Abstract
Loop–loop tertiary interactions play a key role in the folding and catalytic activity of natural hammerhead ribozymes. Using a combination of NMR spectroscopy, site-directed mutagenesis and kinetic and infectivity analyses, we have examined the structure and function of loops 1 and 2 of the (+) and (–) hammerheads of chrysanthemum chlorotic mottle viroid RNA. In both hammerheads, loop 1 is a heptanucleotide hairpin loop containing an exposed U at its 5′ side and an extrahelical U at its 3′-side critical for the catalytic activity of the ribozyme in vitro and for viroid infectivity in vivo, whereas loop 2 has a key opened A at its 3′-side. These structural features promote a specific loop–loop interaction motif across the major groove. The essential features of this tertiary structure element, base pairing between the 5′ U of loop 1 and the 3′ A of loop 2, and interaction of the extrahelical pyrimidine of loop 1 with loop 2, are likely shared by a significant fraction of natural hammerheads.
INTRODUCTION
Hammerhead ribozymes are small catalytic RNAs first discovered in viroids and satellite RNAs, where they mediate processing of the multimeric strands generated during replication (1,2), and more recently identified in transcripts of the genomes of certain animal and plant species (3,4–6). They catalyze specific self-cleavage of the RNA backbone via a transesterification reaction that releases 5′ hydroxyl and 2′,3′-cyclic phosphodiester termini. Hammerheads have potential therapeutic interest as catalytic antisense agents, because they can trans-hybridize to a complementary target substrate, cleave that substrate, and bind subsequently to a new target molecule (7). In this respect, a number of engineered hammerheads are currently under clinical trials (8,9).
Natural hammerheads are composed of a highly conserved catalytic core flanked by three stems (helices I to III) usually capped by short loops (1–3) (Figure 1A). Interactions between loops 1 and 2 greatly increase the catalytic activity of the ribozymes (10,11). These tertiary contacts are distal from the active site but promote the adoption by the central core of a catalytically active conformation, as shown recently by the X-ray structures of the Schistosoma mansoni (12) and tobacco ring spot virus satellite RNA (sTRSV) (13) full-length hammerheads. These structures have reconciled structural and biochemical views on the catalytic mechanism of hammerheads (14), and have also provided important information on how the major grooves of loops 1 and 2 contact each other. However, the sequence and secondary structure of loops 1 and 2 of natural hammerheads do not appear to follow common patterns (15) and, in spite of the likely relevance on therapeutic ribozyme design, a systematic study of the structure and interactions of loops 1 and 2 in hammerhead ribozymes remains to be carried out.
Figure 1.
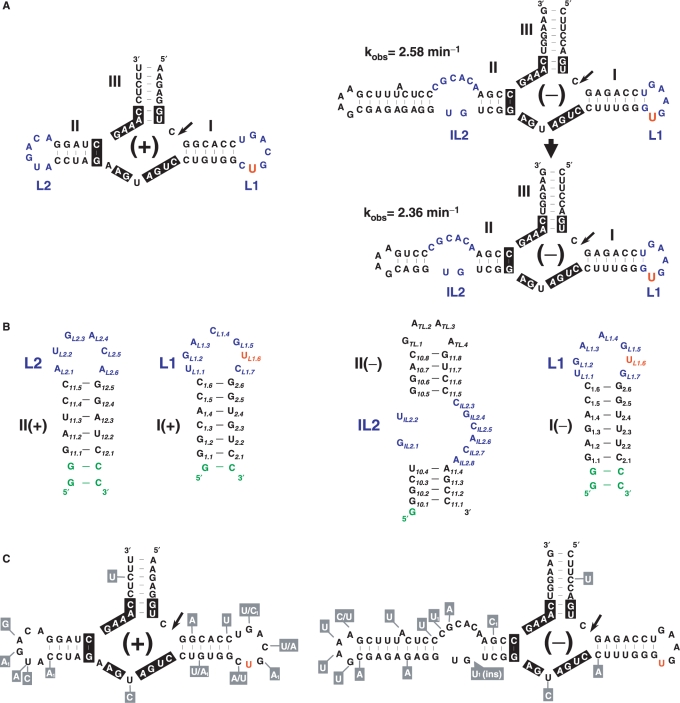
Primary and secondary structure analysis of the CChMVd (+) and (–) hammerheads. (A) Wild-type hammerheads and secondary structure of a CChMVd (–) hammerhead with a shortened but functional domain II as revealed by the similar self-cleavage rate-constants. Active site nucleotides highly or strictly conserved in most natural hammerheads are indicated with a black background. (B) Secondary structures of the I– and II– (right), and I+ and II+ (left) domains studied by NMR spectroscopy. Additional nucleotides are shown in green. (C) Natural sequence variability found in CChMVd (+) and (–) hammerheads, indicated within boxes with gray background. Variants labeled with subscript 1 have only been detected once in the databases. In (A), (B) and (C), the extrahelical U of domain I is shown in red.
The present study focuses on the structure and functional impact of the helix I-loop 1 and helix II-loop 2 domains (hereafter called domains I and II for simplicity) of the hammerheads formed by the (+) and (–) RNAs of chrysanthemum chlorotic mottle viroid (CChMVd) (16,17), the infectivity of which can be assessed with a relatively fast bioassay. In the CChMVd (+) hammerhead, domains I (I+) and II (II+) most likely interact through their hairpin loops, as in most natural hammerheads (4), whereas in the CChMVd (–) hammerhead, the hairpin loop of domain I (I-) presumably interacts with an internal loop of domain II (II-), hereafter referred to as internal loop 2 of this hammerhead (Figure 1A). A tertiary contact of this rather unusual class has also been observed in the Schistosoma hammerhead but with opposite loop topology, because in this case an internal loop of domain I interacts with the apical loop of domain II (12).
Using a combination of NMR spectroscopy, site-directed mutagenesis, kinetics analysis and infectivity bioassays, we show that loops 1 of the CChMVd (+) and (–) hammerheads fold into related three-dimensional structures, and that the same holds for loops 2 of these hammerheads. Mutations that disrupt the structures or change the identity of key nucleotides involved in loop–loop contacts have a strong effect on both catalytic activity in vitro and viroid replication in vivo. Mutagenesis data, together with sequence and structure comparisons with other hammerheads, indicate that the three-dimensional structures of loops 1 and 2 are conserved, giving rise to a tertiary interaction motif embedded within the loop–loop complexes that is likely shared by a significant fraction of natural hammerheads. Since these loop 1–loop 2 contacts are critical for hammerhead activity under the low magnesium levels existing in vivo (10,11), the findings reported here are expected to contribute to a better understanding of the principles underlying hammerhead architecture and to the rational design of improved ribozymes of this class.
MATERIALS AND METHODS
Site-directed mutagenesis
PCR-based protocols reported previously (18–20) were applied to plasmid pCM20 containing a monomeric insert of CChMVd (16). The resulting plasmids with the specific point mutations were used as template for amplifying their monomeric inserts with adjacent 5′-phosphorylated primers, and the PCR products were gel-purified and ligated to obtain dimeric head-to-tail cDNAs that were cloned and sequenced to confirm the presence of the desired mutations in both units.
In vivo analysis of viroid viability and genetic stability
For each bioassay, which was repeated at least once, blocks of four chrysanthemum plants (Dendranthema grandiflora Tzvelez, cv. ‘Bonnie Jean’) were mechanically inoculated with buffer (mock-inoculated control) or with 40 ng per plant of the monomeric RNAs resulting from self-cleavage during in vitro transcription of linearized recombinant plasmids containing dimeric head-to-tail inserts of the CChMVd 20 variant (wild-type) and the L1(–).U6 mutants. The primary transcripts and their self-cleavage products were separated by denaturing PAGE in 5% gels, and the monomeric CChMVd RNAs were eluted and quantified. Infectivity was assessed by symptom expression and northern-blot hybridization, and genetic stability by RT-PCR amplification, cloning and sequencing of the resulting progenies (20).
In vitro analysis of catalytic activity
Self-cleavage rate constants were determined as reported previously (10). The uncleaved primary transcripts corresponding to the CChMVd (+) and (–) hammerheads (and their mutants), flanked by two Gs in 5′ to enhance transcription and 4–5 nt in 3′ for proper restriction, were incubated in 50 mM PIPES-NaOH pH 6.5 for 1 min at 95°C and slowly cooled to 25°C for 15 min. After taking a zero-time aliquot, reactions were triggered by adding MgCl2 to 10 mM. Aliquots were removed at time intervals and quenched with a 5-fold excess of stop solution at 0°C, with the primary transcripts and their cleavage products being separated by denaturing PAGE in 15% gels. The product fraction at different times Ft was assessed by quantitative scanning and fitted to the equation Ft = Fo + F∞ (1-e–kt), where Fo and F∞ are the product fractions at zero time and at the reaction endpoint, respectively, and k is the first-order cleavage rate constant (21).
NMR RNA sample preparation
Unlabeled (all), 15N-labeled (domains I− and II−) and 13C/15N-labeled (domains I− and II+) RNA oligonucleotides encompassing either domain I or II of the CChMVd (–) and (+) hammerheads (Figure 1B) were prepared by T7-driven in vitro transcription from oligonucleotide DNA templates. 15N-labeled and 13C/15N-labeled NTPs were prepared from cellular RNA as described (22). After a purification procedure based on gel electrophoresis, all samples were microdialyzed in aqueous solutions containing 10 mM sodium phosphate (pH 6.0) and 0.1 mM EDTA. The final RNA concentration in the NMR samples ranged from 0.5 to 1.0 mM. The I− and II− samples were also analyzed in the same buffer supplemented with 5 mM MgCl2 to evaluate the effect of Mg2+ on the structures. The I+ sample was additionally examined in 0.5 mM sodium phosphate (pH 6.0) to reduce formation of a competing duplex.
NMR spectroscopy
NMR spectra were acquired on Bruker 600 and 700 MHz spectrometers, processed with Topspin 1.3 (Bruker Biospin) and analyzed using Sparky 3.110 (Goddard and Kneller, University of California, San Francisco). For the unlabeled samples, spectra recorded in D2O were acquired at several temperatures (typically 24°C and 34°C), and included series of 2D NOESY (mixing times 80, 120 and 250 ms), DQF-COSY, TOCSY (60 ms), ROESY and 1H-31P HETCOR experiments. NOESY spectra were also recorded in H2O at 8°C with 150 ms mixing time. The relaxation delay was 2 s for all experiments with unlabeled samples. For the 15N-labeled or 13C/15N-labeled domain I−, II− and II+ samples, 2D 1H-15N HSQC and HNN COSY experiments were recorded in H2O at 27°C to evaluate hydrogen bonding between bases via two bond N-N couplings. For the 13C/15N-labeled domain I− and II+ samples, 2D 1H-13C HSQC, 3D 13C-edited NOESY-HMQC (100 and 200 ms mixing times), 3D HCCH E-COSY, 2D HCCH-TOCSY, 3D HCP, 2D quantitative HCP, 3D 13C-edited 1H-31P HETCOR and 2D quantitative Γ−HCNCH (23) spectra were acquired at 27°C in D2O. Together, these data allowed for the assignment of all relevant nuclei contained in these sequences, as well as for the extraction of complete sets of distance and dihedral restraints for structure calculation.
Constraints for domain I− and II+ structure determination
Distance constraints were semi-quantitatively estimated from the 2D NOE intensities at 80, 120 and 250 ms mixing times. Cross-peaks corresponding to covalently constrained intra-base and intra-sugar distances were used as a reference for constraint calibration. Constraints involving ambiguous or overlapped cross-peaks were obtained from the 13C-edited NOESY-HMQC spectra at 100 and 200 ms mixing times. Based on the absence of NOE interactions, lower limit distance constraints were imposed on several proton pairs. With the exception of hydrogen bonding restraints, all distance restraints had a minimum width of 1 Å. Hydrogen-bonding distance constraints for Watson–Crick A:U, G:C and G:U (wobble) pairs were introduced based on the observation of HNN-COSY couplings, and/or patterns of NOEs and chemical shifts observed in H2O NOESY experiments. With the exception of the U9:G15 wobble pair in domain I−, no hydrogen bonding restraints were imposed on loop nucleotides. Sugar conformation was inferred from the analysis of DQF-COSY, TOCSY and 3D HCCH-E-COSY spectra, and constrained using the backbone dihedral angle δ. Backbone angles β, γ and ε were also constrained using a combination of observations and measurements from 3D HCCH-E-COSY, HCP and 13C-edited 1H-31P HETCOR, and 2D quantitative HCP experiments.
Domains I− and II+ structure calculation
The distance and dihedral constraints specified above were then used to determine the three-dimensional structure of domains I− and II+. 100 (I−) or 60 (II+) structures were iteratively refined by high-temperature restrained molecular dynamics using the ff99 force field of Amber 8.0 (Case et al., University of California, San Francisco). We set up a protocol in which each structure undergoes two simulated annealing stages of 35 and 20 ps. In the first stage, the structure is kept at 7000 K with covalent, van der Waals, and restraint terms initially reduced 10 times and then gradually allowed to return to their standard values during 10 ps. The temperature is then slowly reduced to 1 K while the relative weighting of the electrostatic and hydrogen-bonding terms is progressively increased from 0 to 1. No cut-off is used for the nonbonded interactions, and the solvent is mimicked with a distance-dependent dielectric constant. In the second (refinement) stage, a Generalized Born model is used to simulate the presence of solvent. The structure is heated to 600 K, slowly cooled to 1 K and minimized. The relative weighting of the constraint terms is initially decreased 10 times and then slowly returned to 1 K before cooling. For each refinement round, the initial structures are disordered, high-temperature snapshots obtained from the 7000 K stage of a previous iteration. After several refinement rounds, the final sets of 29 I− and 33 II+ converged structures were selected solely on the basis of total energy and constraint violation energy, and used to generate the refinement statistics (Table 1).
Table 1.
Experimental constraints and refinement analysis for domains I− and II+ of the CChMVd hammerheads
| I− | II+ | |
|---|---|---|
| Experimental constraintsa | ||
| NOE distance constraints | 436 (19) | 360 (18) |
| Intraresidue | 142 (6) | 121 (6) |
| Interresidue | 294 (13) | 239 (12) |
| Hydrogen bonding distance constraints | 23 | 19 |
| Dihedral constraints | 84 (4) | 74 (4) |
| Dihedral constraints (chirality) | 92 (4) | 80 (4) |
| Refinement analysisb | ||
| NOE violations >0.25 Å | 0 | 0 |
| Largest NOE violation (Å) | 0.23 ± 0.01 | 0.13 ± 0.01 |
| Distance constraint violation energyc (Kcal mol−1) | 10.96 ± 0.19 | 3.46 ± 0.24 |
| Dihedral violations >10° | 0 | 0 |
| Largest dihedral violation (°) | 9.21 ± 0.06 | 1.40 ± 1.02 |
| Dihedral constraint violation energyd (Kcal mol−1) | 2.48 ± 0.03 | 0.13 ± 0.10 |
| R.m.s deviation from ideal bond lengths (Å) | 0.01 ± 0.00 | 0.01 ± 0.00 |
| R.m.s deviation from ideal bond angles (°) | 2.56 ± 0.01 | 2.49 ± 0.01 |
| Heavy atom pair-wise r.m.s deviation (Å): | 1.05 ± 0.31 | 1.02 ± 0.29 |
| Heavy atom r.m.s deviation from average structure (Å): | 0.74 ± 0.19 | 0.71 ± 0.20 |
aAverage number of constraints per residue indicated in parenthesis.
bAverage and standard deviation values obtained from 29 (I−) and 33 (II+) converged structures.
cForce constant K = 30 Kcal mol−1 Å−2.
dForce constant K = 90 Kcal mol−1 rad−2.
CChMVd (+) full-length hammerhead model building
After superimposing NMR structures of CChMVd domains I− and II+ on domains I and II of the crystal structure of the Schistosoma hammerhead (12), we removed the crystallographic domain I and II atoms and inserted the unpaired A11 nucleotide at the base of stem II (Figure 1A) in a stacked conformation, using PDB structure 1GID (24) as a model. After performing the appropriate base changes in stem III and domain I to obtain the CChMVd (+) sequence, the position of the loop 1 and loop 2 residues was modified to accomplish the tertiary interactions revealed by site-directed mutagenesis, taking care to set backbone dihedral angles to acceptable values (25). The structure was then progressively energy-minimized using the ff99 force field of Amber 8.0 (Case et al., University of California, San Francisco) with intra- and inter-domain base pair hydrogen-bonding restraints, and a Generalized Born model to mimic solvent effects.
RESULTS AND DISCUSSION
NMR spectroscopy
We have examined by NMR spectroscopy the solution structure of domains I and II of the CChMVd (+) and (–) hammerheads (Figures 1 and 2 and Supplementary Figures S1 and S2): domains I− and II+ were studied by 2D and 3D methods using unlabeled and 13C/15N-labeled samples, domain II− by 2D methods using unlabeled and 15N-labeled samples and domain I+ by 2D methods using an unlabeled sample. To facilitate the NMR analyses, we focused on a smaller version of domain II− in which the corresponding helix was shortened, after verifying that this artificial ribozyme retained most of the catalytic activity of its wild-type counterpart (Figure 1A).
Figure 2.
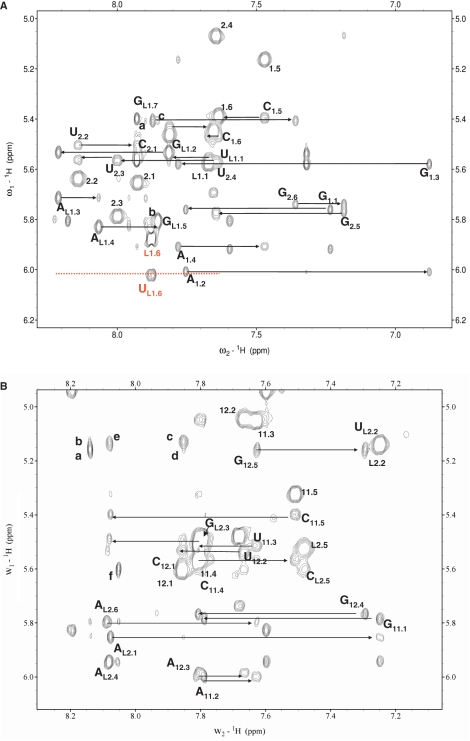
NMR spectra of domains I and II of the CChMVd (+) and (–) hammerheads. The assignments of the H2/H6/H8–H1′/H5 NOESY regions (250 ms mixing time) are shown for domains I– (A) and II+ (B). Intraresidue H1′-H6/H8 cross-peaks are labeled with residue name and number, intraresidue H5-H6 crosspeaks are labeled with residue number, and sequential NOE connectivities are indicated with horizontal arrows. In (A), cross-peaks (a) to (c) are assigned as follows: a, L1.G7 H1′-L1.A4 H2; b, L1.G5 H1′-L1.G7 H8; c, L1.G7 H1′-L1.G5 H8. The assignments of the extrahelical L1.U6 residue are shown in red, and the discontinuous line indicates the absence of any significant sequential interactions. In (B), cross-peaks (a) to (f) are: a, G12.5 H1′-L2.A1 H2; b, L2.U2 H1′- L2.A1 H2; c, L2.U2 H1′- L2.A6 H2; d, G12.5 H1’-L2.A6 H2; e, L2.U2 H1’- L2.A4 H8; f, L2.C5 H1′-L2.A4 H2. The assignments of the terminal base pairs of I– and II+ (shown in green in Figure 1B) have been omitted for clarity.
Three-dimensional structure of domain I of the CChMVd (–) hammerhead
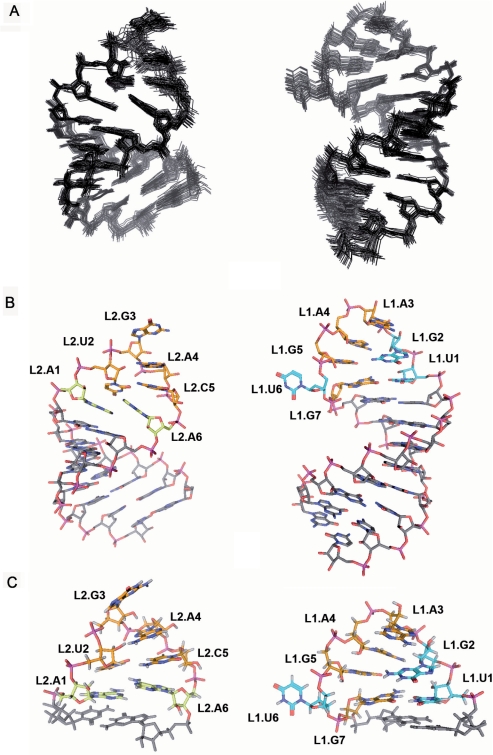
Twenty-nine converged structures with low total and restraint-violation energies compose the conformational ensemble representing the solution structure of domain I− (Figure 3A and Table 1). This domain forms a hairpin capped by an apical UGAAGUG heptaloop (Figure 3B). The first residue of the heptaloop, L1.U1, establishes a G:U wobble pair with L1.G7, as indicated by HNN-COSY data (data not shown). These 2 nt closing the loop adopt standard C3′-endo anti conformations, and are connected by sequential interactions with their neighbor nucleotides in the stem (Figure 2A). In contrast, the remaining loop nucleotides adopt C2′-endo sugar conformations and their glycosidic angles are in the high anti range, as indicated by TOCSY, COSY and ΓHCNCH (23) data. L1.G2 is stacked on L1.U1 as revealed by sequential NOE interactions, and its base is turned towards the major groove side of the loop and located opposite to L1.G5, with which it establishes a weak L1.G2 amino to L1.G5 N7 hydrogen bond (Figure 3C). The next two loop residues, L1.A3 and L1.A4, make the backbone turn of the loop and their bases are located at opposite sides of the loop. L1.A3 is on top of the loop, with its sugar partially stacked on the L1.A4 base and its base located in the major groove above L1.G2; L1.A4 is located in the minor groove below L1.A3, with both its sugar and base partially stacked on the L1.G5 base. Like the L1.A4 base, the L1.G5 base is turned toward the minor groove of the loop, leaving its Hoogsteen edge in the vicinity of the L1.G2 amino group (Figure 3C). L1.U6 is outside the helix and in the major groove of the loop, allowing the L1.G5 base to partially stack on the L1.G7 nucleotide closing the heptaloop (Figure 3B and C). This is supported by weaker or absent sequential NOEs between L1.U6 and its neighbors, and by the observation of NOE contacts between L1.G5 and L1.G7 (Figure 2A, peaks b and c).
Figure 3.
Three-dimensional structures of domains I– and II+ of the CChMVd hammerheads determined by NMR spectroscopy. In (A)–(C) domain I− structures are shown on the right, seen from the major groove of loop 1, and domain II+ structures are shown on the left, seen from the minor groove of loop 2. (A) View of the superimposed 33 and 29 converged structures of domains II+ and I–, respectively. (B) View of converged structures of domains II+ and I–. (C) Close view of the AUGACA hexaloop 2 and UGAAGUG heptaloop 1. In (B) and (C) the carbon atoms of L1.U1, L1.G2 and L1.U6 (domain I–) and L2.A1 and L2.A6 (domain II+) are colored in blue and yellow, respectively, those of other loop nucleotides in orange, and those of stem residues in gray. In (A) and (B), hydrogen atoms have been omitted for clarity.
Overall, the UGAAGUG heptaloop of domain I− forms a fairly compact although rather open structure stabilized by both base–base and base–sugar hydrogen-bonding and stacking interactions. The lower part of the loop contains two noncanonical L1.U1:G7 and L1.G2:G5 base pairs that stack on each other, looping out the penultimate L1.U6 nucleotide (Figure 3B and C). The U:G and G:G base pairing interactions force the Watson–Crick edges of L1.U1 and L1.G2 to face the major groove, as also do the L1.A3 base and the L1.U6 nucleotide. In contrast, the L1.G7 and L1.G5 bases, together with L1.A4, are oriented toward the minor groove of the loop (Figure 3B and C).
The NMR spectra of domain I of the CChMVd (+) hammerhead show that loop 1 is structurally conserved in both hammerheads
Since domains I of the two CChMVd hammerheads show high sequence identity (16) (Figure 1), we next examined whether the structure of loops 1 is also conserved. The NMR data indicated that hairpin I+ is capped by an apical UGACGUC heptaloop structurally similar to the UGAAGUG apical loop of domain I−. This conclusion is supported by related chemical shift, TOCSY and NOE patterns (Supplementary Figure S1) indicating that the stacking and pairing of nucleotides are similar in both loops, and that L1.U6 of domain I+ also holds an extrahelical location.
Natural sequence variability correlates with the structural orientation of residues in loop 1
Sequence comparisons of numerous natural CChMVd variants (16,26,27) show that nucleotides L1.U1 and L1.U6 of the heptaloop capping domain I+ are strictly conserved, with the sequence variability mapping predominantly at positions L1.4 and L1.7 (Figure 1C). This conservation pattern is correlated with the orientation of residues observed in the three-dimensional structure of domain I− (Figure 3C), for which essentially no sequence variability has been reported: residues facing the major groove of the heptaloop, where the presumed contacts with domain II take place, are significantly more conserved than residues facing the minor groove.
The first and the extrahelical U residues of domain I have a strong impact on catalytic activity
The first L1.U1 and penultimate L1.U6 nucleotides of loop 1 are strictly conserved (Figure 1C), and face the major groove side of domain I− (Figure 3C). The available crystallographic data indicate that tertiary contacts between loops 1 and 2 are established through the major groove (12,13). To determine whether these nucleotides play a significant role on loops 1–2 interactions, we tested the effects of mutations on the self-cleavage activity of the CChMVd (+) and (–) hammerheads in vitro. The four possible mutations of L1.U6 in the (–) hammerhead, L1(–).U6A, L1(–).U6G, L1(–).U6C and L1(–).U6Δ, caused a reduction of the catalytic constant of one to two orders of magnitude (Table 2). The two purine mutants were particularly detrimental, with L1(–).U6A almost eliminating the catalytic activity of the ribozyme. In the (+) hammerhead, the same L1(+).U6A mutant caused a similar reduction in the self-cleavage rate. Mutation of L1.U1 also caused a large effect on ribozyme activity in both polarities, as also did both L1.G2A mutants. In contrast, the L1(–).A4G mutation, involving one of the residues facing the minor groove, did not affect significantly the self-cleavage rate constant (Table 2).
Table 2.
Self-cleavage rate constants of the wild-type CChMVd (−) and (+) hammerheads and some single and double mutants
| Variant | kobs (min−1) | |
|---|---|---|
| (−) | Wild-type | 2.58 ± 0.11 |
| IIa | 2.36 ± 0.28 | |
| L1.U6Aa | 0.05 ± 0.01 | |
| L1.U6Ga | 0.16 ± 0.02 | |
| L1.U6Ca | 0.38 ± 0.06 | |
| L1.U6Δa | 0.34 ± 0.05 | |
| L1.A4Ga | 2.25 ± 0.41 | |
| L1.U1Aa | 0.13 ± 0.02 | |
| IL2.A8Ua | 0.066 ± 0.002 | |
| L1.U1A/IL2.A8Ua | 1.09 ± 0.07 | |
| L1.G2Aa | 0.90 ± 0.19 | |
| IL2.C7Ua | 0.59 ± 0.01 | |
| L1.G2A/IL2.C7Ua | 0.84 ± 0.04 | |
| (+) | Wild-type | 3.33 ± 0.80 |
| L1.U6A | 0.049 ± 0.014 | |
| L1.U1C | 0.91 ± 0.08 | |
| L2.A6G | 2.76 ± 0.72 | |
| L1.U1C/L2.A6G | 0.042 ± 0.003 | |
| L1.G2A | 0.79 ± 0.09 | |
| L2.C5U | 2.18 ± 0.16 | |
| L1.G2A/L2.C5U | 0.28 ± 0.07 |
Single mutants with kobs ≤ 0.1 min−1 are shown in bold.
aSmaller version of the (−) hammerhead in which the domain II helix was shortened (Figure 1A).
The extrahelical U residue of domain I- is essential for viroid replication
To further assess the role of the extrahelical L1.U6 nucleotide in vivo, we inoculated chrysanthemum plants with full-length CChMVd RNAs containing each of the four mutations of the CChMVd (–) L1.U6 residue. The effects of these mutations on viroid replication were noteworthy in all cases (Table 3): plants inoculated with L1.U6A and L1.U6▵ RNAs did not express symptoms, and northern-blot hybridizations with a specific riboprobe failed to detect the viroid RNA, whereas plants inoculated with L1.U6G and L1.U6C RNAs showed a delay in symptom expression with respect to the wild-type RNA; these plants did contain CChMVd RNA, but RT-PCR amplification, cloning and sequencing revealed that both mutations had reverted to the original L1.U6 in the resulting progeny. These results are consistent with L1.U6 playing a critical role in the catalytic activity of the hammerhead although, given the high informational density of viroid RNAs, the involvement of this residue in critical function(s) other than self-cleavage cannot be excluded (17).
Table 3.
Bioassay of CChMVd RNAs with mutations affecting the L1.U6 residue of the (−) hammerhead
| Mutant | Symptomsa | Hybridization | Progeny |
|---|---|---|---|
| Wild-type | 8/8 (4/4 + 4/4) | Yes | |
| L1.U6A | 0/8 (0/4 + 0/4) | No | NAb |
| L1.U6G | 6/8 (2/4 + 4/4) | Yes | Reversion |
| L1.U6C | 8/8 (4/4 + 4/4) | Yes | Reversion |
| L1.U6Δ | 0/8 (0/4 + 0/4) | No | NA |
aNumber of plants with symptoms with respect to those inoculated in two independent experiments with four plants each.
bNot applicable.
Solution structure of domain II of the CChMVd (+) and (−) hammerheads
To get further insights on how the conserved heptaloop of domain I interacts with domain II, we have also analyzed the solution structure of domains II+ and II− by NMR spectroscopy. The NMR data show that domain II− forms a hairpin capped by a GAAA tetraloop and containing an asymmetric 2:6 GU:CGCACA internal loop 2, as depicted in Figure 1B. Broadening of the internal loop resonances (Supplementary Figure S2) indicated that this loop exchanges between two or more conformations in solution, and prevented detailed NMR analyses of this domain. In contrast, domain II+ forms a more stable hairpin closed by an apical AUGACA hexaloop. The solution conformation of hairpin II+ is represented by 33 structures with very low total- and restraint- violation energies (Figure 3A; Table 1). All six loop nucleotides are predominantly anti and C3′-endo, with the exception of L2.G3 that is mostly C2′-endo and, strikingly, none is base-paired. L2.A6 is intercalated between L2.A1, which is stacked on the adjacent C11.5:G12.5 stem pair, and L2.U2 (Figure 3B and C). This is clearly indicated by the relative intensities of the NOEs between the L2.A1 and L2.A6 H2s and the H1′ protons of G12.5 and L1.U2 (Figure 2B, peaks a–d). L2.U2 and L2.C5 are located relatively far from each other with their Watson–Crick edges oriented towards the major and the minor groove side of the loop, respectively, as revealed by a medium-intensity L1.U2 H1′-L2.C5 H5 NOE interaction (data not shown). The loop backbone turns at the L2.U2-G3 step, and the L2.A4 base is turned toward the minor groove side of the loop and anchored to the L2.U2 sugar via hydrogen bonds between the L2.U2 2′OH and the N7 and exocyclic amino group of L2.A4 (Figure 3C); this is supported by nonsequential NOEs between L2.U2 H1′ and H2′ sugar protons and L2.A4 H8 (Figure 2B, peak e). In contrast, the positions of the next two loop nucleotides, L2.C5 and L2.A6, are rather unstable: the sugar-phosphate backbone undergoes a large change in orientation at the L2.C5-A6 step, and the stacking of both the C5 sugar and base with the neighboring residues is limited (Figure 3B and C), as indicated by broadening of the L2.C5 resonances by conformational exchange (Figure 2B) and by significantly weakened or nonobservable sequential interactions between L2.C5 and its two neighbors, L2.A4 and L2.A6 (Figure 2B). In addition, intercalation of the L2.A6 base between L2.A1 and L2.U2 leaves a significant space between this nucleotide and the C11.5:G12.5 stem pair, in front of the L2.A1 base. This is supported by weakened L2.A6-G12.5 sequential interactions (Figure 2B).
Overall, none of the loop nucleotides is base-paired, and only the L2.U2 base is turned toward the major groove side of the loop. The L2.G3, L2.A4, L2.C5 and L2.A6 bases are stacked on the 3′-side of the loop and mostly oriented toward the minor groove. A significant change in orientation occurs for the L2.A6, L2.C5 and L2.A4 backbone relative to the double-helical stem, brought about by the simultaneous unwinding and opening toward the minor groove of these nucleotides. This backbone conformation is accommodated by trans γ angles in the L2.A6 and G12.5 nucleotides closing the 3′-end of the loop, and is stabilized by hydrogen bonding between the L2.U2 sugar and the Hoogsteen edge of the L2.A4 base. Due to this backbone conformation and to L2.A6 intercalation between L2.A1 and L2.U2, the L2.C5 and L2.A6 bases are solvent-exposed and/or exhibit weak stacking interactions with their neighbors (Figure 3B and C).
The conserved R:A opposition at the bottom of loop 2 is essential for the catalytic activity of CChMVd hammerheads
The last two C and A nucleotides of loops 2 are strictly conserved in CChMVd (+) (L2, AUGACA) and (–) (IL2, GU:CGCACA) hammerheads, whereas the first is always a purine (Figure 1C). We have tested how mutations in these residues modulate the catalytic activity of the CChMVd hammerheads. Self-cleavage was almost abolished in the IL2(–).A8U mutant (Table 2), in which impairment of the 5′-R:3′-A opposition most likely produces major changes in the structure of loop 2 (see above). In contrast, substitution of the penultimate C residue by U caused smaller effects on the catalytic activity of both ribozymes, as also did the conservative L2(+).A6G substitution.
Comparative analyses indicate that the three-dimensional structures of loops 1 and 2 are conserved in a significant fraction of natural hammerheads
Loop 1
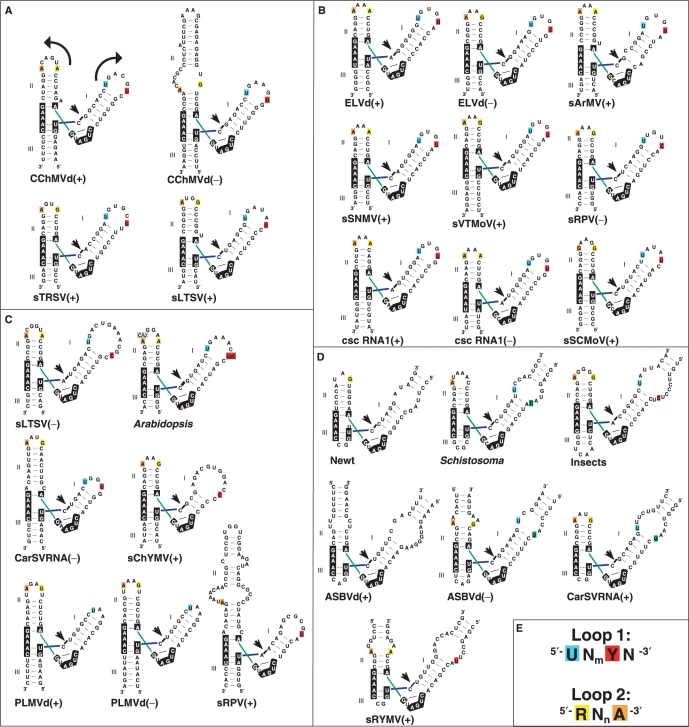
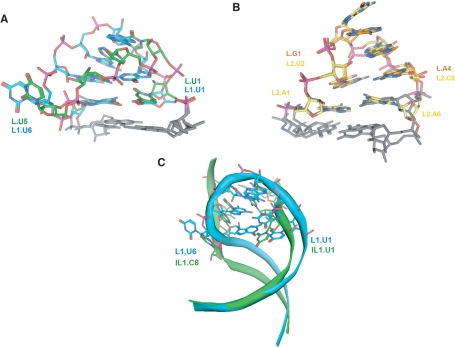
In the CChMVd (+) and (–) hammerheads, loop 1 contains an extrahelical L1.U6 nucleotide, and L1.U1, L1.G2 and L1.U6 face the major groove in the three-dimensional structure of domain I− (Figure 3C). A comparative analysis of 27 natural hammerheads (4) reveals that 17 conform in sequence to a conserved UNmYN loop 1 motif, where Y is a pyrimidine and m is usually 3 or 4 in apical loops (Figure 4). The closest relatives to CChMVd hammerheads are those of the (+) strands of sTRSV and the satellite RNA of lucerne transient streak virus (sLTSV), in which loop 1 is also a heptaloop with L1.U1, L1.G2 and L1.U6 residues (Figure 4A). Some hammerheads with loops 1 of more than seven nucleotides, like those of Arabidopsis thaliana and the (–) strand of sLTSV also match the conserved motif, as does the UGGUG pentaloop of the hammerhead of the (–) strand of the carnation small viroid-like RNA (CarSV) and the internal loop of the cave cricket hammerhead (Figure 4). However, the most common form of loops 1 are UN3YN hairpin hexaloops, which also match the loop 1 motif and are present in nine natural hammerheads (Figure 4B). The structure of these loops is unknown, but a SCOR (28) search revealed that, in the sequence-related 23S rRNA UCAAUG (29) and 16S rRNA UUAAUU, UUAACU and UUAAAU (30), UCAAUU (31) and UUAAGU (32) hexaloops, the 5′ U faces the major groove and the penultimate residue (equivalent to L1.U6 in heptaloops) is always located outside the helix and in the major groove side of the loop. Indeed, these hexaloops superpose quite well with the CChMVd (–) UGAAGUG heptaloop (Figure 5A).
Figure 4.
Sequence and secondary structure comparative analysis of loops 1 and 2 in natural hammerheads. In (A–D), secondary structures of all known 27 natural hammerheads are organized according to loop 1 type: (A) heptaloops; (B) hexaloops; (C) other apical loops; (D) internal loops. Nucleotides matching the UNmYN loop 1 and RNnA loop 2 conserved motifs are color-coded as indicated in scheme (E). Arrows in the first hammerhead indicate strand orientation. In (B), five UN3UA hexaloops were considered to match the UNmYN motif because U:A and U:G pairs are isosteric (35) and the U:A opposition closing the loop may not form a base-pair, as observed in the UUUAGA and UGAAA loops contained in PDB structures 1NBS (39) and 1NJP (29), respectively. In (D), pyrimidine residues located at the last rather than at the penultimate position of internal loop 1 are green-colored.
Figure 5.
Three-dimensional structure conservation of loops 1 and 2 in natural hammerheads. (A) Superposition of the UGAAGUG CChMVd(–) heptaloop 1 with a U1C2A3A4U5G6 hexaloop from 23S rRNA (PDB code 1NJP) (29). UCAAUG-related hexaloops are the most frequently found loop 1 class of natural hammerheads (Figure 4B). (B) Superposition of the AUGACA CChMVd(+) hexaloop 2 (yellow) with a G1A2A3A4 tetraloop (orange). GNRA tetraloops are the most frequently found loop 2 class of natural hammerheads (Figure 4A–D). (C) Superposition of the UGAAGUG CChMVd(–) heptaloop 1 with the Schistosoma mansoni hammerhead U1C2C3A4:G5U6A7C8 internal loop 1 (12). In (A) and (C) the carbon atoms of the UGAAGUG heptaloop are blue-colored, whereas those of the two other loops are green and adjacent stem residues are grey. Superpositions (A–C) show that RNA loops with different sequence and topology, including those that do not match the conserved sequence motifs shown in Figure 4E, adopt conserved structures containing key 3D features: an exposed U1 and an extrahelical pyrimidine at the 5′ side and 3′ side of the major groove of loop 1, respectively, and an unpaired A at the 3′ side of loop 2.
Loop 2
In the CChMVd (+) hammerhead, loop 2 is an AUGACA hexaloop, where L2.A6 is stacked on L2.A1, and L2.A4, L2.C5 and L2.A6 are partially opened toward the minor groove (Figure 3B and C); in the CChMVd (–) hammerhead, loop 2 is replaced by a GU:CGCACA internal loop. An analysis of the 27 natural hammerheads reveals that 22 match a conserved purine-rich RNnA loop 2 motif, where the 5′ residue is a purine, the 3′ residue is A, and n is usually 2 or 4 in apical loops (Figure 4). The hexaloops contained in the sLTSV (–), Arabidopsis and cave cricket hammerheads are very similar in sequence to the AUGACA loop of CChMVd (+), but the most common loops 2 are GNRA and AAAA apical tetraloops (n = 2), which are present in 12 hammerheads (Figure 4). The structure of the AAAA tetraloop is unknown, but GNRA tetraloops form the most abundant family of RNA hairpin loops (28). Strikingly, there is high similarity between the three-dimensional structures of GNRA tetraloops and the CChMVd (+) AUGACA hexaloop, which superpose quite well (Figure 5B). In GNRA loops, the 2-amino and 2′-OH groups of L2.G1 are hydrogen-bonded to the L2.A4 phosphate and to the Hoogsteen edge of the preceding L2.R3, respectively. These interactions promote opening of L2.R3 and L2.A4 toward the minor groove, resembling the situation observed in the AUGACA hexaloop where the opening of L2.C5 and L2.A6 is accommodated with similar backbone angles and stabilized by hydrogen bonds between the L2.U2 2′OH sugar and the Hoogsteen edge of L2.A4. GNRA tetraloops commonly establish tertiary interactions through the minor groove, using the three exposed residues of their 3′-side (24,33,34). However, since interactions between hammerhead domains I and II occur through the major groove (12,13), a conformational change involving the loop 2 nucleotides is likely coupled to the loop–loop interaction (see below).
In summary, of the 27 natural hammerheads, 16 match both loop sequence motifs, seven match either loop 1 or loop 2 motif, three show partial matches and only the hammerhead of the (+) strand of avocado sunblotch viroid (ASBVd) does not show any match to either sequence motif (Figure 4). Although the sequence motifs are not always matched, sequence conservation is not the only requirement for function in the context of the interactions between the globular loop structures studied here, which are quite different from the interactions between linear strands predominantly stabilized by canonical base pairing. Loops with different sequence and length can superpose closely in space (Figure 5), and different nucleotides can give rise to equivalent noncanonical base-base interactions (35). Indeed, conserved three-dimensional structure rather than sequence has been shown to often determine RNA function (36). In this respect, L1.U1 of 23S rRNA UAA triloops—which are similar to loops 1 of peach latent mosaic viroid (PLMVd) hammerheads not matching the proposed sequence motifs (Figure 4C)—faces the major groove side of the loop (37), whereas the last A of UGAGAU loops—similar to loops 2 of the same hammerheads—is turned toward the minor groove, exposed and highly mobile (38). Likewise, the internal loops of the Schistosoma, ASBVd (–) and CarSV (+) hammerheads do not match the loop 1 motif either (Figure 4D). However, in the X-ray structure of the Schistosoma hammerhead the backbone of internal loop 1 forms an elbow at the IL1.C8 pyrimidine nucleotide (shown in green in Figure 4D). This residue is extrahelical and protrudes from the major groove of domain I into the CAAAUA loop 2, where the 3′ A is also turned toward the minor groove (12). A superposition of domains I of the Schistosoma and CChMVd (−) hammerheads reveals that, despite the sequence, length and topology differences between the two loops, the extrahelical CChMVd L1.U6 and Schistosoma IL1.C8 residues are close in space and induce similar backbone kinks (Figure 5C). Furthermore, the CChMVd (+) and (–) hammerheads, both matching the proposed sequence motifs, share with the atypical Schistosoma hammerhead a similar tertiary interaction motif embedded within their loop–loop complexes (see below).
A loop–loop interaction motif shared by most natural hammerheads
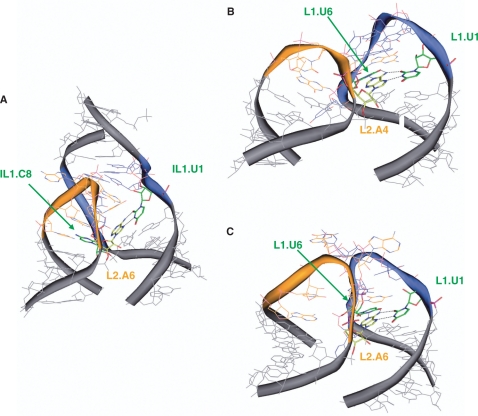
The structures of CChMVd domains I− and II+ (Figure 3) and NMR analyses of domains I+ and II− (Supplementary Figures S1 and S2), together with the effects of site-directed mutagenesis (Table 2), sequence and structure conservation patterns of natural hammerheads (Figures 4 and 5), and crystal structures of the Schistosoma (12) and sTRSV (+) (13) hammerheads, indicate that the contacts between loops 1 and 2 of most natural hammerheads take place across the major groove, and that they present two distinctive features relevant for hammerhead function: (i) turning of the 3′ A of loop 2 to establish a base-pair with the 5′ U of loop 1 and (ii) interaction of the extrahelical pyrimidine residue of loop 1 with the 3′-side of loop 2 (Figure 6).
Figure 6.
Loop 1–loop 2 complexes in natural hammerhead ribozymes. (A) and (B) are from the crystallographic structures of the Schistosoma (12) and sTRSV (+) hammerheads, respectively, and (C) is from a mutagenesis-supported model of the CChMVd (+) hammerhead (Materials and methods section). The views are from the minor groove of domain II and the major groove of domain I. The carbon atoms of loop 1 nucleotides are in blue, those of loop 2 in orange, and stem nucleotides in gray. The highlighted (green- and yellow-colored carbon atoms) contacts between IL1.U1, IL1.C8 and L2.A6 (A), L1.U1, L1.U6 and L2.A4 (B) and L1.U1, L1.U6, and L2.A6 (C) form the proposed tertiary interaction motif likely conserved in most natural hammerheads. Hydrogen atoms have been omitted for clarity.
A reverse Hoogsteen pair between the 3′ A of loop 2 and the 5′ U of loop 1 is observed in the X-ray structures of the Schistosoma and sTRSV (+) hammerheads (12,13) (Figure 6A and B), which contain highly dissimilar loops (Figure 4). This base pair is also supported by our mutagenesis analysis of the CChMVd (+) and (–) hammerheads (Table 2). We evaluated possible base-pair interactions between L1.U1 and L2.A6/IL2.A8, and L1.G2 and L2.C5/IL2.C7 by designing two double mutants for each polarity and comparing their effects with the corresponding single mutants. Partial restoration of the catalytic activity by L1(–).U1A/IL2(–).A8U indicated a direct base-pair interaction between L1.U1 and the 3′ A of loop 2. The other double mutants did not support Watson–Crick interactions between L1.U1 and L2.A6/IL2.A8 or L1.G2 and L2.C5/IL2.C7. Altogether, the effects of single and double mutants (Table 2) were consistent with a reverse Hoogsteen pair between L1.U1 and L2.A6/IL2.A8, and with intercalation of L2.C5/IL2.C7 between L1.U1 and L1.G2 to interact with the L1.U1 sugar (Figure 6C).
The observed base-pairing between the 3′ A of loop 2 and L1.U1 partly explains the sequence conservation patterns: these two nucleotides are strictly conserved in the CChMVd (+) and (–) hammerheads (Figure 1C), and are part of the UNmYN loop 1 and RNnA loop 2 motifs conserved in other natural hammerheads (Figure 4). Furthermore, the 5′-R:3′-A opposition at the bottom of loop 2 [or some alternative unstable pairing involving the 3′ A, like the C:A opposition in the Schistosoma and sRPV (+) hammerheads, see Figure 4], may facilitate the opening of the 3′ A to pair with L1.U1 across the major groove (Figure 6). In agreement with this view, mutations disrupting the L2.R1:L2.A6/IL2.A8 opposition, like IL2(–).A8U, or affecting the 5′ U of loop 1, such as L1(–).U1A, caused major detrimental effects on the catalytic activity of the CChMVd hammerheads (Table 2). In contrast, more conservative changes, like L2(+).A6G or L1(+).U1C, produced smaller (but still significant) effects, as did mutations affecting the C preceding L2.A6/IL2.A8 in loop 2 and the G following L1.U1 in loop 1 (Table 2).
In the sTRSV (+) hammerhead, loop 1 is a UGUGCUU heptaloop related to loops 1 of the CChMVd (+) and (–) hammerheads and matching the UNmYN motif, and loop 2 is a GUGA tetraloop matching the RNnA motif (Figure 4A). In the recently published X-ray structure of this hammerhead, the penultimate U of loop 1 (L1.U6) establishes a Watson–Crick pair with the 3′ A of loop 2, giving rise to a base triple formed by these 2 nt and L1.U1 (13) (Figure 6B). L1.U6 holds an extrahelical location in the undocked loop 1 of both CChMVd (+) and (–) hammerheads (Figure 3 and Supplementary Figure S1). Our mutagenesis data indicate that this nucleotide plays a major role on catalysis, supporting an interaction (Figure 6C) analogous to that observed in the sTRSV hammerhead X-ray structure: the L1.U6A mutant (which would impede pairing with the 3′ A of loop 2) almost abolished self-cleavage in the CChMVd hammerheads (Table 2) and completely blocked viroid replication in vivo (Table 3), with the L1(–).U6G mutant, and to a lesser extent L1(–).U6Δ and L1(–).U6C, also diminishing significantly the catalytic activity. The contacts involving the penultimate U of loop 1 explain the remaining sequence conservation patterns: this nucleotide is strictly conserved in natural variants of the CChMVd (+) hammerhead (Figure 1C), and is also part of the UNmYN loop 1 motif conserved in other natural hammerheads (Figure 4).
Since loops 1 of the sTRSV (+) and CChMVd (+) and (–) hammerheads are related, and the undocked conformations of the sTRSV (+) GUGA and CChMVd (+) AUGACA loops 2 are similar (Figure 5B), we can have a glimpse of loops 1 and 2 before and after the loop–loop docking interaction by comparing the isolated CChMVd loop structures (Figures 3 and 5) with the bound loop structures in the sTRSV (+) crystal (13) (Figure 6B) and in the CChMVd (+) mutagenesis-supported model (Figure 6C). The U1 and unpaired U6 residues of loop 1 face the major groove in the undocked loop structures (Figures 3, and 5A and C), and form a base triple across the major groove with the 3′A of loop 2 in the loop–loop complexes (Figure 6B and C). The last two residues of the undocked GNRA and AUGACA loops 2 are dynamic and/or partially opened towards the minor groove (Figures 3 and 5B). These two residues turn further toward the minor groove in the loop–loop complexes, so that the 3′A pairs with L1.U1, and the preceding base protrudes into the rather open loop 1 structure (Figure 3), intercalating between L1.U1 and L1.G2 (Figure 6).
A previous mutagenesis analysis of the sTRSV (+) hammerhead (11) provides further support for the structural conservation of loops 1 and 2 among natural hammerheads and for the importance of the proposed loop–loop interaction motif. In a series of 11 mutants substituting each of the residues of loops 1 and 2 with C, mutations affecting L1.U6, L1.U1 and L1.G4 of loop 1, and L2.A4, L2.G3 and L2.G1 of loop 2, produced the greatest reductions in self-cleavage activity, while the L1.C5, L1.U7 and L2.U2 mutations, compatible with the consensus structures of loops 1 and 2, increased the activity (11). In agreement with these findings, L1.U1, L1.U6, L2.G1, L2.G3 and L2.A4 would either be required for maintaining the appropriate undocked conformations of the loops (Figure 5A and B), or directly involved in key tertiary interactions (13) (Figure 6B).
In the Schistosoma hammerhead the consensus motifs are only partly matched, and the 3′ pyrimidine of internal loop 1 (IL1.C8) occupies the last rather than the penultimate position of the loop, as in the ASBVd (–) and CarSV (+) hammerheads (see green-colored residues in Figure 4D). Nevertheless, this pyrimidine is extrahelical and holds a position similar to L1.U6 in the undocked CChMVd (–) loop 1 (Figure 5C). In the crystal structure, IL1.C8 protrudes into loop 2, stacking on stem II in front of L2.C1 (12), and facilitating the displacement of the 3′ A of loop 2 to pair across the major groove with L1.U1 (Figure 6A).
Although induced-fit effects occur upon the establishment of the loop–loop tertiary contacts, a number of essential structural and dynamic features are present in the undocked domains of the CChMVd (+) and (–) hammerheads. They include the extrahelical location of L1.U6 and the availability of the Watson–Crick edge of L1.U1 in the major groove of loop 1, together with the partial opening and flexibility of the last 2 nt of loop 2 (Figure 3C). These structural signatures are conserved (Figures 4 and 5), thus highlighting the relevance of the undocked loop conformations in triggering the loop–loop contact motif that probably leads to the adoption of a functional conformation by the hammerhead. This motif is consistent with our mutagenesis analysis of the CChMVd (+) and (–) hammerheads (Table 2) and with a previous mutagenesis study of the sTRSV (+) hammerhead (11); it has been observed in the X-ray structures of the Schistosoma (12) and sTRSV (+) (13) hammerheads, and is likely shared by a significant fraction of the natural hammerheads.
CONCLUSIONS
We have characterized two novel hairpin loop RNA structures, an UGAAGUG heptaloop and an AUGACA hexaloop that cap domains I and II of the hammerheads embedded in the CChMVd (–) and (+) strands, respectively. Analysis of these rather unusual structures has led us to identify loop 1 and 2 three-dimensional foldings that are presumably conserved in a large fraction of natural hammerheads: loop 1 contains an exposed U in its 5′-side and an extrahelical pyrimidine in its 3′-side, while loop 2 has an unpaired and opened A in its 3′-side. Mutations that disrupt the loop shapes or modify the identity of these nucleotides have strong effects on the catalytic activity of the ribozyme in vitro and on the infectivity of the viroid in vivo. The results presented here provide new structural insights on how natural hammerheads operate and may help to enhance loop–loop interactions in artificial trans-acting hammerheads designed against specific RNAs.
Coordinates
The coordinates of domains I− and II+ have been deposited in the Protein Data Bank (accession codes 2RO2 and 2RPK, respectively).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Educación y Ciencia of Spain (BFU2004-07274/BMC, SAF2007-60243 and Ramón y Cajal contract to J.G.); (BFU2005-06808/BMC and BFU2008-03154/BMC to R.F.); (Ramón y Cajal contract to M.D.P.); the CIPF (predoctoral fellowship to D.D.); the Human Frontier Science Organization (long-term fellowship to M.D.P.); the European Union (EU-NMR RII3-026145). Funding for open access charge: Ministerio de Educación y Ciencia of Spain.
Conflict of interest statement. None declared.
This article is dedicated to the memory of Ángel Ramírez Ortiz.
Supplementary Material
ACKNOWLEDGEMENTS
31P and Γ–HCNCH experiments were carried out at BMRZ, Frankfurt, Germany, with the help of C. Ritcher. We are grateful to L. Pérez and A. Ahuir for excellent technical assistance.
REFERENCES
- 1.Hutchins CJ, Rathjen PD, Forster AC, Symons RH. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986;14:3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 3.Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores R, Hernandez C, De la Peña M, Vera A, Daros JA. Hammerhead ribozyme structure and function in plant RNA replication. Methods Enzymol. 2001;341:540–552. doi: 10.1016/s0076-6879(01)41175-x. [DOI] [PubMed] [Google Scholar]
- 5.Scott WG. Ribozymes. Curr. Opin. Struct. Biol. 2007;17:280–286. doi: 10.1016/j.sbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Westhof E. A tale in molecular recognition: the hammerhead ribozyme. J. Mol. Recognit. 2007;20:1–3. doi: 10.1002/jmr.806. [DOI] [PubMed] [Google Scholar]
- 7.Uhlenbeck OC. A small catalytic oligoribonucleotide. Nature. 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- 8.Tafech A, Bassett T, Sparanese D, Lee CH. Destroying RNA as a therapeutic approach. Curr. Med. Chem. 2006;13:863–881. doi: 10.2174/092986706776361021. [DOI] [PubMed] [Google Scholar]
- 9.Drude I, Dombos V, Vauleon S, Muller S. Drugs made of RNA: development and application of engineered RNAs for gene therapy. Mini Rev. Med. Chem. 2007;7:912–931. doi: 10.2174/138955707781662609. [DOI] [PubMed] [Google Scholar]
- 10.De la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 12.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi YI, Martick M, Lares M, Kim R, Scott WG, Kim SH. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 2008;6:e234. doi: 10.1371/journal.pbio.0060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson JA, Uhlenbeck OC. When to believe what you see. Mol. Cell. 2006;23:447–450. doi: 10.1016/j.molcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Shepotinovskaya IV, Uhlenbeck OC. Catalytic diversity of extended hammerhead ribozymes. Biochemistry. 2008;47:7034–7042. doi: 10.1021/bi7025358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarro B, Flores R. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of self-cleaving viroids with hammerhead ribozymes. Proc. Natl Acad. Sci. USA. 1997;94:11262–11267. doi: 10.1073/pnas.94.21.11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De la Peña M, Flores R. An extra nucleotide in the consensus catalytic core of a viroid hammerhead ribozyme: implications for the design of more efficient ribozymes. J. Biol. Chem. 2001;276:34586–34593. doi: 10.1074/jbc.M103867200. [DOI] [PubMed] [Google Scholar]
- 18.Weiner MP, Costa GL, Schoettlin W, Cline J, Mathur E, Bauer JC. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene. 1994;151:119–123. doi: 10.1016/0378-1119(94)90641-6. [DOI] [PubMed] [Google Scholar]
- 19.Byrappa S, Gavin DK, Gupta KC. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 1995;5:404–407. doi: 10.1101/gr.5.4.404. [DOI] [PubMed] [Google Scholar]
- 20.Gago S, De la Peña M, Flores R. A kissing-loop interaction in a hammerhead viroid RNA critical for its in vitro folding and in vivo viability. RNA. 2005;11:1073–1083. doi: 10.1261/rna.2230605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stage-Zimmermann TK, Uhlenbeck OC. Hammerhead ribozyme kinetics. RNA. 1998;4:875–889. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price SR, Oubridge C, Varani G, Nagai K. Preparation of RNA-protein complexes for X-ray crystallography and NMR. In: Smith C, editor. RNA-Protein Interaction: Practical approach. New York: Oxford University Press; 1998. pp. 37–74. [Google Scholar]
- 23.Rinnenthal J, Richter C, Ferner J, Duchardt E, Schwalbe H. Quantitative Gamma-HCNCH: determination of the glycosidic torsion angle chi in RNA oligonucleotides from the analysis of CH dipolar cross-correlated relaxation by solution NMR spectroscopy. J. Biomol. NMR. 2007;39:17–29. doi: 10.1007/s10858-007-9167-5. [DOI] [PubMed] [Google Scholar]
- 24.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 25.Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Headd JJ, Richardson DC, Ham D, Hershkovits E, Williams LD, et al. RNA backbone: consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De la Peña M, Navarro B, Flores R. Mapping the molecular determinant of pathogenicity in a hammerhead viroid: a tetraloop within the in vivo branched RNA conformation. Proc. Natl Acad. Sci. USA. 1999;96:9960–9965. doi: 10.1073/pnas.96.17.9960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De la Peña M, Flores R. Chrysanthemum chlorotic mottle viroid RNA: dissection of the pathogenicity determinant and comparative fitness of symptomatic and non-symptomatic variants. J. Mol. Biol. 2002;321:411–421. doi: 10.1016/s0022-2836(02)00629-0. [DOI] [PubMed] [Google Scholar]
- 28.Klosterman PS, Tamura M, Holbrook SR, Brenner SE. SCOR: a structural classification of RNA database. Nucleic Acids Res. 2002;30:392–394. doi: 10.1093/nar/30.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Culyba M, Volkman H, Krugh TR. 2003. RCSB PDB accession codes 1HS1, 1HS3, 1HS4. [Google Scholar]
- 31.Zhang H, Ricart B, Huck M, Krugh TR. 2003. RCSB PDB accession code 1HS8. [Google Scholar]
- 32.Zhang H, Fountain MA, Krugh TR. Structural characterization of a six nucleotide RNA hairpin loop found in E. coli: r(UUAAGU) Biochemistry. 2001;40:9879–9886. doi: 10.1021/bi011226x. [DOI] [PubMed] [Google Scholar]
- 33.Oubridge C, Kuglstatter A, Jovine L, Nagai K. Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle. Mol. Cell. 2002;9:1251–1261. doi: 10.1016/s1097-2765(02)00530-0. [DOI] [PubMed] [Google Scholar]
- 34.Hainzl T, Huang S, Sauer-Eriksson AE. Structure of the SRP19 RNA complex and implications for signal recognition particle assembly. Nature. 2002;417:767–771. doi: 10.1038/nature00768. [DOI] [PubMed] [Google Scholar]
- 35.Leontis NB, Stombaugh J, Westhof E. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collier AJ, Gallego J, Klinck R, Cole PT, Harris S, Harrison GP, Aboul-ela F, Varani G, Walker S. A conserved RNA structure within the HCV IRES eIF3 binding site. Nat. Struct. Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- 37.Wimberly BT, Guymon R, McCutcheon JP, White SW, Ramakrishnan V. A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 38.Leeper TC, Martin MB, Kim H, Cox S, Semenchenko V, Schmidt FJ, Van Doren SR. Structure of the UGAGAU hexaloop that braces Bacillus RNase P for action. Nat. Struct. Biol. 2002;9:397–403. doi: 10.1038/nsb775. [DOI] [PubMed] [Google Scholar]
- 39.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.