Abstract
Two BODIPY-labeled colchicine derivatives were synthesized and shown to bind to tubulin but only partially inhibit tubulin polymerization in the presence of GTP. Cytotoxicity studies were carried out in HeLa, HepG2, Raji and Vero cells. Apoptosis-inducing properties were determined by caspase 3/7 activity and flow cytometry and interactions between the derivatives and tubulin were verified by fluorescence microscopy of living cells.
Colchicine, a natural product extracted from Colchicum autumnale, is used, despite serious side effects, as treatment of gout (Figure 1). Colchicine can bind to tubulin,1,2 initiating a conformational change of tubulin subunits that leads to rapid depolymerization of microtubules.3 In contrast, paclitaxel stabilizes the α,β-tubulin dimer by occupying a different binding site than colchicine.4
Figure 1.
Colchicine.
We desired to take advantage of the differential binding of colchicine to un-polymerized tubulin in order to track sub-cellular localization of tubulin dimers and small oligimers in living, non-permeablized cells. We were particularly interested in the flux of tubulin in neurons and the flagella of Chlamydomona; and in understanding the role of sequestration in normal function of tubulin structures in these cells.
Meeting the project goals requires a colchicine analog that is cell permeable, non-cytotoxic, tubulin binding, stable for extended microscopy studies (non-bleaching), excited at wavelengths compatible with living cells (green to red, not blue), of good quantum yield (to detect low abundance dimers), insensitive to environment, and minimally inhibiting of tubulin polymerization. This is a demanding set of requirements.
Prior approaches have used anti-tubulin antibodies or fluorescein-labeled colchicine (FC) to study the localization of tubulin in fixed cells.5–7 Unfortunately, spectrofluorometry experiments indicated that the anionic nature of FC resulted in binding properties different from those of unlabeled colchicine.8 Additionally, FC is sensitive to photo-bleaching, is highly pH sensitive, and has limited cellular permeability. These properties indicate FC is of low utility for long term microscopy studies. Both dansyllabeled colchicine (DC)9 and NBD-labeled colcemid (NBC)10,11 have been reported. Unfortunately both DC and NBC have low quantum yields and are highly environmentally sensitive. Additionally, DC has an excitation wavelength in the deep blue region. Together these properties render these two reagents of low utility for microscopy studies on living cells. Hence, there is a need for optimized fluorescent colchicine analogues.
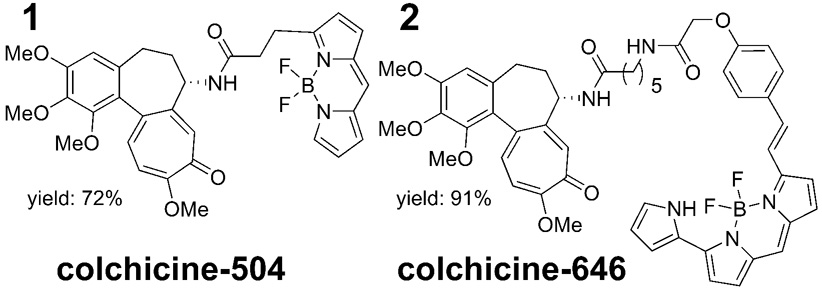
Herein, we report the synthesis and characterization of two pH-independent BODIPY-labeled colchicine derivatives, designated colchicine-504 and colchicine-646 designed to meet our requirements (Figure 2).
Figure 2.
BODIPY labeled colchicines.
The synthesis of both probes proceeded as planned. Deacetyl colchicine was prepared from colchicine using a published procedure.12,13 Deacetyl colchicine was then coupled with the corresponding BODIPY succinimidyl esters (Molecular Probes D2184 and D10001) in dimethylformamide, followed by purification using normal phase chromatography. Structure and purity were determined by NMR and LC-MS.14,15
Each conjugate exhibited a fluorescence spectra that was minimally perturbed from the originating dye and insensitive to solvent and pH.16 The dyes were chosen for minimal overlap between absorbance and emission spectra to give a low background signal for fluorescence based assays. Examples are shown in Figure 5 and Figure 6 (fluorescence polarization) and Figure 6 (fluorescence microscopy). Standard broadband filters were sufficient for visualization by the latter technique. Additionally both conjugates retained their high quantum yields and were photo-stable. Overall, the photophysical properties of each conjugate meet the project goals.
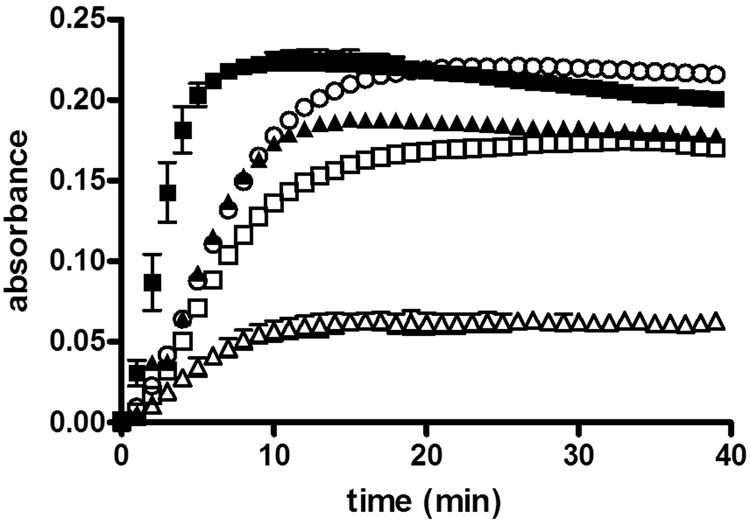
Figure 5.
Microtubule polymerization inhibition by colchicine-504 or colchicine-646.a
a Purified tubulin (7.2 µM) was treated with Paclitaxel (10 µM) ■, or colchicine (5 µM)△, or colchicine-504 (5 µM)□, or colchicine-646 ▲ (5 µM), or DMSO control O at 37°C in the presence of buffer (10 mM, GTP, 80 mM PIPES, 1 mM EGTA, 1 mM MgCl2, pH 6.8, total DMSO (0.5%)). The formation of microtubules was followed by measuring the absorbance at 340 nm; normalized values are the mean of two independent experiments.
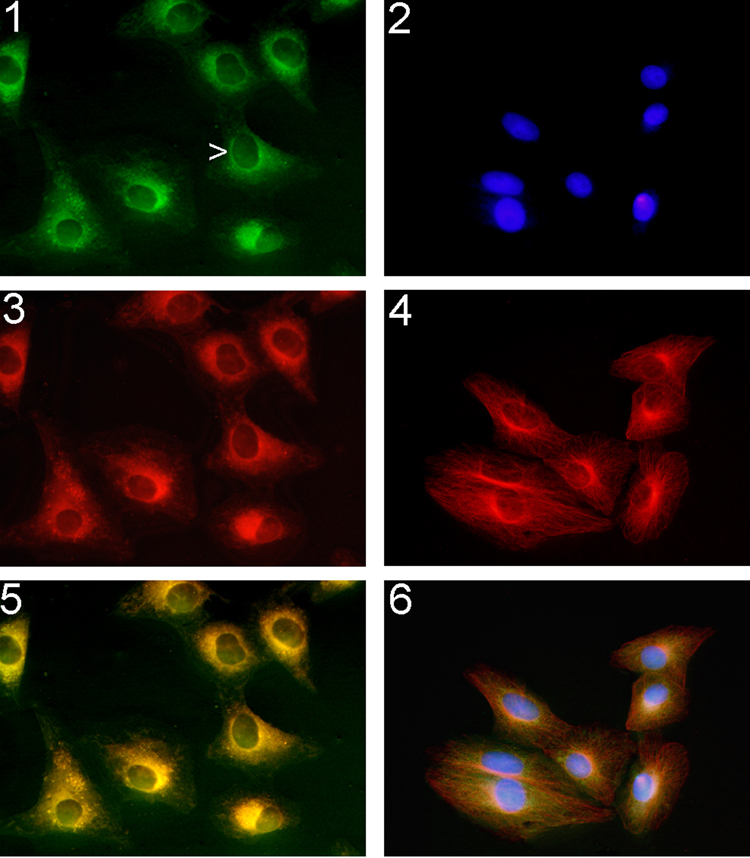
Figure 6.
1, 2, and 3: Living Vero cells treated colchicine analogs for 3h and visualized by fluorescence microscopy. 1) colchicine-504 (1 µM); 3) colchicine-646 (1 µM); 5) merge of 1) and 2); Staining was found in the perinuclear region of the Vero cell (arrow) and extended to the cell periphery. 2, 4, and 6: Fixed and permeabilized Vero cells were incubated with 2) 4',6-diamidino-2-phenylindole; 4) anti-α-tubulin antibody followed by a Texas-Red conjugated goat anti-mouse antibody; 6) colchicine-504 (1 µM) merge with 2) and 4).
Binding of colchicine-504 and colchicine-646 to tubulin was measured by fluorescence polarization (FP) (Figure 5). First, we determined the solubility of BODIPY-labeled compounds in PBS to ensure that both compounds were soluble under the FP conditions used. In contrast to colchicine, which is highly soluble in PBS + 5% DMSO, colchicine-504 and colchicine-646 were soluble at concentration of 32 µM and <10 µM, respectively (Table 1). The FP assay was carried out under the same conditions reported for tubulin competitive binding SPA assays.17,18
Table 1.
Cytotoxicity (EC50), permeability, and solubility of colchicine, colchicine-504, and colchicine-646.
| HeLa EC50 (µM)a |
HepG2 EC50 (µM)a |
Raji EC50 (µM)a |
Vero EC50 (µM)a |
Perme- abilityb (10− 6cm/s) |
Solu- bilityc (µM) |
|
|---|---|---|---|---|---|---|
| Col- chicine |
0.039 ±0.006 |
0.031 ±0.01 |
0.014 ±0.002 |
0.53 ±0.09 |
415 ±47 |
>250 |
| 504 | 0.36 ±0.07 |
0.18 ±0.09 |
0.138 ±0.02 |
2.3 (±0.9) |
108 ±24 |
32 ±15) |
| 646 | 1.9 ±0.7 |
1.2 ±0.6 |
0.65 ±0.19 |
>10 | <10 | <10 ±2 |
Viability was assessed in a 96 well format. Cells were plated at 2000 cells/well, and drugs were added after 12h. After 48 h, CellTiter-Glow solution was added, and cell viability was measured after 10 min. The EC50 values were obtained by fitting data to the following equation (y = min + (max − min)/1 + (x/Kd) Hill slope). Values reported are the means of two independent experiments done in triplicate.
Permeability (PAMPA) was measured as previously reported.23
Solubility (PBS + 5% DMSO) was assessed using Multiscreen Solubility assay (Millipore, 96 well plate). Briefly, each compound was serial diluted in PBS:acetonitrile (1:1) and absorbance was measured at 350 nm (calibration). Next, 5 µL of a 10 mM solution of colchicine, colchicine-504 or colchicine-646 was added to 95 µL PBS in a filter plate and agitated for 2h at room temperature. After filtration, absorbance at 350 nm was measured, and concentration was determined on the basis of calibration. Two independent experiments were done in duplicate.
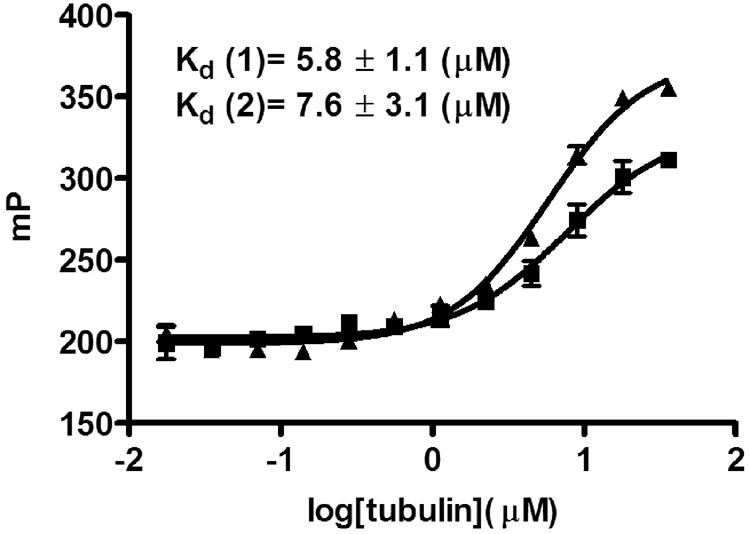
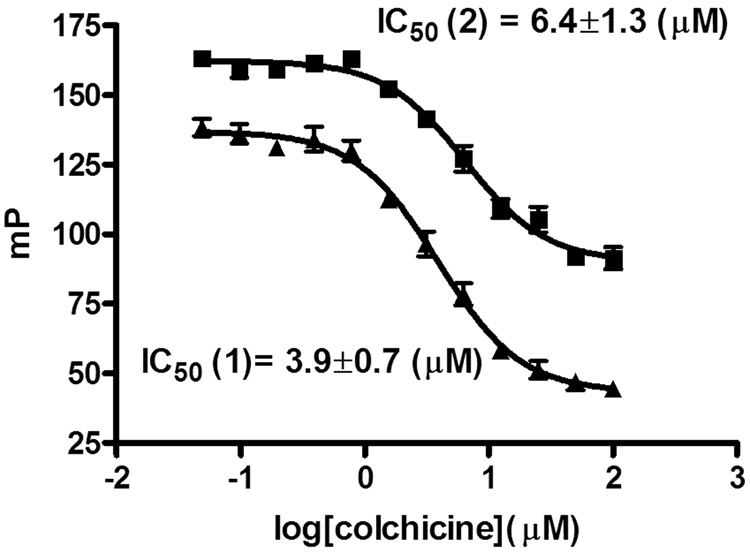
For both compounds saturable binding was measured with Kd values of 5.8 µM, (colchicine-504) and 7.6 µM (colchicine-646), respectively (Figure 3). These binding affinities were slightly higher than the reported values for FC5 (14.5 µM) and DC9 (10–20 µM). A binding study with colchicine-504 and BSA, used in the same concentration as tubulin, showed no unspecific binding of colchicine-504 (data not shown). A 10-fold higher concentration of colchicine-646 was used in respect colchicine-504 because of the relative lower quantum yield (Figure 4). The competition FP assay was carried out in the presence of 10 µM tubulin (roughly 2 × Kd). Both compounds showed reversible binding using colchicine as competitor. IC50 values of 3.9 µM (colchicine-504) and 6.4 µM (colchicine-646) were measured for colchicine under assembly conditions (1 mM GTP, 37°C) as well as under non-assembly conditions (no GTP, rt) verifying binding to tubulin and not preformed microtubles.19 IC50 values for colchicine between 90 nM and 6.5 µM were reported using [3H] colchicine using various amounts of tubulin.5,18 Recently, a tubulin-based FP for the paclitaxel binding site was reported20 but to our knowledge this is the first developed tubulin-based FP assay for the colchicine binding site.
Figure 3.
Direct binding assay with colchicine-504 ▲and colchicine-646 ■ to tubulin (fluorescence polarization).a
aAssay was carried out in black 384 well plate; tubulin was serially diluted from 72 µM to 0.035 µM in the presence of 20 µl buffer (10 mM, GTP19, 80 mM PIPES, 1 mM EGTA, 1 mM MgCl2, pH 6.8, 5% total DMSO) and colchicine-504 or colchicine-646 (10 nM or 100 nM, respectively). The binding was measured after 2h at 37°C19 using fluorescence polarization. The Kd values were obtained by fitting data to the following equation (y = min + (max − min)/1 + (x/Kd) Hill slope). Values are means of two experiments carried out in triplicate.
Figure 4.
Competition binding assay with colchicine and colchicine-504 ▲ and colchicine-646 ■ to tubulin (fluorescence polarization).a
a Assay was carried out in black 384 well plate; colchicine was serially diluted from 100 µM to 0.04 µM in the presence of 20 µl buffer (10 mM, GTP19, 80 mM PIPES, 1 mM EGTA, 1 mM MgCl2, pH 6.8), tubulin (10 µM), and colchicine-504 or colchicine-646 (10 nM or 100 nM, respectively). The binding was measured after 2h at 37°C19 using fluorescence polarization. The Kd values were obtained by fitting data to the following equation (y = min + (max − min)/1 + (x/Kd) Hill slope). Values are means of three experiments carried out in triplicate.
The biochemical activity of the colchicine-504 and colchicine-646 in comparison with colchicine was analyzed by studying depolymerization of microtubules in the presence of these drugs using the CytoDYNAMIX™ assay (Cytoskeleton) (Figure 5), following the manufacturer’s recommended protocol. Purified tubulin was treated in the presence of GTP with paclitaxel, colchicine, and colchicine-504 and colchicine-646. The degree of polymerization was followed over time, measuring light scattering at 340 nm, which is proportional to the concentration of microtubule polymer.21,22
The polymerization reaction of tubulin showed nucleation, growth, and steady state. In the presence of paclitaxel, microtubules formation was enhanced in comparison with untreated tubulin, due to the stabilization of theα,β-tubulin dimer. For colchicine derivatives, we observed a weak inhibition of tubulin polymerization for colchicine-646 and a stronger inhibition using compound colchicine-504 at 5 µM. Colchicine itself exhibited a very strong inhibition at this concentration. Because this is a low DMSO content assay (0.5%) solubility could have influenced the degree of inhibition of tubulin polymerization observed.
Colchicine and its derivatives were incubated at various concentrations with cultured HeLa, HepG2, Raji, and Vero cells to determine cell viability after 48 h of exposure (Table 1). A commercially available luminescence cell viability assay, CellTiter-Glo (Promega), was used to quantify the ATP in viable cells.
Colchicine is toxic in a low nanomolar concentration to HeLa, HepG2, and Raji cell lines. Interestingly, the EC50 value found for Vero cells was 530 nM. The cytotoxicity of the colchicine-504 and colchicine-646 was significantly less than that of colchicine. Both compounds were less toxic to Vero cells than to other cell lines tested. PAMPA studies showed both derivatives were less permeable than colchicine itself (Table 1). The decreased permeability of the colchicine analogues might contribute to their lower cytotoxicity values. Because we intend to apply these agents in living cells, we chose Vero, the line that exhibited the lowest overall toxicity for further characterization.
Colchicine is known to activate caspase 3/7 in various cells.24 We investigated the induction of apoptosis by measuring caspase 3/7 activity in Vero cells in the presence of colchicine and its derivatives (Table 2).
Table 2.
Cell cycle and apoptosis analysis of Vero cells treated with colchicine, colchicine-504 and colchicine-646.
| Compound | EC50a,b(µM) [Apoptosis] |
EC50a,b(µM) [Cell cycle arrest] |
|---|---|---|
| Colchicine | 0.39 ±0.13 | 0.27 ±0.09 |
| 504 | 2.6 ±0.7 | 0.96 ±0.28 |
| 646 | 10.5 ±2.3 | 6.8 ±3.1 |
Vero cells were treated with compounds in a dose-response manner and analyzed after 18 hours.
Cells were stained with propidium iodide (PI) using a CycleTEST Plus DNA reagent kit (Beckton Dickinson). EC50 values were determined by cell-cycle analysis representing the concentration for 50% cells arrested in G2/M phase. The EC50 values were obtained by fitting data to the following equation (y = min + (max − min)/1 + (x/Kd) Hill slope). Values are means of two independent experiments.
Cells were treated with Caspase-Glo (Promega) and measured after 1 hour. The EC50 values were obtained by fitting data to the following equation (y = min + (max − min)/1 + (x/Kd) Hill slope). Values are means of two independent experiments carried out in duplicate.
The EC50 (cytotoxicity) determined for colchicine and its fluorescent analogues using Vero cells was similar to the EC50 determined for the activation of caspase 3/7. For all compounds we observed a 3- to 4-fold increase in caspase 3/7 activity at higher concentrations. The apoptosis-inducing activity of colchicine and its derivatives was also characterized by cell cycle analysis. Vero cells were treated with compounds at various concentrations, incubated for 18 h, stained with propidium iodide, and analyzed by flow cytometry. An increase in the G2/M DNA content in cells treated with colchicine and its analogues was observed (Table 2). The compound concentrations necessary to arrest 50% of the cells in the G2/M phase was in the same range as the EC50 values determined for cytotoxicity and apoptosis.
To illustrate the permeability and selectivity of these analogues, we treated Vero cells with colchicine-504 and colchicine-646 for 3 h and visualized them by fluorescence microscopy. (Figure 6, 1, 3, and 5.)
Both compounds penetrated the cell membranes of living Vero cells and stained the perinuclear regions, a region known to have high concentrations of both tubulin and microtubules.6 Additional staining was observed on the endoplasmic reticulum and in the cytosol.5 The tubulin skeleton of fixed and permeabilized Vero cells were stained with 4',6-diamidino-2-phenylindole (Figure 6, 2), anti-α-tubulin antibody followed by a Texas-Red conjugated goat anti-mouse antibody (Figure 6, 4), and colchicine-504 merged with 1 and 4 (Figure 6, 6). Selective tubulin labeling of colchicine-504 was demonstrated by colocalization with antibody stain. Clearly both compounds are permeable and could functionally stain tubulin rich compartments at low concentrations.
In conclusion, we introduced two new BODIPY-labeled colchicine analogs with biochemical properties similar to those of colchicine. Because of their photo-stable and pH-independent fluorescent properties, these probes are be superior to FC for the analysis of the cellular distribution of tubulin. Currently work is ongoing in the application of these compounds to monitoring the dynamics of tubulin flux.
Supplementary Material
Acknowledgements
This work was supported by the American Lebanese and Syrian Associated Charities (ALSAC), and St. Jude Children’s Research Hospital. We thank Michele Connelly for culturing the cell lines used.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Hastie SB. Pharmacol Ther. 1991;51:377–401. doi: 10.1016/0163-7258(91)90067-v. [DOI] [PubMed] [Google Scholar]
- 2.Weisenberg RC, Borisy GG, Taylor EW. Biochemistry. 1968;7:4466–4479. doi: 10.1021/bi00852a043. [DOI] [PubMed] [Google Scholar]
- 3.Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 4.Nogales E, Wolf SG, Downing KH. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 5.Moll E, Manz B, Mocikat S, Zimmermann HP. Exp Cell Res. 1982;141:211–220. doi: 10.1016/0014-4827(82)90083-0. [DOI] [PubMed] [Google Scholar]
- 6.Albertini DF, Clark JI. Cell Biol Int Rep. 1981;5:387–397. doi: 10.1016/0309-1651(81)90009-6. [DOI] [PubMed] [Google Scholar]
- 7.Clark JI, Garland D. J Cell Biol. 1978;76:619–627. doi: 10.1083/jcb.76.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Craenenbroeck E, Engelborghs Y. Biochemistry. 1999;38:5082–5088. doi: 10.1021/bi9821925. [DOI] [PubMed] [Google Scholar]
- 9.Das L, Datta AB, Gupta S, Poddar A, Sengupta S, Janik ME, Bhattacharyya B. Biochemistry. 2005;44:3249–3258. doi: 10.1021/bi048211u. [DOI] [PubMed] [Google Scholar]
- 10.Hiratsuka T, Kato T. J Biol Chem. 1987;262:6318–6322. [PubMed] [Google Scholar]
- 11.Sengupta S, Puri KD, Surolia A, Roy S, Bhattacharyya B. Eur J Biochem. 1993;212:387–393. doi: 10.1111/j.1432-1033.1993.tb17673.x. [DOI] [PubMed] [Google Scholar]
- 12.Ducray P, Lebeau L, Mioskowski C. Helv. Chim. Acta. 1996;79:2346–2352. [Google Scholar]
- 13.Lagnoux D, Darbre T, Schmitz ML, Reymond JL. Chem. Eur. J. 2005;11:3941–3950. doi: 10.1002/chem.200401294. [DOI] [PubMed] [Google Scholar]
- 14.colchicine-504: 1H-NMR (400 MHz) δ= 7.22 (d, J. H., 1H), 7.17 (s, 1H), 7.03 (s, 1H), 6.95 (m, 1H), 6.75 (d, J = 10.7 Hz, 1H), 6.49 (m, 1H), 6.20 (m, 1H), 6.11 (s, 1H), 4.55 (m, 1H), 3.96 (s, 3H), 3.93 (s, 3H), 3.88 (s, 3H), 3.65 (s, 3H), 2.68-2.54 (m, 2H), 2.55 (m, 6H), 2.36-2.22 (m, 4H); MS calcd for C34H36BF2N3O6 (+H) 632.47, found 632.50.
- 15.colchicine-646: 1H-NMR (400 MHz) δ= 10.43 (bs, H., NH), 8.03 (bs, 1H, NH), 7.63-7.52 (m, 2H), 7.32-7.26 (m, 2H), 7.21-7.18 (m, 1H), 7.06-7.01 (m, 2H), 6.89-6.92 (m, 4H), 6.90-6.86 (m, 3H), 6.80 (d, J = 10.8 Hz, 1H), 6.47 (m, 1H), 6.38 (m, 1H), 6.13 (m, 1H), 5.30 (m, 1H), 3.97 (s, 3H), 3.92 (s, 3H), 3.89 (s, 3H), 3.63 (s, 3H), 3.47-3.41 (m, 1H), 3.26-3.18 (m, 1H), 4.58-4.51 (m, 3H), 2.61-2.30 (m, 3H), 2.21-2.06 (m, 3H), 1.48-1.71 (m, 4H), 1.23 (m, 2H); MS calcd for C49H50BF2N5O8 (+H) 886.76, found 886.69.
- 16.see supplemental information.
- 17.Tahir SK, Han EK, Credo B, Jae HS, Pietenpol JA, Scatena CD, Wu-Wong JR, Frost D, Sham H, Rosenberg SH, Ng SC. Cancer Res. 2001;61:5480–5485. [PubMed] [Google Scholar]
- 18.Zhang LH, Wu L, Raymon HK, Chen RS, Corral L, Shirley MA, Narla RK, Gamez J, Muller GW, Stirling DI, Bartlett JB, Schafer PH, Payvandi F. Cancer Res. 2006;66:951–959. doi: 10.1158/0008-5472.CAN-05-2083. [DOI] [PubMed] [Google Scholar]
- 19.No difference was observed for experiments carried out in the presence and absence of GTP and at rt or 37°C (data not shown).
- 20.Morais S, O'Malley S, Chen W, Mulchandani A. Anal Biochem. 2003;321:44–49. doi: 10.1016/s0003-2697(03)00436-6. [DOI] [PubMed] [Google Scholar]
- 21.Shelanski ML, Gaskin F, Cantor CR. Proc Natl Acad Sci U S A. 1973;70:765–768. doi: 10.1073/pnas.70.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JC, Timasheff SN. Biochemistry. 1977;16:1754–1764. doi: 10.1021/bi00627a037. [DOI] [PubMed] [Google Scholar]
- 23.Ruell JA, Tsinman KL, Avdeef A. European Journal of Pharmaceutical Sciences. 2003;20:393–402. doi: 10.1016/j.ejps.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Lam MS, Birkeland A, Riffel A, Montana L, Sullivan ME, Post JM. J Pharmacol Toxicol Methods. 2006;54:313–319. doi: 10.1016/j.vascn.2006.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.