Abstract
Oryza (23 species; 10 genome types) contains the world's most important food crop — rice. Although the rice genome serves as an essential tool for biological research, little is known about the evolution of the other Oryza genome types. They contain a historical record of genomic changes that led to diversification of this genus around the world as well as an untapped reservoir of agriculturally important traits. To investigate the evolution of the collective Oryza genome, we sequenced and compared nine orthologous genomic regions encompassing the Adh1-Adh2 genes (from six diploid genome types) with the rice reference sequence. Our analysis revealed the architectural complexities and dynamic evolution of this region that have occurred over the past ∼15 million years. Of the 46 intact genes and four pseudogenes in the japonica genome, 38 (76%) fell into eight multigene families. Analysis of the evolutionary history of each family revealed independent and lineage-specific gain and loss of gene family members as frequent causes of synteny disruption. Transposable elements were shown to mediate massive replacement of intergenic space (>95%), gene disruption, and gene/gene fragment movement. Three cases of long-range structural variation (inversions/deletions) spanning several hundred kilobases were identified that contributed significantly to genome diversification.
INTRODUCTION
Comparative analysis of plant genomes has provided important insights into genome organization, shared ancestral gene order (synteny), and mechanisms underlying their conservation and disruption (reviewed in Bennetzen, 2007; Tang et al., 2008). However, these studies lacked the phylogenetic breadth to thoroughly elucidate the mechanisms, rates, or directionality of genome evolution. An exciting and emerging paradigm for studying genome evolution is deep comparative analysis of closely related species (Ma and Bennetzen, 2004; Ammiraju et al., 2006, 2007; Hawkins et al., 2006; Clark et al., 2007; Grover et al., 2007, 2008). This comparative phylogenomic approach blends a new dimension of phylogenetic inference with structural genomic knowledge and thereby offers the precise resolution needed to understand specifics of genome evolution.
The genus Oryza is an ideal model system of exceptional global importance (The Rice Chromosome 3 Sequencing Consortium, 2005; Wing et al., 2005). More than half of humanity relies on domesticated rice for daily caloric needs. Also, it constitutes the central comparative genomics species for monocots (International Rice Genome Sequencing Project, 2005; Wing et al., 2005; Paterson, 2006; Bennetzen, 2007; Jung et al., 2008). Furthermore, the genus has diversified across a broad ecological range (Vaughan et al., 2003) within a narrow evolutionary time scale (∼15 million years [MY]) with several closely spaced speciation events, constituting an almost stepwise historical genomic record.
The 23 species of Oryza have been classified into 10 distinct genome types, represented by six diploids (A, B, C, E, F, and G) and four allotetraploids (BBCC, CCDD, HHJJ, and HHKK) (Nayar, 1973; Aggarwal et al., 1997; Ge et al., 1999), have a genome size variation of 3.6-fold, and are a rich source of unique allelic variation for rice improvement (Brar and Khush, 1997). The phylogenetic relationships among these genome types are also largely resolved (Ge et al., 1999; Zou et al., 2008). A major comparative genomics consortium under the auspices of the Oryza Map Alignment Project (www.omap.org; Wing et al., 2005; Ammiraju et al., 2006; Kim et al., 2008) was assembled to understand the code and context of genome evolution in this genus at two different resolutions: macro (chromosomal) and micro (sequence; ranging from a few orthologous genomic regions to the level of complete chromosome arms). As part of these efforts, extensive genus-wide and genome-scale public resources have been generated, including 16 BAC libraries and their associated BAC end sequence/SNaPshot fingerprint physical maps (Ammiraju et al., 2006; Kim et al., 2008). These resources, coupled with enormous genetic and functional experimental resources already in place for O. sativa (Jung et al., 2008), make Oryza uniquely suited for connecting the power of model system research with its surrounding ecological dynamics.
As an initial microlevel foray into the collective Oryza genome, we conducted a genus-wide, large-scale sequence analysis of a single orthologous genomic region (Adh1-Adh2) from representative species spanning all six diploid genome types. Previous studies indicated that Oryza has experienced several rounds of rapid diversification associated with speciation events in a short evolutionary time span (those for A genome species, C genome species, and for F-G genome species; Zhu and Ge, 2005; Zhang and Ge, 2007; Zhu et al., 2007; Zou et al., 2008). By comparing these genomes, our goal is to understand the organization, evolutionary origins, and complexity of genomic repertoires in diploid Oryza. In addition, this study illuminates the underlying dynamics of Oryza genome evolution.
RESULTS
A Genus-Wide Vertical Comparative Sequence Data Set
We isolated and sequenced a set of orthologous BACs harboring the Adh1 locus from four AA genome species and one species for each of the five remaining diploid genome types, BB through GG (Table 1), comprising >2 Mb of sequence. This genus-wide vertical comparative sequence data set was compared with a 600,118-bp reference sequence containing the Adh1 locus (OsAdh1RefSeq) from the short arm of chromosome 11 of O. sativa ssp japonica (pseudo molecule build 4; Rice Annotation Project, 2008). The Adh1 region was selected because it has been used as a model locus for comparative genomics across many plant species (Gaut and Clegg, 1991, 1993; Avramova et al., 1996; Sang et al., 1997; Tikhonov et al., 1999; Tarchini et al., 2000; Hass et al., 2003; Ilic et al., 2003; Grover et al., 2007).
Table 1.
BAC Clones from Divergent Oryza Lineages Constituting the Genus-Wide Adh1 Vertical Comparative Sequence Data Set
| Species | Genome | Clone Address | Insert Size (bp) | Total Length of the Sequenced Regiona |
|---|---|---|---|---|
| O. sativa ssp indica | AA | OSI9Ba083O10 | 142,000 | 299,037 |
| OSI9Ba092B13 | 170,000 | |||
| O. glaberrima | AA | OG__Ba066K08 | 120,493 | 120,493 |
| O. rufipogon | AA | OR_CBa141L10 | 155,003 | 155,003 |
| O. nivara | AA | OR_BBa102H20 | 204,633 | 204,633 |
| O. punctata | BB | OP__Ba004F03 | 128,102 | 128,102 |
| O. officinalis | CC | OO__Ba194G19 | 164,731 | 164,731 |
| O. australiensis | EE | OA_CBa016E12 | 251,568 | 251,568 |
| OA_CBa062H21 | 214,351 | 214,351 | ||
| O. brachyantha | FF | OB__Ba045I08 | 216,000 | 216,000 |
| O. granulata | GG | OG_ABa077F15 | 149,708 | 285,707 |
| OG_ABa032P05 | 176,311 |
Size of the sequence after trimming the overlapping regions. For O. australiensis (EE), the two BAC sequences are nonoverlapping.
Genomic Architecture of Oryza Genomes at the Adh1 Locus
Gene and Transposable Element Content
To achieve consistency in comparisons across the diploid Oryza species, we reannotated the OsAdh1RefSeq using a set of stringent criteria (see Methods; Ma et al., 2005). With some exceptions, the reannotated genes largely agreed with previous annotation results (Tarchini et al., 2000; TIGR rice annotation release V5; see Supplemental Table 1 online). We excluded 12 of the 61 annotated genes (TIGR V5) because they did not meet our gene confirmation criteria (see Methods) and corrected four gene and four transposable element (TE) models (e.g., merging of split genes; separating genes that were integrated into TE models and identification of pseudogenes that contained TEs). The excluded and corrected genes are listed in Supplemental Tables 2 and 3 online along with the reasons for exclusion or correction. For simplicity, when discussing genes and TEs for each species, we used the following nomenclature prefixes: J, japonica; I, indica; N, O. nivara; R, O. rufipogon; G, O. glaberrima; P, O. punctata; O, O. officinalis; A, O. australiensis; B, O. brachyantha; and GR, O. granulata.
The final reannotated data set for OsAdh1RefSeq resulted in the identification of 46 intact genes (including gene 12 that is embedded within a Pack-MULE TE, a mutator like element [MULE] that carries a gene or gene fragments [Jiang et al., 2004]) and four apparent pseudo (ψ) genes (originating by frameshift mutations [J13-2 and J13-3] and TE insertion [J11-9 and J11-12]). The structures of 36 genes were supported by full-length cDNA (Fl-cDNA) or Fl-assembled EST evidence and were classified as expressed, whereas the 10 remaining genes were classified as hypothetical (see Supplemental Table 1 online). An interesting feature of OsAdh1RefSeq was the frequent occurrence of duplicated genes (Tarchini et al., 2000). Seventy-six percent (38 out of 50, including ψ genes) of the annotated genes were organized into eight different gene families, many of them organized in tandemly arrayed clusters. These included genes J2, J5, J6, J11, J13, J14, J18, and J20 (see Supplemental Table 1 online; Figure 1). Copy numbers for the individual gene families varied from 2 to 14 (Figure 1; see Supplemental Table 1 online). Some of the gene families belonged to much larger superfamilies, such as F-box (Jain et al., 2007), nucleotide binding site–leucine-rich repeat (NBS-LRR) (Zhou et al., 2004; Yang et al., 2008), protein kinase (Yang et al., 2006), and receptor kinase genes (Zhang et al., 2005, 2007). Many of these gene families have been shown to be associated with biotic and abiotic stress responses. Supplemental Table 1 online summarizes the properties of the final OsAdh1RefSeq gene set and their putative functional classifications based on similarity to known plant genes.
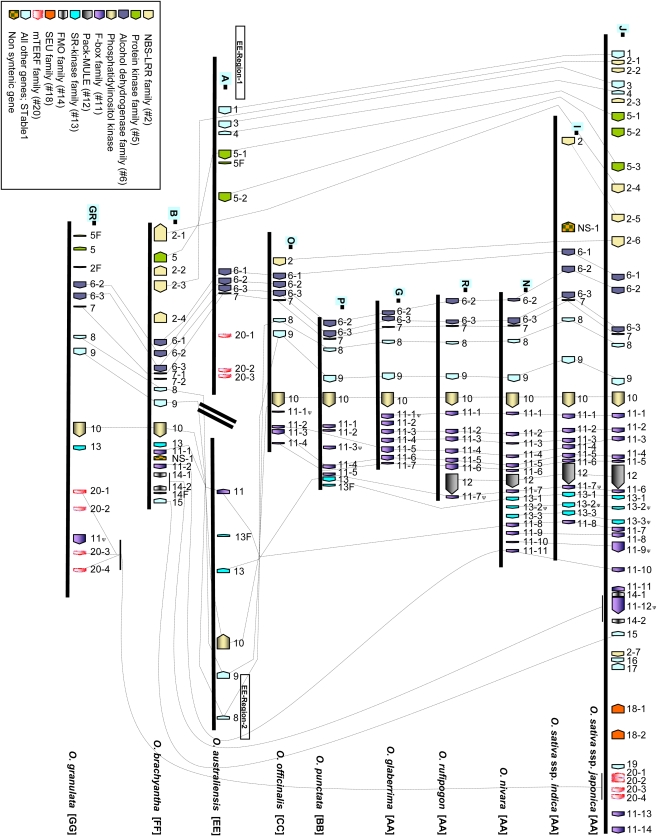
Figure 1.
Comparative Phylogenomic View of the Orthologous Adh1 Vertical Data Set across the Genus Oryza.
Genes from each Oryza species are denoted by the first letter of each species (i.e., J, japonica; I, indica; N, nivara; R, rufipogon; G, glaberrima, P, punctata; O, officinalis; A, australiensis, B, brachyantha; GR, granulata). Each gene family is color coded to depict dynamics of gene family evolution.
A total of 178 genes, plus 14 putative ψ genes and eight TE-embedded genes or gene fragments, were identified in the analyzed Oryza regions (see Supplemental Table 4 online; Figure 1). Gene density in the OsAdh1RefSeq was one gene every 13.3 kb (Table 3), slightly lower than the genomic average of 12.1 kb (International Rice Genome Sequencing Project, 2005; Rice Annotation Project, 2007). For the remaining diploid Oryza regions, gene density was highest in O. glaberrima (AA), with one gene every 10 kb, and lowest in O. australiensis (EE), with 1 gene per 27 kb (Table 3). For the purposes of estimating gene densities, only intact genes were considered, and this value would, in some cases, be higher if the ψ genes and TE-embedded genes were included. Variation in gene density was mainly due to differences in the physical length of intergenic space across the Oryza lineages (Figure 1). Such variations were observed even for the closely related and recently radiated A genome species.
Table 3.
General Genomic Features of the Genus-Wide Adh1 Vertical Comparative Sequence Data Set
| Genomic Feature | O. sativa ssp japonica (AA) | O. sativa ssp indica (AA) | O. nivara (AA) | O. rufipogon (AA) | O. glaberrima (AA) | O. punctata (BB) | O. officinalis (CC) | O. australiensis (EE) | O. brachyantha (FF) | O. granulata (GG) |
|---|---|---|---|---|---|---|---|---|---|---|
| Genome size (Mb) | 397 | 397 | 448 | 439 | 357 | 425 | 651 | 965 | 362 | 882 |
| Sequence size (kb) | 600 | 299 | 204 | 155 | 120 | 128 | 164 | 466 | 213 | 285 |
| Number of apparent intact genesa | 45 | 18 | 19 | 12 | 12 | 11 | 11 | 17 | 20 | 12 |
| Gene density (kb/gene) | 13.3 | 16.6 | 10.7 | 12.9 | 10.0 | 11.6 | 14.9 | 27.4 | 10.7 | 23.8 |
| Number of apparent pseudogenesb | 4.0 | 2.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | NO | NO | 3 |
| Number of genes carried by transposons | 1.0 | 2.0 | 2.0 | 1.0 | 1.0 | NO | 1.0 | NO | NO | 1 |
| Percentage of BAC sequence occupied by gene sequences (including introns) | 31.3 | 21.2 | 28.0 | 24.0 | 29.4 | 26.0 | 18.6 | 11.6 | 40.0 | 11.1 |
| Percentage of BAC sequences occupied by TE sequences | 41.2 | 53.7 | 47.6 | 48.6 | 39.0 | 46.5 | 56.4 | 58.8 | 28.3 | 66.2 |
| GC content (%) | 41.0 | 42.7 | 41.5 | 41.4 | 41.9 | 41.6 | 42.4 | 44.0 | 39.4 | 46.4 |
Genes that are partial due to their position at the end of a BAC sequence are included as intact.
Genes that have in-frame shifts leading to premature stop codons or that have TE insertions.
Table 2summarizes the compositional diversity and relative nucleotide contribution of different TE classes across the Oryza Adh1 regions. The OsAdh1RefSeq was found to be populated by all known classes and types of TEs. The lowest overall TE content in base pairs was found in O. brachyantha (FF) (28%), whereas O. granulata (GG) contained the highest (66%). Long terminal repeat (LTR) retrotransposons constituted the single largest TE class in terms of size contribution, whereas miniature inverted repeat transposable elements (MITEs) outnumbered all other TE classes in the compact Oryza genomes, including O. brachyantha (FF), one of the basal species. Surprisingly, the contributions of class II (DNA) elements exceeded or were comparable with those of the Class I (RNA) elements across Oryza, with the exception of the BB, EE, and GG genomes. This bias most likely reflects the fact that the vertical data set was gene rich. Previous studies have shown that DNA elements are more concentrated in gene-rich regions, whereas RNA elements are enriched in heterochromatic regions of the rice genome (International Rice Genome Sequencing Project, 2005; Zhang et al., 2007).
Table 2.
Composition and Sequence Contribution of TEs across the Genus-Wide Adh1 Vertical Comparative Sequence Data Set
| TEs | O. sativa ssp japonica (AA) | O. sativa ssp indica (AA) | O. nivara (AA) | O. rufipogon (AA) | O. glaberrima (AA) | O. punctata (BB) | O. officinalis (CC) | O. australiensis (EE) | O. brachyantha (FF) | O. granulata (GG) |
|---|---|---|---|---|---|---|---|---|---|---|
| Class I: retroelement
| ||||||||||
| LTR | ||||||||||
| Ty1/copia | 4.8 | 6.0 | 6.1 | 5.7 | 3.2 | NO | 2.0 | 16.0 | 9.9 | 13.0 |
| Ty3/gypsy | 4.6 | 9.9 | NO | NO | NO | 2.0 | 11.8 | 11.4 | 0.1 | 15.8 |
| Unclassified LTR | 0.7 | 0.6 | NO | NO | NO | 5.3 | 6.6 | 7.7 | 0.5 | 16.7 |
| LTR/TRIM | 0.1 | NO | NO | NO | NO | NO | NO | 0.1 | 0.1 | NO |
| SoloLTR | 1.1 | 5.8 | 2.0 | 2.6 | 0.8 | 13.4 | 2.7 | 5.0 | 1.1 | 3.6 |
| Non-LTR | ||||||||||
| LINE | 0.5 | 0.6 | NO | NO | NO | 3.5 | 1.9 | 2.8 | 0.2 | 0.2 |
| SINE | 0.4 | 0.2 | 0.1 | 0.1 | 0.1 | 0.3 | 0.3 | 0.4 | NO | 0.5 |
| Total Class I (%) | 12.1 | 23.0 | 8.2 | 8.4 | 4.2 | 24.5 | 25.3 | 43.4 | 12.0 | 49.7 |
| Class II: DNA transposon
| ||||||||||
| CACTA | 5.6 | 5.3 | 5.9 | 7.8 | 10.1 | 0.1 | 13.2 | 3.6 | 0.2 | NO |
| hAT | 2.4 | 1.2 | 2.3 | 1.9 | 1.4 | 3.1 | 2.2 | 2.2 | 0.5 | 3.3 |
| Helitron | 3.3 | 6.2 | 4.6 | 7.2 | 3.3 | 5.3 | 1.1 | 2.3 | 0.2 | 1.1 |
| PILE | 0.4 | 0.4 | 0.6 | 0.8 | 0.9 | NO | 3.4 | 0.1 | 0.4 | 3.6 |
| TC1 | 0.9 | 0.3 | 0.1 | NO | NO | 0.1 | NO | NO | NO | NO |
| MULE | 5.9 | 5.8 | 10.2 | 8.7 | 7.4 | 5.9 | 6.6 | 5.5 | 4.4 | 4.4 |
| Pack-MULE | 1.5 | 2.2 | 5.4 | 4.5 | 0.9 | NO | NO | NO | NO | NO |
| MITE/stowaway | 3.0 | 2.5 | 3.1 | 2.9 | 3.3 | 1.1 | 1.4 | 0.5 | 1.7 | 1.1 |
| MITE/tourist | 3.4 | 2.6 | 3.9 | 3.5 | 2.5 | 2.6 | 1.4 | 0.5 | 5.7 | 1.0 |
| Other class II | 2.9 | 4.3 | 3.3 | 2.9 | 5.0 | 3.7 | 1.4 | 0.6 | 3.2 | 2.0 |
| Total class II (%) | 29.1 | 30.7 | 39.5 | 40.2 | 34.8 | 22.0 | 31.1 | 15.4 | 16.3 | 16.4 |
| Total identified TEs (%) | 41.2 | 53.7 | 47.6 | 48.6 | 39.0 | 46.5 | 56.4 | 58.8 | 28.3 | 66.2 |
| Genome size (Mb) | 397 | 397 | 448 | 439 | 357 | 425 | 651 | 965 | 362 | 882 |
Values represent the percentage of each genome represented by the particular class of TE. NO, none observed.
Our data indicated that the majority of TE lineages could be identified using homology to known Oryza TEs, suggesting that most TE lineages have been present throughout Oryza evolution. However, these TE lineages differed in their degree of divergence, abundance, and genome size contribution (Table 2) in species-specific ways, as shown previously (Piegu et al., 2006; Ammiraju et al., 2007; Zuccolo et al., 2007, 2008; Kim et al., 2008).
Table 3 summarizes the general composition of the targeted genomic Oryza sequences in terms of genic, TE, and GC content. A positive correlation between GC content, TE content, and genome size was observed for these species (in each pairwise comparison, r2 value is >0.62). No major differences in the average length of genes, exons, and introns were found that could have had an impact on genome size among the corresponding Oryza lineages.
Divergence in Coding Regions, Phylogenetic Relationships, and Tentative Timing of Genus Radiation
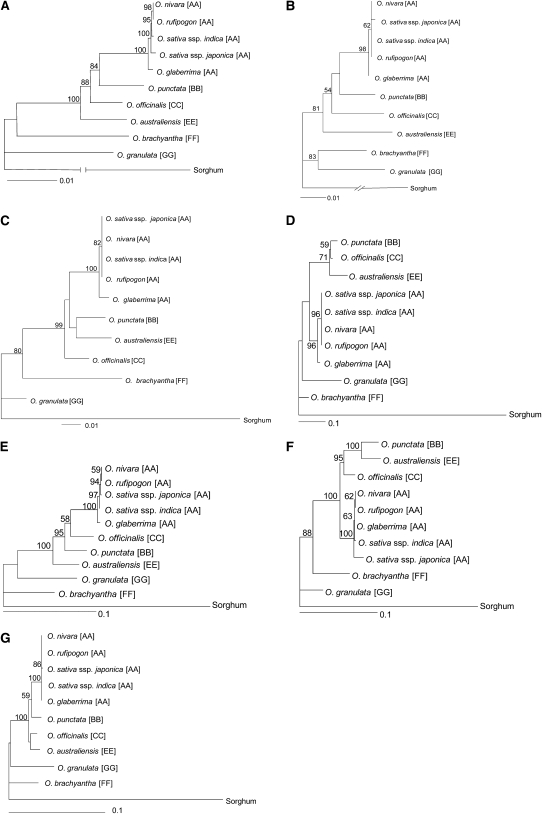
To interpret various rearrangements, their lineage specificity, and timing in a evolutionary framework, we determined the phylogenetic relationships and tentative molecular timing of respective Oryza speciation events using a core set of six genes (6-2, 6-3, 7, 8, 9-exon2, and 10) for which there was complete sequence coverage across the genus-wide vertical data set. Coding sequences from this core set were aligned to produce both individual and consensus phylogenetic trees (Figures 2B to 2G; see Supplemental Data Set 1 online). The topological relationships of the consensus tree (e.g., Figure 2A) were in agreement with previously published reports (Ge et al., 1999; Zou et al., 2008).
Figure 2.
Phylogenetic Relationships of Individual Conserved Core Genes across Oryza and a Consensus Tree Depicting All Topologies.
(A) Consensus diploid tree with all six core genes.
(B) Oryza Adh1 gene species tree.
(C) Oryza Adh2 species tree.
(D) Oryza RZ53 gene species tree.
(E) Oryza NifS gene species tree.
(F) Oryza Peroxidase gene species tree.
(G) Oryza PIK gene species tree.
To estimate the extent of sequence divergence, and, consequently, tentative species divergence times within Oryza, we calculated the synonymous (Ks) and nonsynonymous substitution (Ka) rate variation for the core gene set. Varying levels of Ks and Ka were observed among the genes (see Supplemental Table 5 online). All core genes appeared to be under purifying selection as indicated by Ka/Ks values of <1 (see Supplemental Table 5 online), except for gene 10 in O. nivara and O. rufipogon, and all core genes produced phylogenetic trees of similar topology. Two out of the six core genes showed elevated Ks values in one or more Oryza species (i.e., B6-2 [Adh2] in O. brachyantha [FF], and A7 and GR7 in O .australiensis [EE] and O. granulata [GG]). Previous studies have shown that the summing of overall synonymous divergence from a reasonable number of genes can reduce inaccuracies of exceptional rate variation in calibrating molecular clocks (Zhang et al., 2002; Ma and Bennetzen, 2004). Because the core gene set is small, and to avoid introducing a bias in the molecular clock estimate, we excluded Ks values for genes 6-2 and 7 in deriving an average molecular divergence time. Using a mutation rate of 6.5 × 10−9 mutations per synonymous site per year (Gaut et al., 1996) and a combined average of four core genes, we estimated that the genus Oryza diversified from a common ancestor ∼15 million years ago (MYA) (see Supplemental Table 6 online). It should be noted, however, that the molecular divergence calculated for O. nivara and O. rufipogon is from a single gene (10).
Our results suggest that the A genome lineages diversified∼0.58 MYA, whereas the BB, CC, and EE genome lineages diversified around 5.7 to 7.5 MYA (all within a 2-MY range). The FF and GG genomes, the most distantly related species, last shared a common ancestor with the AA-BB-CC-EE clade ∼13 to 14.6 MYA (within a 1-MY range).
Divergence in the Intergenic Regions and the Extent of Sequence Flux
Recent intra- and interspecies (Ma and Bennetzen, 2004; Ma et al., 2004) comparisons among three AA genome lineages of Oryza revealed rapid and ongoing sequence flux in intergenic regions that affected the overall size of these genomes. To our knowledge, no information is available with respect to the degree of intergenic sequence divergence in the remaining AA genome species and the nine other Oryza genome types relative to the O. sativa Refseq. To investigate this question, pairwise global sequence alignments were conducted between individual Oryza species with a core segment (6-2 to 10 interval; Figure 1) of the OsAdh1RefSeq that had complete sequence coverage in all Oryza species. The results indicated a massive replacement of intergenic space that ranged from ∼71 to ∼95% in the BB thru the GG genome types, respectively, relative to the OsAdh1Refseq, through independent and lineage-specific insertions and deletions (see Supplemental Table 7 online). These results are consistent with previous cross-genus studies among grasses (i.e., sorghum-maize and wheat-barley) that have shown nearly complete divergence within a 9 to 14 MY time frame (Tikhonov et al., 1999; Ramakrishna et al., 2002c). In the AA genomes, ∼15 to 30% of intergenic sequence has been replaced since their divergence from a common ancestor.
Specificities and Chronology of LTR Retrotransposon Accumulation, Retention, and Elimination
Across the genus, we identified a total of 35 intact LTR retrotransposons (defined as a retrotransposable element containing two intact LTR sequences) and 60 solo LTRs, which are by-products of unequal homologous recombination between paired LTRs from the same or related LTR retrotransposons. Supplemental Table 8 online tallies the number, size, and coordinates of intact LTR retrotransposons and solo LTRs in the compared regions from each Oryza species. They ranged from zero in O. glaberrima (AA) to 11 in O. australiensis (EE) and 10 in O. granulata (GG). We estimated the insertion time of each intact element using the level of sequence divergence between the two LTRs and a substitution rate of 1.3 × 10−8 (Ma and Bennetzen, 2004) as described by SanMiguel et al. (1998). The insertion time estimates ranged from 0.01 to 10 MYA.
LTR Retrotransposon Dynamics in the AA Genome Lineage
Analysis of the japonica (OsAdh1RefSeq) and indica subspecies of cultivated rice revealed the presence of nine intact LTR retrotransposons (five in ssp japonica and four in ssp indica) and 12 solo LTRs (three in japonica and nine in indica) with only one intact element (J-ILTR1) and one solo LTR (J-sLTR5) shared between them. The average insertion time estimated for the three unshared intact elements in indica was 0.13 MYA, which is much lower than the estimated divergence time of 0.22 to 0.44 MYA for indica and japonica haplotypes (this study; Ma and Bennetzen, 2004). On the other hand, only two out of the four unshared intact elements in the japonica region were dated to be younger than the species divergence time (Table 4; see Supplemental Figure 1 online). Collectively, these five intact elements and 10 solo LTRs represent independent insertion and deletion events in the two lineages after divergence from a last common ancestor (LCA) (see Supplemental Figure 1 online). The other two unshared intact LTR retroelements identified in the OsAdh1RefSeq were estimated to have inserted ∼1.4 MYA (J-ILTR3) and 1.18 MYA (J-ILTR4), thus predating A genome radiation (see above), suggesting their retention only in the japonica region and deletion in the other A genome species. In fact, a solo LTR with an intact target site duplication (TSD) was identified at the same syntenic position in all other A genome species for J-ILTR3, thereby supporting this hypothesis (see Supplemental Figure 1 online).
Table 4.
Intact LTR Retrotransposons and Solo LTRs in the Aligned Adh1 Regions
| Age (MYA)
|
||||||
|---|---|---|---|---|---|---|
| Species | Species Comparison | Intact | Solo LTR | Range | Average | Intact:Solo LTRa |
| O. sativa ssp indica (AA) | Present only in japonica | 4 | 2 | 0.01–1.4 | 0.69 | 1.67:1 |
| Present only in indica | 3 | 8 | 0.03–0.96 | 0.13 | 0.44:1 | |
| Common to both | 1 | 1 | 0.96 | – | – | |
| O. nivara (AA) | Present only in japonica | 4 | None | 0.01–1.4 | 0.69 | 5:0 |
| Present only in nivara | None | 2 | – | – | 1:3 | |
| Common to both | 1 | 1 | 1.12 | – | – | |
| O. rufipogon (AA) | Present only in japonica | 2 | None | 0.01–1.4 | 0.71 | 3:1 |
| Present only in rufipogon | None | 2 | – | – | 1:3 | |
| Common to both | 1 | 1 | 1.12 | – | – | |
| O. glaberrima (AA) | Present only in japonica | 3 | None | 0.01–1.4 | 0.79 | 3:1 |
| Present only in glaberrima | None | 1 | – | – | 0:2 | |
| Common to both | – | 1 | – | – | – | |
| O. punctata (BB) | Present only in japonica | 3 | 1 | 0.01–1.4 | 0.79 | 3:1 |
| Present only in punctata | 1 | 9 | 1.05 | – | 1:9 | |
| Common to both | – | – | – | – | – | |
| O. officinalis (CC) | Present only in japonica | 3 | 3 | 0.01–1.4 | 0.79 | 1:1 |
| Present only in officinalis | None | 2 | – | – | 0:2 | |
| Common to both | – | – | – | – | – | |
| O. australiensis (EE)b | Present only in japonica | 4 | 3 | 0.01–1.4 | 0.89 | 1:0.75 |
| Present only in australiensis | 11 | 13 | 0.05–3.6 | 0.96 | 1:1.18 | |
| Common to both | – | – | – | – | – | |
| O. brachyantha (FF) | Present only in japonica | 6 | 3 | 0.01–1.4 | 0.62 | 2:1 |
| Present only in brachyantha | 1 | 6 | 2.85 | – | 1:6 | |
| Common to both | – | – | – | – | – | |
| O. granulata (GG) | Present only in japonica | 6 | 3 | 0.01–1.4 | 0.62 | 2:1 |
| Present only in granulata | 10 | 5 | 0.11–10 | 2.84 | 2:1 | |
| Common to both | – | – | – | – | – | |
From the aligned regions only.
From both BACs of O. australiensis.
The single shared intact element (J-ILTR1) between indica and japonica was also shared with O. rufipogon (AA) and O. nivara (AA) but was absent in O. glaberrima (AA). Since the insertion time of J-ILTR1 was estimated to be older (0.96 MYA) than the radiation time of O. glaberrima from the LCA of O. sativa (0.58 [this study] to 0.67 MYA; Ma and Bennetzen, 2004), it is likely that J-ILTR1 was either present in O. glaberrima and has since been deleted, or it inserted at its present syntenic position immediately after divergence of O. glaberrima from the other AA genome lineages. Similarly, a solo LTR (J-sLTR5) was found in all A genome species but absent at the orthologous position in O. punctata (BB), whereas another solo LTR (I-sLTR7) was shared among indica, O. rufipogon, and O. nivara regions but was absent in the other AA genomes, suggesting differential accumulation and retention of LTR retrotransposons independently and in a lineage-specific fashion across these species.
LTR Retrotransposable Elements Are Not Conserved between the Oryza Genome Types at the Adh1 Region
We were unable to identify orthologous intact LTR retrotransposons or solo LTRs beyond the AA genome species. The majority of intact LTR retrotransposons that differentiate these genomes appear to have inserted quite recently and independently within the last 5 MY (50% within 1 MY and 75% within 2 MY) after speciation (see Supplemental Table 8 online). Our inability to detect LTR retrotransposon insertions that occurred prior to speciation is consistent with the unstable nature of LTR retrotransposons and their rapid turnover in plants, mainly due to illegitimate or unequal recombination (Ma and Bennetzen, 2004).
To assess the variation in the rate of unequal recombination and its impact on independent compressions and expansions, we calculated the ratio of intact elements to solo LTRs (Table 4). Aligned orthologous regions from pairwise comparisons of each Oryza species with OsAdh1Refseq were used for this purpose. The general trend observed was a recent expansion of OsAdh1RefSeq relative to most of the other orthologous regions (Table 4) due to recent independent insertions of LTR retrotransposons and also by elevated rates of unequal recombination in the other species. These results further confirm the findings of Ma and Bennetzen (2004) and extend them to a genus-wide perspective. Two species in particular, O. punctata (BB) and O. brachyantha (FF), exhibited elevated rates of unequal recombination, leading to recent compressions of their respective genomic regions (Table 4). It should be noted that the O. brachyantha (FF) genome is one of the smallest in the genus, whereas O. punctata (BB) has the smallest genome in the recently and rapidly radiated O. officinalis complex (Miyabayashi et al., 2007; Zhang and Ge, 2007). This complex includes five diploid species with CC, BB, and EE genomes and four allotetraploid species having a BBCC or CCDD genomic constitution. Two other species, O. glaberrima (AA) and O. officinalis (CC), had no recent insertions in the Adh1 genomic region.
Microsynteny
Despite the rapid evolutionary divergence detected in the Adh1 intergenic regions of the diploid Oryza, gene order and orientation were largely maintained. However, several structural rearrangements were observed that differentiate these genomic regions. The evolutionary origins and underlying molecular mechanisms of these rearrangements are described in the following sections.
Asymmetric Evolution of Gene Families Has Caused Frequent Perturbations in Synteny
A large proportion of unshared genes was found in the regions flanking the 6-2 to 10 core genic interval and was found to increase with phylogenetic distance within the genus (see Supplemental Table 9 online). We observed several synteny perturbations associated with complex and dynamic patterns of tandemely duplicated genes belonging to different families. These patterns included lineage-specific gene gain (duplication), loss (pseudogenization or elimination), and other small or large structural rearrangements. Pertinent results for each gene family are described in detail below.
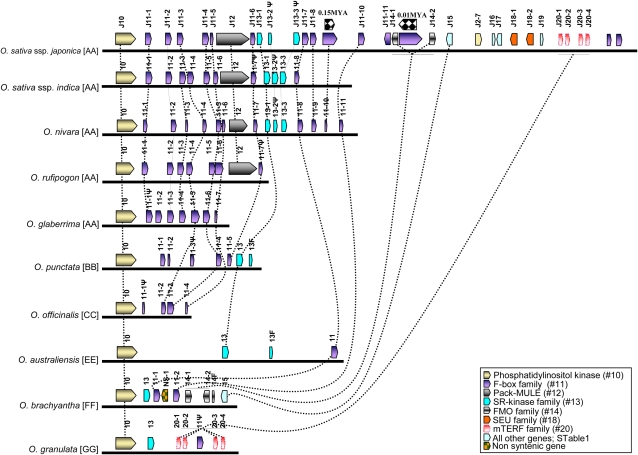
F-box Gene Family
The F-box gene family constitutes one of the largest families in the rice genome, with nearly 687 members that are distributed across all 12 chromosomes (Jain et al., 2007). The F-box family expanded quite recently in the rice genome, predominantly by localized tandem duplications (Jain et al., 2007). Annotation of OsAdh1RefSeq revealed the presence of 14 F-box or F-box–related genes (Figures 1 and 3) designated J11-1 to J11-14. Ten are transcriptionally active, as supported by Fl-cDNA or Fl-assembled EST evidence (see Supplemental Table 1 online; Figures 1 and 3), two were classified as hypothetical genes (i.e., J11-5 contains an F-box domain; and J11-14 is phylogenetically related to all F-box genes in this region [see Supplemental Table 1 online; Figure 4]), and two (J11-9 and J11-12) contained LTR retrotransposon insertions and were classified as ψ genes. All 14 were organized in the same transcriptional orientation, except gene J11-11, and were interspersed with unrelated genes in this region. We used a phylogenetic approach, using nucleotide and protein sequences, to determine orthologous and paralogous relationships among these genes (Figure 4; see Supplemental Data Set 2 online). This analysis revealed (Figures 1, 3, and 4) patterns of rapid divergence (clades that were not well supported by bootstrap values) and variation in copy number due to lineage-specific gene loss and gain. A single F-box gene (I11-3; Figures 1 and 3) was identified that was conserved in all AA genome species except OsAdh1RefSeq, suggesting that it is a recent and lineage-specific deletion in ssp japonica that occurred after divergence from the LCA. Furthermore, gene 11-6 was found to be intact in OsAdh1RefSeq and O. nivara (AA) but contained frameshift mutations causing premature stop codons in both the O. rufipogon (AA) and the indica subspecies. In addition, for one of the two OsAdh1RefSeq F-box pseudogenes (J11-9ψ) containing TE insertions, an ortholog that contained an intact open reading frame (ORF) was identified in O. nivara, suggesting that it is a recent pseudogenization specifically in ssp japonica after its divergence from a common progenitor. Dating of the LTR retrotransposon insertions into genes J11-9ψ and J11-12ψ at ∼0.15 and ∼0.01 MYA, respectively, confirmed that pseudogenization was recent.
Figure 3.
Expanded View of Figure 1 That Depicts F-box Gene Family Dynamics.
Figure 4.
Unrooted Phylogenetic Tree Demonstrating the Evolutionary Origin and Diversification of F-box or F-box–Like Genes in the Genus-Wide Adh1 Vertical Sequence Data Set. Bootstrap values (.50%) are displayed on branches.
Interestingly, a subcluster of F-box genes (J11-1, J11-2, J11-3, J11-4, and J11-5; Figures 1 and 3) was found to span the genomic interval between OsAdh1RefSeq genes J10 and J12 (Pack-MULE) in all AA, BB, and CC genome species but was completely absent in the EE, FF, and GG genomes. Although these latter species did not contain F-box genes in this genomic interval, we did find evidence for the presence of this gene class in the orthologous regions of the allopolyploids O. ridleyi (HHJJ) and O. coarctata (HHKK) (see Supplemental Figure 2 online). These data suggest that an F-box gene or gene(s) resided in this location, between genes 10 and 12, before the divergence of the Oryza from a LCA and that the F-box gene(s) underwent lineage-specific deletion, amplification, and pseudogenization during the course of Oryza evolution.
S-Receptor Kinase Family
We identified an orthologous family of putative Ser/Thr receptor-like kinase genes (OsAdh1RefSeq gene family J13; Figure 1; see Supplemental Table 1 online) that could be traced from the AA through the GG genome types (see Supplemental Table 10 online; Figures 1 and 3). For the three AA genome species where full sequence coverage was available (i.e., OsAdh1RefSeq, O. sativa ssp indica, and O. nivara), this gene family consisted of three family members (13-1, 13-2, and 13-3). Orthologs (Figures 1 and 3) for J13-1 had intact ORFs in all AA genomes, whereas all three orthologs for J13-2 contained frameshift mutations and were therefore classified as ψ genes. Interestingly, the three J13-3 orthologs had intact ORFs in both O. sativa ssp indica and O. nivara but had frameshift mutations in OsAdh1RefSeq, suggesting a recent pseudogenization of this copy. Although, this gene family was not found in O. rufipogon (AA), O. glaberrima, and O. officinalis (CC), due to lack of sequence coverage in the region, it is highly likely that this family is still (or was) present in these lineages as well, since it was detected in all other Oryza genome types.
The remaining genome types either had full (EE, FF, and GG), partial (BB), or no sequence coverage (CC). A single member of this family was present in the BB, EE, FF, and GG genomes as well as two gene fragments in the BB (P13F) and EE (A13F) genomes. Phylogenetic relationships among these gene family members were determined and are shown in Supplemental Figure 3B and Supplemental Data Set 3 online, except for the highly divergent single family members found in the FF and GG genomes (no bootstrap support; Figure 1; see Supplemental Figure 3B online). Variation in copy number of gene family 13 suggests that the common ancestor of Oryza contained three copies of gene family 13 that were retained in the AA genome lineage but were deleted independently in the BB, EE, FF, and GG lineages. Alternatively, multiple gene copies found in the EE, BB, and AA genome types arose in these lineages via gene duplication. Two lines of evidence suggest that the first case is more likely. First, the identification of two fragments of this family in orthologous positions in the BB and EE genomes provides evidence that functional genes may have resided in these locations but have since been partially deleted and rearranged. Second, the estimated timing of duplication of the three paralogs (J13-1, J13-2, and J13-3) at 23 to 33 MYA suggests an ancestral origin of the organization of gene family 13 (see Supplemental Table 13A online).
Flavin Monooxygenase Gene Family
We identified a flavin monooxygenase (FMO) gene family (J14-1 and J14-2; Figure 1; see Supplemental Table 1 online) in OsAdh1RefSeq that could be traced to the FF genome of O. brachyantha but was not found in O. granulata (GG). No sequence coverage was available for the remaining AA, BB, CC, and EE genome types to determine if these genes were present. In O. brachyantha (FF), the FMO gene family was composed of two intact genes and one truncated (ψ) gene (Figure 1). Phylogenetic analysis revealed species-specific grouping with two FMO genes (J14-1 and J14-2) from O. sativa forming one clade and those from O. brachyantha forming a second clade (see Supplemental Figure 3A online), suggesting lineage-specific evolution/divergence of this family. These data also suggest that the origin of the FMO genes in their present location near Adh1 began in the FF genome lineage, probably by movement from another region in a genome ancestral to the O. brachyantha genome. Alternatively, the FMO family originated near Adh1 in a common ancestor of the FF and GG genomes, as hypothesized above, but has been deleted from the GG genome lineage.
Mitochondrial Transcription Termination Factor Gene Family
Four copies of the mitochondrial transcription termination factor (mTERF) family (OsAdh1RefSeq gene family J20; Figure 1; see Supplemental Table 1 online) were found in both O. sativa subspecies. The Adh1-linked location of this family appeared to be ancient since we could identify family members in orthologous positions in both O. australiensis (EE) and O. granulata (GG). Phylogenetic analysis revealed a clustering of all four mTERF members of O. granulata (GG) in a single monophyletic group (see Supplemental Figure 3F and Supplemental Data Set 3 online). Two of the four genes appear to have originated by recent duplication events (GR20-1-GR20-3 duplication ∼0.4 MYA; GR20-2-GR20-4 duplication ∼5.4 MYA; see Supplemental Figure 3F online), suggesting independent and lineage-specific evolution of this family in the GG genome. A20-3 strongly clustered with 20-1 of the japonica and indica genomes, whereas A20-1 and A20-2 clustered strongly with 20-2 of indica and japonica, indicating either a putative inversion in O. australiensis (EE) relative to O. sativa involving this segment (see Supplemental Figures 3F and 5 online) or a duplication in an inverse orientation. As in the case of O. granulata (GG), the lineage-specific birth of a new mTERF member was also found in O. australiensis (EE) in the form of an A20-1 to A20-2 duplication that occurred ∼1.7 MYA.
Protein Kinase Gene Family
We identified an ancient protein kinase family (OsAdh1RefSeq gene family members J5-1, J5-2, and J5-3; Figure 1; see Supplemental Table 1 online) that could be traced across orthologous positions in the AA, EE, FF, and GG genomes. Two intact genes (A5-1 and A5-2) and one gene fragment (A5F) from this family were found in the orthologous region of O. australiensis (EE). One member of this family was found in each of the O. brachyantha (FF) and O. granulata (GG) genomes but was found in opposite transcriptional orientation relative to the AA and EE genomic regions. Phylogenetic analysis indicated that A5-2 is orthologous to O. sativa J5-3/I5-3 and that A5-1 is tightly grouped with O. sativa J5-1 and J5-2 (see Supplemental Figure 3C and Supplemental Data Set 3 online). We dated the J5-1/5-2 duplication to ∼10 MYA based on the synonymous substitution rate, which is higher than the species divergence time with O. australiensis, thereby suggesting its deletion in the O. australiensis genome subsequent to speciation. The single member of this family (GR5) in O. granulata (GG) was present in the same transcriptional orientation relative to O. brachyantha (FF) but in opposite orientation relative to OsAdh1Refseq, indica, and O. australiensis (EE) (Figure 1). In addition, GR5 was highly divergent in comparison to its homologs in the rest of the Oryza species, as indicated by its basal position, and no cladistic support in the phylogenetic tree (see Supplemental Figure 3C online).
Alcohol Dehydrogenase (Adh) Gene Family
We identified three members of the alcohol dehydrogenase gene family on OsAdh1RefSeq (J6-1, J6-2, and J6-3) that could be traced across all diploid Oryza genomes where sequence coverage was available, including the CC, EE, FF, and GG genome types. Although only a small gene fragment of the Adh3 gene could be identified in O. granulata (GG), its presence confirms that this triplication predates the divergence of these Oryza lineages.
Previous analysis indicated that the Adh1 and Adh2 duplication predates the grass radiation (∼65 MYA; Gaut et al., 1999). To test whether Adh3 was also part of such an ancient duplication or was recently duplicated in Oryza only, we estimated the synonymous substitution rate (Ks) through pairwise alignments with Adh1 and Adh2 coding sequences and calculated the timing of duplication to be >53 MYA, indicating that Adh3 was also an ancient duplication and that its duplication partner was Adh2 (see Supplemental Table 13B online). Furthermore, pairwise comparisons of the three paralogs demonstrated that, like the Adh1 and Adh2 genes of O. sativa, Adh3 is also under strong functional constraint, as indicated by a Ka/Ks ratio <0.5 (see Supplemental Table 13B online).
NBS-LRR Gene Family
We identified a NBS/LRR gene family (J2; Figure 1; see Supplemental Table 1 online) in OsAdh1RefSeq that could be tracked in the orthologous positions in the AA (O. sativa ssp indica), CC, and FF genome types but was absent in the EE genome type. Although partial sequence coverage in this region was available for the GG genome, we were unable to precisely determine the presence or absence of this family at the orthologous position. Although no orthologous sequences were available for three AA genomes (O. nivara, O. rufipogon, and O. glaberrima) and O. punctata (BB), the fact that this family was found in the FF genome suggests that it originated before the FF genome. Alternatively, it could have originated in a common ancestor with the GG genome or earlier in evolutionary time but has since been deleted.
In OsAdh1RefSeq, the NBS/LLR family is composed of seven genes (NB-ARC [two copies: J2-1 and J2-7], LRR [three copies: J2-2, J2-3, and J2-5], and NBS-LRR [NBARC-LRR domains; two copies: J2-4 and J2-6) (Figure 1; see Supplemental Table 1 online). The orthologous indica sequence retained all orthologous copies except gene J2-6. Although we only had partial sequence coverage for the CC and FF genome types, we were able to identify one and four members of this family, respectively. Further phylogenetic analysis of the FF genome NBS-LRR genes with the same from OsAdh1Refseq revealed a complex set of rearrangements (see Supplemental Figures 3E and 4 and Supplemental Data Set 3 online) that included (1) an inversion involving a segment between B2-1 and B2-3 of O. brachyantha with J2-1 to J2-4, (2) a deletion of two protein kinase genes within this inverted segment of O. brachyantha, and (3) a duplication of B2-2 to B2-4. Because the single protein kinase gene B5 (Figure 1; see Supplemental Figure 4 online) was located in the opposite orientation relative to OsAdh1RefSeq, O. australiensis (EE), and O. coarctata (HHKK) genomes, this inversion appears to be specific to the FF genome.
RZ53
Gene J7 (Figure 1; see Supplemental Table 1 online) was classified as an expressed gene in OsAdh1Refseq and is one of the six core genes that traverse the diploid Oryza phylogeny. It was present as a single copy in all Oryza species, except in O. brachyantha (FF), where an additional copy was found adjacent to the first copy (Figure 1). Like in the case of the first copy, the second copy was also under purifying selection (Ka/Ks = <0.5; see Supplemental Table 13C online). However, the gene duplication was estimated to have happened >20 MYA, based on the Ks value between the two paralogs (see Supplemental Table 13C online). These data suggest that the duplication of the RZ53 gene in the FF genome occurred early after the separate descent of this lineage from the other Oryza and has since diverged quite rapidly relative to the parental gene. Alternatively, the duplication may be more ancient but has since been retained only in O. brachyantha (FF).
Lineage-Specific Structural Rearrangements
A 350-kb Inversion in O. australiensis (EE)
An inversion/deletion spanning >350 kb (relative to O. sativa ssp japonica RefSeq sequence) was identified in the O. australiensis (EE) genome by comparative sequence and physical map analysis (Figure 1; see Supplemental Figures 5 and 6 online). Analysis showed that, with the exception of gene copy number variation and the absence of gene family 2, the order and orientation of genes A1 to A7 was preserved on a single 251-kb BAC clone from O. australiensis (OA_CBa0016E12). However, instead of genes A8, A9, A10, A11, and A13 immediately following A7 in O. australiensis (EE), gene family 20 was located next to gene A7, which is situated 350 kb away from gene J7 in the rice Refseq, suggesting that a large inversion/deletion occurred between genes A7 and A20 in O. australiensis. To determine whether a deletion/inversion occurred between genes A7 and A20 in the EE genome, we screened the O. australiensis BAC library with gene-specific probes for genes 8, 9, and 10, thereby identifying a set of clones that fell into a single contig the could be placed on the O. australiensis physical map very close to the O. australiensis Adh1 BAC (OA_CBa016E12; region-1; see Supplemental Figure 6 online) containing genes A1 to A7. A single BAC clone was selected from this contig (OA_CBa062H21; region-2), sequenced and annotated, and was shown to contain genes A8, A9, and A10 (see Supplemental Figures 5 and 6 online), thereby supporting the hypothesis that a large inversion and not a deletion occurred between A7 and A20.
Because gene order (specifically from genes 7 through 10) was found to be contiguous in this region for all the diploid Oryza species, and there were no large genomic rearrangements with the exception of O. australiensis, it is likely that this inversion is specific to the EE genome. The orthologous region of a distantly (>50 MYA) related grass, Sorghum bicolor (http://www.phytozome.net/sorghum), retained the same order of genes from 6-3 to 10, thereby confirming its ancestral nature and further supporting the conclusion that a large-scale inversion occurred in the Adh1 region in the lineage leading to the O. australiensis genome.
An ∼200-kb Deletion/Inversion in O. granulata (GG)
A large orthologous segment of ∼219 kb, containing ∼13 genes and 3ψ genes between J13-1 and J20-1, appears to have been deleted in O. granulata (GG) relative to OsAdh1RefSeq (Figure 1; see Supplemental Figure 7 online). Because this orthologous region was found intact in O. brachyantha (FF) (based on the presence of genes B13-1, B14-1, B14-2, and B15), the GG genome rearrangement appears to have occurred specifically in the O. granulata lineage. To validate this hypothesis, we examined the orthologous region of the S. bicolor genome for the presence or absence of the ∼219-kb region and were able to identify genes 6-3, 8, 9, 10, and 14 in the same order and orientation as OsAdh1RefSeq and the FF genome. The presence or absence of gene family 13 and 11 could not be precisely determined because of the incompleteness of the S. bicolor draft sequence in this region. These results suggest that the large rearrangement (either a deletion or an inversion) detected in O. granulata occurred specifically in the GG lineage.
Gene Movement
We identified a total of four genes or gene fragments that appear to have originated in the Adh1 region by relocation; movement of two was TE mediated, and the other two moved by unknown mechanisms.
Pack-MULE Mediated Gene Movement
OsAdh1RefSeq gene J12 was found to be transcriptionally active (as indicated by Fl-cDNA support) and encodes a putative LRR kinase with 54% protein sequence similarity to the bacterial blight resistance gene Xa21 (Song et al., 1995; Tarchini et al., 2000). This gene was conserved only in the AA genome lineages, for which full sequence coverage was available (i.e., OsAdh1RefSeq, ssp indica, O. nivara, and O. rufipogon). Although the coding sequence and total predicted protein length was similar to Xa21, the single intron in gene 12 varied in size from 6.5 kb (japonica, indica, and O. rufipogon) to 0.1 kb (O. nivara) (see Supplemental Figure 8 online). Careful examination of this conserved gene showed that it was embedded within a Pack-MULE, of which the terminal inverted repeat (TIR) was previously described (Os0874 family; Jiang et al., 2004). All four orthologous Pack-MULEs contained intact TSDs (see Supplemental Figure 8 online), with the exception of a degraded TIR in OsAdh1RefSeq. No putative parental gene of J12 was identified in the IRGSP RefSeq.
To determine the origin of this Pack-MULE, we examined the BB genome of O. punctata (BB) and found a MULE, at the same location with intact TSDs (see Supplemental Figure 8 online). This MULE was absent at the syntenic positions in EE, FF, and GG genomes. Although no sequence coverage was available for the CC genome of O. officinalis, examination of orthologous sequences in the BB and CC subgenomes of O. minuta (BBCC) revealed the presence of the identical MULE in the BB subgenome only. Thus, our data demonstrate that a single MULE invaded the genome of a common ancestor of the AA and BB genomes prior to polyploidization >5.7 MYA. It is likely that this ancestral MULE was a Pack-MULE that had already acquired the LRR kinase gene homology. In this model, a presumably early event in the derivation of the BB lineage was the deletion of most of the internal sequences of this Pack-MULE in a common ancestor of the O. punctata and O. minuta BB chromosomes. Multiple independent indels would explain the current variation in the AA genome LRR kinase regions inside this Pack-MULE. Alternatively, the LRR kinase homology may have been acquired within an early AA genome progenitor, but this would require that the MULE became a Pack-MULE by an internal ectopic conversion without transposition of the element.
A second case of TE-mediated gene movement was found by the identification of a Pack-MULE (Os0053 family; Jiang et al., 2004) with identical TSDs in all AA genome lineages where sequence coverage was available, except for OsAdh1RefSeq. The Pack-MULE contains a single gene fragment that putatively originated from gene dbj|BAD27916.1 located on rice chromosome 2. The absence of this Pack-MULE along with the gene fragment at the syntenic location of OsAdh1RefSeq, but the presence of a single intact TSD followed by ∼68 bp of unrelated sequence suggests lineage-specific excision of this Pack-MULE followed by gap repair.
Gene Movement by Unknown Mechanisms
A noncolinear, non-TE-related hypothetical gene (INS-1; Figure 1) was identified in the indica subspecies only (Figure 1). This candidate gene has significant homology to tubulin-specific chaperon binding cofactor genes (pfam domain 07986). Similarity searches detected the presence of two intact copies of this gene in the rice RefSeq (LOC_Os06g41110 on chromosome 6 and LOC_Os02g10130 on chromosome 2, and the structures of both were supported by Fl-cDNA gi|37990492|dbj|AK120869.1). These two RefSeq copies (chromosomes 6 and 2) are also found at the syntenic locations in the indica subspecies (contig002647 and contig002647, respectively). No flanking sequences of these two RefSeq genes could be found in the corresponding Adh1 region of indica, suggesting that (INS-1) most likely originated in its present location in ssp indica by insertion and not by deletion in the rice RefSeq.
Lastly, a single nonsyntenic gene was identified in O. brachyantha (FF) (BNS-1; Figure 1). Two homologs of this gene were found in a recently duplicated region of chromosome 11 and 12 of the RefSeq (LOC_Os11g03200 and LOC_Os12g02950; both have Fl-cDNA support), although it is predicted to be 100 amino acids shorter in protein length than the rice RefSeq homologs. The precise mechanism of this single gene or gene fragment movement is not known.
DISCUSSION
Here, we report the generation and analysis of a unique genus-wide vertical comparative sequence data set, encompassing all diploid genome types of Oryza, using a single biologically important genomic region. It constitutes the largest comparative genomic sequence layout available for any plant genus. The power of this data set lies in the resolution it offers. With representation of a broad ecological selection history over a short evolutionary period (∼15 MY; this study) and an outgroup species at every phylogenetic node, this data set finds comparative resolution only in studies of the genera Drosophila (Clark et al., 2007) and Gossypium (Grover et al., 2007, 2008) among higher eukaryotes.
We chose the Adh1-Adh2 region for our comparative analysis for several reasons. This region has long been the subject of intense genetic, evolutionary, and functional investigation across many plant lineages. Furthermore, it was one of the first local chromosomal segments developed as a comparative exploratory model for microsynteny among the cereals. The rice Adh1-Adh2 region on chromosome 11 is microsyntenic to that of maize (Zea mays) chromosome 4 (Tikhonov et al., 1999; Tarchini et al., 2000). However, this colinearity is punctuated by several exceptions; a significant interruption is mediated by the Adh1 gene relocation in a common ancestor of these grasses to chromosome 1 in maize and a colinear region on linkage group C in sorghum (Tikhonov et al., 1999; Ilic et al., 2003). Beyond these results, the long-term evolution of this region is largely unknown. Here, we extend the study of the evolutionary history of this region to a broad array of closely related Oryza species.
Overall, our analyses unveiled several significant insights into the history and tempo of Oryza evolution. First, our results indicated that the Adh1-Adh2 region has undergone a number of physical changes in a relatively short evolutionary time frame; second, a large number of these changes are very recent (and thus narrowly lineage specific), and at least some are frequent (e.g., duplications). We discuss here three major forces contributing to genomic instability: (1) gene family variation, (2) TE action, and (3) other short- and long-range DNA rearrangements. However, given the rapid and lineage-specific diversification coupled with a partially overlapping sequence data set, it was not surprising that we were able to decipher the exact order and timing of most, but not all, of these major rearrangement events.
Much of the decline in the number of shared genes was observed in the members of various gene families of the sequenced region (Figure 1; see Supplemental Table 9 online). Specific features observed are (1) remarkable plasticity in the copy number of individual gene families, (2) continuous lineage-specific gain and loss of individual members or entire gene clusters throughout the evolution of Oryza at every ancestral node, (3) rapid evolution relative to many neighboring genes, and (4) an apparent ongoing trend for the steady increase of genes via duplication in the AA genomes (most gene families have higher copy numbers in O. sativa or the other A genomes than in the B, C, E, F, and G genomes). Furthermore, the observed association between phylogenetic and physical clustering of members of different families (i.e., members from a tightly linked tandem array are also phylogenetically close) suggest unequal crossing over as the major mechanism for their amplification.
In general, large multigene families, such as disease resistance genes and genes involved in reproduction or morphological complexity, evolve rapidly (Leister et al., 1998; Meyers et al., 1999; Song et al., 2001; Ramakrishna et al., 2002a, 2002b; Song and Messing, 2002; Fiebig et al., 2004; Schein et al., 2004). The frequent gain and loss of gene family members observed in this region follows the birth-and-death model of gene evolution (Ohno, 1970; Nei et al., 1997; Michelmore and Meyers, 1998; Nei and Rooney, 2005). This model posits that genomes gain new genes by continuous duplication, with some acquiring new functions through random mutation followed by natural selection. While a few duplicated genes persist through vertical descent, most are lost either by inactivation (and subsequent slow sequence degeneration through indels and point mutations) or by rapid elimination, usually through unequal homologous recombination.
This dynamic of gain and loss in tandem gene families can lead to complex networks of synteny in cross-species comparisons. Approximately 53 and 68%, respectively, of the total gene complements of rice and Arabidopsis thaliana are composed of paralogous gene families (Lin et al., 2008). Nearly 30% of these are arranged as tandem clusters. Many rice gene families have experienced expansions relative to Arabidopsis (Lin et al., 2008). However, little is known about the evolutionary dynamics of individual gene families at the genus level in any set of species, including Oryza. Therefore, our study provides a unique glimpse into the dynamics of these processes across a broad range of recently diverged lineages.
The heterogeneous evolution of Oryza genomes is particularly well demonstrated by TEs. First, TE content correlated with genome size in this study, as in previous investigations (Ammiraju et al., 2006, 2007; Piegu et al., 2006; Zuccolo et al., 2007). Our analyses indicated that intergenic regions have been in rapid and constant flux, to the extent of >95% replacement, mainly mediated by LTR retrotransposons within ∼15 MY. In particular, several independent expansions (O. granulata [GG], O. australiensis [EE], O. officinalis [CC], and O. sativa [AA]) and apparent contractions (O. glaberrima [AA], O. brachyantha [FF], and O. punctata [BB]) indicate that genome size has probably not evolved in a step-wise fashion, assuming that this region mirrors the dynamics of each genome in total. An ancestral retrotransposon family, RWG, discovered in the analysis of the O. granulata Adh1 region, was shown to occupy up to one-quarter of the EE and GG nuclear genomes (Piegu et al., 2006; Ammiraju et al., 2007). Therefore, it would be interesting to determine if the elevated unequal recombination that appears to have contributed to a contraction of the genomes in the BB, FF, and O. glaberrima (AA) lineages in the Adh1 region will hold up at the whole-genome level. However, because >77% of the solo LTRs identified in this analysis have TSDs (see Supplemental Tables 8, 11, and 12 online), it appears that unequal intraelement and intrastrand recombination is the primary mechanism employed to remove LTR retrotransposons. This mechanism only slows down but cannot reverse the expansion of a genome caused by LTR retrotransposon activity (Devos et al., 2002; Ma et al., 2004; Bennetzen et al., 2005).
Still, it is astounding that these apparent rates of unequal homologous recombination could vary so dramatically within the genus Oryza. Although an earlier study predicted such variation across angiosperms (Vitte and Bennetzen, 2006), the flowering plant variation could have been generated in >200 million years of independent evolution. Moreover, the differences observed by Vitte and Bennetzen (2006) were not across orthologous regions, so a trivial explanation could have been that some genomes yielded a higher percentage of solo LTRs merely because they had smaller genomes with a higher percentage of recombinationally active euchromatin. In this study, orthologous gene-rich regions were compared, so all should be euchromatic. If unequal recombination rates can vary twofold or more within a 15 million year period, then this indicates that quantitative activity and/or the pairing requirements of recombination are themselves highly unstable. It is not known whether this variation is associated with unequal recombination in meiosis or in the DNA repair associated with somatic cell growth, but it is surprising that either of these processes would have shown such high levels of interspecies variability.
Two other fundamental sources of diversity created by TEs are the generation of presence/absence polymorphisms by gene or gene fragment movement and the disruption of genes by insertion. Further studies are needed to assess whether functions have been acquired by any of these mobilized gene components or by gene-TE (e.g., transposase) fusions. These observed mechanisms (i.e., gene movement [TE or non-TE mediated] and gene disruption) appear to be active in all phylogenetic branches of Oryza. An intriguing case was that of a Pack-MULE mediated relocation of gene J12 in AA genomes. Previous work has shown that MULEs acquire small gene fragments, in the order of 1 kb or less, and some are even transcribed (Jiang et al., 2004). Although the mechanism of MULE gene/gene fragment acquisition is still unknown, there is no evidence to date that a MULE can capture genomic sequences after it has inserted into the genome. Interestingly, the size of the J12 gene (9.8 kb) is dramatically larger than any known sequence acquisition by a Pack-MULE so far described (Jiang et al., 2004). Although we were unable to identify the parental gene for J12 in the IRGSP RefSeq, our data suggest that MULEs may be capable of acquiring much larger gene/gene fragments than previously known. The evolutionary implications of this gene/fragment relocation to the Adh1 region are not known. It is possible that the reorganization of this apparently functional gene may confer a selective advantage.
Despite the central importance of structural rearrangements in chromosomal and organismal evolution, little has been learned about the molecular processes underlying their origin and maintenance, especially in plants. Our analyses suggest that the unstable architectural complexity observed in the Adh1-Adh2 genomic region was significantly fashioned by several small (10 to 100 kb) and medium (100 to 500 kb) size rearrangement polymorphisms. These include a 350-kb inversion polymorphism seen in O. australiensis, a 250-kb deletion/inversion polymorphism found in O. granulata, and a 73-kb inversion polymorphism detected in O. brachyantha. We established, in all three cases, a rearrangement-free orientation as the ancestral state.
We have further presented evidence for their recent and unique origin, leading to species-specific rearrangement polymorphisms. For all these structural rearrangements the break points map in close vicinity to duplicated sequences (gene family members or TEs; see Supplemental Figures 4, 5, and 7 online). Although we expect that duplication-rich regions are features found along all Oryza chromosomes, these results nevertheless suggest that ectopic recombination between duplicated sequences in direct or inverted orientation at nonhomologous chromosomal sites is a common phenomenon over an evolutionary timescale. The significant frequency of deletions, including losses of all tandem members of some gene families, suggests that DNA loss by mechanisms that don't require homologous recombination (e.g., illegitimate recombination; Devos et al., 2002; Ma et al., 2004) is also quite frequent. The biological consequences, if any, of the great majority of these rearrangements are unknown.
However, it has been shown in the case of some insects, for instance, that such structural rearrangements have the potential to create new avenues for speciation and adaptations to new ecological niches (Coghlan et al., 2005). Similarly, small changes in the expression of regulatory genes, a frequent outcome of TE insertion and other chromosomal rearrangements, have been found to have profound effects on plant architecture and development (Doebley et al., 2006). Further studies are needed to investigate the frequency, commonality, and molecular nature of such biologically significant changes, and comparative analysis of genome structure and evolution will continue to provide a leading avenue into this research (Bennetzen and Chen, 2008).
METHODS
Isolation and Sequencing of Adh1 Orthologous Regions
Adh1-containing BAC clones were isolated by screening seven published (Oryza nivara [AA], O. rufipogon [AA], O. glaberrima [AA], O. punctata [BB], O. officinalis [CC], O. brachyantha [FF], and O. granulata [GG]) and two unpublished (O. sativa ssp indica cultivar 93-11 and O. australiensis [EE]; www.genome.arizona.edu) BAC libraries with a radiolabeled Adh1 probe using previously described methods (Ammiraju et al., 2006). Validated clones were sequenced to phase III quality as previously described (International Rice Genome Sequencing Project, 2005), followed by sequence submission to GenBank. A total of 600,118 bp of sequence (coordinates 5319784 to 5919902) from chromosome 11 of the IRGSP RefSeq (build 4) was used as the OsAdh1RefSeq for our comparative analysis.
Sequence Analysis
TEs were annotated using both structure- and homology-based methods. Known repeats were identified by homology searches using RepeatMasker (http://www.repeatmasker.org) and then manually curated using a custom rice TE library (Jiang et al., 2003). The structure-based annotation involved independent annotation of TEs based on the identification of TE signatures, such as TIR, LTR, TSD, PBS, and PPT sequences using Cross_match, Dotter (Sonhammer and Durbin, 1995), and CONSED (Gordon et al., 1998). LTR retrotransposon insertion dates were calculated as previously described (SanMiguel et al., 1998), using a rate of 1.3 × 10−8 mutations per site per year (Ma and Bennetzen, 2004).
Gene Identification
Putative genes were identified by similarity searches against transcript (Blat; Kent, 2002; BlastN), protein (BlastX and BlastP), and conserved domain databases at the National Center for Biotechnology Information, and FGENESH (trained for monocots; http://www.softberry.com/) ab-initio gene prediction. Alignments of Oryza sequences were used to comparatively predict and further refine gene annotations. An identified gene candidate was only considered a gene if (1) it was not transposon related, (2) it contained a known and conserved functional domain, and (3) it had an Arabidopsis thaliana or cereal genome homolog outside of the Oryza genus. All tiers of annotation were overlaid on individual BAC sequences and were visualized and edited using Apollo (Lewis et al., 2002). Some exceptions to the previously available annotations are listed in Supplemental Tables 2 and 3 online.
Calculation of Divergence
Pairwise global alignments of individual Oryza genomic regions were conducted using MLAGAN (Brudno et al., 2003) against the 6-2 to 10 interval of OsAdh1RefSeq using default settings. MLAGAN is a global alignment tool that first recursively identifies colinear anchors of maximum local sequence identity and then generates a global alignment. Consequently, a few rearranged sequences (i.e., those that span inversion breakpoints) were not included in the analysis. Conserved noncoding sequences, defined as those 10 bp or greater in length with 90% or greater nucleotide identity, were identified in pairwise alignments using mVISTA (Frazer et al., 2004; http://genome.lbl.gov/vista/mvista/submit.shtml). These parameters were chosen to capture the evolutionary divergence in the genus Oryza. Gene and TE annotations of OsAdh1Refseq and individual Oryza lineages were used to classify sequences as coding or intergenic and used to calculate percent divergence as described by Dubcovsky and Dvorak (2007).
Phylogenetic Analysis
The genus-wide phylogenetic relationships of different Oryza lineages were estimated using six conserved core genes (6-2, 6-3, 7, 8, 9, and 10). DNA and protein sequences of these genes were aligned using ClustalX (Thompson et al., 1994) and MUSCLE (Edgar, 2004) and adjusted manually. Phylogenetic analyses of DNA sequences were conducted using maximum likelihood methods incorporating the best fit model of sequence evolution for each gene using Modeltest (Posada and Crandall, 1998) as implemented in PAUP* 4.0b10 (Swofford, 2002). Heuristic searches were performed with 100 replicates of random taxon addition. Phylogenetic analysis of paralogous gene family protein sequences was conducted using the maximum likelihood method with the JTT model (Jones et al., 1992) and a gamma distribution as implemented in PHYLIP3.67 (http://evolution.genetics.washington.edu/phylip.html). Bootstrap analysis with 10,000 replicates (Felsenstein, 1985) was used to evaluate clade support. In cases where no coverage was found in the sequenced region of the indica subspecies, orthologous gene family members were retrieved from the syntenic region of the 93-11 whole-genome draft sequence assembly (Yu et al., 2005). The rates of synonymous and nonsynonymous substitutions for coding sequences of the core genes were estimated as described by Ma and Bennetzen (2004). Dating individual gene duplications was conducted using the synonymous substitution rate of Adh1 and Adh2 genes from grasses (Gaut et al., 1999).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ266019 to FJ266028. The accession number for the region containing the 600,118 bp reference sequence containing the Adh1 locus (OsAdh1RefSeq) from the short arm of chromosome 11 of O. sativa ssp japonica is Rice Annotation Project, 2008. Clone addresses for each species used in this analysis are provided in Table 1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenomic View of the Genus-Wide Adh1 Vertical Sequence Data Set Depicting Both Gene and Transposon Dynamics.
Supplemental Figure 2. The Presence of F-Box Gene Family in the Polyploid Oryza Species at Orthologous Adh1 regions.
Supplemental Figure 3. Evolutionary Dynamics of Different Gene Families in Their Orthologous Regions.
Supplemental Figure 4. Complex Rearrangements in O. brachyantha (FF) Relative to OsAdh1Refseq.
Supplemental Figure 5. Putative Inversion in O. australiensis (EE) Relative to OsAdh1RefSeq.
Supplemental Figure 6. Putative Inversion in O. australiensis (EE) Relative to OsAdh1Refseq and Validation of the Inversion by Physical Mapping.
Supplemental Figure 7. Large Rearrangement in O. granulata (GG) Relative to OsAdh1Refseq.
Supplemental Figure 8. Pack-MULE–Mediated Gene Movement/Acquisition in the AA Genome Lineages.
Supplemental Table 1. List of Genes in OsAdh1Refseq.
Supplemental Table 2. Previously Identified Putative Gene Models from OsAdh1Refseq That Were Omitted from the Comparative Analysis.
Supplemental Table 3. Previously Identified Gene Models That Were Corrected in This Study.
Supplemental Table 4. Total Number of Genes (Intact, Pseudo, TE Embedded, and Gene Fragments) Identified in the Adh1 Genus-Wide Vertical Sequence Data Set.
Supplemental Table 5. Synonymous (Ks) and Nonsynonymous (Ka) Values for Six Core Genes Spanning the Adh1 Genus-Wide Vertical Sequence Data Set.
Supplemental Table 6. Molecular Divergence Times of Individual Oryza Species Using Four OsAdh1Refseq Core Genes to Derive an Approximate Molecular Clock.
Supplemental Table 7. Sequence Conservation in Coding and Intergenic Regions Derived from Pair-Wise Comparisons Between a Core Segment (J6-2 to J10 Interval) of the Adh1 Genus-Wide Vertical Sequence Data Set.
Supplemental Table 8. List of Intact LTR Retrotransposons and Solo LTRs: Presence or Absence of TSDs, and Their Coordinates.
Supplemental Table 9. Summary of Comparisons of the Number of Shared and Unshared Genes in the Adh1 Genus-Wide Vertical Sequence Data Set.
Supplemental Table 10. List of Shared or Unshared Genes or Gene Families across the Adh1 Genus-Wide Vertical Sequence Data Set.
Supplemental Table 11. Summary of Intact LTR Retrotransposons and Solo LTRs Based on the Presence or Absence of Target Site Duplications.
Supplemental Table 12. Detailed Breakdown of Intact LTR Retrotransposons and Solo LTRs Based on the Presence or Absence of Target Site Duplications.
Supplemental Table 13. Molecular Timing of Various Duplication Events.
Supplemental Data Set 1. Alignments of Core Genes Used to Derive Phylogenetic Relationships Depicted in Figure 2.
Supplemental Data Set 2 Alignments of F-box Gene Family Members Used to Derive Phylogenetic Relationships Used in Figure 4.
Supplemental Data Set 3. Alignments of Various Gene Family Members Used to Derive Phylogenetic Relationships Described in the Supplemental Figure 3.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation (Grant DBI-0321678 to R.A.W. and S.J.), the Bud Antle Endowed Chair (to R.A.W.), and the Georgia Research Alliance (to J.L.B.). We thank members of the Arizona Genomics Institute's BAC/EST Resource, Sequencing, Finishing, and Bioinformatics groups for generating and processing the high-quality sequence data used in this analysis. We also thank other members of the Oryza Map Alignment Project research team and advisory committee for fruitful discussions.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Rod A. Wing (rwing@ag.arizona.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aggarwal, R.K., Brar, D.S., and Khush, G.S. (1997). Two new genomes in the Oryza complex identified on the basis of molecular divergence analysis using total genomic DNA hybridization. Mol. Gen. Genet. 254 1–12. [DOI] [PubMed] [Google Scholar]
- Ammiraju, J., et al. (2006). The Oryza bacterial artificial chromosome library resource, construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 16 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammiraju, J., et al. (2007). Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J. 52 342–351. [DOI] [PubMed] [Google Scholar]
- Avramova, Z., Tikhonov, A., SanMiguel, P., Jin, Y.K., Liu, C.N., Woo, S.S., Wing, R.A., and Bennetzen, J.L. (1996). Gene identification in a complex chromosomal continuum by local genomic cross-referencing. Plant J. 10 1163–1168. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2007). Patterns in grass genome evolution. Curr. Opin. Plant Biol. 10 176–181. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., and Chen, M. (2008). Grass genomic synteny illuminates plant genome function and evolution. Rice. 1 109–118. [Google Scholar]
- Bennetzen, J.L., Ma, J., and Devos, K. (2005). Mechanisms of recent genome size variation in flowering plants. Ann. Bot. (Lond.) 95 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar, D.S., and Khush, G.S. (1997). Alien introgression in rice. Plant Mol. Biol. 35 35–47. [PubMed] [Google Scholar]
- Brudno, M., Do, C.B., Cooper, G.M., Kim, M.F., Davydov, E., Green, E.D., Sidow, A., Batzoglou, S., and Progra, N.C.S. (2003). LAGAN and Multi-LAGAN, Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A.G., et al. (2007). Evolution of genes and genomes on the Drosophila phylogeny. Nature 8 203–218. [DOI] [PubMed] [Google Scholar]
- Coghlan, A., Eichler, E.E., Oliver, S.G., Paterson, A.H., and Stein, L. (2005). Chromosome evolution in eukaryotes, a multi-kingdom perspective. Trends Genet. 21 673–682. [DOI] [PubMed] [Google Scholar]
- Devos, K.M., Brown, J.K.M., and Bennetzen, J.L. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J.F., Gaut, B.S., and Smith, B.D. (2006). The molecular genetics of crop domestication. Cell 127 1309–1321. [DOI] [PubMed] [Google Scholar]
- Dubcovsky, J., and Dvorak, J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316 1862–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence-limits on phylogenies - An approach using the bootstrap. Evolution Int. J. Org. Evolution 39 783–791. [DOI] [PubMed] [Google Scholar]
- Fiebig, A., Kimport, R., and Preuss, D. (2004). Comparisons of pollen coat genes across Brassicaceae species reveal rapid evolution by repeat expansion and diversification. Proc. Natl. Acad. Sci. USA 101 3286–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer, K.A., Pachter, L., Poliakov, A., Rubin, E.M., and Dubchak, I. (2004). VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32 W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., and Clegg, M.T. (1991). Molecular evolution of alcohol dehydrogenase 1 in members of the grass family. Proc. Natl. Acad. Sci. USA 88 2060–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., and Clegg, M.T. (1993). Molecular evolution of the Adh1 locus in the genus Zea. Proc. Natl. Acad. Sci. USA 90 5095–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., Morton, B.R., McCaig, B.C., and Clegg, M.T. (1996). Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., Peek, A.S., Morton, B.R., and Clegg, M.T. (1999). Patterns of genetic diversification within the Adh gene family in the grasses (Poaceae). Mol. Biol. Evol. 16 1086–1097. [DOI] [PubMed] [Google Scholar]
- Ge, S., Sang, T., Lu, B.R., and Hong, D.Y. (1999). Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc. Natl. Acad. Sci. USA 96 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. (1998). Consed: A graphical tool for sequence finishing. Genome Res. 8 195–202. [DOI] [PubMed] [Google Scholar]
- Grover, C.E., Kim, H., Wing, R.A., Paterson, A.H., and Wendel, J.F. (2007). Microcolinearity and genome evolution in the AdhA region of diploid and polyploid cotton (Gossypium). Plant J. 50 995–1006. [DOI] [PubMed] [Google Scholar]
- Grover, C.E., Yu, Y., Wing, R.A., Paterson, A.H., and Wendel, J.F. (2008). A phylogenetic analysis of indel dynamics in the cotton genus. Mol. Biol. Evol. 25 1415–1428. [DOI] [PubMed] [Google Scholar]
- Hass, B.L., Pires, J.C., Porter, R., Phillips, R.L., and Jackson, S.A. (2003). Comparative genetics at the gene and chromosome levels between rice (Oryza sativa) and wild rice (Zizania palustris). Theor. Appl. Genet. 107 773–782. [DOI] [PubMed] [Google Scholar]
- Hawkins, J.S., Kim, H., Nason, J.D., Wing, R.A., and Wendel, J.F. (2006). Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 16 1252–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic, K., SanMiguel, P.J., and Bennetzen, J.L. (2003). A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice genomes. Proc. Natl. Acad. Sci. USA 100 12265–12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Jain, M., Nijhawan, A., Arora, R., Agarwal, P., Ray, S., Sharma, P., Kapoor, S., Tyagi, A.K., and Khurana, J.P. (2007). F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N., Bao, Z., Zhang, X., Eddy, S.R., and Wessler, S.R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431 569–573. [DOI] [PubMed] [Google Scholar]
- Jiang, N., Bao, Z., Zhang, X., Hirochika, H., Eddy, S.R., McCouch, S.R., and Wessler, S.R. (2003). An active DNA transposon family in rice. Nature 421 163–167. [DOI] [PubMed] [Google Scholar]
- Jones, D.T., Taylor, W.R., and Thornton, J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8 275–282. [DOI] [PubMed] [Google Scholar]
- Jung, K.H., An, G.H., and Ronald, P.C. (2008). Towards a better bowl of rice, assigning function to tens of thousands of rice genes. Nat. Rev. Genet. 9 91–101. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. (2002). BLAT - The BLAST-like alignment tool. Genome Res. 12 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H., et al. (2008). Construction, alignment and analysis of twelve framework physical maps that represent the ten genome types of the genus Oryza. Genome Biol. 9 R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister, D., Kurth, J., Laurie, D.A., Yano, M., Sasaki, T., Devos, K., Graner, A., and Schulze-Lefert, P. (1998). Rapid reorganization of resistance gene homologues in cereal genomes. Proc. Natl. Acad. Sci. USA 95 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S.E., et al. (2002). Apollo: A sequence annotation editor. Genome Biol. 3 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H.N., Ouyang, S., Egan, A., Nobuta, K., Haas, B.J., Zhu, W., Gu, X., Silva, J.C., Meyers, B.C., and Buell, C.R. (2008). Characterization of paralogous protein families in rice. BMC Plant Biol. 8 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and Bennetzen, J.L. (2004). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Devos, K.M., and Bennetzen, J.L. (2004). Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., SanMiguel, P., Lai, J.S., Messing, J., and Bennetzen, J.L. (2005). DNA rearrangement in orthologous Orp regions of the maize, rice and sorghum genomes. Genetics 170 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Dickerman, A.W., Michelmore, R.W., Sivaramakrishnan, S., Sobral, B.W., and Young, N.D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20 317–332. [DOI] [PubMed] [Google Scholar]
- Michelmore, R.W., and Meyers, B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8 1113–1130. [DOI] [PubMed] [Google Scholar]
- Miyabayashi, T., Nonomura, K.I., Morishima, H., and Kurata, N. (2007). Genome size of twenty wild species of Oryza determined by flow cytometric and chromosome analyses. Breed. Sci. 57 73–78. [Google Scholar]
- Nayar, N.M. (1973). Origin and cytogenetics of rice. Adv. Genet. 17 153–292. [Google Scholar]
- Nei, M., Gu, X., and Sitnikova, T. (1997). Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. USA 94 7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., and Rooney, A.P. (2005). Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. (1970). Evolution by Gene Duplication. (London: George Allen and Unwin).
- Paterson, A.H. (2006). Leafing through the genomes of our major crop plants, strategies for capturing unique information. Nat. Rev. Genet. 7 174–184. [DOI] [PubMed] [Google Scholar]
- Piegu, B., Guyot, R., Picault, N., Roulin, A., Saniyal, A., Kim, H., Collura, K., Brar, D.S., Jackson, S., Wing, R.A., and Panaud, O. (2006). Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 16 1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada, D., and Crandall, K.A. (1998). MODELTEST: Testing the model of DNA substitution. Bioinformatics 14 817–818. [DOI] [PubMed] [Google Scholar]
- Ramakrishna, W., Dubcovsky, J., Park, Y.-J., Busso, C., Emberton, J., SanMiguel, P., and Bennetzen, J.L. (2002. c). Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, W., Emberton, J., Ogden, M., SanMiguel, P., and Bennetzen, J.L. (2002. a). Structural analysis of the maize Rp1 complex reveals numerous sites and unexpected mechanisms of local rearrangement. Plant Cell 14 3213–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, W., Emberton, J., SanMiguel, P., Ogden, M., Llaca, V., Messing, J., and Bennetzen, J.L. (2002. b). Comparative sequence analysis of the sorghum Rph region and the maize Rp1 resistance gene complex. Plant Physiol. 130 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Annotation Project (2007). Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 17 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Annotation Project (2008). The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 36(Database issue): D1028–D1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, T., Donoghue, M.J., and Zhang, D.M. (1997). Evolution of alcohol dehydrogenase genes in peonies (Paeonia): Phylogenetic relationships of putative nonhybrid species. Mol. Biol. Evol. 14 994–1007. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20 43–45. [DOI] [PubMed] [Google Scholar]
- Schein, M., Yang, Z.H., Mitchell-Olds, T., and Schmid, K.J. (2004). Rapid evolution of a pollen-specific oleosin-like gene family from Arabidopsis thaliana and closely related species. Mol. Biol. Evol. 21 659–669. [DOI] [PubMed] [Google Scholar]
- Song, R.T., Llaca, V., Linton, E., and Messing, J. (2001). Sequence, regulation, and evolution of the maize 22-kD alpha zein in gene family. Genome Res. 11 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R.T., and Messing, J. (2002). Contiguous genomic DNA sequence comprising the 19-kD zein gene family from maize. Plant Physiol. 130 1626–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.Y., Wang, G.L., Chen, L.L., Kim, H.S., Pi, L.Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.X., Zhu, L.H., Fauquet, C., and Ronald, P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270 1804–1806. [DOI] [PubMed] [Google Scholar]
- Sonhammer, E.L., and Durbin, R. (1995). A dot-matrix program with dynamic threshold control suited for gene, DNA and protein sequencing analysis. Gene 167 1–10. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2002). PAUP, Phylogenetic Analysis Using Parsimony, Version 4.0b10. (Sunderland, MA: Sinauer Associates).
- Tang, H., Bowers, J.E., Wang, X., Ming, R., Alam, M., and Paterson, A.H. (2008). Synteny and collinearity in plant genomes. Science 320 486–488. [DOI] [PubMed] [Google Scholar]
- Tarchini, R., Biddle, P., Wineland, R., Tingey, S., and Rafalski, A. (2000). The complete sequence of 340 kb of DNA around the rice Adh1-Adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell 12 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Rice Chromosome 3 Sequencing Consortium (2005). Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res. 15: 1284–1291. [DOI] [PMC free article] [PubMed]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A.P., SanMiguel, P.J., Nakajima, Y., Gorenstein, N.M., Bennetzen, J.L., and Avramova, Z. (1999). Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, D.A., Morishima, H., and Kadowaki, K. (2003). Diversity in the Oryza genus. Curr. Opin. Plant Biol. 6 139–146. [DOI] [PubMed] [Google Scholar]
- Vitte, C., and Bennetzen, J.L. (2006). Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. USA 103 17638–17643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing, R.A., et al. (2005). The Oryza Map Alignment Project: The golden path to unlocking the genetic potential of wild rice species. Plant Mol. Biol. 59 53–62. [DOI] [PubMed] [Google Scholar]
- Yang, S., Gu, T., Pan, C., Feng, Z., Ding, J., Hang, Y., Chen, J.Q., and Tian, D. (2008). Genetic variation of NBS-LRR class resistance genes in rice lines. Theor. Appl. Genet. 116 165–177. [DOI] [PubMed] [Google Scholar]
- Yang, S.H., Feng, Z.M., Zhang, X.Y., Jiang, K., Jin, X.Q., Hang, Y.Y., Chen, J.Q., and Tian, D.C. (2006). Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol. Biol. 62 181–193. [DOI] [PubMed] [Google Scholar]
- Yu, J., et al. (2005). The genomes of Oryza sativa, a history of duplications. PLoS Biol. 3 e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Vision, T.J., and Gaut, B.S. (2002). Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol. Biol. Evol. 219 1464–1473. [DOI] [PubMed] [Google Scholar]
- Zhang, L.B., and Ge, S. (2007). Multilocus analysis of nucleotide variation and speciation in Oryza officinalis and its close relatives. Mol. Biol. Evol. 24 769–783. [DOI] [PubMed] [Google Scholar]
- Zhang, S., Chen, C., Li, L., Meng, L., Singh, J., Jiang, N., Deng, X.W., He, Z.H., and Lemaux, P.G. (2005). Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 139 1107–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Gu, Y.Q., Singh, J., Coleman-Derr, D., Brar, D.S., Jiang, N., and Lemaux, P.G. (2007). New insights into Oryza genome evolution, high gene colinearity and differential retrotransposon amplification. Plant Mol. Biol. 64 589–600. [DOI] [PubMed] [Google Scholar]
- Zhou, T., Wang, Y., Chen, J.Q., Araki, H., Jing, Z., Jiang, K., Shen, J., and Tian, D. (2004). Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics 271 402–415. [DOI] [PubMed] [Google Scholar]
- Zhu, Q., and Ge, S. (2005). Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 167 249–265. [DOI] [PubMed] [Google Scholar]
- Zhu, Q.H., Zheng, X.M., Luo, J.C., Gaut, B.S., and Ge, S. (2007). Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: Severe bottleneck during domestication of rice. Mol. Biol. Evol. 24 875–888. [DOI] [PubMed] [Google Scholar]
- Zou, X.H., Zhang, F.M., Zhang, J.G., Zang, L.L., Tang, L., Wang, J., Sang, T., and Ge, S. (2008). Analysis of 142 genes resolves the rapid diversification of the rice genus. Genome Biol. 9 R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo, A., Ammiraju, J., Kim, H., Sanyal, A., Jackson, S., and Wing, R.A. (2008). Rapid and differential proliferation of the Ty3-Gypsy LTR retrotransposon Atlantys in the genus Oryza. Rice 1 85–99. [Google Scholar]
- Zuccolo, A., Sebastian, A., Talag, J., Yu, Y., Kim, H., Collura, K., Kudrna, D., and Wing, R.A. (2007). Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evol. Biol. 7 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.