Abstract
Substances containing a phenolic moiety are often metabolized to quinones whose high reactivity makes them difficult to study. Some of these precursors have clear health benefits, and some quinones themselves are used in cancer therapy, whereas others are deleterious. For example, dietary intake of phytoestrogen, genistein (Gen), seems to play a preventive role in breast cancer (BC) whereas prolonged exposure to chemically similar mammalian estrogens is clearly associated with elevated incidence of BC. Although both can be metabolized to reactive quinones, the catechol estrogen quinones (CEQs) modify DNA by redox cycling and/or depurination via a Michael addition. Here, we report an investigation of the chemical reactivity of Gen and estrone quinones to determine the chemical differences in of these two biologically important molecules. The catechol genistein quinone (CGenQ), has a half life of 4 ± 1 s under physiological condition, as determined by glutathione trapping. It disappears by reacting with H2O to give a dihydrate, CGenQ·(H2O)2, whose structure was proved by NMR. Under reductive conditions, CGenQ·(H2O)2 is readily reduced to reform the catechol genistein (CGen). This reversible oxidation of CGen to CGenQ and the prompt moderation of its reactivity by hydration to CGenQ·(H2O)2 effectively moderates any redox cycling or depurination reaction of CGenQ with DNA. Catechol estrogen quinones, on the other hand, are sufficiently long-lived that they can damage DNA via a Michael addition or by redox cycling. Although the reactivity of CEQ in a nonaqueous solvent is similar to that of CGenQ, its reactivity in aqueous media with the free Ade base is more than 600 times that of CGenQ. These results suggest that rapid hydration of a quinone can moderate its reactivity toward biomolecules, allowing them to express, for example, estrogen-like properties without exhibiting the deleterious properties of redox cycling or directly damaging DNA via depurination reactions.
Introduction
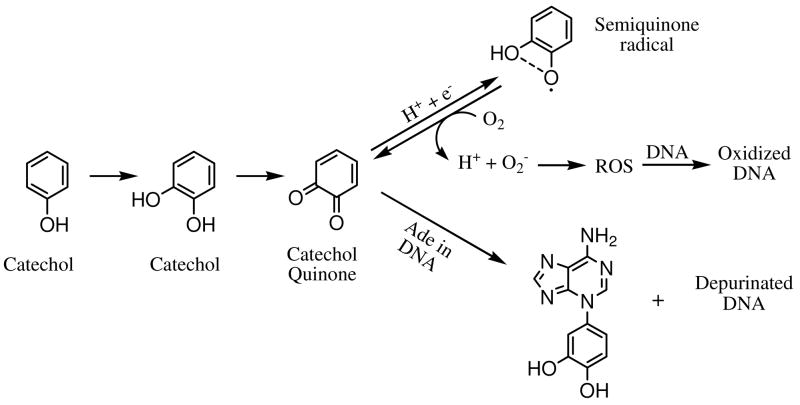
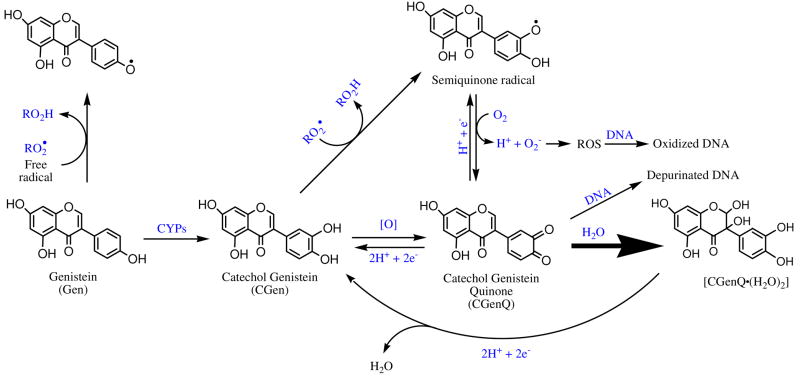
Quinones are commonly produced by oxidation of many natural products (e.g., estrogens, phytoestrogens) and related compounds. The usual substrates, an aromatic compound containing oxygen substituents (e.g. phenols, hydroquinones, catechols), become quinones by the action of monooxygenase or peroxidase.1 Among the quinones to which humans are exposed are those from (1) benzene and poly-aromatic hydrocarbons (PAH),2–9 (2) antioxidants from dietary intake of soy foods, red wine, etc,10–14 (3) xenobiotic drugs for anti-tumor and anti-biotic therapy15–17, and (4) xenoestrogens and endogenous estrogens used in hormone replacement therapy.18–20 Quinones are often toxic, eliciting a variety of cytotoxic and genotoxic effects in vivo.21 For example, quinones are redox-active, forming reactive oxygen species (ROS) via redox cycling and causing oxidative damage to DNA and proteins.22–24 Moreover, quinones are electrophiles and can effect serious cell damage via direct covalent modification of DNA and/or proteins.12, 24–26 To illustrate ROS generation and direct DNA modification, we show in Scheme 1 example reactions involving phenol, catechol, and catechol quinone.
Scheme 1.
Mechanisms for quinone-induced carcinogenesis, either indirectly via ROS or directly via Michael addition and depurination.
Owing to widespread human exposure to quinones and their important implications in carcinogenesis, we hypothesize that the chemical properties of quinone species are a starting point to understand their biological properties. Our assessment makes use of two model quinones, one generated from the phytoestrogen, genistein (Gen), and the other from the mammalian estrogen, estrone (E1). Estrogens, E1 and 17β-estradiol (E2), are potent sex hormones in mammals, whereas Gen is an important member of the isoflavone family and a phytoestrogen, for which there is a some evidence that dietary intake moderates the risk of hormone-related cancers.27 These materials exert biological effects via binding to estrogen receptors (ERs).28–30 Although both have a phenol substructure that can be oxidized to a catechol and ultimately to a potentially toxic catechol quinone31–35, they do exhibit some significant differences in their biological properties. Both Gen and estrogens can serve as antioxidants that scavenge free radicals36–45 presumably by donating hydrogen atoms from the phenolic hydroxyl groups, forming ArO• radicals.46 Although the catechols are good antioxidants47, owing to stabilization of the catecholic ArO• radical by hydrogen bonding with the adjacent hydroxyl group46, the ArO• radical is also a semiquinone radical (see Scheme 1) that can be reduced back to catechol quinones, producing ROS and causing oxidative damage to DNA and proteins.
From the perspective of human health, the balance of chemopreventive and chemopromotive effects of quinones is highly variable, depending on chemical structure. The quinones of Gen and estrogens are appropriate examples. Epidemiologic and experimental evidence reveals that prolonged exposure to estrogens is associated with elevated incidence of breast cancer (BC), leading to significant concerns about hormone replacement therapy.48–50 On the other hand, dietary intake of Gen isoflavones from soy is a useful source of antioxidants and may be linked with the lower BC incidence in Asian women. Nevertheless, its protective effects have not been conclusively established.51–56
Although there is a clear association between estrogen exposure and BC risk, the mechanism whereby estrogens contribute to BC remains to be firmly established. One view is that ER-mediated cell proliferation may account for the correlation.57–62 Another is that a competitive mechanism focused on the genotoxicity of estrogen quinones may actually initiate BC. Extensive cell-culture and animal studies support estrogen-induced mutations through either removal of DNA bases25, 63–68 or generation of ROS via their endogenous catechol estrogen quinone metabolites,21, 26, 58, 64, 69, 70. According to the genotoxic mechanism, endogenous estrogens are oxidized to catechol estrogens (CE) to form either 2-hydroxyestrogen (2-OH-E) or 4-hydroxyestrogen (4-OH-E) via phase I metabolism mediated by P450 enzymes. 2-OH-E and 4-OH-E are subsequently oxidized to catechol estrogen-2,3-quinone (CE-2,3-Q) and 3,4-quinone (CE-3,4-Q), respectively. These CEQs react with nucleophilic adenine (Ade) and/or guanine (Gua) bases by a 1,4-Michael addition, often leading to rapid loss of the purine by glycosyl-bond cleavage and leaving an abasic site in the DNA. Although these sites can be repaired enzymatically, an overabundance or error-prone repair may lead to DNA mutations and ultimately cancer. Do similar oxidative processes occur with Gen, forming catechol genistein (CGen) and ultimately genistein quinones (CGenQ)? As of yet there are no answers to this question.
The stability of the various quinones may explain the balance of salutary and deleterious effects of the oxidation products of Gen and estrone. Our specific hypothesis is that longer-lived, more stable quinones have more time to participate in both direct and indirect DNA damage whereas more reactive quinones react rapidly with solvent. Supporting this idea is the greater half life of the more genotoxic CE-3,4-Q compared to the 2,3-isomer under physiological conditions.71 To test further the hypothesis, we investigated the reactivity of both CGenQ and CE1Qs, employing nuclear magnetic resonance (NMR) and liquid-chromatography-electrospray ionization mass spectrometry (LC/MS). Using the approach of glutathione trapping, first proposed by Bolton and coworkers71, we determined the half life of the quinone from genistein, characterized the products of both its reaction in H2O and its subsequent reduction, and conducted analogous analyses for the quinones of estrone. We also obtained data that allow comparisons of the reactivity of both quinones in reaction with the Ade free base, under aqueous and nonaqueous conditions, and with calf thymus DNA. We are intrigued by the prospect that the deleterious effect of certain quinones are counterbalanced by the salutary effects of others, possibly promoting a “fight fire with fire” strategy whereby one quinone precursor may serve as a sacrificial agent to lower the toxicity of quinones from another.
Experimental Section
Materials
Oxone, 2-iodobenzoic acid (IBA), adenine (Ade), disodium hydrogen phosphate (Na2HPO4), glutathione (GSH), dimethyl formamide (DMF), trifluoacetic acid (TFA, LC/MS grade), formic acid (FA, LC/MS grade), ascorbic acid, Omnisolv acetonitrile (ACN), Omnisolv methanol (MeOH) and ethanol (EtOH) were purchased from Sigma-Aldrich (St. Louis, MO). E1 was purchased from Steraloids (Newport, RI). Gen was purchased from Acros (Morris Plains, NJ). D2O, deuterated d7-DMF and uniformly 15N-labeled (U-15N) adenine 5′-monophosphate (AMP) were from Cambridge Isotope Laboratory (Andover, MA). Calf thymus DNA was purchased from USB Corp (Cleveland, OH). Focus solid-phase extraction (SPE) cartridges (50 mg sorbent, 6 mL load capacity) were from Varian Inc (Palo Alto, CA). Milli-Q water was used for LC/MS and SPE.
The oxidant, 2-iodoxybenzoic acid (IBX), was synthesized from IBA and oxone, as previously described.72 The U-15N-labelled internal standard (IS), 4-hydroxyestrone-1-N3(U-15N)Ade (4-OH-E1-1-N3(U-15N)Ade), was synthesized and characterized, as reported recently.73 Deuterated d7-DMF was used in NMR experiments.
Preparation of Catechol Quinones
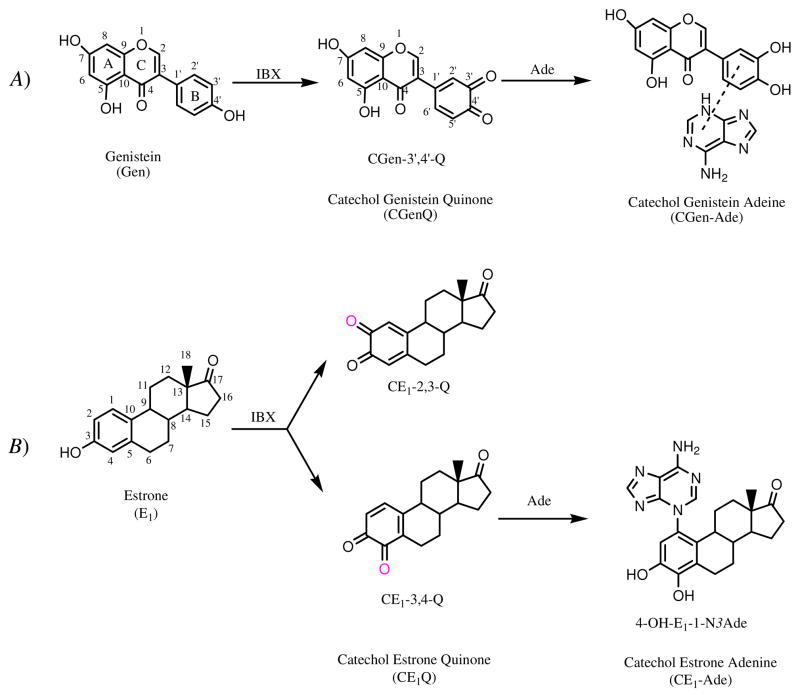
To form the catechol quinone product of Gen, IBX (1 mg) and Gen (1 mg) were mixed in 1 mL of DMF and allowed to react for 30 min. Catechol genistein-3′,4′-quinone (CGen-3′,4′-Q) was obtained and was used as such. An identical procedure was used to prepare simultaneously two catechol estrone quinones: catechol estrone-2,3-quinone (CE1-2,3-Q) and catechol estrone-3,4-quinone (CE1-3,4-Q) (see Scheme 2 for the structures of Gen, E1, CGen-3′,4′-Q, CE1-2,3-Q and CE1-3,4-Q). Given that CGen-3′,4′-Q is the only quinone species derived from Gen, it will be abbreviated simply as CGenQ in this paper.
Scheme 2.
A) Synthesis of CGen-3′,4′-Q and its CGen-Ade adducts from Gen. IBX oxidizes Gen at ring B, forming a single CGenQ product, CGen-3′,4′-Q. CGen-3′,4′-Q reacts with Ade and forms several CGen-Ade adducts. B) Synthesis of CE1-2,3-Q, CE1-3,4-Q and their CE1-Ade adducts from E1. IBX oxidizes E1, forming two CE1Qs: CE1-2,3-Q and CE1-3,4-Q. CE1Qs react with Ade to form 4-OH-E1-1-N3Ade and other CE1-Ade adducts, whose structures are not shown.
Preparation of Catechol Adenine Adducts
To explore the quinone chemistry, a general procedure of synthesizing the quinones and studying their reactivity with adenine was adopted (Scheme 2). (1) To prepare catechol adenine adducts in the absence of H2O, freshly prepared quinones (1 mg Gen or E1 + 1 mg IBX in 1 mL DMF) were mixed with Ade free base (1 mg) and reacted for 3 h. (2) To prepare adducts in predominantly aqueous media, 50 μL of DMF containing freshly prepared quinones was replaced with 100 μL of ACN. Five μL of the quinone(s) in ACN were added to 1 mL of a Na2HPO4 (50 mM, pH 7.4) solution containing Ade free base (1 mg) and incubated at 37 °C overnight. (3) To measure the reactivity with double-stranded DNA, the Ade free base in procedure (2) was replaced with calf thymus DNA (1 mg).
The products obtained from the reaction with the Ade free base were diluted by 500 fold with H2O/ACN/FA (93.5/6.5/0.1%) in preparation for LC/MS analysis. Cleanup steps were carried out for products obtained from the reaction with calf thymus DNA. To precipitate DNA, 2.5 volumes of EtOH were added, and the solution kept overnight at −20 °C. The suspension was centrifuged at 14 k/rpm, 4 °C for 10 min, the supernatant isolated, and the solvent removed under reduced pressure. The analyte was reconstituted in 3 mL of H2O/MeOH (2/1, v/v). The U-15N-labeled internal standard, 4-OH-E1-1-N3(U-15N)Ade (1 pmole), was added, and the solution submitted to SPE cleanup. The SPE cartridge was conditioned with 5 mL MeOH followed by 5 mL H2O. The analyte was loaded and washed with 5 mL H2O followed by 5 mL 10% ACN. The DNA adducts were eluted with 4 × 1 mL MeOH/ACN/H2O/TFA (30/60/10/0.1%). The eluent solvent was removed under reduced pressure, reconstituted in 20 μL of H2O/ACN/FA (93.5/6.5/0.1%) in preparation for LC/MS analysis. Samples in 5 μL were loaded for LC/MS analysis for analysis of the various products.
Kinetic Experiments with GSH Trapping
Freshly prepared quinones from IBX + Gen (or E1) (500 μg in 0.50 mL DMF for 30 min) were added to 9.5 mL Na2HPO4 buffer at 37 °C. Aliquots of 200 μL was removed at various intervals and combined with 200 μL GSH (10 mM in 50 mM Na2HPO4 buffer) at 37 °C. The aliquots were diluted by 100 fold with H2O/ACN/FA (93.5/6.5/0.1%) and analyzed by LC/MS. Gen was used as is an internal standard to quantify the levels of catechol quinones trapped with GSH.
Preparation of Catechol from Catechol Quinones
To obtain catechols from corresponding catechol quinones, five stoichiometric amounts of ascorbic acid were added to freshly prepared quinones in DMF.
Liquid Chromatographic Mass Spectrometric (LC/MS) Analysis
Microflow LC/MS and LC tandem MS (LC/MS/MS) were conducted on an LCQ Deca quadrupole ion-trap mass spectrometer (Thermo-Fisher, San Jose, CA) coupled to a capillary LC (Waters Corp., Milford, MA). An uncoated 360/75 μm OD/ID fused-silica capillary column with 15 μm PicoFrit™ nano tip (New Objective Inc., Woburn, MA) was custom-packed with C18(2) particles (Luna 3 μm 100 Å, Phenomenex, Torrance, CA) to afford a column that was ~ 12 cm in length. The sample solution was loaded and analyzed with solvent A (97% H2O/3% ACN/0.1% FA) and B (3% H2O/97% ACN/0.1% FA) starting with 97% A for 8 min and then a 37 min linear gradient to 100% B. The eluent with a total flow rate of 8 μL/min was split before the column to achieve a flow rate of 270 nL/min at the tip.
Sample ions were introduced with electrospray ionization (ESI) into the mass spectrometer operated in the positive-ion mode. The spray voltage was 1.8–2.5 kV, and no nitrogen sheath gas was used. The maximum time for injection of ions into the trap was 150 ms, and the number of microscans was one. The mass range for the mass spectrometer scan was m/z 100 to 1000 unless otherwise noted. For MS2 experiments, the collision energy was optimized at 40% of the maximum energy available (~ 5 eV).
When accurate mass measurements were needed, LC/MS was carried out with an ion-trap/FT-ICR mass spectrometer (LTQ-FT, Thermo Fisher, San Jose, CA). A resolving power of the FT component was 100,000 (at m/z 400), and the mass range for scanning was from m/z 100–1000.
NMR Analysis
1H NMR measurements were performed on an INOVA-500 (Varian, Inc., Palo Alto, CA) at 25 °C. The 1H NMR assignments were established by Correlation Spectroscopy (COSY), gradient Heteronuclear Multiple Quantum Coherence (gHMQC) and gradient Heteronuclear Multiple Bond Coherence (gHMBC). 1H and 13C results were referenced to d7-DMF. The NMR characterization of CGenQ was performed after 30 min of reaction of 1 mg Gen and 1 mg IBX in 0.5 mL d7-DMF. The two quinone products (CE1-2,3-Q and CE1-3,4-Q) were prepared similarly. For the characterization of CGenQ·(H2O)2 adduct, 1.6 mg Gen and 3.2 mg IBX were reacted in 0.4 mL d7-DMF for 3 h, 0.2 mL D2O was added, and the solution vortexed. For the kinetic measurements for the formation of CGenQ or CE1Q, 1H NMR spectra were collected under quantitative conditions by using 8 transients, a 70° flip angle pulse width and a spectral width of 4820 Hz. The acquisition started after mixing 1 mg Gen (or E1) and 1 mg IBX in 1 mL d7-DMF and was repeated at various intervals at constant temperature (25 °C).
Results and Discussion
(1) Formation and Stability of CGenQ or CE1Q
The product formed in the IBX oxidation of genistein (Gen) in DMF is strikingly simple (Table 1 summarizes the 1H and 13C chemical shifts for the product). The oxidation occurs at the 3′ and 4′ positions of ring B, resulting in the formation of CGen-3′,4′-Q (Scheme 2A). No detectable oxidation of the A ring occurs. Given the deleterious effects of long-term exposure to estrogens, we reinvestigated the formation of CE1Qs, using IBX oxidation to prepare the materials.74 The products of the reaction are CE1-2,3-Q and CE1-3,4-Q at a ratio of 60:40, according to NMR (Scheme 2B, see Figure S1 in Supporting Information for the NMR data). Both classes of quinones, CGenQ and CE1Qs, are relatively stable in DMF. The disappearance of both Gen and E1 quinones follow 2nd-order kinetics (Figure S2 in Supporting Information) with rate constants of 3.3×10−3 and 6.4×10−3 M−1 s−1, respectively. The smaller rate constant for the Gen-quinone reaction accounts for the lower amount of CGenQ, approximately 40% of the total CE1Qs (CE1-2,3-Q + CE1-3,4-Q), after 30 min of reaction in DMF.
Table 1.
1H and 13C NMR assignment for CGen-3′,4′-Q (structure in Scheme 3).
| Position | 1Ha | 13Cb | 1H-13Cc | |
|---|---|---|---|---|
| Chemical shift (ppm) | Coupling Constant (Hz) | Chemical shift (ppm) | 13C Chemical shift (ppm) and corresponding position | |
| 2 | 8.79 (s) | - | 154.5 | 116.2 (3), 140.4 (1′), 154.8 (9), 176.7 (4) |
| 6 | 6.37 (d) | 2.1 | 96.7 | 91.3 (8), 101.5 (10), 159.5 (5), 162.5 (7) |
| 8 | 6.52 (d) | 2.1 | 91.3 | 96.7 (6), 101.5 (10), 154.8 (9), 162.5 (7) |
| 2′ | 6.84 (d) | 2.2 | 124.9 | 116.2 (3), 137.7 (6′), 176.9 (4′) |
| 5′ | 6.48 (d) | 10.2 | 126.2 | 140.4 (1′), 176.5 (3′) |
| 6′ | 7.59 (dd) | 10.3, 2.2 | 137.7 | 124.9 (2′), 176.9 (4′) |
1H peak splitting (s, singlet; d, doublet; dd: doublet of doublet).
Determined by gradient Heteronuclear Multiple Quantum Coherence (gHMQC).
Determined by gradient Heteronuclear Multiple Bond Coherence (gHMBC).
(2) Half Life of Catechol Genistein Quinone in water
Unlike in DMF, quinones are labile in water under physiological conditions. Bolton and coworkers75 demonstrated that glutathione (GSH) trapping can be used for measuring the half lives of quinones; specifically, CE-2,3-Q, CE-3,4-Q71 and raloxifene, in H2O. GSH is a strong nucleophile that reacts with electrophilic catechol quinones via a Michael addition to form covalently bound adduct(s). We adopted this GSH-trapping protocol to determine the half life of CGenQ; GSH binds to CGenQ to form predominantly two mono GSH adducts. Analogous to the reaction of estrogens, the binding sites are the 2′ and 5′ position of CGenQ, giving 3′-hydroxygenistein-2′-SG (3′-OH-Gen-2′-SG) and 3′-hydroxygenistein-5′-SG (3′-OH-Gen-5′-SG). In addition to the mono GSH adducts, a small fraction of CGenQ reacts to add two GSH molecules whereby the second GSH molecule binds to the 2′ or 5′ position of CGenQ to afford 3′-hydroxygenistein-2′,5′-diSG (3′-OH-Gen-2′,5′-diSG). Simple addition occurs to conjugate the quinones in the first step of reaction with GSH, yielding presumably 3′-OH-Gen-2′-SG and 3′-OH-Gen-5′-SG of M.W. 591, as determined by ESI MS. The 3′-OH-Gen-2′,5′-diSG conjugate has a molecular weight of 896, two mass units less than would expected from a simple addition of a second GSH. To add a second GSH, we propose that 3′-OH-Gen-2′-SG and 3′-OH-Gen-5′-SG are re-oxidized to catechol quinones, CGen-3′,4′-Q-2′-SG and CGen-3′,4′-Q-5′-SG (M.W. 589), respectively. We confirmed the formation of these MW-589 species with LC/MS analyses, supporting that conjugation of these intermediate quinones with a second GSH to afford 3′-OH-Gen-2′,5′-diSG.
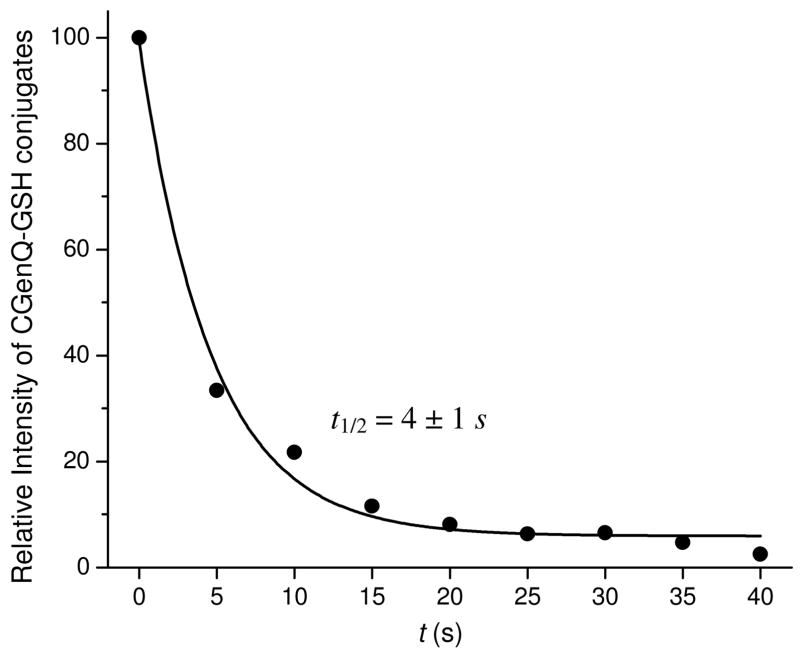
To measure the half life of CGen-3′,4′-Q, we followed the time course for 3′-OH-Gen-2′-SG and 3′-OH-Gen-5′-SG by using their mass spectral signal intensities, summing and normalizing them to 100% at t = 0. Diluting the final CGenQ/GSH mixture by 100 fold prior to analysis prevented additional oxidation of residual Gen by IBX during the LC/MS analysis. To quantify the GSH adducts, the signal for unreacted Gen was used as an IS (see Figure 1 for the reaction of CGenQ in H2O trapped by GSH). The data, when fit with a single exponential function, reveal that the half life of CGenQ is 4 ± 1 s at 37 °C, pH 7.4. In contrast, the half lives of CE1-2,3-Q and CE1-3,4-Q are considerably longer at 42 s and 730 s (12.2 min), respectively.71
Figure 1.
Measurement of the half lives of CGen-3′,4′-Q with GSH trapping. Quinones were deactivated in H2O at 37 °C, pH 7.4. Aliquots were removed and combined with GSH at various time. Data were fitted with an exponential function.
(3) Products of the reaction of Quinones in H2O
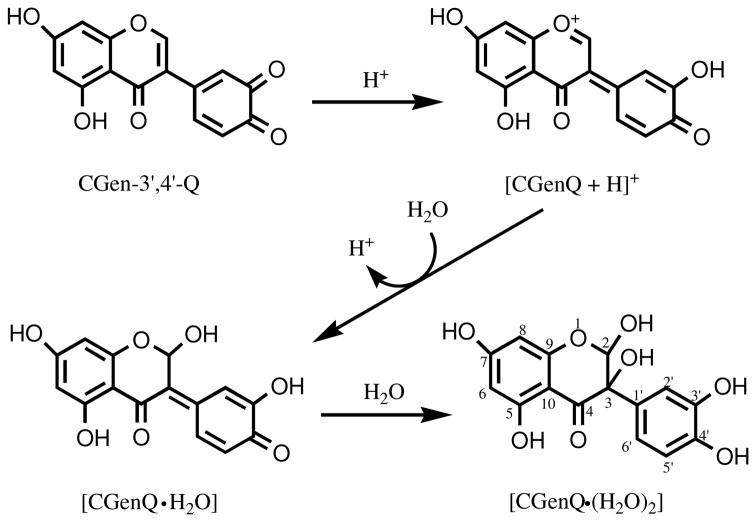
To characterize the decay product(s) of CGenQ in H2O, we mixed d7-DMF containing freshly prepared CGenQ with D2O (v/v, 2:1) and monitored the products by NMR (for a summary of the 1H and 13C chemical shifts for the product of CGenQ reacting with water, see Table 2). The NMR results are consistent with CGenQ being converted to a dihydrate CGenQ·(H2O)2 (see Scheme 3 for structures). CGenQ·(H2O)2 is stable in H2O, and no additional reaction of the dihydrate occurs, even for a period of 48 h. A mechanism for converting CGenQ to CGenQ·(H2O)2 (Scheme 3) shows that the O1 ether and the C2-C3 double bond in the C ring of genistein provide a driving force for facile protonation of CGenQ, whose conjugate acid is stabilized as an intermediate oxonium ion with a high degree of conjugation. The oxonium ion undergoes subsequent hydration to CGenQ·H2O. This hydration reaction and the overall stabilization it provides may be characteristics of many quinones that are formed from isoflavones and other health-promoting materials containing phenolic moieties.
Table 2.
1H, 13C NMR assignment for CGenQ·(H2O)2 (structure in Scheme 3).
| Position | 1H a | 13C b | 1H-13C c | |
|---|---|---|---|---|
| Chemical shift (ppm) | Coupling Constant (Hz) | Chemical shift (ppm) | 13C Chemical shift (ppm) and corresponding position | |
| 2 | 5.46 (s) | - | 91.6 | 92.9 (3) |
| 6 | 5.97 (d) | 1.8 | 95.9 | 90.1 (8), 101.9 (10), 157.6 (5/7), 167.9 (9) |
| 8 | 6.18 (d) | 1.8 | 90.1 | 95.9 (6), 101.9 (10), 167.9 (9), 174.5 (4) |
| 2′ | 7.06 (d) | 2.1 | 112.5 | 92.9 (3), 116.3 (6′), 144.4 (4′) |
| 5′ | 6.78 (d) | 8.5 | 114.9 | 127.3 (1′), 144.5 (3′) |
| 6′ | 6.89 (dd) | 8.5, 2.1 | 116.3 | 112.5 (2′), 144.4 (4′) |
1H peak splitting (s, singlet; d, doublet; dd: doublet of doublet).
Determined by gradient Heteronuclear Multiple Quantum Coherence (gHMQC).
Determined by gradient Heteronuclear Multiple Bond Coherence (gHMBC).
Scheme 3.
Proposed mechanism for the reaction of CGenQ, Gen-3′,4′-Q, with H2O to form CGenQ·(H2O)2.
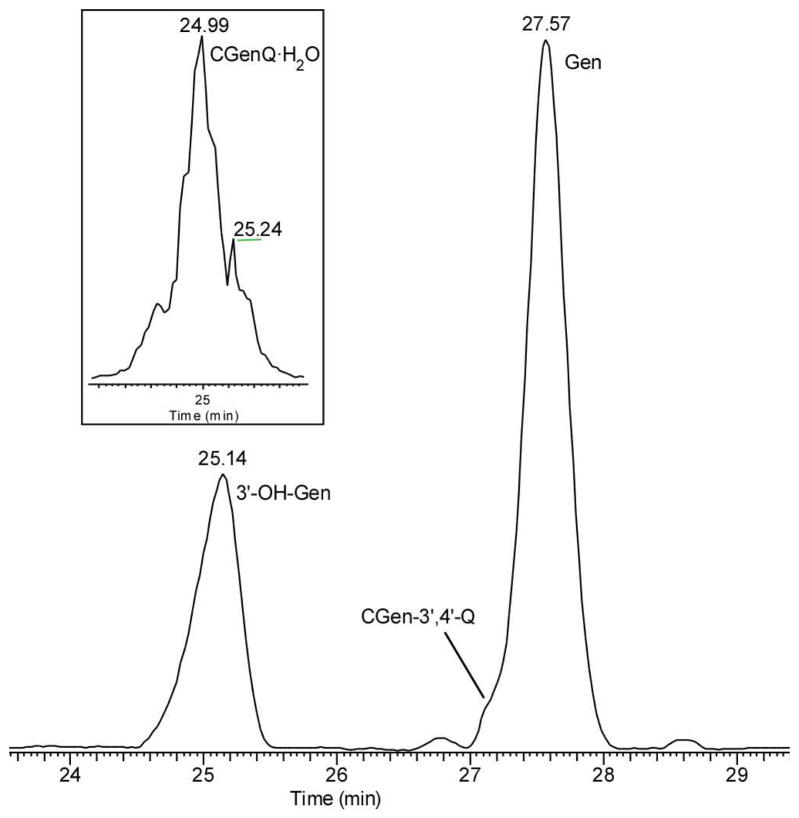
We also investigated the hydration of CGenQ by using ESI and accurate-mass analysis (LTQ-FT) (see Figure 2 for the LC trace of the products generated from the reaction of CGenQ in H2O). CGenQ reacts rapidly in H2O, making the detection of unhydrated CGenQ difficult by LC/MS. CGenQ·(H2O)2 is expected to lose water molecule(s) readily after protonation as the dihydrate carries several aliphatic hydroxyl groups. The dominant product is seen, however, as protonated CGen of m/z 287, as confirmed by accurate mass measurements. At first thought, this is inconsistent with the NMR results, which indicate formation of the dihydrate (expected m/z 321 for the protonated species, [CGenQ·(H2O)2 + H]+). To characterize further the species of m/z 287 seen by MS, we prepared 3′-OH-Gen by the reduction of CGen-3′,4′-Q in DMF. Mixing an equal amount of the synthetic 3′-OH-Gen with the unknown and analyzing again by LC/MS show that the unknown co-elutes with synthetic 3′-OH-Gen, and both are seen as ions of m/z 287 in the mass spectrum. The product-ion mass spectra (MS/MS) of the [M + H]+ of the two compounds are nearly identical (see Table S1 in Supporting Information), establishing that the material arising from the dihydrate during the LC/MS analysis is 3′-OH-Gen.
Figure 2.
LC/MS analysis of CGen-3′,4′-Q. The identity of [CGen-3′,4′-Q + H2O] conjugate (tR = 24.99 min) is consistent with an accurate mass measurement.
We can rationalize the results from MS and NMR by invoking reduction of the CGenQ·(H2O)2 to CGen in the LC/MS solution. The CGenQ·(H2O)2 presumably has a high reduction potential, allowing ready conversion back to CGen with H2O release. The likely reducing agent is formic acid (FA), which we add as a modifier for the LC/MS analysis. We are unable to test easily this proposal because, without the addition of acid in the LC experiment, we are unable to detect any products. The presence of FA, even an unintended amount, leads to rapid reduction of CGenQ·(H2O)2 to CGen.
During the trapping experiment, GSH reacts with CGenQ to form exclusively GenQ-GSH conjugates at t = 0; no CGen is detectable. The observation reveals that, in reaction with GSH, conjugation is preferred over reduction of the GenQ. CGen, however, forms later, as seen in the LC/MS of GenQ trapped by GSH after incubating GenQ in H2O. We propose that GSH cannot form a conjugate with CGenQ·(H2O)2; rather the reduction by GSH to form CGen becomes the dominant path. This facile conversion obviates detection of any major amount of CGenQ·(H2O)2 in the LC/MS analysis. We do clearly see, however, a residual amount of mono-hydrated CGenQ, CGenQ·H2O, as shown in the reconstructed ion chromatogram in the inset of Figure 2. The window for mass selection was centered at the theoretical mass of [CGenQ·H2O + H]+ with a width of ± 0.001 mass units (m/z 303.0499 ± 0.0010). The reconstructed ion chromatogram shows that the monohydrate, CGenQ·H2O, elutes at 24.99 min whereas CGen elutes at 25.14 min. Although the retention time difference is small, it is reproducible in replicates done even over many days, indicating that that the hydration is not caused by the ESI process. The difference indicates that a more hydrophilic species is formed by the addition of H2O to CGenQ. The mono-hydrate, CGenQ·H2O, may be a product of in-source fragmentation of CGenQ·(H2O)2 or an intermediate in the di-hydration reaction (Scheme 3). In negative-ion ESI, where formic acid can be avoided and the water lose attenuated, accurate mass measurements show that both the dihydrate CGenQ·(H2O)2 and monohydrate CGenQ·H2O can be observed at m/z 319.0458 (~ 3 ppm) and 301.0352 (~ 3 ppm), respectively. The abundance ratio of the two ions is 1:4, respectively.
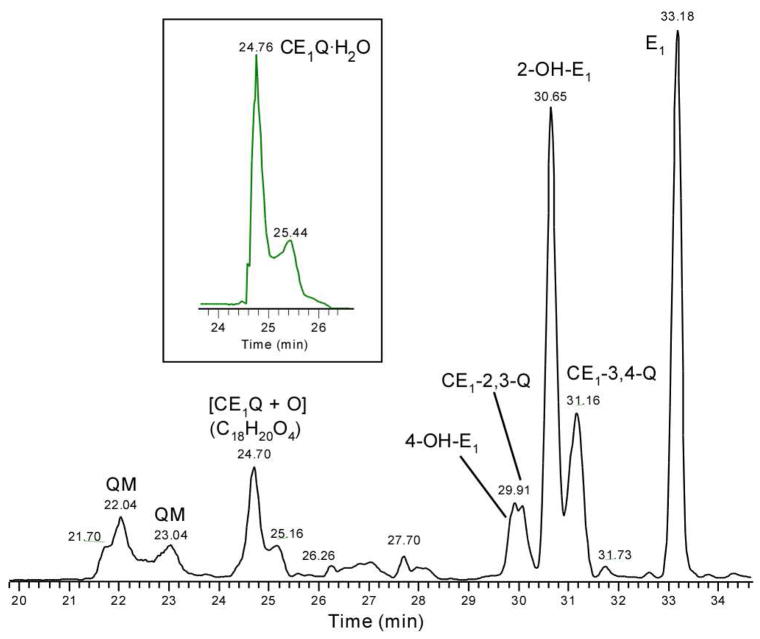
For the estrone quinones, NMR analysis indicates that CE1-2,3-Q is formed in higher abundance than CE1-3,4-Q. Owing to its faster reaction in H2O, CE1-2,3-Q is of lower abundance as seen by LC/MS analysis (Figure 3). In addition to the two quinones, there are two other species whose [M + H]+ ions are of m/z 285 and that elute at 22.04 and 23.04 min, respectively, both of which have the formulae of CE1Q as established by accurate mass measurements. These new species are probably quinone methides (QMs); a previous study revealed that CE1Qs isomerize to their QMs via a process assisted by H2O.26, 71, 76 Interestingly, we found no QMs in the reactions of CGenQ. A plausible explanation is that the presence of O1 and the absence of OH at C2 in CGenQ prevent the formation of a QM.
Figure 3.
LC/MS analysis of CE1Qs. The identities of two [CE1Q + H2O] conjugates (tR = 24.76 and 25.44 min) are consistent with accurate mass measurement.
Analogous to the products from Gen, we also see two species of m/z 287 in the LC/MS analysis of the reaction products of CE1Q. They coelute with authentic 2,3-OH-E1 and 3,4-OH-E1, supporting their assignments as catechol estrones. The product pattern that results from the reaction of CE1Qs is more complicated than for the genistein quinone. Analogous to the reactions of the latter species, estrone quinones also produce a monohydrate, CE1Q·H2O (see inset in Figure 3 for the accurate-mass selective ion chromatogram for CE1Q·H2O). To achieve high specificity, we set the window for mass selection at the theoretical mass of [CE1Q·H2O + H]+ with a variability of ± 0.001 mass units (i.e., m/z 303.1591 ± 0.0010). Two different hydrates elute, presumably CE1-2,3-Q·H2O and CE1-3,4-Q·H2O.71 Furthermore, two new species elute at 24.70 and 25.16 min, respectively. Accurate mass measurements reveal that each has the formula [CE1Q + O] (m/z 301 for a protonated species). Given that the further oxidation of CE1Qs is not of major interest here, we did not investigate them further.
(4) Reactivity of Quinones with Ade and DNA
(4–1) Formation of Ade Adducts in Aqueous solution
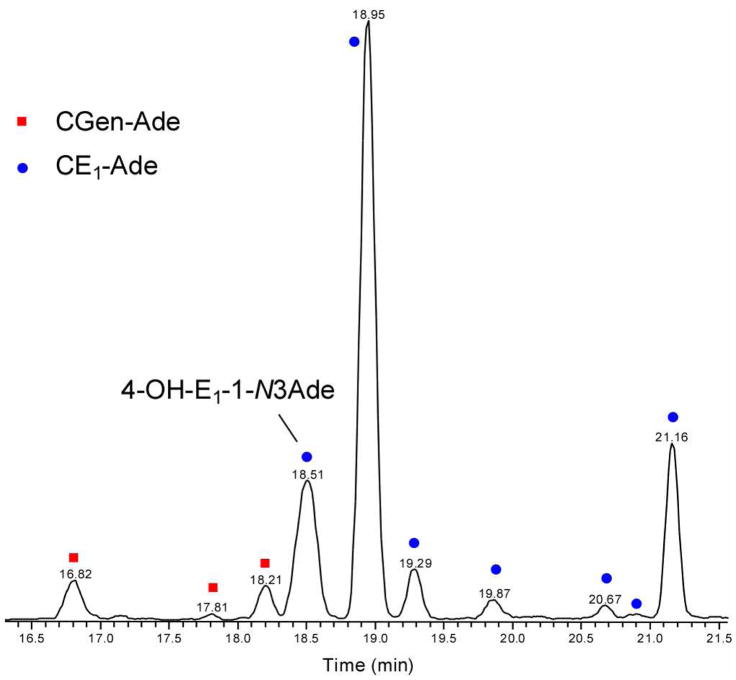
The rapid reaction of the genistein quinone in water suggests that the probability for the quinone to react with DNA bases is low. To test, we compared the ratio of modified Ade species formed from the genistein quinone reacting with Ade to those of estrone quinones also reacting with Ade. We added CGenQ or CE1Q in ACN (5 μL) to 1 mL of Na2HPO4 solution (50 mM, pH 7.4) containing the Ade free base and incubated the solution overnight at 37 °C. Any Ade adducts, formed under these identical conditions, were combined in a 50:1 (Gen:E1) ratio, in anticipation that the diminished reactivity of CGenQ with respect to CE1Q would make difficult a quantitative measurement over a large concentration range, and the solution was diluted with 50 mM Na2HPO4. This strategy allowed us to measure accurately the relative amount of Ade modified by catechol genistein quinone (CGen-Ade) compared to the amount of Ade modified by the estrone quinones (CE1-Ade). An LC/MS trace (Figure 4) shows that the Gen-modified Ade species elute from 16.8–18.2 min, whereas CE1-Ade species elute from 18.5–21.2 min. A reconstructed ion chromatogram shows the elution of CGen-Ade (m/z 420.1) and CE1-Ade adducts (m/z 420.2); we confirmed the assignments by accurate mass measurements. Product-ion mass spectra (MS/MS) provide additional support for these adduct assignments. One of the characteristic fragmentations of the CGen-Ade adducts is a through-ring cleavage in ring C to generate product ions of m/z 153 and/or a complementary ion of m/z 268 (see Figure S3 in Supporting Information). Putative structures of the CGen-Ade adducts, along with their major cleavages induced by MS/MS are given in Figure S4 in Supporting Information.
Figure 4.
Reconstructed ion chromatogram showing the formation of CGen-Ade adducts and CE1-Ade adducts. Analysis is of a 50:1 mixture of CGenQ + Ade and CE1Q + Ade (37 °C, pH 7.4). The observed ratio of CGen-Ade adducts to CE1-Ade adducts is 1:620.
In accord with the half lives of quinones, the high propensity of CGenQ to undergo hydration diminishes its reactivity with the nucleobase such that only trace amounts of CGen-Ade adducts can form compared to CE1-Ade adducts. Considering that 50 times more CGenQ-Ade adducts were injected, we found that integration of peak area in LC shows that the total CE1-Ade adducts are 620 ± 60 times more abundant than the CGen-Ade adducts.
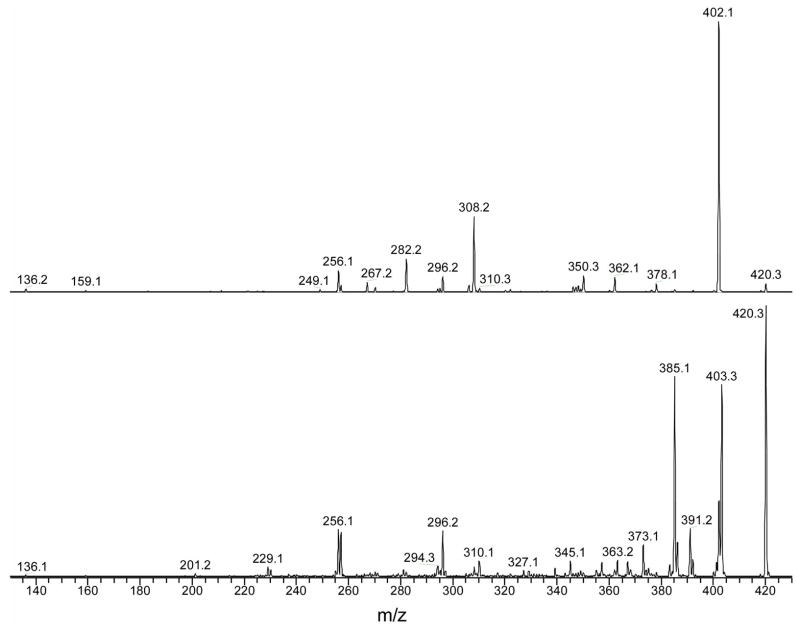
The most abundant CE1-Ade adduct, eluting at 18.95 min, is probably weakly bound and dissociates rapidly during electrospray to form the fragment ion of m/z 136, [Ade + H]+, and its complement, the ion of m/z 285, [CE1Q + H]+. In addition to the weakly-bound species, there are two major and more stable CE1-Ade adducts that elute at 18.51 min and 21.16 min. The early eluter is the well characterized 4-OH-E1-1-N3Ade (see Figure 5, bottom panel, for the product-ion mass spectrum taken in an MS/MS experiment), which is a putative biomarker for breast cancer.77, 78 The nearly equally abundant CE1-Ade eluting at 21.16 min gives an unusual product-ion mass spectrum (top panel in Figure 5). Missing in this spectrum are ions formed by the losses of NH3 and CH2NH from [4-OH-E1-1-N3Ade + H]+. These ions are formed by migration of H-atom(s) from the estrone to the Ade moiety73 and are characteristic of a nucleobase that adds to the C1 position of the A ring of the steroid. The unknown CE1-Ade adduct likely has a structure in which the migration of H atom(s) to Ade is prohibited. A full structural study is beyond the scope of this article.
Figure 5.
MS/MS product-ion spectra of the two CE1-Ade adducts, 4-OH-E1-1-N3Ade at tR = 18.51 min (bottom panel) and unknown at tR = 21.16 min (top panel) from Figure 4.
(4–2) Formation of Ade Adducts in DMF
Both CGenQ and CE1Qs, however, are relatively stable in DMF. In this nonaqueous solvent, the reactivities of the quinones are more “intrinsic” (not significantly determined by the properties of the solvent). In this medium, the ratio of CGen-Ade adducts to CE1-Ade adducts increases remarkably to 1:3 (Figure S5 in Supporting Information, also see Table S2 for accurate mass measurements for CGen-Ade adducts). Given that the amount of CGenQ is ~ 40% of the total CE1Qs, we conclude that the “intrinsic reactivities” to modify Ade are similar for the two quinones. The significantly shorter half life of CGenQ in H2O accounts for its greatly reduced reactivity with Ade in aqueous solution (i.e., under physiological conditions).
MS/MS product-ion analyses reveal that the two CGen-Ade adducts formed in DMF and eluting at 17.95 and 19.38 min are the same species formed in aqueous solution and eluting at 16.82 and 18.21 min, respectively, as determined at a different time. Another product eluting at 18.38 min in the reaction with DMF, however, is barely detectable in the reaction in H2O. Clearly, the solvent affects the formation not only of the CGen-Ade adducts but also the CE1-Ade adducts. The species eluting at 21.22 and 21.32 min are 4-OH-E1-1-N3Ade and the weakly-bound CE1-Ade, respectively. Unlike in H2O, where the weakly bound species is ~ 4 times as abundant as 4-OH-E1-1-N3Ade, these two species are produced in DMF in nearly equal amounts. The species produced in DMF and eluting at 23.67 min corresponds to that eluting at 21.16 min from the reaction in water, but its abundance is considerably less than 4-OH-E1-1-N3Ade when formed in DMF.
(4–3) Formation of DNA Adducts with Calf Thymus DNA
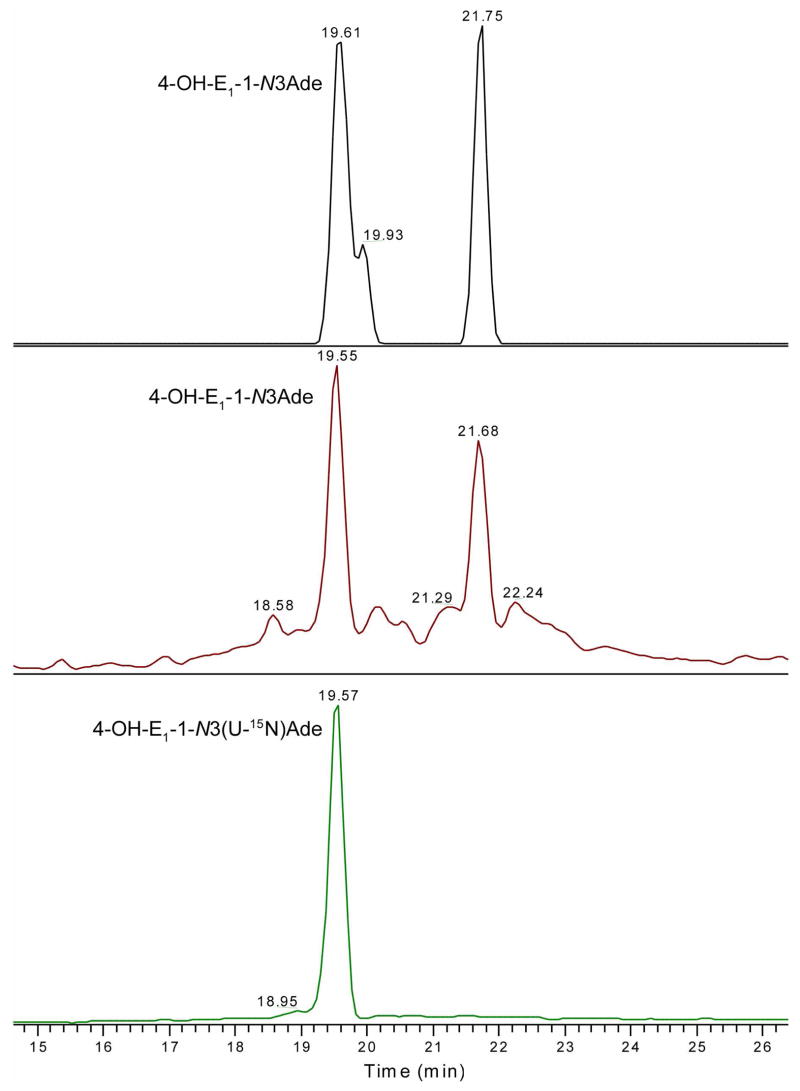
We extended the reactivity comparison of the two quinones by carrying out a competitive reaction with calf thymus DNA. The reaction involved approximately 60 nmole of CE1-2,3-Q and 40 nmole of CE1-3,4-Q in 5 μL ACN, to which we added 1 mg calf thymus DNA in 1 mL of H2O (50 mM Na2HPO4, pH 7.4). After overnight incubation at 37 °C, we added the internal standard, U-15N labeled IS, 4-OH-E1-1-N3(U-15N)Ade (1 pmole), to assure good quantification and submitted the mixture to SPE cleanup. The total ion chromatogram (TIC) obtained by monitoring the elution of the IS, 4-OH-E1-1-N3(U-15N)Ade (tR = 19.57 min), which was obtained in a LC/MS analysis (see the LC traces in Figure 6), allows the identification of one of the CE1-Ade adducts and allows for quantification of all of them. A TIC produced by integrating the product-ion intensities generated by MS/MS of the m/z 420.2 [M + H]+ ions show two species that elute at tR of 19.55 and 21.68 min. One coelutes with the U-15N labeled 4-OH-E1-1-N3Ade, indicating that the species at tR of 19.55 min is 4-OH-E1-1-N3Ade. Comparing the MS/MS product-ion spectra (see Figure S6 in Supporting Information) with those of the CE1-Ade adducts formed from the reaction with Ade free base in H2O, we conclude that the two species formed from the reaction with calf thymus DNA correspond to those eluting at tR 18.51 and 21.16 min, respectively, formed in the reaction in water (Figure 5). (Retention times cannot be precisely compared because the analyses were done at different times and under slightly different conditions.)
Figure 6.
Product mixture from reaction of CE1Q and calf thymus DNA at 37 °C, pH 7.4, as analyzed with the ion-trap/FT-ICR mass spectrometer. Accurate mass measurement in top panel shows two and possibly three major CE1-Ade adducts. The middle panel show the corresponding TIC measured in ion trap. Bottom panel shows the TIC of internal standard, 4-OH-E1-1-N3 (U-15N)Ade in ion trap.
The product-ion spectra of 4-OH-E1-1-N3Ade with U-15N labeled IS are characteristic of the assigned structure, as was discussed in a recent publication.73 The identities of these CE1-Ade adducts are also consistent with accurate mass measurements (top panel in Figure 6). The mass tolerance is ±0.0015 mass units (±3.6 ppm) from the theoretical mass, and all signals within that tolerance and produced within the mass range of 200–2000 comprise the chromatograms in the figure. Two CE1-Ade adducts are clearly characterized and a third eluting at 19.93 min, is missed in the TIC generated from the product ions (middle panel, Figure 6). The shoulder species may correspond to the weakly bound adduct (species of tR 18.95 in Figure 4 and tR 21.32 min in Figure S5), which dissociates promptly during the precursor-ion isolation step of the MS/MS sequence, and thus cannot be seen in the MS/MS mode as displayed in the middle panel of Figure 6. The use of U-15N labeled IS allows us to quantify the amount of 4-OH-E1-1-N3Ade formed by depurinating of the double-stranded DNA to be 190 ± 40 fmol (formed in the incubation of 60 nmole of CE1-2,3-Q and 40 nmole of CE1-3,4-Q with 1 mg calf thymus DNA under physiological conditions). In addition, there is a second CE1-Ade adduct of approximately equal abundance. We were unable to detect any of modified guanine bases, which is in accord with previous observations that catecholestrogen-modified Gua depurinates from DNA at a slower rate than does modified Ade. The amount of a modified quinone is less than 30% of the adenine. The more facile release of CE-modified Ade suggests that there will be a higher probability in identifying them in tissue extracts or biological fluids for biomarker discovery. In contrast, CE-Gua adducts may be a more appropriate target for exploring biomarkers that remain in the double-stranded DNA.
The same procedure with CGenQ at either the same level or 20 times of the amount used for CE1Qs produced, not surprisingly, no detectable CGen-modified Ade species (detection limit is 50 fmol). The lower reactivity is in accord with more rapid reaction of CGenQ in H2O.
(5) Metabolism of Genistein and its Quinones
The observations of the rapid conversion of CGenQ to its H2O conjugate and the facile reduction of CGenQ·(H2O)2 to CGen allow us to propose a mechanism whereby the ability of certain quinones (e.g., that from genistein) to damage macromolecules is considerably diminished by a fast reaction with H2O (Scheme 4). Thus, although Gen and related quinones can be metabolized to catechols and to quinones, the quinones do not participate significantly in redox cycling or add directly to purine bases of DNA because they are rapidly and covalently hydrated. The structure of CGenQ·(H2O)2 is one that would be expected to be incapable of generating ROS and/or attacking nucleophilic sites in DNA. This stable dihydrate can be reduced back to CGen, 3′-OH-Gen. The short “dwell time” of Gen in its quinone state should lead to low cytotoxicity and/or genotoxicity, as must be proved in other studies. Instead, we expect that Gen would act primarily as an antioxidant in vivo.
Scheme 4.
Proposed mechanism for the antioxidant activities of Gen and its catechol and the “detoxification” of its quinone metabolite.
In comparison, CE1Qs, CE1-3,4-Q in particular, react with H2O at substantially slower rates. These longer-lived quinones are available to participate in redox cycling and/or to damage DNA by direct reaction followed by depurination. Once CE1Q hydrates are formed, they can be reduced back to their catechols, CE1s and then reoxidized generating again relatively long-lived quinones available again for redox cycling and depurinating reactions. If Gen and similar materials are present along with estrogens, however, oxidative phase I metabolism of Gen may absorb the oxidizing equivalents in vivo, preventing oxidation of estrogens and preventing its untoward biological effects.
Conclusions
The quinone from genistein is much less reactive with Ade and with DNA, suggesting that it is less biologically deleterious than the CEQs. The diminished reactivity of CGenQ is due to its rapid reaction with water, converting it to a dihydrated species, CGenQ·(H2O)2, under physiological conditions. Under mild reductive conditions, the dihydrate is reduced back to CGen. The cycling of CGenQ, CGenQ·(H2O)2 and CGen in water is an effective means to diminish any potential toxicity. The rapid reaction with water exhibited by the Gen quinone suggests to us that Gen exerts primarily anti-oxidation activities as radical scavenger. In addition, by competing for quinone formation, the oxidative metabolism of Gen may reduce ROS generation by other more deleterious quinone species. Simple chemical reactivities, of course, are not the only predictors of genotoxicity; other factors (e.g., bioavailability) may also play a role. Nevertheless, the chemical properties of estrogen and estrogen-like materials are so compellingly different that we expect those differences to have an impact on biological and toxicological properties.
Supplementary Material
LC/MS, MS/MS, accurate mass, NMR and additional mass spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
The work was supported by the National Centers for Research Resources of the NIH under Grant No. P41RR000954. We are grateful to Prof. K. Moeller and Dr. V. Birman for insightful discussions on the NMR structure characterization of the hydrated product of catechol genistein quinone.
References
- 1.Monks TJ, Hanzlik RP, Cohen GM, Ross D, Graham DG. Toxicol Appl Pharmacol. 1992;112:2–16. doi: 10.1016/0041-008x(92)90273-u. [DOI] [PubMed] [Google Scholar]
- 2.Cavalieri EL, Rogan EG, Devanesan PD, Cremonesi P, Cerny RL, Gross ML, Bodell WJ. Biochemistry. 1990;29:4820–4827. doi: 10.1021/bi00472a011. [DOI] [PubMed] [Google Scholar]
- 3.Casale GP, Singhal M, Bhattacharya S, RamaNathan R, Roberts KP, Barbacci DC, Zhao J, Jankowiak R, Gross ML, Cavalieri EL, Small GJ, Rennard SI, Mumford JL, Shen ML. Chem Res Toxicol. 2001;14:192–201. doi: 10.1021/tx000012y. [DOI] [PubMed] [Google Scholar]
- 4.FlowersGeary L, Bleczinski W, Harvey RG, Penning TM. Chem-Biol Interact. 1996;99:55–72. doi: 10.1016/0009-2797(95)03660-1. [DOI] [PubMed] [Google Scholar]
- 5.Van de Water RW, Pettus TRR. Tetrahedron. 2002;58:5367–5405. [Google Scholar]
- 6.Sorensen M, Autrup H, Moller P, Hertel O, Jensen SS, Vinzents P, Knudsen LE, Loft S. Mutat Res-Rev Mutat Res. 2003;544:255–271. doi: 10.1016/j.mrrev.2003.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Xue WL, Warshawsky D. Toxicol Appl Pharmacol. 2005;206:73–93. doi: 10.1016/j.taap.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Shimada T. Drug Metab Pharmacokinet. 2006;21:257–276. doi: 10.2133/dmpk.21.257. [DOI] [PubMed] [Google Scholar]
- 9.Barrett-Connor E, Mosca L, Collins P, Geiger MJ, Grady D, Kornitzer M, McNabb MA, Wenger NK. N Engl J Med. 2006;355:125–137. doi: 10.1056/NEJMoa062462. [DOI] [PubMed] [Google Scholar]
- 10.Galati G, Sabzevari O, Wilson JX, O’Brien PJ. Toxicology. 2002;177:91–104. doi: 10.1016/s0300-483x(02)00198-1. [DOI] [PubMed] [Google Scholar]
- 11.Azam S, Hadi N, Khan NU, Hadi SM. Toxicol Vitro. 2004;18:555–561. doi: 10.1016/j.tiv.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Rietjens I, Boersma MG, van der Woude H, Jeurissen SMF, Schutte ME, Alink GM. Mutat Res-Fundam Mol Mech Mutagen. 2005;574:124–138. doi: 10.1016/j.mrfmmm.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 13.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, De Cabo R, Sinclair DA. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baur JA, Sinclair DA. Nature Reviews Drug Discovery. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 15.Powis G. Free Radic Biol Med. 1989;6:63–101. doi: 10.1016/0891-5849(89)90162-7. [DOI] [PubMed] [Google Scholar]
- 16.Gutierrez PL. Free Radic Biol Med. 2000;29:263–275. doi: 10.1016/s0891-5849(00)00314-2. [DOI] [PubMed] [Google Scholar]
- 17.Asche C. Mini-Rev Med Chem. 2005;5:449–467. doi: 10.2174/1389557053765556. [DOI] [PubMed] [Google Scholar]
- 18.Grodstein F, Stampfer MJ, Colditz GA, Willett WC, Manson JE, Joffe M, Rosner B, Fuchs C, Hankinson SE, Hunter DJ, Hennekens CH, Speizer FE. N Engl J Med. 1997;336:1769–1775. doi: 10.1056/NEJM199706193362501. [DOI] [PubMed] [Google Scholar]
- 19.Rossouw JE, Anderson GL, Prentice RL, LaCroix AZ, Kooperberg C, Stefanick ML, Jackson RD, Beresford SAA, Howard BV, Johnson KC, Kotchen M, Ockene J. JAMA-J Am Med Assoc. 2002;288:321–333. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- 20.Grodstein F, Clarkson TB, Manson JE. N Engl J Med. 2003;348:645–650. doi: 10.1056/NEJMsb022365. [DOI] [PubMed] [Google Scholar]
- 21.Bolton JL, Trush MA, Penning TM, Dryhurst G, Monks TJ. Chem Res Toxicol. 2000;13:135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- 22.Obrien PJ. Chem-Biol Interact. 1991;80:1–41. doi: 10.1016/0009-2797(91)90029-7. [DOI] [PubMed] [Google Scholar]
- 23.Davies MJ. BBA-Proteins Proteomics. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Pfohl-Leszkowicz A, Manderville RA. Mol Nutr Food Res. 2007;51:61–99. doi: 10.1002/mnfr.200600137. [DOI] [PubMed] [Google Scholar]
- 25.Cavalieri EL, Stack DE, Devanesan PD, Todorovic R, Dwivedy I, Higginbotham S, Johansson SL, Patil KD, Gross ML, Gooden JK, Ramanathan R, Cerny RL, Rogan EG. Proc Natl Acad Sci U S A. 1997;94:10937–10942. doi: 10.1073/pnas.94.20.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolton JL, Thatcher GRJ. Chem Res Toxicol. 2008;21:93–101. doi: 10.1021/tx700191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lof M, Weiderpass E. Nutrition Research. 2006;26:609–619. [Google Scholar]
- 28.Kuiper G, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, van der Burg P, Gustafsson JA. Endocrinology. 1998;139:4252–4263. doi: 10.1210/endo.139.10.6216. [DOI] [PubMed] [Google Scholar]
- 29.Nikov GN, Hopkins NE, Boue S, Alworth WL. Environ Health Perspect. 2000;108:867–872. doi: 10.1289/ehp.00108867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verheus M, van Gils CH, Keinan-Boker L, Grace PB, Bingham SA, Peeters PHM. Journal Of Clinical Oncology. 2007;25:648–655. doi: 10.1200/JCO.2006.06.0244. [DOI] [PubMed] [Google Scholar]
- 31.Kulling SE, Honig DM, Simat TJ, Metzler M. J Agric Food Chem. 2000;48:4963–4972. doi: 10.1021/jf000524i. [DOI] [PubMed] [Google Scholar]
- 32.Kulling SE, Honig DM, Metzler M. J Agric Food Chem. 2001;49:3024–3033. doi: 10.1021/jf0012695. [DOI] [PubMed] [Google Scholar]
- 33.Kulling SE, Lehmann L, Metzler M. J Chromatogr B. 2002;777:211–218. doi: 10.1016/s1570-0232(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 34.Awad HM, Boersma MG, Vervoort J, Rietjens I. Arch Biochem Biophys. 2000;378:224–233. doi: 10.1006/abbi.2000.1832. [DOI] [PubMed] [Google Scholar]
- 35.Awad HM, Boersma MG, Boeren S, van Bladeren PJ, Vervoort J, Rietjens I. Chem Res Toxicol. 2001;14:398–408. doi: 10.1021/tx000216e. [DOI] [PubMed] [Google Scholar]
- 36.Hodgson JM, Croft KD, Puddey IB, Mori TA, Beilin LJ. J Nutr Biochem. 1996;7:664–669. [Google Scholar]
- 37.Kapiotis S, Hermann M, Held I, Seelos C, Ehringer H, Gmeiner BMK. Arterioscler Thromb Vasc Biol. 1997;17:2868–2874. doi: 10.1161/01.atv.17.11.2868. [DOI] [PubMed] [Google Scholar]
- 38.RuizLarrea MB, Mohan AR, Paganga G, Miller NJ, Bolwell GP, RiceEvans CA. Free Radic Res. 1997;26:63–70. doi: 10.3109/10715769709097785. [DOI] [PubMed] [Google Scholar]
- 39.Heim KE, Tagliaferro AR, Bobilya DJ. J Nutr Biochem. 2002;13:572–584. doi: 10.1016/s0955-2863(02)00208-5. [DOI] [PubMed] [Google Scholar]
- 40.Bolton JL. Toxicology. 2002;177:55–65. doi: 10.1016/s0300-483x(02)00195-6. [DOI] [PubMed] [Google Scholar]
- 41.Tang MY, Abplanalp W, Ayres S, Subbiah MTR. Metab-Clin Exp. 1996;45:411–414. doi: 10.1016/s0026-0495(96)90212-7. [DOI] [PubMed] [Google Scholar]
- 42.RuizLarrea MB, Leal AM, Martin C, Martinez R, Lacort M. J Physiol Biochem. 1997;53:225–229. [PubMed] [Google Scholar]
- 43.Ayres S, Abplanalp W, Liu JH, Subbiah MTR. Am J Physiol-Endocrinol Metab. 1998;37:E1002–E1008. doi: 10.1152/ajpendo.1998.274.6.E1002. [DOI] [PubMed] [Google Scholar]
- 44.Prokai L, Prokai-Tatrai K, Perjesi P, Zharikova AD, Perez EJ, Liu R, Simpkins JW. Proc Natl Acad Sci U S A. 2003;100:11741–11746. doi: 10.1073/pnas.2032621100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moosmann B, Behl C. Proc Natl Acad Sci U S A. 1999;96:8867–8872. doi: 10.1073/pnas.96.16.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright JS, Johnson ER, DiLabio GA. J Am Chem Soc. 2001;123:1173–1183. doi: 10.1021/ja002455u. [DOI] [PubMed] [Google Scholar]
- 47.Ruiz-Larrea MB, Martin C, Martinez R, Navarro R, Lacort M, Miller MJ. Chem Phys Lipids. 2000;105:179–188. doi: 10.1016/s0009-3084(00)00120-1. [DOI] [PubMed] [Google Scholar]
- 48.Colditz GA, Hankinson SE, Hunter DJ, Willett WC, Manson JE, Stampfer MJ, Hennekens C, Rosner B, Speizer FE. N Engl J Med. 1995;332:1589–1593. doi: 10.1056/NEJM199506153322401. [DOI] [PubMed] [Google Scholar]
- 49.Henderson BE, Feigelson HS. Carcinogenesis. 2000;21:427–433. doi: 10.1093/carcin/21.3.427. [DOI] [PubMed] [Google Scholar]
- 50.Ravdin PM, Cronin KA, Howlader N, Berg CD, Chlebowski RT, Feuer EJ, Edwards BK, Berry DA. N Engl J Med. 2007;356:1670–1674. doi: 10.1056/NEJMsr070105. [DOI] [PubMed] [Google Scholar]
- 51.Messina M, Barnes S. J Natl Cancer Inst. 1991;83:541–546. doi: 10.1093/jnci/83.8.541. [DOI] [PubMed] [Google Scholar]
- 52.Setchell KDR, Cassidy A. J Nutr. 1999;129:758S–767S. doi: 10.1093/jn/129.3.758S. [DOI] [PubMed] [Google Scholar]
- 53.Shu XO, Jin F, Dai Q, Wen WQ, Potter JD, Kushi LH, Ruan ZX, Gao YT, Zheng W. Cancer Epidemiol Biomarkers Prev. 2001;10:483–488. [PubMed] [Google Scholar]
- 54.Moon YJ, Wang XD, Morris ME. Toxicol Vitro. 2006;20:187–210. doi: 10.1016/j.tiv.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 55.Messina M, McCaskill-Stevens W, Lampe JW. J Natl Cancer Inst. 2006;98:1275–1284. doi: 10.1093/jnci/djj356. [DOI] [PubMed] [Google Scholar]
- 56.Fink BN, Steck SE, Wolff MS, Britton JA, Kabat GC, Schroeder JC, Teitelbaum SL, Neugut AI, Gammon MD. Am J Epidemiol. 2007;165:514–523. doi: 10.1093/aje/kwk033. [DOI] [PubMed] [Google Scholar]
- 57.Nandi S, Guzman RC, Yang J. Proc Natl Acad Sci U S A. 1995;92:3650–3657. doi: 10.1073/pnas.92.9.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yager JD, Liehr JG. Annu Rev Pharmacol Toxicol. 1996;36:203–232. doi: 10.1146/annurev.pa.36.040196.001223. [DOI] [PubMed] [Google Scholar]
- 59.Flototto T, Djahansouzi S, Glaser M, Hanstein B, Niederacher D, Brumm C, Beckmann MW. Horm Metab Res. 2001;33:451–457. doi: 10.1055/s-2001-16936. [DOI] [PubMed] [Google Scholar]
- 60.Clemons M, Goss P. N Engl J Med. 2001;344:276–285. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- 61.Santen RJ. J Clin Endocrinol Metab. 2002;87:3007–3012. doi: 10.1210/jcem.87.7.8589. [DOI] [PubMed] [Google Scholar]
- 62.Platet N, Cathiard AM, Gleizes M, Garcia M. Crit Rev Oncol/Hematol. 2004;51:55–67. doi: 10.1016/j.critrevonc.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 63.Liehr JG, Avitts TA, Randerath E, Randerath K. Proc Natl Acad Sci U S A. 1986;83:5301–5. doi: 10.1073/pnas.83.14.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liehr JG. Endocr Rev. 2000;21:40–54. doi: 10.1210/edrv.21.1.0386. [DOI] [PubMed] [Google Scholar]
- 65.Liehr JG. Regul Toxicol Pharmacol. 2000;32:276–282. doi: 10.1006/rtph.2000.1432. [DOI] [PubMed] [Google Scholar]
- 66.Yue W, Santen RJ, Wang JP, Li Y, Verderame MF, Bocchinfuso WR, Korach KS, Devanesan P, Todorovic R, Rogan EG, Cavalieri EL. J Steroid Biochem Mol Biol. 2003;86:477–486. doi: 10.1016/s0960-0760(03)00377-7. [DOI] [PubMed] [Google Scholar]
- 67.Li KM, Todorovic R, Devanesan P, Higginbotham S, Kofeler H, Ramanathan R, Gross ML, Rogan EG, Cavalieri EL. Carcinogenesis. 2004;25:289–297. doi: 10.1093/carcin/bgg191. [DOI] [PubMed] [Google Scholar]
- 68.Yager JD, Davidson NE. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 69.Liehr JG, Roy D. Free Radic Biol Med. 1990;8:415–423. doi: 10.1016/0891-5849(90)90108-u. [DOI] [PubMed] [Google Scholar]
- 70.Han XL, Liehr JG. Cancer Research. 1994;54:5515–5517. [PubMed] [Google Scholar]
- 71.Iverson SL, Shen L, Anlar N, Bolton JL. Chem Res Toxicol. 1996;9:492–499. doi: 10.1021/tx950178c. [DOI] [PubMed] [Google Scholar]
- 72.Frigerio M, Santagostino M, Sputore S. J Org Chem. 1999;64:4537–4538. [Google Scholar]
- 73.Zhang Q, Gross ML. Chem Res Toxicol. 2008;21:1244–1252. doi: 10.1021/tx800067s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saeed M, Zahid M, Rogan E, Cavalieri E. Steroids. 2005;70:173–178. doi: 10.1016/j.steroids.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 75.Yu LN, Liu H, Li WK, Zhang FG, Luckie C, van Breemen RB, Thatcher GRJ, Bolton JL. Chem Res Toxicol. 2004;17:879–888. doi: 10.1021/tx0342722. [DOI] [PubMed] [Google Scholar]
- 76.Bolton JL, Shen L. Carcinogenesis. 1996;17:925–929. doi: 10.1093/carcin/17.5.925. [DOI] [PubMed] [Google Scholar]
- 77.Gaikwad NW, Yang L, Muti P, Meza JL, Pruthi S, Ingle JN, Rogan EG, Cavalieri EL. Int J Cancer. 2008;122:1949–1957. doi: 10.1002/ijc.23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Q, Aft RL, Gross ML. Chem Res Toxicol. 2008;21:1509–1513. doi: 10.1021/tx8001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LC/MS, MS/MS, accurate mass, NMR and additional mass spectra. This material is available free of charge via the Internet at http://pubs.acs.org.