Abstract
Rad51 is the central catalyst of homologous recombination in eukaryotes and is thus critical for maintaining genomic integrity. Recent crystal structures of filaments formed by Rad51 and the closely related archeal RadA and eubacterial RecA proteins place the ATPase site at the protomeric interface. To test the relevance of this feature, we mutated conserved residues at this interface and examined their effects on key activities of Rad51: ssDNA-stimulated ATP hydrolysis, DNA binding, polymerization on DNA substrates and catalysis of strand-exchange reactions. Our results show that the interface seen in the crystal structures is very important for nucleoprotein filament formation. H352 and R357 of yeast Rad51 are essential for assembling the catalytically competent form of the enzyme on DNA substrates and coordinating its activities. However, contrary to some previous suggestions, neither of these residues is critical for ATP hydrolysis.
INTRODUCTION
Rad51 is central to the homologous recombination pathway for the accurate repair of DNA double-strand breaks. The main function of Rad51 is to align a broken DNA end with a homologous sequence on an intact sister chromatid or homologous chromosome. It does so by forming a filament that coats a single strand (formed by other enzymes that process the broken ends), then searching for homologous sequences in duplex DNA and catalyzing the exchange of the matching strands (1).
Yeast and human Rad51 proteins have been characterized extensively. In vitro they bind single- and double-stranded DNA in an ATP-dependent manner (2,3). In the presence of ATP or nonhydrolyzable ATP analogs they can catalyze strand-exchange reactions between homologous DNA substrates (4–6). DNA binding as well as high salt concentrations stimulate the ATPase activity of Rad51 (7,8). These properties parallel those of eubacterial RecA, which performs a similar biological role and shares a conserved core domain with Rad51 (9,10). However, some dissimilarities between the biochemical properties of RecA and Rad51 have been described: they include differences in the ATP hydrolysis rates, in the apparent cooperativity of binding to DNA, and, under certain conditions, the polarity of the strand-exchange reaction (4,11–13).
Electron microscopy studies established that the filaments formed by RecA and Rad51 on DNA are right-handed, helical structures with ∼6 protein protomers per turn and that the DNA is bound near the filaments axis (14–16). The pitch of the filaments is remarkably heterogeneous. One factor influencing this heterogeneity is the nucleotide cofactor: filaments formed with nonhydrolyzable ATP analogs (and thus most likely to represent the active state for homology searching and strand exchange) appear more elongated with a helical pitch ranging between 90 and 130 Å, while others formed with ADP or without a nucleotide cofactor are more compressed with a pitch of 60–80 Å. Thus, the switch between the two forms is likely modulated in response to ATP binding and/or hydrolysis, but the details of this allosteric mechanism are not currently understood.
Notably, Rad51 and its homologs from other kingdoms share a structurally conserved ATPase core domain with many other oligomeric ATPases, including many helicases and the F1 ATPase (17). In many of these systems, residues from one protomer contact the ATP bound to its neighbor. Some such residues, usually arginines, actively participate in ATP hydrolysis and are often called ‘arginine fingers’ (17,18). Others act as ‘sensors’ that couple nucleotide binding to changes in the relative orientation of domains or entire protomers. Recent crystal structures of homologous recombinases identified conserved residues that appeared likely to play similar roles.
Several crystal structures of RecA, Rad51 and their archeal homolog RadA have been determined (19–24). Their overall architecture is conserved, with a common core domain, containing both the ATPase and DNA-binding activities, preceded by a polymerization motif. The latter is a single β strand that adds onto the central β sheet of another protomer's core domain. This ‘trans’ β strand is followed by a flexible linker, giving significant structural flexibility to the overall polymer. Rad51 and RadA have an additional helix–hairpin–helix containing N-terminal domain not found in RecA that may serve as a transient docking spot for duplex DNA (25). Instead, RecA has a small C-terminal domain that may interact with duplex DNA but differs in structure and may also be regulatory (26).
Most RecA crystal structures were determined in the absence of DNA, and regardless of the nucleotide cofactor bound, correspond to the inactive or compressed form, with pitches between 72 and 83 Å (19,20,27–30). However, the structure of a RecA filament bound to DNA and a nonhydrolyzable ATP analog has recently been reported, with an extended pitch of 94 Å (24). The major difference between the low- and high-pitch filaments is the relative orientation of the ATPase domains. In the compressed RecA forms, the ATPase site is largely exposed to solvent. In contrast, the ATPase site is buried within the interface between adjacent core domains (Figure 1) in the extended form. This interface agrees well with previous biochemical and EM data, and places a series of conserved residues from one protomer in close proximity to (or in direct contact with) the ATP bound to its neighbor (31–34).
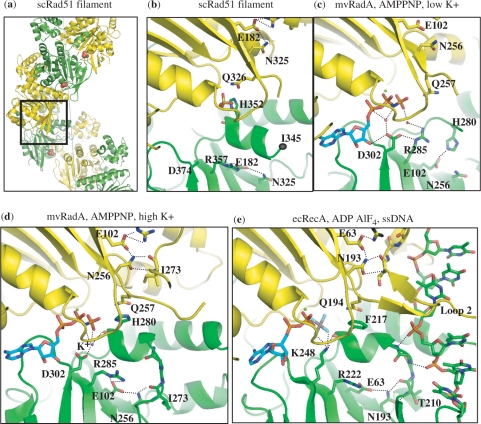
Figure 1.
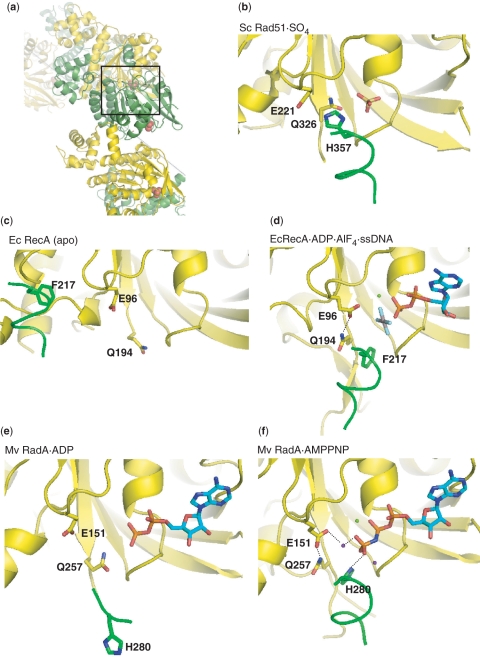
A network of interactions conserved across three kingdoms. (a) Crystal structure of the scRad51 filament (PDBid 1szp) (21); alternate protomers are colored yellow and green. The ATPase site is at the interface and here contains a sulfate ion in the position where the β phosphate of ATP would bind (shown in space-filling mode). The dotted gray line denotes the filament axis. (b) Enlarged and cut-away view of the boxed portion of part a. The phosphate-binding ‘P-loop’ of one protomer abuts the H352-bearing helix of the neighboring subunit. This loop and the helix following it are oriented similarly in (b–d). DNA-binding loop 2, which is disordered, ends near residue 345. The sulfate ion, and several highly conserved residues discussed in the text are shown. The side chains of T345 and D374 were disordered in this structure. Some of the hydrogen-bonding network is shown as dotted lines, although the resolution of this structure renders some of the details uncertain. (c) The interface seen in the mvRadA structure in the presence of AMPPNP (cyan carbons) but without K+ (PDBid 1t4g) (23). The helix following Loop 2 is unfolded, and the conserved R makes a water-mediated contact (red spheres) to the ATP analog (23). (d) mvRadA in the presence of AMPPNP (cyan) and high [K+] (purple dot) (PDBid 2fpm)(36). Loop 2 is visible, albeit rather poorly ordered. The conserved H contacts the 3rd phosphate group of the ATP analog. A K+ ion also binds this phosphate, as well as D302 and the carbonyl of H280. Although high K+ is required for mvRadA activity, the conservation of this aspartate implies that similar ion-mediated interactions are probably important in eukarytoic Rad51 as well. (E) EcRecA in the presence of ADP AlF4 (cyan and CPK coloring) and ssDNA (PDBid 3cmw) (24). Figures 1 and 7 were made with PYMOL (DeLano Scientific LLC).
There are still no structures available for the eukaryotic and archeal proteins with ordered DNA present. However, scRad51 and mvRadA formed filaments in the crystal with extended pitches, from 104–130 Å, regardless of the nucleotide cofactor (21,23). Although the extended forms vary in detail, like in the case of RecA-DNA filament, the ATPase site is placed between the protomers and conserved residues at this interface contact the nucleotides cofactors; such residues are likely candidates for the roles of sensors or fingers similar to other ATPases.
Here we examine how mutation of some of the conserved interface residues in scRad51 affects the formation of nucleoprotein filaments, in vitro strand-exchange reactions and DNA-stimulated ATPase activity. We focused on H352 and R357, which are situated across from the P-loop in the extended filament structures (Figure 1). These residues participate in a highly conserved network of interactions that connects the two adjacent protomers, the ATPase site, and the DNA. H352 is conserved in most Rad51 and RadA proteins, but replaced by F in eubacterial RecAs, whereas R357 is strictly conserved across family members in all three kingdoms. We also examined the behavior of scRad51 I345T, originally identified in a screen for gain-of-function mutations that bypass the in vivo requirement for the accessory factors Rad55 and Rad57 (35). This mutant was shown to bind DNA more avidly in the presence of ATP. I345 is located at the C-terminal end of loop 2, and its backbone also participates in the network shown in Figure 1. It is conserved as I in most eukaryotic and archeal family members but as T in RecAs.
Some structures have implied that H352 and R357 could play a direct role in catalyzing ATP hydrolysis, while others imply that they may be conserved for other reasons. In some structures of archaeal mvRadA with AMPPNP bound, H280, the equilvalent of H352, extends from one protomer into the neighbor's ATPase site, directly contacting the 3rd phosphate group (36). However, this region appears quite flexible. In the scRad51 filament structure, H352 also interacts the neighboring protomer's ATPase site, but the contact details differ from those seen in mvRadA and between alternating protomer–protomer interfaces within the same scRad51 crystal (21). In other structures, the helix bearing this histidine is unfolded: those of mvRadA filaments bound to ADP or in the presence of low [K+] and AMPPNP, and the core domain of human rad51 (which lacked the polymerization motif and thus did not form a filament) (22,37,38). In the RecA case, the equivalent residue is a phenylalanine, and thus more likely to be involved in interprotomer communication than catalysis. In the presence of an ATP analog and DNA, but not in other structures, this residue also interacts with the neighboring protomer's ATPase site (24). Although most changes to this F destroy RecA function, one mutation, F217Y, greatly increases ATP-mediated cooperativity (33).
In most structures, R357 (or its equivalent) forms an ion pair with another strictly conserved residue, E182, and packs against the helix that contains H352. However, in the mvRadA structures where that helix is unfolded, the arginine comes into close contact with the neighboring protomer's nucleotide cofactor, raising the possibility that it could act as a more traditional arginine finger.
We find that this interface is indeed functionally important in yeast Rad51. Several substitutions of H352 and R357 in scRad51 strongly affect the ability of the protein to form filaments on single and double-stranded DNA and impair the catalysis of strand-exchange reactions in vitro. We also find that R357M and I345T mutants show enhanced ATP hydrolysis in the absence of ssDNA, and altered responses of ssDNA binding to nucleotide cofactor. However, H352 and R357 are not critical catalytic components of the ATPase site. Our data indicate that they are more analogous to the sensors of other related ATPases than to arginine fingers.
MATERIALS AND METHODS
Protein expression and purification
ScRad51 was purified as described previously, with minor modification (21). Briefly, Rad51 was expressed in the Rosetta (DE3) pLysS cell line (Novagen) using a PET3 vector (21). After centrifugation, (NH4)2SO4 was stirred slowly into the clarified lysate (0.24 g ml–1) and the solution was centrifuged. The pellet (containing Rad51), was re-dissolved and dialyzed into buffer consisting of 20 mM Tris–HCl, pH 7.5, 0.5 mM EDTA, 5% (v/v) glycerol, 50 mM NaCl, 1 M urea, 1 mM DTT. Using an AKTA FPLC (Pharmacia) the protein prep was subjected to the following columns: Q-Sepharose, Hi-Trap Blue, Heparin and Mono-Q (all from GE healthcare). The purified Rad51 protein was dialyzed into 20 mM Tris–HCl, pH 7.5, 30% (v/v) glycerol, 0.5 mM EDTA, 50 mM NaCl, 1 mM DTT and concentrated. All purification proceeded at 4°C. All mutant proteins were expressed and purified similarly to WT scRad51; the purity was estimated to be at least 95% for each of the proteins used in this study.
Site-directed mutagenesis
Site-directed mutations were introduced using the Stratagene Quikchange Kit and verified by sequencing.
DNA oligonucleotides
DNA oligonucleotides were purchased from IDT and PAGE-purified. The following oligonucleotides were used:
#1: 5′-ACAGCACCAGATTCAGCAATTAAGCTCTAAGCCATCCGCAAAAATGACCTCAAAACAAAAGGA
#2: 5′-TCCTTTTGTTTTGAGGTCATTTTTGCGGATGG
#3: 5′-ACAGCACCA GATTCAGCAATTAAGCTCTAAG
#4: 5′-CTTAGAGCTTAATTGCTGAATCTGGTGCTGT
#5: 5′-TCCTTTTGTTTTGAGGTCATTTTTGCGGATGGCTTAGAGCTTAATTGCTGAATCTGGTGCTGT
Strand-exchange assay
Our assay was based on that of Mazin et al. (5). Duplex DNA with a 31-nt 5′ single-stranded tail was obtained by annealing oligos #1 and #2. Rad51-DNA filaments were pre-formed on this substrate by incubating 7 μM Rad51 protein with 21 μM (nt) DNA at 30°C in a reaction containing 30 mM HEPES, pH 7.0, 20 mM KCl, 1 mM DTT, 6.2 mM ATP and 2 mM Mg(OAc)2; after 5 min the [Mg(OAc)2] was increased to 18 mM and incubation was continued for another 15 min. The strand-exchange reaction was started by adding a 31-mer dsDNA (oligos # 3 and 4) substrate in which the strand homologous to the ssDNA tail was 32P labeled. Aliquots were removed at 0, 15, 45 and 60 min; to stop the strand-exchange reaction EDTA (50 mM), SDS (1%) and proteinase K (500 mg/ml) were added to each. Five-microliter aliquots were loaded onto a 9% polyacrylamide gel and electrophoresed for 2 h at 120 V. Bands were visualized with a Molecular Dynamics Phosphor Imager, and their intensities were measured using the Image Quant software (Molecular Dynamics). The background reaction level was determined from control reactions using Rad51 storage buffer in place of protein. Each point in Figure 3c is the average of three independent experiments.
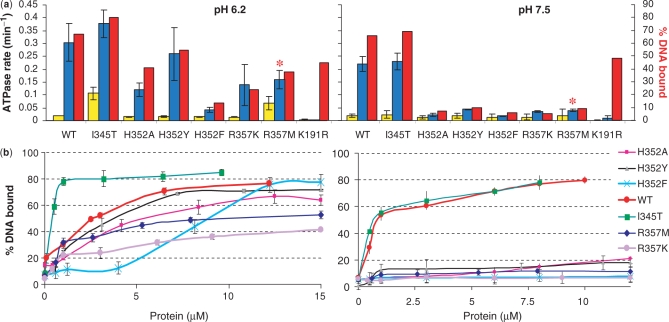
Figure 3.
Correlation of ATPase with ssDNA-binding activities. (a) ATPase activity of WT scRad51 and mutant forms in the absence (yellow bars) and presence (blue bars) of ssDNA, and %DNA bound under the same conditions (red bars; extrapolated for 5 μM Rad51 from the plots in part B).Five micromolar Rad51 and 15 μM (in nt.) of an ss63-mer, 5 mM ATP and 15 mM Mg++ were used. For the R357M mutant the rates in the presence of DNA (*) represent initial rates (see text for more details). (b) Binding to ssDNA in the presence of 5 mM ATP and 15 mM Mg++; titrations of 63-mer ssDNA oligos with Rad51 proteins. Fifteen-micromolar 63-mer ssDNA (nucleotides) was incubated with Rad51 proteins under conditions used in the ATPase and passed through alkali-treated nitrocellulose and DEAE filters. The percentage bound represents the amount of DNA retained on nitrocellulose compared with the total DNA. Results shown are an average of values obtained by quantitation of three different filter pairs, to which drops from the same sample were applied.
DNA-binding assay
A double filter binding assay was used (35,39). Nitrocellulose filters (0.45 μm HAWP, Millipore) and DEAE membranes (Whatman) were prepared as described (39). Binding reactions were carried out in 20 mM MES pH6.2, or 20 mM Tris–acetate, pH7.5, 10 mM Mg(OAc)2, 1 mM DTT, 7.5 μg/ml BSA. 0.6 μM (nt or bp) 32P-labeled 63-mer DNA (either oligo #1 or the duplex of #1 and #5) was titrated with Rad51. For comparison to the ATPase assays (i.e. the experiments in Figure 3), the conditions were 50 mM MES pH 6.2 or 30 mM Tris–acetate, pH 7.5, 15 mM MgAc, 15 μM (nt) ssDNA. All reactions were equilibrated at 30°C.
After incubation, samples were filtered through a vacuum manifold apparatus with a nitrocellulose filter on top a DEAE one, supported by Whatman 5 qualitative filter paper. Five micro-liter drops from the reaction mixtures were applied on the filters; for each reaction triplicate aliquots were filtered. The filters were washed with buffer and dried on paper. Binding of the 32P-labeled DNA to the filters was quantified as in the strand-exchange assays. The binding efficiency was calculated as the ratio of signal on the nitrocellulose filter to the sum of the signal on both (nitrocellulose and DEAE) filters.
ATPase assay
ATP hydrolysis rates were measured in a coupled ATP- regenerating and NADH-oxidation system (40–42). Reaction contained 30 mM Tris–acetate pH 7.5 or 50 mM MES pH 6.2, 50 mM KCl, 5 mM ATP, 15 mM MgAc2,10 U/ml pyruvate kinase, 10 U/ml lactate dehydrogenase, 0.3 mM phosphoenolpyruvate, 50 μg/ml BSA, 1 mM DTT, 250 μM NADH and 5 μM Rad51. After 15 min incubation at 30°C, each reaction was divided into two 118 μl aliquots. Two microliters of ssDNA 63-mer (oligo #1) was added to one such that the final DNA: protein ratio was 3-nt/protomer; these samples were equilibrated for an additional 30 min at 30°C to allow the formation of protein–DNA filaments. Two microliters of water was added to the other aliquot, followed by a 5 equilibration. After equilibration, the decrease in absorbance at 340 nm (resulting from NADH oxidation) was monitored for at least 20 min at 30°C using the kinetics mode of a Beckman Coulter DU640B spectrophotometer. For each protein variant control experiments were performed with longer timescales (1 h) in order to check if the rates remained stable during this time interval. Each result reported in Figure 3a is the average of at least three independent experiments.
The ATPase rate was calculated from the following equation:
where Kpath is the molar absorption coefficient for NADH for a given optical path length (42). The rates were corrected for background NADH decomposition of controls containing no ATPase.
SFM
The dsDNA used in the SFM experiments was made by linearizing plasmid pDERI1 with ScaI (43). The resulting 1821-bp blunt-end linear double-stranded DNA was deproteinized by phenol:chloroform:iso-amyl alcohol (25:24:1) extraction, precipitated with ethanol and dissolved in H2O.
Rad51 nucleoprotein filaments were formed in a 10 μl reaction volume containing 25 mM HEPES-KOH (pH7.5), 10 mM MgCl2, 2 mM cofactor (ATP or AMPPNP), 7.5 μM DNA (concentration in nucleotides), 2.5 μM Rad51 and 30 mM KCl. Reactions were carried out at 37°C for 30 min and then placed on ice. For imaging, reaction mixtures were diluted 15-fold in deposition buffer (10 mM HEPES-KOH pH 7.5, 10 mM MgCl2) and deposited on freshly cleaved mica. After 20 s, the mica was washed with water (glass distilled; SIGMA) and dried with a stream of filtered air. Images were obtained on a NanoScope IIIa and a NanoScope IV (Veeco Instruments) with a type E scanner, operating in tapping mode in air. Silicon tips (Nanoprobes) were obtained from NanoWorld.
The filament lengths were measured by manual tracing from NanoScope images imported into IMAGE SXM 1.62 (NIH IMAGE version modified by Steve Barrett, Surface Science Research Centre, University of Liverpool, Liverpool, UK).
RESULTS
Mutation of H352 or R357 strongly impairs strand-exchange activity
WT and mutant scRad51 were tested for their ability to catalyze a DNA strand-exchange reaction. In addition to H352A, H352Y, H352F, R357M and R357K we also tested two control mutants, I345T and K191R. The I345T scRad51 functions as a positive control in our assays, since it is fully active in vitro in binding ssDNA, hydrolyzing ATP and catalyzing strand-exchange reactions (21,35). K191R, in the P-loop, has been shown in both Rad51 (44–46) and RecA (47,48) to drastically reduce ATP hydrolysis without destroying binding. These studies have also shown that K to R mutants are partially active in catalyzing strand-exchange in vitro and, for scRad51, in vivo (49).
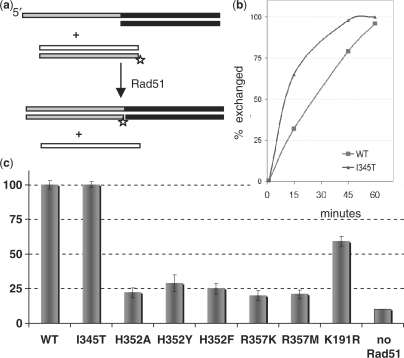
We assayed strand exchange between oligonucleotide substrates (5) (Figure 2). Filaments were pre-formed on tailed substrates and the reaction started by adding 32P-labeled homologous dsDNA. The reactants and the products were differentiated by gel electrophoresis. All the mutations at H352 and R357 severely impaired strand exchange, as shown in Figure 2; the most active, H352Y, exchanged only 30% of the substrate in 60 min, whereas the WT and I345T proteins were able to complete the strand exchange-reaction in 60 min (I345T was slightly faster than WT). For the P-loop mutant K191R a moderate decrease in reaction efficiency was observed (strand exchange was ∼50% complete after 60 min), in accordance with previous in vitro analysis (44). Thus, even relatively conservative substitutions of the interface residues H352 and R357 severely impact the strand-exchange activity of Rad51.
Figure 2.
H352 and R357 mutants are deficient in strand exchange. (a) In vitro strand-exchange assay. Rad51-DNA filaments are pre-formed on tailed duplex substrates, after which a 32P-labeled 31-mer dsDNA substrate homologous to the ssDNA tail is added. The progress of the strand-exchange reaction is followed by monitoring the size of the labeled fragment on a nondenaturing gel. (b) The kinetics of the strand-exchange reactions promoted by WT Rad51 protein and mutant I345T. (c) Percent of the label found in the strand-exchange product after 60min for WT and mutant Rad51 proteins. For each protein variant the value shown in the bar graph represents the average of three separate experiments.
ATP hydrolysis is impaired but not abrogated by mutation of H352 or R357
ATPase activity was measured in an NADH coupled assay (41,42). To determine the stimulation of ATP hydrolysis by DNA, rates were measured in the absence and in the presence of an ssDNA oligo (oligo #1). Control experiments determined that under identical solution conditions this 63 mer stimulated the ATPase activity of WT Rad51 similarly to longer ssDNA substrates such as poly(dT)96 or PhiX174 (data not shown). As the formation and stability of Rad51 filaments on oligonucleotides can be influenced by pH (50), we performed both the ATPase and the DNA-binding assays at two pH values, 6.2 and 7.5. In the absence of DNA, WT Rad51 hydrolyzed ATP very slowly (∼0.018 min–1). After ssDNA 63 mer was added and the protein–DNA filaments allowed to form, the hydrolysis rates increased 7- and 15-fold at pH 7.5 and pH 6.2, respectively.
As expected, I345T showed ATPase activity comparable to or slightly higher than the WT protein (Figure 3a) (35). The ATPase rate of this variant was also elevated in the absence of DNA. On the timescale of our assay, K191R was inactive: under no conditions were its ATP hydrolysis rates distinguishable from controls with no Rad51 added. This confirms that K191 is a key catalytic residue and shows that our protein preparations are not contaminated with other ATPases.
The mutations at H352 and R357 did not abolish ATP hydrolysis activity, or its stimulation by ssDNA. However, in all cases, particularly at pH 7.5, the rate of ssDNA-stimulated ATP hydrolysis was significantly lower than for WT protein. The sharp contrast between the behavior of the H352/R357 mutants and the K191R mutant argues that, unlike K191, H352 and R357 are not essential for ATP hydrolysis. Nevertheless, the impact of point substitutions of these highly conserved residues on the ssDNA stimulated ATPase activity is quite large, and was even larger in initial assays carried out at lower protein and DNA concentrations. We hypothesized that the decreased levels of stimulation are related to the deficiency of these mutants in forming filaments on ssDNA. Our DNA-binding studies described below support this interpretation (Figure 3a and b).
One of the variants, R357M, is unique in that upon DNA addition its hydrolytic activity is initially slightly stimulated, but subsequently becomes 0 (Figure S1). Under our reaction conditions the initial stimulation lasts for about 30 min and the calculated amount of ATP hydrolyzed during this time interval is substoichiometric with respect to the protein. We do not understand the molecular origins of this effect but suspect that it might be related to our finding (discussed below) that ATP inhibits ssDNA binding by R357M. It is not simply the result of exonuclease contamination, because control experiments showed no degradation of a 32P-labeled 48-nt ssDNA after a 5-h incubation in the presence of 28 µM Rad51 and 10 mM Mg++ (data not shown).
ssDNA binding correlates with ssDNA-stimulated ATPase activity
To examine if any correlation exists between the ssDNA stimulated ATPase activity and the efficiency of ssDNA binding, we performed DNA-binding assays under conditions virtually identical to those used in the ATPase assays. Binding efficiency was determined by a filter binding assay (35,39).
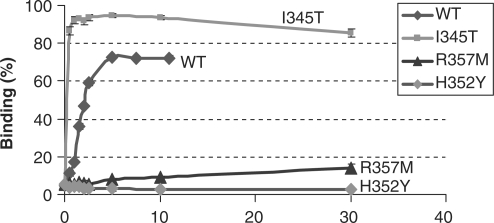
Figure 3b shows titrations of a 32P-labeled 63-mer ssDNA with protein in the presence of the same concentrations of ATP and Mg2+ as those used in the ATPase assays. All of the mutants at H352 and R357 bound ssDNA poorly at pH 7.5, although some (H352Y, H352A) bound reasonably well at pH 6.2. Rad51-I345T bound ssDNA with higher affinity than WT [as previously reported (35)]; this was especially striking at low pH, where ∼9-fold less of the I345T mutant was required to achieve half maximal binding, and we were probably in a stoichiometric binding regime.
For each protein variant, we interpolated the DNA-binding curves at the protein concentration where the ATPase assays were performed and plotted the DNA-binding efficiency (Figure 3a). This highlights the correlation between the efficiency of ssDNA binding and the stimulation of ATP hydrolysis by ssDNA. The notable exception is P-loop mutant K191R which, although it binds DNA efficiently, cannot detectably hydrolyze ATP. By comparison point mutations in position R352 and R357 show measurable rates of hydrolysis that generally reflect their deficiency in ssDNA binding.
I345T and R357M substitutions alter the coupling between binding of ssDNA and nucleotide cofactor
Two variants, I345T and R357M, hydrolyzed ATP faster than the WT protein in the absence of DNA at pH 6.2. The relative stimulation of ATPase activity by ssDNA was lower for these mutants than for the WT. This was especially striking for R357M which showed only a 2-fold stimulation and—as previously mentioned—even this effect is short-lived.
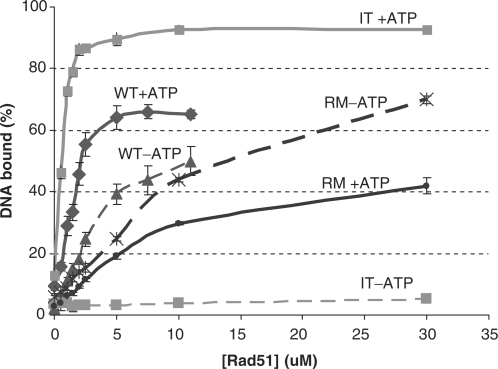
Further experiments showed that for both I345T and R357M the normal coupling between DNA and ATP binding is altered. When ATP-dependent and ATP-independent ssDNA binding are directly compared (Figure 4), WT Rad51 binds ssDNA in the absence of ATP, although not as well as in the presence, as expected (2). However, binding of R357M to DNA appears to be inhibited by the presence of ATP while—surprisingly—for I345T no DNA binding in the absence of ATP could be detected.
Figure 4.
I345T and R357M show altered ATP–DNA coupling. ssDNA binding by Rad51 WT, I345T and R357M in the absence (dashed lines) and presence (solid lines) of ATP. Double filter binding assay as in Figure 3b, with 0.6 μM (in nt) 63-mer ssDNA and with or without 5 mM ATP, 10 mM MgOAc at pH 6.2.
SFM shows impaired filament formation by H352A and R357M
The effects of these mutations on filament formation were visualized by scanning force microscopy analysis of unfixed samples. This technique allows visualization of irregular structures, implying possible dynamic rearrangements (51).
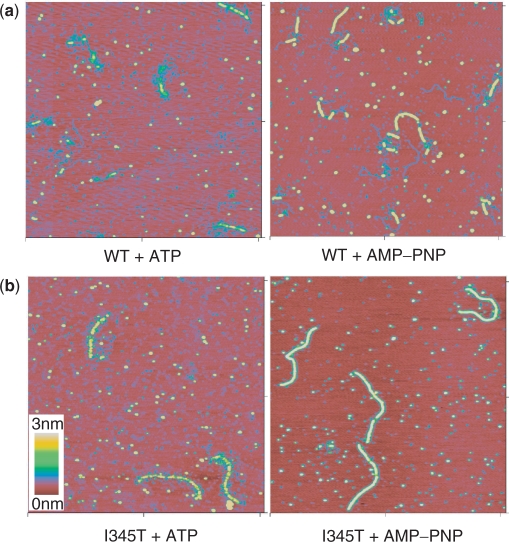
None of the proteins tested showed significant filament formation on dsDNA in the absence of nucleotide cofactor. WT scRad51 formed irregular filaments on dsDNA in the presence of ATP and Mg2+ (Figure 5a). At a protein : DNA ratio of 1 protein to 3 nt, scRad51 only partially covered the DNA substrate. It formed more regular filaments on dsDNA in the presence of the nonhydrolyzable ATP analog AMP-PNP and Mg2+. Here also, the DNA was only partially covered: each DNA molecule had multiple separate filament segments of different lengths. (Because of the variable amount of DNA covered the contour length of these partial filaments was not analyzed.)
Figure 5.
DNA WT and I345T form filaments on dsDNA. SFM images of complexes formed by WT (a) and I345T (b) scRad51 on double-stranded DNA in the presence of ATP or AMPPNP. All images are 2 × 2 μM and height is represented by color as shown by color bar. See Figure S2 for similar images with H352A and R357M.
I345T behaved similarly to WT with respect to filament structure. On dsDNA irregular filaments were formed in the presence of ATP and regular filaments were formed in the presence of AMP-PNP (Figure 5b). However, in contrast to WT, I345T covered the whole DNA substrate at the same protein:DNA ratio. That suggests enhanced and/or more cooperative dsDNA binding by the I345T protein, which correlates with the filter binding assays of both ss- and dsDNA binding (Figures 3 and 6).
Figure 6.
dsDNA binding. Filter binding assays showing the relative binding efficiency of WT scRad51 and I345T, R357M and H352A variants to double-stranded 63-mer DNAs in the presence of ATP at pH 7.5.
To estimate DNA extension in the I345T filaments, their contour length was measured and compared to that of bare DNA. The average length of the nucleoprotein filaments protein formed by I345T with AMPPNP was 0.754 μm (±0.006, n = 35), while that of bare DNA was 0.530 μm (±0.001, n = 120). This DNA extension of 42% is somewhat less than the 50% extension of DNA within hRad51 filaments determined previously in the same way, perhaps due to small unbound DNA segments that were not resolved in these images (51).
The H352A and R357M variants did not form filaments on dsDNA in any conditions tested (±ATP or AMPPNP; Figure S2). Both proteins formed ill-defined complexes that only occasionally bound double-stranded DNA. Filter binding assays also showed very poor dsDNA binding by these proteins, even in the presence of ATP (Figure 6). Poor binding to the duplex product provides an additional explanation for their poor strand-exchange activity.
DISCUSSION
We found that residues H352 and R357, at the interprotomer interface, are critically important for Rad51 function. Substitutions in these positions significantly impaired the ability of Rad51 to catalyze in vitro strand-exchange reactions. Our DNA binding, ATPase and SFM data indicate that the H352 and R357 mutants were deficient in their ability to undergo ATP-dependent polymerization on DNA—a critical step for assembling the catalytically competent form of the enzyme. In the absence of DNA, Rad51 mutants H352A/Y/F and R357M/K hydrolyze ATP similarly or—in some cases—slightly better than the WT protein. This result indicates that (i) the observed functional deficiency of these variants is not caused by misfolding or nonspecific aggregation and (ii) neither H352 nor R357 is critical for hydrolysis per se. Under conditions where their DNA-binding handicap is at least partially overcome (e.g. high concentrations), the mutant proteins can, indeed, hydrolyze ATP in a DNA-dependent manner. Notably, ATP binding and not hydrolysis is critical for function of homologous recombinases (47–49). Like some other AAA+ superfamily members, Rad51 may use ATP to control a conformational switch, becoming activated by cofactor binding and resetting upon hydrolysis and ADP + Pi release (17). Although our data show that H352 and R357 are not analogous to the trans-acting catalytic arginine fingers often found in NTPases, they are good candidates for trans-acting sensors of ATP presence. Recent work suggests that D374 of scRad51, conserved as K in RecAs and D in Rad51/RadA proteins, may aid catalysis in trans: a cation bound by the equivalent D302 of RadA interacts with the ATP's 3rd phosphate, as does the lysine of RecA (Figure 1d and e) (24,52,53). However, this residue's main role may also be in sensing the presence of ATP (and thus properly organizing the interface) rather than in directly catalyzing its hydrolysis.
The similar behavior of the H352 and R357 mutants indicates that they probably disrupt the same cooperative pathway of interactions required for the ATP-dependent assembly of active Rad51-DNA filaments. H352 is ideally located to promote communication between filament subunits: it protrudes from an α helix and can dock into the neighboring protomer's ATPase site. The two structures most likely to reflect the active state for homology searching and ATPase activity are those of RecA with ADP AlF4, and mvRadA with AMPPNP and high K+ (which activates this archaeal enzyme's activity). In both these cases, this conserved H/F stacks against the side chain of an invariant glutamine (326 of scRad51; 194 of RecA). This glutamine, which has long been proposed to be involved in ATP–DNA crosstalk (30,54,55), appears to position an invariant glutamate, which in turn positions the attacking water molecule in the ATPase site (Figure 7). The helix bearing this conserved H/F structurally links the 3rd phosphate of the ATP analog to the DNA-binding end of loop 2. At the N-terminus of the helix, the DNA's phosphate backbone contacts the backbone NH groups of two highly conserved glycines, while at the C-terminal end, the carbonyl of the H/F interacts with a positive charge that bridges it to the cofactor's 3rd phosphate: the tip of K248 in RecA and a K+ ion bound to D302 in RadA. These highly conserved charged interactions may also stabilize the helix by interacting with its dipole moment. Since this helix is unfolded and/or poorly ordered in several archaeal structures that do not include all the cofactors and/or form high-pitch right-handed filaments (37,38,56,57), its dynamics may play an important regulatory role. R357 lies on a β strand that packs against the H352-bearing helix. In the mvRadA filament in the presence of ADP or AMPPNP but the absence of the high K+ required to activate this particular enzyme, the unfolding of this helix allows R357, normally occluded by the helix, to make a water-mediated contact with ATP bound to the neighboring protomer (22,23). The conformational metastability of this protein segment may be important in propagating changes associated with ligand and cofactor binding.
Figure 7.
Coupling of the intersubunit interface to the ATPase site. In each panel, only one helix from the green protomer is shown, with the backbone drawn as a thin tube, for clarity. In the active forms, docking of the F/H that extends from this helix into the neighbor's ATPase site appears to orient the conserved glutamine, which in turn orients the glutamate that is proposed to activate an attacking water molecule for ATP hydrolysis. Green spheres are Mg++, purple, K+. Only the most relevant hydrogen bonds are drawn. (a) Overall view of the yeast Rad51 filament, from the same view as (b–e). The region that is expanded (with most of the green protomer cut away) in part b is boxed. In each panel, the yellow protomer's P-loop and the helix following it are similarly oriented. (B)Yeast Rad51 with sulfate (PDBid 1szp) (21). (c) E. coli RecA, in the absence of substrates (PDBid 2reb) (19). (d) E. coli RecA with ADP AlF4 and ssDNA (PDBid 3cmw) (24). (e) Methanococcus voltae RadA with ADP (PDBid 2fpk) (22). (f) M. voltae RadA with AMPPNP (PDBid 2fpm) (22).
The importance of this segment following loop 2 has also been noted in prokaryotic RecA proteins, where it was termed a ‘RecA signature sequence’ (58). Mutagenesis studies suggested that F217 and R222 of Escherichia coli RecA (analogous to scRad51 H352 and R357, respectively) are involved in propagating ATP-mediated allosteric transitions between protein subunits in the filament (31–33). Our data suggest that the Rad51 and RadA proteins posses a similar ‘signature motif’ in this region with a similar role in filament formation and activation.
Some of our results with the I345T scRad51 variant were quite unexpected. We initially employed I345T as a positive control, since it had been previously characterized in vivo and in vitro. Our data agreed that it has increased affinity for DNA and elevated ATPase rates (35). In addition, the SFM images showed that it coats dsDNA substrates better than the WT protein. Since the I345T substitution is in DNA binding loop 2, a simple explanation for these results would be that it increases Rad51's affinity for all DNA. However, this hypothesis cannot explain the fact that at pH 6.2 the I345T mutant displays an increased ATP hydrolysis rate in the absence of DNA as well as an absolute requirement for the ATP cofactor in DNA binding—two new results reported here. I345T's absolute requirement for ATP in DNA binding at low pH was quite unexpected. WT Rad51 is known to bind ss- and dsDNA in the absence of ATP, although the ATP-independent binding mode is unlikely to be physiologically relevant (2). In vitro, however, the two binding modes are not interconvertible and indirect evidence suggests that they are mechanistically different. I345T should be useful for addressing the differences between these two binding modes in future studies.
The I345T mutant is more sensitive than WT to the presence of cofactor, and in this sense, more RecA-like. Exactly why, however, is not immediately clear from the available static crystal structures. In RecA, this residue is conserved as T in the WT protein, and it does not contact the DNA at all. However, it lies within the critical loop 2, and the two glycines immediately following it bind the DNA (Figure 1). The backbone carbonyl and nitrogen of this T are hydrogen bonded to the strictly conserved N193, which immediately precedes the allosteric affector Q194. In mvRadA, the equivalent residue is I273, and even in the absence of DNA it makes similar interactions. Loop 2 is in a similar, although not identical conformation, to that seen in RecA when bound to DNA. It may be that isoleucine in this position stabilizes a conformation of loop 2 that is competent, but suboptimal for DNA binding. This might explain why WT Rad51 is less dependent on ATP for DNA binding than the I345T mutant. Threonine at this postion may enhance DNA binding in the presence of ATP because when the suboptimal conformation is no longer stabilized, ATP binding can more easily drive a conformational change to the optimal configuration.
In sum, these data establish the functional significance of the intermolecular interface seen in the scRad51 and the RadA crystal structures. The conserved residues chosen for mutation (H352, R357 and I345) are not critical for catalysis of ATP hydrolysis but rather play important roles in coordinating cofactor binding to functionally important conformational changes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM058827 to P.A.R.); Susan G. Komen fellowship (to A.A.G.); Dutch Cancer Society scholarship (to J.H.A.V.); Association of International Cancer Research (to C.W.) and NCI program project (CA92584 to C.W.). Funding for open access charge: National Institutes of Health (GM058827).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lorraine Symington and Doug Bishop for helpful conversations and Yuen-Ling Chan for invaluable technical assistance.
REFERENCES
- 1.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 2.Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1999;274:2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- 3.Namsaraev EA, Berg P. Binding of Rad51p to DNA. Interaction of Rad51p with single- and double-stranded DNA. J. Biol. Chem. 1998;273:6177–6182. doi: 10.1074/jbc.273.11.6177. [DOI] [PubMed] [Google Scholar]
- 4.Namsaraev E, Berg P. Characterization of strand exchange activity of yeast Rad51 protein. Mol. Cell Biol. 1997;17:5359–5368. doi: 10.1128/mcb.17.9.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazin AV, Zaitseva E, Sung P, Kowalczykowski SC. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Stasiak AZ, Masson JY, McIlwraith MJ, Stasiak A, West SC. Conformational changes modulate the activity of human RAD51 protein. J. Mol. Biol. 2004;337:817–827. doi: 10.1016/j.jmb.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 8.Shim KS, Schmutte C, Yoder K, Fishel R. Defining the salt effect on human RAD51 activities. DNA Repair (Amst.) 2006;5:718–730. doi: 10.1016/j.dnarep.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 10.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta RC, Bazemore LR, Golub EI, Radding CM. Activities of human recombination protein Rad51. Proc. Natl Acad. Sci. USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 13.Vispe S, Defais M. Mammalian Rad51 protein: a RecA homologue with pleiotropic functions. Biochimie. 1997;79:587–592. doi: 10.1016/s0300-9084(97)82007-x. [DOI] [PubMed] [Google Scholar]
- 14.Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 15.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl Acad. Sci. USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egelman EH, Yu X. The location of DNA in RecA-DNA helical filaments. Science. 1989;245:404–407. doi: 10.1126/science.2667137. [DOI] [PubMed] [Google Scholar]
- 17.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 18.Scheffzek K, Lautwein A, Kabsch W, Ahmadian MR, Wittinghofer A. Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras. Nature. 1996;384:591–596. doi: 10.1038/384591a0. [DOI] [PubMed] [Google Scholar]
- 19.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 20.Datta S, Ganesh N, Chandra NR, Muniyappa K, Vijayan M. Structural studies on MtRecA-nucleotide complexes: insights into DNA and nucleotide binding and the structural signature of NTP recognition. Proteins. 2003;50:474–485. doi: 10.1002/prot.10315. [DOI] [PubMed] [Google Scholar]
- 21.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 22.Qian X, Wu Y, He Y, Luo Y. Crystal structure of Methanococcus voltae RadA in complex with ADP: hydrolysis-induced conformational change. Biochemistry. 2005;44:13753–13761. doi: 10.1021/bi051222i. [DOI] [PubMed] [Google Scholar]
- 23.Wu Y, He Y, Moya IA, Qian X, Luo Y. Crystal structure of archaeal recombinase RADA: a snapshot of its extended conformation. Mol. Cell. 2004;15:423–435. doi: 10.1016/j.molcel.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 24.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 25.Aihara H, Ito Y, Kurumizaka H, Yokoyama S, Shibata T. The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J. Mol. Biol. 1999;290:495–504. doi: 10.1006/jmbi.1999.2904. [DOI] [PubMed] [Google Scholar]
- 26.Cox MM. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 27.Xing X, Bell CE. Crystal structures of Escherichia coli RecA in a compressed helical filament. J. Mol. Biol. 2004;342:1471–1485. doi: 10.1016/j.jmb.2004.07.091. [DOI] [PubMed] [Google Scholar]
- 28.Rajan R, Bell CE. Crystal structure of RecA from Deinococcus radiodurans: insights into the structural basis of extreme radioresistance. J. Mol. Biol. 2004;344:951–963. doi: 10.1016/j.jmb.2004.09.087. [DOI] [PubMed] [Google Scholar]
- 29.Xing X, Bell CE. Crystal structures of Escherichia coli RecA in complex with MgADP and MnAMP-PNP. Biochemistry. 2004;43:16142–16152. doi: 10.1021/bi048165y. [DOI] [PubMed] [Google Scholar]
- 30.Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 31.Skiba MC, Knight KL. Functionally important residues at a subunit interface site in the RecA protein from Escherichia coli. J. Biol. Chem. 1994;269:3823–3828. [PubMed] [Google Scholar]
- 32.McGrew DA, Knight KL. Molecular design and functional organization of the RecA protein. Crit. Rev. Biochem. Mol. Biol. 2003;38:385–432. doi: 10.1080/10409230390242489. [DOI] [PubMed] [Google Scholar]
- 33.Kelley De Zutter J, Forget AL, Logan KM, Knight KL. Phe217 regulates the transfer of allosteric information across the subunit interface of the RecA protein filament. Structure (Camb.) 2001;9:47–55. doi: 10.1016/s0969-2126(00)00552-9. [DOI] [PubMed] [Google Scholar]
- 34.VanLoock MS, Yu X, Yang S, Lai AL, Low C, Campbell MJ, Egelman EH. ATP-mediated conformational changes in the RecA filament. Structure (Camb.) 2003;11:187–196. doi: 10.1016/s0969-2126(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 35.Fortin GS, Symington LS. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 2002;21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y, Qian X, He Y, Moya IA, Luo Y. Crystal structure of an ATPase-active form of Rad51 homolog from Methanococcus voltae. Insights into potassium dependence. J. Biol. Chem. 2005;280:722–728. doi: 10.1074/jbc.M411093200. [DOI] [PubMed] [Google Scholar]
- 37.Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 39.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl Acad. Sci. USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreuzer KN, Jongeneel CV. Escherichia coli phage T4 topoisomerase. Methods Enzymol. 1983;100:144–160. doi: 10.1016/0076-6879(83)00051-8. [DOI] [PubMed] [Google Scholar]
- 41.Kowalczykowski SC, Krupp RA. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein. Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J. Mol. Biol. 1987;193:97–113. doi: 10.1016/0022-2836(87)90630-9. [DOI] [PubMed] [Google Scholar]
- 42.Kiianitsa K, Solinger JA, Heyer WD. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem. 2003;321:266–271. doi: 10.1016/s0003-2697(03)00461-5. [DOI] [PubMed] [Google Scholar]
- 43.Ristic D, Wyman C, Paulusma C, Kanaar R. The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc. Natl Acad. Sci. USA. 2001;98:8454–8460. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sung P, Stratton SA. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- 45.Morrison C, Shinohara A, Sonoda E, Yamaguchi-Iwai Y, Takata M, Weichselbaum RR, Takeda S. The essential functions of human Rad51 are independent of ATP hydrolysis. Mol. Cell Biol. 1999;19:6891–6897. doi: 10.1128/mcb.19.10.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst.) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Rehrauer WM, Kowalczykowski SC. Alteration of the nucleoside triphosphate (NTP) catalytic domain within Escherichia coli recA protein attenuates NTP hydrolysis but not joint molecule formation. J. Biol. Chem. 1993;268:1292–1297. [PubMed] [Google Scholar]
- 48.Shan Q, Cox MM, Inman RB. DNA strand exchange promoted by RecA K72R. Two reaction phases with different Mg2+ requirements. J. Biol. Chem. 1996;271:5712–5724. doi: 10.1074/jbc.271.10.5712. [DOI] [PubMed] [Google Scholar]
- 49.Morgan EA, Shah N, Symington LS. The requirement for ATP hydrolysis by Saccharomyces cerevisiae Rad51 is bypassed by mating-type heterozygosity or RAD54 in high copy. Mol. Cell Biol. 2002;22:6336–6343. doi: 10.1128/MCB.22.18.6336-6343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim JM, Maraboeuf F, Kim SK, Shinohara A, Takahashi M. Effect of ions and nucleotides on the interactions of yeast Rad51 protein with single-stranded oligonucleotides. J. Biochem. (Tokyo) 2001;129:469–475. doi: 10.1093/oxfordjournals.jbchem.a002879. [DOI] [PubMed] [Google Scholar]
- 51.Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qian X, He Y, Wu Y, Luo Y. Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae. J. Mol. Biol. 2006;360:537–547. doi: 10.1016/j.jmb.2006.05.058. [DOI] [PubMed] [Google Scholar]
- 53.Cox JM, Abbott SN, Chitteni-Pattu S, Inman RB, Cox MM. Complementation of one RecA protein point mutation by another. Evidence for trans catalysis of ATP hydrolysis. J. Biol. Chem. 2006;281:12968–12975. doi: 10.1074/jbc.M513736200. [DOI] [PubMed] [Google Scholar]
- 54.Voloshin ON, Wang L, Camerini-Otero RD. The homologous pairing domain of RecA also mediates the allosteric regulation of DNA binding and ATP hydrolysis: a remarkable concentration of functional residues. J. Mol. Biol. 2000;303:709–720. doi: 10.1006/jmbi.2000.4163. [DOI] [PubMed] [Google Scholar]
- 55.Hortnagel K, Voloshin ON, Kinal HH, Ma N, Schaffer-Judge C, Camerini-Otero RD. Saturation mutagenesis of the E. coli RecA loop L2 homologous DNA pairing region reveals residues essential for recombination and recombinational repair. J. Mol. Biol. 1999;286:1097–1106. doi: 10.1006/jmbi.1998.2515. [DOI] [PubMed] [Google Scholar]
- 56.Ariza A, Richard DJ, White MF, Bond CS. Conformational flexibility revealed by the crystal structure of a crenarchaeal RadA. Nucleic Acids Res. 2005;33:1465–1473. doi: 10.1093/nar/gki288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen LT, Ko TP, Chang YC, Lin KA, Chang CS, Wang AH, Wang TF. Crystal structure of the left-handed archaeal RadA helical filament: identification of a functional motif for controlling quaternary structures and enzymatic functions of RecA family proteins. Nucleic Acids Res. 2007;35:1787–1801. doi: 10.1093/nar/gkl1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roca AI, Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Prog. Nucleic Acid. Res. Mol. Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.