Abstract
Genes co-expressed may be under similar promoter-based and/or position-based regulation. Although data on expression, position and function of human genes are available, their true integration still represents a challenge for computational biology, hampering the identification of regulatory mechanisms. We carried out an integrative analysis of genomic position, functional annotation and promoters of genes expressed in myeloid cells. Promoter analysis was conducted by a novel multi-step method for discovering putative regulatory elements, i.e. over-represented motifs, in a selected set of promoters, as compared with a background model. The combination of transcriptional, structural and functional data allowed the identification of sets of promoters pertaining to groups of genes co-expressed and co-localized in regions of the human genome. The application of motif discovery to 26 groups of genes co-expressed in myeloid cells differentiation and co-localized in the genome showed that there are more over-represented motifs in promoters of co-expressed and co-localized genes than in promoters of simply co-expressed genes (CEG). Motifs, which are similar to the binding sequences of known transcription factors, non-uniformly distributed along promoter sequences and/or occurring in highly co-expressed subset of genes were identified. Co-expressed and co-localized gene sets were grouped in two co-expressed genomic meta-regions, putatively representing functional domains of a high-level expression regulation.
INTRODUCTION
Co-expression is essential to sustain normal function of cells and tissues. Genes can be co-expressed because they are co-regulated, have similar promoters, share combinations of functional regulatory sequence motifs binding transcription factors (TF), and/or they are co-localized. Genes could be co-localized because they are close to each other on a linear chromosome, thus being under the influence of the same regulators (e.g. enhancers acting locally on a limited chromosomal region) and/or under the effect of local control, possibly based on specific chromatin modifications. Genes could be considered co-localized also because they are preferentially located in a given functional district of the three-dimensional interphase nucleus, i.e. chromosome territories, thus being exposed to a particularly concentrated mixture of regulatory proteins (1). On the other hand, part of co-expressed genes (CEG) are functionally related and tend to be under the control of similar gene circuits (2–5). Thus, the integrated study of co-expression, co-regulation, co-localization and functional similarity may help in understanding basic and general rules governing genomic expression and may allow identifying mechanisms and specific switches of expression regulation in considered biological processes (6,7).
Haematopoiesis is an ideal biological model for studying regulation of gene expression in cellular differentiation since it represents a plastic process where multipotent stem cells gradually limit their differentiation potential, generating different precursor cells that finally evolve in eight distinct types of terminally differentiated cells. Myelopoiesis is the part of haematopoiesis leading to differentiation of myeloid cell lineages (erythrocytes, megakaryocytes, granulocytes and mono/macrophages). In a recent study on myelopoiesis (8), the analysis of the correlations between expression patterns of genes, their biological roles and their physical position in the human genome led to the identification of (i) chromatin domains containing clusters of genes relevant for specific myeloid lineages and (ii) chromosomal regions with low transcriptional activity that partially overlap genomic clusters related to non-haematopoietic functions.
In this article, we address the study on gene expression regulation by integrating analyses performed at multiple levels. Specifically, the analysis of gene co-expression and co-localization is integrated with the analysis of promoter sequences.
The search for DNA motifs in gene promoters is a challenging problem that has been of longstanding interest in computational biology (9–11). Numerous pattern discovery programs, which are based on different algorithms and methodologies, have been proposed and coded in stand-alone or web applications [e.g. MEME (12), GibbsSampler (13), Weeder (14), WordSpy (15), COOP (16), MOST (17) and RSAT (18); for a critical review see Tompa et al. (19)].
We introduce a novel methodology for the identification of putatively relevant motifs in gene promoters. The method is composed of a cascade of non-standard analytical procedures, and it is conducted on top of a MySQL database organizing different levels of interconnected data. In particular, this approach allows:
Finding, in a selected set of promoter sequences, motifs over-represented as compared with a biologically meaningful background model.
Analyzing promoter sequences by regions. Since several functional motifs have strong position-related prevalence (20,21), accounting for positional distribution in promoter sequences could refine the search for biologically meaningful motifs.
Integrating motif over-representation with additional, biologically relevant properties of motifs, such as similarity with known-functional sequences.
The application of this computational approach to the analysis of expression regulation during myeloid cell differentiation allowed identifying a greater number of significantly over-represented motifs in the promoters of co-expressed and co-localized genes when compared with those of simply co-expressed genes. All results are stored in a dedicated website where details of promoter sequences analyses can be browsed, along with gene transcriptional and functional characteristics (http://compgen.bio.unipd.it/MoDi/).
MATERIALS AND METHODS
Myelopoiesis gene expression data
The myelopoiesis data set consists of gene expression data for 24 samples generated from 8 different types of myeloid cells. Specifically, RNA from CD34 + , haematopoietic stem/progenitor cells (HSC), myeloid precursors (myeloblasts, monoblasts, erythroblasts and megakaryoblasts) and terminally differentiated cells (monocytes, neutrophils and eosinophils) was analysed using Affymetrix GeneChip HG-U133A, as described in (8). Robust multi-array average (RMA) procedure was applied to raw signals (i.e. CEL files) in order to background adjust and normalize microarray intensities and to generate gene expression values. Raw data of 24 considered samples are publicly available as a GEO series (GSE12837). Probe–gene relationships were obtained using GeneAnnot-based custom Chip Definition Files with a total of 11 446 unique custom probesets (gahgu133a_1.1.1.cdf; www.xlab.unimo.it/GA_CDF/) (22). For each of eight cell types, RMA expression values in corresponding replicate samples were averaged, and then, the data matrix was standardized per gene, obtaining a vector of eight expression values for each gene.
Reference set of human genes: genomic localization and promoter sequences
A reference set of human genes was collected by selecting all EntrezGene human entries corresponding to trustable RefSeq nuclear genes with Known, Reviewed or Validated Status and excluding Mitochondria, Plasmids, Plastids and Pseudogenes. Each EntrezGene ID was matched to the corresponding mRNA and EST sequences of UCSC Genome Browser. The genomic region including all of these sequences was selected as the reference gene locus and used to predict the exact Transcription Start Site (TSS) position. Gene loci whose genomic position could not be unambiguously determined according to this procedure were discarded. Then, the promoter sequences were retrieved, each spanning from 1000 bp upstream to 100 bp downstream of the predicted TSS (−1000, +100). These sequences constituted the reference set of gene promoters (REFGP).
QT clustering of myelopoiesis expression data: groups of Co-Expressed Genes (CEG)
In order to identify sets of co-expressed human genes (CEG) along myelopoiesis, we first selected genes varying in tissue/cell type-dependent manner, and then quality threshold (QT) clustering was used to group CEG. The Shannon entropy (H) was adopted as measure of expression variability (23) and used to rank and filter genes. The genes selected as variably expressed in myeloid cells were then grouped by the similarity of expression profile. Spearman correlation was adopted as a similarity measure and cluster analysis was performed by the QT clustering (24) of TMEV software (http://www.tm4.org/mev.html). QT clustering was adopted since it allows setting a priori thresholds for cluster quality, such as minimum values for the correlation between gene pairs within the cluster and for the number of genes per cluster.
Local Correlation Score (LCS): Co-Expressed chromosomal Regions (CER)
We searched for chromosomal regions including CEG, which could represent functional domains of higher-level gene expression regulation. The search for co-expressed chromosomal regions (CER) was based on the Local Correlation Score (LCS), a statistic for local correlation of gene expression patterns. The significance of local enrichment in CEG was evaluated using locally adaptive procedure (LAP) (25). The correlation between expression patterns of genes localized in a region (LCS) was computed using a sliding window whose width was set equal to twice μid,c + 2σid,c, where μid,c and σid,c are, respectively, the mean and standard deviation of log-transformed intergenic distances, and are computed independently on each chromosome c. In details, for each gene contained in the gene expression data matrix and located at specific position j on chromosome c, Spearman correlation was computed pairwise for all of the neighbouring genes located in the window j ± nc, with nc equal to μid,c + 2σid,c. The LCS was defined as the median correlation among all of the pairwise correlation coefficients. If no gene was contained in the window except the central gene at position j, the LCS was set equal to zero. Then, the significance of positive or negative local peaks in LCS values was evaluated using LAP. LAP procedure consists of three main steps: (i) adoption of a statistic for each gene contained into the gene expression data matrix; (ii) adaptive bandwidth smoothing of the statistic after sorting the statistical scores according to the chromosomal position of the corresponding genes and (iii) application of a permutation test to identify chromosomal regions with significant positive or negative peaks of the selected statistic, with a q-value correction for multiple tests. The LAP procedure was applied to LCS statistic and allowed the identification of CER with significantly high (+CER) or significantly low (–CER) levels of local correlation among gene expression patterns. These genomic regions include groups of genes co-expressed and co-localized that were considered for subsequent analyses.
Inter Regional Correlation Score (IRCS): Co-expressed Chromosomal Meta-Regions (CEMR)
Distinct chromosome arms and chromatin domains may occupy discrete territories in the cell nucleus, whose topological characteristics are essential for gene regulation (26). Therefore, we searched for groups of CER showing similar expression patterns. Previously selected +CER were clustered according to the similarity of expression, quantified in terms of IRCS. IRCS between two regions CERA and CERB was defined as the median value of all pairwise Spearman correlations between the n genes of CERA and the m genes of CERB. QT clustering based on IRCS was used to group different CER into CEMR. These CEMR may be indicative of functional domains characterized by a high-level expression regulation.
Framework for motif discovery in promoters sequences
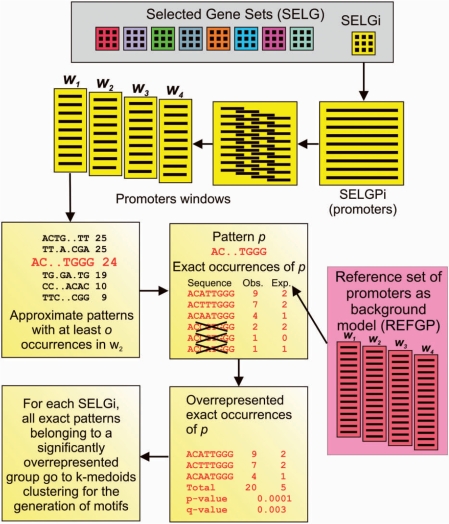
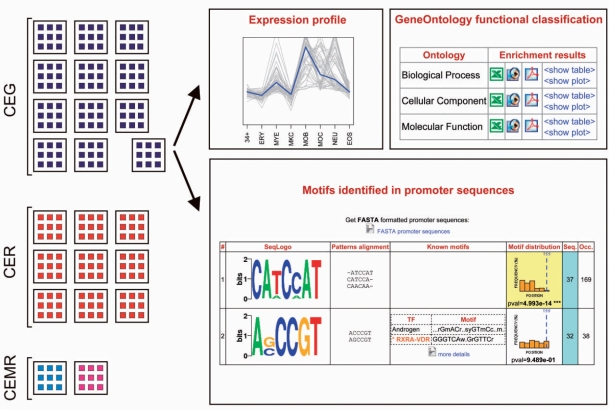
A computational framework was developed for identifying putative regulatory motifs in promoter sequences of selected groups of genes (SELGPi) as compared with REFGP. The framework comprises methodologies and software for completing a number of analysis steps, including (i) approximate patterns enumeration, (ii) significance scoring of over-representation, (iii) generation of motifs and (iv) their comparison with known regulatory sequences (Figure 1). Data obtained from different levels of analysis are recorded in a MySQL database and integrated with expression data.
Figure 1.
Scoring exact patterns over-representation in a selected set of gene promoters. For each SELGi, the corresponding SELGPi is divided in overlapping sequence windows. Approximate patterns occurring in at least 30% of the considered promoters are used to select the corresponding group of exact patterns. Then, within the groups of exact patterns, only patterns showing more occurrences than expected by chance: their total number of occurrences is used to compute over-representation P-value of the whole group of exact patterns. If the P-value is significant after Benjamini FDR correction, they are clustered together with other over-represented patterns that identify over-represented motifs corresponding to the specific SELGPi.
Groups of exact patterns over-represented in promoter windows
Full-length promoter sequences of the REFGP are divided into overlapping sequence windows, with window width and overlap defined by the user. For each window, sequence patterns of a given length are examined. The occurrences (in both strands) of each pattern and the sequences that contain each pattern are counted and stored in MySQL database tables, thus providing an estimate of expected frequencies in the REFGP.
Then, as shown in Figure 1, for each of the selected groups of gene promoters (SELGPi), a specific sequence window is considered, and approximate patterns occurring in at least s-sequences are identified with SPEXS (27). We considered ungapped patterns with two, not lateral, variable positions, in which any of the four nucleotides is allowed, e.g. ANATGNTCGT, N={A,C,T,G}). However, the evaluation of over-representation of the approximate patterns may produce many false positive and false negative results. In fact, approximate patterns can match both over-represented and under-represented exact patterns, thus biasing the over-representation statistic. Therefore, each approximate pattern is first associated to the group of corresponding exact patterns, which are subsequently filtered to select only those occurring in at least two different sequences and being more represented in SELGPi than expected by chance, according to the estimated frequency in the REFGP. Thus, each approximate pattern is associated to the group X containing a set of h exact patterns satisfying these conditions and the total occurrences n of these exact patterns in SELGPi are compared to the total expected frequencies, according to REFGP. Finally, the probability of observing more than n occurrences, for the group X within the SELGPi, is evaluated using the binomial distribution as described in (28). The false discovery rate (FDR) is then used to control false positive results due to multiple statistical tests (29).
In this way, for each SELGPi (and for each combination of pattern length and sequence window), a given number of significantly over-represented groups of exact patterns is identified.
k-medoids clustering of significant patterns generates motifs
For each SELGPi, all the exact patterns belonging to any over-represented group X are then compared and assembled into motifs with k-medoids clustering (30), using the TAMO package (31). In order to assess the pairwise distances between patterns, we adopted the end-space free alignment algorithm, whose peculiarity is avoiding penalization for mismatched overlapping prefixes and/or suffixes of aligned sequences. Since k-medoids clustering requires an a priori determined number of clusters k and is a heuristic process to find the optimal value of k (i.e. the minimum number of clusters producing a sufficiently good pattern partitioning), the clustering analysis was repeated with increasing values of k until partitioning optimality was less or equal to a user-defined threshold. Partitioning optimality (dk) was defined as the maximum of the distances between cluster medoids and individual cluster members. To stabilize results, for each incremental value of k, the k-medoids clustering was performed 100 times. Then the partitioning having the minimum dk was selected as the best among the 100 trials.
Finally, to obtain a single over-represented motif corresponding to a cluster of over-represented patterns, all patterns within a cluster were multialigned using ClustalW (32) with a stringent gap open penalty (100) to avoid gaps in the resulting consensus sequence. A matrix of nucleotide frequencies in motif positions was computed from patterns alignment result and visualized with a sequence logo. Over-represented motifs occurring in <30% of SELGPi were not included in final results.
Comparison with known TFBS
A set of known binding sequences for human transcription factors (TFs) was collected from JASPAR and TESS databases. In total 142 sequence motifs or variant of motifs, each known to be recognized by a TF were collected. This group also included motifs corresponding to 21 TFs for which a specific involvement in myeloid cell differentiation is well known (Supplementary Data file 1). For each TF, all available binding sequences were grouped with k-medoids clustering, as above described, to generate one motif and the corresponding matrix of nucleotide frequencies.
Significantly over-represented motifs were compared with known-transcription factor binding sequences (TFBS) using the ‘scan’ function of the TAMO library (31) to evaluate the similarity of the corresponding groups of sequences. Two motifs were considered similar if the match among their corresponding sets of sequences exceeds 70% of the best possible score that could be obtained, given the number and length of sequences.
The scripts constituting our motif discovery framework are freely available at the URL http://compgen.bio.unipd.it/MoDi/ along with a copy of the promoter database used for the analyses and associated documentation.
Uniformity analysis of motifs distribution along promoter sequences
Uniformity analysis was performed to identify motifs with occurrences non-homogeneously distributed along promoter sequences. The occurrences of each motif (Mi) in each promoter sequence of the SELGPi are compared with uniform distribution: Chi-squared test is used to evaluate the significance of the differences between the observed and the expected occurrences of Mi, in a set of non-overlapping windows of promoter sequences, as described in (21).
RESULTS
Datasets
Myelopoiesis gene expression data
As detailed in Materials and methods section, the gene expression dataset was contained in a data matrix with 11 446 genes/custom probesets and 24 samples for eight different cell types of the human myeloid lineage (Supplementary Data file 2: RMA gene expression data matrix).
Reference set of human genes: promoter, position
A reference set of human genes was collected, as the complete set of EntrezGeneIDs corresponding to trustable nuclear genes. For 15 138 of these genes, the genomic position was precisely defined and promoter sequences retrieved. Thus, 15 138 promoter sequences, each encompassing 1000 bp upstream and 100 bp downstream of the predicted gene TSS (–1000, +100), constituted the REFGP (freely available at the URL http://compgen.bio.unipd.it/MoDi/).
Integrated dataset
The intersection of 11 446 genes/probesets of the expression data matrix with the reference set of 15 138 gene/promoters comprised 9716 genes for which genomic localization, promoter sequence, and expression data in myeloid cells were available. For each gene in the integrated dataset, Gene Ontology functional annotations were retrieved from the EntrezGene database.
Sets of CEG in myeloid cells
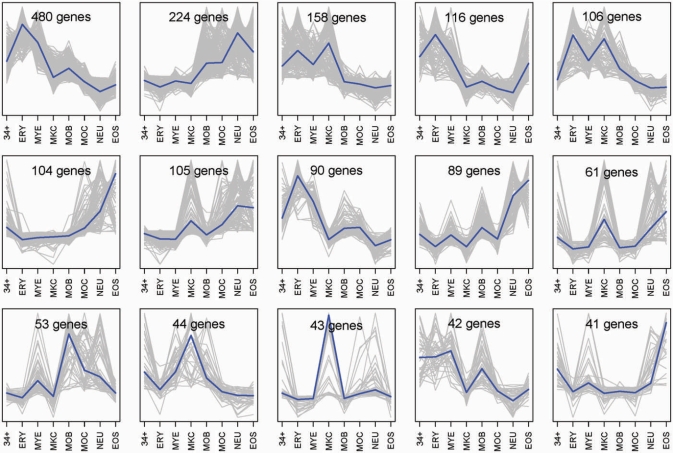
Sets of genes showing similar expression patterns constitute the first most intuitive candidates for sharing regulatory motifs. Genes with variable expression were selected and grouped according to their expression patterns into sets of CEG (Figure 2, yellow panel, and Figure 3). A set of 2796 (29%) genes with highly to moderately variable expression were selected (Shannon entropy, H ⩽ 2.8) and then grouped by similarity of expression using QT clustering. Setting maximum cluster diameter to 0.25 (minimum correlation of 0.75) with at least 15 genes per cluster, we obtained 44 gene clusters, including a total of 2455 genes. Each cluster represents a group of human genes co-expressed during myelopoiesis (CEG) (Supplementary Data file 3: sets of human genes co-expressed during myelopoiesis). In Figure 3, expression plots of 15 CEG sets with at least 40 genes per set are reported, whereas all plots are available in the Supplementary Data file 3. Functional GO terms enrichment was tested on each considered CEG detecting significantly enriched terms (hypergeometric test with a P-value ⩽ 0.05 and at least 10% of geneset genes, or five genes, in each category). Results are available online (http://compgen.bio.unipd.it/MoDi/).
Figure 2.
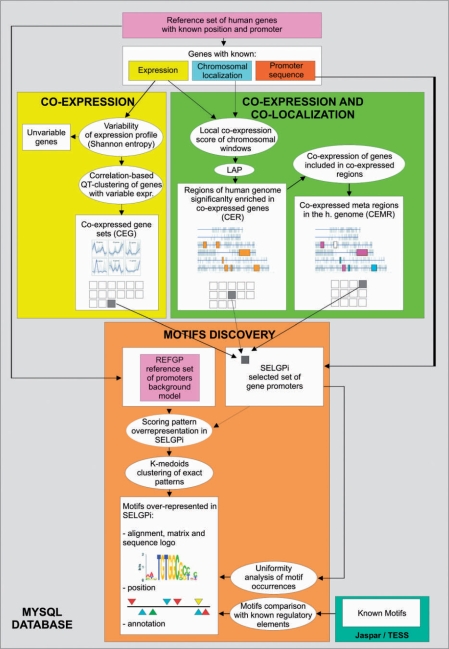
Experimental schema. Genomic databases with information on gene localization and sequences are used to generate the database of reference promoter sequences. Information on genes including expression data, chromosomal localization and promoter sequences are then taken into account by the integrated analytical framework. CEG sets are selected by the mere analysis of expression data. Then, expression data and genomic information are combined to select co-expressed and co-localized genes, thus identifying CER, which are subsequently grouped in CEMR, according to the similarity of the associated expression patterns. All of these set of genes are then used to select the corresponding sets of SELGPi, which are analysed with our motif discovery procedure. The motif discovery analysis combines the use of approximate patterns for grouping of underlying exact patterns, binomial distribution with FDR correction, in order to select over-represented exact patterns with an adequate statistical significance. Then, clustering of over-represented patterns leads to the identification of significantly over-represented motifs. Posterior analyses of over-represented motifs allow their further annotations with likely biologically relevant characteristics, including their non-uniform distribution along promoters, their occurrences in SELGPi, their matches with known TFBS and their occurrence in a subset of highly correlated genes.
Figure 3.
Expression plots of 15 co-expressed gene sets with at least 40 genes per set. QT clustering analysis allowed identifying 44 groups of highly correlated genes: CEG sets. The plot shown in grey expression patterns corresponds to the 15 CEG including more than 40 genes; the blue-bold line represents the median of the expression profiles in each gene set. (34+: CD34+ HSC; ERY: Erythroblasts; MYE: Myeloblasts; MKC: Megakaryoblasts; MOB: Monoblasts; MOC: Monocytes; NEU: Neutrophils; EOS: Eosinophils).
CER, sets of neighbouring genes similarly expressed during myelopoiesis
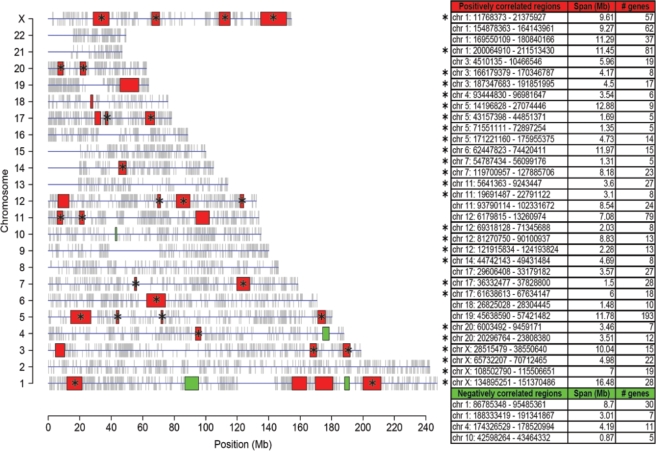
A number of evidences support the existence of mechanisms for positional regulation of gene expression, influencing transcription within specific chromosomal regions (7). This high level of gene expression regulation was taken into account as well, and we looked for CER along myelopoiesis (Figure 2, green panel). The analysis of CER was carried out using the LCS statistic. LCS is computed for each gene position, considering the correlation with neighbouring genes within a specific window, as described in Materials and methods section. Window width was selected independently for each chromosome, taking into account the different gene density of chromosomes. The average window width was 3.66 Mb, with values ranging from 0.72 Mb (chr 19) to 5.69 Mb (chr 13), thus including on average 5.7 genes per window. Then, by applying LAP to LCS statistic (q-value ⩽ 0.01), we identified chromosomal regions, including at least five genes per region, with significantly high (positively correlated regions, +CER) or significantly low correlation among gene expression patterns (negatively correlated regions, –CER). We identified 34 +CER, including a total of 922 genes covering 211.84 Mb (7% of the human genome), and 4 –CER, with a total of 53 genes covering 16.77 Mb (0.54% of the human genome) (Figure 4).
Figure 4.
Genomic positions of co-expressed regions in the human chromosomes. The analysis of CER was carried out using the LCS statistic, with a sliding window approach applied to the human genome. Regions harbouring genes with highly positive and negative correlation of expression in myelopoiesis are shown in red and green, respectively. The table on the right reports details on CER localization, span, and the number of genes from the region, which is present in the gene expression data matrix. An asterisk is used to mark positively correlated CER, which were selected for subsequent analyses, including the CEMR and motif discovery analyses.
It could be noticed that the number and the span of negatively correlated regions are considerably lower than those of positively correlated even if the correlation coefficients between genes are symmetrically distributed around zero, within the genomic windows used for computing LCS (Supplementary Data file 4, panel A). Therefore, we also investigated the relationship between the physical distance and the correlation of adjacent genes. This analysis showed an apparent inverse correlation between the distance of adjacent gene pairs and the correlation coefficient of corresponding expression patterns. Positively correlated genes tend to be closer to each other, whereas negatively correlated genes tend to be separated by larger intergenic regions (Supplementary Data file 4, panel B).
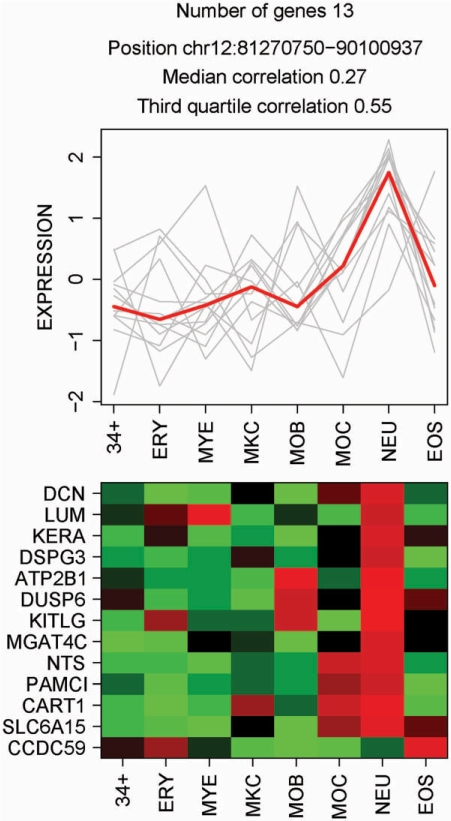
For the subsequent analyses, we focused only on positively correlated regions. Since the widths of +CER (6.23 Mb in average, min 1.31 Mb, max 16.48 Mb) exceed those of the original windows used for LCS computation, we verified the actual level of correlation between genes included into +CER. The distribution of pairwise Spearman correlations between genes of each +CER was evaluated and 26 CER with high correlation between corresponding genes (third quartile of pairwise correlations ⩾0.5) were selected for further analyses, as reliable groups of co-localized genes and co-expressed genes. Figure 5 reports the expression plot and heatmap, with genes ordered by genomic position, for a +CER including 13 genes localized in chromosome 12 (81 270 750 − 90 100 937); plots and heatmaps for the complete set of 26 +CER are in Supplementary Data file 5. The information about CER is also available using distributed annotation system (DAS) (33) (http://compgen.bio.unipd.it/Annotations/das/). Functional GO terms enrichment of CER was conducted as described for CEG.
Figure 5.
Example of a co-expressed region. Expression plot and heatmap, with genes ordered by genomic position, describing a region of chromosome 12 (81 270 750 − 90 100 937) harbouring 13 co-expressed genes. The red-bold line represents the median of expression profiles in the gene set. (34+: CD34+HSC; ERY: Erythroblasts; MYE: Myeloblasts; MKC: Megakaryoblasts; MOB: Monoblasts; MOC: Monocytes; NEU: Neutrophils; EOS: Eosinophils).
CEMR
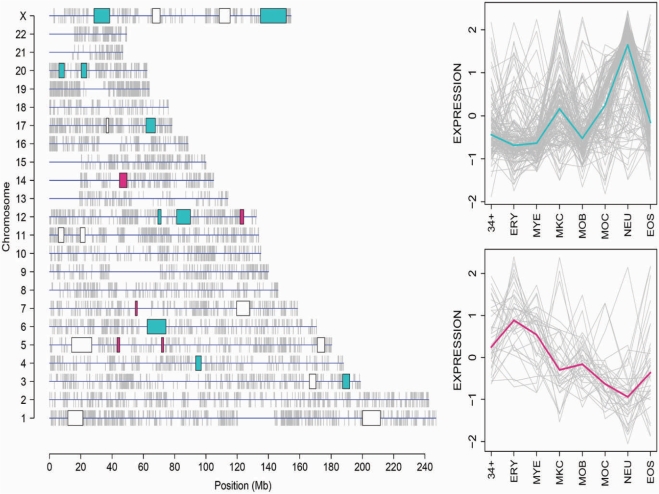
We reasoned that the number of possible variants of gene expression profiles, calculated on eight different cell types, is expected to be limited and that it would be possible to find similarity among pairs or groups of profiles corresponding to different +CER. Moreover, since distinct chromosome portions may occupy discrete territories in the cell nucleus (26,34), +CER with similar expression profiles might constitute chromatin domains with a specific functional role and a peculiar localization within the nucleus (CEMR; Figure 2, green panel). Therefore, as detailed in Materials and methods section, IRCS as similarity measure and QT clustering (with maximum distance set to 0.7) were used for grouping +CER into meta-regions (CEMR). Fifteen out of 26 selected +CER were grouped into two CEMR: one CEMR includes 10 +CER, distributed in seven different chromosomes, whereas the other is composed of five +CER located in four different chromosomes. The remaining 11 +CER cannot be grouped into CEMR according to the selected thresholds. The genes belonging to each CEMR clearly show a specific expression profile and have median correlation among them ⩾0.4 (Figure 6). Figure 6 reports the position of the 26 original +CER in human chromosomes. Magenta and light blue colours indicate +CER belonging to the two selected CEMR, for which the expression profiles are also given, whereas white blocks represent +CER, which cannot be grouped into CEMR. CEMR information is available as DAS annotation (33). Functional GO terms enrichment of CEMR was conducted, as described for CEG.
Figure 6.
Genomic localization of CEMR. Genomic positions of 26 original positively correlated CER in the human chromosomes and their grouping into CEMR are shown. The two CEMR are marked in magenta and light blue colours both in the chromosomes plot on the left and in the expression profiles on the right. White blocks on the chromosomes plot on the left represent CER which can't be grouped into CEMR according to the selected thresholds used for the analysis. (34+: CD34+ HSC; ERY: Erythroblasts; MYE: Myeloblasts; MKC: Megakaryoblasts; MOB: Monoblasts; MOC: Monocytes; NEU: Neutrophils; EOS: Eosinophils).
Identification of motifs over-represented in promoters of CEG sets, CER or CEMR, with putative regulatory role
As above described, 44 CEG sets were identified by classical analysis of gene expression data (see also Supplementary Data file 3 and Figure 3), whereas integrated analysis of gene expression and chromosomal localization allowed identifying 26 sets of genes co-expressed and co-localized (CER), and two sets of genes included in CEMR of the human genome (CEMR). For each of these gene sets, the corresponding group of gene promoters was considered and analysed to discover significantly over-represented motifs in SELGPi, as compared with a large group of 15 138 promoters (REFGP, being the background model), in order to identify putative regulatory elements (Figure 2).
Each selected set of genes is related to a specific expression pattern and likely to a specific biological role in a given differentiation context. Thus, significant motifs identified for each considered gene set are expected to play a functional role in a specific program of cellular differentiation.
Incidentally, since each considered promoter sequence was defined separately, we checked for overlapping promoter sequences belonging to gene pairs included in the same gene set: only 0.17% of considered sequences overlap in 13 (mostly very small) regions. Among all the considered genes, only two promoters, belonging to genes included in the same gene set, show an overlap of a number of nucleotides close to 1100, which is the length of considered promoter sequences: this is a pair of divergent genes with a unique bidirectional promoter (PDCD10, programmed cell death 10 and SERPINI1, neuroserpin precursor). This finding is in accordance with previous data on co-expression of genes with bidirectional promoters: PDCD10 and SERPINI1 were included in the same set of CEG (35).
From each considered 1100 bp promoter, five sequence windows of 300 bp in width and overlapping each other 100 bp, were extracted. For each sequence window, we selected approximate patterns of six and eight nucleotides, occurring in at least 30% of promoter sequences in the SELGPi. Since a previous systematic survey of known regulatory sites and motifs, available in TRANSFAC database, enlightened the over-representation of even-length functional motifs (16), we focused on even-length patterns as well. Among exact sequences matched by a given approximate pattern, a group of exact patterns was selected as over-represented (with FDR set to 0.05) in the specific SELGPi sequence window, as detailed in Materials and methods section. Thus, for each of the considered windows and for each pattern length, groups of over-represented exact patterns were identified, and subsequently clustered to obtain over-represented sequence motifs, each of them defined by a motif consensus sequence (represented as sequence logo) and a matrix of nucleotide frequencies in motif positions. Each detected motif is associated to the following attributes (Figure 7, and dedicated website: http://compgen.bio.unipd.it/MoDi/): (i) matrix of nucleotides frequencies in motif positions and sequence logo; (ii) alignment of patterns belonging to the motif; (iii) similarity with known TFBS and possible involvement in myelopoiesis; (iv) motif distribution along promoter sequences and uniformity test P-value; (v) number of sequences in which the motifs was found and total number of occurrences; and (vi) occurrence in subgroup of promoters belonging to highly co-expressed genes.
Figure 7.
Results of motif discovery analysis. The results of motif discovery analysis on promoters of each selected set of genes are integrated with gene expression data, functional information about genes, and more details concerning motifs characteristics in a dedicated website (http://compgen.bio.unipd.it/MoDi/). The dedicated website allows simultaneous exploration of information concerning the selected sets of genes and the over-represented motifs.
A total of 5325 significantly over-represented motifs were identified in 59 out of the 72 considered groups of gene promoters (Table 1).
Table 1.
Statistics on over-represented motifs
| Genes | Motifs |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All | Similar to known TFBS |
Similar to myeloid TFBS |
Non-uniform (NU) |
Occurring in highly correlated subgroup of genes (HC) |
NU and HC |
|||||||
| N | N | N | Percentage | N | Percentage | N | Percentage | N | Percentage | N | Percentage | |
| Gene sets (59 out of 72) | ||||||||||||
| Total | 1532.0 | 5325.0 | 1009.0 | 187.0 | 1772.0 | 1881.0 | 578.0 | |||||
| Average | 26.0 | 90.3 | 17.1 | 18.9 | 3.2 | 3.5 | 30.0 | 33.3 | 31.9 | 35.3 | 9.8 | 10.9 |
| CEG (32 out of 44) | ||||||||||||
| Total | 967.0 | 1943.0 | 367.0 | 70.0 | 651.0 | 645.0 | 210.0 | |||||
| Average | 30.2 | 60.7 | 11.5 | 18.9 | 2.2 | 3.6 | 20.3 | 33.5 | 20.2 | 33.2 | 6.6 | 10.8 |
| CER (25 out of 26) | ||||||||||||
| Total | 390.0 | 3369.0 | 637.0 | 117.0 | 1109.0 | 1227.0 | 359.0 | |||||
| Average | 15.6 | 134.8 | 49.0 | 36.4 | 4.7 | 3.5 | 85.3 | 63.3 | 94.4 | 70.0 | 27.6 | 20.5 |
| CEMR (2 out of 2) | ||||||||||||
| Total | 175.0 | 13.0 | 5.0 | 0.0 | 12.0 | 9.0 | 9.0 | |||||
| Average | 87.5 | 6.5 | 2.5 | 38.5 | 0.0 | 0.0 | 6.0 | 92.3 | 4.5 | 69.2 | 4.5 | 69.2 |
The table reports information on the over-represented motifs that where found in the whole set of selected gene sets and in the three distinct types of gene sets (CEG, CER and CEMR). Reported mean values are calculated only considering those gene sets with over-represented motifs: the number of gene sets with over-represented motifs is shown in the first column, together with the total number of gene sets for each group. Then for each group of gene sets, statistics on the total number of genes are shown along with statistics on the total number of over-represented motifs (All), as well as with statistics on the number of over-represented motifs matching specific characteristics: including motifs matching known TFBS (known TFBS), including motifs matching known TFBS with a known role in myelopoiesis, motifs with non-uniform distribution along the promoters (non-uniform), motifs occurring in a subgroup of highly correlated genes and motifs satisfying the latter two characteristics.
One-third of discovered motifs are non-uniformly distributed along promoters. Since different evidences indicate non-uniform distribution of predicted motifs as a feature supporting their functional role, spatial distribution of over-represented motifs along promoters was also considered. Motifs with occurrences non-homogeneously distributed along promoter sequences were identified. Thus, each motif is associated to a bar graph representing the number of motif occurrences in six promoter regions and to a P-value from uniformity test. About one-third of discovered motifs (1772) resulted to be significantly non-uniformly distributed along the promoter sequence, with P-value ⩽ 0.05.
Presence of specific motifs in promoters may have a stronger effect on genes co-expression
As previously seen, each motif was identified because over-represented in a specific set of gene promoters and occurring in at least 30% of them. Expression patterns of genes whose promoters actually contain a specific motif were compared computing pairwise correlation coefficients. In this way, we were able to detect those motifs that occur in genes actually having correlations higher than the original gene set to which they belong (comparing the third quartiles of pairwise correlations), and that are supposed to be enriched in sequence elements truly functional in controlling gene expression. Thirty-five percent of discovered motifs (1881) fall in this category.
In addition, 11% of discovered motifs (578 in total) were found to have both properties of being non-uniformly distributed and occurring in highly correlated subgroups of genes. This, numerically manageable, fraction of discovered motifs may be envisaged as the group of best candidates for being functional regulatory elements.
Functional role in myeloid differentiation of discovered motifs is supported by previous knowledge
Motif-describing matrices were compared with those describing 142 known motifs (representing sequence elements binding known transcription factors, 29 of which correspond to 21 TF that are specifically involved in myeloid cell differentiation). Among the 5325 identified motifs, 19% (1009) are similar to at least one of the 142 known TFBS, retrieved from public databases and 187 of them are similar to binding sites for 21 selected TF relevant for myelopoiesis. On the other hand, 118 of the 142 known TFBS (83%), and 17 of the 21 binding sites of TF known to play a role in myeloid cells differentiation (81%) have a match with significantly over-represented motifs.
Comparison of motifs discovery results in CEG, CER, and CEMR gene promoters
In Supplementary Data file 6, the number of discovered motifs in CEG, CER and CEMR is shown as a bar plot. On average, there are 60.7 motifs in CEG, 134.8 in CER and 6.5 in CEMR sets. Apparently, more motifs are found in the promoters of genes co-expressed and co-localized (CER) than in groups of simply co-expressed genes (CEG). This observation also applies to subsets of motifs with biologically meaningful characteristics: i.e. those non-uniformly distributed, those occurring in highly correlated subgroups of genes, and those matching known TFBS. The number of motifs in the two considered CEMR is the lowest for all motif types.
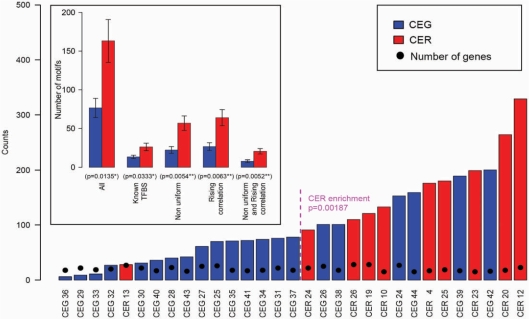
In order to assess the significance of the difference between the number of over-represented motifs in CEG and CER, we considered a subset of CEG and CER with similar number of genes, so as to exclude any possible bias related to the number of genes in each gene set because a non-negligible difference exists between the number of genes in CEG (mean 30.2) and CER (mean 15.6). Thus, we focused on 21 CEG and 10 CER (ranging from 15 to 29 genes each) and, after sorting gene sets according to the number of over-represented motifs, we found a significant enrichment in CER among the gene sets with higher number of motifs (9 CER out of the 15 gene sets, P-value 0.00187; Figure 8, main panel). Moreover, the number of motifs identified in CEG and CER is significantly different also considering t-tests results: P-values are significant at α = 0.05 both considering all motifs and considering subsets of motifs with specific biological meaningful characteristics (Figure 8, small panel).
Figure 8.
Comparison of the number of significant motifs found in CEG and CER. The number of significantly over-represented motifs within promoters of 21 CEG (blue bars) and 10 CER (red bars) of similar cardinality (from 15 to 29 genes each, as indicated by black points in the main panel) was compared. The significance of the difference was tested with t-test by considering all motifs and subgroup of motifs satisfying different characteristics (small panel). In addition, in the main panel, the selected sets of genes are sorted according to the number of over-represented motifs discovered in each gene set. The relative enrichment in CER (9 out of 15), in the first half of gene sets with higher number of motifs, is significant according to hypergeometric test (P-value 0.00187).
Motif discovery results in a pathways-oriented view
KEGG (http://www.genome.ad.jp/kegg) and Biocarta (http://www.biocarta.com) databases provide pathway maps representing knowledge on molecular interactions and networks involved in metabolism as well as genetic or environmental regulation of cellular processes. Ingenuity pathway analysis (IPA; http://www.ingenuity.com) is a commercial software for modeling biological systems. We integrated results of motifs discovery in SELGPi (e.g. CEG sets) with biologically meaningful annotations on gene relationships such as KEGG and Biocarta information, and with IPA-supported charts of functional relationships. In particular, we considered gene sets associated to over-represented motifs possibly recognized by a TF known to have a role in myelopoiesis. These sets of genes, together with the putative regulator(s) (i.e. the known TF binding the discovered motif), were mapped to KEGG and Biocarta pathways, and analysed by IPA. This allowed finding interesting examples of regulators and regulated gene products, which are involved in specific pathways or regulatory circuits.
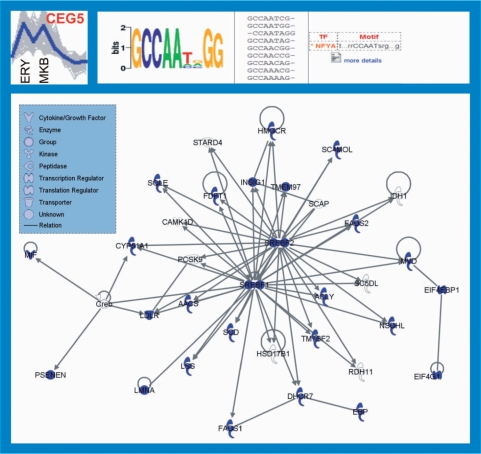
For instance, CEG5 genes are highly expressed in erythroblasts and megakaryoblasts, and at least in part regulated by NFYA, according to motif discovery results, which are known to be involved in myeloid cells differentiation and to be expressed in erythroid and myeloid progenitors. In addition, many genes included in CEG5 show different types of previously reported interactions, which can be enlightened using the IPA software (Figure 9); this software allows identifying network of relationships among selected sets of genes according to the literature reports. This interaction network between the genes included in CEG5, by means of IPA, constitutes a significant finding and confirms the existence of biologically meaningful relationships, in addition to the sharing of sequence motifs, among the identified genes.
Figure 9.
Expression profile, example of motif discovery results and network of interactions between genes included in CEG5. The top left panel reports expression plot for CEG5 genes, with marked expression peaks corresponding to erythroblasts and megakaryoblasts cell contexts. The top right panel contains a screenshot from the website with supplementary results (http://compgen.bio.unipd.it/MoDi/) showing one of the over-represented sequence motifs matching a known binding site for NFYA. In details, this panel reports the sequence logo of the discovered motif; the sequences are composed of the discovered motif and the known motif associated to NFYA binding site. The bottom panel represents the network of interactions obtained from IPA analysis on the CEG5 genes. Within the network, nodes represent gene products, whereas edges represent biologically meaningful interactions supported by literature findings. Shapes of different nodes represent different molecular functions, as detailed in the legend. Arcs (arrows) represent regulatory relationships (gene product A activates expression of gene B or regulates the activity of protein B), whereas edges represent protein–protein interactions.
CEG44 genes are highly expressed in megakaryoblasts, and a motif similar to GATA1 binding site is present in half of their promoters. Two of these genes are included into the KEGG pathway ‘Arachidonic acid metabolism: prostaglandin and leukotriene metabolism’ (Supplementary Data file 7, panel A). Platelets contain thromboxanes that are derived from arachidonic acid and are relevant for platelets function, including their aggregation and activation. The same gene set also includes pro-platelet basic protein (PPBP) that is involved in platelet production and other genes involved in other haematopoietic cells differentiation, such as Bruton agammaglobulinemia tyrosine kinase (BTK), that is also expressed in platelets, and monocyte to macrophage differentiation-associated (MMD).
Finally, CEG33 genes that are characterized by expression peaking in erythroblasts, and the putative regulators GATA1 and LMO2, identified according with motif discovery results, were mapped on Biocarta pathways. GATA1 and AHSP [also named erythroid-associated factor (ERAF)] are included in the gene set and are known to interact in the pathway of the Haemoglobin's Chaperone. This gene set with an erythroid-specific pattern of expression includes as well Glycophorin E, an erythroid antigen related to the M blood group and Adducin-2 (ADD2) that is involved in regulating erythrocytes’ precursors proliferation: the figure in panel B of Supplementary Data file 7 shows a simplified version of Haemoglobin's Chaperone chart derived from Biocarta. Furthermore, the gene set includes other genes involved in the differentiation of other haematopoietic cells such as PRKDC (protein kinase, DNA-activated, catalytic polypeptide) that is relevant for B cells differentiation and is also expressed in myeloid cells or DLK1 (delta-like 1 homolog) that is involved in T cells differentiation.
DISCUSSION
In the present work, we considered different levels of gene expression regulation in human myelopoiesis, integrating the study of promoter-based and position-related control of transcription, of ∼10 000 genes, in myeloid cells and identifying sequence elements in gene promoters with putative regulatory function along differentiation.
A novel methodology for the identification of putative regulatory elements in promoter sequences was developed, aiming at identifying motifs over-represented in a selected set of promoters, as compared with a background model built according to a reference set of promoters. Different lines of evidence support the fact that promoter regulatory elements are functional in a given biological context according to their position relative to the TSS. Tabach and colleagues (20) proved that positional-dependent TFBS tend to be located in the region close to and upstream the TSS. Thus, promoter sequences were divided in partially overlapping windows, which were considered individually for the selection of over-represented patterns.
The framework for the identification of regulatory motifs integrates a number of steps, including approximate patterns enumeration, calculation of exact patterns over-representation, and generation of motifs by clustering sequences of exact patterns. The first step of the analysis is the enumeration of approximate patterns found in the selected group of promoters. Biologically, functional binding sites are degenerated and the over-representation of exact patterns is a subtle signal, often too difficult to identify with statistical techniques. The adopted statistical measure that was described by Van Helden et al. (28) was based on the binomial distribution, allowing associating a P-value to the observed occurrences of patterns or motifs. We adopted the FDR to guarantee a global control over false positives. One improvement of our framework relies on the fact that the over-representation significance was calculated neither for a single exact pattern, nor for a simple approximate one. Instead, we considered the subgroup of exact patterns matching a given approximate one, capable of maximizing the significance of global over-representation of the group in the selected set of gene promoters, as compared with the background model. This approach was finally chosen considering that: (i) the over-representation signal of each exact pattern is subtle and strongly dependent from expected number of occurrences, in turn dependent from pattern length; (ii) the over-representation of a pattern including fully variable positions is not informative because it may match exact patterns both over- and under-represented, the latter obscuring likely biologically meaningful signals. In the last phase of the analytical pipeline, clustering of all exact patterns belonging to over-represented groups gives rise to over-represented motifs, which are eventually compared with known TFBS and further analysed. Thus, identified motifs are associated to different attributes, such as non-uniform distribution along promoters, accounting for position-dependent function, and occurrence in promoters of highly CEG, possibly indicating that the presence of the motifs is crucial for determining specific expression behaviour. Non-uniformly distributed motifs occurring in promoters of highly CEG are probably the most interesting candidates for a functional role along myelopoiesis.
A very large set of putative promoter sequences (∼15 000, i.e. a good representation of the whole set of human promoters) was used as background model for computing patterns over-representation. Using real promoter sequences as background model allows identifying motifs specifically associated to a given set of promoters (of genes sharing expression, positional or functional characteristics) avoiding motifs that are simply highly frequent in human promoter regions. This approach reduces the number of false positives or uninteresting results, i.e. repetitive sequence elements and structural elements constitutively present in most human promoters.
Following a classic assumption that ‘genes co-expression implies, at least in part, co-regulation’, 44 groups of promoters of highly CEG were identified and associated each to a specific expression pattern in myeloid cells. Then, combining expression and positional information, chromosomal regions with significantly high positive LCS were identified (CER), comprising ∼10% of the genes and 7% of the genome. These could represent functional domains of high-level gene expression regulation.
It is worthwhile noting that regions including genes with negative correlation of expression are fewer and smaller than positively correlated regions. Moreover, an apparent inverse correlation exists between the distance of adjacent gene pairs and the correlation coefficient of corresponding expression vectors. Confirming the previous reports (36), in the whole human genome, genes positively correlated tend to be close to each other, whereas negatively correlated genes tend to be separated by larger intergenic regions.
The results of the motifs discovery analyses are available in a dedicated website representing the first collection of putative regulatory motifs acting during myeloid cells differentiation. Expression profiles of genes belonging to a specific gene set are displayed together with information regarding gene annotation, gene-ontology, and promoter sequences. Motifs found in the selected gene set are shown together with all the information on attributes derived from the post-processing of motif discovery results. Results about CERs and CEMRs are also available as a resource based on the DAS, and can be accessed by querying the MyDas server. This represents an expandable framework in which additional results of novel analyses on extended myeloid cells datasets may be easily integrated.
Significantly, over-represented motifs were identified in 80% of considered gene sets, with 90 motifs per gene set in average. About 20% of over-represented motifs are similar to known TFBS, and all of the considered motifs binding TF, with a known role in myeloid cells differentiation, were found over-represented in at least one gene set. About one-third of identified motifs are non-uniformly distributed along promoters and another one-third is represented in promoters of genes with highly correlated expression profiles; the intersection of these categories accounts for 11% of all over-represented motifs.
Motifs discovery results detailed in the website could be profitably used to gain biological interpretation since the various characteristics of each motif are shown together with gene expression and function information. For example, focusing on CEGs, with at least one motif non-uniformly distributed and occurring in a subset of genes with high correlation, it could be noticed that in the promoters of CEG5 three over-represented motifs were found, all similar to the known binding site of nuclear transcription factor Y alpha (NFYA), which was previously reported to be involved in myeloid cells differentiation and to be expressed in erythroid and myeloid progenitors (37–39). The expression pattern of CEG5, peaking in erythroblasts, and megakaryoblasts is coherent with previous reports. In addition, the analysis of Gene Ontology functional classes, and IPA analysis also highlighted the existence of biologically meaningful relationships among CEG5 genes as well as the enrichment in biological process categories related to the synthesis of thromboxanes, and therefore, essential for platelets function. Furthermore, CEG11 shows the highest expression level in monoblasts, and is enriched in genes involved in immune, defence and inflammatory response, as inferred from Gene Ontology functional classification. Among its over-represented motifs that are as well non-uniformly distributed and occurring in highly CEG, we found a motif similar to the binding site for CEBPB, i.e. a well known transcription factor playing a role in the differentiation and functionality of mono/macrophages and also granulocytes (40–42). From another point of view, motifs discovery results can help identifying possible regulatory circuits in myeloid differentiation, involving specific transcription factors and/or combination of them. For example, GATA1 (GATA binding protein 1/globin transcription factor 1) has an established role in the differentiation of megakaryocytes, erythrocytes and also eosinophils (43,44). Motifs possibly recognized by GATA1 are present among over-represented motifs of CEG33 and CEG42, including genes with, respectively, erythroblasts- and eosinophils-specific expression patterns, as well as among motifs of CEG34 and CEG44, comprising genes with megakaryoblasts-specific expression patterns. Moreover, among these gene sets, CEG33 and CEG44 share predicted binding sequences for LMO2 (LIM domain only 2), a transcription factor with a role in haematopoiesis, with previously described interactions with GATA1 (45). In particular, it can interact with GATA1 and is involved in erythroid differentiation (43,46). It is worth noticing that CEG44 includes a limited number of genes, and therefore over-represented Gene Ontology functional classes cannot be identified with statistical significance. Nevertheless, the mapping of CEG44 genes on KEGG pathways and additional functional information allowed enlightening biologically meaningful relationships between the selected genes. Then, focusing on SPI1 (spleen focus forming virus (SFFV) proviral integration oncogene spi1) (43), which has a role in influencing myeloid cells differentiation towards granulocytes (neutrophils), monocytes and also eosinophils cell lineages, we found over-represented motifs corresponding to its known-binding sites in many CEG and CER gene sets. Among them, there are CEG20, CEG24 and CEG42, with an eosinophils-specific expression pattern, as well as different CER including genes with high expression in neutrophils (such as CER3, CER4, CER5, CER15, CER20, CER22 and CER24). Furthermore, these CER are enriched in genes involved in development and immune system related functional categories according to the analysis of Gene Ontology functional classes. In addition, motif discovery results were integrated with biologically relevant information concerning regulatory and metabolic pathways as well as with databases describing molecular interactions such as IPA. The integration of computational results and biological data allowed verifying that complex relationships can be enlightened in the selected gene sets as described in the Results section.
When observing the variety of expression profiles associated to the different CEG sets and to positive CER gene sets, it is evident that the CER profiles can be fitted to a smaller number of expression patterns, showing peculiar expression behaviours in differentiation lineages, and that their cross comparison allows the identification of only two groups of CER gene sets, i.e. the CEMR. These findings allow supposing that mechanisms controlling expression of specific chromosomal regions, which may involve epigenetic modifications, could be particularly important in specific differentiation lineages. The expression patterns of the two CEMR can indeed be related to cell contexts representing early and late differentiation stages, and gene co-expression in CER and CEMR gene sets might be related to different levels of epigenetic regulation.
Genes adjacent each other on a chromosome can be under the influence of the same, locally acting, regulators and/or under the effect of local epigenetic control, based on specific chromatin modifications. Moreover, additional levels of epigenetic regulation may act on genes located on a given functional district of the three-dimensional interphase nucleus (26,34). Indeed, the resultant gene expression derives from different layers of transcriptional regulation, mediated by epigenetic mechanisms and by specific combinations of transcription factors binding sequences, within gene promoters. In this view, we performed the comparative study of promoters of CEG, CER and CEMR with the final purposes of identifying putatively functional regulatory elements, involved in specific differentiation switches and lineage choices in myeloid cells, and formulating hypotheses on the relative role of genetic and epigenetic regulation of transcription during myelopoiesis.
More motifs were found in the groups of promoters of CER gene sets than those of promoters of CEG with comparable cardinality. However, since CERs share basically two expression profiles, we wondered if the total number of identified motifs in CER gene sets could have been overestimated and if the same motifs were actually found over-represented, by parallel analyses, in different CER gene sets. To assess whether the higher number of motifs found in CERs was due to a high number of shared motifs between CERs, we built a list of non-redundant motifs found in the two supersets containing all results of CERs and all CEGs by pairwise comparison of motifs IUPAC consensus sequences. In the two supersets, non-redundant motifs were 16% fewer than original motifs (1653 and 2754 non-redundant motifs were identified in CEG and CER, respectively) and the ratio between motifs found in CER and CEG gene sets remains unchanged if non-redundant motifs are considered. Thus, conversely, the number of non-redundant motifs found over-represented in CER gene promoters remains considerably higher than that found in CEG gene promoters. This result is in accordance with the fact that few significantly over-represented motifs were found in CEMR gene sets: the gene promoters of different CER sets, grouped by similarity of expression in a CEMR set, do not share numerous similar motifs.
The motifs discovery approach is characterized by the independent analysis of different promoter regions, and by the posterior analysis of distribution of over-represented motifs, accounting for the role of binding site position in determining its function. The identification of motifs found in subgroups of genes highly co-expressed and comparison of detected motifs with known TFBS is expected to increase the completeness of results. The statistical analysis adopted for identifying putative biologically relevant motifs was both stringent, adopting false discovery rate, and capable of identifying subtle over-representation signals. Indeed, the analysis is conducted on subgroups of exact patterns for which the significance of global over-representation is maximized in the selected set of gene promoters, as compared with the background model based on a numerous set of real gene promoter sequences.
In conclusion, the integrated analysis of co-expressed and/or co-localized sets of genes proved to be effective in enlightening biologically relevant results, including findings related to the differentiation of various myeloid lineages, which are coherent with previous knowledge of the considered, complex biological system. This work constitutes an important improvement in the methodologies for characterizing gene expression regulation of entire genomic regions biologically relevant for a specific process.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Padova (CPDA065788/06, CPDR074285/07); Fondazione CARIPARO (Progetti Eccellenza 2006); MIUR (2007CHSMEB_002 to S.F and S.Bo); Italian Association for Biology and Genetics (mobility fellowships program 2007 to F.F.); Consorzio Interuniversitario per le Biotecnologie (mobility fellowships program 2007 to F.F.). Funding for open access charge: Fondazione CARIPARO (Progetti Eccellenza 2006).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
This article is dedicated to the memory of our friend and colleague Prof. Stefano Ferrari (1951–2008).
REFERENCES
- 1.Kosak ST, Scalzo D, Alworth SV, Li F, Palmer S, Enver T, Lee JS, Groudine M. Coordinate gene regulation during hematopoiesis is related to genomic organization. PLoS. Biol. 2007;5:e309. doi: 10.1371/journal.pbio.0050309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caron H, van Schaik B, van der MM, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, Voute PA, et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 3.Lercher MJ, Urrutia AO, Hurst LD. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 2002;31:180–183. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
- 4.Vogel JH, von Heydebreck A, Purmann A, Sperling S. Chromosomal clustering of a human transcriptome reveals regulatory background. BMC Bioinformatics. 2005;6:230. doi: 10.1186/1471-2105-6-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purmann A, Toedling J, Schueler M, Carninci P, Lehrach H, Hayashizaki Y, Huber W, Sperling S. Genomic organization of transcriptomes in mammals: coregulation and cofunctionality. Genomics. 2007;89:580–587. doi: 10.1016/j.ygeno.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Coppe A, Danieli GA, Bortoluzzi S. REEF: searching REgionally Enriched Features in genomes. BMC Bioinformatics. 2006;7:453. doi: 10.1186/1471-2105-7-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics. 2008;91:243–248. doi: 10.1016/j.ygeno.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Ferrari F, Bortoluzzi S, Coppe A, Basso D, Bicciato S, Zini R, Gemelli C, Danieli GA, Ferrari S. Genomic expression during human myelopoiesis. BMC Genomics. 2007;8:264. doi: 10.1186/1471-2164-8-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 10.Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- 11.Pevzner PA, Sze SH. Combinatorial approaches to finding subtle signals in DNA sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:269–278. [PubMed] [Google Scholar]
- 12.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 13.Thompson W, Rouchka EC, Lawrence CE. Gibbs Recursive Sampler: finding transcription factor binding sites. Nucleic Acids Res. 2003;31:3580–3585. doi: 10.1093/nar/gkg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G, Yu T, Zhang W. WordSpy: identifying transcription factor binding motifs by building a dictionary and learning a grammar. Nucleic Acids Res. 2005;33:W412–W416. doi: 10.1093/nar/gki492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bortoluzzi S, Coppe A, Bisognin A, Pizzi C, Danieli GA. A multistep bioinformatic approach detects putative regulatory elements in gene promoters. BMC Bioinformatics. 2005;6:121. doi: 10.1186/1471-2105-6-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pizzi C, Bortoluzzi S, Bisognin A, Coppe A, Danieli GA. Detecting seeded motifs in DNA sequences. Nucleic Acids Res. 2005;33:e135. doi: 10.1093/nar/gni131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Brohee S, van Helden J. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 20.Tabach Y, Brosh R, Buganim Y, Reiner A, Zuk O, Yitzhaky A, Koudritsky M, Rotter V, Domany E. Wide-scale analysis of human functional transcription factor binding reveals a strong bias towards the transcription start site. PLoS. ONE. 2007;2:e807. doi: 10.1371/journal.pone.0000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casimiro AC, Vinga S, Freitas AT, Oliveira AL. An analysis of the positional distribution of DNA motifs in promoter regions and its biological relevance. BMC Bioinformatics. 2008;9:89. doi: 10.1186/1471-2105-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrari F, Bortoluzzi S, Coppe A, Sirota A, Safran M, Shmoish M, Ferrari S, Lancet D, Danieli GA, Bicciato S. Novel definition files for human GeneChips based on GeneAnnot. BMC Bioinformatics. 2007;8:446. doi: 10.1186/1471-2105-8-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert C.J., Jr Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heyer LJ, Kruglyak S, Yooseph S. Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callegaro A, Basso D, Bicciato S. A locally adaptive statistical procedure (LAP) to identify differentially expressed chromosomal regions. Bioinformatics. 2006;22:2658–2666. doi: 10.1093/bioinformatics/btl455. [DOI] [PubMed] [Google Scholar]
- 26.Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories – a functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Brazma A, Jonassen I, Vilo J, Ukkonen E. Predicting gene regulatory elements in silico on a genomic scale. Genome Res. 1998;8:1202–1215. doi: 10.1101/gr.8.11.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Helden J, Andre B, Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B – Methodol. 1995;57:289–300. [Google Scholar]
- 30.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning; DataMining, Inference and Prediction. New York: Springer; 2001. [Google Scholar]
- 31.Gordon DB, Nekludova L, McCallum S, Fraenkel E. TAMO: a flexible, object-oriented framework for analyzing transcriptional regulation using DNA-sequence motifs. Bioinformatics. 2005;21:3164–3165. doi: 10.1093/bioinformatics/bti481. [DOI] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature. 2007;445:379–781. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- 35.Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuoka Y, Inaoka H, Kohane IS. Inter-species differences of co-expression of neighboring genes in eukaryotic genomes. BMC Genomics. 2004;5:4. doi: 10.1186/1471-2164-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marziali G, Perrotti E, Ilari R, Coccia EM, Mantovani R, Testa U, Battistini A. The activity of the CCAAT-box binding factor NF-Y is modulated through the regulated expression of its A subunit during monocyte to macrophage differentiation: regulation of tissue-specific genes through a ubiquitous transcription factor. Blood. 1999;93:519–526. [PubMed] [Google Scholar]
- 38.Sjin RM, Krishnaraju K, Hoffman B, Liebermann DA. Transcriptional regulation of myeloid differentiation primary response (MyD) genes during myeloid differentiation is mediated by nuclear factor Y. Blood. 2002;100:80–88. doi: 10.1182/blood.v100.1.80. [DOI] [PubMed] [Google Scholar]
- 39.Miller JD, Stacy T, Liu PP, Speck NA. Core-binding factor beta (CBFbeta), but not CBFbeta-smooth muscle myosin heavy chain, rescues definitive hematopoiesis in CBFbeta-deficient embryonic stem cells. Blood. 2001;97:2248–2256. doi: 10.1182/blood.v97.8.2248. [DOI] [PubMed] [Google Scholar]
- 40.Pham TH, Langmann S, Schwarzfischer L, El Chartouni C, Lichtinger M, Klug M, Krause SW, Rehli M. CCAAT enhancer-binding protein beta regulates constitutive gene expression during late stages of monocyte to macrophage differentiation. J. Biol. Chem. 2007;282:21924–21933. doi: 10.1074/jbc.M611618200. [DOI] [PubMed] [Google Scholar]
- 41.Duprez E, Wagner K, Koch H, Tenen DG. C/EBPbeta: a major PML-RARA-responsive gene in retinoic acid-induced differentiation of APL cells. EMBO J. 2003;22:5806–5816. doi: 10.1093/emboj/cdg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nerlov C, McNagny KM, Doderlein G, Kowenz-Leutz E, Graf T. Distinct C/EBP functions are required for eosinophil lineage commitment and maturation. Genes Dev. 1998;12:2413–2423. doi: 10.1101/gad.12.15.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 44.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osada H, Grutz G, Axelson H, Forster A, Rabbitts TH. Association of erythroid transcription factors: complexes involving the LIM protein RBTN2 and the zinc-finger protein GATA1. Proc. Natl Acad. Sci. USA. 1995;92:9585–9589. doi: 10.1073/pnas.92.21.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perry C, Soreq H. Transcriptional regulation of erythropoiesis. Fine tuning of combinatorial multi-domain elements. Eur. J. Biochem. 2002;269:3607–3618. doi: 10.1046/j.1432-1033.2002.02999.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.