Summary
The C. elegans DAF-2 insulin-like signaling pathway, which regulates lifespan and stress resistance, has also been implicated in resistance to bacterial pathogens. Loss-of-function daf-2 and age-1 mutants have increased lifespans (Kenyon et al. 1993; Larsen et al. 1995; Lithgow et al. 1995) and are resistant to a variety of bacterial pathogens (Garsin et al. 2003). This raises the possibility that the increased longevity and the pathogen resistance of insulin-like signaling pathway mutants are reflections of the same underlying mechanism (Lithgow 2003; Bolm et al. 2004). Here we report that regulation of lifespan and resistance to the bacterial pathogen Pseudomonas aeruginosa is mediated by both shared and genetically distinguishable mechanisms. We find that loss of germline proliferation enhances pathogen resistance and this effect requires daf-16, similar to the regulation of lifespan. In contrast, the regulation of pathogen resistance and lifespan is decoupled within the DAF-2 pathway. Long-lived mutants of genes downstream of daf-2, such as pdk-1 and sgk-1 (Paradis et al. 1999; Hertweck et al. 2004), show wildtype resistance to pathogens. However, mutants of akt-1 and akt-2, which we find to individually have modest effects on lifespan, show enhanced resistance to pathogens. We also demonstrate that pathogen resistance of daf-2, akt-1, and akt-2 mutants is associated with restricted bacterial colonization and that daf-2 mutants are better able to clear an infection after challenge with P. aeruginosa. Moreover, we find that pathogen resistance among insulin-like signaling mutants is associated with increased expression of immunity genes during infection. Other processes that affect organismal longevity, including Jun kinase signaling and caloric restriction, do not affect resistance to bacterial pathogens, further establishing that aging and innate immunity are regulated by genetically distinct mechanisms.
Introduction
The genetic regulation of both the rate of aging and resistance to bacterial pathogens are conserved aspects of animal physiology that have been well studied in the nematode Caenorhabditis elegans (Kurz & Tan 2004; Ewbank 2006; Antebi 2007). Genes affecting many processes, including neuroendocrine signaling, caloric restriction, and mitochondrial function, have been found to modulate the lifespan of C. elegans. The DAF-2 insulin-like signaling pathway regulates lifespan in C. elegans (Kenyon et al. 1993), and homologous neuroendrocine signaling pathways regulate lifespan in other animals, including mice (Bluher et al. 2003; Holzenberger et al. 2003; Tatar et al. 2003). A number of pathways have been found to regulate innate immunity in C. elegans, including p38 MAPK and DAF-2 insulin-like signaling (Ewbank 2006). Loss-of-function mutants of daf-2 and age-1 enhance resistance to a variety of bacterial pathogens in C. elegans (Garsin et al. 2003), and insulin-like signaling appears to function in parallel to p38 MAPK in the regulation of innate immunity (Troemel et al. 2006).
The insulin-like signaling pathway also regulates dauer formation and resistance to a variety of abiotic stresses in C. elegans (Riddle et al. 1997). The phenotypes associated with loss-of-function mutations in daf-2 are suppressed by loss-of-function mutations in daf-16, which encodes a FOXO transcription factor. DAF-2 regulates DAF-16 at least in part through the activation of phosphoinositide 3-kinase (PI3-kinase), encoded by age-1 and aap-1. PI3-kinase potentiates the activity of four serine threonine kinases: the PI3K-dependent kinase homolog PDK-1, the Akt/PKB homologs AKT-1 and AKT-2, and the serum- and glucocorticoid-inducible kinase homolog, SGK-1. These kinases appear to have distinct roles in C. elegans physiology. Dauer formation is regulated by akt-1, akt-2, and pdk-1, but not sgk-1, and oxidative stress resistance is regulated predominately by sgk-1 and pdk-1 (Hertweck et al. 2004). In contrast, both pdk-1 and sgk-1 are individually required for normal lifespan, whereas akt-1 and akt-2 appear to function together to regulate lifespan (Hertweck et al. 2004; Oh et al. 2005; Baumeister et al. 2006).
The C. elegans germline also regulates lifespan. Worms which lack a germline, either due to mutations which result in loss of germline proliferation or due to laser ablation of the germline, are longer lived (Hsin & Kenyon 1999; Arantes-Oliveira et al. 2002). This lifespan extension is DAF-16-dependent, but appears to be regulated in parallel to DAF-2. Some sterile worms are resistant to bacterial pathogens, and this resistance is also DAF-16 dependent (Miyata et al. 2008). In contrast to the regulation of lifespan by the germline, which appears to be mediated by signals from the germline stem cells, resistance to bacterial pathogens in these sterile animals appears to be mediated by signals from the embryos.
Extensive analyses using transcriptional profiling have identified target genes whose expression is regulated by DAF-16 and DAF-2 either directly or indirectly (Murphy et al. 2003; McElwee et al. 2004; McElwee et al. 2006). These genes fall into at least two functional groups. The first group includes stress responsive and detoxifying genes, including genes that encode enzymes which neutralize reactive oxygen species (ROS). The second group includes a diverse classes of confirmed and putative antimicrobial effector molecules, including lysozymes (such as lys-7), saposin-like genes, such as spp-1 (Kato et al. 2002), glycine/tyrosine-rich antimicrobial peptides, such as nlp-31 (Couillault et al. 2004), ASABF-type antimicrobial peptides, such as abf-2 (Banyai & Patthy 1998), and thaumatins (such as thn-2).
Infection-mediated killing of C. elegans by the Gram negative pathogen Pseudomonas aeruginosa (hereafter PA14) is associated with the accumulation of bacteria within the intestine (Tan et al. 1999), which is accelerated in mutants with enhanced susceptibility to killing, such as the p38 MAPKK mutant sek-1(km4) (Kim et al. 2002). Aging in C. elegans is associated with bacterial accumulation and packing in the intestine. Worms fed heat-killed or UV-killed E. coli OP50 have longer lifespans than those fed live OP50 (Gems & Riddle 2000; Garigan et al. 2002). Thus, resistance to bacterial infection could enhance lifespan in C. elegans. The co-regulation of aging and innate immunity by the DAF-2 insulin-like signaling pathway raises the possibility that the increased longevity and pathogen resistance of insulin-like signaling pathway mutants are reflections of the same underlying mechanism (Lithgow 2003; Bolm et al. 2004). Here we present evidence that regulation of lifespan and resistance to PA14 is mediated by both shared and genetically distinguishable mechanisms.
Results
Germline proliferation negatively regulates pathogen resistance through a daf-16-dependent mechanism
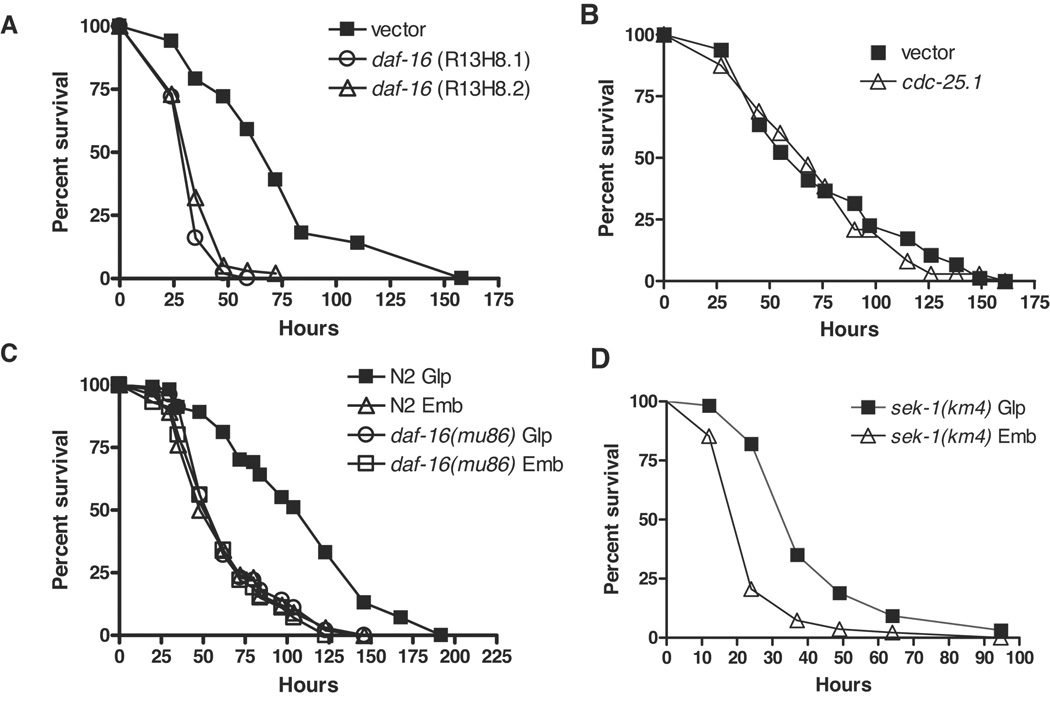
In fertile animals, daf-16 is required for the extended lifespan and enhanced pathogen resistance of daf-2 mutants. daf-16 mutants, while shorter lived (Lin et al. 1997), were reported not to have enhanced susceptibility to E. faecalis, S. aureus, and P. aeruginosa (Garsin et al. 2003; Kerry et al. 2006; Troemel et al. 2006). Some bacterial infections impair egg-laying, resulting in the hatching of eggs within infected animals, a phenotype termed “bagging”, which contributes to killing the worm and thereby complicating the interpretation of survival assays (Shapira & Tan 2008). To eliminate bagging, we therefore reassessed the requirement for daf-16 in immune function using sterile animals. We observed that knockdown of daf-16 by RNA interference (RNAi) in rrf-3(pk1426);glp-4(bn2) animals rendered these animals significantly more susceptible to PA14 (Figure 1A). The rrf-3(pk1426) mutation sensitized the animals to RNAi (Simmer et al. 2002), and the glp-4(bn2) mutation resulted in the development of sterile adults lacking germline proliferation (Beanan & Strome 1992). Similar results were observed with daf-16 RNAi knockdown in glp-4(bn2) worms (Supplemental Table S1). By contrast, daf-16 RNAi knockdown in sterile animals with intact germline proliferation, pha-1(e2123), resulted in survival kinetics that was indistinguishable from control (Supplemental Table S1). pha-1(e2123) animals have intact germline proliferation but are sterile because viable progeny do not develop at non-permissive temperature (Schnabel & Schnabel 1990). Given that glp-4(bn2) animals have extended life span (Arantes-Oliveira et al. 2002) and the C. elegans germline has been shown to negatively regulate adult lifespan in a daf-16-dependent manner, such that ablation of the germline increases lifespan only in daf-16(+) worms (Hsin & Kenyon 1999; Lin et al. 2001), we wondered whether the C. elegans germline could also modulate immune function and whether this is dependent on DAF-16. We investigated the effect of loss of germline proliferation on resistance to PA14 by performing C. elegans survival assays using worms in which cdc-25.1 has been knocked down by RNAi (Shapira et al. 2006; Shapira & Tan 2008). RNAi knockdown of cdc-25.1 generates two distinct populations of sterile worms: one with and another without proliferating germlines (hereafter referred to as Emb and Glp, respectively; Supplemental Text). First, we showed that cdc-25.1 did not affect pathogen resistance beyond that observed in other Glp animals; glp-4(bn2) germline-deficient animals treated with cdc-25.1 dsRNA had similar survival kinetics on PA14 compared to those treated with vector control (Figure 1B). We then compared the survival of Emb and Glp subpopulations of N2 and daf-16(mu86) null mutants (Supplemental Table S1) on PA14. The ability of daf-16(mu86) and N2 animals to survive infection by PA14 was not significantly different among sterile Emb populations. This corroborates the previous reports (Garsin et al. 2003; Troemel et al. 2006) that in animals with intact germline proliferation, daf-16 is dispensable for resistance to bacterial pathogens. In the N2 strain, resistance to PA14 was enhanced by 75% in Glp animals as compared to Emb animals (Figure 1C). However, the enhanced resistance of Glp animals to PA14 was completely suppressed in daf-16(mu86) animals, where no significant difference between the survival of Emb and Glp animals could be detected. In contrast, the pathogen sensitive p38 MAPKK mutant sek-1(km4) Glp animals were more resistant to PA14 than sek-1(km4) Emb animals, indicating that failure to observe a difference between daf-16(mu86) Emb and Glp animals is not a result of limited statistical power (Figure 1D). Based on these results, we conclude that germline proliferation negatively regulates resistance to PA14, either directly or indirectly, in a daf-16-dependent but sek-1-independent manner. This observation is consistent with a previous report that sterile animals, such as glp-1(e2141) and fer-15(b26);fem-1(hc17), are resistant to bacterial pathogens in a daf-16-dependent manner (Miyata et al. 2008). In animals with proliferating germlines, daf-16 is required for lifespan regulation but not for resistance to PA14. These observations suggest that despite an overlap in the role of the daf-2/daf-16 pathway in affecting lifespan and pathogen resistance, some aspects of the regulation could be molecularly distinct.
Figure 1. Germline proliferation regulates pathogen resistance through a daf-16-dependent mechanism.
Survival of worms on PA14 was monitored over time at 25°C. (A) RNAi knockdown of daf-16 causes enhanced susceptibility to PA14 in worms without a proliferating germline. Enhanced susceptibility of rrf-3(pk1426);glp-4(bn2) worms to pathogen was observed with two independent RNAi constructs compared to vector control (logrank test, p < 0.0001): R13H8.1, which targets all isoforms of daf-16, and R13H8.2, which targets the most broadly-expressed isoforms (Supplemental Figure S1). Survival of worms treated with R13H8.1 and R13H8.2 were indistinguishable (logrank, p = 0.04). Similar results are observed with daf-16 RNAi knockdown in glp-4(bn2) worms (Supplemental Table S1). (B) RNAi knockdown of cdc-25.1 does not affect the ability of glp-4(bn2) worms to survive infection by PA14 (logrank, p = 0.26, compared to vector control). (C) Enhanced resistance to PA14 due to loss of germline proliferation is daf-16-dependent, but daf-16 is dispensable for resistance to pathogen in animals with intact germline proliferation. In N2 populations, Glp worms were more resistant to PA14 infection than Emb worms (logrank test p < 0.0001), but in daf-16(mu86) populations Emb and Glp were not significantly different (logrank test p = 0.08). In comparison to N2 Emb, daf-16 Emb was not significantly different (logrank test p = 0.07). (D) The enhanced pathogen resistance due to loss of germline proliferation does not require sek-1. Compared to sek-1(km4) Emb, the mean survival of sek-1(km4) Glp is significantly enhanced (logrank, p < 0.0001). Representative experiments shown. Complete supporting data are presented in Table S1.
Lifespan and pathogen resistance are distinctly regulated by components of the insulin-like signaling pathway
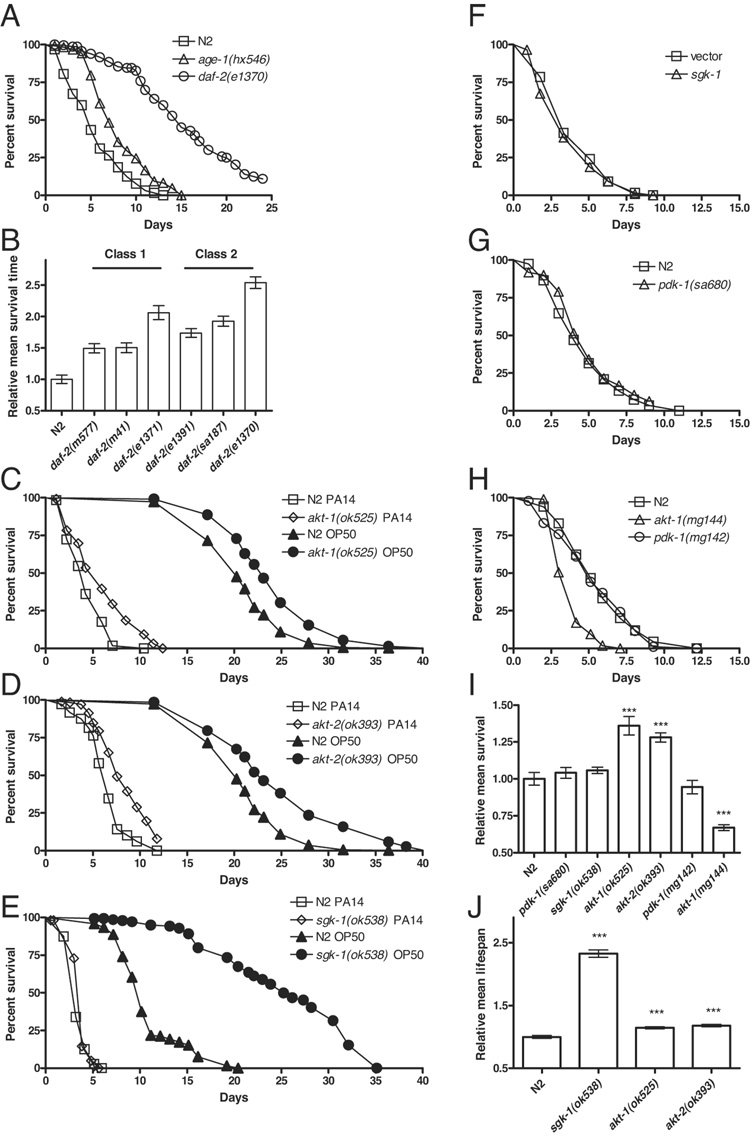
To investigate the possible distinct molecular nature of the regulation of lifespan and immunity, we examined additional components of the daf-2/daf-16 pathway. Previously, using fertile animals, hypomorphic alleles of daf-2 and age-1 were reported to be resistant to pathogenic bacteria (Garsin et al. 2003). The strength of these mutants varied considerably, with the strongest resistance observed in daf-2(e1370) animals. daf-2(e1370) mutants however, exhibit other pleiotropic effects, such as reduced brood size and increased incidence of internal hatching (Gems et al. 1998; Patel et al. 2008) that, as noted earlier, could confound the interpretation of pathogen resistance assays that are based on survival (Shapira & Tan 2008). Using worms rendered Glp by cdc-25.1 RNAi, we confirmed that daf-2(e1370) and age-1(hx546) were resistant to PA14 and noted that daf-2(e1370) was significantly more resistant to PA14 than age-1(hx546) (Figure 2A). Moreover, we found that daf-2(e1370) was significantly more resistant to PA14 than other daf-2 alleles, and pathogen resistance varied more within than between the two phenotypic classes of daf-2 alleles (Figure 2B). We also observed that aap-1(ok282), a deletion mutant of the p50/p55 PI3K regulatory subunit (Wolkow et al. 2002), was resistant to PA14 (Supplemental Table S1). Like age-1, aap-1 loss-of-function mutants are long lived and arrest as dauers when raised at high temperatures (Wolkow et al. 2002). Thus, daf-2, age-1, and aap-1 are each required for the regulation of both lifespan and resistance to bacterial pathogens.
Figure 2. Aging and pathogen resistance are distinctly regulated by components of the insulin-like signaling pathway.
(A) The enhanced pathogen resistance of long-lived daf-2 and age-1 mutants is observed in sterile animals. Survival of sterile daf-2(e1370), age-1(hx546), and N2 adults on PA14 over time is shown. The survival of each strain was significantly different in pairwise comparisons (logrank, p < 0.0001). (B) Hypomorphic daf-2 mutants were resistant to PA14. daf-2(m577), daf-2(m41), daf-2(1371), daf-2(e1391), daf-2(sa187), and daf-2(e1370) were more resistant to pathogen than N2 (logrank, p < 0.0001). Of the daf-2 mutants tested, daf-2(e1370) was reproducibly the most resistant (logrank, p < 0.0001 in pairwise comparisons). Note that pathogen resistance varied more within than between phenotypic classes among daf-2 alleles (Gems et al. 1998). (C) akt-1(ok525) mutants were resistant to PA14 (logrank test, p < 0.0001; open symbols). They were also slightly, but significantly longer lived, with a mean lifespan of 115% compared to N2 (logrank p < 0.0001; closed symbols) when fed with UV-killed OP50 as 1-day old adults. (D) akt-2(ok393) mutants were resistant to PA14 (logrank test, p < 0.0001; open symbols). They were only slightly but significantly longer lived, with a mean lifespan of 118% compared to N2 (logrank p < 0.0001; closed symbols) when fed with UV-killed OP50 as 1-day old adults. (E) sgk-1(ok583) mutants were not significantly different than N2 in resistance to PA14 (logrank test, p = 0.11). sgk-1(ok538) mutants exhibited a mean lifespan of 230% compared to N2 (logrank, p < 0.0001). Lifespan of Glp N2 and sgk-1(ok538) fed with UV-killed OP50 starting as 1-day old adults is plotted. (F) The sgk-1(ok538) mutant has a strong developmental delay, which is substantially weaker with sgk-1 RNAi knockdown. Thus, we used RNAi knockdown to assess the role of sgk-1 in pathogen resistance without developmental timing as a confounding factor. RNAi knockdown of cdc-25.1 and sgk-1 was performed serially. cdc-25.1 RNAi knockdown was performed maternally followed by egg lay to undiluted sgk-1 RNAi bacteria. RNAi knockdown of sgk-1 did not enhance pathogen resistance (logrank, p = 0.27). The efficacy of the induction of RNAi was confirmed by RNAi knockdown of daf-16, which enhanced susceptibility to pathogen (Table S1). Moreover, RNAi knockdown of sgk-1 has been reported to extend lifespan to 160% of control levels (Hertweck et al. 2004). (G) pdk-1(sa680) mutants were also not significantly different than N2 in resistance to PA14 (logrank test, p = 0.12). (H) The pathogen resistance of gain-of-function mutants in akt-1 and pdk-1 mirrored the loss-of-function mutants. akt-1(mg144) was sensitive compared to N2 (logrank p < 0.0001), whereas pdk-1(mg142) was not significantly different from N2 (logrank p = 0.6). (I) Relative mean survival on PA14 for experiments shown in Figure 2. (J) Relative mean lifespan on UV-killed OP50 for experiments shown in Figure 2. All experiments used cdc-25.1 RNAi Glp animals. Representative experiments shown. In panels C–E, representative lifespan and pathogen resistance assays were combined in the same plot to ease visual comparison. Complete supporting data are presented in Supplementary Table S1.
Downstream of PI3K are four serine threonine kinases: PDK-1, AKT-1, AKT-2, and SGK-1. Loss-of-function mutations in pdk-1 and sgk-1 cause lifespan extension (Paradis et al. 1999; Hertweck et al. 2004). However, there are conflicting reports on the requirement for akt-1 and akt-2 in the regulation of lifespan. While the deletion mutants akt-1(ok525) and akt-2(ok393) are reported not to affect lifespan (Hertweck et al. 2004), others have reported that RNAi knockdown of akt-1 significantly increased mean lifespan (Hamilton et al. 2005; Hansen et al. 2005). In addition, the akt-1(ok525);akt-2(ok393) double mutant is reported to form constitutive dauers at all temperatures, which can be bypassed by daf-16 RNAi during development, and to have significantly increased lifespan (Oh et al. 2005). We wondered if the specificity of these kinases extends to the relationship between pathogen resistance and lifespan. It has been reported that E. coli OP50 could grow and proliferate in older worms and contribute to a more rapid death compared to worms grown on dead or nonproliferating bacteria (Garigan et al. 2002). This raises the possibility that results obtained from lifespan assays that used live E. coli OP50 could be affected by normal lifespan and immune function. We therefore determined the adult lifespans of akt-1(ok525) and akt-2(ok393) using UV-killed E. coli as a food source and monitored survival of these strains on PA14 at 25°C. All assays used worms rendered Glp by cdc-25.1 RNAi. The akt-1(ok525) and akt-2(ok393) mutants exhibited a small but significant increase in mean lifespan on UV-killed E. coli (Figure 2C and 2D, closed symbols) and a proportionately larger increase in resistance to PA14 (Figure 2C and 2D, open symbols). We speculated that akt-1 and akt-2 function partially redundantly for the regulation of pathogen resistance, as has been found for the regulation of dauer formation (Hertweck et al. 2004). We were unable to generate akt-1(ok525);akt-2(ok393) double mutants, which appeared to be embryonic or early-larval lethal in our experiments. However, we found that double RNAi knockdown of both akt-1 and akt-2 enhanced pathogen resistance to a greater extent than either single RNAi (Supplemental Table S1), consistent with a model of partial redundancy for the regulation of pathogen resistance.
Knockdown of sgk-1 by RNAi increased mean lifespan to more than 160% relative to controls (Hertweck et al. 2004). Corroborating this result, we observed that the deletion mutant sgk-1(ok538) had a mean lifespan on UV-killed E. coli of 230% relative to N2 (Figure 2E, closed symbols). Yet, despite this large lifespan extension, sgk-1(ok538) animals were indistinguishable from N2 animals in their ability to survive PA14 infection (Figure 2E, open sybmols). All assays used worms rendered Glp by cdc-25.1 RNAi. Similarly, we observed that PA14 resistance of animals fed with sgk-1 dsRNA was indistinguishable from the vector control (Figure 2F). For this assay, cdc-25.1 RNAi knockdown was achieved by maternal exposure to cdc-25.1 dsRNA-expressing bacteria followed by egg lay on sgk-1 RNAi bacteria, thus maximizing the effective concentration of sgk-1 dsRNA. The long-lived loss-of-function mutant pdk-1(sa680) (Paradis et al. 1999) also had wildtype-like survival on PA14 in a cdc-25.1 RNAi Glp background (Figure 2G). pdk-1(sa680) carries a missense mutation and thus may possess residual function. To further reduce PDK-1 function, we knocked down pdk-1 expression using RNAi in a pdk-1(sa680) background. Consistent with the suggestion that pdk-1(sa680) is a strong loss-of-function mutation (Paradis et al. 1999), knockdown of pdk-1 did not enhance the pathogen resistance of pdk-1(sa680) animals (Figure S3).
To complement our observations with the loss-of-function mutants we tested gain-of-function mutants, akt-1(mg144) and pdk-1(mg142) for their ability to survive PA14 infection in a cdc-25.1 RNAi Glp background. akt-1(mg144) showed increased susceptibility to PA14, whereas pdk-1(mg142) was indistinguishable from wildtype (Figure 2H) despite having a decreased lifespan (Paradis et al. 1999). These results indicate that pathogen resistance, like lifespan and dauer formation, is regulated by a distinct subset of the serine threonine kinases downstream of AGE-1 (Figure 2I and 2J). We conclude that both longevity and resistance to bacterial pathogens are regulated by AKT-1 and AKT-2. However, despite their strong effects on lifespan, mutants in sgk-1 and pdk-1 do not affect immune function.
Regulation of lifespan and pathogen resistance by DAF-16
The mechanism by which PDK-1, AKT-1, AKT-2 and SGK-1 differentially regulate resistance to pathogen and lifespan is unclear. Genetic and biochemical experiments in worms and mammals indicate that AKT and SGK directly interact with DAF-16 and its mammalian homolog at least in part by the phosphorylation of overlapping but distinct residues on DAF-16 and its mammalian homologs (Kobayashi & Cohen 1999; Brunet et al. 2001; Henderson & Johnson 2001). Mutation of these phosphorylation sites results in constitutive nuclear localization of DAF-16, and may be sufficient to cause constitutive dauer formation (Lee et al. 2001), but has no detectable effect on lifespan (Lee et al. 2001; Lin et al. 2001). Using the same transgenic worm strains in which the consensus AKT and SGK phosphorylation sites have been mutated (Lin et al. 2001), we likewise did not observe an effect on resistance to PA14 (Supplementary Table S2). Animals without germline have enhanced DAF-16 nuclear localization in their intestine, and this appears to be associated with increased longevity and resistance to pathogen (Lin et al. 2001). However, it appears that nuclear localization regulated by AKT and SGK phosphorylation sites is not sufficient to confer increased longevity or enhanced resistance to pathogens.
We also wondered whether lifespan and resistance to bacterial pathogens could be distinctly regulated by the differential activity of DAF-16 in different tissues. DAF-16 has detectable roles in neurons and intestine in affecting lifespan (Wolkow et al. 2000; Libina et al. 2003; Iser et al. 2007). Using the same transgenic worm strains (Libina et al. 2003), we examined the role of DAF-16 in regulating pathogen resistance in neurons and intestine. When daf-16 was expressed as a transgene under its native promoter in daf-16(mu86); daf-2(e1370) animals, pathogen resistance was strongly restored (Supplemental Figure S4A,D). In contrast, when expression of daf-16 was restricted to either the neurons or the intestine, only partial rescue of the resistance phenotype of daf-2(e1370) was observed (Supplemental Figure S4B–D). This pattern of rescue is not substantially different from the pattern that was previously reported for lifespan with these strains (Libina et al. 2003). This suggests that a complex interaction between tissues, including neurons and intestine, characterizes the regulation of lifespan and pathogen resistance, but this does not distinguish the regulation of lifespan and pathogen resistance.
Resistance of insulin-like signaling mutants is associated with reduced bacterial colonization, enhanced bacterial clearance, and increased expression of antimicrobial genes
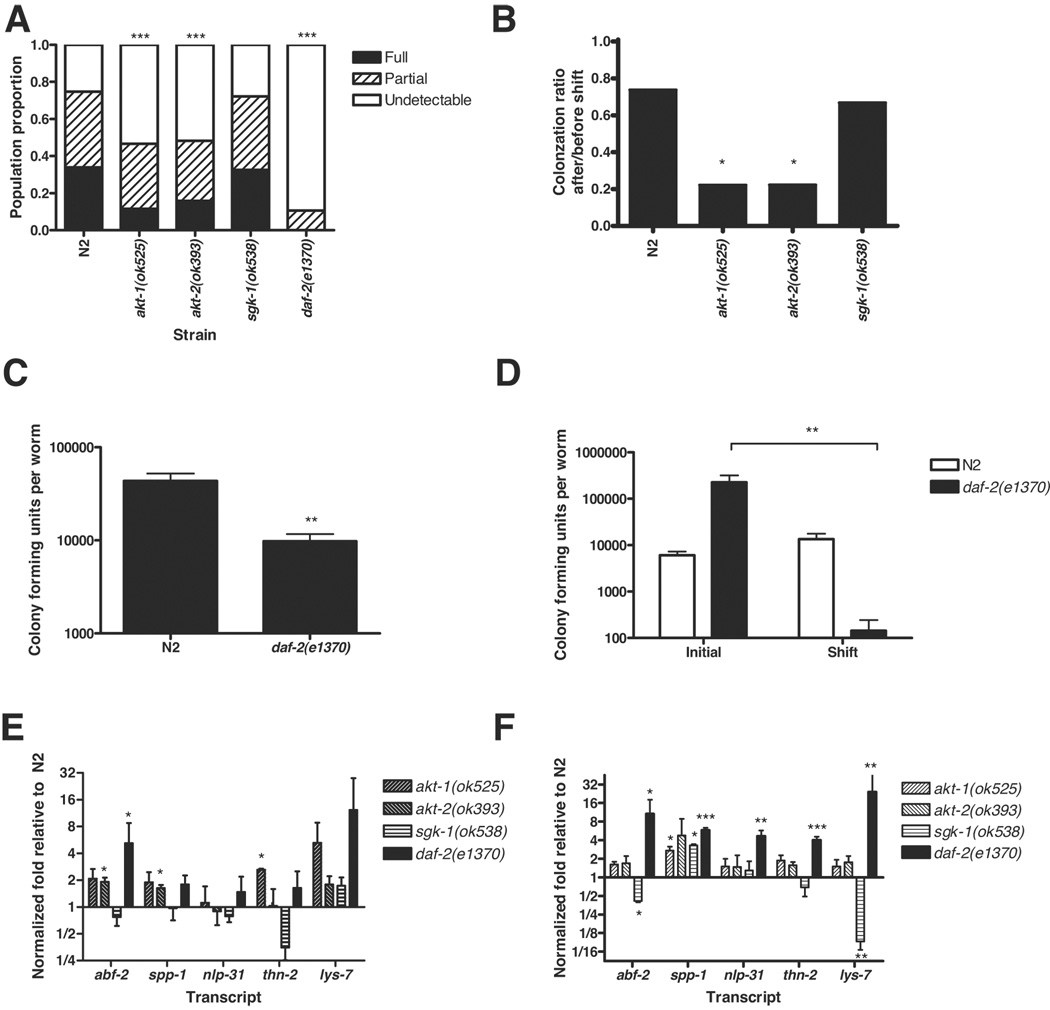
Killing of C. elegans by PA14 is associated with the accumulation of bacteria within the intestine (Tan et al. 1999), which is accelerated in mutants with enhanced susceptibility to killing, such as sek-1(km4) (Kim et al. 2002). We hypothesized that pathogen resistant mutants, such as akt-1, akt-2 and daf-2 mutants, should accumulate PA14 more slowly than N2. To test this hypothesis, we exposed 1-day-old adult N2, akt-1(ok525), akt-2(ok393), sgk-1(ok538), and daf-2(e1370) worms to GFP-expressing PA14 for 24 h (Tan et al. 1999). The extent of PA14 accumulation was assessed by the presence of GFP fluorescence in the worm intestine. A semi-quantitative scoring scale was used to compare the degree of colonization between populations (Figure 3A). The extent of bacterial colonization in sgk-1(ok538) animals was indistinguishable from N2, consistent with wildtype-like pathogen resistance of sgk-1(ok538) animals. In contrast, within the akt-1(ok525) and akt-2(ok393) populations fewer worms were fully or partially colonized and more had undetectable colonization compared to N2. The daf-2(e1370) population had further reduced colonization, in accord with its stronger pathogen resistance phenotype in the PA14 survival assays. Differences in feeding and defecation might influence the extent of bacterial colonization. daf-2(e1370) animals exhibit reduced pumping (Kenyon et al. 1993; Gems et al. 1998). Mutations in eat-2 also result in decreased pharyngeal pumping but no obvious defects in pharyngeal anatomy (Avery 1993). However, eat-2(ad465) did not exhibit reduced colonization after 24 h exposure to PA14-GFP and instead exhibited slightly increased bacterial accumulation (Figure S5). This suggests that reduced pharyngeal pumping does not substantially affect bacterial accumulation as measured by PA14-GFP colonization. daf-2(e1370) animals also exhibited reduced rates of defecation (Figure S6). Reduced defecation results in increased sensitivity to PA14 (Shapira & Tan 2008), presumptively due to a reduced ability to clear pathogenic bacteria. Thus, reduced defecation in daf-2(e1370) cannot explain the reduced colonization of daf-2(e1370) mutants.
Figure 3. Resistance of daf-2 pathway mutants is associated with reduced bacterial colonization, enhanced bacterial clearance, and increased immunity gene expression.
(A) akt-1(ok525), akt-2(ok393), and daf-2(e1370) animals accumulated significantly less PA14-GFP in their intestine than N2 following a 24 h exposure (pairwise comparison Chi-square, p < 0.0001). No significant difference in bacterial accumulation was observed between sgk-1(ok538) and N2 populations (Chi-square, p = 0.86). For each strain, population proportion by degree of bacterial accumulation in the intestine was categorized as fully colonized, partially colonized, or undetectable colonization corresponding to averages of 4.22 × 104, 2.45 × 104, and 3.48 × 103 colony forming units of PA14-GFP, respectively. The pooled sample sizes are N=336 N2, 343 akt-1(ok525), 328 akt-2(ok393), 126 sgk-1(ok538), and 28 daf-2(e1370). (B) Pathogen clearance was enhanced in akt-1(ok525) and akt-2(ok393) compared to N2. The proportion of colonized worms before and after shift from PA14-GFP to E. coli OP50 was quantified, and the ratio of colonization after/before was plotted. The extent of clearance was significantly greater in akt-1(ok525) and akt-2(ok393) compared to N2 (log-linear analysis, p < 0.05) but not significantly different in sgk-1(ok538). (C) Quantification of live bacteria from whole worm lysates of daf-2(e1370) and N2 following exposure to PA14 for 24 h. The CFU counts were significantly less in daf-2 than N2 (t-test, p = 0.009). (D) Pathogen clearance was enhanced in daf-2(e1370) compared to N2. The initial PA14 CFUs obtained from daf-2 and N2 worms exposed to PA14 for 24 h and 18 h, respectively, were compared to PA14 CFUs from worms obtained 24 h after the shift to OP50-1. While the CFUs from initial and shifted N2 populations were not significantly different (t-test, p = 0.2), the CFUs were reduced more than 1000 fold in daf-2 worms after shift (t-test, p = 0.01). Use of equal initial exposure time produced comparable results after shift (data not shown). (E–F) Mean normalized expression level of immunity genes in akt-1(ok525), akt-2(ok393), sgk-1(ok538), and daf-2(e1370) exposed to (E) OP50 or (F) PA14, plotted as fold relative to expression level in N2 exposed to PA14 on a log2 scale. Transcript levels were measured by quantitative RT-PCR and normalized to the average of 3 control genes. Mean and standard error of the mean from 2 to 4 independent experiments is shown. Pairwise comparisons to N2 were performed. (E) Antimicrobial gene expression is elevated in akt-1(ok525), akt-2(ok393), and daf-2(e1370) but not sgk-1(ok538). (F) Each antimicrobial gene was expressed at significantly higher levels in daf-2(e1370) than N2 under infected conditions. Taken as a whole, the set of antimicrobials was expressed at higher levels in daf-2(e1370), akt-1(ok525), and akt-2(ok393) than N2 (binomial test, p = 0.03). Antimicrobial gene expression was mixed in sgk-1(ok538) mutants (binomial test, p = 0.3) with significantly lower levels of abf-2 and lys-7 but higher levels of spp-1. Error bars indicate standard error of the mean. * p < 0.05, ** p < 0.01, *** p < 0.001
We further assessed the immune competence of daf-2 pathway mutants by determining their ability to clear an infection. For the clearance assay, animals were initially exposed to PA14-GFP for 24 h and then shifted to an E. coli food source for 24 h to recover. The extent of colonization was assayed before and after the shift in two categories: detectable and undetectable colonization. The ratio of colonization before and after shift was used as a metric of bacterial clearance (Figure 3B). For each strain tested, colonization was significantly reduced after the 24 h shift to E. coli. However, the magnitude of the drop in colonization was significantly larger for akt-1(ok525) and akt-2(ok393) than N2 (log-linear analysis, p < 0.05) whereas N2 and sgk-1(ok538) were not significantly different in their ability to clear PA14-GFP. The extent of PA14-GFP colonization in daf-2(e1370) was too low to measure accurately in the PA14-GFP clearance assay. To overcome this problem, the magnitude of the effect of daf-2(e1370) on the accumulation of PA14 was further quantified by enumerating live intestinal PA14 as colony forming units (CFUs). Following 24 h exposure of N2 and daf-2(e1370) to PA14, worms were disrupted and CFUs of PA14 were quantified from the worm lysates. daf-2 worms had significantly lower CFUs than N2 animals (Figure 3C), confirming that they were colonized to a lesser extent. For the clearance assay, daf-2 and N2 animals were initially exposed to PA14 for a predetermined period and then shifted to an E. coli food source. The number of live bacteria within each worm strain was determined immediately before and 24 h after the shift to E. coli. Because equal exposure time resulted in unequal initial colonization between daf-2 and N2 animals (Figure 3C), we exposed daf-2 and N2 worms to PA14 for 24 h and 18 h, respectively. Despite higher PA14 inoculums in daf-2 than N2 animals (Figure 3D, initial), daf-2 worms were able to reduce intestinal PA14 by more than 1000 fold following the shift to E. coli (Figure 3D, shift). In contrast, the PA14 load in N2 did not change significantly 24 h after shift to E. coli (Figure 3D, shift). In animals that were shifted to PA14 for 24 h, PA14 load did not decrease in either strain (data not shown), indicating that the decrease in live PA14 in daf-2(e1370) animals is a function of its increased ability to clear the pathogen from the intestine. Thus, the enhanced resistance of daf-2(e1370) worms to PA14 is associated with a corresponding enhanced ability to restrict bacterial colonization and to clear pathogens.
A microarray analysis of insulin-like pathway mutants identified putative antimicrobial genes as potential DAF-16 transcriptional targets (Murphy et al. 2003). Thus, a plausible mechanism for the enhanced ability of daf-2 animals to restrict bacterial colonization is the constitutively elevated expression of proteins with antibacterial functions. We have observed that several genes that are up-regulated in daf-2 mutants, including thn-2, spp-1, and lys-7, are necessary to restrict colonization by PA14 (T. Kawli and M.-W. Tan, unpublished data). To demonstrate that resistance of daf-2 pathway mutants is correlated with the expression of antimicrobials, we compared the expression levels of five candidate antimicrobial genes in N2 and daf-2-pathway worms that were exposed to OP50 or PA14 for 12 h using quantitative RT-PCR. The criteria for selecting these genes are as follows. First, they are differentially regulated in daf-2 mutants ((Murphy et al. 2003) and E. Evans, S. Slutz and M.-W. Tan, unpublished data) or respond to PA14 infection (Shapira et al. 2006; Troemel et al. 2006). Second, they either have demonstrated antimicrobial activity (the defensin-like gene abf-2 (Banyai & Patthy 1998), the saposin gene spp-1 (Kato et al. 2002), and the neuropeptide-like protein nlp-31 (Couillault et al. 2004)) or are homologous to proteins shown to have antimicrobial activity and when knocked down resulted in increased sensitivity to PA14 without affecting normal lifespan (thn-2 (O'Rourke et al. 2006; Shapira et al. 2006) lys-7 (T. Kawli and M.-W Tan, unpublished)). spp-1 and abf-2 expressions were also induced during Salmonella typhimurium infection and each is required to prevent colonization by this enteric pathogen (Alegado & Tan 2008). Following 12 h exposure to OP50, antimicrobial gene expression was elevated in akt-1(ok525), akt-2(ok393) and daf-2(e1370) but not sgk-1(ok538) (Figure 3E). Expression of abf-2 was significantly higher in akt-2(ok393) and daf-2(e1370), spp-1 was expressed at higher levels in akt-2(ok393) and thn-2 was expressed at higher levels in akt-1(ok525). To confirm that antimicrobial gene expression remained elevated during PA14 infection, we examined gene expression following a 12 h exposure to PA14. The expression levels of abf-2, spp-1, nlp-31, thn-2 and lys-7 were each significantly higher in daf-2(e1370) worms than in N2 worms (Figure 3F). The average expression of each of the five candidate immunity genes was also higher in both akt-1(ok525) and akt-2(ok393) compared to N2 (Figure 3F), consistent with their pathogen resistance phenotypes. In contrast, the expression of these immunity genes in sgk-1(ok538) was mixed between up and down-regulation. While spp-1 was expressed at significantly higher levels, abf-2 and lys-7 expression were significantly lower in sgk-1(ok538), and the overall pattern of expression of antimicrobial genes in sgk-1(ok538) was not significantly different than N2 (Figure 3F), consistent with the wildtype-like pathogen resistance of sgk-1(ok538) mutants. These findings indicate that the expression of these immunity genes as an aggregate during infection is predictive of bacterial colonization and survival, and may underlie the differences between AKT and SGK in immune modulation.
Lifespan extension is uncoupled from pathogen resistance in known aging mutants
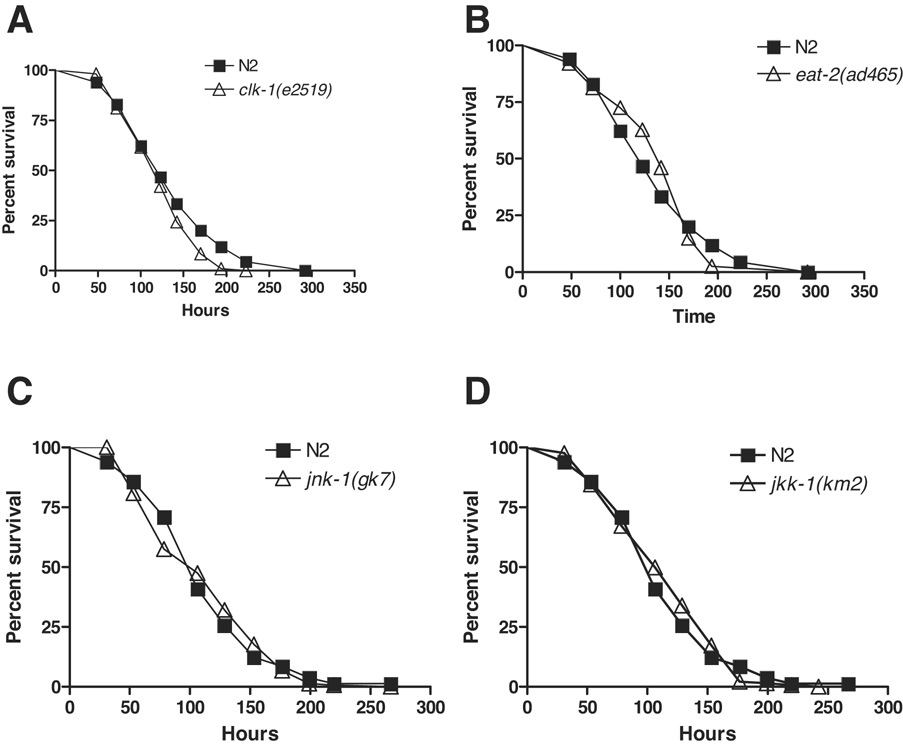
To further dissociate the genetic basis for lifespan and immune regulation, we tested additional well-characterized aging mutants for resistance to PA14. Mutations in clk-1, a gene necessary for ubiquinone synthesis, alter developmental and physiological timing and extend lifespan in C. elegans (Lakowski & Hekimi 1996). Survival of clk-1(e2519) on PA14 was not significantly different than N2 in a cdc-25.1 RNAi Glp background. (Figure 4A). Mutations in eat-2 affect the rate of pharyngeal pumping, and eat-2 is widely used as a genetic model for lifespan extension due to caloric restriction (Lakowski & Hekimi 1998). As with clk-1, we did not observe increased resistance to PA14 in eat-2(ad465) animals in a cdc-25.1 RNAi Glp background (Figure 4B). This finding is not unexpected because we previously found that an eat-1 mutant, which also exhibits a reduced rate of pharyngeal pumping (Avery 1993) and increased lifespan (Lakowski & Hekimi 1998), was indistinguishable from wildtype for resistance to PA14 (Tan et al. 1999). Both clk-1 and eat-2 are reported to function independently of daf-2 insulin-like signaling to increase lifespan (Lakowski & Hekimi 1996; Lakowski & Hekimi 1998; Houthoofd et al. 2003). Consistent with this observation, neither gene appears to be required for the regulation of resistance to bacterial pathogens.
Figure 4. Pathogen resistance is not linked with lifespan extension in aging mutants.
Other long and short-lived mutants do not have increased or decreased pathogen resistance. Neither clk-1 (A) nor eat-2 (B) were more resistant than N2 (logrank test, p = 0.02 and 0.7 respectively). clk-1(e2519) was not more sensitive than N2 as replicate experiments yield mean times to death from 92% to 104% relative to N2 (Supplemental Table S1). Likewise, neither jnk-1(gk7) (C) nor jkk-1(km2) (D) was more sensitive than N2 (logrank, p = 0.91 and 0.71 respectively). All experiments used cdc-25.1 RNAi Glp animals. Representative experiments shown. Complete supporting data are presented in Supplementary Table S1.
The c-Jun N-terminal kinase pathway positively regulates lifespan in parallel to the DAF-2/AKT-1/AKT-2 pathway but both converge on DAF-16 (Oh et al. 2005). Loss-of-function jnk-1(gk7) and jkk-1(km2) mutants have shortened lifespans (Oh et al. 2005). However, unlike daf-16(mu86), neither mutant was more sensitive than N2 as cdc-25.1 RNAi Glp animals to PA14 (Figure 4C–D). Complementing these results, the absence of a pathogen sensitivity phenotype for a jnk-1 mutant with intact germline was previously reported (Kim et al. 2002). Our analysis with Glp animals further shows that neither jnk-1 nor jkk-1 is required for germline-regulated pathogen defense.
Discussion
We showed that resistance to bacterial pathogens and lifespan can be uncoupled within the daf-2 pathway (Figure 1–Figure 3) and in a variety of aging mutants (Figure 4). Pathogen resistance and lifespan are concordantly regulated by the DAF-2 insulin-like receptor and by AGE-1/AAP-1 PI3 kinase signaling (Figure 2A and Supplemental Table S1). Downstream of DAF-2 and AGE-1, four serine theonine kinases affect the activity and localization of the forkhead transcription factor DAF-16, which directly or indirectly regulates the expression of genes involved in the regulation of life span and immunity. We showed that these serine threonine kinases differentially regulate pathogen resistance and lifespan (Figure 5). These observations support two direct conclusions regarding the regulation of lifespan and pathogen resistance. First, by showing that deletion mutations in akt-1 or akt-2 have increased lifespan on UV-killed E. coli, an assay that eliminated the possible contribution of immune function for survival on live E. coli, we can conclude that AKT-1 and AKT-2 regulate lifespan at least in part independently of their regulation of pathogen resistance. Second, we conclude that SGK-1 and PDK-1 regulate lifespan independently of pathogen resistance because loss-of-function mutations in sgk-1 and pdk-1 enhanced lifespan without affecting pathogen resistance. Taken together, these findings indicate that lifespan and pathogen resistance are regulated by genetically distinguishable mechanisms.
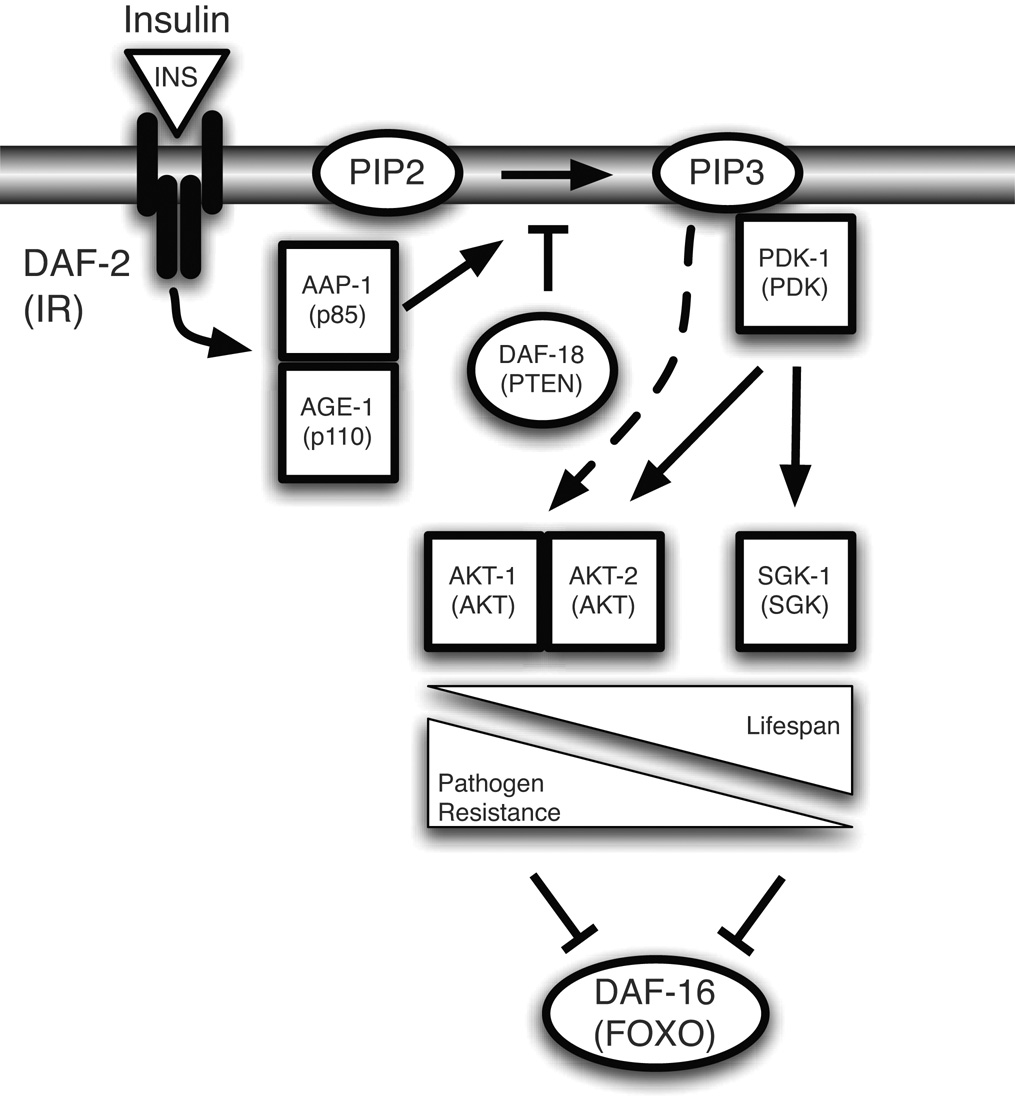
Figure 5. Model of the regulation of pathogen resistance and lifespan by insulin-like signaling.
Activation of DAF-2 potentiates the generation of PIP3 by AGE-1, which in turn activates PDK-1. Activated PDK-1 leads to the phosphorylation of DAF-16 by AKT-1, AKT-2, and SGK-1. AKT-1 and AKT-2 contribute to the regulation of pathogen resistance and lifespan, whereas SGK-1 regulates lifespan but not pathogen resistance. A hypothetical PDK-1-independent input to AKT-1 and AKT-2 is also indicated. Model adapted from (Hertweck et al. 2004) with additional data from this report.
The interpretation of the wildtype-like pathogen resistance of the pdk-1(sa680) mutant depends on whether pdk-1(sa680) retain retains residual PDK-1 activity. Biochemical evidence indicates that PDK-1 functions upstream of AKT-1, AKT-2, and SGK-1. However, the developmental requirement for pdk-1 can be bypassed by overexpression of akt-1 or akt-2 but not sgk-1 in a pdk-1(sa680) background (Hertweck et al. 2004). We observed that RNAi knockdown of pdk-1 in a pdk-1(sa680) background does not enhance pathogen resistance, consistent with pdk-1(sa680) being a strong loss-of-function or null mutant. If pdk-1(sa680) is a null allele, then the pathogen resistance phenotypes of akt-1, akt-2, sgk-1, and pdk-1 loss-of-function mutants (Figure 2) suggest that AKT-1 and AKT-2 can act independently of PDK-1 in the regulation of pathogen resistance. Alternatively, pdk-1(sa680) may retain residual signaling to AKT-1 and AKT-2 but not SGK-1. Resolution of this issue requires the identification of a pdk-1 null allele. These observations illustrate that pathogen resistance provides an alternative phenotype with which to delineate insulin-like signaling.
DAF-2 and DAF-16 regulate worm physiology as part of an intricate signaling pathway with contributions from tissue-specific and yet-unidentified components. We failed to find evidence that regulation of pathogen resistance and lifespan are distinguished by the AKT/SGK phosphorylation sites on DAF-16 or by the tissue specific function of DAF-16 in insulin-like signaling. However, we could not rule out the possibility that one or both of these mechanisms contributes to the differential regulation of pathogen resistance and lifespan. An alternative hypothesis which has been examined elsewhere (Paradis & Ruvkun 1998; Lin et al. 2001; Hertweck et al. 2004) is that AKT-1/AKT-2/SGK-1 competes with an alternative parallel branch of the DAF-2 pathway. Activation of a parallel pathway that converges on DAF-16 could account for the superlative pathogen resistance phenotypes of daf-2(e1370) mutants we observed.
We have also found evidence of a role for DAF-16 in pathogen resistance as a target of signaling regulated by germline proliferation in addition to its role downstream DAF-2. Our results indicate that daf-16 is necessary for the enhancement of resistance in germline-deficient animals, which parallels the observation that lifespan is regulated by germline signaling (Hsin & Kenyon 1999; Lin et al. 2001). In animals with proliferating germlines, however, daf-16 is required for lifespan regulation but not for resistance to PA14. In contrast, sek-1, jnk-1, and jkk-1 do not appear to suppress the pathogen resistance of Glp animals. The observation that a daf-16 null mutant completely suppresses the pathogen resistance associated with loss of germline proliferation suggests that the effect of eliminating germline proliferation on pathogen resistance is mediated by activation of DAF-16.
Recently, analyses of several mutants identified from an enhanced pathogen resistance (Epr) screen suggest that sterility contributes to pathogen resistance in a DAF-16-dependent manner (Miyata et al. 2008). In contrast to the regulation of lifespan by the germline, which appears to be mediated by signals from the germline stem cells, the authors suggest that resistance to PA14 in these sterile animals is mediated by signals from the embryos. It will be interesting to determine what distinguishes sterile strains in which PA14 susceptibility is daf-16-dependent, such as glp-1(e2141) and fer-15(b26);fem-1(hc17), from those strains in which PA14 susceptibility is daf-16 independent, such as pha-1(e2123) and cdc-25.1 Emb (Figure 1 and Table S1). One possible explanation is that pha-1(e2123) and cdc-25.1 Emb animals possess fertilized embryos, whereas fertilization does not occur in any of the sterile animals in which daf-16-dependent pathogen resistance is observed. If the embryo-signal hypothesis of sterile pathogen resistance is confirmed, it would represent another instance of the distinguishable regulation of pathogen resistance and lifespan. Like daf-2 insulin-like signaling, the pathogen resistance of sterile mutants represents the regulation of pathogen resistance and lifespan by convergent signals acting on DAF-16.
We examined two mechanisms that are associated with pathogen resistance in insulin-like signaling mutants. We have shown that the pathogen resistance of insulin-like signaling mutants is associated with reduced bacterial colonization and enhanced expression of antimicrobial genes. The enhanced resistance of daf-2 pathway worms to PA14 (Figure 2) is associated with a corresponding enhanced ability to restrict bacterial colonization (Figure 3A,C) and to clear pathogens (Figure 3B,D). The expression of putative antimicrobial genes during infection is predictive of bacterial colonization and survival and may underlie the differences between akt-1, akt-2, and sgk-1 in immune modulation (Figure 3E,F). Our data suggest a model wherein pathogen resistance of daf-2 pathway mutants is caused in part by reduced bacterial colonization due to enhanced expression of immunity genes.
An ROS resistance model has also been proposed to explain the pathogen resistance of daf-2 mutants, such that resistance to oxidative stress is partially required for resistance to pathogen (Chavez et al. 2007). It has been reported that sgk-1 and pdk-1 mutants have enhanced ROS resistance, but akt-1 and akt-2 mutants do not, which may account for the effects of these mutants on lifespan (Hertweck et al. 2004). We speculate that enhanced ROS resistance alone is insufficient for enhanced resistance to P. aeruginosa and that enhanced expression of antimicrobials may be necessary. One possibility is that the superlative resistance of daf-2(e1370) is mediated by enhancements of both ROS resistance and antimicrobials. Thus, the regulation of both ROS resistance and antimicrobial gene expression by the daf-2 pathway may explain the convergence of pathogen resistance and lifespan regulation.
Our results indicate that merely being long lived is not sufficient for resistance to bacterial pathogens, nor are short-lived mutants necessarily sensitive to pathogens (Figure 4). In addition, several examples of pathogen-sensitive or pathogen-resistant mutants with normal lifespans have been reported. Loss of the GATA transcription factor elt-2 in adults enhances sensitivity to pathogen without shortening lifespan (Shapira et al. 2006), and a relatively normal lifespan is reported for the pathogen-sensitive sek-1(km4) and pmk-1(km25) mutants (Kim et al. 2002; Troemel et al. 2006). Also, the necrosis-deficient mutants vha-12(n2915) and unc-32(e189) are resistant to two Gram negative bacteria but have normal lifespan (Wong et al. 2007).
The C. elegans model of aging and innate immunity may provide insights into the regulation of these processes in other organisms, including humans. The human gut is colonized by hundreds of distinguishable bacterial strains (Gill et al. 2006), and the composition of this intestinal microbiota varies with genotype, age, diet, and health (Lupp & Finlay 2005; Turnbaugh et al. 2006). Moreover, decline in immune function is an important facet of human aging (Ginaldi et al. 2001) and pathogenesis is a major cause of mortality in older adults (Heron 2007). In C. elegans, aging is also associated with a decline in pathogen resistance (Kurz & Tan 2004), and post-reproductive worms are compromised in their ability to respond transcriptionally to PA14 exposure (E. A. Evans and M.-W. Tan, unpublished observation).
Aspects of the molecular regulation of aging and innate immunity are also conserved from worms to human. Insulin-like signaling is an evolutionarily conserved regulator of lifespan, and reduction of insulin-like signaling increases lifespan in flies (Clancy et al. 2001; Clancy et al. 2002) and mice (Bluher et al. 2003; Holzenberger et al. 2003; Tatar et al. 2003). Reduced insulin-like signaling also increases resistance to bacterial pathogens in Drosophila (Libert et al. 2008). However, in contrast to C. elegans, canonical antimicrobial peptides do not appear to be expressed at higher levels in Drosophila insulin-like signaling mutants (Libert et al. 2008). Also, Akt and FOXO, the fly homologues of AKT-1/2 and DAF-16, play a causal role in a bacterial infection-induced wasting (Dionne et al. 2006). In mammals, insulin and IGF-1 signaling have broad effects on the innate and adaptive immune systems (Heemskerk et al. 1999; Kelley et al. 2007). Dietary restriction is another conserved regulator of lifespan. We observed no enhancement of pathogen resistance in worms subjected to a genetic form of dietary restriction (Figure 4B). Similarly, dietary restriction does not enhance pathogen resistance in Drosophila (Libert et al. 2008). In mammals, including humans, reports of the effects of dietary restriction on immunity are mixed (Jolly 2007) but suggest either no effect (McFarlin et al. 2006), a marginally positive effect (Rankin et al. 2006), or negative effects (Gardner 2005) on immune function. Because many aspects of the molecular regulation of aging and innate immunity are conserved from worms to mammals, it will be interesting to determine if our finding that the genetic regulation of lifespan and pathogen resistance in C. elegans are distinguishable may apply to other organisms as well.
Experimental Procedures
Strains
Caenorhabditis elegans strains were obtained from the Caenorhabditis Genetics Center (CGC) unless otherwise mentioned. sgk-1(ok538), akt-1(ok525) and akt-2(ok393) were backcrossed to N2 4x and an independent backcrossed line of each was gratefully received from M. Hertweck and R. Baumeister of the University of Freiburg (Hertweck et al. 2004). Within experiments, worm strains were grown at matched temperatures. daf-2(e1370) were grown at 20°C. rrf-3(pk1426);glp-4(bn2), glp-4(bn2), and pha-1(e2123) were grown at 15°C and shifted to 25°C by L4. age-1(hx546) and pdk-1(sa680) were grown at 15°C until adulthood. sgk-1(ok538), akt-1(ok525), akt-2(ok393), clk-1(e2519) and other strains were grown at 25°C unless matched with strains grown at lower temperatures.
Pseudomonas aeruginosa strains included the clinical isolate PA14 and PA14-GFP, an isogenic GFP-expressing strain (Tan et al. 1999). Escherichia coli was a streptomycin-resistant derivative of OP50, OP50-1, obtained from the CGC. Bacteria expressing dsRNA directed against daf-16, akt-1, and cdc-25.1 were part of a C. elegans RNAi library expressed in E. coli (Geneservice, Cambridge, U.K.). Bacteria expressing dsRNA directed against akt-2 and sgk-1 were part of a C. elegans RNAi library expressed in E. coli (Open Biosystems, Huntsville, Alabama). Bacterial expressing dsRNA directed against pdk-1 was a gift from W. Iser and C. Wolkow (Gami et al. 2006). All bacterial strains were cultured by shaking at 37°C.
cdc-25.1 RNAi knockdown
RNAi knockdown of cdc-25.1 in egg-laying adults was used to produce a mixed population of animals with or without a proliferating germline (Glp), which could be clearly distinguished at the 1-day old adult stage using a stereomicroscope. Those animals with a germline are fertile, but the eggs they lay will not hatch (Emb). The distinction between Glp and Emb animals are confirmed by DIC microscopy. Glp animals are readily confirmed by the absence of the gonads and germline and by the absence of eggs laid on plates containing only Glp animals. Emb animals are recognizable as young adults by the presence of embryos in the uterus of Emb worms. Within the population of cdc-25.1(RNAi) animals, a small percentage of animals have ambiguous classification as Glp or Emb, and these animals are not used in any of the experiments we performed. Two sequential 4 h egg lays on RNAi plates at 25°C were used as described (Shapira et al. 2006). Unless noted, Glp animals were selected for the C. elegans survival assays.
C. elegans survival assays
Assays to determine the ability of C. elegans to survive PA14 infection were performed as described (Shapira & Tan 2008). Briefly, PA14 was grown overnight in King’s Broth containing 100 mg/ml rifampicin at 37°C. 10 µl was spread on modified nematode growth (NG) media and grown for 24 h at 37°C. Worms were infected at 25°C by feeding on PA14 lawns. Kaplan-Meier survival analysis was performed using StatView 5.0.1. The Mantel-Cox logrank test was used to assess statistical significance of differences in survival. Only p-values < 0.01 were considered significant to minimize type I errors. Mean time to death and standard error of the mean was calculated in StatView and then normalized to N2 for graphical comparison (Table S1–2).
For lifespan assays, UV-killed bacteria was prepared as described (Gems & Riddle 2000). Briefly, OP50-1 was grown in LB with 100 mg/ml streptomycin, concentrated 20X, and 80µl was spread onto NG plates. Plates were placed at ~23°C for 16h, then exposed to UV in a Stratagene Stratalinker 2400 for 4 min.
Bacterial accumulation assay
Young adult worms were exposed to PA14-GFP for 24h at 25°C, and then examined at 40X magnification for intestinal accumulation of GFP using a Leica MZFLIII stereomicroscope. Individual worms were categorized as fully colonized, partially colonized, or undetectable colonization. These categories have been validated against colony-forming unit counts (CFUs). Specifically, 6 worms from each category were individually enumerated for CFUs; with fully colonized, partially colonized, or undetectable colonization corresponding to averages of 4.22 × 104, 2.45 × 104 and 3.48 × 103 colony forming units of PA14-GFP, respectively (data not shown). Population proportions of these categories were compared by Chi-square test using Statview 5.0.1. Experimental replicates were pooled for graphical display and a summary statistical analysis.
Colony forming unit measurement
Pools of ten worms were picked into M9 containing 25 mM levamisole, washed twice in M9 containing 25 mM levamisole, then exposed to 25 mM levamisole, 1mg/ml ampicillin, and 1mg/ml gentamycin in M9 for 1 h to kill external bacteria. Worms were washed again in M9 containing 25 mM levamisole and disrupted in M9 containing 1% Triton-X100 using a motorized pestle. Serial dilutions of worm lysate were grown overnight on LB containing 100µg/ml Rifampicin at 37°C and colony forming units were quantified. At least three replicates for each condition were processed in parallel. For shift experiments, worms were moved to OP50-1 lawns on NG agar containing 300µg/ml streptomycin.
RNA extraction and qRT-PCR
Worms were grown until young adults at 20°C on OP50-1, then moved to 6–8 modified NG plates containing 10 µl PA14 grown overnight. After a 12 h exposure at 25°C, worms were resuspended in M9, washed 3 times, resuspended in 1 ml of Trizol, and stored at −80C until use. Total RNA was extracted as described (Reinke et al. 2000) and DNase treated using TurboDNAse (Ambion, Austin TX). RT-PCR was performed with the Bio-Rad iScript One-Step RT-PCR Kit with SYBR Green in 25 µl reactions using a Bio-Rad iCycler according to the manufacturer’s instructions (BioRad Laboratories, Hercules, CA). The average cycle threshold of three primer pairs (ama-1, F44B9.5, and pan actin (act-1,3,4)) was used as a loading control correction. Summary statistics and statistical tests were calculated from N2-normalized cycle threshold values prior to conversion to relative fold change. Calculations were performed with Microsoft Excel 2003.
Primers
Whenever possible, primers were designed to amplify a sequence found in spliced cDNA but not genomic DNA by having one of the primer pairs overlap an exon junction. Primer design was aided with the program AutoPrime (Wrobel et al. 2004). Primer sequences are available from the authors upon request.
Supplementary Material
Acknowledgments
We thank A. Brunet, A. Fire, and the members of the Tan lab, for critical comments, discussions and technical advice; S. Slutz for assistance with RT-PCR; T. Kawli for assistance with microscopy; B. Hamlin for assistance with survival assays; the Caenorhabditis Genetics Center, the C. elegans Gene Knockout Consortium, M. Hertweck, and R. Baumeister for worm strains; W. Iser and C. Walkow for RNAi clones. This work was supported by grants from the National Institutes of Health (M.-W.T.). E.A.E. was supported by a Stanford Graduate Fellowship and a National Science Foundation Graduate Research Fellowship. W.C.C. was supported by a Stanford Graduate Fellowship and a Howard Hughes Medical Institute Graduate Fellowship.
References
- Alegado RA, Tan MW. Resistance to antimicrobial peptides contributes to persistence of Salmonella typhimurium in the C. elegans intestine. Cell Microbiol. 2008;10:1259–1273. doi: 10.1111/j.1462-5822.2008.01124.x. [DOI] [PubMed] [Google Scholar]
- Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:1565–1571. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133:897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyai L, Patthy L. Amoebapore homologs of Caenorhabditis elegans. Biochim Biophys Acta. 1998;1429:259–264. doi: 10.1016/s0167-4838(98)00237-4. [DOI] [PubMed] [Google Scholar]
- Baumeister R, Schaffitzel E, Hertweck M. Endocrine signaling in Caenorhabditis elegans controls stress response and longevity. J Endocrinol. 2006;190:191–202. doi: 10.1677/joe.1.06856. [DOI] [PubMed] [Google Scholar]
- Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Bolm M, Chhatwal GS, Jansen WT. Bacterial resistance of daf-2 mutants. Science. 2004;303:1976. doi: 10.1126/science.303.5666.1976a. [DOI] [PubMed] [Google Scholar]
- Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol Cell Biol. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez V, Mohri-Shiomi A, Maadani A, Vega LA, Garsin DA. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics. 2007;176:1567–1577. doi: 10.1534/genetics.107.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Hafen E, Leevers SJ, Partridge L. Dietary restriction in long-lived dwarf flies. Science. 2002;296:319. doi: 10.1126/science.1069366. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Dionne MS, Pham LN, Shirasu-Hiza M, Schneider DS. Akt and FOXO dysregulation contribute to infection-induced wasting in Drosophila. Curr Biol. 2006;16:1977–1985. doi: 10.1016/j.cub.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Ewbank JJ. Signaling in the immune response. WormBook. 2006:1–12. doi: 10.1895/wormbook.1.83.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gami MS, Iser WB, Hanselman KB, Wolkow CA. Activated AKT/PKB signaling in C. elegans uncouples temporally distinct outputs of DAF-2/insulin-like signaling. BMC Dev Biol. 2006;6:45. doi: 10.1186/1471-213X-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner E. Caloric restriction decreases survival of aged mice in response to primary influenza infection. J Gerontol A Biol Sci Med Sci. 2005;60:688–694. doi: 10.1093/gerona/60.6.688. [DOI] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- Gems D, Riddle DL. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics. 2000;154:1597–1610. doi: 10.1093/genetics/154.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginaldi L, Loreto MF, Corsi MP, Modesti M, De Martinis M. Immunosenescence and infectious diseases. Microbes Infect. 2001;3:851–857. doi: 10.1016/s1286-4579(01)01443-5. [DOI] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk VH, Daemen MA, Buurman WA. Insulin-like growth factor-1 (IGF-1) and growth hormone (GH) in immunity and inflammation. Cytokine Growth Factor Rev. 1999;10:5–14. doi: 10.1016/s1359-6101(98)00022-7. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Heron M. Deaths: leading causes for 2004. Natl Vital Stat Rep. 2007;56:1–95. [PubMed] [Google Scholar]
- Hertweck M, Gobel C, Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp Gerontol. 2003;38:947–954. doi: 10.1016/s0531-5565(03)00161-x. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Iser WB, Gami MS, Wolkow CA. Insulin signaling in Caenorhabditis elegans regulates both endocrine-like and cell-autonomous outputs. Dev Biol. 2007;303:434–447. doi: 10.1016/j.ydbio.2006.04.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly CA. Is dietary restriction beneficial for human health, such as for immune function? Curr Opin Lipidol. 2007;18:53–57. doi: 10.1097/MOL.0b013e3280115416. [DOI] [PubMed] [Google Scholar]
- Kato Y, Aizawa T, Hoshino H, Kawano K, Nitta K, Zhang H. abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem J. 2002;361:221–230. doi: 10.1042/0264-6021:3610221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley KW, Weigent DA, Kooijman R. Protein hormones and immunity. Brain Behav Immun. 2007;21:384–392. doi: 10.1016/j.bbi.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kerry S, TeKippe M, Gaddis NC, Aballay A. GATA transcription factor required for immunity to bacterial and fungal pathogens. PLoS ONE. 2006;1:e77. doi: 10.1371/journal.pone.0000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan MW, Ausubel FM. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem J. 1999;339(Pt 2):319–328. [PMC free article] [PubMed] [Google Scholar]
- Kurz CL, Tan MW. Regulation of aging and innate immunity in C. elegans. Aging Cell. 2004;3:185–193. doi: 10.1111/j.1474-9728.2004.00108.x. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- Libert S, Chao Y, Zwiener J, Pletcher SD. Realized immune response is enhanced in long-lived puc and chico mutants but is unaffected by dietary restriction. Mol Immunol. 2008;45:810–817. doi: 10.1016/j.molimm.2007.06.353. [DOI] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ. Does anti-aging equal anti-microbial? Sci Aging Knowl Environ. 2003;2003:PE16. doi: 10.1126/sageke.2003.25.pe16. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Finlay BB. Intestinal microbiota. Curr Biol. 2005;15:R235–R236. doi: 10.1016/j.cub.2005.03.032. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thornton J, Gems D. Diapause-associated metabolic traits reiterated in long-lived daf-2 mutants in the nematode Caenorhabditis elegans. Mech Ageing Dev. 2006;127:458–472. doi: 10.1016/j.mad.2006.01.006. [DOI] [PubMed] [Google Scholar]
- McFarlin BK, Flynn MG, Mahon AK, Stewart LK, Timmerman KL, Lyle RM, Campbell WW. Energy restriction with different protein quantities and source: implications for innate immunity. Obesity. 2006;14:1211–1218. doi: 10.1038/oby.2006.138. [DOI] [PubMed] [Google Scholar]
- Miyata S, Begun J, Troemel ER, Ausubel FM. DAF-16-Dependent Suppression of Immunity during Reproduction in C. elegans. Genetics. 2008 doi: 10.1534/genetics.107.083923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003 doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- O'Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Ailion M, Toker A, Thomas JH, Ruvkun G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 1999;13:1438–1452. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DS, Garcia-Garza A, Nanji M, McElwee JJ, Ackerman D, Driscoll PC, Gems D. Clustering of Genetically Defined Allele Classes in the Caenorhabditis elegans DAF-2 Insulin/IGF-1 Receptor. Genetics. 2008 doi: 10.1534/genetics.107.070813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin J, Shute M, Heffron S, Sake K. Energy restriction but not protein source affects antioxidant capacity in athletes. Free Radic Biol Med. 2006;41:1001–1009. doi: 10.1016/j.freeradbiomed.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, Kim SK. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Myer BJ, Priess JR. C. elegans II. Plainsview, New York: Cold Spring Harbor Labortory Press; 1997. [Google Scholar]
- Schnabel H, Schnabel R. An Organ-Specific Differentiation Gene, pha-1, from Caenorhabditis elegans. Science. 1990;250:686–688. doi: 10.1126/science.250.4981.686. [DOI] [PubMed] [Google Scholar]
- Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, Tan MW. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci U S A. 2006;103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M, Tan MW. Genetic analysis of Caenorhabditis elegans innate immunity. Methods Mol Biol. 2008;415:429–442. doi: 10.1007/978-1-59745-570-1_25. [DOI] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- Wolkow CA, Munoz MJ, Riddle DL, Ruvkun G. Insulin receptor substrate and p55 orthologous adaptor proteins function in the Caenorhabditis elegans daf-2/insulin-like signaling pathway. J Biol Chem. 2002;277:49591–49597. doi: 10.1074/jbc.M207866200. [DOI] [PubMed] [Google Scholar]
- Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel G, Kokocinski F, Lichter P. AutoPrime: selecting primers for expressed sequences. Genome Biology. 2004;5:P11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.