Abstract
To elucidate the function of the transcriptional coregulator PRMT1 (protein arginine methyltranferase 1) in interferon (IFN) signaling, we investigated the expression of STAT1 (signal transducer and activator of transcription) target genes in PRMT1-depleted cells. We show here that PRMT1 represses a subset of IFNγ-inducible STAT1 target genes in a methyltransferase-dependent manner. These genes are also regulated by the STAT1 inhibitor PIAS1 (protein inhibitor of activated STAT1). PIAS1 is arginine methylated by PRMT1 in vitro as well as in vivo upon IFN treatment. Mutational and mass spectrometric analysis of PIAS1 identifies Arg 303 as the single methylation site. Using both methylation-deficient and methylation-mimicking mutants, we find that arginine methylation of PIAS1 is essential for the repressive function of PRMT1 in IFN-dependent transcription and for the recruitment of PIAS1 to STAT1 target gene promoters in the late phase of the IFN response. Methylation-dependent promoter recruitment of PIAS1 results in the release of STAT1 and coincides with the decline of STAT1-activated transcription. Accordingly, knockdown of PRMT1 or PIAS1 enhances the anti-proliferative effect of IFNγ. Our findings identify PRMT1 as a novel and crucial negative regulator of STAT1 activation that controls PIAS1-mediated repression by arginine methylation.
Keywords: Transcriptional repression, interferon signaling, STAT1, PIAS1, PRMT1, arginine methylation
Interferons (IFNs) constitute a family of secreted autocrine and paracrine proteins that regulate innate and adaptive immunity in response to viral and microbial infections and modulate proliferation, survival, and apoptosis of normal cells as well as tumor cells (Borden et al. 2007). IFNs exert their effects by binding to cell surface receptors, which triggers receptor dimerization/oligomerization, allowing transphosphorylation and activation of intracellular receptor-associated tyrosine kinases of the JAK (Janus kinase) family. Activated JAKs phosphorylate critical tyrosine residues on the receptor, which results in the recruitment of a family of nucleo–cytoplasmic shuttling transcription factors, the STAT (signal transducer and activator of transcription) proteins (Levy and Darnell 2002; Schindler et al. 2007). In the following, receptor-bound STATs are activated by phosphorylation of a single tyrosine residue within the C terminus leading to STAT dimerization via a reciprocal phosphotyrosine-SH2 (src homology 2) interaction. The STAT dimers are retained in the nucleus, where they bind specific DNA sequences in promoter regions of target genes and induce gene transcription (Meyer and Vinkemeier 2004). Among the seven mammalian STAT proteins STAT1 and STAT2 are mediators of the IFN signaling pathway, whereas the remaining STAT family members respond to distinct cytokines. Type I IFNs, such as IFNα and IFNβ, activate STAT1/STAT2 heterodimers, and type II IFNγ mainly signals via STAT1 homodimers (Borden et al. 2007).
Proper duration and magnitude of IFN-induced gene activation is crucial, as abnormal and imbalanced JAK–STAT activation is associated with immune disorders and cancer (Darnell 2002; Levy and Darnell 2002). Therefore, cells have evolved mechanisms to allow rapid and transient activation and to prevent excess responses to IFNs and other cytokines. Restriction and inactivation of the STAT pathway is accomplished by regulators such as protein tyrosine phosphatases, which inactivate JAKs and STATs in the cytoplasm or in the nucleus (Levy and Darnell 2002). In addition, SOCS (suppressors of cytokine signaling) proteins switch off the activity of JAKs by acting as pseudosubstrates or by recruiting the ubiquitination machinery (Alexander and Hilton 2004). The PIAS (protein inhibitor of activated STAT) protein family operates in the nucleus and the family member PIAS1 interacts with activated STAT1 and suppresses the DNA-binding ability of the transcription factor (Liu et al. 1998; Shuai and Liu 2005). PIAS1 depletion causes increased expression of a subset of STAT1 target genes (Liu et al. 2004). In agreement with this observation, PIAS1 knockout mice reveal enhanced protection against pathogenic infections (Liu et al. 2004). Whether the Sumo E3 ligase activity of PIAS1 and sumoylation of STAT1 is required for its repressive activity on IFN-inducible gene expression is controversial in the literature (Rogers et al. 2003; Ungureanu et al. 2003, 2005; Song et al. 2006). Until now, little was known about the molecular mechanism leading to activation of PIAS1, which then restrains STAT1 activity, and how PIAS1 interferes with the chromatin binding of STAT1.
Arginine methylation is catalyzed by a family of enzymes, called PRMTs (protein arginine methyltransferases), consisting of 11 members (Boisvert et al. 2005; Pal and Sif 2007). These enzymes share a central highly conserved catalytic domain and perform mono- and dimethylation of the guanidino group of the arginine residues using S-adenosylmethionine (SAM) as methyl donor (Bedford and Richard 2005). PRMTs regulate various cellular processes such as RNA processing, nucleo–cytoplasmic transport, DNA repair, transcription, and signal transduction. Several reports implicate arginine methylation in the regulation of IFN signaling: PRMT1 interacts with the cytoplasmic domain of the IFNα receptor, and its depletion alters IFNα-induced growth arrest (Abramovich et al. 1997; Altschuler et al. 1999). Tyrosine phosphorylation, transcriptional activity, and protein stability of STAT transcription factors were suggested to be influenced by arginine methylation (Mowen et al. 2001; Zhu et al. 2002; Chen et al. 2004). However, arginine methylation of STAT transcription factors itself was not confirmed by other groups, and the nonspecific methyltransferase inhibitor MTA, which was commonly used to study the in vivo relevance of arginine methylation in this pathway, executes strong side effects and inhibits IFN-induced phosphorylation and activation of STATs (Meissner et al. 2004; Komyod et al. 2005). Consequently, how exactly PRMTs and arginine methylation might regulate IFN signaling, is so far unclear.
Here, we investigated the influence of PRMT1 on IFN signaling and found that PRMT1 represses a subset of STAT1 target genes that overlaps with genes regulated by PIAS1. PRMT1 methylates PIAS1 at Arg 303 (R303) in vitro and in vivo. Methylation-deficient and methylation-mimicking PIAS1 mutants revealed that PIAS1 methylation is essential for the repressive function of PRMT1 in IFN-inducible transcription and allows the recruitment of PIAS1 to STAT1 target gene promoters in the late phase of the IFN response. Our findings identify PRMT1 as a novel inhibitor of IFN signaling that is involved in turning-off STAT1 activation by regulating the repressive function of PIAS1.
Results
PRMT1 coregulates PIAS1-dependent STAT1 target genes
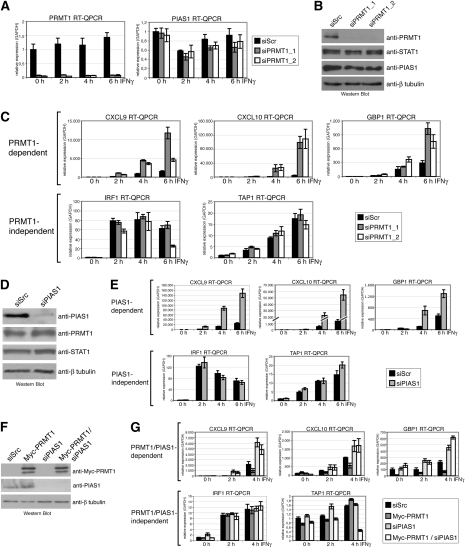
Since the role of arginine methylation in IFN signaling is controversial (Abramovich et al. 1997; Mowen et al. 2001; Meissner et al. 2004; Komyod et al. 2005), we investigated the potential role of PRMT1 in this pathway. We depleted PRMT1 in HeLa cells using transient transfection of two different siRNAs directed against the PRMT1 transcript (siPRMT1_1 and siPRMT1_2). The efficient depletion of endogenous PRMT1 by the two alternative siRNAs in comparison with the control siRNA (siScr) was determined on RNA (Fig. 1A) and protein level (Fig. 1B). The expression of STAT1 or PIAS1 was not altered by knockdown of PRMT1 (Fig. 1A,B). To examine the effect of PRMT1 on IFNγ-inducible STAT1-dependent transcription we performed RT-QPCR of known STAT1 target genes (Ramana et al. 2002) in control (siScr) and PRMT1-depleted HeLa cells either unstimulated (0 h) or stimulated with IFNγ for 2, 4, and 6 h. As shown in Figure 1C, the IFNγ-inducible transcription of several STAT1 targets, including CXCL9 (Cxc chemokine ligand 9), CXCL10 (Cxc chemokine ligand 10), and GBP1 (guanylate-binding protein 1), was enhanced threefold to 10-fold upon knockdown of PRMT1 using either of the two siRNAs against PRMT1. In contrast, the basal transcript level of these genes was not affected by PRMT1 depletion. IFN induction of other STAT1 target genes, like IRF1 (IFN response factor 1) and TAP1 (transporter associated with anti-gene presentation 1), was not significantly influenced by PRMT1 knockdown, suggesting that transcriptional activation of only a subset of STAT1 target genes is repressed by PRMT1 (Fig. 1C). The PRMT1-dependent STAT1 targets identified here appeared to match the subset of IFN-induced genes selectively controlled by the inhibitor of STAT1 signaling, PIAS1 (Liu et al. 2004). We asked whether siRNA-mediated depletion of the PIAS1 protein would affect the same subset of IFNγ-dependent STAT1 target genes as PRMT1 depletion does. Indeed, we found that PIAS1 knockdown, which was verified by Western blot (Fig. 1D), enhanced the IFN-inducible transcription of the PRMT1-dependent STAT1 targets CXCL9, CXCL10, and GBP1, whereas PRMT1-independent genes, like IRF1 and TAP1, were not impaired by suppression of PIAS1 (Fig. 1E). We obtained similar results for the IFN-responsive A375 melanoma (Supplemental Fig. S1) and HEK293 cells (data not shown). Taken together, these data identify PRMT1 and PIAS1 as negative regulators of the same STAT1 target genes.
Figure 1.
PRMT1 and PIAS1 regulate the same subset of STAT1 target genes. (A–C) HeLa cells were transfected with control siRNA (siScramble, siScr) or with two alternative siRNAs against PRMT1 (siPRMT1_1 and siPRMT1_2). After 48 h, cells were induced with 5 ng/mL IFNγ for the indicated time points. Subsequently cells were harvested and total RNA or protein extracts were prepared. (A) RT-QPCR was performed for detection of transcript levels of PRMT1 or PIAS1, respectively. Results were normalized using GAPDH mRNA level as a reference. Transcript levels in uninduced control cells were set to 1. (B) For detection of knockdown efficiency, protein levels of PRMT1, STAT1, and β-tubulin were analyzed by Western blot from 20 μg of protein extract (not induced with IFNγ). For detection of PIAS1, 80 μg of extract were used. (C) Total RNA was analyzed for transcript levels of CXCL9, CXCL10, GBP1, IRF1, and TAP1 normalized to GAPDH. Transcript levels in uninduced control cells were set to 1. (D,E) siRNA-mediated control (siScr) and PIAS1 knockdown (siPIAS1) was performed in HeLa cells. IFNγ induction was conducted as in A–C. RNA and protein extracts were prepared. (D) PIAS1 knockdown was determined by Western blot from uninduced extracts. (E) Transcript levels of STAT1 target genes were analyzed by RT-QPCR as described in C. (F,G) HEK293 cells were transfected with control siRNA (siScr) or siRNA against PIAS1, and as indicated, after 24 h cells were transfected with Myc-tagged PRMT1. After 48 h, cells were induced with 5 ng/mL IFNγ for the indicated time points. Subsequently, cells were harvested for the preparation of total RNA or protein extracts. (F) Efficiency of PRMT1 overexpression and PIAS1 knockdown was analyzed by Western blot for Myc-tag, PIAS1, and β-tubulin in uninduced extracts. (G) Transcript levels of STAT1 target genes were analyzed by RT-QPCR as described in C.
PRMT1 acts upstream of PIAS1 in STAT1 signaling
In the following, we studied whether PRMT1 and PIAS1 synergize in their repressive function on STAT1-mediated transcription. We performed single and double knockdown of PRMT1 and PIAS1 in HeLa cells (Supplemental Fig. S2A). Using RT-QPCR we found that the single and double depletion results in a similar increase in IFN-induced transcription of the PRMT1/PIAS1-dependent STAT1 targets. Double depletion of PRMT1 and PIAS1 did not lead to an additive effect compared with the effects of single depletion of PRMT1 or PIAS1 (Supplemental Fig. S2B). As expected, the PRMT1/PIAS1-independent targets were not influenced by the double depletion (Supplemental Fig. S2B). These observations suggest that PRMT1 and PIAS1 functionally depend on each other in the STAT1 pathway.
To further dissect the relationship between PRMT1 and PIAS1 we expressed Myc-tagged PRMT1 by transient transfection in HEK293 cells (Fig. 1F) and investigated IFN-induced transcription. Overexpression of PRMT1 diminished transcriptional induction of specifically the PRMT1/PIAS1-dependent subgroup of STAT1 target genes (Fig. 1G), reinforcing our findings in PRMT1-depleted cells (Fig. 1C). When we depleted PIAS1 in cells overexpressing PRMT1 (Fig. 1F), the repressive effect of exogenous PRMT1 was abrogated and transcriptional induction of the PRMT1/PIAS1-dependent STAT1 target genes was enhanced to a similar extent as with the sole depletion of PIAS1 (Fig. 1G). These results indicate that PRMT1 operates upstream of PIAS1 in this pathway and requires PIAS1 to inhibit IFNγ-dependent transcription.
Endogenous PRMT1 and PIAS1 proteins interact and are recruited to STAT1 target genes in the late phase of the IFN response
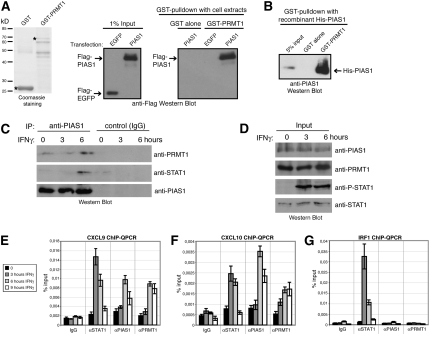
Next, we investigated whether the functional cooperation between PRMT1 and PIAS1 could reflect interaction of both proteins. We performed GST pulldown experiments of Flag-EGFP- (control) or Flag-PIAS1-overexpressing HEK293 cell extracts with GST alone and GST-PRMT1 (Fig. 2A). Pulldowns were analyzed by anti-Flag Western blot and revealed a specific interaction between PRMT1 and PIAS1 in vitro (Fig. 2A). Using recombinant His-tagged PIAS1 protein we demonstrated in further pulldown assays with GST-PRMT1 that the two proteins interact directly (Fig. 2B).
Figure 2.
PRMT1 interacts with PIAS1 and binds simultaneously with PIAS1 to STAT1 target gene promoters in the late phase of the IFNγ response. (A) Protein lysates of Flag-EGFP- (control) or Flag-PIAS1-overexpressing HEK293 cells were subjected to GST pulldown experiments. (Left panel) Equal amounts of GST alone and GST-PRMT1 protein, as indicated in the Coomassie-stained SDS gel (full-length protein bands marked with asterisks), coupled to glutathione beads were incubated with 250 μg of protein lysates. (Right panel) Bound PIAS1 protein was visualized by Western blot using Flag antibody. (Middle panel) One percent input (2.5 μg) of HEK293 cell extract is shown. (B) GST pulldown with recombinant His-tagged PIAS1 purified from Sf9 cells were performed using GST and GST-PRMT1 protein. Bound PIAS1 protein was detected by anti-His-tag Western blot. Five percent input of recombinant PIAS1 is shown. (C,D) Co-IPs of endogenous PIAS1, PRMT1, and STAT1 were performed. HeLa cells were induced with IFNγ for the indicated time points and nuclear extracts were prepared. (C) IP of PIAS1 (anti-PIAS1) or control IP (IgG) was carried out from 1 mg of nuclear extract and examined by Western blot using antibodies against PRMT1, STAT1, and PIAS1. (D) Input Western blots corresponding to CoIP in C were stained for PIAS1, PRMT1, P-STAT1, and STAT1. (E–G) Recruitment of STAT1, PIAS1, and PRMT1 to STAT1 target gene promoters was investigated by ChIP. HeLa cells were treated in a time course experiment with IFNγ (5 ng/mL). Subsequently, cells were harvested and subjected to ChIP analysis using control antibodies (IgG) and antibodies against STAT1, PIAS1, and PRMT1. Immunoprecipitated DNA was analyzed in triplicates by QPCR with primers for the CXCL9 (E), CXCL10 (F), and IRF1 (G) gene promoters and displayed as percentage input. Standard deviation was calculated from the triplicates, and error bars are indicated accordingly. Representative figure legend is show in E.
We then determined whether the two proteins bind also endogenously to each other and how such an interaction might be influenced in the course of the IFN response. HeLa cells were left untreated or treated for 3 and 6 h with IFNγ. Nuclear extracts of these cells were subjected to anti-PIAS1 immunoprecipitation (IP). Immunoprecipitates were probed for the presence of PRMT1 by Western blot analysis. PRMT1 und PIAS1 associated very weakly in the uninduced state and at the 3-h induction time point, whereas the interaction of the two proteins was enhanced after 6 h of IFNγ treatment (Fig. 2C). When we examined the PIAS1 precipitates for the presence of STAT1, PIAS1 was found to associate with STAT1 after 6 h of IFNγ treatment in agreement with other reports, which also showed an IFN-dependent interaction (Liu et al. 1998). Essentially the same results were obtained, when we included ethidium bromide in the IP (Lai and Herr 1992) indicating that the interaction between PIAS1, PRMT1, and STAT1 was not DNA-mediated (data not shown). IFNγ stimulation did not affect the protein levels of the three proteins, as shown by Western blot analysis of the corresponding nuclear extracts (Fig. 2D). These data strongly suggest that PIAS1 preferentially interacts with PRMT1 and STAT1 in the late phase of the IFN response.
Subsequently, we explored whether PRMT1 and PIAS1 are recruited to STAT1 target gene promoters. We performed chromatin IP (ChIP) analysis in either uninduced cells or 3, 6, and 9 h after addition of IFNγ using antibodies against STAT1, PIAS1, and PRMT1 and assayed for the enrichment of the PRMT1/PIAS1-dependent STAT1 target genes CXCL9 and CXCL10 and the PRMT1/PIAS1-independent target IRF1. STAT1 binding to the CXCL9 and CXCL10 promoter peaked at 3 h IFNγ treatment, gradually declined between 6 and 9 h and reached the basal (uninduced) level of recruitment at the 9 h time point (Fig. 2E,F). Consistent with this detachment of STAT1 from the CXCL9 and CXCL10 promoter and the decrease in transcriptional induction in the late phase of IFN treatment (Supplemental Fig. S3), PIAS1 and PRMT1 bound to both promoters after 6 and 9 h of IFN induction (Fig. 2E,F). In contrast, the promoter of the PRMT1/PIAS1-independent STAT1 target gene IRF1 was never occupied by PIAS1 or PRMT1 throughout the whole IFN time course, but STAT1 was strongly enriched after 3 and 6 h of IFNγ stimulation (Fig. 2G). These results indicate that both PRMT1 and PIAS1 are simultaneously recruited to the PRMT1/PIAS1-dependent genes in the late phase of IFNγ signaling, which coincides with the detachment of STAT1 and with the gradual decline of transcriptional activation.
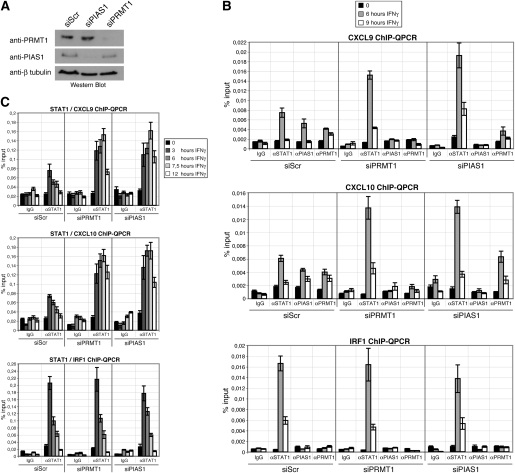
PRMT1 is required for promoter recruitment of PIAS1 and promoter detachment of STAT1
To test whether PRMT1 influences the promoter recruitment of PIAS1 we conducted ChIP analysis in PRMT1-depleted cells and control (siScr) cells, which were either untreated or IFN-treated. PRMT1 depletion in the corresponding chromatin samples was verified by Western blot (Fig. 3A). In control cells STAT1, PIAS1, and PRMT1 binding to the CXCL9 and CXCL10 promoter overlapped after 6 h of IFNγ stimulation (Fig. 3B, top and middle panels), in agreement with Figure 2, E and F. Depletion of PRMT1 resulted in a loss of PIAS1 recruitment and in an enhanced and prolonged recruitment of STAT1 to the CXCL9 and CXCL10 promoter during the IFN response (Fig. 3B, top and middle panels). In contrast association of STAT1 with the PRMT1/PIAS1-independent IRF1 promoter was not influenced by PRMT1 suppression (Fig. 3B, bottom panel). A similar effect on STAT1 binding was also observed in PIAS1-depleted cells, in which IFN-induced STAT1 binding of the CXCL9 and CXCL10 promoter was increased (Fig. 3A,B, top and middle panels), but binding of the IRF1 promoter was unchanged (Fig. 3B, bottom panel). PIAS1 knockdown did not affect the recruitment of PRMT1 to the CXCL9 and CXCL10 gene promoter (Fig. 3B, top and middle panels) corroborating our observation that PRMT1 acts upstream of PIAS1 in this pathway (Fig. 1G). However, PRMT1 promoter recruitment requires IFNγ-induced binding of STAT1 to its target genes, as ChIP analysis in STAT1-deficient U3A cells revealed (Supplemental Fig. S4). To strengthen our observation that PRMT1 and PIAS1 regulate the magnitude and kinetic of STAT1 promoter binding we performed ChIP analysis for STAT1 in a more detailed IFN induction time course including additional time points. As shown in Figure 3C, depletion of PRMT1 or PIAS1 resulted in an increased and prolonged STAT1 binding with a clearly delayed peak of STAT1 recruitment to a subset of target genes. These results indicate that PRMT1 regulates PIAS1-mediated repression of STAT1 target genes at the chromatin level by allowing promoter recruitment of PIAS1, which subsequently diminishes promoter association of STAT1.
Figure 3.
PIAS1 and STAT1 promoter recruitment depends on PRMT1. (A–C) HeLa cells were transfected with control siRNA or siRNA against PRMT1 and PIAS1, respectively. After 48 h, cells were induced with IFNγ (5 ng/mL) for the indicated time points and subjected to lysis for ChIP analysis. Depletion of PRMT1 and PIAS1 was determined by Western blot of the corresponding chromatin samples (A). Promoter recruitment of STAT1, PIAS1, and PRMT1 to STAT1 target gene promoters (CXCL9, CXCL10, and IRF1) was determined by ChIP-QPCR in PRMT1 or PIAS1 knockdown cells (B,C). Immunoprecipitated DNA was analyzed in triplicates by QPCR and results were displayed as percentage input.
Recent studies suggested a role of PIAS1-mediated sumoylation of STAT1 in the repression of STAT1 signaling (Ungureanu et al. 2003, 2005). We asked whether PRMT1 has an impact on the sumoylation of STAT1. PRMT1 depletion did not change the levels of sumoylated STAT1 (Supplemental Fig. S5) indicating that PRMT1 does not control the sumoylation activity of PIAS1. Moreover, overexpression of the sumoylation-deficient PIAS1 mutant W372A (Liu et al. 2007) revealed a similar repressive effect on the IFN induction of the same STAT1 target genes as wild-type PIAS1 (Supplemental Fig. S6). These results imply that sumoylation activity by PIAS1 is not essential for transcriptional inhibition of the STAT1 target genes investigated here.
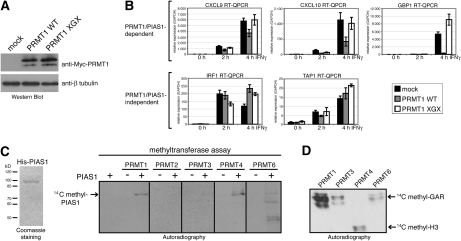
The methyltransferase activity of PRMT1 is required for its repressive function in STAT1 signaling
As PRMT1 possesses arginine methyltransferase activity toward histones and other nuclear proteins (Wang et al. 2001; Kwak et al. 2003; Barrero and Malik 2006) we studied in the following whether the catalytic activity of PRMT1 is required for its repressive function in STAT1-dependent transcription. Therefore, we established transient overexpression of wild-type PRMT1 (WT) and the catalytically inactive PRMT1 mutant XGX (Balint et al. 2005), which were equally well expressed (Fig. 4A). As expected IFNγ-induced expression of CXCL9, CXCL10, and GBP1 was diminished in the presence of exogenous wild-type PRMT1 compared with control cells (Fig. 4B). In contrast, the catalytically inactive PRMT1 mutant lost its repressive capacity and transcriptional induction of CXCL9, CXCL10, and GBP1 occurred to a similar extent as in control cells (Fig. 4B) leading to the conclusion that the catalytic activity of PRMT1 is required for its inhibitory effect on STAT1-mediated transcription.
Figure 4.
The arginine methyltransferase activity of PRMT1 is required for its repressive function in IFN-dependent transcription, and PIAS1 is arginine methylated in vitro. (A,B) HeLa cells were transfected either with PRMT1 wild type (WT) or PRMT1 catalytic inactive mutant (XGX). After 48 h, cells were induced with 5 ng/mL IFNγ for the indicated time points. Subsequently cells were harvested for the preparation of total RNA or protein extracts. (A) Overexpression of PRMT1 WT and PRMT1 XGX in uninduced cell extracts was monitored by Western blot using anti-Myc antibody. (B) Transcript levels of the indicated STAT1 target genes were analyzed with RT-QPCR and normalized to GAPDH. Transcript levels in uninduced control cells were set 1. (C) In vitro methylation assays of His-tagged PIAS1 purified from Sf9 cells (as indicated in the Coomassie-stained gel in the left panel) were performed with recombinant GST-PRMTs in the presence of methyl-14C-labeled SAM. Methylation products were resolved on SDS-PAGE, blotted, and visualized by autoradiography. (D) As positive control for the enzymatic activity of the GST-PRMT preparations in C, in vitro methylation assays were perfomed using established substrates of PRMTs, like GST-GAR protein (glycine–arginine-rich domain of fibrillarin) or bulk histones, and analyzed by autoradiography.
We next asked whether PRMT1 and other PRMTs would possess methyltransferase activity toward PIAS1. Therefore, we employed an in vitro methylation assay using recombinant baculovirus-expressed His-tagged PIAS1, GST-PRMTs, or immunopreciptitated PRMT5 from cell extracts as enzyme source and radiolabeled methyl-donor SAM (Fig. 4C; data not shown). This assay revealed that PIAS1 is in vitro methylated by PRMT1, PRMT4, and PRMT6 (Fig. 4C). The catalytic activity of the various enzyme preparation was verified using established substrates in an in vitro methylation assay (Fig. 4D; data not shown).
To address the question whether PRMT4 and PRMT6, which methylate PIAS1 in vitro as well, influence IFN-induced STAT1 target gene expression in a similar way as PRMT1, we depleted PRMT4 and PRMT6, respectively, using transient transfection of siRNAs in HeLa cells (Supplemental Fig. S7A,C). Transcriptional induction of the PRMT1/PIAS1-dependent STAT1 target genes CXCL9 and GBP1 was not impaired by knockdown of neither PRMT4 (Supplemental Fig. S7B) nor PRMT6 (Supplemental Fig. S7D). These data demonstrate that among the PRMTs capable of methylating PIAS1 in vitro, PRMT1 is the only enzyme that cooperates with PIAS1-dependent repression in a methyltransferase-dependent manner in vivo.
Arg 303 in PIAS1 is methylated by PRMT1 in vivo
To investigate the occurrence of in vivo methylation of PIAS1 we performed metabolical labeling of HeLa cells with L-[methyl-3H]-methionine. Cells were transfected with Flag-tagged PIAS1 and additionally with control siRNA (siScr) or with siRNA targeting PRMT1 (siPRMT1). Subsequently, cells were stimulated with IFNγ or not and simultaneously metabolical labeling was performed for 3 h in the presence of translational inhibitors. Flag-PIAS1 protein was immunopurified from the cell extracts, resolved by SDS-PAGE and methylation was detected by fluorography. PIAS1 was methylated in response to IFNγ treatment in the control cells, whereas knockdown of PRMT1 resulted in the loss of PIAS1 methylation (Fig. 5A). The efficiency of PIAS1 overexpression, PRMT1 depletion, and IFNγ stimulated tyrosine 701 phosphorylation of STAT1 was verified by Western blot analysis (Fig. 5A). To confirm that translational inhibition avoided incorporation of the radiolabeled methionine by de novo protein biosynthesis (Liu and Dreyfuss 1995), cells were incubated with 35S-methionine and protein synthesis inhibitors. No label was detectable in total protein lysate from these cells by fluorography (Supplemental Fig. S8) indicating that radiolabeling of PIAS1 was indeed due to post-translational modification. Similar results for in vivo methylation of the endogenous PIAS1 protein were obtained (Fig. 5B), whereas methylation of STAT1, which was reported by Mowen et al. (2001), was not detectable in this assay (Fig. 5C). These data indicate that in vivo methylation of PIAS1 occurs in response to IFNγ treatment and that PRMT1 is the responsible enzyme for PIAS1 methylation in this pathway.
Figure 5.
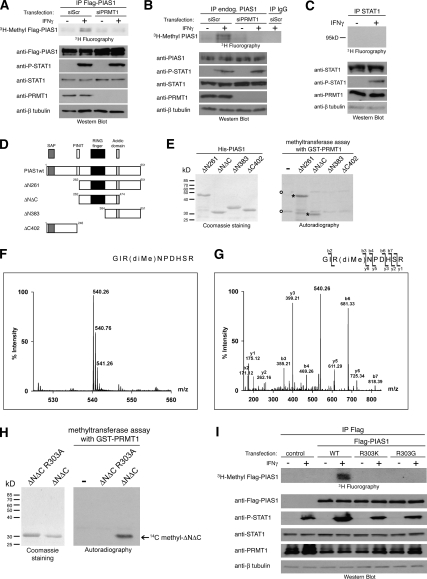
PRMT1 methylates R303 in PIAS1 in vivo. (A) HeLa cells were first transfected with control siRNA (siScr) or siRNA against PRMT1 (siPRMT1_1 was used). After 24 h cells were transfected with Flag-tagged PIAS1, and 48 h later cells were induced with IFNγ for 3 h (+) or not (−) and simultaneously labeled with L-(methyl-3H) methionine in the presence of translation inhibitors. (Top panel) Subsequently, Flag-tagged PIAS1 was immunoprecipitated from cell lysates and analyzed by SDS-PAGE followed by fluorography. (Bottom panels) Effective knockdown of PRMT1 and overexpression of PIAS1 was checked by Western blot. (B) HeLa cells were transfected with control siRNA (siScr) or siRNA against PRMT1 (siPRMT1_1 was used). After 72 h cells were induced with IFNγ for 3 h (+) or not (−) and simultaneously labeled with L-(methyl-3H) methionine in the presence of translation inhibitors. (Top panel) Subsequently, endogenous PIAS1 was immunoprecipitated from cell lysates and analyzed by SDS-PAGE followed by fluorography. (Bottom panels) Efficient knockdown of PRMT1 and endogenous PIAS1 and STAT1 levels were checked by Western blot. (C) HeLa cells were induced with IFNγ for 3 h (+) or not (−) and simultaneously labeled with L-(methyl-3H) methionine in the presence of translation inhibitors. (Top panel) Subsequently, endogenous STAT1 was immunoprecipitated from cell lysates and analyzed by SDS-PAGE followed by fluorography. (Bottom panels) The endogenous STAT1 and PRMT1 levels were checked by Western blot. (D) Schematic representation of PIAS1 deletion constructs used in E. Conserved domains are indicated: The N-terminal SAP box (Saf-A/B, Acinus, PIAS) is required for DNA-binding, PINIT motif is involved in nuclear retention of certain family members, RING finger-like domain is essential for Sumo ligase activity and the acidic domain is important for protein–protein interactions (Duval et al. 2003; Shuai and Liu 2005). (E) His-tagged PIAS1 deletion constructs were purified from E. coli, and equal amounts were used for in vitro methylation with recombinant GST-PRMT1 in the presence of methyl-14C-labeled SAM. Coomassie staining illustrates the protein amounts used for the methylation assay (left panel). Methylation products were resolved on SDS-PAGE, blotted, and visualized by autoradiography (right panel). PIAS1-specific methylation products are marked with asterisks and nonspecific methylation products with a circle. (F,G) PIAS1 ΔNΔC was in vitro methylated by recombinant PRMT1 and subsequently analyzed by mass spectrometry. (F) The NanoESI-MS spectrum of the tryptic fragment 301GIRNPDHSR309 revealed a peak with m/z 540.26 (calculated m/z 540.29) corresponding to the doubly charged ion of the dimethylated sequence. (G) Fragment ion (MS/MS) spectrum resulting from the doubly charged precursor with m/z 540.26 produced y ions and the b ions by consecutive fragmentation reactions. Relevant ions were labeled according to the accepted nomenclature. Dimethylation of Arg 303 was detected in the mass of the b3, b4, b6, and b7 ions. (H, right panel) In vitro methylation of PIAS1 ΔNΔC or the mutant PIAS1 ΔNΔC R303A by GST-PRMT1 was monitored by autoradiography. (Left panel) The Coomassie staining reveals that equal amounts of both PIAS1 deletion proteins were used for the methylation assay. (I) HeLa cells were transiently transfected with Flag-tagged EGFP (control), Flag-tagged PIAS1 wild type (WT), PIAS1 R303K mutant, and PIAS1 R303G mutant, respectively. Metabolic labeling with L-(methyl-3H) methionine, anti-Flag IP, fluorography (top panel), and corresponding Western blot (bottom panels) were performed as in A.
In order to map the methylation site(s) in PIAS1, we expressed several N- and C-terminal deletion constructs of PIAS1 (Fig. 5D) as His-tagged fusions in bacteria (Fig. 5E). In vitro methylation in the presence of GST-PRMT1 revealed that the PIAS1 deletion ΔNΔC is the smallest fragment still methylated by PRMT1 (Fig. 5E). This indicates that the methylation site(s) of PRMT1 in PIAS1 are located between amino acid 262 and 384, which encompass the RING finger domain and the acidic domain.
To identify the exact site(s) of PRMT1 methylation in PIAS1 we methylated PIAS1 ΔNΔC in the presence of GST-PRMT1 in vitro. The ESI mass spectrum revealed a doubly charged peak at m/z 540.26 (calculated m/z 540.29) corresponding to the dimethylated sequence of the tryptic PIAS1 fragment GIRNPDHSR (amino acids 301–309) (Fig. 5F). Fragment ion (MS/MS) spectrum resulting from this doubly charged precursor clearly indicated that the first arginine within the peptide (R303 in the full-length context of PIAS1) is dimethylated (Fig. 5G). Mutation of the R303 to alanine (R303A) confirmed this arginine, which is localized between the PINIT motif and the RING finger domain, as the in vitro methylation site of PIAS1 ΔNΔC protein by PRMT1 (Fig. 5H).
To validate R303 also as the in vivo methylation site of PIAS1 by PRMT1, we mutated R303 in the full-length PIAS1 to lysine (R303K) or glycine (R303G). Subsequently, we performed overexpression and metabolical labeling of both Flag-tagged mutants or wild-type Flag-tagged PIAS1 (WT) in unstimulated and IFNγ-stimulated HeLa cells. Immunoprecipitates of PIAS1 wild type revealed in vivo methylation of PIAS1 upon IFNγ stimulation, whereas both PIAS1 mutants, R303K and R303G, were not methylated in the absence or in the presence of IFNγ (Fig. 5I). These results indicate that PIAS1 is methylated upon IFNγ stimulation at a single arginine, R303, in vivo.
Methylation of R303 in PIAS1 is required for its repressive effect on STAT1 and for its recruitment to STAT1 target gene promoters
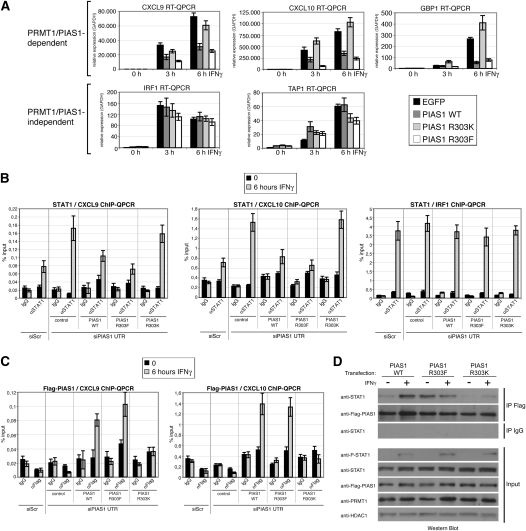
To investigate the impact of PIAS1 methylation on its repressive activity in STAT1-activated transcription we explored overexpression of PIAS1 wild type, a methylation-deficient PIAS1 mutant (R303K), and a methylation-mimicking mutant, in which the arginine was mutated to phenylalanine (R303F) (Mostaqul Huq et al. 2006). The mutations of R303 did not affect the sumoylation activity of PIAS1 (Supplemental Fig. S9), its nuclear localization, or the cellular localization of STAT1, and its IFN-induced shuttling (Supplemental Fig. S10). Subsequent to transfection of PIAS1 wild type and the two mutants, IFNγ treatment was performed and STAT1 target gene expression was analyzed by RT-QPCR. Overexpression of PIAS1 wild type and the methylation-mimicking PIAS1 R303F mutant decreased the IFN-induced transcription of PRMT1/PIAS1-dependent targets compared with control transfected cells (Fig. 6A). In contrast, the methylation-deficient PIAS1 R303K mutant exhibited a relief of this repression and resulted in a transcriptional induction of the PRMT1/PIAS1-dependent targets comparable with control cells (Fig. 6A). Expression of the different PIAS1 proteins did not affect the IFN-induced transcriptional activation of PRMT1/PIAS1-independent STAT1 target genes (Fig. 6A). These findings demonstrate that PRMT1-mediated methylation of R303 in PIAS1 is crucial for repression of STAT1-activated transcription.
Figure 6.
Methylation of R303 in PIAS1 is crucial for PIAS1-dependent repression of STAT1 signaling and for PIAS1 promoter recruitment. (A) HeLa cells transiently overexpressing either Flag-EGFP, Flag-PIAS1 wild type (WT), methylation-deficient Flag-PIAS1 R303K mutant or methylation-mimicking Flag-PIAS1 R303F mutant were induced with IFNγ (5 ng/mL) for the indicated time points. Subsequently, RNA was prepared and analyzed by RT-QPCR for transcript levels of STAT1 target genes. Results were normalized to GAPDH and transcript levels in uninduced control cells were set to 1. (B,C) HeLa cells were transfected with control siRNA (siScr) or siRNA against the 3′UTR of PIAS1 (siPIAS1 UTR = siPIAS1 UTR1 + 2). After 24 h, the siPIAS1 UTR-treated cells were transfected with empty vector (control), Flag-tagged PIAS1 wild-type (WT), R303K mutant, or R303F mutant constructs. After 48 h of this second transfection, cells were treated with IFN for 6 h and subjected to ChIP analysis. Recruitment of STAT1 (B) and Flag-PIAS1 (C) to the indicated gene promoters was determined by QPCR. Results were displayed as percentage input. (D) HeLa cells were transfected with Flag-tagged PIAS1 wild-type (WT), R303K mutant, or R303F mutant constructs. After 48 h, cells were treated with IFN for 6 h and subjected to co-IP analysis. IP of Flag-PIAS1 (anti-Flag) or control IP (IgG) was carried out from 500-μg nuclear extract and examined by Western blot using antibodies against STAT1 and Flag-PIAS1. Input Western blots of the nuclear extracts corresponding to the co-IP were stained for P-STAT1, STAT1, Flag-PIAS1, PRMT1, and HDAC1 (the latter served as loading control).
To determine the impact of R303 methylation on the promoter recruitment of STAT1 and PIAS1, and to avoid at the same time an interference of the endogenous PIAS1 protein in this assay, we first depleted the endogenous PIAS1 expression in HeLa cells by using siRNAs targeting the 3′ untranslated region (UTR) of PIAS1 (Supplemental Fig. S11A). Subsequently, we transiently transfected the PIAS1-depleted cells with empty vector (control), Flag-PIAS1 wild type, methylation-deficient Flag-PIAS1 R303K mutant, and methylation-mimicking Flag-PIAS1 R303F mutant. The exogenous PIAS1 expression was not affected by the siPIAS1 targeting the 3′UTR, and the expression levels of the different PIAS1 constructs were equal (Supplemental Fig. S11A). When we subjected these cells to IFNγ treatment, we found that similar to the siRNA targeting the PIAS1 ORF (Fig. 1E), the siPIAS1 UTR resulted in an increased transcriptional activation of the specific subset of STAT1 targets in comparison with siScr transfected cells (Supplemental Fig. S11B). Overexpression of PIAS1 wild type and R303F mutant rescued this effect, whereas PIAS1 R303K was not able to perform the rescue (Supplemental Fig. S11B). ChIP analysis of these cells revealed that IFN-induced promoter recruitment of STAT1 was enhanced in the siPIAS1 UTR-transfected cells compared with siScr-transfected cells, in agreement with Figure 3B. Further, overexpression of PIAS1 wild type and methylation-mimicking R303F mutant rescued this effect; i.e., both PIAS1 proteins decreased the STAT1 recruitment back to the level detected in siScr-transfected cells (Fig. 6B; Supplemental Fig. S12A). In contrast, the methylation-deficient PIAS1 R303K mutant was unable to substitute for the function of the endogenous PIAS1 protein in the PIAS1-depleted cells and the STAT1 recruitment remained increased (Fig. 6B; Supplemental Fig. S12A). These observations apply to the PRMT1/PIAS1-dependent STAT1 targets, and are consistent with the transcriptional effects of PIAS1 wild type and the mutants on this particular subset of genes (Fig. 6A; Supplemental Fig. 11B). In agreement with the PRMT1/PIAS1-independent transcriptional regulation of IRF1, PIAS1 depletion as well as overexpression of the different PIAS1 constructs did not influence STAT1 recruitment to this promoter (Fig. 6B). When we analyzed the promoter recruitment of PIAS1 in these cells, we noticed that the binding of PIAS1 R303K to the PRMT1/PIAS1-dependent STAT1 targets was completely lost, whereas the binding of methylation-mimicking PIAS1 R303F mutant was similar to PIAS1 wild type (Fig. 6C; Supplemental Fig. S12B). In agreement with the PRMT1/PIAS1-independent transcriptional regulation of IRF1, the different PIAS1 constructs did not associate with this promoter (Supplemental Fig. S12B). These results show that PRMT1-mediated methylation of R303 in PIAS1 is essential for the recruitment of PIAS1, for the release of STAT1 from target gene promoters, and for turning off of STAT1-activated transcription.
To test whether the methylation of R303 itself contributes to the interaction between PIAS1 and STAT1, we performed co-IP experiments from cells overexpressing either Flag-PIAS1 wild type, Flag-PIAS1 R303F, or Flag-PIAS1 R303K in the absence or presence of IFNγ stimulation. Nuclear extracts were subjected to anti-Flag IP and, subsequently, immunoprecipitates were stained for the presence of endogenous STAT1. As expected, the interaction between PIAS1 wild type and STAT1 was enhanced upon addition of IFNγ (Fig. 6D). The methylation-mimicking PIAS1 R303F mutant revealed already association with STAT1 without IFNγ stimulation, which was not further increased upon treatment with IFNγ. In contrast, the methylation-deficient PIAS1 R303K mutant exhibited reduced binding to STAT1, under uninduced as well as induced conditions (Fig. 6D). These data suggest that the PRMT1-mediated methylation of R303 in PIAS1 facilitates its interaction with STAT1, and in this way might allow promoter recruitment of PIAS1.
Knockdown of PRMT1 or PIAS1 enhances the anti-proliferative effect of IFNγ
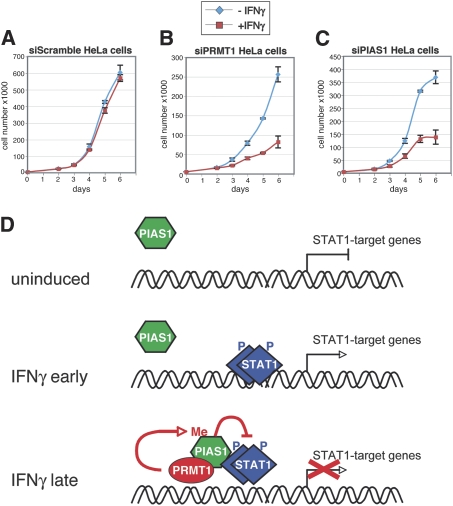
Finally, we elucidated the impact of PRMT1-regulated repression by PIAS1 on the biological outcome of IFN signaling and STAT1 function. Therefore we measured the growth/proliferation of HeLa cells in the presence of different concentrations of IFNγ. A concentration of 15 ng IFNγ/mL resulted in a growth arrest, whereas at lower concentration of 7.5 ng IFNγ/mL the anti-proliferative effect of IFN was not detectable (Supplemental Fig. S13) (Bromberg et al. 1996). In the following, HeLa cells were transfected with control siRNA (siScr), siPRMT1, or siPIAS1 and subjected to growth curve analysis in the presence of IFNγ. Depletion of endogenous PRMT1 and PIAS1, respectively, was monitored and verified by Western blot analysis (Supplemental Fig. S14). Relative to cells transfected with siScr, depletion of PRMT1 or PIAS1 resulted in the occurrence of anti-proliferative activity at low doses of IFNγ (Fig. 7A–C) and enhanced anti-proliferative activity at high doses of IFNγ (data not shown). These results demonstrate that PRMT1/PIAS1-mediated repression of STAT1 activity is important for the biological effects of IFN.
Figure 7.
Knockdown of PRMT1 or PIAS1 enhances the anti-proliferative effect of IFNγ. (A–C) HeLa cells were transfected with control siRNA, siRNA against PRMT1 (siPRMT1_1 was used) or against PIAS1. After 48 h, cells were trypsinized and 10,000 cells per well were seeded and after 4 h induced with 7.5 ng/mL IFNγ. At the indicated time points, cells were harvested and cell number was determined. For each condition and time point triplicates were counted. Error bars are depicted accordingly. (D) Model for the regulation of PIAS1-mediated repression of STAT1 signaling by PRMT1.
Discussion
Previous reports suggested that arginine methylation influences IFN signaling, since PRMT1 was shown to interact with the cytoplasmic domain of the IFNα receptor (Abramovich et al. 1997) and its depletion interfered with IFNα-induced growth arrest (Altschuler et al. 1999). PRMT1 was reported to methylate the transcription factor STAT1, and this was shown to inhibit the association of STAT1 and PIAS1 and consequently to enhance transcriptional activation of IFN responsive genes (Mowen et al. 2001; Nien et al. 2007). However, arginine methylation of STAT1 was not confirmed by others (Meissner et al. 2004) and also not in the present work (Fig. 5C). Furthermore, most studies investigating the impact of arginine methylation in IFN signaling used nonspecific methyltransferase inhibitors, like MTA, which execute strong side effects and inhibit various cellular kinase cascades (p38 and ERK) and the IFN-induced phosphorylation of STAT1 and STAT3 (Meissner et al. 2004; Komyod et al. 2005). Therefore, we set out in the present study to elucidate the role of arginine methylation in STAT1 signaling and identified the PIAS1 protein as an important target of PRMT1 in this pathway.
Our findings reveal a novel mechanism how the nuclear inhibitor of STAT1 signaling, PIAS1, is activated in the course of the IFN response to restrain STAT1 function and thus how duration and magnitude of IFN signaling is balanced. We demonstrate that arginine methylation by PRMT1 influences the transcriptional activity of STAT1 negatively by regulating the interaction between STAT1 and it repressor protein PIAS1. Interestingly, in the literature PRMT1 has been reported to act as transcriptional coactivator (Wang et al. 2001; An et al. 2004; Wagner et al. 2006) as well as corepressor (Kwak et al. 2003; Yu et al. 2006). Our model for the cooperation between PRMT1 and PIAS1 in STAT1 signaling is summarized in Figure 7D. We find that PRMT1 represses a subset of IFNγ-activated STAT1 target genes, which overlaps with genes regulated by PIAS1 (Liu et al. 2004). PRMT1 mediates its function in STAT1 signaling through PIAS1, as depletion of PIAS1 abrogates the inhibitory activity of PRMT1. In search of the molecular mechanism of this cooperation, we discover that PRMT1 dimethylates PIAS1 at R303. Association between PRMT1 and PIAS1 and methylation of R303 is induced in the late phase of the IFNγ response and enables gene-specific recruitment of PIAS1. Therefore, depletion of PRMT1 results in the loss of PIAS1 promoter binding. Consistently, transcriptional repression and promoter recruitment of a methylation-deficient PIAS1 mutant is abrogated, reinforcing that PRMT1-dependent methylation of PIAS1 is important for its repressive function. The promoter-associated methylation of PIAS1 then triggers the interaction between STAT1 and PIAS1 and enforces the release of STAT1 from its target genes, which subsequently results in the decline of transcriptional activation. This interpretation is supported by our findings that the methylation-deficient PIAS1 mutant exhibits a decreased interaction with STAT1 in co-IP, and that depletion of PRMT1 or PIAS1 results in an enhanced and prolonged recruitment of STAT1 to a subset of its target genes.
Regulation of the repressive activity of PIAS1 is necessary, as the protein is predominantly localized in the nucleus. A constitutively active form of PIAS1 would straight away inhibit activated STAT1 dimers, which enter the nucleus upon stimulus. But initially, STAT1 dimers need to perform their transactivation function and to enable a proinflammatory and anti-proliferative gene expression pattern. Therefore, mechanisms must exist to allow a targeted interaction between STAT1 and its nuclear repressor protein in the late phase of the immune response. Here, we describe such a mechanism: R303 methylation catalyzed by PRMT1 facilitates the STAT1–PIAS1 interaction, which was reported to be a direct interaction (Liao et al. 2000). R303 is localized in a conserved but functionally not assigned part between the PINIT motif and the RING finger domain of PIAS1. Although the C-terminal portion of PIAS1 (not encompassing the R303) is responsible for the direct interaction between PIAS1 and STAT1, the N-terminal part of PIAS1 including R303 modulates the association between the two proteins. This N-terminal domain restricts the interaction of PIAS1 to STAT1 dimers by inhibiting its interaction with STAT1 monomers (Liao et al. 2000) supporting our findings that methylation of R303 is important for the PIAS1–STAT1 interaction. Interestingly, the methylation site is not embedded in a consensus methylation sequence of PRMT1, which typically harbors the methylated arginine in a RGG or RXR context (Smith et al. 1999). In general, methylation of the arginine residue is thought to regulate protein–protein and protein–nucleic acid interactions potentially by increasing its “hydrophobic” character (Boisvert et al. 2005). Methylation of arginine residues occurs also in other chromatin proteins involved in cytokine signaling, like the PRMT1-driven methylation of the arginine–glycine rich N terminus of the coactivator NIP45, which promotes the association between NIP45 and the transcription factor NFAT and results in an augmented activity of the IL-4 and IFNγ gene promoter (Mowen et al. 2004).
Mechanistically, it was suggested that the interaction between PIAS1 and STAT1 inhibits the DNA-binding ability of STAT1 and causes the reduction of transcriptional activity (Liu et al. 1998; Shuai and Liu 2005). Similar data were also reported for other PIAS family members; for example, PIAS3 interacts with STAT3 and suppresses the interaction of STAT3 with the DNA (Chung et al. 1997). Given that R303 is conserved also in other PIAS family members, arginine methylation of PIAS proteins might play a more general role in cytokine-dependent gene regulation. PIAS1 knockout mice reveal enhanced protection against pathogenic infections, although PIAS1 represses only a subset of 9% of the IFN-induced STAT target genes (Liu et al. 2004), indicating that PIAS1 is a physiologically important negative regulator. A common characteristic of genes regulated by PIAS1 and also PRMT1, like the GBP1, CXCL9, and CXCL10 gene, is that they contain low-affinity binding sites of STAT1, whereas the high-affinity binding site of STAT1 in the IRF1 gene promoter does not depend on PIAS1 as well as PRMT1 (Liu et al. 2004).
Methylation by PRMT1 seems to be not the only mechanism for how PIAS1 function is regulated. It was reported recently that PIAS1 is phosphorylated at Ser 90 by the IKKα kinase particularly in response to TNFα and LPS stimulation, and that this modification is required to repress the NF-κB/STAT1 activity (Liu et al. 2007). Similar to the regulation of PIAS1 by PRMT1, S90 phosphorylation was needed to allow promoter recruitment of PIAS1 in NF-κB-activated transcription and resulted in the detachment of NF-κB from its targets. In contrast to the regulation by PRMT1, the Sumo E3-ligase activity was necessary for the repressive function of PIAS1, since a sumoylation-deficient PIAS1 mutant was defective in S90 phosphorylation (Liu et al. 2007). Interestingly, we did not find phosphorylated PIAS1 species in response to IFNγ treatment (data not shown). Thus, the mechanism of PIAS1 regulation might differ depending on the stimulus, transcription factor, target genes, and cell type. In the future it will be important to also study the role of arginine methylation of PIAS1 in its STAT1-independent functions.
In summary, our results identify PIAS1 as the crucial target of arginine methylation and PRMT1 in IFN signaling. PRMT1 is involved in turning off transcriptional activity of STAT1 and represses the transcription factor by regulating the interaction between STAT1 and it repressor protein PIAS1 in a methylation-dependent manner. Hence, PRMT1 is a very interesting target in therapies of viral infections and also deregulated STAT signal transduction. How PRMT1 itself is regulated in the course of IFN stimulation, needs to be addressed in the future.
Materials and methods
Plasmids
The following plasmids were used: pcDNA3.1-PRMT1 wild-type and pcDNA3.1-PRMT1-Myc XGX mutant were published recently (Balint et al. 2005). pGex2T-GAR and pGex2T-PRMT3 were described previously (Tang et al. 1998). pGex4T-1-PRMT1 and pGex4T-1-PRMT2 were published (Scott et al. 1998). Murine pGex4T-1-PRMT4 (Chen et al. 1999) and human pGex2T-PRMT6 (Hyllus et al. 2007) were described.
PIAS1 full-length and deletion constructs were generated by PCR amplification from the pBK-CMV-PIAS1 plasmid (Sapetschnig et al. 2002) introducing at the 5′-end of a XhoI site and at the 3′-end of a Hind III site and cloned via these sites in the pFastBac HT vector or the pRSETA vector (Invitrogen). The following constructs were generated: pFastBac HT-Pias1 full-length (amino acids 1–651), pRSETA-PIAS1 ΔN45 (amino acids 46–651), ΔN261 (amino acids 262–651), ΔN383 (amino acids 384–651), ΔC402 (amino acids 1–253), and ΔNΔC (amino acids 262–474). Furthermore, pRSETA-PIAS1 ΔNΔC was used for site-directed mutagenesis (Stratagene QuickChange Kit) of Arg 303 to alanine. The pN3-3xFlag PIAS1 (full-length) construct was generated by PCR amplification introducing at the 5′-end of an EcoRI site and at the 3′-end of a SalI site and cloned into a modified pEGFP-N3 vector (Clontech; EGFP tag replaced by triple Flag tag). pN3-3xFlag PIAS1 R303 mutants were generated by site-directed mutagenesis (Stratagene Quickchange Kit). R303 was exchanged to K, G, and F, respectively.
Antibodies
The following antibodies were employed: anti-Flag (F3165) from Sigma, anti-HA (sc-805) from Santa Cruz Biotechnologies, anti-5-His (34,660) from Qiagen, anti-STAT1 (sc-346) from Santa Cruz Biotechologies, anti-STAT1pY701 (9171) from Cell Signaling, anti-β-tubulin (MAB 3408) from Chemicon, anti-HDAC1 (sc-7872) from Santa Cruz Biotechnologies, anti-PRMT6 (Wagner et al. 2006), anti-PRMT4 (Kleinschmidt et al. 2008), anti-PRMT1 (07-404) from Upstate Biotechnologies for Western blot, anti-PRMT1 (Kleinschmidt et al. 2008) for ChIP, and anti-PIAS1 (sc-8152) from Santa Cruz Biotechnologies for Western blot/IP. For ChIP analysis we generated a polyclonal rabbit antibody against the C terminus of human PIAS1 (amino acids 384–651). The Myc (9E10) antibody was a kind gift from Martin Eilers. As control antibodies in ChIP and IP we used anti-p15 (sc-1429) from Santa Cruz Biotechnologies or rabbit IgG from Pierce (31,235) and from Sigma (I5006), respectively.
Cell culture and transfections
HeLa and HEK293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). A375 cells were cultured in RPMI supplemented with 10% FBS. IFNγ (Strathmann Biotec) was used at concentrations between 5 and 15 ng/mL for the indicated time points. Transfection of HeLa and HEK293 cells was carried out using the Ca-Phosphat method.
siRNA oligonucleotide duplexes were purchased from Dharmacon for targeting the human PRMT1, PIAS1, PRMT4, and PRMT6, respectively. The transfections were carried out with Oligofectamine (Invitrogen). siRNA duplexes were transfected at a final concentration of 100 nM in HeLa, HEK293, or A375 cells. Afer 2 d cells were collected for protein or RNA preparation. The siRNA sequences are listed in the Supplemental Material.
RT-QPCR
Total RNA was isolated using PeqGold total RNA Kit (PeqLab). Two micrograms of RNA were applied to reverse transcription by incubation with 0.5 μg of oligo(dT)17 primer and 100 U of M-MLV reverse transcriptase (Invitrogen). cDNA was analyzed by QPCR, which was performed using ABsolute QPCR SybrGreen Mix (Abgene) and the Mx3000P real-time detection system (Agilent). The primers used in RT-QPCR are indicated in the Supplemental Material. All amplifications were performed in triplicate using 0.5 μL of cDNA per reaction. The triplicate mean values were calculated according to the ΔΔCt quantification method (Pfaffl 2001) using the GAPDH gene transcription as reference for normalization. Relative mRNA levels in uninduced control cells were equated to 1 and the other values were expressed relative to this. The RT-QPCR results were reproduced at least three times in independent experiments and representative data sets are shown.
ChIP
HeLa cells were cross-linked in the presence of 1% formaldehyde for 10 min at room temperature. Subsequent ChIP analysis was performed as described previously (Wagner et al. 2006). Immunoprecipitated and eluted DNA was purified with QIAquick columns (Qiagen) and amplified by QPCR. The ChIP-QPCR primers are indicated in the Supplemental Material. All amplifications were performed in triplicate using 0.6 μL of DNA per reaction. The triplicate mean values were displayed as percentage input and standard deviation was calculated. The presented results were reproduced several times in independent experiments.
Preparation of protein extracts and IP
Whole-cell extract were prepared from HeLa or HEK293, respectively. Cells were rinsed in PBS and lysed in IPH buffer (50 mM Tris at pH 8, 150 mM NaCl, 5 mM EDTA, 0.5% NP40, 1 mM DTT, 150 mM NaF, 2 mM Na3VO4). After lysis the extracts were cleared by centrifugation and supernatant was used for Western blot analysis.
Nuclear extract were prepared from HeLa cells. Cells were washed with PBS and subsequently lysed in buffer A1 (10 mM HEPES KOH at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.04% NP40, 2 mM Na3VO4) for 5 min on ice. After centrifugation cytoplasmic supernatant was removed and nuclei were washed in buffer A2 (10 mM HEPES KOH at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 2 mM Na3VO4) to exclude cytoplasmatic contamination. The remaining nuclear pellet was lysed in IPH buffer, and thereafter a benzonase digestion was performed. For IP, 1–2 μg of the indicated antibodies were incubated with 500 μg of nuclear extract as described previously (Kleinschmidt et al. 2008).
Recombinant proteins and GST pulldown experiments
GST fusion proteins and His tag proteins were prepared from Escherichia coli BL21 according to standard procedures. His-tagged PIAS1 (full length) was expressed and purified from Sf9 cells according to the manufacturer's instructions for the Bac-to-Bac Baculovirus Expression System (Invitrogen). GST and GST-PRMT1 immobilized on glutathione beads were blocked with 10% goat serum in HEGN buffer (30 mM HEPES-KOH at pH 7.6, 125 mM KCl, 0.16 mM EDTA, 16% glycerol, 0.16% NP-40, 1 mM DTT) for 10 min at room temperature. Precleared whole-cell extract (250 μg) (Kleinschmidt et al. 2008) or recombinant His-tagged PIAS1 (0.5 μg) was incubated with the blocked beads for 1 h at room temperature. Subsequently, beads were washed five times with IPH buffer and examined by Western blot analysis.
In vitro methyltransferase assay
Eluted and dialyzed GST-PRMTs (0.5 μg) were incubated with approximatly 2–4 μg of substrate in the presence of [14C-methyl]-SAM (58.3 mCi/mM; GE Healthcare Life Science) for 2 h at 37°C. Reaction was stopped by addition of SDS-PAGE sample buffer and analyzed by SDS-PAGE, blotting and autoradiography.
In vivo methylation assay
Transfected Hela cells were pretreated with cycloheximid and chloramphenicol in the presences or absence of IFNγ (5 ng/mL). After a 30-min pretreatment, L-[methyl-3H]-methionine (70 Ci/mM; GE Healthcare Life Science) was added and cells were cultured for another 3 h (Liu and Dreyfuss 1995). Whole-cell extracts were prepared and employed for IP of Flag-tagged PIAS1 or endogenous PIAS1 as well as STAT1. Immunoprecipitates were separated by SDS-PAGE. The gel was incubated in Enlight solution (MoBiTec) according to manufacturer instructions and 3H-labeled proteins were visualized by fluorography.
Mass spectrometric analysis
Gel-excised methylated PIAS1 protein (ΔNΔC) bands were washed with 50% (v/v) acetonitrile in 25 mM ammonium bicarbonate, shrunk by dehydration in acetonitrile, and dried in a vacuum centrifuge. Reduction/alkylation of disulfide bonds and enzymatic in-gel digestion was performed as described (Bauer and Krause 2005). MS and MS/MS experiments were carried out on a quadrupole orthogonal acceleration time-of-flight mass spectrometer Q-Tof Ultima equipped with a CapLC liquid chromatography system (Micromass). Peptides were separated using a capillary column (Atlantis dC18, 3 μm, 100 Å, 150 mm × 75 μm i.d.; Waters GmbH) and an eluent flow rate of 200 nL/min. Mobile phase A was 0.1% formic acid (v/v) in acetonitrile-water (3:97, v/v) and B was 0.1% formic acid in acetonitrile-water (8:2, v/v). Runs were performed using a gradient of 3%–64% B in 100 min. Data-dependent aquisition was used, whereas peptides, which covered the methylation sites, were preferentially selected for MS/MS. The processed MS/MS spectra (MassLynx version 4.0 software) were compared with the theoretical fragment ions of peptides of human PIAS1 protein.
Growth curves
For analysis of IFNγ-induced growth arrest, 10,000 cells per well were seeded and subsequently stimulated with 7.5 or 15 ng/mL IFNγ. Cells were harvested at the indicated time points and cell numbers were determined in triplicates for each condition and time point by using the Neubauer counting chamber. For growth arrest analysis in PRMT1- or PIAS1-depleted cells, we performed transient transfection of the corresponding siRNAs in HeLa cells for 48 h. Subsequently, cells were trypsinized, seeded (10,000 cells per well) and treated with IFNγ. The knockdown was monitored in parallel by Western blot analysis.
Acknowledgments
We thank Hervey Herschman for providing the pGEX2T-GAR plasmid. We thank Alexander Brehm, Martin Eilers, Jean-Francois Naud, and Uwe Vinkemeier for critical reading of the manuscript and valuable suggestions. We thank all members of the U.M.B. laboratory and Alexandra Sapetschnig for their help during work progress, and Inge Pelz for technical assistance. This work was supported by the DFG (Deutsche Forschungsgemeinschaft), by funding from Land Hessen, and from BMBF (Bundesministerium für Bildung und Forschung) to U.M.B.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.489409.
Supplemental material is available at http://www.genesdev.org.
References
- Abramovich C., Yakobson B., Chebath J., Revel M. A protein–arginine methyltransferase binds to the intracytoplasmic domain of the IFNAR1 chain in the type I interferon receptor. EMBO J. 1997;16:260–266. doi: 10.1093/emboj/16.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander W.S., Hilton D.J. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu. Rev. Immunol. 2004;22:503–529. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- Altschuler L., Wook J.O., Gurari D., Chebath J., Revel M. Involvement of receptor-bound protein methyltransferase PRMT1 in antiviral and antiproliferative effects of type I interferons. J. Interferon Cytokine Res. 1999;19:189–195. doi: 10.1089/107999099314333. [DOI] [PubMed] [Google Scholar]
- An W., Kim J., Roeder R.G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Balint B.L., Szanto A., Madi A., Bauer U.M., Gabor P., Benko S., Puskas L.G., Davies P.J., Nagy L. Arginine methylation provides epigenetic transcription memory for retinoid-induced differentiation in myeloid cells. Mol. Cell. Biol. 2005;25:5648–5663. doi: 10.1128/MCB.25.13.5648-5663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero M.J., Malik S. Two functional modes of a nuclear receptor-recruited arginine methyltransferase in transcriptional activation. Mol. Cell. 2006;24:233–243. doi: 10.1016/j.molcel.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P.J., Krause E. Accessibility of cysteines in the native bovine rod cGMP-gated channel. Biochemistry. 2005;44:1624–1634. doi: 10.1021/bi0478749. [DOI] [PubMed] [Google Scholar]
- Bedford M.T., Richard S. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Boisvert F.M., Chenard C.A., Richard S. Protein interfaces in signaling regulated by arginine methylation. Sci. STKE. 2005;271:re2. doi: 10.1126/stke.2712005re2. [DOI] [PubMed] [Google Scholar]
- Borden E.C., Sen G.C., Uze G., Silverman R.H., Ransohoff R.M., Foster G.R., Stark G.R. Interferons at age 50: Past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg J.F., Horvath C.M., Wen Z., Schreiber R.D., Darnell J.E., Jr Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon α and interferon γ. Proc. Natl. Acad. Sci. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Ma H., Hong H., Koh S.S., Huang S.M., Schurter B.T., Aswad D.W., Stallcup M.R. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Chen W., Daines M.O., Hershey G.K. Methylation of STAT6 modulates STAT6 phosphorylation, nuclear translocation, and DNA-binding activity. J. Immunol. 2004;172:6744–6750. doi: 10.4049/jimmunol.172.11.6744. [DOI] [PubMed] [Google Scholar]
- Chung C.D., Liao J., Liu B., Rao X., Jay P., Berta P., Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- Darnell J.E., Jr Transcription factors as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- Duval D., Duval G., Kedinger C., Poch O., Boeuf H. The “PINIT” motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 2003;554:111–118. doi: 10.1016/s0014-5793(03)01116-5. [DOI] [PubMed] [Google Scholar]
- Hyllus D., Stein C., Schnabel K., Schiltz E., Imhof A., Dou Y., Hsieh J., Bauer U.M. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes & Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt M.A., Streubel G., Samans B., Krause M., Bauer U.M. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 2008;36:3202–3213. doi: 10.1093/nar/gkn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komyod W., Bauer U.M., Heinrich P.C., Haan S., Behrmann I. Are STATS arginine-methylated? J. Biol. Chem. 2005;280:21700–21705. doi: 10.1074/jbc.C400606200. [DOI] [PubMed] [Google Scholar]
- Kwak Y.T., Guo J., Prajapati S., Park K.J., Surabhi R.M., Miller B., Gehrig P., Gaynor R.B. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell. 2003;11:1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- Lai J.S., Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.E., Darnell J.E., Jr Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Liao J., Fu Y., Shuai K. Distinct roles of the NH2- and COOH-terminal domains of the protein inhibitor of activated signal transducer and activator of transcription (STAT) 1 (PIAS1) in cytokine-induced PIAS1–Stat1 interaction. Proc. Natl. Acad. Sci. 2000;97:5267–5272. doi: 10.1073/pnas.97.10.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Liao J., Rao X., Kushner S.A., Chung C.D., Chang D.D., Shuai K. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Mink S., Wong K.A., Stein N., Getman C., Dempsey P.W., Wu H., Shuai K. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat. Immunol. 2004;5:891–898. doi: 10.1038/ni1104. [DOI] [PubMed] [Google Scholar]
- Liu B., Yang Y., Chernishof V., Loo R.R., Jang H., Tahk S., Yang R., Mink S., Shultz D., Bellone C.J.,., et al. Proinflammatory stimuli induce IKKα-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell. 2007;129:903–914. doi: 10.1016/j.cell.2007.03.056. [DOI] [PubMed] [Google Scholar]
- Meissner T., Krause E., Lodige I., Vinkemeier U. Arginine methylation of STAT1: A reassessment. Cell. 2004;119:587–589. doi: 10.1016/j.cell.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Meyer T., Vinkemeier U. Nucleocytoplasmic shuttling of STAT transcription factors. Eur. J. Biochem. 2004;271:4606–4612. doi: 10.1111/j.1432-1033.2004.04423.x. [DOI] [PubMed] [Google Scholar]
- Mostaqul Huq M.D., Gupta P., Tsai N.P., White R., Parker M.G., Wei L.N. Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 2006;25:5094–5104. doi: 10.1038/sj.emboj.7601389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen K.A., Tang J., Zhu W., Schurter B.T., Shuai K., Herschman H.R., David M. Arginine methylation of STAT1 modulates IFNα/β-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- Mowen K.A., Schurter B.T., Fathman J.W., David M., Glimcher L.H. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol. Cell. 2004;15:559–571. doi: 10.1016/j.molcel.2004.06.042. [DOI] [PubMed] [Google Scholar]
- Nien W.L., Dauphinee S.M., Moffat L.D., Too C.K. Overexpression of the mTOR α4 phosphoprotein activates protein phosphatase 2A and increases Stat1α binding to PIAS1. Mol. Cell. Endocrinol. 2007;263:10–17. doi: 10.1016/j.mce.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Pal S., Sif S. Interplay between chromatin remodelers and protein arginine methyltransferases. J. Cell. Physiol. 2007;213:306–315. doi: 10.1002/jcp.21180. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana C.V., Gil M.P., Schreiber R.D., Stark G.R. Stat1-dependent and -independent pathways in IFN-γ-dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- Rogers R.S., Horvath C.M., Matunis M.J. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J. Biol. Chem. 2003;278:30091–30097. doi: 10.1074/jbc.M301344200. [DOI] [PubMed] [Google Scholar]
- Sapetschnig A., Rischitor G., Braun H., Doll A., Schergaut M., Melchior F., Suske G. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 2002;21:5206–5215. doi: 10.1093/emboj/cdf510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C., Levy D.E., Decker T. JAK–STAT signaling: From interferons to cytokines. J. Biol. Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- Scott H.S., Antonarakis S.E., Lalioti M.D., Rossier C., Silver P.A., Henry M.F. Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2) Genomics. 1998;48:330–340. doi: 10.1006/geno.1997.5190. [DOI] [PubMed] [Google Scholar]
- Shuai K., Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- Smith J.J., Rucknagel K.P., Schierhorn A., Tang J., Nemeth A., Linder M., Herschman H.R., Wahle E. Unusual sites of arginine methylation in Poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J. Biol. Chem. 1999;274:13229–13234. doi: 10.1074/jbc.274.19.13229. [DOI] [PubMed] [Google Scholar]
- Song L., Bhattacharya S., Yunus A.A., Lima C.D., Schindler C. Stat1 and SUMO modification. Blood. 2006;108:3237–3244. doi: 10.1182/blood-2006-04-020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Gary J.D., Clarke S., Herschman H.R. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J. Biol. Chem. 1998;273:16935–16945. doi: 10.1074/jbc.273.27.16935. [DOI] [PubMed] [Google Scholar]
- Ungureanu D., Vanhatupa S., Kotaja N., Yang J., Aittomaki S., Janne O.A., Palvimo J.J., Silvennoinen O. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood. 2003;102:3311–3313. doi: 10.1182/blood-2002-12-3816. [DOI] [PubMed] [Google Scholar]
- Ungureanu D., Vanhatupa S., Gronholm J., Palvimo J.J., Silvennoinen O. SUMO-1 conjugation selectively modulates STAT1-mediated gene responses. Blood. 2005;106:224–226. doi: 10.1182/blood-2004-11-4514. [DOI] [PubMed] [Google Scholar]
- Wagner S., Weber S., Kleinschmidt M.A., Nagata K., Bauer U.M. SET-mediated promoter hypoacetylation is a prerequisite for coactivation of the estrogen-responsive pS2 gene by PRMT1. J. Biol. Chem. 2006;281:27242–27250. doi: 10.1074/jbc.M605172200. [DOI] [PubMed] [Google Scholar]
- Wang H., Huang Z.Q., Xia L., Feng Q., Erdjument-Bromage H., Strahl B.D., Briggs S.D., Allis C.D., Wong J., Tempst P,., et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Yu M.C., Lamming D.W., Eskin J.A., Sinclair D.A., Silver P.A. The role of protein arginine methylation in the formation of silent chromatin. Genes & Dev. 2006;20:3249–3254. doi: 10.1101/gad.1495206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Mustelin T., David M. Arginine methylation of STAT1 regulates its dephosphorylation by T cell protein tyrosine phosphatase. J. Biol. Chem. 2002;277:35787–35790. doi: 10.1074/jbc.C200346200. [DOI] [PubMed] [Google Scholar]