Abstract
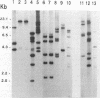
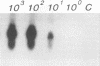
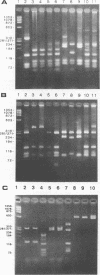
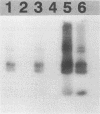
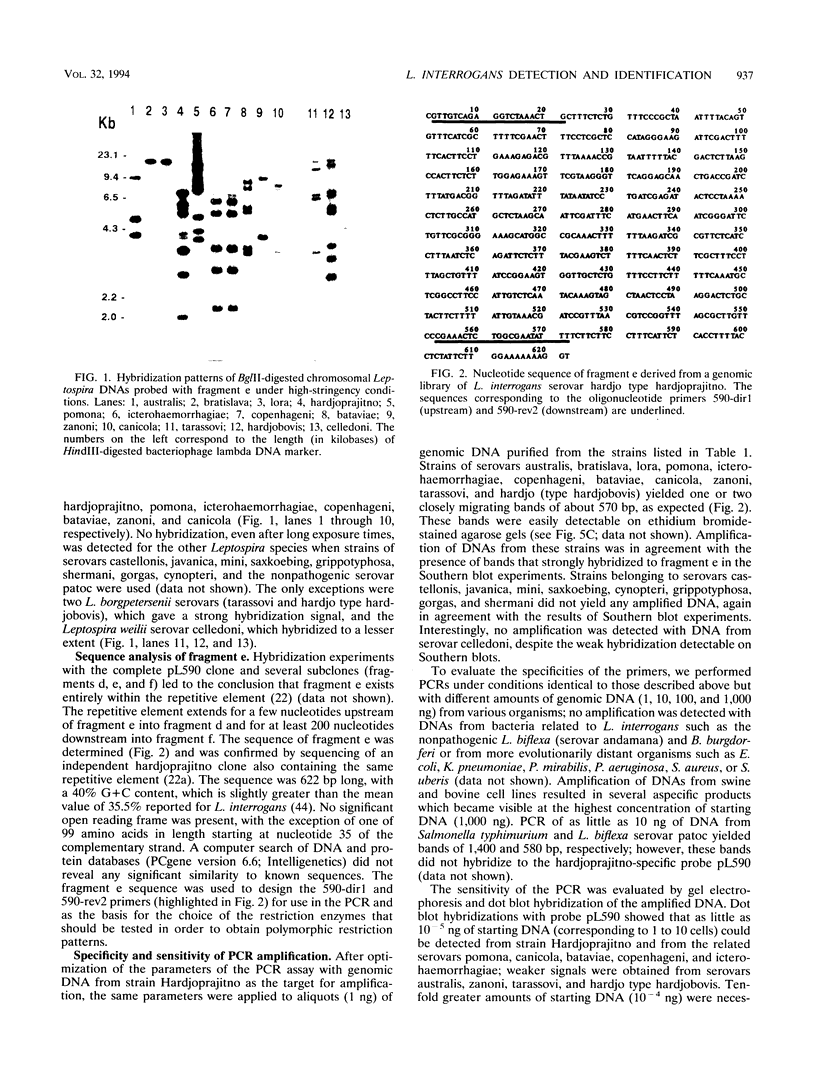
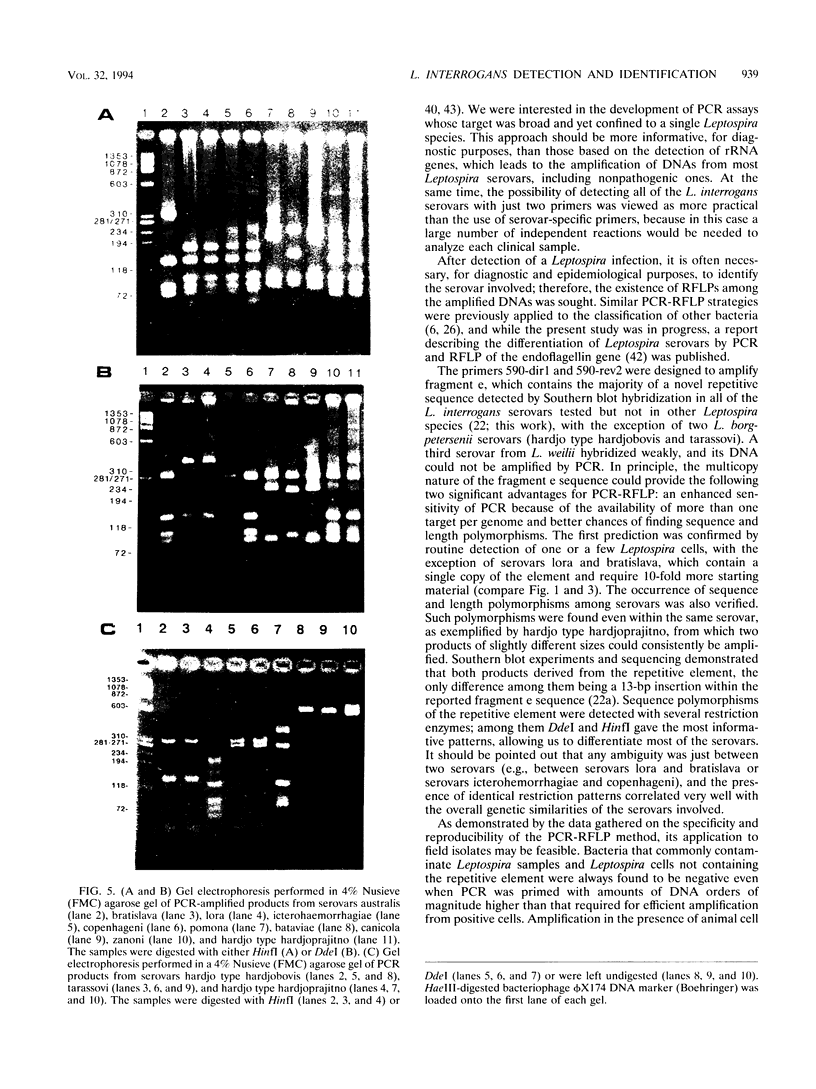
Primers for PCR were selected from a sequenced fragment of clone pL590, which contains a repetitive element present in the genome of Leptospira interrogans serovar hardjo type hardjoprajitno (M. L. Pacciarini, M. L. Savio, S. Tagliabue, and C. Rossi, J. Clin. Microbiol. 30:1243-1249, 1992). A specific DNA fragment was amplified from the genomic DNAs of serovar hardjo type hardjoprajitno and nine serovars also belonging to L. interrogans as a consequence of the spread of the same or a closely related repetitive element within this species (Pacciarini et al., J. Clin. Microbiol. 30:1243-1249, 1992). In addition, specific amplification was obtained from two Leptospira borgpetersenii serovars (tarassovi and hardjo type hardjobovis). Negative PCR results were observed with all of the other Leptospira serovars tested, including nonpathogenic ones (serovars patoc and andamana), another spirochete (Borrelia burgdorferi), bacteria commonly found in biological samples, and swine and bovine cell lines. Direct PCR on biological samples such as kidney samples demonstrated that preliminary isolation and culture of Leptospira cells are not required for efficient detection. Furthermore, digestion of the amplified DNA with the enzymes HinfI and DdeI yielded specific polymorphic patterns, allowing discrimination among the majority of the serovars. These methods were applied to 25 field isolates of serovar pomona, leading to the conclusion that they were suitable for the simple and rapid detection of L. interrogans and for serovar identification.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbour A. G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984 Jul-Aug;57(4):521–525. [PMC free article] [PubMed] [Google Scholar]
- Belák S., Ballagi-Pordány A., Flensburg J., Virtanen A. Detection of pseudorabies virus DNA sequences by the polymerase chain reaction. Arch Virol. 1989;108(3-4):279–286. doi: 10.1007/BF01310940. [DOI] [PubMed] [Google Scholar]
- Demmler G. J., Buffone G. J., Schimbor C. M., May R. A. Detection of cytomegalovirus in urine from newborns by using polymerase chain reaction DNA amplification. J Infect Dis. 1988 Dec;158(6):1177–1184. doi: 10.1093/infdis/158.6.1177. [DOI] [PubMed] [Google Scholar]
- Drancourt M., Kelly P. J., Regnery R., Raoult D. Identification of spotted fever group rickettsiae using polymerase chain reaction and restriction-endonuclease length polymorphism analysis. Acta Virol. 1992 Jan;36(1):1–6. [PubMed] [Google Scholar]
- Ellis W. A., Montgomery J. M., Thiermann A. B. Restriction endonuclease analysis as a taxonomic tool in the study of pig isolates belonging to the Australis serogroup of Leptospira interrogans. J Clin Microbiol. 1991 May;29(5):957–961. doi: 10.1128/jcm.29.5.957-961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J. L., Baril C., Bellenger E., Perolat P., Baranton G., Saint Girons I. Genome conservation in isolates of Leptospira interrogans. J Bacteriol. 1991 Dec;173(23):7582–7588. doi: 10.1128/jb.173.23.7582-7588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J. L., Bellenger E., Perolat P., Baranton G., Saint Girons I. Pulsed-field gel electrophoresis of NotI digests of leptospiral DNA: a new rapid method of serovar identification. J Clin Microbiol. 1992 Jul;30(7):1696–1702. doi: 10.1128/jcm.30.7.1696-1702.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Harris V. G. Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J Bacteriol. 1967 Jul;94(1):27–31. doi: 10.1128/jb.94.1.27-31.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafatos F. C., Jones C. W., Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979 Nov 24;7(6):1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Febvre R. B., Thiermann A. B. DNA homology studies of leptospires of serogroups Sejroe and Pomona from cattle and swine. Am J Vet Res. 1986 Apr;47(4):959–963. [PubMed] [Google Scholar]
- Marshall R. B., Wilton B. E., Robinson A. J. Identification of Leptospira serovars by restriction-endonuclease analysis. J Med Microbiol. 1981 Feb;14(1):163–166. doi: 10.1099/00222615-14-1-163. [DOI] [PubMed] [Google Scholar]
- Michna S. W. Leptospirosis. Vet Rec. 1970 Apr 25;86(17):484–496. doi: 10.1136/vr.86.17.484. [DOI] [PubMed] [Google Scholar]
- Millar B. D., Chappel R. J., Adler B. Detection of leptospires in biological fluids using DNA hybridisation. Vet Microbiol. 1987 Oct;15(1-2):71–78. doi: 10.1016/0378-1135(87)90131-3. [DOI] [PubMed] [Google Scholar]
- Mérien F., Amouriaux P., Perolat P., Baranton G., Saint Girons I. Polymerase chain reaction for detection of Leptospira spp. in clinical samples. J Clin Microbiol. 1992 Sep;30(9):2219–2224. doi: 10.1128/jcm.30.9.2219-2224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. N., Armstrong C. H., Nielsen N. C. Relationship among selected Leptospira interrogans serogroups as determined by nucleic acid hybridization. J Clin Microbiol. 1989 Dec;27(12):2724–2729. doi: 10.1128/jcm.27.12.2724-2729.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou C. Y., Kwok S., Mitchell S. W., Mack D. H., Sninsky J. J., Krebs J. W., Feorino P., Warfield D., Schochetman G. DNA amplification for direct detection of HIV-1 in DNA of peripheral blood mononuclear cells. Science. 1988 Jan 15;239(4837):295–297. doi: 10.1126/science.3336784. [DOI] [PubMed] [Google Scholar]
- Pacciarini M. L., Savio M. L., Tagliabue S., Rossi C. Repetitive sequences cloned from Leptospira interrogans serovar hardjo genotype hardjoprajitno and their application to serovar identification. J Clin Microbiol. 1992 May;30(5):1243–1249. doi: 10.1128/jcm.30.5.1243-1249.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph D., McClelland M., Welsh J., Baranton G., Perolat P. Leptospira species categorized by arbitrarily primed polymerase chain reaction (PCR) and by mapped restriction polymorphisms in PCR-amplified rRNA genes. J Bacteriol. 1993 Feb;175(4):973–981. doi: 10.1128/jb.175.4.973-981.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadass P., Jarvis B. D., Corner R. J., Penny D., Marshall R. B. Genetic characterization of pathogenic Leptospira species by DNA hybridization. Int J Syst Bacteriol. 1992 Apr;42(2):215–219. doi: 10.1099/00207713-42-2-215. [DOI] [PubMed] [Google Scholar]
- Regnery R. L., Spruill C. L., Plikaytis B. D. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J Bacteriol. 1991 Mar;173(5):1576–1589. doi: 10.1128/jb.173.5.1576-1589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. J., Ramadass P., Lee A., Marshall R. B. Differentiation of subtypes within Leptospira interrogans serovars Hardjo, Balcanica and Tarassovi, by bacterial restriction-endonuclease DNA analysis (BRENDA). J Med Microbiol. 1982 Aug;15(3):331–338. doi: 10.1099/00222615-15-3-331. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savio M. L., Pacciarini M. L., Cinco M., Tagliabue S. Identification of Leptospira interrogans strains by monoclonal antibodies and genomic analysis. New Microbiol. 1993 Oct;16(4):315–321. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tamai T., Sada E., Kobayashi Y. Restriction endonuclease DNA analysis of Leptospira interrogans serovars Icterohaemorrhagiae and Copenhageni. Microbiol Immunol. 1988;32(9):887–894. doi: 10.1111/j.1348-0421.1988.tb01450.x. [DOI] [PubMed] [Google Scholar]
- Terpstra W. J., Schoone G. J., Ligthart G. S., ter Schegget J. Detection of Leptospira interrogans in clinical specimens by in situ hybridization using biotin-labelled DNA probes. J Gen Microbiol. 1987 Apr;133(4):911–914. doi: 10.1099/00221287-133-4-911. [DOI] [PubMed] [Google Scholar]
- Terpstra W. J., Schoone G. J., ter Schegget J. Detection of leptospiral DNA by nucleic acid hybridisation with 32P- and biotin-labelled probes. J Med Microbiol. 1986 Aug;22(1):23–28. doi: 10.1099/00222615-22-1-23. [DOI] [PubMed] [Google Scholar]
- Thiermann A. B., Handsaker A. L., Moseley S. L., Kingscote B. New method for classification of leptospiral isolates belonging to serogroup pomona by restriction endonuclease analysis: serovar kennewicki. J Clin Microbiol. 1985 Apr;21(4):585–587. doi: 10.1128/jcm.21.4.585-587.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eys G. J., Gerritsen M. J., Korver H., Schoone G. J., Kroon C. C., Terpstra W. J. Characterization of serovars of the genus Leptospira by DNA hybridization with hardjobovis and icterohaemorrhagiae recombinant probes with special attention to serogroup sejroe. J Clin Microbiol. 1991 May;29(5):1042–1048. doi: 10.1128/jcm.29.5.1042-1048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eys G. J., Gravekamp C., Gerritsen M. J., Quint W., Cornelissen M. T., Schegget J. T., Terpstra W. J. Detection of leptospires in urine by polymerase chain reaction. J Clin Microbiol. 1989 Oct;27(10):2258–2262. doi: 10.1128/jcm.27.10.2258-2262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eys G. J., Zaal J., Schoone G. J., Terpstra W. J. DNA hybridization with hardjobovis-specific recombinant probes as a method for type discrimination of Leptospira interrogans serovar hardjo. J Gen Microbiol. 1988 Mar;134(3):567–574. doi: 10.1099/00221287-134-3-567. [DOI] [PubMed] [Google Scholar]
- Woodward M. J., Redstone J. S. Differentiation of leptospira serovars by the polymerase chain reaction and restriction fragment length polymorphism. Vet Rec. 1993 Mar 27;132(13):325–326. doi: 10.1136/vr.132.13.325. [DOI] [PubMed] [Google Scholar]
- Zuerner R. L., Bolin C. A. Nucleic acid probe characterizes Leptospira interrogans serovars by restriction fragment length polymorphisms. Vet Microbiol. 1990 Sep;24(3-4):355–366. doi: 10.1016/0378-1135(90)90183-v. [DOI] [PubMed] [Google Scholar]
- Zuerner R. L., Bolin C. A. Repetitive sequence element cloned from Leptospira interrogans serovar hardjo type hardjo-bovis provides a sensitive diagnostic probe for bovine leptospirosis. J Clin Microbiol. 1988 Dec;26(12):2495–2500. doi: 10.1128/jcm.26.12.2495-2500.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]