Abstract
Transmissible spongiform encephalopathies are associated with the conformational conversion of the prion protein from the cellular form (PrPC) to the scrapie form. This process could be disrupted by stabilizing the PrPC conformation, using a specific ligand identified as a chemical chaperone. To discover such compounds, we employed an in silico screen that was based on the nuclear magnetic resonance structure of PrPC. In combination, we performed ex vivo screening using the Fukuoka-1 strain-infected neuronal mouse cell line at a compound concentration of 10 μM and surface plasmon resonance. Initially, we selected 590 compounds according to the calculated docked energy and finally discovered 24 efficient antiprion compounds, whose chemical structures are quite diverse. Surface plasmon resonance studies showed that the binding affinities of compounds for PrPC roughly correlated with the compounds' antiprion activities, indicating that the identification of chemical chaperones that bind to the PrPC structure and stabilize it is one efficient strategy for antiprion drug discovery. However, some compounds possessed antiprion activities with low affinities for PrPC, indicating a mechanism involving additional modulation factors. We classified the compounds roughly into five categories: (i) binding and effective, (ii) low binding and effective, (iii) binding and not effective, (iv) low binding and not effective, and (v) acceleration. In conclusion, we found a spectrum of compounds, many of which are able to modulate the pathogenic conversion reaction. The appropriate categorization of these diverse compounds would facilitate antiprion drug discovery and help to elucidate the pathogenic conversion mechanism.
Transmissible spongiform encephalopathies (TSEs) are neurodegenerative diseases that include Creutzfeldt-Jakob disease, chronic wasting disease, scrapie, and bovine spongiform encephalopathy. These diseases are characterized by the accumulation of the scrapie form of the prion protein (PrPSc) in the central nervous system (4, 23). The conversion of PrP from the normal cellular form (PrPC) to the PrPSc form is a characteristic feature of the diseases, although a detailed structure of PrPSc is still unknown. The occurrence of TSEs is associated with specific mutations in PrP, inoculation with infectious material, or apparently spontaneous onset. Since there are currently no established therapies for TSEs, it is important to identify compounds with therapeutic or prophylactic activity against these diseases.
The purpose of this study was to characterize the effects of small compounds (molecular weight of 150 to 350) on the prion's pathogenic conversion reaction and to investigate the structure-activity relationship among a broader spectrum of compounds. The structure-activity relationship would also give us a clue for designing a chemical chaperone (15). This work applied an in silico screening method using AutoDock software (19) to target a wider area around the major PrP pocket, and a larger compound database was utilized for screening. During pathogenic conversion, various regions may be involved, including the factor X-binding site (11) and the hot spots (15). Small compounds that are able to interact with those regions may modulate the conversion reaction (2, 26, 27).
Currently, it is known that various docking programs are generally not accurate in predicting binding affinities or binding modes (28). Although the antiprion effects of compounds can also be characterized by these two parameters, the difficulties described above can be somewhat diminished because pathogenic conversion involves various sites. This conformational change in the protein is quite distinct from the enzymatic reaction in which only the active site is involved. Because PrP contains broadly distributed binding sites, in silico screening could provide a broad spectrum of candidates as antiprion ligands, even if predicted binding sites or binding affinities are not very accurate (21). However, the resulting broad spectrum of effective small compounds finally examined by the ex vivo screening could provide a clue regarding how to optimize the compound structure and a potential hint for elucidating the mechanism of the conversion reaction.
Here, neuronal mouse (GT1-7) cells chronically infected with the Fukuoka-1 strain (18) have been used as a model for examining antiprion activities. The Fukuoka-1 strain was isolated from a case of human Gerstmann-Sträussler-Scheinker syndrome. This cell line could provide a useful model for screening compounds with antiprion activity because it produces PrPSc quite stably. Additionally, this cell line allows for the examination of cellular processes associated with the production of PrPSc. To examine the antiprion activity of approximately 200 compounds selected by using an in silico method, we used a Western blotting technique described previously (15). Results indicated that a variety of compounds could inhibit PrPSc formation when added to the medium of these cells. Compounds that were able to accelerate the formation of PrPSc were seen as well.
To further elucidate the mechanisms of action, we measured the compound binding affinities by using surface plasmon resonance (SPR) and investigated the correlation between antiprion activities and binding affinities. The possible mechanisms behind the effects of small compounds on the prion's pathogenic conversion reaction are discussed below, and we provide direction for further optimization.
MATERIALS AND METHODS
Database and preparation of compounds.
A subset composed of approximately 600,000 compounds from a free compound database named ZINC (10) was used for docking simulations. ZINC is provided by the Shoichet laboratory in the Department of Pharmaceutical Chemistry at the University of California, San Francisco. ZINC5 (2005 version of the database) was used in this study. The flexibility of compounds was determined automatically using AUTOTORS, which is a program in AutoDock software (19), and the united-atom model for the compound was also created with AUTOTORS. Nonpolar hydrogen atoms in the compounds were removed after their partial charges were moved to the covalently bonded carbon atoms, whose charge information was given in the ZINC database.
Preparation for protein and grid space.
The nuclear magnetic resonance structure of mouse PrPC (PDB code 1ag2) was used for the target structure of PrPC. AutoDockTools was used to obtain charge information (Kollman united-atom charge) and to build a united-atom model of PrPC. The solvation parameters were prepared automatically using ADDSOL, which is a program in the AutoDock software package. The dimensions of the grids for the docking analysis were 120 by 120 by 80 points (45 Å by 45 Å by 30 Å), with a grid point spacing of 0.375 Å. The center position was set around the center of the major binding pocket of PrPC, which is the putative GN8 binding site previously reported (15) between helix-2 and the loop between helix-1 and β-strand S2.
Automated docking.
AutoDock version 3.05 was used for automated docking simulation (19). Standard parameters, which were created with mkdpf3, a script program in the AutoDock software package, were used. Parameters were as follows: population size, 50; random starting position and conformation; maximal mutation in translation, 2 Å; mutation in rotation, 50 degrees; elitism, 1; mutation rate, 0.02; crossover rate, 0.8; runs, 10 times; energy evaluations, 250,000; and the Lamarckian genetic search algorithm was selected. Calculations were performed with a 26-node PC cluster in which each node has a dual-core central processing unit (2.8 GHz, Pentium-D; Intel). To select compounds for examination, we conducted in silico screening with AutoDock. We made a list of about 600,000 compounds by ranking the compounds according to the docked energy calculated by AutoDock.
Compounds.
We selected compounds depending on the lowest docked energy among 10 runs. In total, 205 compounds were purchased from Maybridge (Cornwall, United Kingdom), Asinex (Moscow, Russia), and Enamine (Kiev, Ukraine). All compounds were tested for antiprion activity.
Cell culture and antibodies.
We used the immortalized neuronal mouse cell line that was either uninfected or persistently infected with the human TSE agent (Fukuoka-1 strain). The former (uninfected) cell line is known as GT1-7, while the latter (infected) line is designated GT+FK (18). These cells were grown and maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Equitech-bio, Kerrville, TX), 50 U/ml penicillin G sodium, and 50 μg/ml streptomycin sulfate (Invitrogen, Carlsbad, CA). The GT+FK cells were maintained for more than 2 years in our laboratory. Stock solutions of compounds were prepared fresh in 100% dimethyl sulfoxide (DMSO) at 10 mM and stored at 4°C. Before being used, compounds were diluted with medium as indicated in Fig. 1A. Control cells were treated with medium containing solvent (0.1%) alone. Approximately 1.5 × 105 cells were plated in each well of a six-well plate, and compound treatment was started 15 h later. After 72 h of treatment, cells were lysed in 150 μl of 1× Triton X-100-deoxycholate lysis buffer (150 mM NaCl, 0.5% Triton X-100, 0.5% sodium deoxycholate, 50 mM Tris-HCl [pH 7.5]) (20), and the total protein concentration of the sample was adjusted to 2 mg of protein per milliliter. Western blotting for PrPSc was done as described previously (20). For the primary antibody, PrP M-20 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used to detect PrPSc and PrPC. The signals were visualized by SuperSignal (Pierce Biotechnology, Rockford, IL) and scanned using a LAS-1000 UV mini analyzer (Fujifilm, Tokyo, Japan).
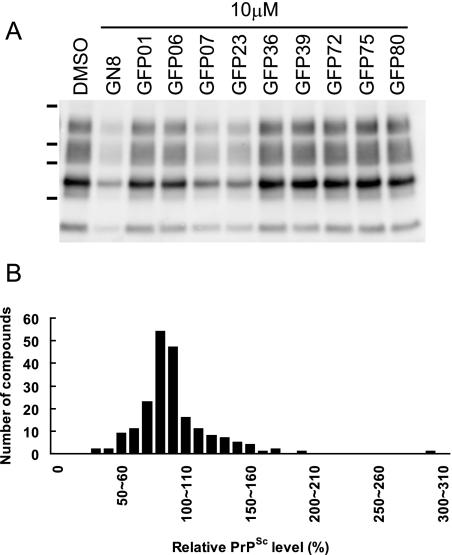
FIG. 1.
Ex vivo screening. (A) Western blotting of PrPSc in GT+FK cells after treatment with different compounds identified through in silico screening. The cells treated with GFP07 or GFP23 showed a significant reduction of PrPSc. Lane DMSO, 0.1% DMSO; lane GN8, 2-pyrrolidin-1-yl-N-[4-[4-(2-pyrrolidin-1-yl-acetylamino)-benzyl]-phenyl]-acetamide (15). Dashes at left represent molecular mass markers (37, 25, 20, and 15 kDa). (B) A histogram shows the number of compounds as a function of the relative PrPSc level. The relative PrPSc levels were divided into subsets of 10%, and the number of compounds contained in each subset is shown. The distribution of the relative PrPSc levels is close to the normal distribution, and intriguingly, some compounds increased the relative PrPSc levels to more than 150% of the control.
The density of PrPSc derived from GT+FK cells in each solution was measured and compared with that of the control treated with medium containing solvent. Compounds were judged to be effective if levels of PrPSc were reduced to less than 70% of the control, as reported previously (8). At least two independent experiments were performed to determine the levels of PrPSc.
Recombinant mouse PrP(121-231).
The expression plasmid for mouse PrP(121-231) (the PrP region from amino acids 121 to 231) was a kind gift from Kurt Wüthrich and Simone Hornemann. The recombinant PrP was prepared for SPR measurements as described previously (9, 15). The concentration of mouse PrP was estimated by using the specific absorbance of ɛ at 280 nm of 1.49 (mg/ml)−1 cm−1.
SPR measurements.
Interactions between the PrP and the compounds were analyzed using a Biacore T100 system. A recombinant mouse PrP(121-231) was immobilized on a sensor chip (product CM5) according to the manufacturer's instructions. Various concentrations of compounds were injected into running buffer (0.1% surfactant P20 in 0.01 M HEPES [pH 7.4]-0.15 M NaCl [product HBS-N; Biacore] containing 5% DMSO [pH 7.4]) for 1 min at a flow rate of 30 μl/min. Then, running buffer without compounds was injected for 10 min at the same flow rate. Data were corrected using a blank sensor chip as a control. The dissociation constant (Kd) was calculated from the analyte concentration dependence of the sensorgram response.
RESULTS
In silico screening.
We first picked up the top 590 compounds from the list, all of which showed a docked energy level lower than −11.45 kcal/mol (Table 1). The differences in the docked energies of these compounds were within 1.87 kcal/mol. From these 590 compounds, we selected 205 commercially available compounds and purchased them from Maybridge, Asinex, or Enamine.
TABLE 1.
Summary of the ligand-screening test in silico and ex vivoa
| Method | Parameter | Number | Hit rate (%)b |
|---|---|---|---|
| Compounds tested in silico | Compounds with a binding score of less than −11.45 kcal/mol | 590 | |
| High-ranking and available compounds (out of 590) | 205 | ||
| Compounds tested ex vivo | Cytotoxic compound | 1 | |
| Not effective (relative PrPSc level of >70%) | 180 | ||
| Effective (relative PrPSc level of <70%) | 24 | 11.7 | |
| More effective (relative PrPSc level of <50%) | 4 | 2.0 |
The numbers of compounds (total n = ∼600,000) in in silico and ex vivo tests are summarized.
The hit rate was calculated from 205 compounds tested ex vivo.
Ex vivo screening.
To evaluate the effect of the selected compounds on the PrP conversion process, we conducted ex vivo screening. We used GT+FK cells (18) to test the above-listed 205 compounds. From among them, 24 compounds that significantly inhibited production of PrPSc in the GT+FK cells at a concentration of 10 μM were identified. The other compounds were almost ineffective, and one compound was highly toxic to the cells at 10 μM. Of the 24 effective noncytotoxic compounds, four compounds reduced PrPSc levels to less than 50% of that of the control (Fig. 1A), and the effects of these compounds were dose dependent (data not shown). Previously identified inhibitors, such as quinacrine (6, 13), also inhibited PrPSc production in the ex vivo assay using GT+FK cells (data not shown). On the other hand, some other compounds increased the levels of PrPSc. After GT+FK cells were treated with these nine compounds, levels of PrPSc were more than 150% higher than that of the control (Fig. 1B).
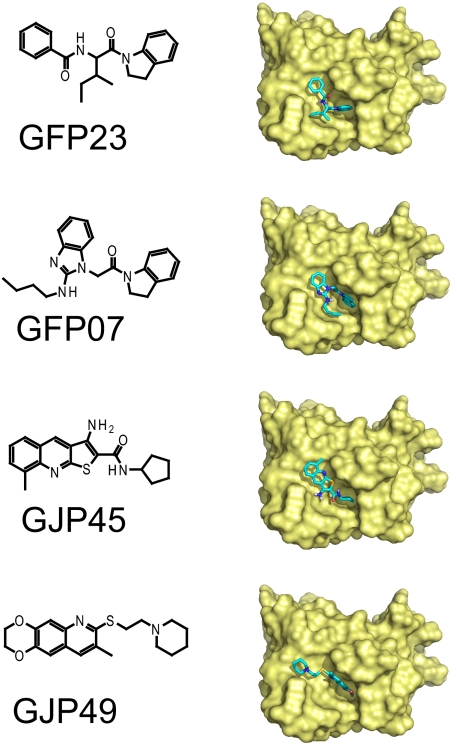
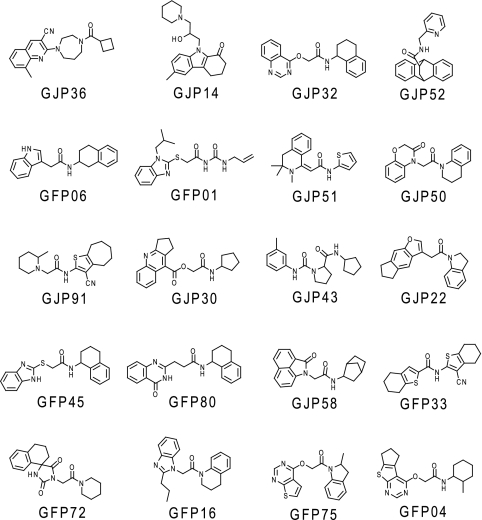
Figure 2 shows four of the hit compounds (effective compounds [relative PrPSc levels of <70%]) and their best docking modes, as calculated with AutoDock. Common chemical moieties were not obvious among these compounds. In the best docking modes, these compounds were commonly located in the major pocket or the putative GN8 binding site. For these compounds, other docking modes were also obtained, with docked energies similar to the lowest ones among 10 docking runs. In addition to the four highly effective compounds listed in Fig. 2, we also obtained a variety of moderately effective compounds (Fig. 3). Any common characteristics among these effective compounds were not observed at this stage.
FIG. 2.
Chemical structures of effective compounds against PrPSc accumulation. The structures of compounds and their predicted modes of docking with PrPC are shown for the top four compounds: GFP23, GFP07, GJP45, and GJP49.
FIG. 3.
Chemical structures of moderately effective compounds against PrPSc accumulation. Compounds with a rank order from 5th to 24th are shown, along with the relative level of PrPSc, from 50% to 70%, in the antiprion ex vivo assay.
Determination of the Kd between representative compounds and PrPC, using the SPR technique.
To evaluate the direct binding affinity of the compounds for PrPC, we used SPR measurements. For the top 13 effective compounds and the 3 most ineffective compounds from ex vivo experiments, we determined Kd values by using SPR sensorgram responses (Fig. 4 and Table 2).
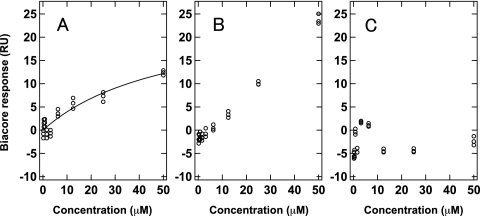
FIG. 4.
Examples of the concentration dependence of the Biacore response for (A) GFP23 as a single and specific binding compound, (B) GJP36 as a multiple or nonspecific binding compound, and (C) GJP82 as a nonbinding compound to mouse PrPC.
TABLE 2.
Results of in silico and ex vivo screenings and Kd valuesa
| Compound | Docked energy (kcal/mol) | Mean level of PrPSc ± SD (%) | Kd (μM) ± SD for PrPC |
|---|---|---|---|
| GFP23 | −11.68 | 36.78 ± 0.63 | 51.2 ± 12.2 |
| GFP07 | −11.78 | 37.33 ± 4.23 | 20.0 ± 17.6 |
| GJP45 | −11.46 | 41.85 ± 18.82 | 86.1 ± 4.6 |
| GJP49 | −11.48 | 47.40 ± 19.89 | 50.8 ± 11.1 |
| GJP36 | −11.93 | 50.62 ± 21.26 | NDb |
| GJP14 | −12.38 | 51.32 ± 7.76 | NDb |
| GJP32 | −12.46 | 53.05 ± 10.72 | NDb |
| GJP52 | −12.00 | 54.71 ± 9.21 | NDc |
| GFP06 | −11.78 | 55.33 ± 12.35 | 48.6 ± 20.3 |
| GFP01 | −11.82 | 56.73 ± 13.15 | No binding |
| GJP51 | −11.56 | 57.44 ± 1.62 | NDd |
| GJP50 | −11.67 | 57.55 ± 4.88 | NDb |
| GJP91 | −11.86 | 58.78 ± 5.89 | 21.9 ± 16.4 |
| GJP30 | −12.29 | 62.89 ± 3.93 | —e |
| GJP43 | −11.47 | 63.16 ± 7.91 | — |
| GJP22 | −12.16 | 63.70 ± 5.23 | — |
| GFP45 | −11.60 | 64.86 ± 16.55 | — |
| GFP80 | −11.51 | 65.01 ± 35.45 | — |
| GJP58 | −11.71 | 65.03 ± 4.59 | — |
| GFP33 | −11.64 | 66.94 ± 0.11 | — |
| GFP72 | −11.54 | 67.11 ± 33.65 | — |
| GFP16 | −11.74 | 67.24 ± 1.24 | — |
| GFP75 | −11.52 | 67.43 ± 23.22 | — |
| GFP04 | −11.81 | 68.77 ± 11.7 | — |
| GFP66 | −11.54 | 93.68 ± 13.69 | No binding |
| GJP82 | −11.83 | 94.61 ± 8.17 | No binding |
| GJP61 | −11.50 | 95.70 ± 0.74 | No binding |
The 24 effective and 3 noneffective compounds in ex vivo tests are shown. Values are the means ± standard deviations (SD) of two to four independent experiments.
Kd was not determined (ND) because of an unlimited increase in Biacore responses.
Kd was not determined because of the low solubility of the compound.
Kd was not determined because of the slow binding kinetics.
—, experiments were not performed.
SPR sensorgram response curves were roughly categorized into three types of curves. The first type was typically observed for compounds that exhibited single and specific binding to PrPC (Fig. 4A). Fitting the compound concentration dependence of the sensorgram response enables the estimation of the Kd. The Kd values were estimated to be about 10 to 90 μM for the top four compounds (GFP23, GFP07, GJP45, and GJP49), which were comparable to the antiprion compound GN8. In the second type of sensorgram response curve, a saturation of response was not observed for the compounds (GJP36, GJP14, GJP32, and GJP50), as shown in Fig. 4B. These compounds bound to PrPC, but it was not possible to calculate the Kd by using a single binding model, which strongly suggested multiple or nonspecific binding with PrPC. The third type of response curve was observed for compounds with no binding (GFP01, GFP66, GJP82, and GJP61) to PrPC, at least at the concentrations displayed (Fig. 4C). Table 2 indicates that most compounds with high antiprion activity can bind directly to PrPC and that the ineffective compounds hardly bind to PrPC.
Comparison between antiprion activity and binding affinity.
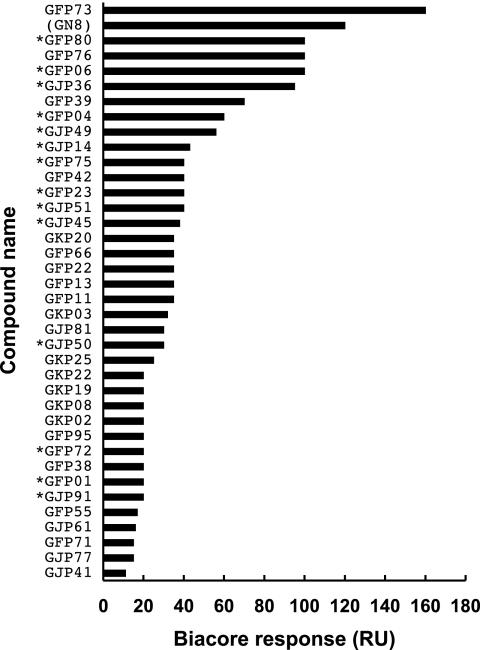
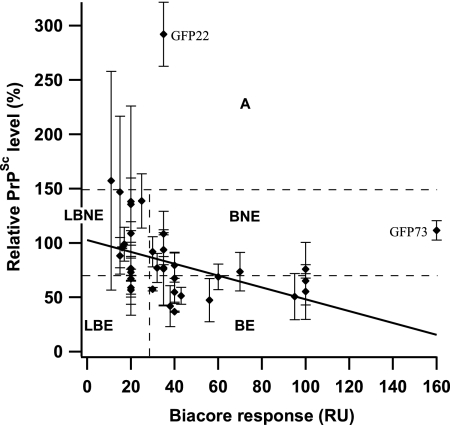
PrPSc levels in the presence of effective or ineffective compounds in ex vivo screening were compared with docked energies obtained by in silico screening or with Kd values derived from SPR responses (Table 2). Docked energies of the listed compounds were similar, from −12.46 to −11.46 kcal/mol, and there were no correlations between PrPSc levels and docked energies. No correlation was seen as well for the 205 compounds selected (data not shown). Meanwhile, a variety of binding patterns was presented in the SPR experiments, but the correlation between PrPSc levels and Kd values was ambiguous. Consequently, to compare the antiprion activities obtained from the ex vivo screening with the binding affinities between compounds and PrPC, we measured SPR responses for 37 compounds at a fixed and relatively high concentration of 100 μM. This analysis included both the effective and the ineffective compounds (Fig. 5). Most effective compounds showed a high Biacore response (>30 response units [RU]), and it was suggested that a compound with antiprion activity might have a high capacity for binding to PrPC. Thus, we tried to confirm the correlation between relative PrPSc levels in the ex vivo screening and Biacore responses in in vitro screening. As shown in Fig. 6, we built a scatter plot from relative PrPSc levels and Biacore responses of these compounds. Excluding two compounds with an extremely high PrPSc level (GFP22) or Biacore response (GFP73), a moderate correlation between PrPSc levels and Biacore responses was observed (r = 0.45) for 35 compounds.
FIG. 5.
In vitro screening using the Biacore system. The affinities of compounds to the mouse PrPC immobilized on a sensor chip were evaluated using Biacore response units (RU). The concentration of each compound was 100 μM. *, Compounds judged to be effective based on comparatively high RU values. GN8 was used as a positive control.
FIG. 6.
The correlation between the relative PrPSc levels and Biacore responses (RU). A scatter plot was built from the relative PrPSc levels and Biacore responses of 37 compounds, including effective and noneffective compounds. Relative PrPSc levels are the means of two to four independent experiments. Error bars represent standard deviations. Based on binding affinities and antiprion activities, these compounds were roughly classified into five categories, i.e., BE, LBE, BNE, LBNE, and A. Compounds GFP22 and GFP73 exhibited high values for levels of PrPSc and Biacore responses, respectively.
Also, we were able to classify the compounds roughly into five categories: (i) binding and effective (BE), (ii) low binding and effective (LBE), (iii) binding and not effective (BNE), (iv) low binding and not effective (LBNE), and (v) acceleration (A). In category BE there were 11 compounds (GJP14, GJP36, GJP45, GJP49, GJP51, GJP52, GFP04, GFP06, GFP23, GFP75, and GFP80) (Fig. 2 and 3); in LBE there were 3 (GJP91, GFP01, and GFP72) (Fig. 3); in BNE there were 10; in LBNE there were 11; and in A there were 2. Chemical structures of compounds belonging to categories BNE through A are listed in Fig. S1 through S3 in the supplemental material.
DISCUSSION
Development of the in silico screening techniques has greatly expedited our search for new, potentially therapeutic inhibitors of PrPSc accumulation (15). Currently, 100,000 compounds per day can be tested via high-throughput screening technology (24), but it is almost impossible to test the same number of compounds in the search for a leading compound against prion diseases because of the complexity of the assay. Although Kocisko et al. have developed a high-throughput screening assay for PrPSc inhibitors, they screened only 2,000 compounds (12). Clearly, one cannot evaluate all compounds in a database such as the Available Chemicals Directory (MDL Information Systems), which includes over 107 compounds.
To date, various in silico screening computer programs have been widely applied for the discovery of small compounds such as enzyme inhibitors, in which well-defined active sites usually serve as the target regions (14). Since PrP has not been proven to have catalytic activity, we used the slow-dynamic information obtained by using Carr-Purcell-Meiboom-Gill relaxation dispersion experiments to assign the appropriate target region, termed “hot spots.” Each compound in the database was evaluated through an analysis of the potential binding sites, and finally, a novel antiprion compound, termed GN8, was discovered (15). GN8 efficiently inhibited the production of PrPSc in ex vivo screening and stabilized the PrPC conformation through specific binding with the major pocket. Thus, we demonstrated that GN8 works as a chemical chaperone. Meanwhile, in vivo experiments showed that GN8 profoundly prolonged the survival time of prion-infected animals when administered peripherally. In that study, we identified merely one effective compound, but that result is likely explained by our strict selection process (15).
In contrast, here, a variety of compounds with a large structural diversity were identified as potent inhibitors, and our study demonstrated that they have therapeutic efficacy against PrPs at a rate of 2%, as shown in Table 1. Based on these data, we expect that the in silico screening would be valuable as the initial screening for potential antiprion drugs from huge compound libraries. Any compounds that simply targeted the major binding pocket of PrPC would be helpful in characterizing the distribution of the active compound structures. Thus, applying in silico screening for use with TSE-infected cell cultures should provide us with a hint regarding the nature of the pathogenic conversion reaction, as well as the way it is regulated by small compounds.
The hit rate of this in silico screening was about 12% (Table 1). It seems that this hit rate is relatively high compared with 1.2% and 5.4% for in silico screens targeting the formylpeptide receptor (7) and AmpC β-lactamase (22), respectively. However, the targets in those screens are different from the PrP. There might be several reasons for the higher hit rate of the in silico screening. One possibility is that the pathogenic conversion from PrPC to PrPSc is a folding phenomenon. The three-dimensional structure of PrPSc is considerably different from that of PrPC, i.e., PrPC is largely α-helical, whereas PrPSc is β-sheet dominant, and the conversion reaction is postulated to be mediated by an active intermediate between PrPC and PrPSc, PrP* (5). Therefore, any ligands interacting with residues that are involved in this process can modulate the reaction. In fact, compared to the active site of an enzyme, hot spots relevant to the pathogenic conversion of the PrP are somewhat broadly distributed (15) and multiple binding sites are possibly involved. Nonspecific binding with the PrP, most likely a hydrophobic interaction, may be somewhat effective in inhibiting the conversion reaction, because this type of nonspecifically bound substance could reduce the fluctuation of protein, thereby decelerating the pathogenic conversion reaction. This phenomenon is represented by the relatively high hit rate and the diverse chemical structures of hit compounds.
The calculated docked-energy differences between compounds listed in Table 2 were quite small and not statistically significant. This is because the evaluation function utilized in the software was not necessarily designed for the interaction between PrPC and small compounds. In view of the pathogenic conversion reaction, it may be necessary to further evaluate the change in the entropy of solvation associated with binding and the change in the chain entropy of the PrP. Rigorous evaluation of these effects can help provide information for the conversion that occurs from PrPC to PrPSc and how the small compounds interact with each substate of the prion.
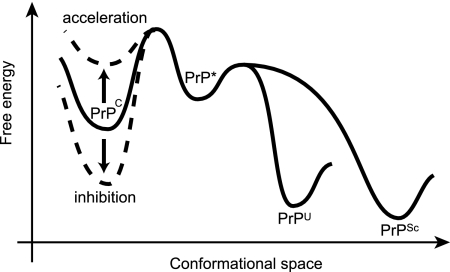
Intriguingly, some compounds could accelerate the conversion, as shown in Fig. 1B and Fig. S3 in the supplemental material. These compounds include GJP41 and GFP22. There are many likely explanations for this result, but one possibility is that the free energy of PrPC could be perturbed by its interaction with small compounds. Some compounds would stabilize the PrPC conformation and inhibit the pathogenic conversion as a chemical chaperone (17), but other compounds could destabilize the protein and accelerate the conversion by decreasing the barrier between PrPC and PrP*, as shown in Fig. 7. However, another possibility is that these compounds could accelerate the conversion process by interacting with molecules other than PrPC, like sodium dodecyl sulfate or Triton X-100, in protein misfolding cyclic amplification reactions (1). However, these compounds do not have the characteristics of a detergent. The common scaffolds of these compounds are not obvious.
FIG. 7.
An illustration of Gibbs free energy as a function of the conformational space of the PrP explains the inhibition (down arrow, dashed line) and acceleration (up arrow, dashed line) mechanism of small compounds bound to PrPC. Compounds in category BE could stabilize the PrPC conformation and reduce the populations of PrP* (an active intermediate), PrP in the unfolded state (PrPU), and PrPSc. On the other hand, compounds in category A could destabilize the PrPC conformation and accelerate the conversion by decreasing the barrier between PrPC and PrP*.
Measurements of the Kd were complicated, as shown in Table 2 and Fig. 4. In SPR measurements, nonspecific binding should be carefully removed. However, for some compounds, such as GJP36, GJP14, GJP32 and GJP50, nonspecificity was rather overwhelming, and it is technically impossible to remove it (Table 2 and Fig. 4B). On the other hand, this nonspecific binding with PrPC is considered a significant contribution to the inhibitory effect, especially for GJP36. Such a nonspecific interaction could also affect the free energy of PrPC shown in Fig. 7. However, this kind of nonspecificity must be avoided because many side effects could result from the nonspecific binding with many physiologically important proteins other than PrPC, which is unfavorable from a pharmacological point of view.
As shown in Fig. 6, we were able to classify the compounds roughly into five categories. Compounds in category BE, including GJP36, GJP14, GJP32, and GJP50, are considered candidates for the chemical chaperones that are capable of stabilizing the PrPC conformation. On the other hand, the existence of compounds belonging to the LBE group indicates the involvement of modulation factors other than direct binding with PrPC. These modulation factors may include factor X (25), detergent-resistant microdomains on the cell membrane (16), the lysosome in which chaperone proteins are degraded, and so on (6). The existence of compounds belonging to category BNE suggests that they bind with regions of PrPC that are less important for the pathogenic conversion reaction. In our previous work, we putatively identified the hot spots in PrPC for pathogenic conversion (15). For example, the compounds that bind to the region far from these spots may be less effective at altering the pathogenic conversion reaction. The compounds belonging to category A would destabilize the PrPC conformation and facilitate the pathogenic conversion reaction (Fig. 7). These small compounds would be potentially hazardous.
Most effective compounds bound to PrPC, whereas the noneffective compounds did not (Table 2), and there was a moderate correlation between binding affinities and antiprion activities (Fig. 6). Therefore, identification of chemical chaperones that bind to the PrPC structure and stabilize it is one efficient strategy for antiprion drug discovery. Additionally, in vitro screening by binding affinity might be useful for antiprion drug discovery. Such compounds should have an advantage over the strain-independent antiprion activity because PrPC, which is the precursor of PrPSc, is identical in all prion strains.
Caughey et al. suggested that well-known anti-TSE compounds such as pentosan polysulfate, certain cyclic tetrapyrroles, and phosphorothioated oligonucleotides attached to the N-terminal region of the PrP and induced the clustering of PrPC to prevent conversion to PrPSc (3). Our results indicated that the inhibitory mechanism of our compounds may be quite distinct from those of known anti-TSE compounds. Our compounds may work as chemical chaperones to stabilize the PrPC conformation through binding with the C-terminal region, because our results are for the binding affinity between the compounds and the PrP lacking the N-terminal region.
Although the common chemical structures of the compounds in each category were not found, compounds belonging to category BE are basically considered to be lead compounds, and thus, each chemical structure could be optimized independently for antiprion activity by referring to the relative levels of PrPSc in the ex vivo screening. Data acquired from this research are also useful for improvement of the docking program and the screening system.
In conclusion, 24 new inhibitors were identified in our screening of 600,000 compounds in the ZINC database. These potent inhibitors have chemical structures that may be easily optimized, and those derivatives might become candidates for a future therapeutic drug. The series of screens has brought a spectrum of compounds that can modulate the pathogenic conversion reaction. These results suggest that some compounds inhibit PrPSc formation through direct interactions with PrPC, whereas other inhibitors may work indirectly. The fact that a number of new inhibitors are classified into the appropriate categories according to their affinities for PrPC distinguishes them from many previously identified PrPSc inhibitors and makes them attractive as potential anti-TSE therapeutic agents.
Supplementary Material
Acknowledgments
We thank Noriyuki Nishida for setting up the ex vivo assay and for valuable technical advice. We thank Tomomi Saeki for technical assistance. We thank Kurt Wüthrich and Simone Hornemann for kindly providing the expression plasmid of mouse PrP(121-231). We thank Kei-ich Yamaguchi, Yosuke Hayano, Tomoharu Matsumoto, and Junko Matsubara for assistance with protein sample preparation.
This study was supported by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation and partly by the Molecular Imaging Project of Japan Society and Technology Agency.
Footnotes
Published ahead of print on 17 November 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Atarashi, R., R. A. Moore, V. L. Sim, A. G. Hughson, D. W. Dorward, H. A. Onwubiko, S. A. Priola, and B. Caughey. 2007. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat. Methods 4:645-650. [DOI] [PubMed] [Google Scholar]
- 2.Barret, A., F. Tagliavini, G. Forloni, C. Bate, M. Salmona, L. Colombo, A. De Luigi, L. Limido, S. Suardi, G. Rossi, F. Auvre, K. T. Adjou, N. Sales, A. Williams, C. Lasmezas, and J. P. Deslys. 2003. Evaluation of quinacrine treatment for prion diseases. J. Virol. 77:8462-8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caughey, B., W. S. Caughey, D. A. Kocisko, K. S. Lee, J. R. Silveira, and J. D. Morrey. 2006. Prions and transmissible spongiform encephalopathy (TSE) chemotherapeutics: a common mechanism for anti-TSE compounds? Acc. Chem. Res. 39:646-653. [DOI] [PubMed] [Google Scholar]
- 4.Chesebro, B. 1999. Prion protein and the transmissible spongiform encephalopathy diseases. Neuron 24:503-506. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, F. E., K. M. Pan, Z. Huang, M. Baldwin, R. J. Fletterick, and S. B. Prusiner. 1994. Structural clues to prion replication. Science 264:530-531. [DOI] [PubMed] [Google Scholar]
- 6.Doh-Ura, K., T. Iwaki, and B. Caughey. 2000. Lysosomotropic agents and cysteine protease inhibitors inhibit scrapie-associated prion protein accumulation. J. Virol. 74:4894-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards, B. S., C. Bologa, S. M. Young, K. V. Balakin, E. R. Prossnitz, N. P. Savchuck, L. A. Sklar, and T. I. Oprea. 2005. Integration of virtual screening with high-throughput flow cytometry to identify novel small molecule formylpeptide receptor antagonists. Mol. Pharmacol. 68:1301-1310. [DOI] [PubMed] [Google Scholar]
- 8.Heal, W., M. J. Thompson, R. Mutter, H. Cope, J. C. Louth, and B. Chen. 2007. Library synthesis and screening: 2,4-diphenylthiazoles and 2,4-diphenyloxazoles as potential novel prion disease therapeutics. J. Med. Chem. 50:1347-1353. [DOI] [PubMed] [Google Scholar]
- 9.Hornemann, S., C. Korth, B. Oesch, R. Riek, G. Wider, K. Wuthrich, and R. Glockshuber. 1997. Recombinant full-length murine prion protein, mPrP(23-231): purification and spectroscopic characterization. FEBS Lett. 413:277-281. [DOI] [PubMed] [Google Scholar]
- 10.Irwin, J. J., and B. K. Shoichet. 2005. ZINC-a free database of commercially available compounds for virtual screening. J. Chem. Infect. Model. 45:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaneko, K., L. Zulianello, M. Scott, C. M. Cooper, A. C. Wallace, T. L. James, F. E. Cohen, and S. B. Prusiner. 1997. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc. Natl. Acad. Sci. USA 94:10069-10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kocisko, D. A., G. S. Baron, R. Rubenstein, J. Chen, S. Kuizon, and B. Caughey. 2003. New inhibitors of scrapie-associated prion protein formation in a library of 2,000 drugs and natural products. J. Virol. 77:10288-10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korth, C., B. C. May, F. E. Cohen, and S. B. Prusiner. 2001. Acridine and phenothiazine derivatives as pharmacotherapeutics for prion disease. Proc. Natl. Acad. Sci. USA 98:9836-9841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuntz, I. D. 1992. Structure-based strategies for drug design and discovery. Science 257:1078-1082. [DOI] [PubMed] [Google Scholar]
- 15.Kuwata, K., N. Nishida, T. Matsumoto, Y. O. Kamatari, J. Hosokawa-Muto, K. Kodama, H. K. Nakamura, K. Kimura, M. Kawasaki, Y. Takakura, S. Shirabe, J. Takata, Y. Kataoka, and S. Katamine. 2007. Hot spots in prion protein for pathogenic conversion. Proc. Natl. Acad. Sci. USA 104:11921-11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangé, A., N. Nishida, O. Milhavet, H. E. McMahon, D. Casanova, and S. Lehmann. 2000. Amphotericin B inhibits the generation of the scrapie isoform of the prion protein in infected cultures. J. Virol. 74:3135-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuda, J., O. Suzuki, A. Oshima, Y. Yamamoto, A. Noguchi, K. Takimoto, M. Itoh, Y. Matsuzaki, Y. Yasuda, S. Ogawa, Y. Sakata, E. Nanba, K. Higaki, Y. Ogawa, L. Tominaga, K. Ohno, H. Iwasaki, H. Watanabe, R. O. Brady, and Y. Suzuki. 2003. Chemical chaperone therapy for brain pathology in G(M1)-gangliosidosis. Proc. Natl. Acad. Sci. USA 100:15912-15917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milhavet, O., H. E. McMahon, W. Rachidi, N. Nishida, S. Katamine, A. Mange, M. Arlotto, D. Casanova, J. Riondel, A. Favier, and S. Lehmann. 2000. Prion infection impairs the cellular response to oxidative stress. Proc. Natl. Acad. Sci. USA 97:13937-13942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew, and A. J. Olson. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19:1639-1662. [Google Scholar]
- 20.Nishida, N., D. A. Harris, D. Vilette, H. Laude, Y. Frobert, J. Grassi, D. Casanova, O. Milhavet, and S. Lehmann. 2000. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J. Virol. 74:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perola, E., W. P. Walters, and P. S. Charifson. 2004. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins 56:235-249. [DOI] [PubMed] [Google Scholar]
- 22.Powers, R. A., F. Morandi, and B. K. Shoichet. 2002. Structure-based discovery of a novel, noncovalent inhibitor of AmpC beta-lactamase. Structure 10:1013-1023. [DOI] [PubMed] [Google Scholar]
- 23.Prusiner, S. B. 1998. Prions. Proc. Natl. Acad. Sci. USA 95:13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, A. 2002. Screening for drug discovery: the leading question. Nature 418:453-459. [DOI] [PubMed] [Google Scholar]
- 25.Telling, G. C., M. Scott, J. Mastrianni, R. Gabizon, M. Torchia, F. E. Cohen, S. J. DeArmond, and S. B. Prusiner. 1995. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell 83:79-90. [DOI] [PubMed] [Google Scholar]
- 26.Touil, F., S. Pratt, R. Mutter, and B. Chen. 2006. Screening a library of potential prion therapeutics against cellular prion proteins and insights into their mode of biological activities by surface plasmon resonance. J. Pharm. Biomed. Anal. 40:822-832. [DOI] [PubMed] [Google Scholar]
- 27.Vogtherr, M., S. Grimme, B. Elshorst, D. M. Jacobs, K. Fiebig, C. Griesinger, and R. Zahn. 2003. Antimalarial drug quinacrine binds to C-terminal helix of cellular prion protein. J. Med. Chem. 46:3563-3564. [DOI] [PubMed] [Google Scholar]
- 28.Warren, G. L., C. W. Andrews, A. M. Capelli, B. Clarke, J. LaLonde, M. H. Lambert, M. Lindvall, N. Nevins, S. F. Semus, S. Senger, G. Tedesco, I. D. Wall, J. M. Woolven, C. E. Peishoff, and M. S. Head. 2006. A critical assessment of docking programs and scoring functions. J. Med. Chem. 49:5912-5931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.