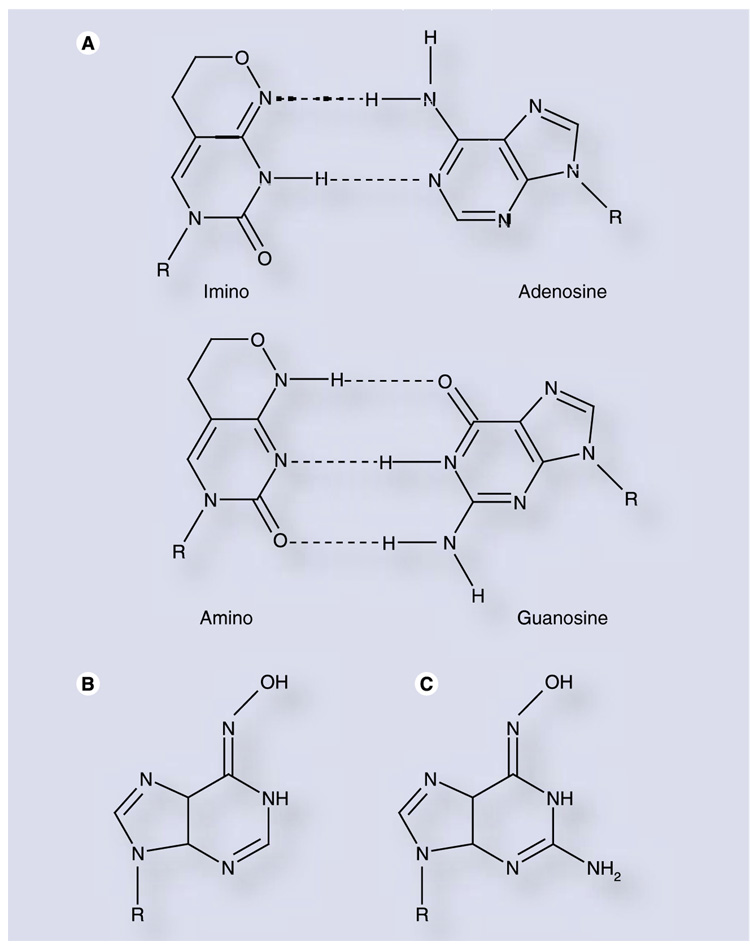
6β-(d-ribofuranosyl)-3,4-dihydro-8H-pyrimido[ 4,5-c][1,2]oxazin-7-one (rP; Figure 8A) is a bicyclic pyrimidine analog which can mimic either cytidine or uridine through tautomerization of the nucleobase. This com-pound was shown to induce transition mutations in Escherichia coli [82] and in an in vitro retroviral replication model [83]. PV genomes synthesized in vitro to contain rP demonstrated substantial reductions in viral fitness, and the nucleotide was an efficient and promiscuous substrate of PV 3Dpol [84]. In spite of these properties, rP did not demonstrate antiviral activity against PV in cell culture, owing to insufficient phosphorylation by cellular kinases [84]. While a prodrug approach may ultimately prove fruitful, this highlights the complexity of designing nucleoside analogs that can induce mutagenesis and, at the same time, maintain activity as substrates for critical cellular enzymes, such as nucleoside and nucleotide kinases required for conversion to the nucleoside triphoshate.
Figure 8. Tautomerization of nucleobase analogs allows for ambiguous base pairing.
(A) Nucleoside P can base pair with either adenosine (top) or guanosine (bottom) through its two tautomers. (B) JA28 and (C) JA30 are N-6-substituted purine analogs that are efficiently and ambiguously incorporated during poliovirus and CVB3 replication [88].
Tautomerization as a strategy for ambiguous basepairing was further advanced with the synthesis of a variety of N-6-modified purine nucleotide analogs [85–87]. The most potent of these analogs, designated JA28 and JA30 (Figure 8B & C, respectively), increased the mutation frequency of PV 35- to 65-fold over a single passage, resulting in 100–1000-fold reductions in viral titer [88]. Similar results were obtained for the picornavirus CVB3. Significantly, these nucleosides were phosphorylated intracellularly, yet did not appear to be substrates for cellular ribonucleotide reductase [88], suggesting that their potential for host genotoxicity may be minimal [89].
Considerations in the design & development of next-generation lethal mutagens
Clearly, one limiting factor for the pharmaceutical exploitation of the lethal mutagenesis phenomenon is the high levels of endogenous nucleosides in normally-functioning cells. All four natural ribonucleosides are found at intracellular concentrations ranging from hundreds of micromolar to low millimolar [65]. Targeting retroviruses with deoxynucleoside analogs can overcome this issue to some degree, as dNTPs are found at much lower concentrations – approximately 40 µM or lower [65].
Another primary concern is the induction of genomic mutation into the host cell (or even into other cells of the body not infected with the target virus). Deoxyribonucleoside analogs clearly have potential for genotoxicity, but ribonucleotides may also be incorporated into cellular DNA if they are converted to the deoxyribonucleotide by cellular ribonucleotide reductase [89]. As the substrate specificity of ribonucleotide reductase is not well-defined, designing analogs resistant to conversion is difficult. Some selectivity to host cell mutation should be achievable owing to the enhanced proofreading activity possessed by cellular replication machinery that is absent in viral replicases [7].
Examination of the nucleoside pools in mammalian cells also provides some direction for the development of antiviral nucleosides. Intracellular pyrimidine concentrations are generally much lower than purine concentrations. Traut compiled approximately 600 published values for nucleoside and nucleotide concentrations [65]. Average concentrations of ATP and GTP in human cells are thought to be approximately 2000 and 300 µM, respectively. Concentrations for the pyrimidines uridine 5′-triphosphate (UTP) and CTP are approximately 250 and 100 µM, respectively. The disparity is due primarily to the extremely high intracellular concentration of ATP, indicative of its essential role as an energy source for myriad cellular processes. Development of pyrimidine analogs thus ensures that intracellular competitors are at a minimum, potentially requiring lower drug concentrations to be effective. Additionally, the purines are intimately involved in essential cellular chemistry beyond DNA and RNA synthesis, being necessary for such diverse processes as metabolic processes, signaling and translation. Thus, focusing on mutagenic pyrimidine analogs may result in fewer adverse cellular effects.
One potential complication in developing mutagenic nucleoside analogs is the variety of targets with which a broad-spectrum antiviral must interact. Although all RdRps are predicted to have a conserved ‘right hand’ structure, consisting of palm, thumb and finger domains, the precise substrate specificity of polymerases from different virus families may vary. It is important to note that, for retroviruses, the target enzyme in the application of ribonucleoside analogs is the host RNA polymerase II. Because a host enzyme is the target, this approach should greatly hinder the development of retroviruses resistant to this class of compounds.
A lofty goal is the development of mutagenic nucleosides that can act as substrates for virus-encoded polymerases but not as substrates for cellular polymerases. This will prove challenging, as the specific structure–function relationships defining substrate specificities for these enzymes are still largely a mystery. Recent work with 2′-modified nucleoside analogs may provide a first step in this direction [90]. These analogs appear to be incorporated by the HCV RdRp and appear to act as chain terminators. Further experimentation demonstrated that 2′-modified nucleoside analogs were inhibitors of the HCV RdRp both in vitro and in vivo [91]. In this case, a mutation conferring resistance was discovered in the RdRp and resistance was directed towards the specific 2′-modification. Importantly, it appears as if these nucleoside analogs are not incorporated by cellular DNA-dependent DNA polymerases or DNA-dependent RNA polymerases [90,91]. This resulted in minimal cellular toxicity at concentrations effective for virus inhibition, and intracellular phosphorylation did not appear to be rate limiting. These promising experiments indicate that modifications to the pentose ring of the nucleoside may allow antiviral activity to be retained while reducing cytotoxicity to the host cell, although differences in substrate selection by various polymerases may render such modifications viral specific. It is important to note, however, that at this time the ability of the mitochondrial transcription machinery to be impacted by these analogs has not been addressed.
Biological implications of lethal mutagenesis & modulation of viral mutation rate
Development of resistance to lethal mutagens
It has often been assumed that application of a mutagen would be unlikely to increase the frequency at which resistant virus variants would arise due to the fact that RNA virus error rates are already extremely high (approximately one mutation per genome per replication cycle). Bull, Sanjuan and Wilke make note that the relevant parameters are difficult to measure and that the ultimate result of increasing mutation frequency cannot be definitively known until more accurate measurements are available [20]. RNA viruses may actually replicate at a suboptimal error rate [20]. Thus, it may be possible that exposure of a virus population to a drug which increases the mutation rate during replication may allow that population to access a peak on the fitness landscape that was unattainable without treatment. This may allow for resistance to the mutagen itself, or to a drug targeting another viral process that is co-administered with the mutagen. There have been reports of increased robustness in viral and subviral pathogens after exposure to a mutagen [92,93], although others have reported no selection of robustness with LCMV in cell culture [94].
Clinical treatment with a lethal mutagen may be unlikely to generate resistant variants, as the high error rate of RNA viruses is necessary for viability and pathogenesis in the host, and a likely mechanism for resistance is the selection of a higher-fidelity polymerase that would restrict quasispecies diversity, countering exogenous increases in mutation rate. Not surprisingly, a ribavirin-resistant variant of PV was identified (PV-G64S) [95,96], whereby resistance is mediated through an increase in polymerase fidelity of approximately threefold. Importantly, although two separate research groups were able to isolate a ribavirin-resistant PV variant, both isolates had exactly the same mutation. This suggests that there may be a very limited number of resistance mechanisms to counter continued exposure to a mutagen. Interestingly, this mutation occurs at a location of the enzyme remote from the catalytic site.
Subsequent work with this resistant variant demonstrated that it had restricted tropism and lessened pathogenesis in a mammalian host [3,4]. This suggests that the error rate is tightly regulated by virus populations to maintain an optimal level of quasispecies diversity. Thus, even if a lethal mutagen were unable to completely extinguish a virus population, changing the error frequency slightly may be sufficient to at least prevent disease or virus spread. Furthermore, application of a mutagen in combination with traditional viral inhibitors may allow for extinction of a virus population under conditions where either treatment alone is insufficient, and may inhibit the evolution of resistance [31].
However, the recent discovery of a ribavirin-resistant FMDV polymerase variant [97] indicates another potential mechanism for resistance to lethal mutagens beyond a global increase in polymerase fidelity. The M296I polymerase incorporates RMP less efficiently than the wild-type polymerase, although quasispecies diversity was not restricted [97]. Biochemical evaluation of the polymerase revealed that this polymerase has lower fidelity (by 2.5-fold) than the wild-type, but with a specific discrimination against ribavirin utilization [arias a et al., submitted manuscript (2008)]. Thus, FMDV can attain resistance by decreasing the efficiency of ribavirin incorporation, although this results in a trade-off of fidelity for normal nucleotide incorporation (specifically, a higher frequency of GMP misincorporation). Interestingly, this is not the first mutator phenotype identified for an RNA virus [98], but it does demonstrate that multiple mechanisms exist to confer resistance to lethal mutagens, and further underscores the importance of a tightly-controlled mutation rate in RNA virus populations.
Alternative approaches to exploiting viral quasispecies
The work described above indicates that changes in virus fidelity can affect not only virus fitness, but pathogenesis and tropism. Modulating fidelity only a few fold in either direction should be sufficient for significant biological effect, whether that be a descent into extinction catastrophe or quasispecies restriction resulting in attenuation.
The discovery of a ribavirin-resistance mutation remote from the active site of PV 3Dpol suggests that mutations in the polymerase active site are not the sole mechanism to modulate fidelity [96]. Changes in overall enzyme conformational dynamics may perturb the active site enough to affect nucleotide incorporation kinetics and thus fidelity. Targeting viral polymerases with small molecule drugs could possibly affect the conformational dynamics required for fidelity of viral replication, thus creating an antiviral effect. However, the interactions necessary to elicit such an effect are unknown, complicating design of such a drug. Furthermore, high-throughput screening approaches to identify small molecules that modulate small changes in fidelity are technically challenging. An additional limitation is that this might require completely distinct molecules to target each individual virus polymerase owing to differences in enzyme structure and activity. Thus, the broad applicability of such a drug may be severely limited.
More interesting is the potential application of high-fidelity viral variants as attenuated vaccine strains. Investigation of the ribavirin resistant G64S PV variant indicates that it demonstrates reduced tissue tropism and pathogenicity [4], and can confer protective immunity to wild-type PV infection in a mouse model [3]. This may provide a broadly applicable strategy for the engineering of live-attenuated vaccines for RNA viruses.
Identifying viruses with increased sensitivity to lethal mutagens
While RNA viruses in general experience heavy fitness loss when exposed to mutagens, the tolerance of RNA viruses to mutagens may vary between viruses, even those that are closely related. We have demonstrated that CVB3 is more sensitive to ribavirin treatment than the closely related picornavirus PV [99]. Surprisingly, this was not due to increased frequency of ribavirin incorporation, as CVB3 3Dpol utilized ribavirin less efficiently than PV 3Dpol in an in vitro primer extension assay [graci jd et al., Manuscript in preparation]. Instead, CVB3 appears to be constrained by a lower extinction threshold, as an equivalent number of mutations caused a more deleterious fitness effect on the CVB3 genome than the PV genome [graci jd et al., Manuscript in preparation (2008)]. This indicates that some viruses may be more susceptible to a lethal mutagenesis-based antiviral approach owing to differences in mutational robustness, possibly due to constraints in the primary protein coding sequence or RNA structural elements which limit the tolerance to mutation.
A measure of the tolerance of an organism to genetic change is the ratio of nonsynonymous to synonymous mutations (dN/dS) [100,101]. Nonsynonymous mutations indicate changes to the codon and its particular amino acid, whereas synonymous mutations indicate a change in the primary nucleotide sequence but with no change in protein coding function, owing to the degeneracy of the genetic code. Thus, the dN/dS ratio can be considered a measure of selective pressure. Viruses exhibiting low dN/dS ratios may therefore display increased sensitivity to lethal mutagenesis, as their genomes are more conserved and therefore may demonstrate reduced mutational robustness. Viruses replicating under heightened selective pressure may therefore be opportune targets for mutagenesis-based antiviral approaches.
Future perspective
Despite extensive investigation of antiviral lethal mutagens in vitro, attempts to exploit this theory for clinical use are just beginning. Although ribavirin has been shown to be a potent mutagen of RNA viruses in vitro, information regarding the clinical importance of this mechanism of action during clinical ribavirin therapy is limited and controversial. In this regard, clinical trials of new antiviral mutagens, such as KP-1212 and KP-1461, will provide critical insight for translating in vitro observations to therapeutic use, as well as providing new avenues for drug development. While a number of nucleoside analogs, particularly those exhibiting ambiguous base pairing properties, have shown sufficient mutagenicity to be potent antivirals in vitro, pharmacological and toxicological liabilities remain. Thus, research leading to improved selectivity and pharmacological properties is required. Understanding the substrate specificity of host cell kinases and ribonucleotide reductases is still in its infancy, but is a field which can have profound implications on the design of future antiviral nucleosides. Further research in this area is likely to provide novel directions for improving the selectivity, pharmacokinetics and pharmacodynamics of nucleoside analogs.
Furthermore, few attempts at studying the implications of lethal mutagenesis in a clinical setting have been undertaken. Emergence of viral resistance, host cell genotoxicity, effects on cellular metabolic processes, and other undesirable off-target interactions will complicate the translation of lethal mutagenesis to the clinic. Clearly, there will be a number of high regulatory hurdles to clear before a highly-mutagenic nucleoside analog can be approved for use in patients. In this regard, the pioneering efforts of Koronis Pharmaceuticals to move KP-1212 and other stealth nucleosides into the clinic will be informative and worth watching in the coming years.
The subject that will likely see the most rapid advances is that of viral resistance to mutagens. It had often been assumed that the primary mechanism of resistance to lethal mutagens would be an overall increase in polymerase fidelity, and this proved true in the case of PV. However, the FMDV ribavirin resistance mutation identified by Sierra et al. demonstrates that other resistance mechanisms are possible, such as polymerase discrimination against specific nucleotides [97]. Investigation of PV and FMDV ribavirin-resistant viruses indicate that viral mutation rate is tightly controlled and likely to be evolutionarily optimized. Thus, further understanding of the interplay between viral fitness and polymerase fidelity will likely have important implications on further development of antiviral drugs, as well as on the generation of effective vaccines.
Executive summary
RNA viruses exist as a quasispecies
Natural RNA virus replication results in the generation of approximately one mutation per genome.
High mutation frequency results in the generation of a viral quasispecies with extensive genetic diversity.
The extinction threshold for a viral population is dependent upon both mutation rate and fecundity.
Viruses may evolve increased mutational robustness to counter the negative effects of high error rate.
An antiviral strategy based on increasing the viral mutation rate (lethal mutagenesis) may allow for effective viral inhibition, with n minimal potential for the generation of resistance.
Nucleoside analogs can induce lethal mutagenesis in virus populations
The antiviral nucleoside ribavirin decreases virus viability in vitro by increasing the viral mutation frequency, although the clinical importance of mutagenesis is still unresolved.
Chemical modificiations to ribavirin have failed to produce more potent mutagens as yet.
‘Universal’, nonhydrogen-bonding nucleosides have shown limited capacity to act as viral mutagens in vitro.
Many promiscuous nucleoside analogs have shown potent induction of viral mutation in vitro with favorable biochemical properties, although only one, KP-1212, has been advanced clinically as a lethal mutagen.
Design of more potent antiviral lethal mutagens will require optimization of structural characteristics influencing interactions with the viral polymerase, as well as with host cell metabolic enzymes.
The mutation frequency of RNA viruses is tightly regulated
Poliovirus can generate resistance to ribavirin by evolving a higher-fidelity polymerase.
A foot-and-mouth disease virus variant displays resistance to ribavirin treatment through specific discrimination against ribavirin while maintaining quasispecies diversity through reducing polymerase fidelity.
Closely-related viruses may differ significantly in their susceptibility to lethal mutagenesis owing to variations in the tolerable error threshold.
Acknowledgements
We thank Jeff Parkins (Koronis Pharmaceuticals) for critical discussion and insightful comments.
Financial & competing interests disclosure
Financial support for the work performed by the authors was provided by the NIH (AI054776 and AI45818 to CEC) and the American Heart Association (established investigator award 0340028N to CEC). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Jason D Graci, PTC Therapeutics, Inc., 100 Corporate Court, South Plainfield, NJ 07080, USA, Tel.: +1 908 912 9249; Fax: +1 908 222 0567; jgraci@ptcbio.com.
Craig E Cameron, The Pennsylvania State University, Department of Biochemistry & Molecular Biology, 201 Althouse Laboratory, University Park, PA 16802, USA, Tel:. +1 814 863 8705; Fax: +1 814 863 7024; cec9@psu.edu.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪ ▪ of considerable interest
- 1.De Clercq E. Antiviral drugs in current clinical use. J. Clin. Virol. 2004;30(2):115–133. doi: 10.1016/j.jcv.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Sidwell RW, Huffman JH, Khare GP, et al. Broad-spectrum antiviral activity of virazole: 1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science. 1972;177(50):705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- 3. Vignuzzi M, Wendt E, Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat. Med. 2008;14(2):154–161. doi: 10.1038/nm1726. Describes the generation of attenuated vaccine virus strains through increasing replication fidelity and restricting viral quasispecies.
- 4. Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439(7074):344–348. doi: 10.1038/nature04388. Small changes in virus fidelity can severely impact viral pathogenesis and tropism.
- 5.Domingo E, Escarmis C, Sevilla N, et al. Basic concepts in RNA virus evolution. FASEB J. 1996;10(8):859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 6.Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc. Natl Acad. Sci. USA. 1999;96(24):13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinhauer DA, Domingo E, Holland JJ. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122(2):281–288. doi: 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- 8.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81(22):12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58(10):465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 10.Domingo E, Martinez-Salas E, Sobrino F, et al. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance: a review. Gene. 1985;40(1):1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- 11.Eigen M, Schuster P. The hypercycle. A principle of natural self-organization. Part A: emergence of the hypercycle. Naturwissenschaften. 1977;64(11):541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 12.Holland JJ, Domingo E, de la Torre JC, Steinhauer DA. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J. Virol. 1990;64(8):3960–3962. doi: 10.1128/jvi.64.8.3960-3962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loeb LA, Essigmann JM, Kazazi F, et al. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl Acad. Sci. USA. 1999;96(4):1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loeb LA, Mullins JI. Lethal mutagenesis of HIV by mutagenic ribonucleoside analogs. AIDS Res. Hum. Retroviruses. 2000;16(1):1–3. doi: 10.1089/088922200309539. [DOI] [PubMed] [Google Scholar]
- 15.Anderson JP, Daifuku R, Loeb LA. Viral error catastrophe by mutagenic nucleosides. Annu. Rev. Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- 16.Holmes EC, Moya A. Is the quasispecies concept relevant to RNA viruses? J. Virol. 2002;76(1):460–465. doi: 10.1128/JVI.76.1.460-462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domingo E. Quasispecies theory in virology. J. Virol. 2002;76(1):463–465. doi: 10.1128/JVI.76.1.463-465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eigen M. Error catastrophe and antiviral strategy. Proc. Natl Acad. Sci. USA. 2002;99(21):13374–13376. doi: 10.1073/pnas.212514799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Summers J, Litwin S. Examining the theory of error catastrophe. J. Virol. 2006;80(1):20–26. doi: 10.1128/JVI.80.1.20-26.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bull JJ, Sanjuan R, Wilke CO. Theory of lethal mutagenesis for viruses. J. Virol. 2007;81(6):2930–2939. doi: 10.1128/JVI.01624-06. Theoretical treatment of lethal mutagenesis.
- 21.Sierra S, Davila M, Lowenstein PR, Domingo E. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J. Virol. 2000;74(18):8316–8323. doi: 10.1128/jvi.74.18.8316-8323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grande-Perez A, Lazaro E, Lowenstein P, Domingo E, Manrubia SC. Suppression of viral infectivity through lethal defection. Proc. Natl Acad. Sci. USA. 2005;102(12):4448–4452. doi: 10.1073/pnas.0408871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilke CO. Quasispecies theory in the context of population genetics. BMC Evol. Biol. 2005;5:44. doi: 10.1186/1471-2148-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilke CO, Wang JL, Ofria C, Lenski RE, Adami C. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature. 2001;412(6844):331–333. doi: 10.1038/35085569. Computational demonstration of mutational robustness.
- 25.Schuster P, Swetina J. Stationary mutant distributions and evolutionary optimization. Bull. Math. Biol. 1988;50(6):635–660. doi: 10.1007/BF02460094. [DOI] [PubMed] [Google Scholar]
- 26.Comas I, Moya A, Gonzalez-Candelas F. Validating viral quasispecies with digital organisms: a re-examination of the critical mutation rate. BMC Evol. Biol. 2005;5(1):5. doi: 10.1186/1471-2148-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Nimwegen E, Crutchfield JP, Huynen M. Neutral evolution of mutational robustness. Proc. Natl Acad. Sci. USA. 1999;96(17):9716–9720. doi: 10.1073/pnas.96.17.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratinier M, Boulant S, Combet C, et al. Transcriptional slippage prompts recoding in alternate reading frames in the hepatitis C virus (HCV) core sequence from strain HCV-1. J. Gen. Virol. 2008;89(Pt 7):1569–1578. doi: 10.1099/vir.0.83614-0. [DOI] [PubMed] [Google Scholar]
- 29.Dinman JD, Ruiz-Echevarria MJ, Peltz SW. Translating old drugs into new treatments: ribosomal frameshifting as a target for antiviral agents. Trends Biotechnol. 1998;16(4):190–196. doi: 10.1016/S0167-7799(97)01167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl Acad. Sci. USA. 2001;98(12):6895–6900. doi: 10.1073/pnas.111085598. Empirical demonstration of lethal mutagenesis of poliovirus using ribavirin.
- 31.Pariente N, Sierra S, Lowenstein PR, Domingo E. Efficient virus extinction by combinations of a mutagen and antiviral inhibitors. J. Virol. 2001;75(20):9723–9730. doi: 10.1128/JVI.75.20.9723-9730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Airaksinen A, Pariente N, Menendez-Arias L, Domingo E. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis. Virology. 2003;311(2):339–349. doi: 10.1016/s0042-6822(03)00144-2. [DOI] [PubMed] [Google Scholar]
- 33.Domingo E, Escarmis C, Lazaro E, Manrubia SC. Quasispecies dynamics and RNA virus extinction. Virus Res. 2005;107(2):129–139. doi: 10.1016/j.virusres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz-Jarabo CM, Ly C, Domingo E, de la Torre JC. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) Virology. 2003;308(1):37–47. doi: 10.1016/s0042-6822(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 35.Daifuku R. Stealth nucleosides: mode of action and potential use in the treatment of viral diseases. BioDrugs. 2003;17(3):169–177. doi: 10.2165/00063030-200317030-00003. [DOI] [PubMed] [Google Scholar]
- 36.Cummings KJ, Lee SM, West ES, et al. Interferon and ribavirin vs interferon alone in the re-treatment of chronic hepatitis C previously nonresponsive to interferon: a meta-ana lysis of randomized trials. JAMA. 2001;285(2):193–199. doi: 10.1001/jama.285.2.193. [DOI] [PubMed] [Google Scholar]
- 37.Davis GL, Esteban-Mur R, Rustgi V, et al. Interferon α-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N. Engl. J. Med. 1998;339(21):1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 38.Lau JY, Tam RC, Liang TJ, Hong Z. Mechanism of action of ribavirin in the combination treatment of chronic HCV infection. Hepatology. 2002;35(5):1002–1009. doi: 10.1053/jhep.2002.32672. [DOI] [PubMed] [Google Scholar]
- 39.Andrei G, De Clercq E. Molecular approaches for the treatment of hemorrhagic fever virus infections. Antiviral Res. 1993;22(1):45–75. doi: 10.1016/0166-3542(93)90085-w. [DOI] [PubMed] [Google Scholar]
- 40.Huggins JW. Prospects for treatment of viral hemorrhagic fevers with ribavirin, a broad-spectrum antiviral drug. Rev. Infect. Dis. 1989;11 Suppl. 4:S750–S761. doi: 10.1093/clinids/11.supplement_4.s750. [DOI] [PubMed] [Google Scholar]
- 41.Krilov LR. Respiratory syncytial virus: update on infection, treatment, and prevention. Curr. Infect. Dis. Rep. 2001;3(3):242–246. doi: 10.1007/s11908-001-0026-3. [DOI] [PubMed] [Google Scholar]
- 42.ICN Pharmaceuticals ICM. 7.5% Virazole® cream approved by the health ministry of the Russian federation. California, CA, USA: 1998. (News release) [Google Scholar]
- 43.Booth CM, Matukas LM, Tomlinson GA, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA. 2003;289(21):2801–2809. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 44.Page T, Connor JD. The metabolism of ribavirin in erythrocytes and nucleated cells. Int. J. Biochem. 1990;22(4):379–383. doi: 10.1016/0020-711x(90)90140-x. [DOI] [PubMed] [Google Scholar]
- 45.Miller JP, Kigwana LJ, Streeter DG, et al. The relationship between the metabolism of ribavirin and its proposed mechanism of action. Ann. NY Acad. Sci. 1977;284:211–229. doi: 10.1111/j.1749-6632.1977.tb21953.x. [DOI] [PubMed] [Google Scholar]
- 46.Graci JD, Cameron CE. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2006;16(1):37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Streeter DG, Witkowski JT, Khare GP, et al. Mechanism of action of 1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide (Virazole), a new broad-spectrum antiviral agent. Proc. Natl Acad. Sci. USA. 1973;70(4):1174–1178. doi: 10.1073/pnas.70.4.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crotty S, Maag D, Arnold JJ, et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000;6(12):1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 49.Contreras AM, Hiasa Y, He W, et al. Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system. J. Virol. 2002;76(17):8505–8517. doi: 10.1128/JVI.76.17.8505-8517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maag D, Castro C, Hong Z, Cameron CE. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J. Biol. Chem. 2001;276(49):46094–46098. doi: 10.1074/jbc.C100349200. [DOI] [PubMed] [Google Scholar]
- 51.Zhou S, Liu R, Baroudy BM, Malcolm BA, Reyes GR. The effect of ribavirin and IMPDH inhibitors on hepatitis C virus subgenomic replicon RNA. Virology. 2003;310(2):333–342. doi: 10.1016/s0042-6822(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 52.Lanford RE, Chavez D, Guerra B, et al. Ribavirin induces error-prone replication of GB virus B in primary tamarin hepatocytes. J. Virol. 2001;75(17):8074–8081. doi: 10.1128/JVI.75.17.8074-8081.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Day CW, Smee DF, Julander JG, et al. Error-prone replication of West Nile virus caused by ribavirin. Antiviral Res. 2005;67(1):38–45. doi: 10.1016/j.antiviral.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 54.Severson WE, Schmaljohn CS, Javadian A, Jonsson CB. Ribavirin causes error catastrophe during Hantaan virus replication. J. Virol. 2003;77(1):481–488. doi: 10.1128/JVI.77.1.481-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chung DH, Sun Y, Parker WB, et al. Ribavirin reveals a lethal threshold of allowable mutation frequency for Hantaan virus. J. Virol. 2007;81(21):11722–11729. doi: 10.1128/JVI.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chevaliez S, Brillet R, Lazaro E, Hezode C, Pawlotsky JM. Analysis of ribavirin mutagenicity in human hepatitis C virus infection. J. Virol. 2007;9:9. doi: 10.1128/JVI.00382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lutchman G, Danehower S, Song BC, et al. Mutation rate of the hepatitis C virus NS5B in patients undergoing treatment with ribavirin monotherapy. Gastroenterology. 2007;132(5):1757–1766. doi: 10.1053/j.gastro.2007.03.035. [DOI] [PubMed] [Google Scholar]
- 58.Perelson AS, Layden TJ. Ribavirin: is it a mutagen for hepatitis C virus? Gastroenterology. 2007;132(5):2050–2052. doi: 10.1053/j.gastro.2007.03.077. [DOI] [PubMed] [Google Scholar]
- 59.Hofmann WP, Polta A, Herrmann E, et al. Mutagenic effect of ribavirin on hepatitis C nonstructural 5B quasispecies in vitro and during antiviral therapy. Gastroenterology. 2007;132(3):921–930. doi: 10.1053/j.gastro.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 60.Lin CC, Xu C, Zhu N, Yeh LT. Absorption, metabolism, and excretion of [14C] viramidine in humans. Antimicrob. Agents Chemother. 2006;50(7):2368–2373. doi: 10.1128/AAC.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu JZ, Yeh LT, Lin CC, Hong Z. Conversion of viramidine to ribavirin in vivo by adenosine deaminase and its inhibition by 2′-deoxycoformycin. Antivir. Chem. Chemother. 2006;17(1):33–39. doi: 10.1177/095632020601700105. [DOI] [PubMed] [Google Scholar]
- 62.Fang C, Srivastava P, Lin CC. Effect of ribavirin, levovirin and viramidine on liver toxicological gene expression in rats. J. Appl. Toxicol. 2003;23(6):453–459. doi: 10.1002/jat.938. [DOI] [PubMed] [Google Scholar]
- 63.Moriyama K, Suzuki T, Negishi T, et al. Effects of introduction of hydrophobic group on ribavirin base on mutation induction and anti-RNA viral activity. J. Med Chem. 2008;51(1):159–166. doi: 10.1021/jm7009952. [DOI] [PubMed] [Google Scholar]
- 64.Graci JD, Cameron CE. Challenges for the development of ribonucleoside analogues as inducers of error catastrophe. Antivir. Chem. Chemother. 2004;15(1):1–13. doi: 10.1177/095632020401500101. [DOI] [PubMed] [Google Scholar]
- 65.Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 1994;140(1):1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 66.Martin P, Jensen DM. Ribavirin in the treatment of chronic hepatitis C. J. Gastroenterol. Hepatol. 2008;23(6):844–855. doi: 10.1111/j.1440-1746.2008.05398.x. [DOI] [PubMed] [Google Scholar]
- 67.Loakes D. Survey and summary: the applications of universal DNA base analogues. Nucleic Acids Res. 2001;29(12):2437–2447. doi: 10.1093/nar/29.12.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loakes D, Brown DM, Linde S, Hill F. 3-nitropyrrole and 5-nitroindole as universal bases in primers for DNA sequencing and PCR. Nucleic Acids Res. 1995;23(13):2361–2366. doi: 10.1093/nar/23.13.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harki DA, Graci JD, Korneeva VS, et al. Synthesis and antiviral evaluation of a mutagenic and non-hydrogen bonding ribonucleoside analogue: 1-β-d-ribofuranosyl-3-nitropyrrole. Biochemistry. 2002;41(29):9026–9033. doi: 10.1021/bi026120w. [DOI] [PubMed] [Google Scholar]
- 70.Nichols R, Andrews PC, Zhang P, Bergstrom DE. A universal nucleoside for use at ambiguous sites in DNA primers. Nature. 1994;369(6480):492–493. doi: 10.1038/369492a0. [DOI] [PubMed] [Google Scholar]
- 71.Harki DA, Graci JD, Edathil JP, et al. Synthesis of a universal 5-nitroindole ribonucleotide and incorporation into RNA by a viral RNA-dependent RNA polymerase. Chembiochem. 2007;8(12):1359–1362. doi: 10.1002/cbic.200700160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loakes D, Brown DM. 5-nitroindole as an universal base analogue. Nucleic Acids Res. 1994;22(20):4039–4043. doi: 10.1093/nar/22.20.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chiaramonte M, Moore CL, Kincaid K, Kuchta RD. Facile polymerization of dNTPs bearing unnatural base analogues by DNA polymerase α and Klenow fragment (DNA polymerase I) Biochemistr. 2003;42(35):10472–10481. doi: 10.1021/bi034763l. [DOI] [PubMed] [Google Scholar]
- 74.Zamyatkin DF, Parra F, Alonso JM, et al. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 2008;283(12):7705–7712. doi: 10.1074/jbc.M709563200. [DOI] [PubMed] [Google Scholar]
- 75.Yu H, Eritja R, Bloom LB, Goodman MF. Ionization of bromouracil and fluorouracil stimulates base mispairing frequencies with guanine. J. Biol. Chem. 1993;268(21):15935–15943. [PubMed] [Google Scholar]
- 76.Suen W, Spiro TG, Sowers LC, Fresco JR. Identification by UV resonance Raman spectroscopy of an imino tautomer of 5-hydroxy-2′-deoxycytidine, a powerful base analog transition mutagen with a much higher unfavored tautomer frequency than that of the natural residue 2′-deoxycytidine. Proc. Natl Acad. Sci. USA. 1999;96(8):4500–4505. doi: 10.1073/pnas.96.8.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harki DA, Graci JD, Galarraga JE, et al. Synthesis and antiviral activity of 5-substituted cytidine analogues: identification of a potent inhibitor of viral RNA-dependent RNA polymerases. J. Med. Chem. 2006;49(21):6166–6169. doi: 10.1021/JM060872x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harris KS, Brabant W, Styrchak S, Gall A, Daifuku R. KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis. Antiviral Res. 2005;67(1):1–9. doi: 10.1016/j.antiviral.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 79.Murakami E, Basavapathruni A, Bradley WD, Anderson KS. Mechanism of action of a novel viral mutagenic covert nucleotide: molecular interactions with HIV-1 reverse transcriptase and host cell DNA polymerases. Antiviral Res. 2005;67(1):10–17. doi: 10.1016/j.antiviral.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 80.Samuels BL, Herndon JE, 2nd, Harmon DC, et al. Dihydro-5-azacytidine and cisplatin in the treatment of malignant mesothelioma: a Phase II study by the Cancer and Leukemia Group B. Cancer. 1998;82(8):1578–1584. [PubMed] [Google Scholar]
- 81.Novel anti-HIV agent enters Phase IIa clinical trial. Expert Rev. Anti Infect. Ther. 2007;5(4):540–541. doi: 10.1586/14787210.5.4.539. No authors listed. [DOI] [PubMed] [Google Scholar]
- 82.Negishi K, Williams DM, Inoue Y, et al. The mechanism of mutation induction by a hydrogen bond ambivalent, bicyclic N4-oxy-2′-deoxycytidine in Escherichia coli. Nucleic Acids Res. 1997;25(8):1548–1552. doi: 10.1093/nar/25.8.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moriyama K, Otsuka C, Loakes D, Negishi K. Highly efficient random mutagenesis in transcription-reverse-transcription cycles by a hydrogen bond ambivalent nucleoside 5′-triphosphate analogue: potential candidates for a selective anti-retroviral therapy. Nucleosides Nucleotides Nucleic Acids. 2001;20(8):1473–1483. doi: 10.1081/NCN-100105242. [DOI] [PubMed] [Google Scholar]
- 84.Graci JD, Harki DA, Korneeva VS, et al. Lethal mutagenesis of poliovirus mediated by a mutagenic pyrimidine analogue. J. Virol. 2007;81(20):11256–11266. doi: 10.1128/JVI.01028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown DM, Lin PK. Synthesis and duplex stability of oligonucleotides containing adenine-guanine analogues. Carbohydr Res. 1991;216:129–139. doi: 10.1016/0008-6215(92)84156-m. [DOI] [PubMed] [Google Scholar]
- 86.Hill F, Williams DM, Loakes D, Brown DM. Comparative mutagenicities of N6-methoxy-2,6-diaminopurine and N6-methoxyaminopurine 2′-deoxyribonucleosides and their 5′-triphosphates. Nucleic Acids Res. 1998;26(5):1144–1149. doi: 10.1093/nar/26.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Too K, Brown DM, Bongard E, et al. Anti-malarial activity of N6-modified purine analogues. Bioorg. Med. Chem. 2007;15(16):5551–5562. doi: 10.1016/j.bmc.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 88.Graci JD, Too K, Smidansky ED, et al. Lethal mutagenesis of picornaviruses with N-6-modified purine nucleoside analogues. Antimicrob. Agents Chemother. 2008;52(3):971–979. doi: 10.1128/AAC.01056-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nordlund P, Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 90.Carroll SS, Tomassini JE, Bosserman M, et al. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 2003;278(14):11979–11984. doi: 10.1074/jbc.M210914200. [DOI] [PubMed] [Google Scholar]
- 91.Migliaccio G, Tomassini JE, Carroll SS, et al. Characterization of resistance to non-obligate chain terminating ribonucleoside analogs which inhibit HCV replication in vitro. J. Biol. Chem. 2003;278(49):49164–49170. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- 92.Sanjuan R, Cuevas JM, Furio V, Holmes EC, Moya A. Selection for robustness in mutagenized RNA viruses. PLoS Genet. 2007;3(6):e93. doi: 10.1371/journal.pgen.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Codoner FM, Daros JA, Sole RV, Elena SF. The fittest versus the flattest: experimental confirmation of the quasispecies effect with subviral pathogens. PLoS Pathog. 2006;2(12):e136. doi: 10.1371/journal.ppat.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martin V, Grande-Perez A, Domingo E. No evidence of selection for mutational robustness during lethal mutagenesis of lymphocytic choriomeningitis virus. Virology. 2008;378(1):185–192. doi: 10.1016/j.virol.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 95. Pfeiffer JK, Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl Acad. Sci. USA. 2003;100(12):7289–7294. doi: 10.1073/pnas.1232294100. Along with [96], identifies a ribavirin-resistant poliovirus and provides characterization of the high-fidelity polymerase that confers resistance.
- 96. Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J. Biol. Chem. 2005;280(27):25706–25716. doi: 10.1074/jbc.M503444200. Along with [95], identifies a ribavirin-resistant poliovirus and provides characterization of the high-fidelity polymerase that confers resistance.
- 97. Sierra M, Airaksinen A, Gonzalez-Lopez C, et al. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J. Virol. 2007;81(4):2012–2024. doi: 10.1128/JVI.01606-06. Identification of a foot-and-mouth disease virus variant with reduced ribavirin incorporation efficiency.
- 98.Korneeva VS, Cameron CE. Structure-function relationships of the viral RNA-dependent RNA polymerase: fidelity, replication speed, and initiation mechanism determined by a residue in the ribose-binding pocket. J. Biol. Chem. 2007;282(22):16135–16145. doi: 10.1074/jbc.M610090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Graci JD, Cameron CE. Lethal mutagenesis: exploiting error-prone replication of riboviruses for antiviral therapy. In: Torrence PF, editor. Antiviral Drug Discovery for Emerging Diseases and Bioterrorism Threats. Hoboken, NJ, USA: Wiley-Interscience; 2005. pp. 203–220. [Google Scholar]
- 100.Holmes EC. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J. Virol. 2003;77(20):11296–11298. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Woelk CH, Holmes EC. Reduced positive selection in vector-borne RNA viruses. Mol. Biol. Evol. 2002;19(12):2333–2336. doi: 10.1093/oxfordjournals.molbev.a004059. [DOI] [PubMed] [Google Scholar]
Websites
- 201.US FDA. www.fda.gov/oashi/aids/virals.html.
- 202.KORONIS. KP-1461 for HIV. www.koronispharma.com/KP1461forHIV.html.