Abstract
The continued evolution of H9N2 and H5N1 viruses and their spread and re-emergence across Eurasia raise concern that prior H9N2 virus infection may limit the detection of subsequent H5N1 infection in gallinaceous poultry by attenuating the severity of disease. We show that H9N2 viruses isolated from Israeli turkeys during 2000–2004 were antigenically and genetically distinguishable. These three H9N2 viruses caused no overt signs of disease in chickens. The 2004 isolate replicated and spread most efficiently, and chickens previously inoculated with this H9N2 virus showed 90%–100% survival after inoculation 1 to 35 days later with lethal H5N1 virus. Chickens that survived did not show signs of disease but did shed lethal H5N1 virus from the cloaca. The modulation of survivability was time-dependent; the effect was maximal five days after H9N2 inoculation. These findings suggest that co-circulation of H9N2 viruses can contribute to the spread of lethal H5N1 viruses.
Keywords: influenza viruses, H9N2, H5N1, lethal challenge, survival, chickens
INTRODUCTION
Avian influenza viruses of the H9N2 subtype have been implicated in the genesis of highly pathogenic (HP) H5N1 strains and in the outcome of HP H5N1-associated disease (Webster et al., 2006). The G1 genotype of the H9N2 virus that circulated in Hong Kong’s live poultry markets in 1997 (A/Quail/HongKong/G1/97) contained six internal genes of the H5N1 virus (A/HongKong/157/97) isolated from humans at that time (Guan et al., 1999, 2000; Lin et al., 2000). The surprisingly low mortality rate of gallinaceous poultry in the Hong Kong markets during that period is explained in part by cross-protective cell-mediated immunity induced by the G1 genotype of the H9N2 virus (Seo and Webster, 2001).
H9N2 viruses of gallinaceous poultry spread from Asia to most Eurasian countries during the 1990s (Aamir et al., 2007; Alexander, 2007). These viruses have circulated widely and cause only low-grade disease in gallinaceous poultry. An unanswered question is whether prior infection with H9N2 viruses reduces the lethality and therefore the detectability of HP H5N1 infection in poultry in the field, thereby facilitating virus spread.
In Israel, H9N2 influenza viruses were first detected in poultry in May 2000, when they caused signs of mild respiratory disease and a drop in egg production, mainly in turkeys. After the initial outbreak, H9N2 viruses were not detected again until December 2001, and they continued to circulate in poultry until April 2003, causing disease outbreaks in chickens, geese, ostriches, and turkeys. A non-homologous inactivated vaccine was widely used to control H9N2 virus spread. H9N2 viruses were introduced a third time in February 2003 and were isolated sporadically through 2006, primarily from chickens.
In March 2006, HP H5N1 virus was detected in Israel. All flocks affected by H5N1 had been vaccinated against H9N2 infection, but evidence of co-infection with H9N2 viruses was observed in the Gaza strip region. As many as one million birds were culled to stop the spread of the H5N1 outbreak (Perk et al., 2007).
The spread of HP H5N1 to poultry in Israel, where H9N2 influenza viruses continued to circulate, allowed us to assess whether the current H9N2 viruses can moderate the lethality of co-circulating HP H5N1 viruses and thus decrease the likelihood of their detection. We antigenically and molecularly characterized three H9N2 viruses isolated from domestic turkeys in Israel and determined experimentally whether these viruses influence the pathogenicity of HP H5N1. We found that some H9N2 influenza viruses modulate the lethality of H5N1 challenge in a time-dependent fashion.
RESULTS
Sequence and phylogenetic analysis of H9N2 influenza viruses
Complete sequencing of the HA and NA genes of the three Israeli H9N2 influenza isolates confirmed that they were of the H9 subtype and were closely inter-related (Fig. 1). There was 96% identity in amino acid sequences between the HA gene of 2004 virus and the 2000 and 2002 viruses, whereas homology between the 2000 and 2002 viruses was close to 99%. The neuraminidase (NA) gene showed 98.5% similarity in the 2000 and 2002 viruses and 96.3% similarity between those two isolates and the 2004 virus. The internal genes differed by an average of 4% (96%–98% similarity of amino acid sequence). Greater divergence in nucleotide sequences (89%–92% similarity) was observed between the internal genes of the H9N2 viruses and the challenge H5N1 virus A/Ck/Israel/364/06 (Table 1). The identity of the surface glycoproteins of challenge viruses A/Ck/Israel/364/06 (H5N1) and A/Ck/Egypt/1C/06 (H5N1) was found to be 99.9% (data not shown).
FIG. 1.
Representation of amino-acid differences between the three H9N2 viruses. Upper sequence in each graph is the sequence of the reference virus, A/ty/Israel/1567/04. Capital letters indicate amino acids. Dark bars indicate changes in the two other H9N2 viruses as compared to the reference sequence; white bars indicate no change. Positions not shown are identical in the three viruses. H9 numbering was used for the HA gene.
TABLE 1.
Percent nucleotide sequence identity of internal proteins of the three Israeli H9N2 virus isolates compared to those of the A/Ck/Israel/364/2006 (H5N1) virus
| Protein | A/ty/Israel/965/00 | A/ty/Israel/90710/02 | A/ty/Israel/1567/04 |
|---|---|---|---|
| NP | 91.1 | 90.6 | 90.4 |
| NS | 91.0 | 91.2 | 90.5 |
| M | 91.4 | 91.2 | 91.4 |
| PA | 91.4 | 91.2 | 91.4 |
| PB1 | 90.1 | 90.1 | 89.3 |
| PB2 | 91.4 | 91.2 | 91.4 |
Phylogenetic analysis of the HA genes of the three Israeli H9N2 isolates showed that they are genetically closely related to A/Quail/Hong Kong/G1/97 (H9N2) and fall into group 1 of the H9N2 influenza virus phylogeny (Perk et al., 2006; Banet-Noach et al., 2007). The HA genes of the three viruses continued to diverge and formed a sister group with the H9N2 viruses isolated in Hong Kong during 1997–1999, indicating their evolution from these viruses (Fig. 2a). The HA gene of the 2004 isolate was more closely related to the G1 reference strain [A/Quail/Hong Kong/G1/97 (H9N2)] than was the HA of the 2000 and 2002 isolates. Phylogenetic analysis of the NA genes showed the same pattern (Fig. 2b), as did analysis of the internal genes (data not shown).
FIG. 2.
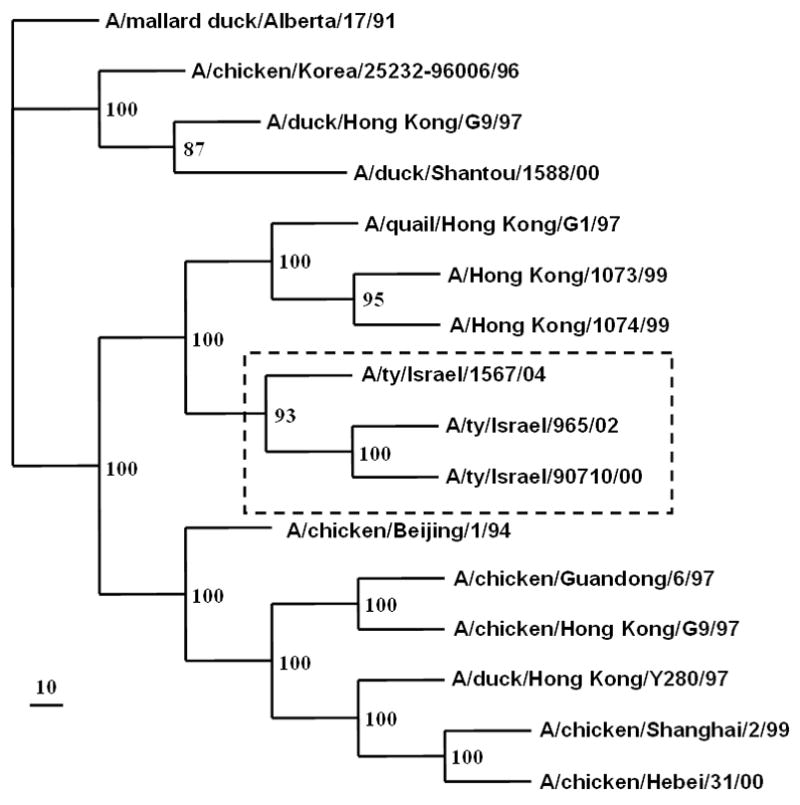
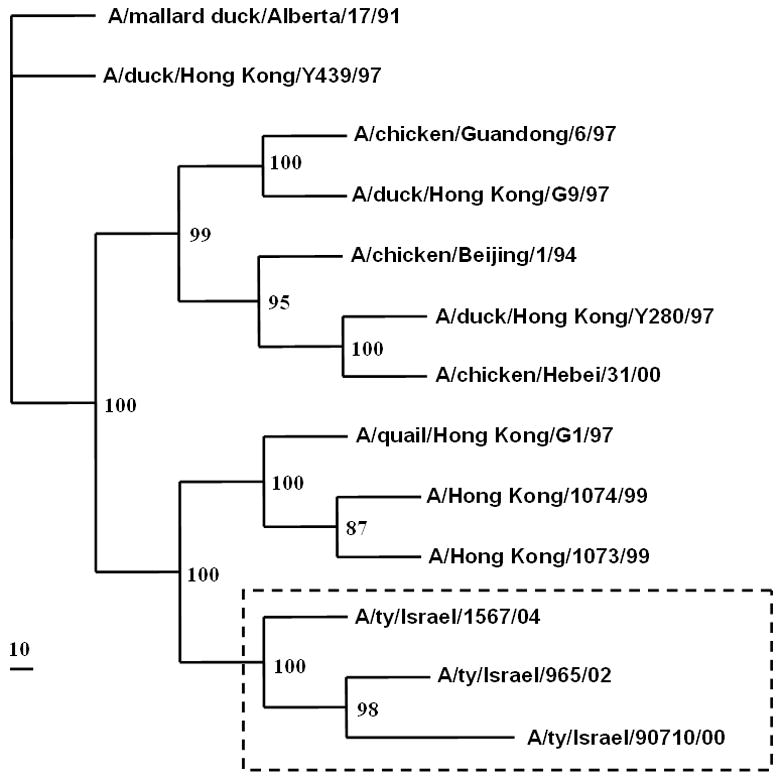
Simplified phylogenetic tree of (a) the HA gene and (b) the NA gene of the Israeli H9N2 viruses.
Growth and antigenic characterization of the H9N2 influenza viruses
The three H9N2 influenza viruses replicated in embryonated chicken eggs and, to a lesser extent, in Madin Darby canine kidney (MDCK) cells (Table 2, Fig. 3). In cell culture, A/ty/Israel/1567/04 (H9N2) replicated to higher titers than the other two strains. HI tests with chicken antisera and a panel of monoclonal antibodies confirmed the H9 subtype of the viruses but showed antigenic drift in comparison to the reference H9N2 viruses isolated in Hong Kong in 1997 (Table 3). The 2004 isolate was most closely related to reference virus A/Qa/HK/G1/97 H9N2. The HA activity of A/ty/Israel/1567/04 (H9N2) was also slightly inhibited by antibodies to the A/Dk/HK/Y280/97 strain, suggesting that the 2004 isolate is antigenically closer to A/Qa/HK/G1/97 (H9N2) than to the other reference H9N2 viruses shown in Table 3 and is antigenically distinguishable from the other two Israeli H9N2 isolates.
TABLE 2.
Replication of H9N2 viruses
| H9N2 virus | Chicken embryos (Log10EID50/ml) | MDCK cells (Log10PFU/ml) | MDCK cells (Log10TCID50/ml) |
|---|---|---|---|
| A/ty/Israel/90710/00 | 9.2 | 7.0 | 3.6 |
| A/ty/Israel/965/02 | 8.8 | 7.7 | 4.7 |
| A/ty/Israel/1567/04 | 8.7 | 8.1 | 6.0 |
FIG. 3.
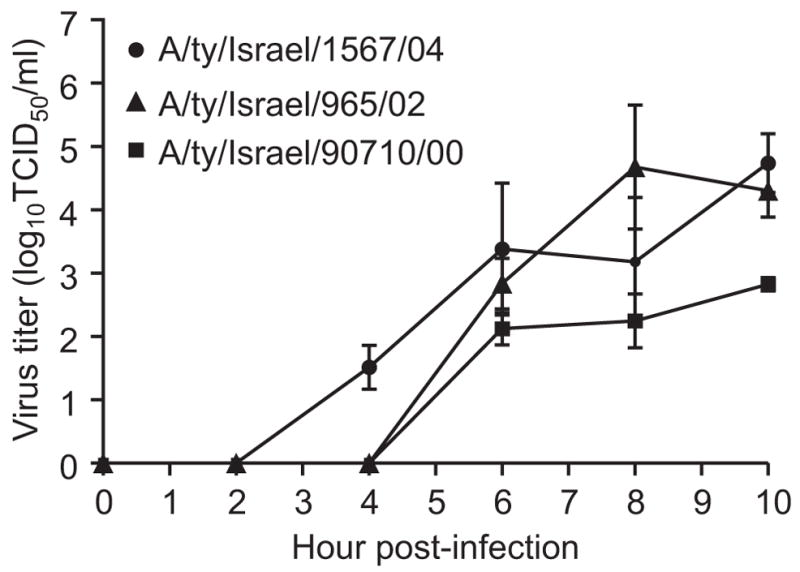
Replication kinetics of the three H9N2 viruses in MDCK cells.
TABLE 3.
Antigenic characterization of viruses by HI assay with anti-H9 monoclonal antibodies and polyclonal chicken antisera
| Polyclonal chicken antiseraa | Anti-H9 monoclonal antibodies | |||||
|---|---|---|---|---|---|---|
| H9N2Virus | aA/ty/Israel/90710/00 | a A/ty/Israel/1567/04 | aA/Qa/HK/G1/97 | aA/Ck/HK/G9/97 | aA/Dk/HK/Y280/97 | |
| G1-26 | G9-6 | 18G4 | 2F4 | |||
| A/ty/Israel/90710/00 | 40 | 160 | 200 | 200 | <b | < |
| A/ty/Israel/965/02 | 80 | 80 | 100 | 800 | < | < |
| A/ty/Israel/1567/04 | 320 | 640 | 12800 | 200 | 100 | 1600 |
| A/Qa/HK/G1/97 | 80 | 320 | 6400 | 3200 | < | < |
| A/Ch/HK/G9/97 | 160 | 1280 | 3200 | 6400 | 3200 | 3200 |
| A/Dk/HK/Y280/97 | 80 | 320 | 800 | 6400 | 25600 | 25600 |
Polyclonal anti-H9 antisera obtained at day 28 p.i. with H9N2 virus. To remove non-specific inhibitors of agglutination, serum was incubated overnight with receptor-destroying enzyme and heated for 30 min at 56°C. The initial dilution was 1:40.
< Indicates a titer less than 1:100 (the initial dilution of the antibodies).
Replication and transmission of H9N2 viruses in chickens
Because the H9N2 influenza viruses affected both chickens and turkeys in Israel, we used chickens—which are smaller and easier to handle in isolators—for in vivo studies. No adaptation of viruses was done prior to inoculation. The A/ty/Israel/965/02 (H9N2) virus replicated to a limited extent in 9-week-old White Leghorn chickens but was not transmitted to contact chickens introduced into the same cage 24 hours p.i. The A/ty/Israel/90710/00 and A/ty/Israel/1567/04 viruses replicated to moderate titers in the tracheas of inoculated chickens, were detected for 7 days, and were transmitted to contact chickens (Table 4); A/ty/Israel/1567/04 was transmitted more effectively than the 2000 isolate at an early stage of infection and replicated more rapidly and to higher titers in contact birds. None of the inoculated or contact birds showed visible signs of disease and all groups continued to gain weight. Only low titers of viruses were isolated from cloacal swabs, and only transiently; virus was isolated at higher titers and with greater frequency from the trachea. Water samples from the feeding trays were collected and analyzed for the 2004 isolate, which was found at relatively high titers up to day 6 (2.3 log10EID50/ml at day 6).
TABLE 4.
Tracheal and cloacal shedding of virus after inoculation with three H9N2 viruses.
| Mean tracheal virus titers* and tracheal shedding (no. shedding/total) | ||||||
|---|---|---|---|---|---|---|
| Inoculated (n=7) | Contact (n=3) | |||||
| Virus | 3 d.p.i. | 5 d.p.i. | 7 d.p.i. | 3 d.p.i | 5 d.p.i | 7 d.p.i. |
| A/ty/Israel/90710/2000 | 3.8 (1/7) | 2 (6/7) | 1.2 (2/7) | 2.5 (1/3) | 3.7 (2/3) | 5( 3/3) |
| A/ty/Israel/965/2002 | 4.5 (3/7) | 3 (7/7) | 2.7 (3/7) | 0 (0/3) | 0 (0/3) | 0 (0/3) |
| A/ty/Israel/1567/2004 | 5.6 (3/7) | 4.4 (7/7) | 3 (6/7) | 5.4 (3/3) | 5.3 (3/3) | 2.9 (3/3) |
|
| ||||||
| Mean cloacal* virus titers and cloacal shedding (no. shedding/total) | ||||||
| A/ty/Israel/90710/2000 | 1.2 (1/7) | 1.5 (1/7) | 0 (0/7) | 0 (0/3) | 0 (0/3) | 2.35 (2/3) |
| A/ty/Israel/965/2002 | 1.2 (2/7) | 0 (0/7) | 0 (0/7) | 0 (0/3) | 0 (0/3) | 0 (0/3) |
| A/ty/Israel/1567/2004 | 1.1 (3/7) | 1.7 (3/7) | 0.5 (2/7) | 1.9 (2.3) | 2.1 (2/3) | 0.5 (1/3) |
Expressed as Log10EID50/ml
Cross-protection studies
Previous studies demonstrated cross protection between the A/Quail/Hong Kong/G1/97 (H9N2) variant isolated in 1997 and the lethal H5N1 virus circulating at that time (Seo and Webster, 2001). Since then, both the H9N2 and H5N1 viruses have continued to evolve and little attention has been paid to the consequences of their co-circulation. To assess the effect of prior infection with the different H9N2 isolates on the course of infection with currently-circulating highly pathogenic H5N1 virus, we inoculated chickens with the three H9N2 isolates and subsequently challenged them with lethal H5N1 virus.
Effect of the H9N2 variants on H5N1 mortality and signs of disease
In the first experiment, groups of four chickens were inoculated with the three H9N2 influenza viruses, respectively, and infection was confirmed via tracheal and cloacal swabs. None of the infected birds showed signs of disease. Thirty-five days after inoculation, the inoculated birds and a non-inoculated control group were challenged with 10 CLD50 of the highly pathogenic H5N1 virus A/Chicken/Egypt/1C/06. The control groups and the groups initially infected with the 2000 and 2002 H9N2 viruses showed signs of severe disease starting 2 to 3 days after challenge, and all birds were dead by the ninth day post-challenge (Fig. 4a).
FIG. 4.
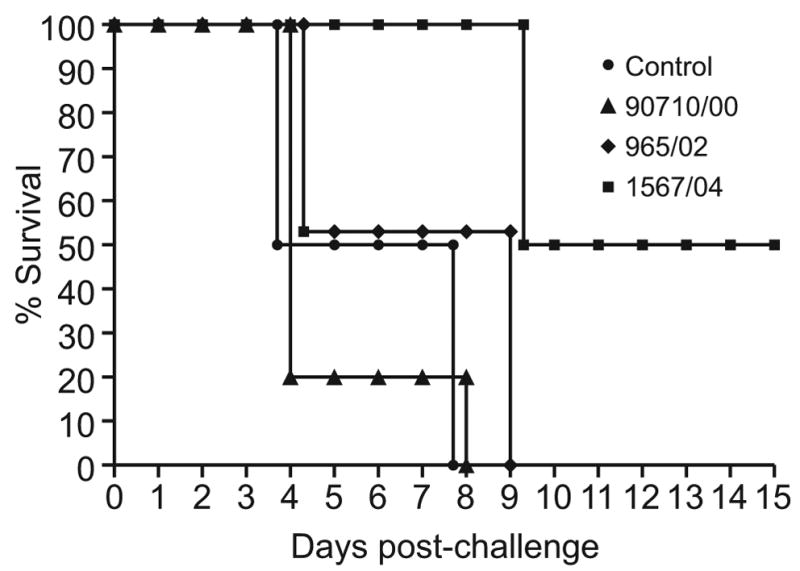
FIG. 4a. Survival of chickens challenged with 10 CLD50 of the highly pathogenic A/Ck/Egypt/1C/06 (H5N1) virus 35 days after inoculation with the three Israeli H9N2 isolates (4 chickens per group).
FIG. 4b. Survival of chickens challenged with 10 CLD50 of the highly pathogenic A/Ck/Israel/364/06 (H5N1) virus 21 days after inoculation with the three Israeli H9N2 isolates (10 chickens per group).
Disease signs and their severity did not differ between the controls and birds previously infected with 2000 (A/ty/Israel/90710/00) and 2002 (A/ty/Israel/965/02) H9N2 viruses, but the latter birds died 1 day later than control birds. Mean survival days for the control group, group inoculated with A/ty/Israel/965/2000, and group inoculated with A/ty/Israel/90710/2002 were 5.2, 5.8 and 6.5, respectively. In contrast, 50% of chickens previously infected with the 2004 H9N2 virus (A/ty/Israel/1567/04) showed no visible signs of disease and survived, while 2 of the 4 died on day 9 after challenge. The birds that died had continued to shed HP H5N1 virus from the trachea and cloaca between days 7 and 9 post-challenge. One chicken that survived shed H5N1 virus until day 5 post-challenge, at a relatively high titer (3.2 log10EID50/ml on day 3).
The same survival pattern was observed when groups of seven birds were inoculated with the same H9N2 isolates. Twenty-four hours after inoculation with the H9N2 viruses, three naïve contact birds were introduced into each group and all birds were challenged simultaneously at day 21 p.i. with 10 CLD50 of the highly pathogenic H5N1 virus A/Chicken/Israel/364/06. All control chickens succumbed to the infection at day 4. All chickens inoculated with 2000 and 2002 H9N2 viruses were dead by days 9 and 12, respectively. The birds that succumbed shed the H5N1 virus from the trachea and cloaca until their deaths. However, only 4 of 10 chickens died in the group previously inoculated with the A/ty/Israel/1567/04 isolate (Fig. 4b). Notably, mortality was observed only within the group of inoculated birds; all contact chickens survived. Birds that survived did not show any virus replication in the trachea and only trace amounts (less than 1.5 log10EID50/ml) of H5N1 virus in the cloaca
Time-dependence of cross-protection
Earlier studies showed that cell-mediated immunity was responsible for cross-protection between H9N2 and HP H5N1 viruses; immunity was transferrable among inbred chickens via immune cells but not serum (Seo et al., 2002). Because circulating cell-mediated immunity is of shorter duration than humoral immunity, protection from HP H5N1 viruses may diminish with increasing time after H9N2 infection.
To test this hypothesis, we inoculated groups of 10 chickens with the 2004 H9N2 Israeli strain (A/ty/Israel/1567/04) and challenged them 5, 20, and 35 days later with 10 CLD50 of highly pathogenic A/Chicken/Israel/364/06 (H5N1) virus. Before challenge, serum samples were collected from all three groups of birds and tested by HI; all samples, including those collected 35 days p.i., were nonreactive with the challenge virus (reciprocal of HI titer was less than 10). As in the previous experiment, all of the control birds died by day 9. None of the chickens inoculated with the H9N2 virus 5 days before challenge showed signs of disease, and all of them survived. Ninety percent (9 of 10) of birds inoculated with H9N2 20 or 35 days before H5N1 challenge survived without signs of disease (Fig. 5).
FIG. 5.
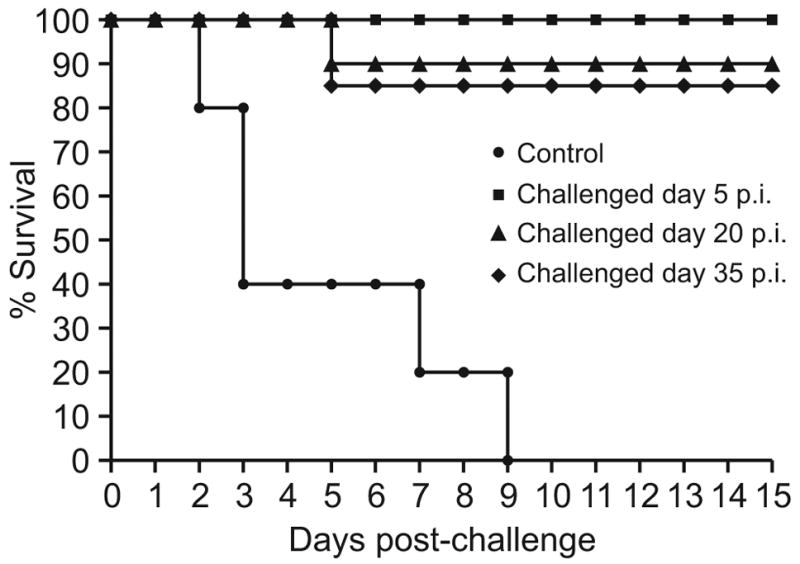
Survival of chickens challenged with 10 CLD50 of the highly pathogenic A/Ck/Israel/364/06 (H5N1) virus 5, 20, or 35 days after inoculation with the A/Ty/Israel/1567/04 (H9N2) virus. (n=10 chickens per group)
To study H9N2-mediated protection at early stages of H9N2 infection, we challenged groups of five birds with 10 CLD50 of the A/Chicken/Israel/364/06 (H5N1) virus 1, 3, or 5 days after inoculation with A/ty/Israel/1567/04 (H9N2) virus (Fig. 6). As in the previous experiment, all birds challenged on day 5 survived without signs of disease. Two of the five birds challenged on day 3 died, and four of the five birds challenged on day 1 died. All of the birds that died showed signs of disease, whereas those that survived did not. Therefore, cross-protection increased progressively from day 1 to day 5 after inoculation.
FIG. 6.
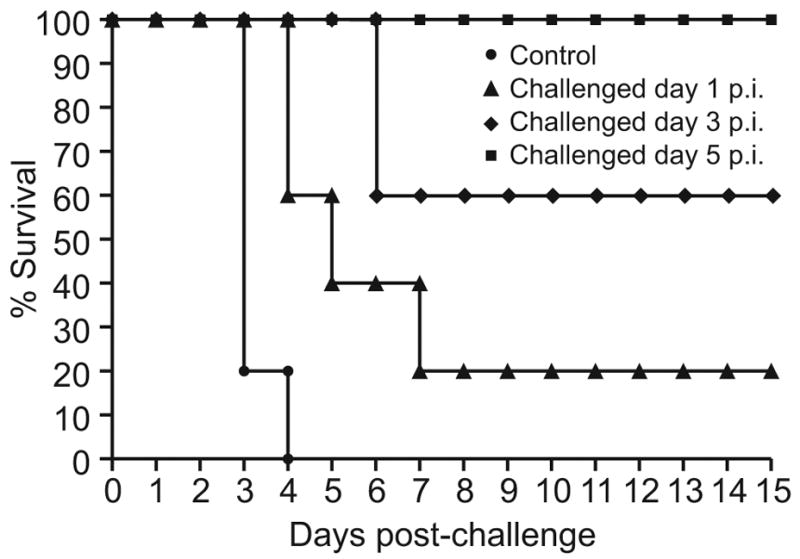
Survival of chickens challenged with 10 CLD50 of the highly pathogenic A/Ck/Israel/364/06 virus 1, 3, or 5 days after inoculation with the A/ty/Israel/04 (H9N2) virus (n=10 chickens per group).
DISCUSSION
H9N2 and HP H5N1 viruses have continuously co-circulated in domestic poultry throughout Eurasia since 1997. The HP H5N1 viruses initially isolated in Hong Kong in 1997 shared internal gene segments with H9N2 viruses isolated from live poultry markets at that time (Guan et al., 1999; Shortridge et al., 1998). However, both the H9N2 and the HP H5N1 viruses have continued to evolve and have formed a number of clades and subclades (WHO, 2007, 2008; Xu et al., 2007). Therefore, it was unknown whether the currently circulating H9N2 viruses limit the detection of subsequent H5N1 infection by reducing its lethality. After large-scale culling was required in Israel to stop the spread of an H5N1 outbreak in poultry flocks, we investigated the potential role of co-circulating H9N2 viruses in the spread of HP H5N1.
We first characterized the H9N2 viruses isolated from turkeys during outbreaks in Israeli poultry in 2000, 2002, and 2004 (Perk et al., 2006). These three viruses were antigenically distinct. Phylogenetic analysis of the HA genes revealed that two distinguishable groups of H9N2 viruses arose from the G1 lineage (A/Quail/HongKong/G1/97) circulating in Hong Kong in 1997. In serological analysis against a panel of anti-H9 monoclonal antibodies, the 2004 isolate was antigenically more closely related to A/Quail/Hong Kong/G1/97 than were the 2000 and 2002 isolates. Phylogenetic analysis of all genes further confirmed this finding. The 2004 H9N2 isolate also replicated more efficiently in chickens than the earlier two isolates and was transmitted more efficiently to contact chickens. Interestingly, we found as much as 2.5 log10EID50/ml of the 2004 isolate in the drinking water of infected chickens on day 5 p.i. (data not shown); this finding suggests how the virus may have been transmitted to naïve birds on poultry farms. None of the three H9N2 isolates caused overt signs of disease.
Only the H9N2 influenza virus that replicated and was transmitted efficiently in chickens modulated disease caused by HP H5N1 virus. Protection against mortality increased progressively between days 1 and 5 after inoculation with A/ty/Israel/1567/04 (H9N2) virus and declined slightly (by approximately 10%) afterward. All chickens challenged five days after H9N2 inoculation survived. After 35 days, 50% of chickens survived a lethal dose of A/Egypt/1C/06 (H5N1) virus and 90% survived a lethal dose of A/Chicken/Israel/364/06 (H5N1) virus. Because the Israeli and Egyptian H5N1 viruses are genetically and biologically closely related (Perk et al., 2006), the difference in survival is likely to reflect variations in the outbred chickens and possibly the 2-week older average age of chickens inoculated with the Israeli H5N1 virus.
A/ty/Israel/1567/04, the H9N2 isolate that modulated H5N1 pathogenicity, differed from the other two isolates in all eight gene segments, although the three isolates had greater than 96% amino acid similarity in all gene segments (Fig. 1). While molecular differences, especially those in the surface genes, are likely to explain the disparate modulation of H5N1 pathogenicity, further studies will be needed to identify the factors responsible.
Our results suggest that A/ty/Israel/1567/04 grows to higher titers in chickens and persists longer than the other two H9N2 viruses studied. Because virus burden is strongly correlated with the T-cell response in non-lethal influenza, it is possible that the 2004 virus exerts a stronger priming effect on cellular immunity. The absence of protective serum antibodies to H5N1 viruses in chickens infected with H9N2 viruses supports a major role for cellular immunity. The contamination of our H9N2 virus stock with H5N1 viruses was ruled out by molecular methods.
Does the modulation of H5N1 pathogenicity by cross-protective immunity to H9N2 help to perpetuate H5N1 viruses in domestic poultry? If chickens are infected with HP H5N1 virus during the “protected window” after H9N2 infection, an initial outbreak of HP H5N1 may go unnoticed while the virus is shed (predominantly in feces) by protected birds and becomes more widespread. Our results suggest that some strains of H9N2 virus, but not others, can modulate the severity of disease caused by HP H5N1 virus and provide partial protection against lethal challenge under the conditions tested. Interestingly, even closely related H9N2 viruses may differ in their protective effect.
The masking of H5N1 pathogenicity is not an issue for countries that use a “stamping out” policy after detection of HP H5N1, as both viruses would be stamped out. However, masking of H5N1 disease signs by H9N2 is a potentially important problem for countries that use H5N1 vaccines and cull flocks only when clinical disease is evident. Because the use of sentinel chickens for detection of HP H5N1 in vaccinated flocks can be compromised by immunity to H9N2 virus, we suggest that prospective virologic surveillance offers the best solution.
The decade-long circulation of HP H5N1 in Southeast Asia, which has remained a reservoir of evolving strains (Kilpatrick et al., 2006), and the circulation of HP H5N1 since 2005 in parts of Africa and Eurasia, have led to a “blame game.” Some countries and authorities consider wild migratory birds to be the ultimate reservoir and spreaders of highly pathogenic viruses. Conversely, wild-bird ecologists and wildlife authorities consider domestic waterfowl to be the current reservoirs of HP H5N1, while migratory species are occasionally infected and may spread HP H5N1 but do not perpetuate the virus. The rate of H9N2 co-infection and the prevalence of H9N2-mediated protection against HP H5N1 virus are unknown but may be contributors to the continued circulation of pockets of H5N1 virus.
The co-circulation of H9N2 and lethal H5N1 viruses may be contributing to the genetic diversity of both subtypes, with the possibility of reassortment of individual genes. While H9N2 viruses cause no signs of disease in experimentally inoculated birds, signs of disease are observed in the field, especially in cases of concurrent stress or co-infection (Banet-Noach et al., 2007). Inactivated H9N2 vaccines are coming into wider use and may help to reduce H9N2 disease severity and the potential masking of infection with highly pathogenic H5N1 viruses.
MATERIALS AND METHODS
Viruses
We characterized three H9N2 viruses isolated from turkeys in Israel in 2000, 2002, and 2004: A/ty/Israel/90710/00, A/ty/GivatHain/965/02, and A/ty/Israel/1567/04. The A/Chicken/Egypt/1C/2006 and A/Chicken/Israel/364/2006 HP H5N1 influenza viruses were used in challenge experiments. All viruses were propagated in the allantoic cavities of 11-day-old embryonated chicken eggs in a biosafety level 3+ (BSL3+) facility approved by the USDA.
Antigenic analysis
We analyzed the cross-reactivity of the H9N2 viruses by hemagglutination inhibition (HI) assay with polyclonal antisera and anti-H9 monoclonal antibodies to three H9N2 viruses representing the Asian lineages in poultry: A/Quail/Hong Kong/G1/97 (A/Qa/HK/G1/97), A/Chicken/Hong Kong/G9/97 (A/Ck/HK/G9/97), and A/Duck/Hong Kong/Y280/97 (A/Dk/HK/Y280/97). Methods were described previously (Palmer et al., 1975).
Genetic and sequence analysis
RNA was extracted from infected allantoic fluid by using the RNeasy Mini Kit (Qiagen, Valencia, CA). Real-time polymerase chain reaction (RT-PCR) was performed with the OneStep RT-PCR Kit (Qiagen, Valencia, CA), and the PCR products were sequenced by using synthetic oligonucleotides produced by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children’s Research Hospital (St. Jude). Sequence results for all genes of the three isolates are available from GenBank under accession numbers: AY738451, EF492273, EF492371, EF492400, EF492342, EF492302, EF492244, DQ683025 (A/ty/Israel/90710/00); AY738452, EF492289, EF492387, EF492416, EF492358, EF492318, EF492271, DQ683035 (A/ty/Israel/965/02); EF492241, EF492297, EF492395, EF492424, EF492366, EF492326, EF492264, DQ683043 (A/ty/Israel/1567/04). HA genes of the 2000 and 2002 isolates are also referred in the NCBI database as A/ty/Neve Ilan/90710/00 and A/ty/Givat Haim/965/02, respectively. Sequence results for all genes of the challenge virus A/Ck/Israel/364/06 are available from GenBank under accession numbers: EF532635, EF532624, EU574919, EU579978, EU579972, EU579966, EU579960, EU579954.
Phylogenetic analysis
Nucleotide and deduced amino acid sequences were aligned and all sequence data were compiled and edited by using the BioEdit program with the ClustalW alignment package (Thompson et al., 1994). Phylogenetic trees were generated and bootstrapped by using the Phylip program, version 3.67.
Tests in animals
Depending on the experiment, four, seven, or ten 6- to 8-week-old White Leghorn chickens were inoculated with 1.0 ml of virus dilution in PBS containing 106 50% egg infectious doses (EID50) per milliliter via a “natural route” (intranasal [0.3 ml], intraocular [0.1 ml], or intratracheal [0.6 ml]). Contact birds were introduced into the inoculated birds’ cages 24 hours post-inoculation (p.i.) to study virus transmission. All birds were observed daily for signs of disease (disheveled feathers, lethargy, fever, or paralysis), mortality, and weight change. Tracheal and cloacal swabs were collected on days 3, 5, 7, and 10 p.i., and virus was titrated in 10-day-old embryonated chicken eggs. Serum samples collected from all birds before inoculation and 4 weeks after inoculation were analyzed via HI assay in microtiter plates with 0.5% chicken red blood cells. To assess cross-protective immunity induced by H9N2 viruses, chickens were then challenged via natural route with 10 chicken lethal doses (CLD50) of A/Chicken/Egypt/1C/2006 or A/Chicken/Israel/364/2006 HP H5N1 virus and observed daily for signs of disease and mortality. All animal experiments using H5N1 and H9N2 viruses were done in USDA-approved BSL3+ facilities. All studies were conducted under applicable laws and guidelines and after approval from the Animal Care and Use Committee at St. Jude.
Acknowledgments
This project was funded by Contract No. HHSN266200700005C with the National Institute of Allergy and Infectious Diseases of the US National Institutes of Health and supported by the American Lebanese Syrian Associated Charities (ALSAC).
We thank Dr. U. Aamir for assistant with the first BSL-3 experiment, Sharon Naron and Donald Samulack, PhD for critical review and scientific editing of the manuscript, James Knowles for manuscript preparation, and Patrick Seiler and Cedric Proctor for technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aamir UB, Wernery U, Ilyushina N, Webster RG. Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000 to 2003. Virology. 2007;361:45–55. doi: 10.1016/j.virol.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DJ. Summary of avian influenza activity in Europe, Asia, Africa, and Australasia, 2002–2006. Avian Dis. 2007;51(1 Suppl):161–166. doi: 10.1637/7602-041306R.1. [DOI] [PubMed] [Google Scholar]
- Banet-Noach C, Perk S, Simanov L, Grebenyuk N, Rozenblut E, Pokamunski S, Pirak M, Tendler Y, Panshin A. H9N2 viruses from Israeli poultry: a five-year outbreak. Avian Dis. 2007;51(1 Suppl):290–296. doi: 10.1637/7590-040206R1.1. [DOI] [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: Were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Chin PS, Dyrting KC, Ellis TM, Webster RG, Peiris M. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J Virol. 2000;74:9372–9380. doi: 10.1128/jvi.74.20.9372-9380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, Chmura AA, Gibbons DW, Fleischer RC, Marra PP, Daszak P. Predicting the global spread of H5N1 avian influenza. Proc Natl Acad Sci USA. 2006;103:19368–19373. doi: 10.1073/pnas.0609227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A. Avian-to-human transmission of H9N2 subtype influenza A viruses: Relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci USA. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DF, Dowdle WR, Coleman MT, Schild GC. Advanced Laboratory Techniques for Influenza Diagnosis. U.S. Department of Health, Education and Welfare Immunology Series, No. 6. Centers for Disease Control; Atlanta, GA: 1975. [Google Scholar]
- Perk S, Banet-Noach C, Shihmanter E, Pokamunski S, Pirak M, Lipkind M, Panshin A. Genetic characterization of the H9N2 influenza viruses circulated in the poultry population in Israel. Comp Immunol Microbiol Infect Dis. 2006;29:207–223. doi: 10.1016/j.cimid.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Perk S, Banet-Noach C, Golender N, Simanov L, Rozenblut E, Nagar S, Pokamunski S, Pirak M, Tendler Y, García M, Panshin A. Molecular characterization of the glycoprotein genes of H5N1 influenza A viruses isolated in Israel and the Gaza Strip during 2006 outbreaks. Virus Genes. 2007;35:497–502. doi: 10.1007/s11262-007-0120-1. [DOI] [PubMed] [Google Scholar]
- Seo HS, Webster RG. Cross-reactive, cell-mediated immunity and protection of chickens from lethal H5N1 influenza virus infection in Hong Kong poultry markets. J Virol. 2001;75:2516–2525. doi: 10.1128/JVI.75.6.2516-2525.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo SH, Hoffman E, Webster RG. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med. 2002;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- Shortridge KF, Zhou NN, Guan Y, Gao P, Ito T, Kawaoka Y, Kodihalli S, Krauss S, Markwell D, Murti KG, Norwood M, Senne D, Sims L, Takada A, Webster RG. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology. 1998;252:331–342. doi: 10.1006/viro.1998.9488. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Peiris M, Chen H, Guan Y. H5N1 outbreaks and enzootic influenza. Emerg Infect Dis. 2006;12:3–8. doi: 10.3201/eid1201.051024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Antigenic and genetic characteristics of H5N1 viruses and candidate H5N1 vaccine viruses developed for potential use as pre-pandemic vaccines. Wkly Epidemiol Rec. 2007;82:164–167. [PubMed] [Google Scholar]
- Writing Committee of the Second World Health Organization Consultation on Clinical Aspects of Human Infection with Avian Influenza A (H5N1) Virus. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- Xu KM, Smith GJD, Bahl J, Duan L, Tai H, Vijaykrishna D, Wang J, Zhang JX, Li KS, Fan XH, Webster RG, Chen H, Peiris JSM, Guan Y. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J Virol. 2007;81:10389–10401. doi: 10.1128/JVI.00979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]