Abstract
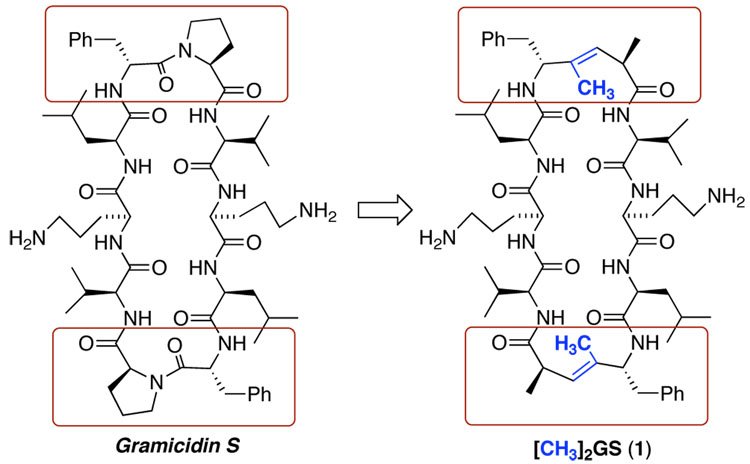
A concise synthesis of a gramicidin S analogue with trisubstituted (E)-alkene dipeptide isostere (TEADI) replacements at both DPhe-Pro positions was realized. Conformational analysis demonstrated that TEADIs can serve as type II β-turn promoters in a cyclic scaffold and successfully mimic a proline residue.
Peptides demonstrate a wide range of diverse physiological properties as hormones, enzyme inhibitors, growth promoters, signaling pathways modulators, antimicrobial agents, etc.1 In order to overcome the limited bioavailability of peptides,2 we are studying the synthesis and pharmacological evaluation of bioisosteric replacements of the amide bond.3 The relatively rigid trisubstituted (E)-alkene dipeptide isosteres (ψ[(E)-C(R)=CH], R≠H, TEADIs) maintain ω-angle planarity and represent useful structural surrogates of hydrolytically labile amide bonds.4 In addition, TEADIs were found to have potential as β-turn promoters in acyclic sequences in our previous studies.5 In a continuation of this work, we have now started to introduce these building blocks into biologically active cyclic peptide sequences as surrogates of the powerful turn-inducing DPhe-Pro sequence6 and evaluate their potential as β-turn promoters as well modulators of biological and metabolic properties.7
Since its discovery in 1942,8 the cyclodecapeptide antibiotic Gramicidin S (GS, cyclo[(Val-Orn-Leu-DPhe-Pro-)2]) has served as an inspiration for the design of antibacterial agents and antimicrobial peptides, as well as a model system for conformational mimicry.9 GS is therefore a particularly significant target for the evaluation of alkene peptide isosteres as surrogates for the type II’ β-turn inducing sequence.
The DPhe-Pro reverse turn is a critical feature of the rigid amphipathic antiparallel β-pleated sheet conformation of GS.10,11 In earlier work,7 we were able to replace the Leu-DPhe peptide bond in GS with a ψ[(E)-C(CF3)=CH] isostere with minimal perturbation of secondary structure and biological activity, while the corresponding ψ[(E)-C(CH3)=CH] isostere failed to maintain the β-pleated sheet conformation according to CD and NMR analyses. A different situation presents itself when the DPhe-Pro peptide bond is replaced. Since the DPhe carbonyl group is not involved in intramolecular H-bonding or dipolar interactions, a ψ[(E)-C(CH3)=CH] surrogate should be as effective as a trifluoromethylated congener in conformationally preorganizing the chain. The restricted backbone rotation imposed by the A1,3-strain across the trisubstituted alkene, and, to a lesser extent, the A1,2-strain experienced by substituents attached to the alkene, should be sufficient for both methyl and trifluoromethyl groups to impose the reverse turn. Furthermore, neither alkene isostere provides an NH hydrogen bond donor group that can lead to the stabilization of γ-turns or other competitive backbone folding patterns that would interfere with the desired β-turn motif. These properties lead, theoretically, to a close match of isostere and DPhe-Pro features (Figure 1)5b We now report an experimental confirmation of this hypothesis.
Figure 1.
GS and analogue with ψ[(E)-C(CH3)=CH] peptide bond surrogates.
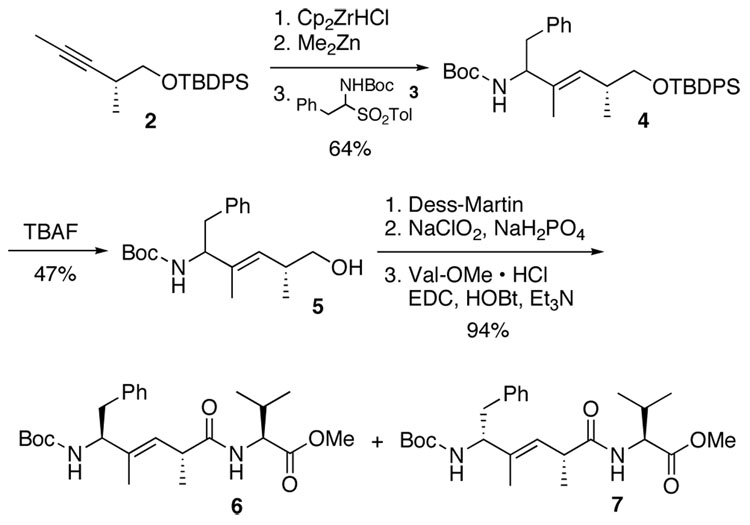
Due to the lability of phenylacetaldimines, the sulfinyl adduct 312 was employed in the organometallic allylation reaction (Scheme 1).13 Using our hydrozirconation/Zr➩Zn transmetalation methodology,14,15 the alkenylzinc species derived from the chiral internal alkyne 213b was added to 3, affording the allylic amide 4 in 64% yield as a ~1:1 mixture of diastereomers. Deprotection of the TBDPS group with TBAF provided the primary alcohol 5. The two diastereomers could not be separated after conversion of 5 to the corresponding acetates;3f,7 however, a two-step oxidation with Dess-Martin periodinane,16,17 followed by coupling with valine methyl ester provided pseudo-tripeptides 6 and 7 which were separated by preparative C18 RP HPLC.18
Scheme 1.
Synthesis of Boc-DPhe-ψ[(E)-C(CH3)=CH]-Pro-Val-OMe
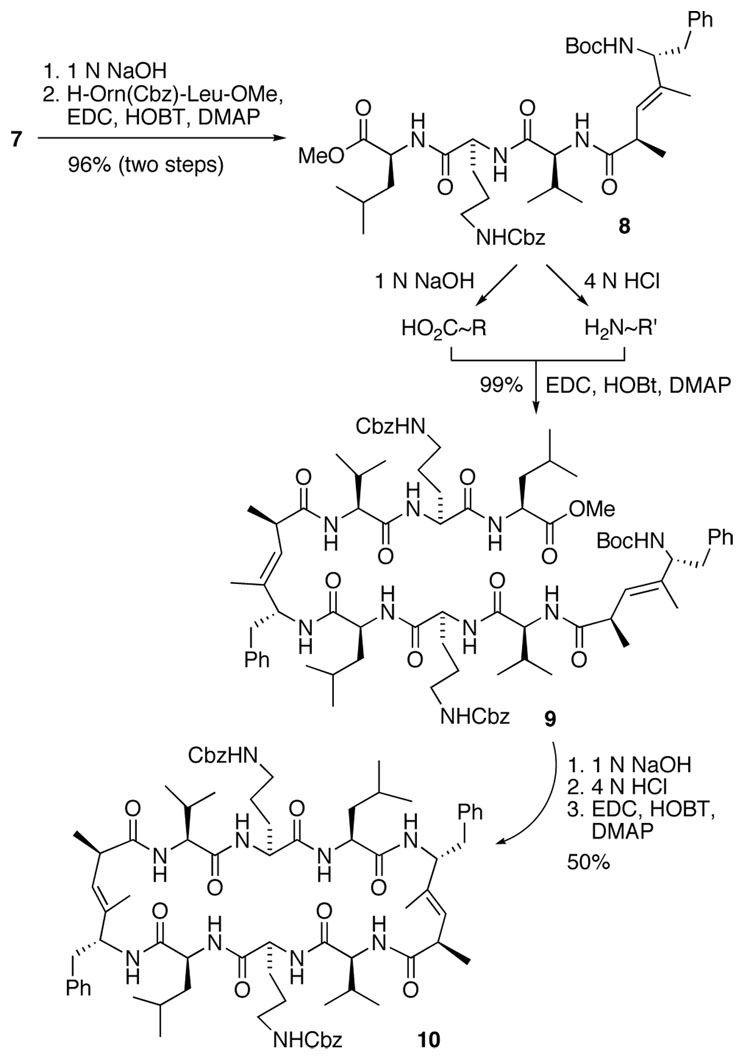
Saponification of pseudo-tripeptide 7 followed by fragment coupling with dipeptide H-Orn(Cbz)-Leu-OMe in the presence of EDC as a coupling reagent afforded the pseudo-pentapeptide 8 in 96% yield over two steps. We initially envisioned a one-pot dimerization-cyclization of 8; however, this approach resulted exclusively in the formation of cyclized pseudo-pentapeptide. In contrast, the stepwise coupling proceeded smoothly to give the pseudo-decapeptide 9 in excellent yield. Saponification of 9 and stepwise removal of the Boc protecting group followed by macrolactamization afforded the desired bis-Cbz-protected GS analogue 10 in 50% yield after preparative C18 RP HPLC purification (Scheme 2).
Scheme 2.
Fragment condensation and synthesis of pseudo-cyclodecapeptide 10
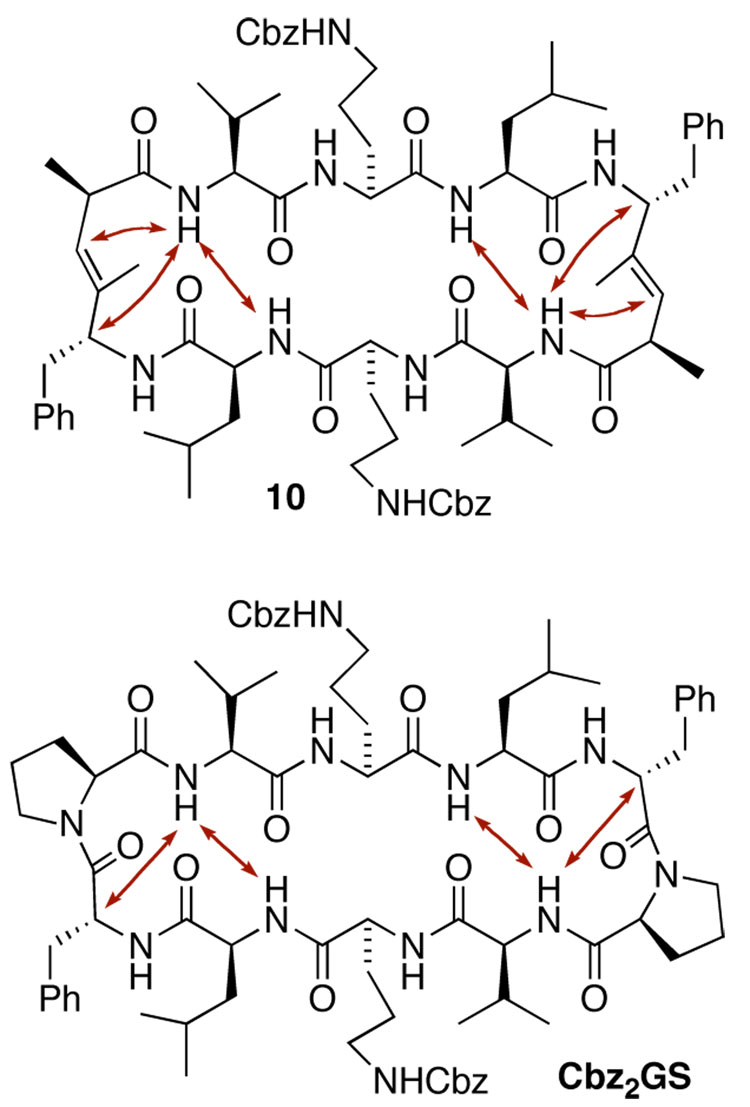
The chemical shifts of all amide protons in 10 were assigned using a combination of COSY, NOESY, HMQC and HMBC data sets collected in DMSO-d6 at 338 K, since some amide 1H NMR signals were obscured at 298 K. Variable temperature NMR was applied to probe the conformation of 10 in solution and determine the level of intramolecular hydrogen bonding (Table 1). Temperature shift coefficients for 10 were in close agreement with the values for bis-Cbz-protected GS (Cbz2GS).7 The NH shifts of Leu and Val residues in 10 showed small temperature coefficients of −1.9 and −1.8 ppb/K, respectively, whereas Orn-NH and DPhe-NH were solvent exposed, thus indicating a hydrogen bonding array typical for an antiparallel β-pleated sheet conformation.19 Furthermore, NOESY spectra showed transannular Leu-NH–Val-NH and Val-NH–DPhe-Hαcontacts for 10 in agreement to what was found for Cbz2GS (Figure 2). The resulting ten-membered intramolecular H-bonding interaction between the valine amide NH and the carbonyl group of the leucine residue is typical for a type II’ β-turn.20
Table 1.
Temperature Shift Coefficients (Δδ/ΔT) of Amide Protons in DMSO-d6 [ppb/K] for 10 and Cbz2GS
| Orn-NH | Leu-NH | Orn-δ-NH | Val-NH | DPhe-NH | |
|---|---|---|---|---|---|
| 10 | −4.8 | −1.9 | −6.3 | −1.8 | −5.9 |
| Cbz2GS | −6.7 | −3.8 | −8.6 | −2.2 | −11.8 |
Figure 2.
Observed NOEs for 10 and Cbz2GS.
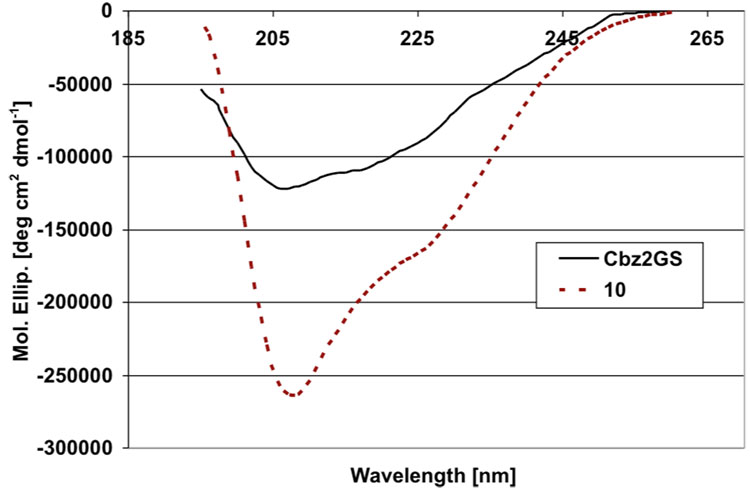
Further confirmation of the close match between the secondary structures of 10 and Cbz2GS was provided by circular dichroism (CD) spectra in EtOH (Figure 3). The strong negative band at ~205–225 nm and a shoulder in the region of 225–235 nm for both 10 and Cbz2GS is consistent with a combination of a type II’ β-turn and a β-sheet conformation in these compounds.21 This result, along with the data from NOESY and variable temperature NMR experiments, further confirmed that the methyl (E)-alkene dipeptide isostere replacements at the former DPhe-Pro positions strongly promoted the archetypical architecture of the parent peptide, gramicidin S. An MMFF-minimized structure for 1 that is in agreement with all experimental data is shown in Figure 4.22
Figure 3.
CD Spectra of 10 and Cbz2GS in EtOH.
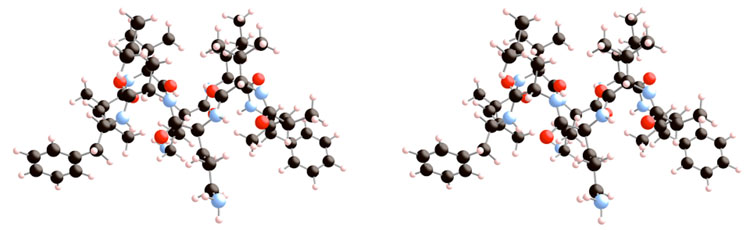
Figure 4.
Stereoview of the minimized structure of 1 derived from an MMFF conformational search algorithm.
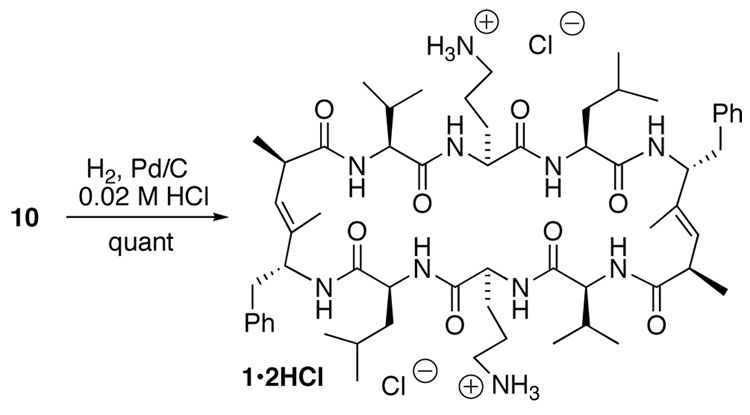
Our previous biological studies showed that free amine functions on the ornithine side chains were necessary to retain the antibacterial, antifungal, and hemolytic activities of GS.7 The Cbz protecting groups in 10 were successfully removed by hydrogenolysis in the presence of 10% Pd/C in a 0.02 M HCl/MeOH solution, without concomitant reduction of the trisubstituted (E)-alkene moieties (Scheme 3). As expected, the hydrochloride salt of 1 also exhibited functional mimicry of the natural product, with an MIC of ~20 µg/mL against Bacillus subtilis, and thus equipotent with GS hydrochloride (MIC ~15 µg/mL in the same assay).
Scheme 3.
Synthesis of the hydrochloride salt of [CH3]2GS (1•2HCl)
The development of proline mimics is of considerable current interest, due to the helix-breaking and unique conformational properties of this amino acid residue.23 We have now demonstrated that a trisubstituted (E)-alkene dipeptide isostere can serve as a bioisosteric replacement for the DPhe-Pro type II’ β-turn in the cyclopeptide antibiotic gramicidin S. The solution conformational analysis and the biological assay of analogues 10 and 1, respectively, provides a strong validation of our design principles. Furthermore, the hydrozirconation/Zr➩Zn transmetalation/imine addition methodology was key to a rapid synthetic access to the target compounds.
Supplementary Material
Experimental procedures and spectral data including copies of 1H and 13C NMR spectra for all new compounds, and 2D NMR spectra of 10 and Cbz2GS. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgment
This work was supported by the NIH/NIGMS P50 program (GM067082). J.X. gratefully acknowledges an Andrew W. Mellon Predoctoral Fellowship for support and Dr. Corey R. J. Stephenson (ETHZ) for helpful discussions.
References
- 1.(a) Kazmierski WM, Kenakin TP, Gudmundssona KS. Chem. Biol. Drug Des. 2006;67:13. doi: 10.1111/j.1747-0285.2005.00319.x. [DOI] [PubMed] [Google Scholar]; (b) Clark RJ, Fischer H, Dempster L, Daly NL, Rosengren KJ, Nevin ST, Meunier FA, Adams DJ, Craik DJ. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13767. doi: 10.1073/pnas.0504613102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ladner RC, Sato AK, Gorzelany J, De Souza M. Drug Disc. Today. 2004;9:525. doi: 10.1016/S1359-6446(04)03104-6. [DOI] [PubMed] [Google Scholar]
- 2.Conradi RA, Hilgers AR, Ho NFH, Burton PS. Pharm. Res. 1992;9:435. doi: 10.1023/a:1015867608405. [DOI] [PubMed] [Google Scholar]
- 3.(a) Wipf P, Fritch PC. J. Org. Chem. 1994;59:4875. [Google Scholar]; (b) Henninger T, Wipf P. In: Methods in Molecular Biology Peptidomimetics. Walker JM, Kazmierski WM, editors. Vol 23. Totowa: Humana Press; 1999. pp. 125–136. [Google Scholar]; (c) Mu Y, Stephenson CRJ, Kendall C, Saini SPS, Toma D, Ren S, Cai H, Strom SC, Day BW, Wipf P, Xie W. Mol. Pharmacol. 2005;68:403. doi: 10.1124/mol.105.013292. [DOI] [PubMed] [Google Scholar]; (d) Wipf P, Werner S, Woo GHC, Stephenson CRJ, Walczak MAA, Coleman CM, Twining LA. Tetrahedron. 2005;61:11488. [Google Scholar]; (e) Levinson N, Hinman R, Patil A, Stephenson CRJ, Werner S, Woo GHC, Xiao J, Wipf P, Lynch KW. RNA. 2006;12:925. doi: 10.1261/rna.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Wipf P, Xiao J, Jiang J, Belikova NA, Tyurin VA, Fink MP, Kagan VE. J. Am. Chem. Soc. 2005;127:12460. doi: 10.1021/ja053679l. [DOI] [PubMed] [Google Scholar]
- 4.(a) Tamamura H, Koh Y, Ueda S, Sasaki Y, Yamasaki T, Aoki M, Maeda K, Watai Y, Arikuni H, Otaka A, Mitsuya H, Fujii N. J. Med. Chem. 2003;46:1764. doi: 10.1021/jm020537i. [DOI] [PubMed] [Google Scholar]; (b) Yang H, Sheng XC, Harrington EM, Ackermann K, Garcia AM, Lewis MD. J. Org. Chem. 1999;64:242. doi: 10.1021/jo981892c. [DOI] [PubMed] [Google Scholar]; (c) Oishi S, Miyamoto K, Niida A, Yamamoto M, Ajito K, Tamamura H, Otaka A, Kuroda Y, Asai A, Fujii N. Tetrahedron. 2006;62:1416. [Google Scholar]
- 5.(a) Wipf P, Henninger TC, Geib SJ. J. Org. Chem. 1998;63:6088. doi: 10.1021/jo981057v. [DOI] [PubMed] [Google Scholar]; (b) Wipf P, Henninger TC. J. Org. Chem. 1997;62:1586. [Google Scholar]
- 6.Wadhwani P, Afonin S, Ieronimo M, Buerck J, Ulrich AS. J. Org. Chem. 2006;71:55. doi: 10.1021/jo051519m. [DOI] [PubMed] [Google Scholar]
- 7.Xiao J, Weisblum B, Wipf P. J. Am. Chem. Soc. 2005;127:5742. doi: 10.1021/ja051002s. [DOI] [PubMed] [Google Scholar]
- 8.Gauze GF, Brazhnikova MG. Am. Rev. Soviet Med. 1944;2:134. [Google Scholar]
- 9.See:Grotenbreg GM, Buizert AEM, Llamas-Saiz AL, Spalburg E, Van Hooft PAV, De Neeling AJ, Noort D, Van Raaij MJ, Van Der Marel GA, Overkleeft HS, Overhand M.J. Am. Chem. Soc 20061287559and references cited therein
- 10.(a) Yamada K, Unno M, Kobayashi K, Oku H, Yamamura H, Araki S, Matsumoto H, Katakai R, Kawai M. J. Am. Chem. Soc. 2002;124:12684. doi: 10.1021/ja020307t. [DOI] [PubMed] [Google Scholar]; (b) Doi M, Fujita S, Katsuya Y, Sasaki M, Taniguchi T, Hasegawa H. Arch. Biochem. Biophys. 2001;395:85. doi: 10.1006/abbi.2001.2567. [DOI] [PubMed] [Google Scholar]
- 11.(a) Kondejewski LH, Farmer SW, Wishart DS, Hancock REW, Hodges RS. Int. J. Pept. Protein Res. 1996;47:460. doi: 10.1111/j.1399-3011.1996.tb01096.x. [DOI] [PubMed] [Google Scholar]; (b) Kondejewski LH, Farmer SW, Wishart D, Kay CM, Hancock REW, Hodges RS. J. Biol. Chem. 1996;271:25261. doi: 10.1074/jbc.271.41.25261. [DOI] [PubMed] [Google Scholar]
- 12.Côté A, Boezio AA, Charette AB. Proc. Nat. Acad. Sci. U. S. A. 2004;101:5405. doi: 10.1073/pnas.0307096101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Wipf P, Xiao J, Geib SJ. Adv. Synth. Catal. 2005;347:1605. [Google Scholar]; (b) Wipf P, Xiao J. Org. Lett. 2005;7:103. doi: 10.1021/ol0477529. [DOI] [PubMed] [Google Scholar]
- 14.(a) Wipf P, Jahn H. Tetrahedron. 1996;52:12853. [Google Scholar]; (b) Wipf P, Kendall C. Top. Organomet. Chem. 2004;8:1. [Google Scholar]
- 15.(a) Wipf P, Kendall C, Stephenson CRJ. J. Am. Chem. Soc. 2001;123:5122. doi: 10.1021/ja0157494. [DOI] [PubMed] [Google Scholar]; (b) Wipf P, Kendall C, Stephenson CRJ. J. Am. Chem. Soc. 2003;125:761. doi: 10.1021/ja028092a. [DOI] [PubMed] [Google Scholar]
- 16.Dess DB, Martin JC. J. Org. Chem. 1983;48:4155. [Google Scholar]
- 17.Wipf P, Kim Y, Goldstein DM. J. Am. Chem. Soc. 1995;117:11106. [Google Scholar]
-
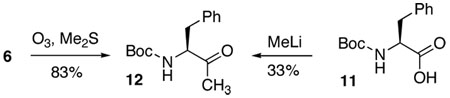
18.The structural assignment of 6 and 7 was based on the ozonolysis product 12, which was identical to the methylation product obtained from Boc-DPhe-OH (11) by HPLC co-injection (Pace RD, Kabalka GW. J. Org. Chem. 1995;60:4838.).
- 19.(a) Wipf P, Fritch PC, Geib SJ, Sefler AM. J. Am. Chem. Soc. 1998;120:4105. [Google Scholar]; (b) Imperiali B, Fisher SL, Moats RA, Prins TJ. J. Am. Chem. Soc. 1992;114:3182. [Google Scholar]; (c) Kessler H. Angew. Chem. Int. Ed. 1982;21:512. [Google Scholar]
- 20.Ball JB, Hughes RA, Alewood PF, Andrews PR. Tetrahedron. 1993;49:3467. [Google Scholar]
- 21.(a) Tamaki M, Akabori S, Muramatsu I. Bull. Chem. Soc. Jpn. 1993;66:3113. [Google Scholar]; (b) Woody RW. Chapter 17. In: Nakanishi K, Berova N, Woody RW, editors. Circular Dichroism. New York: VCH; 1994. pp. 473–496. [Google Scholar]
- 22.A 4300-step conformational equilibrium search was performed with Spartan 04. Irvine, CA: Wavefunction, Inc; [Google Scholar]
- 23.(a) Jenkins CL, Vasbinder MM, Miller SJ, Raines RT. Org. Lett. 2005;7:2619. doi: 10.1021/ol050780m. [DOI] [PubMed] [Google Scholar]; (b) Cordero FM, Pisaneschi F, Batista KM, Valenza S, Machetti F, Brandi A. J. Org. Chem. 2005;70:856. doi: 10.1021/jo0487653. [DOI] [PubMed] [Google Scholar]; (c) Wang XJ, Hart SA, Xu B, Mason MD, Goodell JR, Etzkorn FA. J. Org. Chem. 2003;68:2343. doi: 10.1021/jo026663b. [DOI] [PubMed] [Google Scholar]; (d) Halab L, Lubell WD. J. Am. Chem. Soc. 2002;124:2474. doi: 10.1021/ja012442w. [DOI] [PubMed] [Google Scholar]; (e) Otaka A, Katagiri F, Kinoshita T, Odagaki Y, Oishi S, Tamamura H, Hamanaka N, Fujii N. J. Org. Chem. 2002;67:6152. doi: 10.1021/jo025922u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental procedures and spectral data including copies of 1H and 13C NMR spectra for all new compounds, and 2D NMR spectra of 10 and Cbz2GS. This material is available free of charge via the Internet at http://pubs.acs.org.