Abstract
A synthetic pathway has been constructed for the production of glucuronic and glucaric acids from glucose in Escherichia coli. Coexpression of the genes encoding myo-inositol-1-phosphate synthase (Ino1) from Saccharomyces cerevisiae and myo-inositol oxygenase (MIOX) from mice led to production of glucuronic acid through the intermediate myo-inositol. Glucuronic acid concentrations up to 0.3 g/liter were measured in the culture broth. The activity of MIOX was rate limiting, resulting in the accumulation of both myo-inositol and glucuronic acid as final products, in approximately equal concentrations. Inclusion of a third enzyme, uronate dehydrogenase (Udh) from Pseudomonas syringae, facilitated the conversion of glucuronic acid to glucaric acid. The activity of this recombinant enzyme was more than 2 orders of magnitude higher than that of Ino1 and MIOX and increased overall flux through the pathway such that glucaric acid concentrations in excess of 1 g/liter were observed. This represents a novel microbial system for the biological production of glucaric acid, a “top value-added chemical” from biomass.
The discipline of metabolic engineering was defined more than 15 years ago as “the improvement of cellular activities by manipulations of enzymatic, transport, and regulatory functions of the cell with the use of recombinant DNA technology” (5). It has shown its potential to optimize cellular functions for many purposes including recombinant protein production and pathway engineering for productivity enhancement. More recently, the field has produced several important accomplishments in the area of pathway design for new product generation. We consider a designed pathway as one in which a sequence of conversions is assembled from the individual enzymes of multiple hosts, in a sequence that is not native to any single organism. As examples, we highlight the production of 1,3-propanediol (26), amorphadiene (22), and 1,2,4-butanetriol (27) in Escherichia coli. In these approaches, the conversion steps were designed based on enzyme availability, the recruited enzyme activities from various organisms were identified, and the designed pathways were constructed in E. coli by assembling these enzymatic steps. The general concept incorporated in these metabolic engineering examples is to consider biological components, including enzymes, as interchangeable parts, and the term “synthetic biology” has been used to describe this concept (8, 39). The ability to conceive of and assemble new biosynthetic pathways by combining protein parts from multiple sources leads to increased possibilities for the construction of microbial chemical factories to produce compounds that do not have a microbial origin.
One such compound of interest is d-glucaric acid. d-Glucaric acid is found in fruits, vegetables, and mammals. It is available as a dietary supplement in the form of calcium d-glucarate and has been studied for therapeutic purposes including cholesterol reduction (36) and cancer chemotherapy (33, 34). d-Glucaric acid was also identified as a “top value-added chemical from biomass” in a report of the Pacific Northwest National Laboratory and the National Renewable Energy Laboratory (37), which also described its potential use as a building block for a number of polymers, including new nylons and hyperbranched polyesters. Indeed, d-glucaric acid produced from d-glucose has been successfully utilized to produce a hydroxylated nylon, resulting in a presumably biodegradable fiber (19). d-Glucaric acid is a highly functionalized compound with four chiral carbons and is currently produced by chemical oxidation of glucose, a nonselective and expensive process using nitric acid as the oxidant (37). New catalytic processes using biological systems may lead to higher yield and selectivity.
To construct a biological system for the synthesis of d-glucaric acid, it is reasonable to start with an examination of the naturally existing mammalian pathway, which has been elucidated (20). d-Glucaric acid and l-ascorbic acid are both end products of the d-glucuronic acid pathway. This pathway is a cycle that is initiated with either d-galactose or d-glucose and interacts with the pentose phosphate pathway. d-Glucuronic acid is converted to d-glucaric acid in three successive steps through the intermediates d-glucurono-γ-lactone and d-glucaro-γ-lactone. Therefore, there is a known route for the production of d-glucaric acid from d-glucose. However, this is a lengthy pathway, consisting of more than 10 conversion steps.
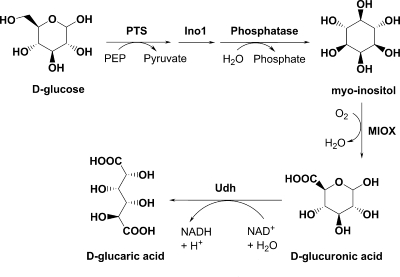
Here we describe the construction of a synthetic pathway for the production of d-glucaric acid in E. coli by combining biological parts from disparate organisms (Fig. 1). myo-Inositol-1-phosphate synthase (Ino1, also known as MIPS), encoded by the INO1 gene of Saccharomyces cerevisiae, is used to produce myo-inositol-1-phosphate from glucose-6-phosphate (14). Ino1 requires NAD+ for activity, but the cofactor is regenerated in the isomerization reaction. The Ino1 substrate is present in E. coli as the result of glucose transport by the phosphoenolpyruvate-dependent phosphotransferase system, which consumes one molecule of phosphoenolpyruvate per molecule of glucose transported (29). myo-Inositol-1-phosphate, the product of the Ino1 reaction, is hydrolytically dephosphorylated to produce myo-inositol by an endogenous phosphatase (23). In yeast, myo-inositol is a constituent of membrane phospholipids, and its derivatives are important for cell signaling. A second recombinant enzyme, myo-inositol oxygenase (MIOX), can convert myo-inositol to d-glucuronic acid using molecular oxygen. This enzyme is present primarily in mammalian sources and represents the first step of myo-inositol catabolism (13). Coexpression of these two enzymes in E. coli enables the production of d-glucuronic acid from d-glucose. As an alternative to the three-step mammalian route for producing d-glucaric acid from d-glucuronic acid, uronate dehydrogenase from Pseudomonas syringae can perform this conversion utilizing NAD+ as a cofactor and a water molecule (7, 35). The udh gene encoding this activity was recently cloned and characterized (40). Expression of this third gene with INO1 and MIOX enables the production of d-glucaric acid from d-glucose.
FIG. 1.
Designed pathway for the production of glucaric acid in E. coli. PTS, phosphoenolpyruvate-dependent phosphotransferase system; phosphatase, SuhB, an endogenous E. coli enzyme (23); PEP, phosphoenolpyruvate.
MATERIALS AND METHODS
Strains, growth media, and plasmids.
E. coli strain DH10B [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ− rpsL (Strr) nupG] was used for all molecular biology manipulations. DH10B and BL21 Star (DE3) [F− ompT hsdSB (rB− mB−) gal dcm rne131 (DE3)] were used as hosts for production of organic acids. Competent cells of both strains were purchased from Invitrogen Corporation (Carlsbad, CA). Cultures were propagated in either Luria-Bertani (LB) or M9 medium. LB (Miller) medium was prepared from dehydrated powder according to the manufacturer's instructions (BD Biosciences, San Jose, CA). M9 was prepared as described previously (32) and consisted of 1× M9 salts (12.8 g/liter Na2HPO4·7H2O, 3 g/liter KH2PO4, 0.5 g/liter NaCl, 1 g/liter NH4Cl), 2 mM MgSO4, 0.1 mM CaCl2, and 10 g/liter (1%) glucose. Leucine was added to a final concentration of 105 μg/ml for DH10B. Kanamycin was added to a final concentration of 20 μg/ml and ampicillin to a final concentration of 100 μg/ml where desired to provide selective pressure for plasmid maintenance.
All molecular biology manipulations were performed according to standard practices (32). The INO1 gene, encoding Ino1, was PCR amplified from a genomic DNA preparation of Saccharomyces cerevisiae with the following primers: forward, 5′-GAATTCATGACAGAAGATAATATTGCTC-3′; reverse, 5′-AAGCTTCTACAACAATCTCTCTTCG-3′. EcoRI and HindIII restriction sites included in the 5′ ends of the primers are underlined. The mouse MIOX gene, encoding myo-inositol oxygenase, was synthesized with codon optimization for expression in E. coli by DNA 2.0 (Menlo Park, CA) based on the sequence with GenBank accession number AF197127. Optimization of the 858-nucleotide (286-codon) sequence was performed by the vendor, with the results summarized as follows (see the supplemental material). A total of 19.2% of the nucleotides were altered, affecting 153 of the 286 codons (53.5%). Among the optimized codons, 144 (94.1%) were altered only at the third nucleotide position. All three nucleotides were changed in three of the codons. The synthetic gene was received as plasmid pJ2-MIOX. EcoRI and HindIII restriction sites were included in the 5′ and 3′ ends of the gene, respectively. Both genes were subcloned into the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible plasmids pMMB206 (25) and pTrc99A (2) to confirm activity of the expressed enzymes. The resulting plasmids were designated pMMB-INO1, pTrc-INO1, pMMB-MIOX, and pTrc-MIOX. For coexpression of both genes, the pRSFDuet-1 vector from Novagen (Gibbstown, NJ), containing two T7 promoters, was used. The INO1 gene was subcloned into the first position by using the EcoRI and HindIII sites, producing plasmid pRSFD-IN. To introduce the MIOX gene into the second position, the HindIII site of pJ2-MIOX was end filled using Klenow enzyme prior to digestion with EcoRI. pRSFD-IN was first digested with XhoI and end filled and then digested with the EcoRI-compatible MfeI prior to ligation with the MIOX gene fragment. The resulting plasmid was designated pRSFD-IN-MI. We previously isolated the udh gene, encoding uronate dehydrogenase, from Pseudomonas syringae (GenBank accession number EU377538) and subcloned the gene into pTrc99A to produce pT1053 (referred to in Table 2 as pTrc-udh) as described previously (40).
TABLE 2.
Production of glucaric acid in BL21 Star (DE3)(pRSFD-IN-MI, pTrc-udh) after 3 days of culturea
| Conditionb | OD600c | Concn (g/liter) of:
|
Yield (%)d | |||
|---|---|---|---|---|---|---|
| Glucose | myo-Inositol | Glucuronic acid | Glucaric acid | |||
| A | 5.10 ± 0.27 | 5.69 ± 0.85 | 0.10 ± 0.02 | NDe | 0.72 ± 0.09 | 17.4 ± 5.1 |
| B | 6.13 ± 0.31 | 1.43 ± 0.81 | 0.18 ± 0.05 | ND | 1.13 ± 0.17 | 13.1 ± 1.0 |
| C | 5.80 ± 0.39 | 2.47 ± 1.00 | 0.23 ± 0.07 | ND | 0.82 ± 0.06 | 11.0 ± 2.4 |
Cultures were grown at 30°C in LB medium supplemented with 10 g/liter glucose and induced with IPTG. Data are the averages and standard deviations of three independent experiments.
Condition A, 0.1 mM IPTG at 0 h; condition B, 0.05 mM IPTG at 0 h; condition C, 0.05 mM IPTG at 0 h and 0.1 mM IPTG at 17.5 h.
OD600, optical density at 600 nm.
Yield = 100 × glucaric acid produced/glucose consumed (mol/mol).
ND, not detectable.
Enzyme assays for Ino1, MIOX, and Udh activity.
Functional expression of the INO1, MIOX, and udh genes was confirmed through in vitro assays of enzyme activity. Crude lysates were prepared by first resuspending cell pellets from 1 to 2 ml culture in 100 to 200 μl 10 mM Tris-Cl (pH 8.0) with 1 mg/ml lysozyme. Cell solutions were lysed by alternating freezing in liquid nitrogen with thawing in 30 to 40°C water for five cycles. The resulting solutions were centrifuged at 14,000 rpm at 4°C for 15 min to remove insolubles. The total protein concentration of lysates was determined using the Bradford method (11).
Assays for myo-inositol-1-phosphate synthase activity were performed as described previously (1, 6). Briefly, glucose-6-phosphate substrate was converted to myo-inositol-1-phosphate in a reaction buffer consisting of 50 mM Tris-acetate (pH 7.5), 0.8 mM NAD+, 14 mM NH4Cl, 5 mM mercaptoethanol, and 5 mM glucose-6-phosphate. Reactions were initiated with the addition of lysate, and reaction mixtures were incubated for 1 h at 37°C. Reactions were terminated with the addition of 0.4 volume 20% trichloroacetic acid. To quantitate the product, inorganic phosphate was removed from the myo-inositol-1-phosphate by oxidation with an equal volume of 0.2 M NaIO4. Excess periodate was destroyed by the addition of an equal volume of 1 M Na2SO3. Control reactions were established without glucose-6-phosphate and without addition of periodate.
Assays for MIOX activity were performed as described previously (4, 30, 31). The reaction buffer consisted of 50 mM Tris-Cl (pH 8.0), 2 mM l-cysteine, 1 mM Fe(NH4)2(SO4)2, and 60 mM myo-inositol. Samples were preincubated without substrate for 10 min at 30°C to activate the MIOX enzyme. Reaction mixtures were incubated for 1 h at 30°C, and then reactions were terminated with the addition of 1/10 volume 30% trichloroacetic acid. The glucuronic acid produced was quantified with an orcinol reagent (13). The reagent consisted of 40 mg orcinol in 10 ml concentrated HCl containing 5.4 mg FeCl3. One volume of sample was mixed with 2 volumes of orcinol reagent, and the mixture was incubated for 30 min in boiling water. After the mixture was cooled to room temperature, absorbance at 670 nm was measured to determine glucuronic acid concentration. Control reactions were established without myo-inositol to account for background.
Assays for uronate dehydrogenase activity were performed by monitoring NADH cofactor generation at 340 nm as described previously (35, 40). The reaction mixture contained 100 mM sodium phosphate buffer (pH 8.0), 2.5 mM glucuronic acid, 0.9 mM NAD+, and bacterial lysate prepared as described above.
Growth conditions for acid production.
Cultures were grown in LB medium supplemented with 10 g/liter glucose and induced with IPTG as indicated in Results. An inoculum was prepared in LB medium, and 1 or 2% (vol/vol) was used to inoculate 250-ml baffled flasks containing 50 or 100 ml of medium. The cultures were incubated at 30°C and 250 rpm, with periodic sampling to determine cell density and product concentration in the culture medium.
Detection and quantification of organic acids.
Metabolites including glucuronic acid and glucaric acid were quantified by high-performance liquid chromatography (HPLC). For glucaric acid assays, samples were pretreated as previously described (28, 40) to separate glucaric acid from other metabolites including glucuronic acid. Briefly, boronic acid affinity gel (Affi-Gel boronate gel; Bio-Rad Laboratories, Hercules, CA), which has an affinity for the coplanar adjacent cis-hydroxyl groups present in glucaric acid (28), was mixed with samples and washed with 80 mM potassium phosphate-20 mM boric acid buffer (pH 7.0). Glucaric acid was eluted with 0.1 M hydrochloric acid. The eluate was neutralized by adding 10 M NaOH and then analyzed by HPLC. HPLC analyses were performed on an Agilent 1100 series instrument equipped with an Aminex HPX-87H column (300 mm by 7.8 mm; Bio-Rad Laboratories, Hercules, CA) and refractive index and diode array detectors under the following conditions: mobile phase, 5 mM sulfuric acid in water; flow rate, 0.5 ml/min; injection volume, 50 μl; temperature, 55°C; UV wavelength, 210 nm.
RESULTS
Verification of recombinant Ino1 and MIOX activities.
The use of myo-inositol-1-phosphate synthase (Ino1) from Saccharomyces cerevisiae to produce high concentrations of myo-inositol through E. coli fermentation has been previously reported (15). Product titers of up to 21 g/liter were obtained under high-cell-density, fed-batch fermentations operated for 54 h. To confirm Ino1 performance in shake flasks, the corresponding gene was amplified, inserted into a compatible vector, and then subcloned into both high- and medium-copy-number plasmids for expression in the common laboratory strain DH10B. Plasmid pTrc-INO1 contains the modified ColE1 replicon, which results in copy numbers of several hundred, while pMMB-INO1 is based on the RSF1010 replicon, with a copy numbers of the order of 10. Two plasmids were evaluated to explore the potential for coexpression of the INO1 and MIOX genes in a single strain using compatible vectors. In vitro activities of 344 nmol/h/mg and 128 nmol/h/mg were measured for cultures harboring pTrc-INO1 and pMMB-INO1, respectively, and incubated at 30°C in LB medium supplemented with 10 g/liter glucose, indicating successful expression of the enzyme. However, only expression from the high-copy-number plasmid resulted in accumulation of measurable quantities of myo-inositol in the culture medium, 0.37 g/liter. Activity was also a strong function of temperature, with none detectable for cultures grown at 37°C. myo-Inositol production was also tested in M9 minimal medium. We postulated that growth in minimal medium with glucose as the only carbon source might increase glucose flux and accordingly increase myo-inositol production. However, only one-half the amount of myo-inositol was produced, suggesting that, while glucose flux may indeed be higher, the Ino1 enzyme expressed under these conditions does not compete as effectively against glycolysis for substrate. Subsequent experiments were conducted in LB medium supplemented with glucose.
MIOX is a protein of primarily eukaryotic origin, and the homologues from humans, mice, rats, and pigs are the ones that have been best characterized (3, 4, 30, 31). MIOX has been functionally expressed in E. coli and purified for characterization of the enzyme's properties; however, to our knowledge, mammalian MIOX has not been used in a whole-cell, recombinant system to produce glucuronic acid. The mouse version of the enzyme had been found to have the most favorable properties upon expression in E. coli (3) and was chosen for investigation. A synthetic version of the gene was purchased from DNA 2.0, with codon optimization for E. coli. This gene was also subcloned into both the high-copy-number and low-copy-number vectors used to evaluate Ino1 activity in DH10B. MIOX activity was initially evaluated at 37°C since the enzyme is of mammalian origin.
The MIOX enzyme is known to require Fe2+ and cysteine for activation in vitro (4). The addition of these compounds to the culture medium did not improve the expression of the enzyme from pTrc-MIOX, as measured in the in vitro assay, but rather resulted in a decrease in activity (Table 1). Glucuronic acid was still measured in the culture medium, though at a lower concentration. The observed decrease in enzyme activity coincided with a significant decrease in cell density, indicating toxicity of these compounds to the host. As reported previously (30, 31), MIOX activity is inhibited by Fe2+ and cysteine at high concentrations. While the extracellular concentrations were set at a level that activates the enzyme in the in vitro assay, the corresponding intracellular concentrations are unknown. It was also reported previously that inclusion of myo-inositol in the culture medium improved soluble expression of MIOX in E. coli (3). We also observed this behavior, noting a sharp decrease in activity of the enzyme when expressed in the absence of myo-inositol supplementation (Table 1). One striking feature of recombinant MIOX is its apparent instability (3). High activity was observed in samples taken during exponential phase (6 h after inoculation), but activity dropped substantially in stationary phase (24 h after inoculation) (Table 1). The background activity of the assay, as measured in control samples containing an empty pTrc99A plasmid, generally increases with time. Note that the high background of the assay results from the nonspecificity of the orcinol reagent, which is known to react with other biological compounds, though to a smaller extent. As a result, the assay may not be reliable for precise quantification of enzyme activity. However, the observed differences between samples with and without myo-inositol and between samples with myo-inositol at early and late time points are sufficiently large that the trends can be considered significant.
TABLE 1.
Activity of recombinant MIOX expressed from high-copy-number pTrc-MIOX in E. coli under various culture conditions
| Culture conditionsa | Activity (nmol/ min/mg) at:
|
Glucuronic acid concn (g/liter) | |
|---|---|---|---|
| 6 h | 12 h | ||
| pTrc99A control | NDb | 82 | ND |
| +MI | 430 | 76 | 0.44 |
| +MI, +Fe, +Cys | 180 | 42 | 0.33 |
| −MI | 28 | 15 | NAc |
Cultures were grown at 37°C in LB medium and induced with 1.0 mM IPTG. Glucuronic acid was measured at 24 h. Supplements: MI, myo-inositol (60 mM, 10.8 g/liter); Fe, Fe(NH4)2(SO4)2 (1 mM); Cys, l-cysteine (2 mM). +, with supplementation; −, without supplementation.
ND, not detectable.
NA, not measured.
Neither in vitro enzyme activity nor in vivo production of glucuronic acid was observed in cultures containing the lower-copy-number pMMB-MIOX construct, suggesting that high expression levels are required to achieve measurable MIOX activity. Because INO1 is actively expressed only at 30°C, the in vivo performance of MIOX when expressed from the high-copy-number plasmid was also evaluated at this temperature. A comparable amount of glucuronic acid, 0.40 g/liter, was produced after 24 h in culture, with titers doubling to 0.78 g/liter after 48 h.
Production of glucuronic acid from glucose.
Production of glucuronic acid from glucose requires the coexpression of both INO1 and MIOX in the same strain. The compatible plasmids pTrc99A and pMMB206 were both investigated, with the expectation that a doubly transformed strain containing either pTrc-INO1 and pMMB-MIOX or pMMB-INO1 and pTrc-MIOX could be used for production. However, our results indicated that reasonable in vivo activities, as determined by accumulation of each desired product in the culture medium, were achievable only with expression of both genes from high-copy-number plasmids. To address this issue, we introduced genes for both enzymes into the high-copy-number pRSFDuet vector, which contains a pair of multicloning sites, each behind a T7 promoter. Enzyme activities were confirmed as described previously, and expression was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). In this manner, an IPTG concentration of 0.1 mM was determined to be preferred. The host strain was also changed from DH10B to BL21 Star (DE3) to enable expression from the T7 promoter. We had previously observed that DH10B was incapable of consuming glucuronic acid for growth (data not shown). BL21 Star (DE3) can metabolize glucuronic acid; however, its consumption appeared to be subject to catabolite repression (data not shown). Therefore, cultivation of the strain in excess glucose prevents consumption of the desired product.
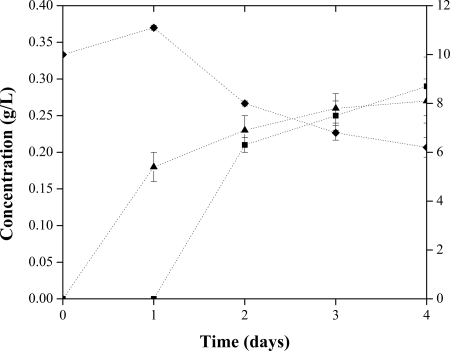
The BL21 Star (DE3) strain carrying pRSFD-IN-MI is capable of producing glucuronic acid from glucose, though to levels of only ∼270 mg/liter (Fig. 2). The culture profile shows that glucuronic acid is present after 24 h with no intermediates detectable, and the concentration increases by 50% in 4 days. However, after 48 h, significant quantities of myo-inositol appear in the culture medium. myo-Inositol continues to accumulate in the medium and is present in concentrations slightly higher than those of the desired end product, glucuronic acid, by the end of the experiment. The final glucuronic acid concentration, 0.27 g/liter, is lower than that observed with direct conversion of myo-inositol in the DH10B(pTrc-MIOX) system above (0.78 g/liter). The accumulation of myo-inositol suggests that MIOX activity is the limiting factor in production of high concentrations of glucuronic acid. In vitro assays confirmed that Ino1 activity is significantly higher than that for the vector-only control throughout the course of the experiment, with only marginal background activity appearing after 3 days (data not shown). In contrast, MIOX activity was only slightly higher than background after 1 day and was subsequently indistinguishable from background. This is consistent with the results summarized previously (Table 1) that indicate that MIOX activity drops sharply after 24 h. Additionally, it is likely that the activity of MIOX in this system is limited by the concentration of myo-inositol produced by Ino1. While an extracellular supplementation of 60 mM (10.8 g/liter) myo-inositol does not mean the intracellular concentration is also this high, it is reasonable to suspect that the intracellular concentrations of myo-inositol that result from Ino1 activity are likely to fall short of the equivalent concentration.
FIG. 2.
Production of glucuronic acid in BL21 Star (DE3)(pRSFD-IN-MI). Cultures were grown in triplicate at 30°C in LB medium supplemented with 10 g/liter glucose and 0.1 mM IPTG. Data points are the averages and standard deviations of the three biological replicates. ▴, glucuronic acid (left axis); ▪, myo-inositol (left axis); ⧫, glucose (right axis).
Production of glucaric acid from glucose.
We recently cloned and characterized the gene encoding uronate dehydrogenase activity from Pseudomonas syringae pv. tomato DC3000 (40). The udh gene was found to be very well expressed in E. coli, resulting in high enzyme activities. For the production of glucaric acid, we utilized a previously constructed vector harboring the udh gene in pTrc99A, which is compatible with pRSFD-IN-MI. Both vectors were introduced into BL21 Star (DE3) to construct an E. coli strain carrying INO1, the MIOX gene, and udh. Productivity of this strain under several different induction conditions was measured (Table 2). To our surprise, up to 1 g/liter of glucaric acid was produced although only 0.27 g/liter of glucuronic acid was previously observed in the system harboring the first two genes. Under induction conditions identical to those previously used for glucuronic acid (Table 2, condition A), 0.72 g/liter of glucaric acid was produced. To further characterize the system, enzyme activities in crude lysates were measured after each day of culture (Fig. 3). Udh activity was highest, more than 2 orders of magnitude higher than Ino1 activity and 3 orders of magnitude higher than MIOX activity. The high activity of Udh thus appears to pull glucose flux through the glucaric acid pathway, leading to a relatively higher titer of glucaric acid. In these samples, MIOX activity does not appear to decrease over time as observed previously; however, the magnitude of the activity remains quite low. Additionally, the first data point here is after 1 day, when MIOX activity was previously shown to have decreased significantly from that observed during exponential growth (Table 1). No glucuronic acid was detected after 3 days of culture time while myo-inositol accumulated, confirming that the MIOX-catalyzed step is limiting.
FIG. 3.
In vitro activity of recombinant Ino1, MIOX, and Udh expressed in BL21 Star (DE3) harboring the corresponding three genes. Cultures were grown at 30°C in LB medium supplemented with 10 g/liter glucose and induced with 0.05 mM IPTG. MIOX activity is presented as net activity to account for background. Data are the averages and standard deviations of three replicates. ▴, Ino1; ▪, MIOX; ⧫, Udh.
The three induction conditions tested resulted in glucaric acid concentrations that ranged from 0.72 to 1.13 g/liter. In general, higher induction levels, i.e., higher IPTG concentrations, resulted in a higher yield of glucaric acid on glucose but lower product concentration (compare, for example, conditions A and B in Table 2). Higher induction levels also led to less glucose consumption and a lower cell density, indicating a metabolic burden associated with higher expression of the three enzymes. However, in the case of lower glucose consumption rate, a higher fraction of glucose flux was directed toward glucaric acid production versus biomass. We also observed that poorer aeration, resulting from doubling the total culture volume from 50 to 100 ml in 250-ml baffled flasks, led to a decrease in the glucaric acid titer by one-half, while growth was not affected (data not shown). This reduced titer is likely attributed to the fact that MIOX, the enzyme for the limiting step, uses molecular oxygen as a cosubstrate (12, 38). Finally, production of glucaric acid in M9 minimal medium was tested; however, a negligible amount of glucaric acid was produced.
DISCUSSION
We have assembled a biosynthetic pathway for the production of glucaric acid using enzymes from three disparate sources: Ino1 from S. cerevisiae, MIOX from mice, and Udh from P. syringae. An endogenous phosphatase also participates in the pathway. The suhB gene product of E. coli has been shown to possess inositol monophosphatase activity in vitro and is therefore a reasonable candidate for this endogenous activity (23). This pathway is attractive from a thermodynamics perspective, since the standard free energy changes (ΔG) for all three steps, as estimated by group contribution theory (21, 24) and considering molecular oxygen as the ultimate oxidant, are all negative: −14.3 kcal/mol for the glucose-to-myo-inositol step; −86.8 kcal/mol for the myo-inositol-to-glucuronic acid step; −55.9 kcal/mol for the glucuronic acid-to-glucaric acid step.
However, as Khosla and Keasling have indicated (18), metabolic engineering is more than simply recruiting various enzymes. It also involves global optimization of metabolic flux when perturbations such as the introduction of new pathways into a host organism are made. Issues of metabolic burden associated with the maintenance of plasmids and expression of plasmid-carried genes are of particular interest in this case (9, 10, 17). In our system, a detectable amount of glucuronic acid was produced in vivo only by high-copy-number plasmids. Glucose-6-phosphate, the first substrate, should not be limiting for central metabolism because LB medium supplemented with excess glucose was used for growth. Therefore, it appears that high expression levels of the recombinant genes are needed in order to compete with the fast and robust glycolysis pathway and to divert glucose-6-phosphate toward glucuronic acid. The result that only small amounts of myo-inositol and no detectable amounts of the organic acids were produced in M9 medium implies that, when glucose is the sole carbon and energy source, almost all of the substrate enters endogenous cellular metabolism. This competition may also explain why the yield of glucaric acid on glucose during the first 2 days of the process, when glucose concentration is higher in the medium, is generally higher than that during the later days, when the concentration is lower (data not shown). The requirement for myo-inositol to achieve high MIOX activity suggests that low productivity from the Ino1 enzyme may ultimately be the limitation for formation of the organic acids in M9 medium. Alternatively, MIOX may be poorly expressed in minimal medium. It should be noted that previous studies with Ino1 that also employed a high-copy-number plasmid for gene expression have demonstrated high levels of myo-inositol production in an alternative chemically defined medium; however, these experiments were conducted in larger-scale, fed-batch fermentations for several days (15). During the initial batch period prior to the onset of glucose feeding (approximately 10 h), the myo-inositol concentration was less than 1 g/liter. Thus, it is worth exploring the extent to which cultivation under fed-batch conditions could improve the productivity of our system.
Plasmid copy number is not the only factor related to expression level that affects the performance of our synthetic system. As shown in Table 2, increasing the inducer concentration to increase expression resulted in lower product concentration. IPTG concentrations below 0.05 mM did not improve glucaric acid production even though glucose consumption rate and growth rate were enhanced due to the reduced metabolic burden (data not shown). E. coli growth is better at 37°C than at 30°C, and the activity of the rate-limiting enzyme MIOX should be higher at 37°C. However, fermentation was performed at 30°C because Ino1 was functionally expressed only at this lower temperature. Considering the reported unusual thermal instability of Udh (7, 35), a temperature lower than 30°C may be better for its activity; however, we observed that the Udh activity at 30°C was much higher than that of either Ino1 or MIOX (Fig. 3) and selected 30°C as the culture temperature to maximize the functional expression of Ino1.
In considering overall limitations on the productivity of this system, potential inhibition by intermediates in the pathway should be examined. MIOX from hog kidney was reported to be inhibited in vitro by d-glucaric acid but not by d-glucuronate and d-glucuronolactone (30, 31). Given that MIOX activity dropped sharply at the stationary phase even in the absence of d-glucaric acid (Table 1), low MIOX activity is more likely due to its intrinsic instability than inhibition by intermediates (3). It should also be noted that we did not overexpress the suhB gene or a phosphatase homologous to its product. However, no myo-inositol-1-phosphate was detected among the culture products, while myo-inositol did accumulate. Therefore, we conclude that the phosphatase activity does not limit flux through the pathway. E. coli also contains the d-glucarate catabolic pathway (16). Indeed, the ability of E. coli to consume d-glucarate as the sole carbon source for growth was used to develop a screen to identify uronate dehydrogenase activity (40). BL21 Star (DE3) can also metabolize d-glucuronic acid. However, the consumption of both organic acids appears to be subject to catabolite repression, preventing the undesirable loss of products in the presence of glucose (data not shown). The theoretical limit of d-glucaric acid titer therefore seems to be determined by the toxicity of the acids and the kinetics of each step. We have observed that E. coli growth and glucose consumption are not affected by the addition of potassium glucarate and sodium glucuronate at concentrations as high as 10 g/liter (data not shown); thus, there is room for improvement of titers by focusing on improving the kinetics of the rate-limiting steps. We are investigating further optimization for enhancing glucose flux to our synthetic pathway by recruiting better enzymes from different sources, engineering these enzymes, and downregulating the competing pathways.
Supplementary Material
Acknowledgments
This work was supported by the Office of Naval Research Young Investigator Program (grant no. N000140510656). S.-H.Y. was supported by a Korea Research Foundation grant funded by the South Korean government (MOEHRD) (KRF-2007-357-D00090). A.M.L. was further supported by a Merck undergraduate research grant (Bioprocess R&D, West Point, PA).
Footnotes
Published ahead of print on 5 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adhikari, J., A. L. Majumder, T. J. Bhaduri, S. DasGupta, and R. L. Majumder. 1987. Chloroplast as a locale of L-myo-inositol-1-phosphate synthase. Plant Physiol. 85:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, E., B. Ochs, and K.-J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 3.Arner, R. J., S. Prabhu, and C. C. Reddy. 2004. Molecular cloning, expression, and characterization of myo-inositol oxygenase from mouse, rat, and human kidney. Biochem. Biophys. Res. Commun. 324:1386-1392. [DOI] [PubMed] [Google Scholar]
- 4.Arner, R. J., S. Prabhu, J. T. Thompson, G. R. Hildenbrandt, A. D. Liken, and C. C. Reddy. 2001. myo-Inositol oxygenase: molecular cloning and expression of a unique enzyme that oxidizes myo-inositol and D-chiro-inositol. Biochem. J. 360:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey, J. E. 1991. Toward a science of metabolic engineering. Science 252:1668-1675. [DOI] [PubMed] [Google Scholar]
- 6.Barnett, J. E. G., R. E. Brice, and D. L. Corina. 1970. A colorimetric determination of inositol monophosphatases as an assay for D-glucose 6-phosphate-1L-myo-inositol 1-phosphate cyclase. Biochem. J. 119:183-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bateman, D. F., T. Kosuge, and W. W. Kilgore. 1970. Purification and properties of uronate dehydrogenase from Pseudomonas syringae. Arch. Biochem. Biophys. 136:97-105. [DOI] [PubMed] [Google Scholar]
- 8.Benner, S. A. 2003. Synthetic biology: act natural. Nature 421:118. [DOI] [PubMed] [Google Scholar]
- 9.Bentley, W. E., N. Mirjalili, D. C. Andersen, R. H. Davis, and D. S. Kompala. 1990. Plasmid-encoded protein: the principal factor in the metabolic burden associated with recombinant bacteria. Biotechnol. Bioeng. 35:668-681. [DOI] [PubMed] [Google Scholar]
- 10.Birnbaum, S., and J. E. Bailey. 1991. Plasmid presence changes the relative levels of many host cell proteins and ribosome components in recombinant Escherichia coli. Biotechnol. Bioeng. 37:736-745. [DOI] [PubMed] [Google Scholar]
- 11.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 12.Charalampous, F. C. 1959. Biochemical studies on inositol. V. Purification and properties of the enzyme that cleaves inositol to D-glucuronic acid. J. Biol. Chem. 234:220-227. [PubMed] [Google Scholar]
- 13.Charalampous, F. C., and C. Lyras. 1957. Biochemical studies on inositol. IV. Conversion of inositol to glucuronic acid by rat kidney extracts. J. Biol. Chem. 228:1-13. [PubMed] [Google Scholar]
- 14.Dean-Johnson, M., and S. A. Henry. 1989. Biosynthesis of inositol in yeast: primary structure of myo-inositol-1-phosphate synthase (EC 5.5.1.4) and functional analysis of its structural gene, the INO1 locus. J. Biol. Chem. 264:1274-1283. [PubMed] [Google Scholar]
- 15.Hansen, C. A., A. B. Dean, K. M. Draths, and J. W. Frost. 1999. Synthesis of 1,2,3,4-tetrahydroxybenzene from D-glucose: exploiting myo-inositol as a precursor to aromatic chemicals. J. Am. Chem. Soc. 121:3799-3800. [Google Scholar]
- 16.Hubbard, B. K., M. Koch, D. R. Palmer, P. C. Babbitt, and J. A. Gerlt. 1998. Evolution of enzymatic activities in the enolase superfamily: characterization of the (D)-glucarate/galactarate catabolic pathway in Escherichia coli. Biochemistry 37:14369-14375. [DOI] [PubMed] [Google Scholar]
- 17.Jones, K. L., S. W. Kim, and J. D. Keasling. 2000. Low-copy plasmids can perform as well as or better than high-copy plasmids for metabolic engineering of bacteria. Metab. Eng. 2:328-338. [DOI] [PubMed] [Google Scholar]
- 18.Khosla, C., and J. D. Keasling. 2003. Metabolic engineering for drug discovery and development. Nat. Rev. Drug Discov. 2:1019-1025. [DOI] [PubMed] [Google Scholar]
- 19.Kiely, D. E., L. Chen, and T. H. Lin. 1994. Simple preparation of hydroxylated nylons—polyamides derived from aldaric acids. Am. Chem. Soc. Symp. Ser. 575:149-158. [Google Scholar]
- 20.Kuellmer, V. April 2001, posting date. Ascorbic acid. In Kirk-Othmer encyclopedia of chemical technology, 4th ed. John Wiley & Sons, Hoboken, NJ. http://mrw.interscience.wiley.com/emrw/9780471238966/home/.
- 21.Li, C., C. S. Henry, M. D. Jankowski, J. A. Ionita, V. Hatzimanikatis, and L. J. Broadbelt. 2004. Computational discovery of biochemical routes to specialty chemicals. Chem. Eng. Sci. 59:5051-5060. [Google Scholar]
- 22.Martin, V. J., D. J. Pitera, S. T. Withers, J. D. Newman, and J. D. Keasling. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21:796-802. [DOI] [PubMed] [Google Scholar]
- 23.Matsuhisa, A., N. Suzuki, T. Noda, and K. Shiba. 1995. Inositol monophosphatase activity from the Escherichia coli suhB gene product. J. Bacteriol. 177:200-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mavrovouniotis, M. L. 1991. Estimation of standard Gibbs energy changes of biotransformations. J. Biol. Chem. 266:14440-14445. [PubMed] [Google Scholar]
- 25.Morales, V. M., A. Bäckman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura, C. E., and G. M. Whited. 2003. Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 14:454-459. [DOI] [PubMed] [Google Scholar]
- 27.Niu, W., M. N. Molefe, and J. W. Frost. 2003. Microbial synthesis of the energetic material precursor 1,2,4-butanetriol. J. Am. Chem. Soc. 125:12998-12999. [DOI] [PubMed] [Google Scholar]
- 28.Poon, R., D. C. Villeneuve, I. Chu, and R. Kinach. 1993. HPLC determination of D-glucaric acid in human urine. J. Anal. Toxicol. 17:146-150. [DOI] [PubMed] [Google Scholar]
- 29.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy, C. C., P. A. Pierzchala, and G. A. Hamilton. 1981. myo-Inositol oxygenase from hog kidney. II. Catalytic properties of the homogeneous enzyme. J. Biol. Chem. 256:8519-8524. [PubMed] [Google Scholar]
- 31.Reddy, C. C., J. S. Swan, and G. A. Hamilton. 1981. myo-Inositol oxygenase from hog kidney. I. Purification and characterization of the oxygenase and of an enzyme containing the oxygenase and D-glucuronate reductase. J. Biol. Chem. 256:8510-8518. [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Singh, J., and K. P. Gupta. 2003. Calcium glucarate prevents tumor formation in mouse skin. Biomed. Environ. Sci. 16:9-16. [PubMed] [Google Scholar]
- 34.Singh, J., and K. P. Gupta. 2007. Induction of apoptosis by calcium D-glucarate in 7,12-dimethyl benz [a] anthracene-exposed mouse skin. J. Environ. Pathol. Toxicol. Oncol. 26:63-73. [DOI] [PubMed] [Google Scholar]
- 35.Wagner, G., and S. Hollman. 1976. Uronic acid dehydrogenase from Pseudomonas syringae: purification and properties. Eur. J. Biochem. 61:589-596. [DOI] [PubMed] [Google Scholar]
- 36.Walaszek, Z., J. Szemraj, M. Hanausek, A. K. Adams, and U. Sherman. 1996. D-glucaric acid content of various fruits and vegetables and cholesterol-lowering effects of dietary D-glucarate in the rat. Nutr. Res. 16:673-681. [Google Scholar]
- 37.Werpy, T., and G. Petersen. 2004. Top. value added chemicals from biomass, volume 1—results of screening for potential candidates from sugars and synthesis gas. U.S. Department of Energy, Washington, DC. http://www.pnl.gov/main/publications/external/technical_reports/PNNL-14808.pdf.
- 38.Xing, G., L. M. Hoffart, Y. Diao, K. S. Prabhu, R. J. Arner, C. C. Reddy, C. Krebs, and J. M. Bollinger, Jr. 2006. A coupled dinuclear iron cluster that is perturbed by substrate binding in myo-inositol oxygenase. Biochemistry 45:5393-5401. [DOI] [PubMed] [Google Scholar]
- 39.Yeh, B. J., and W. A. Lim. 2007. Synthetic biology: lessons from the history of synthetic organic chemistry. Nat. Chem. Biol. 3:521-525. [DOI] [PubMed] [Google Scholar]
- 40.Yoon, S.-H., T. S. Moon, P. Iranpour, A. M. Lanza, and K. J. Prather. Cloning and characterization of uronate dehydrogenases from two pseudomonads and Agrobacterium tumefaciens strain C58. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.