Abstract
MafA is a transcriptional regulator expressed primarily in pancreatic β cells. It binds to the RIPE3b/C1-binding site within the ins gene promoter, which plays a critical role in regulating ins gene expression in response to glucose. Here, we show that MafA is post-translationally modified by the small ubiquitin-related modifiers SUMO-1 and -2. Mutation of a single site in MafA, Lys32, blocks its sumoylation in β cells. Incubation of β cells in low glucose (2 mm) or exposure to hydrogen peroxide increases sumoylation of endogenous MafA. Forced sumoylation of MafA results in reduced transcriptional activity toward the ins gene promoter and increased suppression of the CHOP-10 gene promoter. Sumoylation of MafA has no apparent effect on either its nuclear localization in β cells or its ubiquitin-dependent degradation. This study suggests that modification of MafA by SUMO modulates gene transcription and thereby β cell function.
Insulin is essential for maintaining glucose homeostasis and is synthesized in and secreted from β cells in the islets of Langerhans. Glucose-sensitive cis-regulatory elements on the ins gene promoter are critical in regulating ins gene expression (1). Several transcription factors have been identified that stimulate ins gene transcription through the A and E boxes of the promoter, including Pdx-1 and Beta2/NeuroD1, both MODY (maturity onset diabetes of the young) genes (2). MafA also stimulates ins gene transcription by binding to the RIPE3b/C1 glucose-sensitive element, a Maf recognition element (MARE)3 (3–5). MafA belongs to the family of large Maf proteins, basic leucine zipper transcription factors (6–8). MafA contains a transactivation domain at its N terminus and a DNA-binding domain at its C terminus and homodimerizes through its basic leucine zipper domain. Within pancreatic islets, mafA expression is limited to β cells and is involved in transcription not only of the ins gene but also of other genes involved in β cell-specific functions (9). As might be expected for a regulator of ins gene transcription, MafA-deficient mice display glucose intolerance and develop diabetes, although impaired insulin secretion appears to be the primary defect (10). Additionally, islet structure is abnormal in these mice. Together, these findings indicate that MafA is required for the development and maintenance of mature insulin-producing pancreatic β cells.
Both transcription and post-translational modifications have been implicated in the regulation of MafA under diverse conditions. Transient exposure of β cells to high glucose has been reported to both increase and decrease MafA mRNA and protein (11, 12), whereas chronic exposure to high glucose or lipids has been reported to decrease MafA protein with or without a loss of MafA mRNA (13, 14). Glucose and oxidative stress are reported to regulate mafA expression at the transcriptional level through FoxO1 (15). FoxA2, Nkx2.2, and Pdx-1 modulate mafA expression through conserved sequences in the distal region of the mafA promoter (16). Phosphorylation is thought to be critical for MafA transcriptional activity (17). In vitro kinase assays suggest that MafA may be phosphorylated by ERK2 and p38 mitogen-activated protein kinases (18, 19). Two groups observed that following phosphorylation of Ser65 of MafA, it is sequentially phosphorylated at Ser61, Thr57, Thr53, and Ser49 by GSK3β (glycogen synthase kinase 3β) (17, 20). Phosphorylation at Ser14 has also been reported (18). MafA phosphorylation may also lead to its ubiquitination and degradation by the proteasome (20). These findings suggest that multiple covalent modifications of MafA control its function.
Covalent post-translational modification with SUMO (small ubiquitin-related modifier) regulates diverse cellular processes, including DNA repair, the cell cycle, gene transcription, and nucleocytoplasmic transport (21, 22). Mammals express four SUMO isoforms. Sumoylation with all isoforms occurs in a stepwise process that involves a cascade of SUMO-specific enzymes, an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ligase. SUMO proteases remove SUMO from their targeting proteins. Sumoylation is dynamic, with substrates undergoing rapid conjugation and deconjugation. Only small fractions of the substrates are thought to be subjected to sumoylation at steady state, although the underlying mechanism is still in question. Among the best characterized SUMO substrates are transcription factors, many of which display repressed transcriptional activity upon sumoylation. It has also been proposed that sumoylation of transcription factors contributes to the assembly of promoter complexes and the recruitment of chromatin-modifying enzymes (23).
Several ins gene transcription factors have consensus sumoylation motifs (ΦKXE); these include Pdx-1, C/EBP-β, NFAT, and MafA. Modification of C/EBP-β and NFAT1 by SUMO-1 is important in regulating their transcriptional activity and/or localization (24, 25). However, little is known about the sumoylation of ins gene transcription factors or the possible biological significance of the modification in β cells.
Based on the amino acid sequence, two sumoylation consensus motifs, VK32KE and LK296LE, are present in MafA. Because we are examining mechanisms of glucose-induced changes in transcription in β cells, we tested the idea that SUMO modification of MafA may affect ins gene expression and β cell function. Herein, we provide evidence that MafA is subjected to SUMO-1 or -2 modification in β cells. Hypoglycemia and oxidative stress are potential modulators of MafA sumoylation. Sumoylation regulates MafA transcriptional activity for both ins and CHOP-10 (C/EBP homologous protein 10) gene promoters. These findings demonstrate that MafA is sumoylated in β cells and suggest that sumoylation impacts β cell function.
EXPERIMENTAL PROCEDURES
Cell Culture and Harvest—Early passages of the pancreatic β cell lines rat INS-1 and mouse Min6 were kindly provided by Chris Newgard (Duke University) and Gene Webb (University of Chicago), respectively. Cells were maintained as described (26, 27). Cells at ∼80% confluence were incubated for 2 h in Krebs-Ringer bicarbonate/HEPES buffer containing 2 mm glucose and 0.1% bovine serum albumin (28) before stimulation with 30 mm glucose for Min6 cells and 25 mm glucose for INS-1 cells. After treatment with the reagents indicated in the figure legends, the medium was removed, and cells were washed with cold phosphate-buffered saline (PBS) and harvested in cold lysis buffer (50 mm HEPES, pH 7.5, 0.15 m NaCl, 1% Triton X-100, 0.2 mg/ml phenylmethylsulfonyl fluoride, 0.1 m NaF, 2 mm Na3VO4,10 μg/ml aprotinin, 5 μg/ml pepstatin A, 5 μg/ml leupeptin, and 20 μm N-ethylmaleimide). After 15 min on ice, lysates were sedimented for 15 min at 16,000 × g in a microcentrifuge at 4 °C. Supernatants were stored at -80 °C until further analysis. Protein concentrations were measured using the Bio-Rad Bradford reagent.
Materials—The Dual-Luciferase reporter assay system and passive lysis buffer were purchased from Promega (Madison, WI). The anti-ERK1/2 antibody Y691 was as described (29). The following antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA): c-Maf (M-153), SUMO-1, green fluorescent protein (GFP), rabbit IgG, and Myc. The following reagents were purchased from Sigma: hydrogen peroxide, ionomycin, anisomycin, N-acetyl-l-cysteine, catalase, and N-ethylmaleimide. A vector harboring full-length human mafA was kindly provided by Michael German (University of California, San Francisco). MafA(K32R) was provided by Michael Lawrence. The mafA coding sequence was subcloned into pCMV5 with an N-terminal Myc tag. Vector pCS2 containing N-terminal GFP-SUMO-1, SUMO-1-ΔGG, SUMO-2, hemagglutinin (HA)-SENP2 (SUMO-1/sentrin-specific peptidase 2), FLAG-Ubc9, and Ubc9(C93S) (an interfering mutant) were gifts from Hongtao Yu (University of Texas Southwestern Medical Center). HA-SUMO-1 in pSFFV was a gift from Kim Orth (University of Texas Southwestern Medical Center). Site-directed mutagenesis was performing with the QuikChange kit (Stratagene) according to the manufacturer's instructions. Complementary oligonucleotides from regions -320 to -300 bp of the CHOP-10 gene and A2E1 and MARE of the insI gene were synthesized (Integrated DNA Technologies, Coralville, IA) and blunt end-ligated as 3× repeats into the SmaI site of the pGL3-Luc reporter (Promega). All constructs and mutants were confirmed by sequencing.
Immunoprecipitation and Immunoblotting—Aliquots of cell extracts containing 20 μg of protein were subjected to SDS-PAGE. Proteins were electrotransferred to nitrocellulose membranes (Millipore, Billerica, MA) and blocked with 5% nonfat powdered milk in Tris-buffered saline (20 mm Tris, pH 7.5, and 0.15 m NaCl) containing 0.1% Tween 20 for 2 h at room temperature and incubated with primary antibody overnight at 4 °C, followed by incubation with horseradish peroxidase-conjugated secondary antibody diluted 1:3000 in Tris-buffered saline for 30 min at room temperature. Proteins were detected by enhanced chemiluminescence. In some cases, blots were analyzed by densitometry using a ScanJet 5300C scanner and Multi-Gauge Version 2.3 software. For immunoprecipitation, lysates (0.5–1 mg) were incubated overnight with the indicated antibody and protein A-Sepharose beads at 4 °C. The beads were washed three times with 25 mm Tris, pH 7.4, 0.1 m NaCl, and 0.1% Triton X-100.
Transfections and Reporter Assays—INS-1, Min6, or HEK293 cells were grown in 6-well plates to 80% confluence and cotransfected with pGL3-MARE, pGL3-CHOP-10 (-320 to -300 bp), or pGL3-A2E1, together with pRL-SV40, and the indicated plasmids using the FuGENE HD reagent (Roche Applied Science). After 48 h, the cells were harvested with passive lysis buffer supplemented with 100 mm β-glycerophosphate, 2 mm Na3VO4, 100 mm NaF, 10 μg/ml aprotinin, 5 μg/ml pepstatin A, 5 μg/ml leupeptin, and 0.2 mg/ml phenylmethylsulfonyl fluoride. The lysates were frozen in liquid nitrogen, thawed, and then vortexed for 30 s prior to centrifugation as described above. Promoter activity in the supernatants was assayed with the Dual-Luciferase assay system using a TD-20/20 bioluminometer (Turner Designs), and protein expression was assessed by blotting.
Immunofluorescence—Cells were plated onto coverslips in 12-well dishes and transfected with the indicated plasmids. After 48 h, cells were fixed with ice-cold methanol for 1 h at -20 °C, rinsed three times with PBS, and permeabilized with 0.2% Triton X-100 in PBS for 15 min at 4 °C. Prior to addition of antibodies, cells were incubated in 0.1% Triton X-100 and 4% bovine serum albumin in PBS for 2 h. Primary antibody in the same solution was incubated overnight with cells at 4 °C. After washing, cells were incubated with secondary antibody (1:5000) and 4′,6-diamidino-2-phenylindole (1:10,000) for 1 h. Fluorophores were visualized using a DeltaVision RT deconvolution microscope (Applied Precision, Issaquah, WA). Z-stacks were deconvolved and processed with NIH Image J software.
Subcellular Fractionation—Min6 cells transfected with the indicated plasmids were washed with PBS and harvested in hypotonic lysis buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm dithiothreitol, 1 mm NaF, and 0.5 mm phenylmethylsulfonyl fluoride) with other protease inhibitors as described above. After swelling for 15 min on ice, Nonidet P-40 was added to a final concentration of 0.6%, followed by vortexing for 10 s. A cytosolic fraction was obtained by centrifugation at 5000 × g for 5 min. The pellet was resuspended in hypertonic extraction buffer (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, and 1 mm NaF) plus 1 mm phenylmethylsulfonyl fluoride and other protease inhibitors as described above and incubated on ice for 30 min with vortexing every 5 min. The nuclear extract was obtained by sedimenting the insoluble material at 16,000 × g for 15 min.
Electrophoretic Mobility Shift Assays—Complementary oligonucleotides containing wild-type MARE (5′-TACAGCTTCAGCCCCTCTCGCCATC) and mutant MARE (5′-TACAGCTGACTACCCTCTCGCCATC) consensus sites of the rat insI gene promoter were synthesized (Integrated DNA Technologies). The oligonucleotides were hybridized and end-labeled with T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP. Equal amounts of nuclear extract protein (20 μg) were incubated for 30 min with double-stranded 32P-labeled MARE probe (20,000 cpm) in reaction buffer (10 mm Tris, pH 8.0, 50 mm KCl, 1 mm EDTA, 1 mm dithiothreitol, and 6% glycerol). As indicated, antibodies were added 2 h before incubation with the labeled probe. The reactions were subjected to electrophoresis on 5% polyacrylamide gels, and bands were detected on film by autoradiography.
Statistical Analysis—Results are expressed as means ± S.E. determined from at least three independent experiments. Statistical significance was calculated by analysis of variance as indicated in the figure legends.
RESULTS
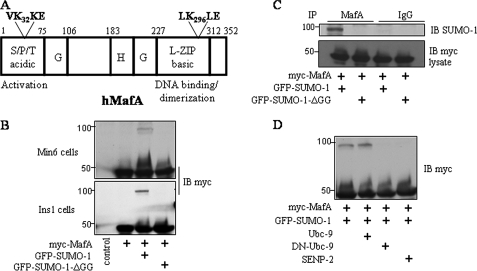
MafA Is Modified by SUMO-1 in Pancreatic β Cells—Previous studies showed that Maf family members MafG and MafB can be modified by SUMO (30, 31). Because there are two sumoylation consensus motifs (VK32KE and LK296LE) in the activation and DNA-binding domains of MafA (Fig. 1A), we investigated whether MafA undergoes sumoylation in pancreatic β cells. In β cells cultured in standard medium, no slowly migrating products, typical of ubiquitin- or SUMO-protein conjugates, were detected by immunoblotting. Because only a relatively small percentage of SUMO substrates are usually found sumoylated in vivo, we transiently expressed epitope-tagged MafA in both Min6 and INS-1 cells. Upon coexpression of Myc-MafA with HA- or GFP-SUMO-1, we observed a Myc-MafA species with an apparent molecular mass increased by the amount expected upon modification by SUMO-1 in β cells (Figs. 1B and 2). Under these conditions, a relatively small amount of MafA was modified. The more slowly migrating band was absent when Myc-MafA was coexpressed with SUMO-1-ΔGG, which lacks the C-terminal diglycine motif and cannot be conjugated to substrates (Fig. 1B). To provide additional evidence that the slowly migrating band is indeed the SUMO-1-modified form of MafA, INS-1 cells were cotransfected with Myc-MafA and GFP-SUMO-1, and MafA was immunoprecipitated with the anti-Maf antibody. Immunoblotting with an antibody to SUMO-1 detected GFP-SUMO-1 linked to MafA in cells transfected with SUMO-1 but not in cells transfected with SUMO-1-ΔGG (Fig. 1C). A comparable profile of MafA sumoylation was observed in HEK293 cells (data not shown). The extent of MafA sumoylation was similar in the presence or absence of overexpression of Ubc9 (ubiquitin-like conjugating enzyme 9), a SUMO-conjugating enzyme (E2) that activates the sumoylation pathway (Fig. 1D). This indicates that Ubc9 is not rate-limiting and suggests that a SUMO E3 ligase would be required to enhance the efficiency of MafA sumoylation. In contrast, coexpression of dominant-negative Ubc9 or the SUMO-specific protease SENP2 reduced detection of sumoylated MafA (Fig. 1D). Thus, SUMO-1 can be covalently coupled to MafA in β cells.
FIGURE 1.
Sumoylation of MafA in pancreatic β cells. A, human MafA (hMafA) with its known domains and functions is shown. VK32KE and LK296LE are two consensus sumoylation motifs. B, Myc-tagged MafA was coexpressed with GFP-SUMO-1 or GFP-SUMO-1-ΔGG in Min6 or INS-1 cells. Cell extracts were immunoblotted (IB) with an antibody to Myc. C, lysates from INS-1 cells expressing Myc-MafA and GFP-SUMO-1 or GFP-SUMO-1-ΔGG were immunoprecipitated (IP) with an anti-c-Maf antibody or control IgG and then detected by immunoblotting with an antibody to SUMO-1. The lower panel shows an anti-Myc immunoblot of the lysates. D, INS-1 cells were transfected with Myc-MafA, GFP-SUMO-1, FLAG-Ubc9, FLAG-dominant-negative (DN) Ubc9, and HA-SENP2 in different combinations as indicated, and cell lysates were immunoblotted with an antibody to Myc. Experiments in each panel were repeated at least three times.
FIGURE 2.
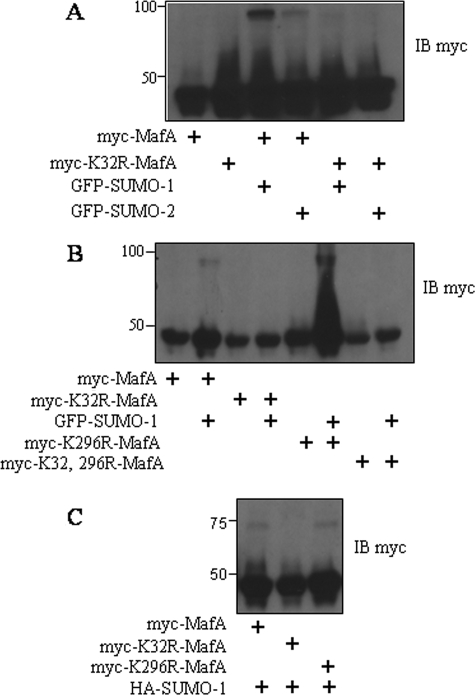
MafA mutants in pancreatic β cells. A–C, immunoblots (IB) with an antibody to Myc of the lysates from INS-1 cells expressing wild-type Myc-MafA, MafA(K32R), MafA(K296R), or MafA(K32R/K296R) and GFP-SUMO-1 or -2 or HA-SUMO-1 as indicated. All experiments were repeated three or more times.
MafA Is Sumoylated at Lys32—SUMO-1 consists of 101 amino acids and shares ∼50% sequence identity with SUMO-2/3. To test whether MafA can be modified by SUMO-2, we coexpressed Myc-MafA and GFP-SUMO-2 in INS-1 cells. Immunoblotting with an antibody to Myc showed that MafA was modified by SUMO-2, but to a lesser extent than by SUMO-1 (Fig. 2A). To identify the sumoylation sites, we made MafA(K32R), MafA(K296R), and the double mutant by site-directed mutagenesis. Sumoylation of MafA(K32R) and MafA(K32R/K296R) was no longer detected in the presence of overexpressed GFP-SUMO-1 or -2. However, MafA(K296R) was still modified (Fig. 2B). Coexpression with HA-SUMO-1 showed similar results (Fig. 2C). These data are consistent with the conclusion that Lys32 is the major site of sumoylation on MafA in pancreatic β cells.
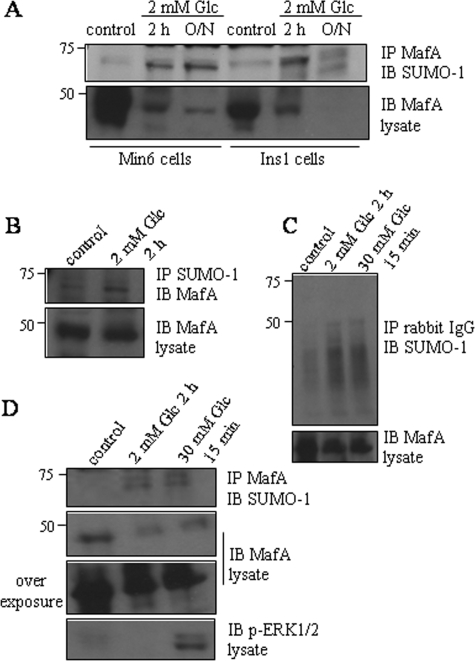
Exposure of Pancreatic β Cells to Low Glucose Increases Sumoylation of Endogenous MafA—To establish the relevance of MafA sumoylation in β cell function, we examined the effect of varying glucose concentrations on endogenous MafA sumoylation. Immunoblotting of immunoprecipitated MafA with anti-SUMO antibodies showed a small amount of endogenous sumoylated MafA in both Min6 and INS-1 cells maintained in standard medium (i.e. in 25 or 11 mm glucose) (Fig. 3A). We confirmed that the 75-kDa MafA species was sumoylated by immunoprecipitating with an anti-SUMO-1 antibody and immunoblotting with an anti-Maf antibody (Fig. 3B). used as a negative control, normal rabbit IgG did not immunoprecipitate these proteins (Fig. 3C). Placing β cells in 2 mm glucose for 2 h enhanced MafA sumoylation. Sumoylation increased further with longer times in low glucose (Fig. 3, A and B). These conditions also dramatically reduced the total amount of MafA, consistent with a previous report (17). Subsequent stimulation of β cells with 30 mm glucose for 15 min following incubation in 2 mm glucose did not decrease the amount of sumoylated MafA (Fig. 3D). These results indicate that varying the glucose concentration can affect the extent of MafA sumoylation in pancreatic β cells.
FIGURE 3.
Effects of glucose on MafA sumoylation in pancreatic β cells. A, lysates from INS-1 or Min6 cells placed in 2 mm glucose (low glucose) for 2 h or overnight (O/N) were immunoprecipitated (IP) with an antibody to c-Maf and immunoblotted (IB) with an antibody to SUMO-1. The lower panel shows an anti-Maf immunoblot of the lysates. B, lysates from Min6 cells starved for 2 h were immunoprecipitated with an antibody to SUMO-1 and immunoblotted with an antibody to c-Maf. The lower panel is an anti-Maf immunoblot of the lysates. C and D, lysates from Min6 cells starved for 2 h or stimulated with 30 mm glucose for 15 min after starvation were immunoprecipitated with an antibody to c-Maf or control rabbit IgG and immunoblotted with an anti-SUMO-1 antibody. The lower panel is an anti-Maf immunoblot of the lysates. All experiments were repeated three times.
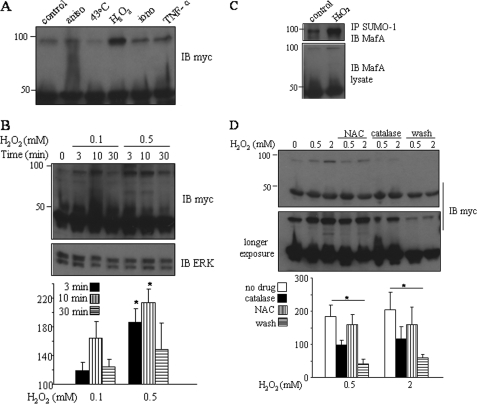
Oxidative Stress Enhances Sumoylation of MafA in Pancreatic β Cells—Protein sumoylation can be induced by heat shock, oxidative stress, electrical stimulation, and ethanol in some systems (32, 33). To identify factors which can affect MafA sumoylation in pancreatic β cells, we exposed β cells to different stimuli, including KCl, anisomycin, ionomycin, hydrogen peroxide, tumor necrosis factor-α, heat shock, and ethanol. Only hydrogen peroxide stimulated the sumoylation of overexpressed MafA to a measurable extent in both Min6 and INS-1 cells (Fig. 4A and data not shown). To explore this finding further, we tested the effects of different concentrations of hydrogen peroxide (0.1, 0.5, 1, and 2 mm) and different times of exposure (3, 10, and 30 min). As little as 0.1 mm hydrogen peroxide increased MafA sumoylation, with the greatest effect observed after 10 min (Fig. 4B). That the band was the sumoylated form of MafA was confirmed by immunoblotting of anti-SUMO-1 immunoprecipitates with an anti-Maf antibody (Fig. 4C). We exposed β cells to catalase and the antioxidant N-acetyl-l-cysteine after hydrogen peroxide treatment. Both agents modestly reduced the amount of sumoylated MafA (Fig. 4D), consistent with a role for oxidative stress. Placing β cells in fresh culture medium for 30 min after hydrogen peroxide treatment more effectively reduced MafA sumoylation (Fig. 4D).
FIGURE 4.
Effects of oxidative stress on MafA sumoylation in pancreatic β cells. A, INS-1 cells were cotransfected with Myc-MafA and GFP-SUMO-1, followed by treatment with 50 μm anisomycin (aniso), 1 μm ionomycin (iono), 100 ng/ml tumor necrosis factor-α (TNF-α), or 0.6 mm H2O2 for 10 min or at 43 °C for 30 min. Lysates were analyzed by immunoblotting (IB) with an antibody to Myc. B, INS-1 cells cotransfected with Myc-MafA and GFP-SUMO-1 were treated with 0.1 or 0.5 mm H2O2 for 3, 10, or 30 min. Lysates were subjected to immunoblotting with an anti-Myc antibody. The lower panel shows an anti-ERK1/2 immunoblot of the lysates. C, lysates from Min6 cells transfected with Myc-MafA and GFP-SUMO-1 and treated with 0.5 mm H2O2 for 10 min were immunoprecipitated (IP) with an anti-SUMO-1 antibody, followed by immunoblotting with an antibody to c-Maf. The lower panel shows an immunoblot of the lysates with an anti-c-Maf antibody. D, INS-1 cells transfected with Myc-MafA and GFP-SUMO-1 were treated with 100 μm N-acetyl-l-cysteine (NAC) for 1 h, followed by 0.5 or 2 mm H2O2 for 10 min; with 1000 units/ml catalase and H2O2 for 10 min; or with H2O2 for 10 min, followed by washing with fresh medium for 30 min. Lysates were immunoblotted with an antibody to Myc. In B and D, the lower panels show results expressed as the means ± S.E. relative to the control set at 100% of three independent experiments. Differences between groups were analyzed using one way analysis of variance, followed by the Holm-Sidak method (*, p < 0.001).
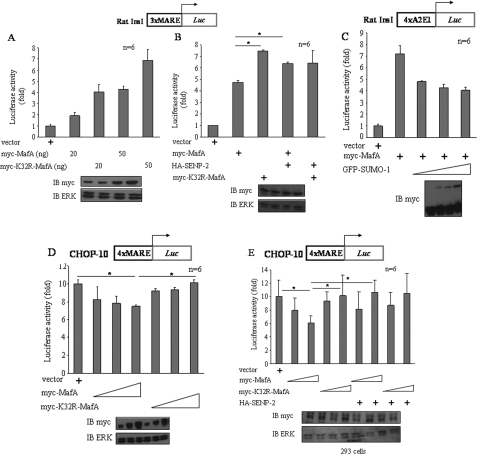
Preventing Sumoylation of MafA Alters Its Transcriptional Activity—To assess the functional consequences of MafA sumoylation, we compared wild-type MafA and MafA(K32R), the mutant lacking the SUMO-1 acceptor site, in transcriptional reporter assays. INS-1 cells were cotransfected with a reporter plasmid containing the rat insI MARE and the MafA constructs. The K32R mutant induced greater reporter activity than an equal amount of wild-type MafA under several conditions, consistent with the idea that sumoylation inhibits MafA activity for the ins gene promoter (Fig. 5, A and B). Further support for this hypothesis came from coexpression with the protease SENP2, which increased reporter activity induced by wild-type MafA, presumably due to reversal of its fractional sumoylation, but had no effect on the activity of MafA(K32R) (Fig. 5B). Min6 cells were transfected with another reporter construct containing the rat insI A2E1 region, which includes MafA- and other transcription factor-binding sites, along with MafA. Increasing amounts of SUMO-1 reduced reporter gene expression (Fig. 5C), also consistent with an inhibitory effect of MafA sumoylation on ins gene transcription.
FIGURE 5.
Effects of sumoylation of MafA on its transcriptional activity in pancreatic β cells. A and B, luciferase assays of rat insI gene promoter region-3×MARE in INS-1 cells overexpressing the indicated plasmids. The lower panels show immunoblots (IB) of the lysates with antibodies to Myc and ERK1/2 (n = 6). C–E, promoter-reporter assays of rat insI gene promoter region-4×A2E1 (C) and CHOP-10 gene promoter region-3×-320/-300 bp (D and E) in Min6 (C), INS-1 (D), and HEK293 (E) cells overexpressing the indicated plasmids. Immunoblots in the lower panels show myc expression. The sumoylated form of MafA is indicated on the blot. Results are expressed as the means ± S.E. of six replicates. Differences between groups were analyzed using one way analysis of variance, followed by Tukey's test in B and D (*, p < 0.05), and using one way analysis of variance, followed by Duncan's method in E (*, p < 0.05).
Disruption of the CHOP-10 gene delays β cell death caused by endoplasmic reticulum stress (34). We showed previously that expression of the CHOP-10 gene is induced in pancreatic β cells maintained in 5.5 mm glucose and repressed by higher concentrations of glucose (27). This is due in part to MafA, which represses CHOP-10 transcription by binding to a MARE-CEB (CAAT enhancer-binding element) site (-320 to -300 bp) in the promoter. We asked whether overexpression of MafA or MafA(K32R) has an effect on the transcriptional activity for the CHOP-10 promoter in pancreatic β cells. Using luciferase linked to the MARE-CEB site from the CHOP-10 promoter, we found that MafA(K32R) had little or no ability to repress reporter gene expression. In contrast, wild-type MafA caused a small but statistically significant repression (Fig. 5D), suggesting that transcriptional repression of this promoter requires modification, presumably sumoylation, of MafA at Lys32. To explore this observation further, wild-type MafA and MafA(K32R) were expressed along with the MARE-CEB site from the CHOP-10 promoter in HEK293 cells, which express no detectable endogenous MafA. Wild-type MafA was able to repress expression of the reporter, whereas MafA(K32R) did not (Fig. 5E). Furthermore, expression of SENP2 reversed repression induced by wild-type MafA. These results support the conclusion that repression of CHOP-10 requires sumoylated MafA.
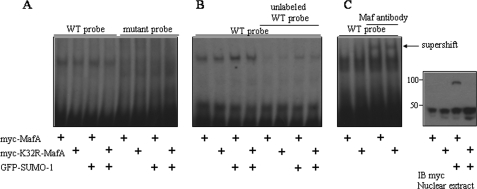
To examine the effects on interaction with the ins gene promoter, wild-type MafA or MafA(K32R) was overexpressed in INS-1 cells or not, and nuclear extracts were prepared for electrophoretic mobility shift assays. Extracts from cells expressing the mutant, which could not be sumoylated, bound DNA as well as did extracts from cells expressing wild-type MafA (Fig. 6, A and B). Binding could be competed by a 50-fold excess of unlabeled wild-type probe. In supershift assays, the anti-Maf antibody bound to both samples, causing a supershift. Expression of SUMO-1 had no detectable impact on the amount of oligonucleotide-bound complex.
FIGURE 6.
Effects of sumoylation of MafA on its DNA binding to the ins gene promoter. A, nuclear extracts of INS-1 cells transfected with the indicated plasmids were incubated with [γ-32P]ATP-labeled wild-type (WT) MARE or mutant probe, followed by electrophoresis. B, electrophoretic mobility shift assay was carried out using INS-1 nuclear extracts with or without preincubation with a 50-fold excess of unlabeled MARE probe. C, electrophoretic mobility shift assay was carried out using INS-1 nuclear extracts incubated without or with an anti-c-Maf antibody. The right panel shows the immunoblot (IB) of nuclear extracts with an anti-Myc antibody. The results in A and B are representative of three experiments and those in C of two experiments.
MafA Sumoylation Has No Detectable Impact on Its Nuclear Localization or Stability in β Cells—Sumoylation has been shown to alter the subcellular localization of certain proteins. To investigate whether MafA sumoylation grossly alters its subcellular localization in β cells, we expressed wild-type MafA, MafA(K32R), and SUMO-1 in different combinations in Min6 cells, followed by indirect immunofluorescence staining. In agreement with previous studies, endogenous MafA was detected in the nucleus of β cells. Overexpressed SUMO-1 was also localized in the nucleus. Expression of SUMO-1 did not detectably alter the localization of endogenous MafA (data not shown). Similar localization patterns were also observed with both wild-type Myc-MafA and the K32R mutant in the presence or absence of coexpressed SUMO-1. Using wild-type MafA plus SUMO-1, the fraction of tagged MafA that was sumoylated should have been greater; using the mutant, the sumoylated species should have been eliminated. In neither case were differences observed. Immunoblotting of SUMO-MafA in subcellular fractions also revealed no differences in its subcellular distribution profile relative to the form that could not be modified (data not shown).
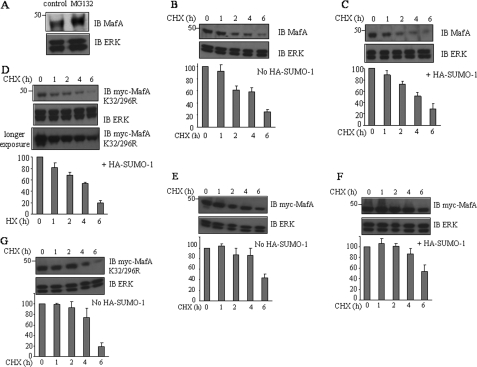
To confirm that MafA is degraded by the proteasome, we treated INS-1 cells with MG132, a proteasome inhibitor. Immunoblotting showed that endogenous MafA protein increased in MG132-treated cells (Fig. 7A). We investigated whether sumoylation might affect ubiquitin-dependent degradation of MafA. Wild-type MafA or the non-sumoylatable K32R or K32R/K296R mutant was expressed in INS-1 cells, and the cells were treated with cycloheximide. Coexpression of SUMO-1 did not greatly affect the half-life of endogenous or overexpressed MafA (Fig. 7, B–G). Wild-type MafA and the double mutant had similar steady-state expression in the presence of the protein synthesis inhibitor (Fig. 7, F and G). These results suggest that SUMO and ubiquitin modifications of MafA do not compete for the same lysine residue.
FIGURE 7.
Effects of sumoylation of MafA on its stability in pancreaticβ cells. A, lysates of INS-1 cells treated with 25 μm MG132 for 6 h were analyzed by immunoblotting with antibodies to c-Maf and ERK1/2. INS-1 cells were either untransfected or transfected with various combinations of GFP-SUMO-1, Myc-MafA, and Myc-MafA(K32R/K296R). After 48 h, cells were treated with 20 μg/ml cycloheximide (CHX) and harvested at 0, 1, 2, 4, or 6 h later. Lysates were subjected to immunoblotting (IB) with antibodies to Myc, c-Maf, and ERK1/2. Experiments were repeated three times. MafA decay curves were plotted from densitometric scans from each experiment. Decay curves for endogenous (B and C) and heterologously expressed (E and F) wild-type MafA and MafA(K32R/K296R) (D and G) are shown from cells also not expressing (B, E, and G) or expressing (C, D, and F) GFP-SUMO-1.
DISCUSSION
We show here that endogenous MafA is sumoylated in pancreatic β cells. To identify the functions associated with this modification, we examined the effects of MafA sumoylation on the ins gene and CHOP-10 gene promoters. Reporter activity driven by the MafA-responsive sequence from the ins gene promoter is enhanced by preventing sumoylation of MafA, leading to the idea that sumoylation of MafA inhibits its ability to stimulate transcription. We found previously that MafA binding to the CHOP-10 promoter is associated with inhibition of CHOP-10 expression (27). Here, we found that blocking sumoylation of MafA prevents transcriptional repression of the CHOP-10 promoter in β cells or in a reconstituted system lacking endogenous MafA, consistent with the conclusion that inhibition of CHOP-10 gene expression by MafA requires its sumoylation. Taken together, these results suggest that sumoylation of MafA regulates important biological activities of this protein that may enhance β cell survival as well as modulate ins gene expression.
Previous chromatin immunoprecipitation experiments demonstrated that MafA is bound to the CHOP-10 promoter under conditions in which CHOP-10 promoter activity is repressed (27). MafA lacking the sumoylation site is not capable of inhibiting CHOP-10 promoter activity in cells lacking endogenous MafA. From these experiments, we conclude that reduced CHOP-10 promoter activity requires binding of sumoylated MafA to the CHOP-10 promoter. Because MafA binds to the promoter under conditions in which it represses transcription and because sumoylated MafA is required for promoter inhibition, it seems most likely that neither altered localization nor degradation plays a substantial role in the functional effects detected on transcription. This hypothesis is supported by the lack of observable changes in these behaviors upon a forced increase in sumoylation.
Transcriptional repression is a function commonly associated with sumoylation (22, 33, 35). The molecular mechanisms by which SUMO modification represses gene transcription may be several. Sumoylation may recruit corepressors or other chromatin modulators, such as histone deacetylases, to promoters. These modulators may assist in initiating the formation of repressive complexes or in retaining the repressive chromatin state, which could account for the substantial effect of low level sumoylated proteins.
Increased sumoylation of MafA is associated with a less robust stimulation of ins promoter activity. For the ins promoter, it is unclear if the decrease in stimulatory activity is due to effects of sumoylated MafA on the promoter complex, such as recruitment of deacetylases, or to lack of sumoylated MafA binding to the promoter. Electrophoretic mobility shift assay experiments suggest that sumoylation does not prevent DNA binding. Further examination of MafA interactions on the ins promoter will be required to define the mechanism underlying its altered function in ins gene transcription.
Mutagenesis indicates that a single site, Lys32, is the predominant, if not only, sumoylation site in MafA, consistent with what was recently reported for MafB (30). Lys32 is in a region containing several reported phosphorylation sites, but is not immediately juxtaposed to these sites. Previous studies have not unambiguously delineated the effects of these putative phosphorylations on MafA activity; thus, predicting how these modifications may interact is not yet possible. Several studies of the ternary complex factor Elk-1 have revealed interplay between phosphorylation and sumoylation that may well serve as a model for the regulation of MafA function. Phosphorylation enhances Elk-1 transcriptional activity, whereas sumoylation inhibits this function (36).
Important questions remaining to be answered concern the mechanisms that determine the extent of MafA sumoylation in β cells and the role of glucose concentration. β cells experience high levels of oxidative stress that are thought to contribute to cell death in response to hyperglycemia (34, 37). The increase in MafA sumoylation by hydrogen peroxide suggested that exposure to elevated glucose might also increase MafA sumoylation; however, the greatest sumoylation of endogenous MafA was detected in cells in low not high glucose. Ongoing studies focus on defining these mechanisms.
Acknowledgments
We thank Hongtao Yu and members of his laboratory for reagents and comments, Michael Lawrence and Eric Wauson for suggestions about this work, Kathy McGlynn for technical assistance, and Dionne Ware for administrative assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant DK55310. This work was also supported by Robert A. Welch Foundation Grant I1243. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MARE, Maf recognition element; PBS, phosphate-buffered saline; GFP, green fluorescent protein; HA, hemagglutinin.
References
- 1.German, M., Ashcroft, S., Docherty, K., Edlund, H., Edlund, T., Goodison, S., Imura, H., Kennedy, G., Madsen, O., Melloul, D., Moss, L., Olson, K., Permutt, M. A., Philippe, J., Robertson, B. P., Rutter, W. J., Serup, P., Stein, R., Steiner, D., Tsai, M. J., and Walker, M. D. (1995) Diabetes 44 1002-1004 [DOI] [PubMed] [Google Scholar]
- 2.Malecki, M. T. (2005) Diabetes Res. Clin. Pract. 68 Suppl. 1, S10-S2115955369 [Google Scholar]
- 3.Olbrot, M., Rud, J., Moss, L. G., and Sharma, A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 6737-6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuoka, T. A., Zhao, L., Artner, I., Jarrett, H. W., Friedman, D., Means, A., and Stein, R. (2003) Mol. Cell. Biol. 23 6049-6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seufert, J., Weir, G. C., and Habener, J. F. (1998) J. Clin. Investig. 101 2528-2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blank, V., and Andrews, N. C. (1997) Trends Biochem. Sci. 22 437-441 [DOI] [PubMed] [Google Scholar]
- 7.Yang, Y., and Cvekl, A. (2007) Einstein J. Biol. Med. 23 2-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motohashi, H., O'Connor, T., Katsuoka, F., Engel, J. D., and Yamamoto, M. (2002) Gene (Amst.) 294 1-12 [DOI] [PubMed] [Google Scholar]
- 9.Aramata, S., Han, S. I., and Kataoka, K. (2007) Endocr. J. 54 659-666 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, C., Moriguchi, T., Kajihara, M., Esaki, R., Harada, A., Shimohata, H., Oishi, H., Hamada, M., Morito, N., Hasegawa, K., Kudo, T., Engel, J. D., Yamamoto, M., and Takahashi, S. (2005) Mol. Cell. Biol. 25 4969-4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao, L., Guo, M., Matsuoka, T. A., Hagman, D. K., Parazzoli, S. D., Poitout, V., and Stein, R. (2005) J. Biol. Chem. 280 11887-11894 [DOI] [PubMed] [Google Scholar]
- 12.Ye, D. Z., Tai, M. H., Linning, K. D., Szabo, C., and Olson, L. K. (2006) Diabetes 55 742-750 [DOI] [PubMed] [Google Scholar]
- 13.Harmon, J. S., Stein, R., and Robertson, R. P. (2005) J. Biol. Chem. 280 11107-11113 [DOI] [PubMed] [Google Scholar]
- 14.Hagman, D. K., Hays, L. B., Parazzoli, S. D., and Poitout, V. (2005) J. Biol. Chem. 280 32413-32418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitamura, Y. I., Kitamura, T., Kruse, J. P., Raum, J. C., Stein, R., Gu, W., and Accili, D. (2005) Cell Metab. 2 153-163 [DOI] [PubMed] [Google Scholar]
- 16.Raum, J. C., Gerrish, K., Artner, I., Henderson, E., Guo, M., Sussel, L., Schisler, J. C., Newgard, C. B., and Stein, R. (2006) Mol. Cell. Biol. 26 5735-5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, S. I., Aramata, S., Yasuda, K., and Kataoka, K. (2007) Mol. Cell. Biol. 27 6593-6605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benkhelifa, S., Provot, S., Nabais, E., Eychene, A., Calothy, G., and Felder-Schmittbuhl, M. P. (2001) Mol. Cell. Biol. 21 4441-4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sii-Felice, K., Pouponnot, C., Gillet, S., Lecoin, L., Girault, J. A., Eychene, A., and Felder-Schmittbuhl, M. P. (2005) FEBS Lett. 579 3547-3554 [DOI] [PubMed] [Google Scholar]
- 20.Rocques, N., Abou, Z. N., Sii-Felice, K., Lecoin, L., Felder-Schmittbuhl, M. P., Eychene, A., and Pouponnot, C. (2007) Mol. Cell 28 584-597 [DOI] [PubMed] [Google Scholar]
- 21.Kerscher, O., Felberbaum, R., and Hochstrasser, M. (2006) Annu. Rev. Cell Dev. Biol. 22 159-180 [DOI] [PubMed] [Google Scholar]
- 22.Gill, G. (2005) Curr. Opin. Genet. Dev. 15 536-541 [DOI] [PubMed] [Google Scholar]
- 23.Yang, X. J., and Gregoire, S. (2006) Mol. Cell 23 779-786 [DOI] [PubMed] [Google Scholar]
- 24.Eaton, E. M., and Sealy, L. (2003) J. Biol. Chem. 278 33416-33421 [DOI] [PubMed] [Google Scholar]
- 25.Terui, Y., Saad, N., Jia, S., McKeon, F., and Yuan, J. (2004) J. Biol. Chem. 279 28257-28265 [DOI] [PubMed] [Google Scholar]
- 26.Lawrence, M. C., McGlynn, K., Park, B. H., and Cobb, M. H. (2005) J. Biol. Chem. 280 26751-26759 [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, M. C., McGlynn, K., Naziruddin, B., Levy, M. F., and Cobb, M. H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11518-11525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khoo, S., and Cobb, M. H. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 5599-5604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boulton, T. G., and Cobb, M. H. (1991) Cell Regul. 2 357-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tillmanns, S., Otto, C., Jaffray, E., Duroure, C., Bakri, Y., Vanhille, L., Sarrazin, S., Hay, R. T., and Sieweke, M. H. (2007) Mol. Cell. Biol. 27 5554-5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motohashi, H., Katsuoka, F., Miyoshi, C., Uchimura, Y., Saitoh, H., Francastel, C., Engel, J. D., and Yamamoto, M. (2006) Mol. Cell. Biol. 26 4652-4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bossis, G., and Melchior, F. (2006) Mol. Cell 21 349-357 [DOI] [PubMed] [Google Scholar]
- 33.Geiss-Friedlander, R., and Melchior, F. (2007) Nat. Rev. Mol. Cell Biol. 8 947-956 [DOI] [PubMed] [Google Scholar]
- 34.Oyadomari, S., Koizumi, A., Takeda, K., Gotoh, T., Akira, S., Araki, E., and Mori, M. (2002) J. Clin. Investig. 109 525-532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, S. H., and Sharrocks, A. D. (2004) Mol. Cell 13 611-617 [DOI] [PubMed] [Google Scholar]
- 36.Yang, S. H., Jaffray, E., Hay, R. T., and Sharrocks, A. D. (2003) Mol. Cell 12 63-74 [DOI] [PubMed] [Google Scholar]
- 37.Poitout, V., and Robertson, R. P. (2002) Endocrinology 143 339-342 [DOI] [PubMed] [Google Scholar]