Abstract
Objective
To compare the global effects of oxidized LDL (oxLDL) and oxLDL-containing immune complexes (oxLDL-IC) on gene expression in human monocytic cells and to identify differentially expressed genes involved with inflammation and survival.
Methods and Results
U937 cells were treated with oxLDL-IC, oxLDL, Keyhole limpet hemocyanin immune complexes (KLH-IC), or vehicle for 4 h. Transcriptome profiling was performed using DNA microarrays. oxLDL-IC uniquely affected the expression of genes involved with pro-survival (RAD54B, RUFY3, SNRPB2, and ZBTB24). oxLDL-IC also regulated many genes in a manner similar to KLH-IC. Functional categorization of these genes revealed that 39% are involved with stress responses, including the unfolded protein response which impacts cell survival, 19% with regulation of transcription, 10% with endocytosis and intracellular transport of protein and lipid, and 16% with inflammatory responses including regulation of I-κB/NF-κB cascade and cytokine activity. One gene in particular, HSP70 6, greatly up-regulated by ox-LDL-IC, was found to be required for the process by which oxLDL-IC augments IL1-β secretion. The study also revealed genes uniquely up-regulated by oxLDL including genes involved with growth inhibition (OKL38, NEK3, and FTH1), oxidoreductase activity (SPXN1 and HMOX1), and transport of amino acids and fatty acids (SLC7A11 and ADFP).
Conclusions
These findings highlight early transcriptional responses elicited by oxLDL-IC that may underlie its cytoprotective and pro-inflammatory effects. Cross-linking of Fcγ receptors appears to be the trigger for most of the transcriptional responses to oxLDL-IC. The findings further strengthen the hypothesis that oxLDL and oxLDL-IC elicit disparate inflammatory responses and play distinct roles in the process of atherosclerosis.
Introduction
Lipid-laden macrophages (foam cells) are the hallmark of the atherosclerotic process and main contributors to progression of cardiovascular disease. Therefore, understanding the processes by which foam cells are formed has been a major objective of atherosclerosis research. It is well established that oxidized LDL (oxLDL) particles are taken up by macrophages leading to accumulation of cholesteryl esters (CE) (1,2). On the other hand, oxLDL is immunogenic and elicits the production of antibodies, predominantly of the proinflammatory IgG1 and IgG3 isotypes (3,4). These antibodies form circulating immune complexes containing oxLDL (oxLDL-IC), and those immune complexes have pro-inflammatory properties (5–7) and are considerably more efficient than oxLDL in the induction of foam cell formation (8–10). While the uptake of both oxLDL and oxLDL-IC produce cells morphologically defined as foam cells, there is evidence for distinct molecular differences. In particular, foam cells formed through oxLDL exposure have reduced survival as compared to foam cells formed through oxLDL-IC exposure (11,12). The basis for this difference in not known. Macrophages exposed to oxLDL-IC have higher levels of CE and display an increased release of cytokines as compared to cells exposed to oxLDL (5,7). These findings have led to the hypothesis that although foam cells generated by exposure to oxLDL and oxLDL-IC appear morphologically similar, they differ in the profile of genes that regulate survival and inflammatory response. To test this hypothesis, we have used DNA microarray analysis and real time quantitative PCR (Q-PCR) to investigate the effects of oxLDL and oxLDL-IC on the transcriptome in human U937 monocytic cells. The results of these studies highlight specific transcriptional responses elicited by oxLDL-IC that may underlie its ability to promote prolonged activation of foam cells.
Materials and Methods
Cells
The human monocytic cell line U937 was obtained from the American Type Culture Collection (ATCC CRL-1593.2) (13). Cells were maintained in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 50μg/ml streptomycin at 37 C, 5% CO2. Cells were seeded at 2×106 cells/2 ml in 6-well plates, and incubated in serum-free medium in the presence of IFN-γ (200 ng/ml) for 18 h prior to addition of experimental treatments. The rationale for IFN-γ treatment is that it is the major cytokine released by activated T cells in macrophage-containing atherosclerotic lesions (14). Thus, the response of IFN-γ primed macrophages to modified lipoproteins is likely to reproduce the conditions at the atheromatous plaque, where the same regions where macrophages and foam cells predominate are heavily infiltrated with CD4+ T cells releasing primarily IFN-γ (15).
Lipoprotein Isolation and Oxidation
LDL (d = 1.019 to 1.063 g/ml) was isolated from plasma of normal volunteers and oxidatively modified using Cu2+ as described previously (16,17). Under the conditions previously reported for copper oxidation of LDL (18) our oxLDL preparations have the following degree of modification: 4 to 7 mmol/mol lysine of malondialdehyde (MDA) (0.4 to 0.7% modification of lysine residues), 0.8 mmol/mol lysine of Nε-(carboxymethyl) lysine (CML) (0.08% modification of lysine residues), and 0.25 mmol/ml lysine of Nε(carboxyethyl)lysine (CEL) (0.025% modification of lysine residues). This degree of LDL modification is associated with formation of auto-antibodies in humans (17,18) and is optimally recognized by the antibody used to form oxLDL-IC (see next section). The endotoxin level in oxLDL preparations was measured using an endotoxin assay kit (Etoxate, Sigma), and found to be below the lower limit of detection (0.015 U/ml).
Preparation of Insoluble Immune Complexes
Soluble immune complexes stimulate macrophages only if carried by red blood cells or immobilized. Immobilization of oxLDL-IC by attachment to matrix proteins is likely to occur in vivo. We used insoluble oxLDL-IC as a form of immobilized immune complexes to stimulate macrophages. Insoluble oxLDL-IC were prepared with human oxLDL and human anti oxLDL antibodies. Anti oxLDL IgG was isolated from whole human serum using a combination of Protein G-Sepharose 4 Fast Flow chromatography (Amersham-Pharmacia Biotech) to isolate IgG, and affinity chromatography in Sepharose-linked human oxLDL to isolate oxLDL antibodies of the IgG isotype, as described previously (3,19). The amount of oxLDL combined with anti-oxLDL IgG to prepare insoluble oxLDL-IC was determined empirically by performing a precipitin curve, constructed by incubating aliquots of the IgG with varying amounts of oxLDL and determining the amount of oxLDL that induced the highest degree of insoluble oxLDL-IC formation as determined by nephelometry. The empirical measurements indicated that a ratio of 150 μg oxLDL protein/500 μg IgG produced peak precipitation. Human Keyhole limpet hemocyanin (KLH) immune complexes (KLH-IC) were prepared with the IgG fraction from anti-KLH human serum isolated using Protein G-Sepharose 4 Fast Flow chromatography (5). The optimal proportion of KLH (endotoxin-free, Calbiochem) to anti KLH IgG for preparation of insoluble human KLH-IC was 200μg/250μg (w/w). All immune complexes were prepared under sterile conditions, washed and re-suspended in PBS.
Measurement of Interleukin 1 beta (IL1-β)
IL1-β levels in conditioned medium were analyzed using two types of ELISA, SearchLight Human Cytokine Array, a multiplex sandwich ELISA (Pierce) and an IL1-β enzyme immunoassay kit from R&D Systems (DLB50; Minneapolis, MN).
Total RNA Preparation and DNA Microarray Hybridization
IFN-γ-treated U937 cells were exposed to oxLDL-IC, KLH-IC, oxLDL (150 μg/ml), or PBS vehicle for 4 h. KLH-IC was used as a control immune complex because KLH has a molecular weight comparable to LDL and because it can engage Fcγ receptors like oxLDL-IC but does not contain lipoproteins. Cells were pelleted by centrifugation at 400×g for 5 min. Total RNA was isolated using Trizol extraction (Invitrogen) and purified using RNeasy Mini kit (Qiagen). RNA quality was assured by using Agilent Bioanalyzer and RNA 6000 nano chip. Total RNA (8 μg) was converted into double-stranded cDNA with a T7-(dT) 24 primer (Genset) and a cDNA synthesis kit (Custom SuperScript; Invitrogen). Biotin-labeled cRNA was synthesized from cDNA by in vitro transcription (Enzo BioArray HighYield RNA Transcript Labeling Kit; Enzo Life Sciences). After purification (RNeasy kit; Qiagen), labeled cRNA was fragmented as recommended by Affymetrix. Hybridization of cRNA samples to Affymetrix HG-U133 plus2 GeneChips, post-hybridization washing, fluorescence staining and scanning were performed at the MUSC DNA Microarray and Bioinformatics Facility. DNA microarray data (raw and normalized) generated by this project are available online through the MUSC DNA Microarray Database (http://proteogenomics.musc.edu/ma/musc_madb.php?page=home&act=manage) and the NCBI Geo (http://www.ncbi.nlm.nih.gov/geo/).
Gene Array Analysis
Hybridization intensity data were normalized using the GCRMA algorithm (20). Identification of differentially expressed genes and hierarchical clustering were performed using dChip software (21). Genes differentially affected by the separate treatments were identified using ANOVA (p<0.001); hierarchical clustering was performed on the resulting genes using the distance metric of 1-Correlation and the Average linkage method. Genes presented here as uniquely affected by either oxLDL-IC or oxLDL were obtained from the resulting heat map. Genes regulated similarly by oxLDL-IC and KLH-IC were identified using the following criteria: 1) FC>2 and p<0.05 (Student’s unpaired t-test) for oxLDL-IC vs PBS and for oxLDL-IC vs oxLDL treatments; 2) FC>2 and p<0.05 (Student’s unpaired t-test) for KLH vs PBS and for KLH vs oxLDL treatments. False discovery rate (FDR) approximated 0.0% as estimated by 50 iterations of randomized sample comparisons. Genes regulated similarly by oxLDL-IC and oxLDL were examined using the same criteria used to identify genes regulated similarly by oxLDL-IC and KLH-IC.
Real Time Quantitative PCR (Q-PCR)
PCR primers were designed using the Beacon Designer 5 software (Primer Biosoft Int., Palo Alto, CA). The forward and reverse primer sequences for the genes examined are shown in Table 1. PCR primers were synthesized by Integrated DNA Technologies, Inc. (Coraville, IA). IFN-γ-treated U937 cells were exposed to oxLDL-IC, oxLDL (150 μg/ml), or the PBS vehicle for 4 h. The RNAeasy mini kit was used to isolate mRNA (Qiagen), and complementary DNA (cDNA) was synthesized using iScript™ cDNA synthesis kit (Bio-Rad). Q-PCR was performed using the iCycler™ real-time detection system (Bio-Rad) with a two-step method using iQ™ SYBR Green Supermix (Bio-Rad). Amplification of glyceraldehydes-3-phosphate dehydrogenase (GAPDH) was performed to standardize the amount of sample RNA. Quantification was performed using the cycle threshold of receptor cDNA relative to that of GAPDH cDNA in the same sample.
Table 1.
PCR primers
| Gene | Accession No. | Symbol | Sequences of primers (5′ to 3′) |
|---|---|---|---|
| Sphingosine kinase 1 | NM_021972 | SPHK1 | Forward: CTGGCAGCTTCCTTGAACCAT
Reverse: TGTGCAGAGACAGCAGGTTCA |
| Heat shock 70kDa protein 6 | NM_002155 | HSPA6 | Forward: CCCTAAGGCTTTCCTCTTGC
Reverse: CATGAAGCCGAGCAGTACAA |
| CD36 | NM_000072 | CD36 | Forward: AAGCAAAGAGGTCCTTATACG
Reverse: TCTGTTCCAACTGATAGTGAAG |
| Low density lipoprotein receptor | NM_000527 | LDLR | Forward: GCAAGGACAAATCTGACGAG
Reverse: TAACGCAGCCAACTTCATC |
| GAPDH | NM_002046 | GAPDH | Forward: CTGAGTACGTCGTGGAGTC
Reverse: AAATGAGCCCCAGCCTTC |
| β-Actin | NM_001101 | ACTB | Forward: TCTAAGAGAATGGCCCAGTC
Reverse: GGCACGAAGGCTCATCATTC |
siRNA Knockdown
U937 cells were transfected with Non-Targeting (control) or HSPA6 (HSP70 6) ON-TARGETplus SMARTpool siRNAs (Dharmacon RNA Technologies, Lafayette, CO) using the Nucleofector™ Device (Amaxa Inc., Gaithersburg, MD) according to manufacturer’s instructions. After 48 h in serum-containing medium, IFN-γ was added and the cells incubated for an additional 18 h. The medium was then replaced with serum-free medium and the cells cultured for 2 h to eliminate all traces of lipoproteins in the serum-supplemented media before addition of the ligand. Longer serum starvation could independently activate the transfected cells and increase the release of cytokines in the media. oxLDL-IC (150μg/ml) was added and the cells incubated for up to 12 h. The conditioned culture medium was removed for IL-1β analysis and RNA extracted for Q-PCR. The extent of siRNA-mediated knockdown of HSP70 6 mRNA levels was confirmed using HSP70 6 and β-actin primers shown in Table I.
Results and Discussion
In this study, DNA microarray transciptomic profiling was performed on RNAs isolated from U937 cells treated with oxLDL-IC, oxLDL, KLH-IC, or PBS vehicle for 4 h. A rationale for choosing 4 h of treatment was based on the finding that between 2 and 6 h post treatment of U937 cells with oxLDL-IC there was an increase in levels of IL-1β released into the conditioned culture medium (Fig. 1). oxLDL did not elicit a similar response (Fig. 1). In light of these findings, changes in gene expression after 4 h of oxLDL-IC exposure could be due to secondary signaling effects of gene products such as IL-1β whose expression was augmented early by oxLDL-IC. Using ANOVA and pair-wise analyses of the resulting microarray hybridization data we sought to determine: 1) genes uniquely regulated by ox-LDL-IC, 2) genes similarly regulated by ox-LDL-IC and KLH-IC but not by oxLDL, 3) genes similarly regulated by ox-LDL-IC and oxLDL, and 4) genes uniquely regulated by ox-LDL.
Figure 1.

Secretion of IL-1β is increased in response to oxLD-IC but not oxLDL. Shown are levels of IL-1β secreted into the conditioned culture medium of U937 cells treated with oxLDL-IC or oxLDL as measured using a multiplex sandwich ELISA. Data are representative of two independent experiments.
Genes uniquely regulated by ox-LDL-IC
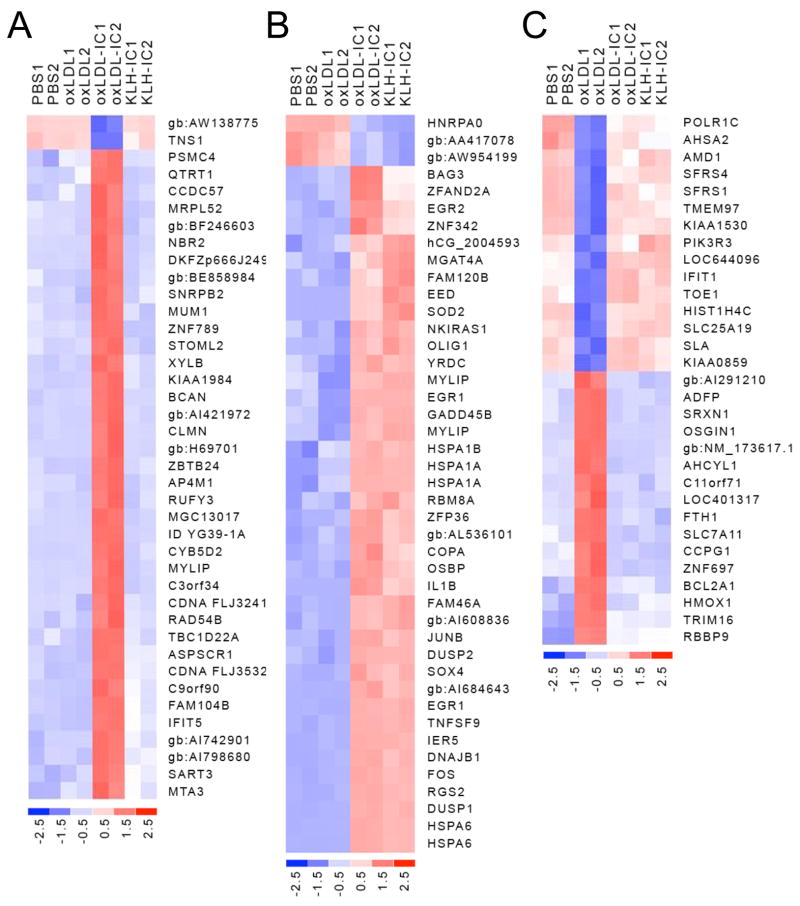
ANOVA analysis revealed that oxLDL-IC uniquely (as compared to the KLH-IC, oxLDL, and PBS treatments) affected the expression of 40 genes (Fig. 2A). From this set of genes, those having a fold change (FC) >2 (4 genes) were subjected to functional categorization. This analysis showed that all four genes (RAD54B, RUFY3, SNRPB2, and ZBTB24) are involved with pro-survival functions (Table 2). These findings concur with previous evidence that oxLDL-IC treatment of U937 cells elicits a survival response (11,12).
Figure 2.
Heat maps depicting expression profiles of regulated genes in U937 cells exposed to oxLDL-IC, KLH-IC, oxLDL, or PBS vehicle. U937 cells were exposed to oxLDL-IC, KLH-IC, oxLDL (150 μg/ml), or PBS vehicle for 4 h. DNA microarray experimentation was performed using Affymetrix U133 Plus 2.0 GeneChips (n = 2). A, Heat map depicting expression profiles of genes uniquely affected by oxLDL-IC. B, Heat map depicting expression profiles of genes regulated similarly by ox-LDL-IC and KLH-IC. C, Heat map depicting expression profiles of genes uniquely affected by oxLDL. Statistical analysis was performed as described under Gene Array Analysis. Colorimetric scaling (Z-standardization) is indicated at the bottom in units of standardization deviation from the mean.
Table 2.
Genes uniquely up-regulated by oxLDL-IC
| Gene | Symbol | Accession No. | Gene ID1 | Function2 | FC3vs PBS | FC vs oxLDL | FC vs KLH-IC |
|---|---|---|---|---|---|---|---|
| Similar to Fibrinogen silencer-binding protein | RAD54B | NM_006550 | 25788 | DNA repair and mitotic activity | 2.10 | 2.06 | 1.93 |
| RUN and FYVE domain containing 3 | RUFY3 | NM_014961 | 22902 | cell division; targets proteins to membrane lipids via interaction with PI3P | 2.12 | 2.18 | 2.17 |
| Small nuclear ribonucleoprotein polypeptide B″ | SNRPB2 | BE466925 | 6629 | RNA splicing, SLE4 autoantigen | 2.71 | 2.95 | 2.65 |
| Zinc finger and BTB domain containing 24 | ZBTB24 | BC036731 | 9841 | regulation of transcription | 3.35 | 3.53 | 3.14 |
NCBI Entrez Gene ID number
based on information from GO biological process annotations and PubMed.
FC, fold change
systemic lupus erythematosus
Genes similarly regulated by ox-LDL-IC and KLH-IC
Differential expression analysis was also conducted to determine whether the regulatory activity of oxLDL-IC is similar to that of KLH-IC, and as such, likely to be mediated via engagement of Fcγ receptors. As a result, 43 genes were identified as being similarly regulated by ox-LDL-IC and KLH-IC (Fig. 2B). Functional categorization of the 31 up-regulated genes from this group (Table 3) revealed that 39% are involved with stress responses, including the unfolded protein response which impacts cell survival. Nineteen percent of the up-regulated genes are involved with regulation of transcription, 10% are involved with endocytosis and intracellular transport of protein and lipid, and 16% affect inflammatory responses including regulation of the I-κB/NF-κB cascade and cytokine activity (Table 3). Genes encoding the cytokines tumor necrosis factor (TNF) (superfamily member 9) and IL-1β were up-regulated 7 fold and 3 fold, respectively. These findings are consistent with evidence showing that human macrophages release significantly higher levels of pro-inflammatory cytokines after incubation with oxLDL-IC as compared to incubation with oxLDL, and support the hypothesis that the mechanisms of activation by oxLDL-IC are similar to those responsible for macrophage activation by KLH-IC, most likely via Fcγ receptor engagement (5).
Table 3.
Genes similarly up-regulated by oxLDL-IC and KLH-IC
| Gene | Symbol | Accession No. | Gene ID1 | Function2 |
|---|---|---|---|---|
| Heat shock 70kDa protein 1A | HSPA1A | NM_005345 | 3303 | stabilizes proteins against aggregation, protein folding, ubiquitin-proteasome pathway |
| Heat shock 70kDa protein 1B | HSPA1B | NM_005346 | 3304 | anti-apoptosis, response to unfolded protein, protein folding |
| Heat shock 70kDa protein 6 (HSP70B′) | HSPA6 | NM_002155 | 3310 | response to unfolded protein |
| DnaJ (Hsp40) homolog, subfamily B, member 1 | DNAJB1 | NM_006145 | 3337 | response to unfolded protein, protein folding, HSP binding |
| BCL2-associated athanogene 3 | BAG3 | NM_004281 | 9531 | anti-apoptosis, protein folding, modulates the activity of HSP70 class |
| Jun B proto-oncogene | JUNB | NM_002229 | 3726 | anti-apoptosis |
| Growth arrest and DNA-damage-inducible, beta | GADD45B | NM_015675 | 4616 | regulation of growth and apoptosis in response to stress |
| Immediate early response 5 | IER5 | NM_016545 | 51278 | mediates cellular response to mitogenic signals |
| SRY (sex determining region Y)-box 4 | SOX4 | BG528420 | 6659 | determination of the cell fate (death/survival) |
| Superoxide dismutase 2, mitochondrial | SOD2 | AL050388 | 6648 | response to oxidative stress |
| Dual specificity phosphatase 1 | DUSP1 | AA530892 | 1843 | response to oxidative stress |
| Mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N- acetylglucosaminyltransferase, isoenzyme A | MGAT4A | BC031487 | 11320 | carbohydrate metabolic process, cell differentiation |
| Coatomer protein complex, subunit alpha | COPA | BC037941 | 1314 | protein transport between endoplasmic reticulum and Golgi |
| yrdC domain containing | YRDC | NM_024640 | 79693 | regulates the activity of a variety of transporters |
| Oxysterol binding protein | OSBP | BC017975 | 5007 | intracellular lipid transport |
| Tumor necrosis factor (ligand) superfamily, member 9 | TNFSF9 | NM_003811 | 8744 | inflammatory response, cytokine activity |
| Interleukin 1, beta | IL1B | NM_000576 | 3553 | inflammatory response |
| v-fos FBJ murine osteosarcoma viral oncogene homolog | FOS | BC004490 | 2353 | inflammatory response, regulator of cell proliferation, differentiation, and transformation. |
| NFKB inhibitor interacting Ras-like 1 | NKIRAS1 | AI970120 | 28512 | inflammatory response, regulates the degradation of IkappaB |
| Ribosomal protein L17 | hCG_2004593 | AA522618 | 6139 | inflammatory response, signal transducer activity, regulation of I-kappaB kinase/NF-kappaB cascade |
| RNA binding motif protein 8A | RBM8A | BC017770 | 9939 | RNA splicing |
| Early growth response 1 | EGR1 | AI459194 | 1958 | regulation of transcription |
| Early growth response 2 | EGR2 | NM_000399 | 1959 | regulation of transcription |
| Oligodendrocyte transcription factor 1 | OLIG1 | AL355743 | 116448 | regulation of transcription |
| Embryonic ectoderm development | EED | AF099032 | 8726 | repression of gene activity, regulator of integrin function |
| Zinc finger protein 342 | ZNF342 | AA761573 | 162979 | regulation of transcription |
| Zinc finger protein 36, C3H type | ZFP36 | NM_003407 | 7538 | mRNA catabolism |
| Zinc finger, AN1-type domain 2A | ZFAND2A | AI984061 | 90637 | binding to metal ions |
| Dual specificity phosphatase 2 | DUSP2 | NM_004418 | 1844 | MAPK signaling pathway |
| Regulator of G-protein signalling 2, 24kDa | RGS2 | NM_002923 | 5997 | cell cycle, regulation of G-protein coupled receptor protein |
| Myosin regulatory light chain interacting protein | MYLIP | NM_013262 | 29116 | cell motility, protein ubiquitination |
NCBI Entrez Gene ID number
based on information from GO biological process annotations and PubMed.
It has been recently shown that immune-complexes activate monocytes/macrophages through the interaction with Fcγ receptors, triggering the secretion of inflammatory cytokines and inhibiting apoptosis (22). The apoptosis protection effects were shown to involve PI3K/Akt, MAPK, and NF-κB pathways (22). Earlier work by our group (11) and by others (12) has shown that oxLDL-IC also promoted survival of monocytes by cross-linking Fcγ receptors. Mechanistically, the pro-survival effects of oxLDL-IC have been shown to involve Akt signaling (12), as well as release of sphingosine kinase 1 (SK1), the enzyme responsible for generating the pro-survival signaling molecule sphingosine-1-phosphate (S1P) (11). Although analysis of the DNA microarray data did not show a statistically significant difference in SK1 expression in response to either oxLDL-IC or KLH-IC as compared to other treatments, Q-PCR analysis showed that SK1 mRNA levels were up-regulated in response to both oxLDL-IC and KLH-IC compared to oxLDL alone (2.3- and 2.2-fold, respectively) (Fig. 3A). Measurement of S1P levels together with SK1 gene expression are necessary since minor increases in the bioactive molecule S1P could be sufficient to transduce signals necessary for cell survival.
Figure 3.
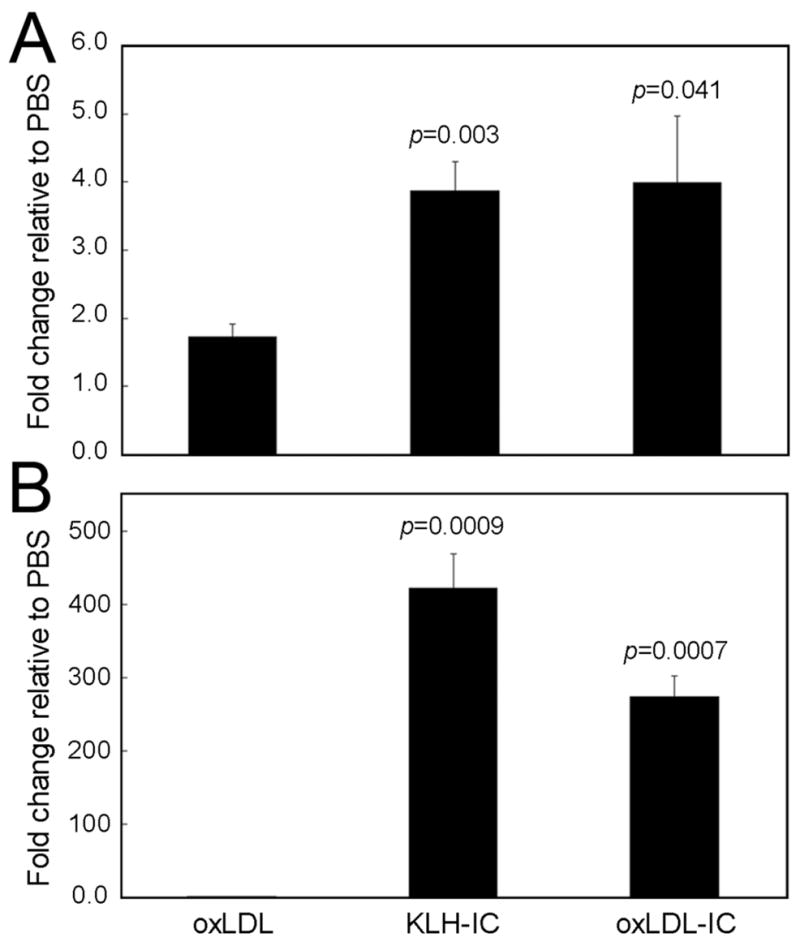
Q-PCR analysis of sphingosine kinase 1 (SK1) and HSP70 6, two genes up-regulated in response to oxLDLIC and KLH-IC. U937 cells were exposed to oxLDL-IC, KLH-IC, oxLDL (150 μg/ml), or PBS vehicle for 4 h. A, Q-PCR analysis of SK1 mRNA levels; and B, Q-PCR analysis of HSP70 6 mRNA levels. Quantification of RNA was performed using the cycle threshold of SK1 and HSP70 6 cDNA relative to that of GAPDH. Values presented in A and B are means of triplicate determinations ± SE; data are representative of two experiments. Indicated p values were derived from a Student’s unpaired t-test of data compared with oxLDL data.
Of those genes similarly regulated by oxLDL-IC and KLH-IC, Superoxide dismutase 2 (SOD2) was up-regulated 71 fold in response to oxLDL-IC and 20 fold in response to KLH-IC both as compared to PBS (Table 3). SOD2, a principal scavenger enzyme in the mitochondrial matrix, protects cells from oxidative stress by detoxifying superoxide generated in mitochondrial respiration by dismutation. High SOD2 expression can enhance fibrosarcoma cell survival in response to apoptotic stimuli via a mechanism that involves regulation of steady state levels of H2O2 (23). Therefore, the augmentation of SOD2 expression that we observed in response to oxLDL-IC could account for the oxLDL-IC-induced cell survival reported previously (11).
Oxysterol binding protein (OSBP), a major sterol-binding protein implicated in lipid metabolism, vesicle transport, and signal transduction was found to be up-regulated in response to oxLDL-IC (4 fold) and KLH-IC (6 fold) compared to PBS (Table 3). Recently, OSBP was found to function as a cholesterol-binding scaffolding protein coordinating the activity of certain phosphatases to control the extracellular signal-regulated kinase (ERK) signaling pathway (24). Furthermore, Perry and Ridgway (25) have shown that when changes in sterol metabolism (oxysterol) induce the translocation of OSBP to the trans-Golgi network, the ceramide transporter (CERT) also translocates to Golgi membranes, transporting more ceramide to sphingomyelin synthase at that site for sphingomyelin synthesis. Thus, up-regulation of OSBP in response to oxLDL-IC could lead to enhanced conversion of ceramide to sphingomyelin. It has been established that cellular accumulations of ceramide, which occur as a result of oxidative stress or in response to inflammatory cytokines, are associated with apoptotic responses (26). We have recently found that indeed short chain ceramides decreased in response to oxLDL-IC compared to controls (data not shown). Therefore, an OSBP-mediated decrease in ceramide levels in response to oxLDL-IC could account for the oxLDL-IC-induced cell survival reported previously (11).
The expression of regulator of G-protein signaling 2 (RGS2) was up-regulated in response to oxLDL-IC (72 fold) and KLH-IC (98 fold) compared to PBS (Table 3). RGS2 has emerged as a multifunctional regulator of G-protein signaling (27). RGS proteins enhance the rate at which certain heterotrimeric G-protein α-subunits hydrolyze GTP to GDP limiting the duration that α-subunits activate downstream effectors (i.e., negatively regulating G protein-coupled receptor signaling) (27). Targeted mutation of RGS2 in mice leads to reduced T cell proliferation and IL-2 production (28). The consequence of RGS2 up-regulation in response to cross-linking of Fcγ receptor on signaling pathways downstream to G protein-coupled receptors in human monocytes/macrophages should now be evaluated.
A member of the heat shock 70 kDa protein (HSP70) family, HSP70 6, displayed the greatest magnitude increase in expression (oxLDL-IC and KLH-IC were 1924-fold and 3516-fold greater, respectively, as compared to PBS) (Table 3). Augmented HSP70 6 expression in response to oxLDL-IC and KLH-IC was also shown by Q-PCR (Fig. 3B). At the present time nothing is known as to the function of HSP70 6. We sought to determine whether the augmented expression of HSP70 6 mRNA could account for oxLDL-IC-induced cytokine release by U937 cells. To address this question we used siRNAs to suppress HSP70 6 expression (Fig. 4A). As shown in Figure 4B, oxLDL-IC-induced secretion of IL1-β was completely inhibited when the expression of HSP70 6 was suppressed by siRNA. In cells treated with vehicle alone, the low level of IL1-β secretion was also decreased by HSP70 6 siRNA (Fig. 4B). The findings indicate that HSP70 6 expression is required in the process by which oxLDL-IC augments the secretion of IL1-β. Using Q-PCR analysis we found that IL1-β mRNA expression was not affected by HSP70 6 siRNA knockdown (data not shown).
Figure 4.

siRNA-mediated suppression of HSP70 6 abrogates oxLDL-IC-induced IL-1β secretion. U937 cells were transfected with HSP70 6 siRNAs and treated with oxLDL-IC as described in Methods. A, The effect on knockdown of HSP70 6 mRNA levels was measured by Q-PCR; quantification of RNA was performed using the cycle threshold of HSP70 6 cDNA relative to that of β-actin. Values presented in A are means of triplicate determinations ± SE; data are representative of three experiments. Indicated p values were derived from a Student’s unpaired t-test of data compared with control siRNA. B, The level of IL1-β secreted into the conditioned culture medium was measured by an IL-1β enzyme immunoassay. Asterisk indicated the undetectable levels of IL-1β in the conditioned culture medium of cells treated with HSP70 6 siRNA. Values presented in B are means of duplicate determinations ± SE; data are representative of two independent experiments.
Genes similarly regulated by ox-LDL-IC and oxLDL
Differential expression analysis of the array data was also conducted to determine whether the regulatory activity of oxLDL-IC is mediated by the oxLDL moiety of the complex. Using the same statistical criteria used to identify genes regulated similarly by oxLDL-IC and KLH-IC, the analysis revealed no genes similarly regulated by oxLDL-IC and oxLDL.
Genes uniquely regulated by ox-LDL
Differential expression analysis was also conducted to determine the transcriptional response of U937 cells to oxLDL. Genes regulated uniquely by oxLDL (31 genes) are shown in the heat map in Figure 2C. From this set of genes, those having a FC > (± 2) were subjected to functional categorization (Table 4). This analysis showed that up-regulated genes are involved with growth inhibition (OSGIN1 and FTH1), oxidoreductase activity (SPXN1 and HMOX1), and transport of amino acids and fatty acids (SLC7A11 and ADFP, respectively) (Table 4).
Table 4.
Genes uniquely regulated by oxLDL
| Gene | Symbol | Accession No. | Gene ID1 | Function2 | FC3vs PBS | FC vs oxLDL-IC | FC vs KLH-IC |
|---|---|---|---|---|---|---|---|
| Interferon-induced protein with tetratricopeptide repeats 1 |
IFIT1 | NM_001548 | 3434 | immune response | −2.58 | −3.77 | −3.25 |
| Src-like-adaptor | SLA | U44403 | 6503 | intracellular signaling cascade | −2.45 | −2.72 | −2.32 |
| Target of EGR1, member 1 (nuclear) | TOE1 | NM_025077 | 114034 | growth inhibitory activity | −1.93 | −2.45 | −2.15 |
| AHA1, activator of heat shock 90kDa protein ATPase |
AHSA2 | AI831431 | 130872 | co-chaperone, response to unfolded protein | −3.20 | −2.35 | −1.87 |
| Phosphoinositide-3-kinase, regulatory subunit 3 (p55, Gamma) |
PIK3R3 | BE622627 | 8503 | modulates the activity of the enzyme phosphatidylinositol 3-kinase activity | −1.89 | −1.76 | −2.31 |
| Zinc finger protein 697 | ZNF697 | AW003092 | 90874 | regulation of transcription | 1.85 | 2.09 | 2.08 |
| Oxidative stress induced growth inhibitor 1 |
OSGIN1 | NM_013370 | 29948 | growth inhibition, cell death | 2.54 | 2.57 | 2.58 |
| Estrogen-responsive B box Protein | TRIM16 | NM_006470 | 10626 | enhancing the alternative secretion pathway of IL-1beta |
4.26 | 2.62 | 2.76 |
| Ferritin, heavy polypeptide 1 | FTH1 | AA083483 | 2495 | inhibition of proliferation, ferric iron binding | 3.23 | 3.40 | 3.92 |
| Sulfiredoxin 1 homolog | SRXN1 | AL121758 | 140809 | oxidoreductase activity | 3.26 | 3.49 | 3.56 |
| Solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 |
SLC7A11 | AB040875 | 23657 | amino acid transport | 3.15 | 4.04 | 3.84 |
| Adipose differentiation-related Protein | ADFP | BC005127 | 123 | long-chain fatty acid transport, sequestering of lipid | 4.20 | 4.42 | 4.42 |
| Heme oxygenase (decycling) 1 | HMOX1 | NM_002133 | 3162 | oxidoreductase activity, iron ion binding | 95.67 | 58.49 | 25.90 |
NCBI Entrez Gene ID number
based on information from GO biological process annotations and PubMed.
FC, fold change
The gene with the greatest level of increase in response to oxLDL was heme oxygenase 1 (HMOX1) (Table 4). Several lines of evidence suggest that HMOX1 expression can be increased several fold by stimuli that induce cellular oxidative stress, including oxidized LDL (29,30). HMOX1 induction is known to lead to an increase in catalytic free iron release. Importantly, HMOX1 also mitigates the cytotoxic effects of iron by mediating the enhancement of intracellular ferritin (31). Consistent with this response, we see that oxLDL augments expression of ferritin, heavy polypeptide 1 (H-ferritin) (Table 4). This is also consistent with other findings showing that oxLDL augments levels of L-ferritin (light-chain ferritin) in THP-1 cells (32,33).
The microarray results also show that the gene encoding adipose differentiation-related protein (ADFP) was uniquely up-regulated in response to oxLDL (Table 4). ADFP is expressed in many cell types and is associated with intracellular neutral lipid droplets (34). The precise relationship between ADFP and lipid droplet biology is still obscure (35,36).
Regulation of receptors involved in modified lipoprotein uptake and foam cell formation
We also examined the effects of oxLDL compared to oxLDL-IC on the expression of several receptors involved in lipoprotein uptake and foam cell formation. LDL receptor (LDLR) expression is characteristically down regulated in differentiated human macrophages (37). However, our group has previously shown that after stimulation with LDL-containing immune complexes, LDLR is up-regulated (38). LDL-containing immune complexes induce both transcriptional and post-transcriptional activation of the LDL-R gene in differentiated monocytes and this induction is independent of the free cholesterol pool of these cells (38). In this study, microarray analysis revealed that LDLR gene expression was induced 2.1 fold (p<0.05) in response to oxLDL-IC compared to oxLDL at 4 h post treatment. The level of expression of genes encoding the scavenger receptors SRB-1 and CD36 were also increased in response to oxLDL-IC treatment (1.6 and 1.7 fold, respectively; p<0.05) as compared to oxLDL. Changes in CD36 and LDLR receptor expression as revealed by DNA microarray analysis were verified using Q-PCR (Fig. 5).
Figure 5.

Q-PCR analysis of the lipoprotein receptors CD36 (A) and LDLR (B). U937 cells were exposed to oxLDL-IC, KLH-IC, oxLDL (150 μg/ml), or PBS vehicle for 4 h. Quantification of RNA was performed using the cycle threshold of receptor cDNA relative to that of GAPDH. Values presented are means of triplicate determinations ± SE; data are representative of two experiments. Indicated p values were derived from a Student’s unpaired t-test of data compared with oxLDL data.
The physiological and pathological significance of LDLR and CD36 upregulation in response to oxLDL-IC remains to be determined. CD36 is an integral membrane protein expressed on monocytes and macrophages and functions as a scavenger receptor for oxidized LDL. It has been shown that minimally oxidized LDL (MM-LDL) pretreatment increases both mRNA levels and protein levels of CD36, whereas “fully” oxLDL pretreatment does not increase expression of CD36 (39). This suggested that MM-LDL primes the macrophage for more effective clearance of oxLDL and foam cell formation (39). Our current data show that CD36 expression was down-regulated in response to oxLDL probably because of “full” oxidation of LDL and/or short incubation time (4 h). oxLDL-IC however induced CD36 upregulation, which could indicate more efficient uptake of existing oxLDL in the plaque.
Furthermore, a number of cytokines and inflammatory agents have been identified as possible mediators involved in regulating expression of scavenger receptors in human monocytes (40). It has been shown that expression of CD36 is upregulated by interleukin-4, monocyte colony-stimulating factor, and phorbol myristate acetate, while lipopolysaccharide and dexamethasone strongly downregulates CD36 mRNA (40); IFN-γ had no effect on CD36 mRNA (40). Thus, it is likely that CD36 upregulation in our study is secondary to the autocrine effect of secreted cytokines induced by oxLDL-IC.
We are currently investigating the possible role of scavenger receptors (class A and class B) and/or lipoprotein receptors in the uptake of oxLDL-IC and whether or not cross-linking of two receptors trigger distinct signaling pathways required to elicit an enhanced macrophage response.
Conclusion
We investigated whether oxidized LDL immune complexes and oxidized LDL differentially affect the expression of genes involved in activation and survival of IFN-γ-treated human monocytes. The findings highlight transcriptional responses elicited by oxLDL-IC that may underlie its reported cytoprotective and pro-inflammatory effects compared to oxLDL. Cross-linking of Fcγ receptor appears to be the principle trigger for macrophage responses to oxLDL-IC. Intriguingly, although oxLDL and oxLDL-IC share the same modified lipid moiety, the two do not regulate genes in common. The findings further strengthen the hypothesis that oxLDL and oxLDL-IC elicit disparate inflammatory responses and thereby play distinct roles in the process of atherosclerosis.
Recent reported findings indicate that there is an important immunological and inflammatory aspect of atherosclerosis. Several autoimmune diseases are associated with accelerated atherosclerosis, increased plasma levels of circulating oxLDL, anti oxLDL antibodies, dyslipidemia, and enhanced inflammation (41,42). Some groups have proposed that the immune response to oxidized LDL may be protective with regard to the development of atherosclerosis (43–45). This postulate is based on experiments carried out with animal models, which have limited relevance to human atherosclerosis (46) and on in vitro data obtained either with Fab fragments (47), which form non-inflammatory complexes due to the lack of the antibody Fc region (48), or with IgM oxLDL antibodies, which are unable to interact with phagocytic cells but represent a minority of the antibody population in humans (4), and have not been shown to confer protection in clinical studies (49). In contrast, human oxLDL-IC are not only pro-inflammatory in vitro (5–7), but are associated with diabetic nephropathy and with increased internal carotid intima-media thickness (50–53).
The striking difference in the cellular effects of oxLDL IC and uncomplexed oxLDL may result from a divergence in the processing of oxLDL and oxLDL-IC, perhaps due to differences in receptor binding, uptake and delivery to lysosomes and/or to lysosomal and post-lysosomal processing. Cross-linking of Fcγ receptor in response to oxLDL-IC is an extremely potent activation signal for human macrophages and is likely to trigger signal transduction pathways that modulate different macrophage functions (54), not likely to be induced as a consequence of scavenger receptor ligation by oxLDL. Our findings may advance efforts to reveal specific targets in the signaling pathway that can have therapeutic implications for blocking cytokine release and to prevent formation of vulnerable plaques.
Acknowledgments
This work was supported by the following NIH grants: HL079274 (SMH), PO1-HL55782 (MLV), HL061873 (WSA), and the South Carolina COBRE in Lipidomics and Pathobiology (P20 RR17677 from NCRR). The work was also supported by an American Heart Association Scientist Development Grant (0435259N) and a grant from the University Research Committee of the Medical University of South Carolina to SMH, and funding from the Department of Veterans Affairs to MLV. We acknowledge the technical assistance of Alena Nareika and personnel of the MUSC DNA Microarray and Bioinformatics Facility including Saurin Jani and Victor Fresco. We thank Charlyne Chassereau for assistance with lipoprotein isolation.
Footnotes
Hammad et al., Genes modulated by oxidized LDL immune complexes
References
- 1.Steinberg D. Low density lipoprotein oxidation and its pathobiological significance. J Biol Chem. 1997;272:20963–6. doi: 10.1074/jbc.272.34.20963. [DOI] [PubMed] [Google Scholar]
- 2.Kunjathoor VV, Febbraio M, Podrez EA, Moore KJ, Andersson L, Koehn S, Rhee JS, Silverstein R, Hoff HF, Freeman MW. Scavenger receptors class A-I/II and CD36 are the principal receptors responsible for the uptake of modified low density lipoprotein leading to lipid loading in macrophages. J Biol Chem. 2002;277:49982–8. doi: 10.1074/jbc.M209649200. [DOI] [PubMed] [Google Scholar]
- 3.Virella G, Koskinen S, Krings G, Onorato JM, Thorpe SR, Lopes-Virella M. Immunochemical characterization of purified human oxidized low-density lipoprotein antibodies. Clin Immunol. 2000;95:135–44. doi: 10.1006/clim.2000.4857. [DOI] [PubMed] [Google Scholar]
- 4.Virella G, Lopes-Virella MF. Lipoprotein autoantibodies: measurement and significance. Clin Diagn Lab Immunol. 2003;10:499–505. doi: 10.1128/CDLI.10.4.499-505.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saad AF, Virella G, Chassereau C, Boackle RJ, Lopes-Virella MF. OxLDL immune complexes activate complement and induce cytokine production by MonoMac 6 cells and human macrophages. J Lipid Res. 2006;47:1975–83. doi: 10.1194/jlr.M600064-JLR200. [DOI] [PubMed] [Google Scholar]
- 6.Virella G, Munoz JF, Galbraith GM, Gissinger C, Chassereau C, Lopes-Virella MF. Activation of human monocyte-derived macrophages by immune complexes containing low-density lipoprotein. Clin Immunol Immunopathol. 1995;75:179–89. doi: 10.1006/clin.1995.1069. [DOI] [PubMed] [Google Scholar]
- 7.Virella G, Atchley D, Koskinen S, Zheng D Lopes-Virella MF; DCCT/EDIC Research Group. Proatherogenic and proinflammatory properties of immune complexes prepared with purified human oxLDL antibodies and human oxLDL. Clin Immunol. 2002;105:81–92. doi: 10.1006/clim.2002.5269. [DOI] [PubMed] [Google Scholar]
- 8.Griffith RL, Virella GT, Stevenson HC, Lopes-Virella MF. Low density lipoprotein metabolism by human macrophages activated with low density lipoprotein immune complexes. A possible mechanism of foam cell formation. J Exp Med. 1988;168:1041–59. doi: 10.1084/jem.168.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klimov AN, Denisenko AD, Vinogradov AG, Nagornev VA, Pivovarova YI, Sitnikova OD, Pleskov VM. Accumulation of cholesteryl esters in macrophages incubated with human lipoprotein-antibody autoimmune complex. Atherosclerosis. 1988;74:41–6. doi: 10.1016/0021-9150(88)90189-x. [DOI] [PubMed] [Google Scholar]
- 10.Libby P. Molecular bases of the acute coronary syndromes. Circulation. 1995;91:2844–50. doi: 10.1161/01.cir.91.11.2844. Review. [DOI] [PubMed] [Google Scholar]
- 11.Hammad SM, Taha TA, Nareika A, Johnson KR, Lopes-Virella MF, Obeid LM. Oxidized LDL immune complexes induce release of sphingosine kinase in human U937 monocytic cells. Prostaglandins Other Lipid Mediat. 2006;79:126–40. doi: 10.1016/j.prostaglandins.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Oksjoki R, Kovanen PT, Lindstedt KA, Jansson B, Pentikainen MO. OxLDL-IgG immune complexes induce survival of human monocytes. Arterioscler Thromb Vasc Biol. 2006;26:576–83. doi: 10.1161/01.ATV.0000201041.14438.8d. [DOI] [PubMed] [Google Scholar]
- 13.Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937) Int J Cancer. 1976;17:565–77. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 14.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–74. doi: 10.1038/nature01323. Review. [DOI] [PubMed] [Google Scholar]
- 15.De Boer OJ, van der Wal AC, Verhagen CE, Becker AE. Cytokin secretion profiles of cloned T cells from human aortic atherosclerotic plaques. J Pathol. 1999;188:174–9. doi: 10.1002/(SICI)1096-9896(199906)188:2<174::AID-PATH333>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Lopes-Virella MF, Koskinen S, Mironova M, Horne D, Klein R, Chassereau C, Enockson C, Virella G. The preparation of copper-oxidized LDL for the measurement of oxidized LDL antibodies by EIA. Atherosclerosis. 2000;152:107–15. doi: 10.1016/s0021-9150(99)00456-6. [DOI] [PubMed] [Google Scholar]
- 17.Virella G, Thorpe SR, Alderson NL, Derrick MB, Chassereau C, Rhett JM, Lopes-Virella MF. Definition of the immunogenic forms of modified human LDL recognized by human autoantibodies and by rabbit hyperimmune antibodies. J Lipid Res. 2004;45:1859–67. doi: 10.1194/jlr.M400095-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Koskinen S, Enockson C, Lopes-Virella MF, Virella G. Preparation of a human standard for determination of the levels of antibodies to oxidatively modified low-density lipoproteins. Clin Diagn Lab Immunol. 1998;5:817–22. doi: 10.1128/cdli.5.6.817-822.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mironova M, Virella G, Lopes-Virella MF. Isolation and characterization of human antioxidized LDL autoantibodies. Arterioscler Thromb Vasc Biol. 1996;16:222–9. doi: 10.1161/01.atv.16.2.222. [DOI] [PubMed] [Google Scholar]
- 20.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. Journal of the American Statistical Association. 2004;99:909–17. [Google Scholar]
- 21.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bianchi G, Montecucco F, Bertolotto M, Dallegri F, Ottonello L. Immune complexes induce monocyte survival through defined intracellular pathways. Ann N Y Acad Sci. 2007;1095:209–19. doi: 10.1196/annals.1397.025. [DOI] [PubMed] [Google Scholar]
- 23.Dasgupta J, Subbaram S, Connor KM, Rodriguez AM, Tirosh O, Beckman JS, Jourd’Heuil D, Melendez JA. Manganese superoxide dismutase protects from TNF-alpha-induced apoptosis by increasing the steady-state production of H2O2. Antioxid Redox Signal. 2006;8:1295–305. doi: 10.1089/ars.2006.8.1295. [DOI] [PubMed] [Google Scholar]
- 24.Wang PY, Weng J, Anderson RG. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 2005;307:1472–6. doi: 10.1126/science.1107710. [DOI] [PubMed] [Google Scholar]
- 25.Perry RJ, Ridgway ND. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol Biol Cell. 2006;17:2604–16. doi: 10.1091/mbc.E06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verheij M, Bose R, Lin XH, Yao B, Jarvis WD, Grant S, Birrer MJ, Szabo E, Zon LI, Kyriakis JM, Haimovitz-Friedman A, Fuks Z, Kolesnick RN. Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature. 1996;380:75–9. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 27.Kehrl JH, Sinnarajah S. RGS2: a multifunctional regulator of G-protein signaling. Int J Biochem Cell Biol. 2002;34:432–8. doi: 10.1016/s1357-2725(01)00141-8. Review. [DOI] [PubMed] [Google Scholar]
- 28.Oliveira-Dos-Santos AJ, Matsumoto G, Snow BE, Bai D, Houston FP, Whishaw IQ, Mariathasan S, Sasaki T, Wakeham A, Ohashi PS, Roder JC, Barnes CA, Siderovski DP, Penninger JM. Regulation of T cell activation, anxiety, and male aggression by RGS2. Proc Natl Acad Sci U S A. 2000;97:12272–7. doi: 10.1073/pnas.220414397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siow RC, Ishii T, Sato H, Taketani S, Leake DS, Sweiry JH, Pearson JD, Bannai S, Mann GE. Induction of the antioxidant stress proteins heme oxygenase-1 and MSP23 by stress agents and oxidised LDL in cultured vascular smooth muscle cells. FEBS Lett. 1995;368:239–42. doi: 10.1016/0014-5793(95)00650-x. [DOI] [PubMed] [Google Scholar]
- 30.Hoekstra KA, Godin DV, Cheng KM. Protective role of heme oxygenase in the blood vessel wall during atherogenesis. Biochem Cell Biol. 2004;82:351–9. doi: 10.1139/o04-006. Review. [DOI] [PubMed] [Google Scholar]
- 31.Vile GF, Basu-Modak S, Waltner C, Tyrrell RM. Heme oxygenase 1 mediates an adaptive response to oxidative stress in human skin fibroblasts. Proc Natl Acad Sci U S A. 1994;91:2607–10. doi: 10.1073/pnas.91.7.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang MK, Choi MS, Park YB. Regulation of ferritin light chain gene expression by oxidized low-density lipoproteins in human monocytic THP-1 cells. Biochem Biophys Res Commun. 1999;265:577–83. doi: 10.1006/bbrc.1999.1725. [DOI] [PubMed] [Google Scholar]
- 33.Kang JH, Kim HT, Choi MS, Lee WH, Huh TL, Park YB, Moon BJ, Kwon OS. Proteome analysis of human monocytic THP-1 cells primed with oxidized low-density lipoproteins. Proteomics. 2006;6:1261–73. doi: 10.1002/pmic.200500290. [DOI] [PubMed] [Google Scholar]
- 34.Heid HW, Moll R, Schwetlick I, Rackwitz HR, Keenan TW. Adipophilin is a specific marker of lipid accumulation in diverse cell types and diseases. Cell Tissue Res. 1998;294:309–21. doi: 10.1007/s004410051181. [DOI] [PubMed] [Google Scholar]
- 35.Londos C, Sztalryd C, Tansey JT, Kimmel AR. Role of PAT proteins in lipid metabolism. Biochimie. 2005;87:45–9. doi: 10.1016/j.biochi.2004.12.010. Review. [DOI] [PubMed] [Google Scholar]
- 36.Jiang HP, Serrero G. Isolation and characterization of a full-length cDNA coding for an adipose differentiation-related protein. Proc Natl Acad Sci U S A. 1992;89:7856–60. doi: 10.1073/pnas.89.17.7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auwerx J. The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia. 1991;47:22–31. doi: 10.1007/BF02041244. Review. [DOI] [PubMed] [Google Scholar]
- 38.Huang Y, Ghosh MJ, Lopes-Virella MF. Transcriptional and post-transcriptional regulation of LDL receptor gene expression in PMA-treated THP-1 cells by LDL-containing immune complexes. J Lipid Res. 1997;38:110–20. [PubMed] [Google Scholar]
- 39.Yoshida H, Quehenberger O, Kondratenko N, Green S, Steinberg D. Minimally oxidized low-density lipoprotein increases expression of scavenger receptor A, CD36, and macrosialin in resident mouse peritoneal macrophages. Arterioscler Thromb Vasc Biol. 1998;18:794–802. doi: 10.1161/01.atv.18.5.794. [DOI] [PubMed] [Google Scholar]
- 40.Yesner LM, Huh HY, Pearce SF, Silverstein RL. Regulation of monocyte CD36 and thrombospondin-1 expression by soluble mediators. Arterioscler Thromb Vasc Biol. 1996;16:1019–25. doi: 10.1161/01.atv.16.8.1019. [DOI] [PubMed] [Google Scholar]
- 41.Frostegard J. Atherosclerosis in patients with autoimmune disorders. Arterioscler Thromb Vasc Biol. 2005;25:1776–85. doi: 10.1161/01.ATV.0000174800.78362.ec. Review. [DOI] [PubMed] [Google Scholar]
- 42.Frostegard J, Svenungsson E, Wu R, Gunnarsson I, Lundberg IE, Klareskog L, Horkko S, Witztum JL. Lipid peroxidation is enhanced in patients with systemic lupus erythematosus and is associated with arterial and renal disease manifestations. Arthritis Rheum. 2005;52:192–200. doi: 10.1002/art.20780. [DOI] [PubMed] [Google Scholar]
- 43.Freigang S, Horkko S, Miller E, Witztum JL, Palinski W. Immunization of LDL receptor-deficient mice with homologous malondialdehyde-modified and native LDL reduces progression of atherosclerosis by mechanisms other than induction of high titers of antibodies to oxidative neoepitopes. Arterioscler Thromb Vasc Biol. 1998;18:1972–82. doi: 10.1161/01.atv.18.12.1972. [DOI] [PubMed] [Google Scholar]
- 44.Palinski W, Miller E, Witztum JL. Immunization of low density lipoprotein (LDL) receptor deficient rabbits with homologous malondialdehyde-modified LDL reduces atherogenesis. Proc Natl Acad Sci USA. 1995;92:821–5. doi: 10.1073/pnas.92.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Binder CJ, Shaw PX, Chang MK, Boullier A, Hartvigsen K, Horkko S, Miller YI, Woelkers DA, Corr M, Witztum JL. The role of natural antibodies in atherogenesis. J Lipid Res. 2005;46:1353–63. doi: 10.1194/jlr.R500005-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Virella G, Lopes-Virella MF. Humoral immunity and atherosclerosis. Nat Med. 2003;9:243–4. doi: 10.1038/nm0303-243. [DOI] [PubMed] [Google Scholar]
- 47.Shaw PX, Horkko S, Tsimikas S, Chang MK, Palinski W, Silverman GJ, Chen PP, Witztum JL. Human-derived anti-oxidized LDL autoantibody blocks uptake of oxidized LDL by macrophages and localizes to atherosclerotic lesions in vivo. Arterioscler Thromb Vasc Biol. 2001;21:1333–9. doi: 10.1161/hq0801.093587. [DOI] [PubMed] [Google Scholar]
- 48.Virella G. Biosynthesis, metabolism and biological properties of immunoglobulins. In: Virella G, editor. Medical Immunology. 6. N.Y. and London: Informa; 2007. pp. 65–72. [Google Scholar]
- 49.Fredrikson GN, Hedblad B, Berglund G, Alm R, Nilsson JA, Schiopu A, Shah PK, Nilsson J. Association between IgM against an aldehyde-modified peptide in apolipoprotein B-100 and progression of carotid disease. Stroke. 2007;38:1495–1500. doi: 10.1161/STROKEAHA.106.474577. [DOI] [PubMed] [Google Scholar]
- 50.Atchley DH, Lopes-Virella MF, Zheng D, Virella G. Oxidized LDL-Anti-Oxidized LDL immune complexes and diabetic nephropathy. Diabetologia. 2002;45:1562–71. doi: 10.1007/s00125-002-0962-y. [DOI] [PubMed] [Google Scholar]
- 51.Lopes-Virella MF, Virella G, Orchard TJ, Koskinen S, Evans RW, Becker DJ, Forrest KY. Antibodies to oxidized LDL and LDL-containing immune complexes as risk factors for coronary artery disease in diabetes mellitus. Clin Immunol. 1999;90:165–72. doi: 10.1006/clim.1998.4631. [DOI] [PubMed] [Google Scholar]
- 52.Lopes-Virella MF, McHenry MB, Lipsitz S, Yim E, Wilson PF, Lackland DT, Lyons T, Jenkins AJ, Virella G. Immune complexes containing modified lipoproteins are related to the progression of internal carotid intima-media thickness in patients with type 1 diabetes. Atherosclerosis. 2007;190:359–69. doi: 10.1016/j.atherosclerosis.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 53.Yishak AA, Costacou T, Virella G, Zgibor J, Fried L, Walsh M, Evans RW, Lopes-Virella M, Kagan VE, Otvos J, Orchard TJ. Novel predictors of overt nephropathy in subjects with type 1 diabetes. A nested case control study from the Pittsburgh Epidemiology of Diabetes Complications cohort. Nephrol Dial Transplant. 2006;21:93–100. doi: 10.1093/ndt/gfi103. [DOI] [PubMed] [Google Scholar]
- 54.Indik ZK, Park JG, Hunter S, Schreiber AD. The molecular dissection of Fc gamma receptor mediated phagocytosis. Blood. 1995;86:4389–99. Review. [PubMed] [Google Scholar]